Abstract
The degradation of human ether-a-go-go-related gene (hERG, KCNH2) transcripts containing premature termination codon (PTC) mutations by nonsense-mediated mRNA decay (NMD) is an important mechanism of long QT syndrome type 2 (LQT2). The mechanisms governing the recognition of PTC-containing hERG transcripts as NMD substrates have not been established. We used a minigene system to study two frameshift mutations, R1032Gfs*25 and D1037Rfs*82. R1032Gfs*25 introduces a PTC in exon 14, whereas D1037Rfs*82 causes a PTC in the last exon (exon 15). We showed that R1032Gfs*25, but not D1037Rfs*82, reduced the level of mutant mRNA compared to the wild-type minigene in an NMD-dependent manner. The deletion of intron 14 prevented degradation of R1032Gfs*25 mRNA indicating that a downstream intron is required for NMD. The recognition and elimination of PTC-containing transcripts by NMD required that the mutation be positioned >54-60 nt upstream of the 3'-most exon-exon junction. Finally, we used a full-length hERG splicing-competent construct to show that inhibition of downstream intron splicing by antisense morpholino oligonucleotides inhibited NMD and rescued the functional expression of a third LQT2 mutation, Y1078*. The present study defines the positional requirements for the susceptibility of LQT2 mutations to NMD and posits that the majority of reported LQT2 nonsense and frameshift mutations are potential targets of NMD.
Keywords: arrhythmia, long QT syndrome, KCNH2, patch-clamp, potassium channels
1. Introduction
The long QT syndrome type 2 (LQT2) is caused by mutations in the human ether-a-go-go-related gene (hERG, KCNH2) (Curran et al., 1995). hERG encodes the pore forming subunit of the rapidly activating delayed rectifier K+ channel in the heart. Over five hundred mutations have been identified in patients with LQT2 (Kapplinger et al., 2009; Lieve et al., 2013; Nagaoka et al., 2008; Napolitano et al., 2005; Splawski et al., 2000; Tester et al., 2005). More than 30% of LQT2 mutations are nonsense or frameshift mutations that introduce premature termination codons (PTCs). We previously reported that PTC-containing hERG mRNAs are degraded by nonsense-mediated mRNA decay (NMD) (Bhuiyan et al., 2008; Gong et al., 2007; Zarraga et al., 2011). NMD is an RNA quality control mechanism that selectively degrades mRNA harboring PTCs (Kuzmiak and Maquat, 2006). NMD eliminates abnormal mRNA transcripts harboring PTCs, and thereby preventing the production of truncated proteins that often have dominant-negative effects. Thus, NMD protects against severe disease phenotypes by converting dominant-negative effects to haploinsufficiency (Khajavi et al., 2006).
The identification of potential NMD targets has important implications in genotype-phenotype correlations in LQT2. The LQT2 mutation R1014* generates truncated hERG channel protein that exhibits a dominant-negative effect on the wild-type (WT) channel in the context of the hERG cDNA construct (Gong et al., 2004). However, when an intron-containing minigene is used, the R1014* mutant mRNA is decreased by NMD (Gong et al., 2007). Therefore, haploinsufficiency rather than dominant-negative effect is the underlying mechanism for the R1014* mutation. Most mutation carriers in this family have a mild clinical phenotype which is consistent with this mechanism (Gong et al., 2007). In some cases, NMD could be detrimental if it prevents the production of truncated proteins that are fully or partially functional. This is typified in the LQT2 Q1070* mutation in which NMD results in a nearly complete elimination of mutant mRNA, precluding the formation of functional, truncated channels (Bhuiyan et al., 2008). Although several LQT2 nonsense and frameshift mutations have been shown to induce NMD, the mechanisms by which the NMD machinery recognizes PTC-containing hERG transcripts have not been established.
There are currently several models that describe the recognition of NMD substrates in mammalian cells. The classic model posits that NMD occurs when translation terminates >50-55 nt upstream of the 3'-most exon-exon junction (Kuzmiak and Maquat, 2006). According to this model NMD is linked to splicing and translation. Pre-mRNA splicing results in deposition of a multi-protein complex, known as exon-junction-complex (EJC), 20-24 nt upstream of each exon-exon junction (Kuzmiak and Maquat, 2006). The EJCs are displaced by the ribosome during the pioneer round of translation. If translation terminates at a PTC that is located 50-55 nt upstream of an exon–exon junction the downstream EJC serves as a binding platform for NMD factors that trigger NMD. Recent studies, however, have challenged the EJC-dependent model of NMD. For instance, PTCs located as close as 8-10 nt upstream of the 3'-most exon-exon junction still elicit NMD in T cell receptor-β and Ig-μ transcripts (Bühler et al., 2006; Carter et al., 1996). Insertion of an intron downstream of β-globin termination codon does not elicit NMD, whereas some PTCs that are located in the last exon or even in intron-less mRNA trigger NMD, suggesting that the presence of a downstream intron is neither sufficient nor required for triggering NMD (Bateman et al., 2005; Bühler et al., 2006; Rajavel and Neufeld, 2001; Singh et al., 2008). These experimental data support an EJC-independent model in which the physical distance between the termination codon and the poly(A)-binding protein C1 is a crucial determinant for recognition of NMD substrates (Eberle et al., 2008). Clearly, the mechanisms that define the susceptibility of PTC-containing mRNA to NMD may vary in different genes. To understand the mechanisms by which the NMD machinery discriminates between premature and normal termination codons in hERG it is necessary to determine the positional requirements associated with NMD-sensitivity and NMD-resistance. In this study we used hERG minigene and full-length hERG splicing-competent constructs to investigate the role of LQT2 PTC position in susceptibility of LQT2 mutations to NMD.
2. Materials and methods
2.1. Plasmid constructs and transfection
The hERG minigene spanning exons 12-15 and the full-length hERG splicing-competent construct composed of hERG cDNA exons 1-10 and hERG genomic DNA from intron 10 to poly(A) site were generated as previously described (Gong et al., 2007; 2011). The full-length splicing competent construct encodes the full-length hERG potassium channel which corresponds to the hERGa isoform (NM_000238.3). The hERG mutations were generated by pAlter mutagenesis system (Promega, Madison, WI). Flp-In HEK293 cells (Invitrogen, Carlsbad, CA) were stably transfected with pcDNA5 hERG minigenes or full-length hERG splicing-competent constructs and selected with 100 μg/ml hygromycin B. The Flp-In HEK293 cells contain the FRT site at a single genomic locus, allowing stable integration of hERG constructs via Flp recombinase-mediated DNA recombination at a specific genomic location in all cell clones.
2.2. RNase protection assay and immunoblot analysis
RNase protection assay (RPA) was performed as previously described (Gong et al., 2011). Briefly, antisense RNA riboprobes were transcribed in vitro in the presence of biotin-14-CTP. The probe contained 277 nt spanning the region of exons 12 and 13. The total length of the probe was 409 nt and contained sequences from the pCRII vector at both ends. The expression level of hygromycin B resistance gene in the pcDNA5 vector was used as an internal control for normalization. The probe for the hygromycin B resistance gene contained 158 nt of the gene and 70 nt from the pGEM vector. Yeast RNA was used as a control for the complete digestion of the probes by RNase. The relative intensity of RPA bands was quantified using ImageJ.
Immunoblot was performed as previously described (Gong et al., 2010). The cell lysates were subjected to SDS-polyacrylamide gel electrophoresis and the blots were probed with an antibody against the N-terminus of the hERG channel protein (H175, Santa Cruz Biotechnology Inc., Santa Cruz, CA). The expression level of hygromycin B phosphotransferase (HPH) encoded by hygromycin B resistance gene in the pcDNA5 vector was used as an internal control for normalization. The polyclonal antibody against HPH was used at 1:1000 dilution as previously described (Gong et al., 2010).
2.3. RNA interference
An UPF1 short hairpin RNA (shRNA), targeting the coding sequence of hUPF1 (5'-GAGAATCGCCTACTTCACT-3'), was used to inhibit expression of UPF1 (Gong et al., 2007). A double stable mammalian expression system was used to express hERG minigenes and UPF1 shRNAs. The design of the Flp-Cre cell line has been previously described (Stump et al., 2012a). Briefly, a loxP2272/loxP target site was introduced into Flp-In HEK293 cells (Invitrogen, Carlsbad, CA) at a single genomic locus. Flp recombinase-mediated recombination and Cre recombinase-mediated cassette exchange reactions were performed to stably introduce a single copy of each gene of interest. The WT hERG and R1032Gfs*25 mutant minigenes were introduced into the Flp-Cre HEK293 cells at the FRT site, whereas the UPF1 and scramble shRNAs were introduced into the Flp-Cre HEK293 cells at the loxP2272/loxP site. The knockdown of the UPF1 protein was analyzed by immunoblot as previously described (Gong et al., 2007).
2.4. Morpholino oligonucleotide treatment
Morpholino oligonucleotides (MO) were synthesized by Gene Tools (Philomath, OR). The antisense MO (5’-AAGCAGGGCTGGAGCTTACCTGAGA-3’) was designed to target the 5’-splice site of intron 14 (Gong et al., 2011). An invert MO with the same sequence but in reverse orientation was used as control. The Endo-Porter method was used to deliver MOs into the cells.
2.5. Patch-clamp recordings
Membrane currents were recorded in whole-cell configuration as previously described (Gong et al., 2004). The bath solution contained (in mM) 137 NaCl, 4 KCl, 1.8 CaCl2, 1 MgCl2, 10 glucose, and 10 HEPES (pH 7.4 with NaOH). The pipette solution contained (in mM) 130 KCl, 1 MgCl2, 5 EGTA, 5 MgATP, and 10 HEPES (pH 7.2 with KOH). hERG channel current was activated by depolarizing steps between −70 and 50 mV from a holding potential of −80 mV. Tail current was recorded following repolarization to −50 mV. All patch-clamp experiments were performed at 22-23 °C.
2.6. Statistical analysis
Data are presented as mean ± standard error of mean. Student’s t-test was used for comparison between two groups. ANOVA with Bonferroni correction was used for comparisons between more than two groups. P<0.05 is considered statistically significant.
3. Results
3.1. LQT2 mutation R1032Gfs*25 but not D1037Rfs*82 leads to NMD
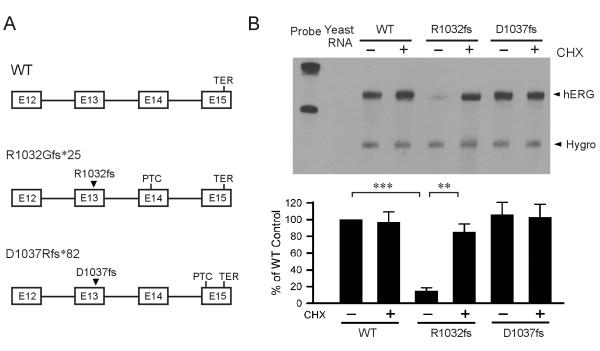
To determine the role of PTC position of LQT2 mutations in susceptibility to NMD, we studied two frameshift LQT2 mutations, R1032Gfs*25 and D1037Rfs*82. The R1032Gfs*25 and D1037Rfs*82 mutations have been previously reported as G1301fs/24 (del3094) and G1036fs/82 (insG3107-3108), respectively (Splawski et al., 2000). We used a minigene containing the hERG genomic sequence spanning exon 12 to exon 15 (Fig. 1A) (Gong et al., 2007). R1032Gfs*25 introduces a PTC in exon 14, and D1037Rfs*82 results in a PTC in the last exon (exon 15) of the hERG gene. We stably expressed the WT and LQT2 mutant minigenes in Flp-In HEK293 cells and performed RPA analysis. As shown in Fig. 1B, R1032Gfs*25, but not D1037Rfs*82, led to a reduced level of mutant mRNA compared to the WT minigene. Since NMD requires protein translation, cycloheximide (CHX) is often used to abrogate NMD. Treatment with CHX had no effect on the levels of WT and D1037Rfs*82 mutant mRNA, but significantly increased the level of R1032Gfs*25 mutant mRNA. These results suggest that position of PTC is important for susceptibility of hERG mutations to NMD.
Fig. 1.
Analysis of the R1032Gfs*25 and D1037Rfs*82 mutations using minigene constructs. (A) Diagram of minigenes showing the positions of WT termination codon (TER) and mutation-induced PTCs. (B) RPA analysis of WT, R1032Gfs*25 and D1037Rfs*82 stably expressed in HEK293 cells. Cells were treated (+) or not treated (−) with 100 μg/ml of CHX for three hours before RNA isolation. The quantitative data after normalization using the level of hygromycin resistance gene (Hygro) are plotted as percentage of WT control (n=4, ***P<0.001, **P<0.01).
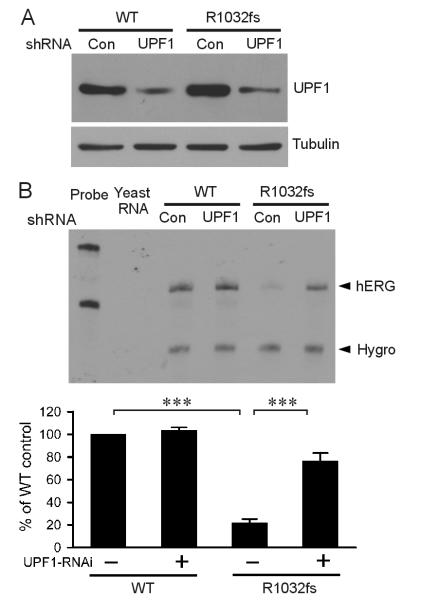
To further test the role of NMD in the reduction of R1032Gfs*25 mutant mRNA, we used RNA interference method to knock down UPF1, a key NMD factor. In these experiments, we stably expressed the hERG minigene and UPF1 shRNA in Flp-Cre HEK293 cells. Flp-Cre cells contain a single copy of the FRT site and of the loxP2272/loxP site, allowing expression of two genes of interest following Flp recombinase-mediated recombination and Cre recombinase-mediated cassette exchange reactions (Stump et al., 2012a). Knockdown of UPF1 protein was confirmed by immunoblot using anti-UPF1 antibody (Fig. 2A). RPA analysis showed that the level of the mutant hERG mRNA was significantly increased in the UPF1 shRNA-transfected cells compared to that observed in scrambled shRNA-transfected cells (Fig. 2B). These results indicated that NMD is responsible for the degradation of R1032Gfs*25 mutant mRNA.
Fig. 2.
Effect of suppression of UPF1 by RNA interference on NMD of R1032Gfs*25. A double stable mammalian expression system was used to express hERG minigenes and either scrambled (Con) or UPF1 (UPF1) shRNAs. (A) Immunoblot analysis of UPF1 protein. (B) RPA analysis of the effect of UPF1 knockdown. RPA signals were quantified, normalized to hygromycin resistance gene (Hygro), and shown as percentage of WT control (n=3, ***P<0.001).
3.2. Downstream intron is required for NMD of R1032Gfs*25
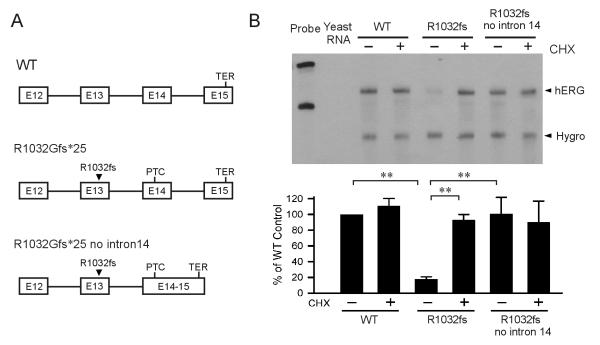
Because the R1032Gfs*25 PTC is found in exon 14 and the D1037Rfs*82 PTC occurs in exon 15, one explanation for the difference in NMD sensitivity of these two mutations is the presence of downstream intron as proposed in the EJC-dependent model. To test whether a downstream intron is required for NMD of R1032Gfs*25, we removed intron 14 from the R1032Gfs*25 minigene construct (Fig. 3A). As shown in Fig. 3B, the removal of intron 14 resulted in the restoration of R1032Gfs*25 mutant mRNA and the treatment with CHX had no effect on the level of mutant mRNA, suggesting that a downstream intron is required for NMD of hERG PTC mutations.
Fig. 3.
Effect of downstream intron on NMD of the R1032Gfs*25 mutation. (A) Diagram of the minigenes of WT, R1032Gfs*25 and R1032Gfs*25 without intron 14. (B) RPA analysis of mRNA from WT, R1032Gfs*25 and R1032Gfs*25 without intron 14 minigenes. Cells were treated (+) or not treated (−) with 100 μg/ml of CHX for three hours before RNA isolation. RPA signals were quantified, normalized to hygromycin resistance gene (Hygro), and shown as percentage of WT control (n=3 or 4, **P<0.01).
3.3. PTC position and susceptibility to NMD
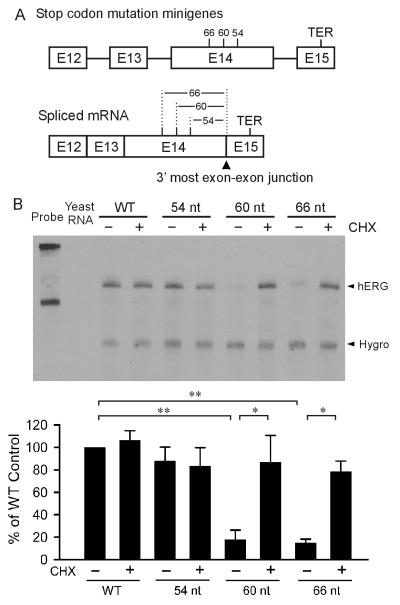
To determine the positional requirement of NMD of hERG mutations, we introduced PTC mutations into the hERG minigene at different distances from the 3’-most exon-exon junction. Three PTC mutations were introduced at the positions of 54 nt (S1092), 60 nt (S1090) and 66 nt (P1088) upstream of the 3’-most exon-exon junction in the spliced minigene (Fig. 4A). As shown in Fig. 4B, PTCs at 60 nt and 66 nt, but not 54 nt upstream of the 3’-most exon-exon junction resulted in a reduced level of mutant mRNA compared to the WT minigene. Treatment with CHX had no effect on the levels of WT and 54 nt mutant mRNA, but significantly increased the level of 60 nt and 66 nt mutant mRNA. These results suggest that PTCs at 60 nt and 66 nt, but not 54 nt upstream of the 3’-most exon-exon junction lead to the degradation of mutant mRNA by NMD.
Fig. 4.
Effect of PTC position on NMD of hERG stop codon mutations. (A) Diagram of the PTC position relative to the 3’-most exon-exon junction in the minigene and the spliced mRNA. (B) RPA analysis of mRNA from HEK293 cells stably expressing WT and mutant minigenes containing PTC mutations 54 nt, 60 nt and 66 nt upstream to the 3’-most exon-exon junction. Cells were treated (+) or not treated (−) with 100 μg/ml of CHX for three hours before RNA isolation. RPA signals were quantified, normalized to hygromycin resistance gene (Hygro), and shown as percentage of WT control (n=3 or 4, **P<0.01, *P<0.05).
3.4. The Y1078* mutation caused NMD leading to a reduction in mutant protein expression and hERG current
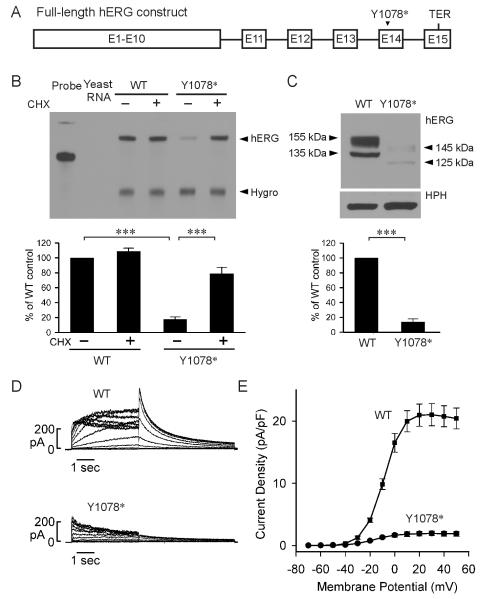
The above mutational studies indicate that the boundary that separates NMD-sensitive and NMD-resistant PTC mutations resides between residues 1090 and 1092 within exon 14, which is between 54 and 60 nt upstream of the 3’-most exon-exon junction. To further test the positional effect of LQT2-associated NMD we characterized the Y1078* mutation which represents the nearest LQT2 PTC mutation upstream of this boundary. Y1078* has been previously reported as A1077fs+X (3234delC, Kapplinger et al., 2009). The mutation introduces a PTC at residue 1078, 34 nt upstream of the boundary or 94 nt upstream of the 3’-most exon-exon junction. In order to study the effect of NMD at the mRNA as well as the protein and functional levels, we introduced the Y1078* mutation into a full-length hERG splicing-competent construct that contained hERG cDNA from exon 1 to exon 10 and hERG genomic DNA from intron 10 to the poly(A) site (Fig. 5A) (Gong et al., 2011; Zarraga et al., 2011). We stably transfected WT and Y1078* full-length hERG constructs into Flp-In HEK293 cells and performed RPA analysis. As shown in Fig. 5B, the mRNA level from Y1078* was significantly lower than the WT construct. Treatment with protein synthesis inhibitor CHX had no effect on the WT mRNA level, but significantly increased the mRNA level of Y1078*, suggesting that the Y1078* mutant mRNA was degraded by NMD.
Fig. 5.
Analysis of the Y1078* mutation in the context of full-length splicing-competent hERG construct. (A) Diagram of the full-length splicing-competent hERG construct. (B) RPA analysis of mRNA from cells expressing WT and Y1078* constructs. Cells were treated (+) or not treated (−) with 100 μg/ml of CHX for three hours before RNA isolation. RPA signals were quantified, normalized to hygromycin resistance gene (Hygro), and shown as percentage of WT control (n=3, ***P<0.001). (C) Immunoblot analysis of WT hERG and Y1078* channel proteins. Cell lysates were subjected to SDS-PAGE and immunoblotted with an antibody against the N-terminus of the hERG channel. The level of protein bands was quantified, normalized to HPH, and shown as percentage of WT control (n=3). (D) Representative currents recorded from HEK293 cells stably expressing WT and Y1078*. (E) I-V plot of tail current density measured at −50 mV following test voltages from −70 to 50 mV for WT and Y1078*.
To determine whether NMD of the Y1078* mutant mRNA would cause a decrease in hERG channel protein and function, we performed immunoblot (Fig. 5C) and patch clamp experiments (Fig. 5D, E). The WT construct expressed two protein bands at 135 kDa and 155 kDa, representing the core-glycosylated immature form and the complex-glycosylated mature form of hERG channel protein, respectively (Gong et al., 2011). The Y1078* mutant generated two protein bands at 125 kDa and 145 kDa, reflecting the deletion of 82 amino acids due to the PTC at position 1078. The level of the Y1078* mutant protein was markedly decreased. In patch clamp experiments, Y1078* caused a significant decrease in hERG current. The maximum tail current densities for WT and Y1078* were 21.0 ± 1.8 pA/pF (n=7) and 1.9 ± 0.4 pA/pF (n=11, P<0.001), respectively.
3.5. Inhibition of downstream intron splicing by antisense MO rescues functional expression of the Y1078* mutant channels
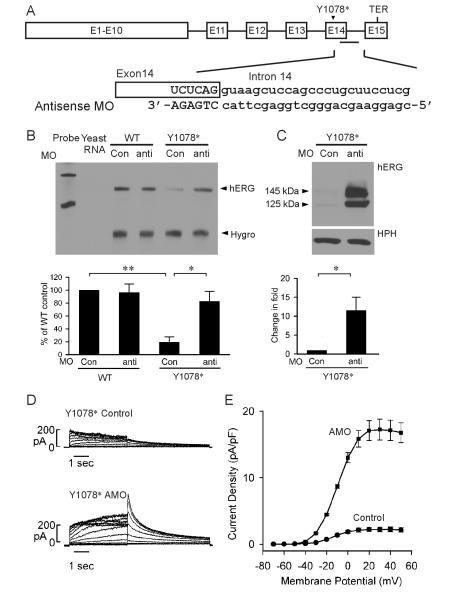
To determine whether the splicing of downstream intron is important in NMD of Y1078*, we used an antisense MO approach. MOs are nonionic DNA analogs that can form RNA-morpholino hybrids and inhibit splicing by blocking the access of splicing factors to splice sites. We used an antisense MO that targets the 5’-splice site of intron 14 to inhibit the splicing of intron 14 (Fig. 6A) (Gong et al., 2011). In these experiments, HEK293 cells stably expressing WT and Y1078* full-length hERG splicing-competent constructs were treated with 5 μM antisense or invert MO for 48 hours. As shown in Fig. 6B, treatment with antisense MO had no effect on the WT mRNA level, but resulted in a significant increase in the level of the Y1078* mutant mRNA compared to the invert MO. These results suggest that inhibition of intron 14 splicing by antisense MO can prevent NMD of the Y1078* mutant mRNA.
Fig. 6.
Inhibition of intron 14 splicing by antisense MO prevents NMD of Y1078*. Cells stably expressing the WT and Y1078* full-length splicing-competent hERG construct were treated with 5 μM invert (Con) or antisense MO for 48 hours. (A) Diagram of the full-length splicing-competent hERG construct and the sequences of antisense MO. (B) RPA analysis of the effect of antisense MO. RPA signals were quantified, normalized to hygromycin resistance gene (Hygro) and shown as percentage of WT control (n=3, **P<0.01, *P<0.05). (C) Immunoblot analysis of the effect of antisense MO. Cell lysates were subjected to SDS-PAGE and immunoblotted with anti-hERG and anti-HPH antibodies. The level of protein bands was quantified, normalized to HPH, and shown as fold changes in the presence of antisense MO (n=4). (D) Representative currents recorded from HEK293 cells stably expressing Y1078* following treatment with 5-μM invert (Control) or antisense MO (AMO) for 48 hours. (E) I-V plot of hERG tail current density measured at -50 mV following test voltages from -70 to 50 mV for WT and Y1078* in the presence of invert or antisense MO.
To demonstrate that inhibition of NMD by antisense MO also results in rescue of Y1078* at protein level, we analyzed hERG protein expression by immunoblot (Fig. 6C). Treatment with antisense MO significantly increased the level of the Y1078* mutant protein compared with the invert MO. These results suggest that inhibition of NMD by antisense MO restores the expression of the Y1078* mutant protein. To study functional rescue of Y1078*, we performed patch clamp recordings (Fig. 6D, E). hERG current was significantly increased by the antisense MO treatment as compared to that observed in invert MO treated cells. The maximum tail current densities for Y1078* treated with invert and antisense MO were 2.2 ± 0.4 pA/pF (n=10) and 17.2 ± 1.6 pA/pF (n=11, P<0.001), respectively. These patch clamp results demonstrate that inhibition of NMD by antisense MO can rescue functional expression of the Y1078* mutation.
4. Discussion
Our present study defines the positional requirements for susceptibility of LQT2 mutations to NMD. Several lines of evidence were found to support an EJC-dependent model for recognition of NMD substrates in hERG PTC mutations. First, NMD occurs in LQT2 mutations that introduce a PTC in the penultimate exon, but not in the last exon. Second, a downstream intron is required for NMD of the R1032Gfs*25 mutation. Third, degradation of PTC-containing mRNA by NMD requires the presence of PTCs located >54-60 nt upstream of the 3'-most exon-exon junction. Fourth, blocking of downstream intron splicing inhibits NMD of the Y1078* mutation. The fact that blocking downstream intron splicing abrogates NMD of Y1078* strongly suggests that the physical distance between the termination codon and the poly(A)-binding protein C1, as proposed by the EJC-independent model, is a not a crucial determinant for NMD of LQT2 mutations. We also show that suppression of NMD by antisense MO leads to the rescue of Y1078* channel expression and hERG current. These findings suggest that EJC-dependent NMD can be prevented by targeting downstream intron splicing using antisense MO, resulting in the rescue of functional expression of mutant proteins.
As >30% of reported LQT2 mutations are nonsense and frameshift mutations, identification of the positional requirements for NMD in hERG is of significant importance in predicting pathogenic mechanism based on the PTC location. Our results suggest that the 3’-boundary for NMD of hERG PTC-containing mRNA is between residues S1090 and P1092 in exon 14. Among the LQT2 PTC mutations identified so far, Y1078* is the nearest PTC mutation upstream of this boundary and D1037Rfs*82 is the nearest PTC mutation downstream of this boundary. We show that Y1078*, but not D1037Rfs*82 is sensitive to NMD. Thus, LQT2 mutations that introduce PTCs upstream of this 3’-boundary are potential targets of NMD, while those harboring a PTC downstream of this boundary are expected to escape NMD.
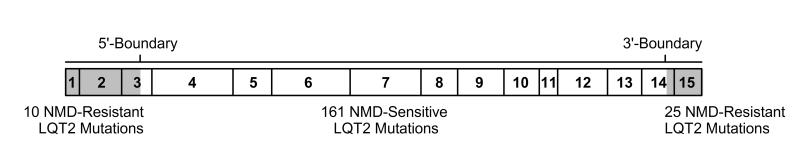
Our recent studies have indicated that LQT2 nonsense mutations near the translational start site of hERG can escape NMD due to translation reinitiation at in-frame, downstream methionine codons (Stump et al., 2012b; 2013). Translation of hERG transcripts containing the C39* or C44* nonsense mutations is reinitiated at Met60 and resulted in the generation of N-terminally truncated, trafficking defective channels. hERG transcripts containing the Q81* mutation were translated from Met124 and resulted in the expression of truncated, channels with accelerated deactivation kinetics. Although the hERG gene contains many in frame methionine residues, only those with a strong Kozak sequence near 5’-region of the transcript may be used for translation reinitiation (Kozak, 2001). We have shown that Met60, Met124, Met133 and Met137 are capable of reinitiating translation (Stump et al., 2012b; 2013). Translation reinitiation is compatible with an EJC-dependent model of NMD in which the EJC-complexes are displaced by the ribosome before they can associate with NMD machinery. The LQT2 frameshift mutation A142Gfs*3, which is located downstream of these methionine residues, fails to reinitiate translation and is sensitive to NMD (Stump et al., 2012b). Thus, the 5’-boundary for NMD of hERG PTC-containing mRNA is likely at M137. Transcripts containing PTCs upstream of this 5’-boundary may be translated into N-terminally truncated hERG channels, while those occurring downstream are predicted be degraded by NMD. In support of the proposed 5’- and 3’-boundaries governing NMD susceptibility, LQT2 PTC mutations located within exons 3, 6, 8, 12, and 13 have previously been shown to be sensitive to NMD (Gao et al., 2013; Gong et al., 2007; Stump et al., 2012b; Sun et al., 2009; Zarraga et al., 2011). A review of the literature revealed that 161 out of 196 reported LQT2 nonsense and frameshift mutations are predicted to be degraded by NMD (Fig. 7, Supplemental Table S1).
Fig. 7.
Schematic of hERG mRNA (exons 1-15) and the 5’ and 3’ boundaries that govern the susceptibility of LQT2 frameshift and nonsense mutations to NMD. The relative position and number of NMD-resistant and NMD-sensitive LQT2 PTC mutations are shown.
In this study, we used the full-length splicing-competent hERG construct to characterize NMD at mRNA, protein and functional levels. We showed that the Y1078* mutant mRNA was degraded leading to a marked decrease in the expression of hERG channel protein and a nearly complete loss of hERG current. Our present work highlights the importance of using splicing-competent hERG constructs in the characterization of LQT2 PTC mutations. To date, several LQT2 nonsense and frameshift mutations have been analyzed using cDNA constructs under the assumption that PTC mutations would generate truncated hERG channels (Choe et al., 2006; Christé et al., 2008; Gong et al., 2004; Li et al., 1997; Nof et al., 2010; Paulussen et al., 2005; Trudeau et al., 2011). However, based on the 3’-boundary identified in the present study, all of these PTC mutations are expected targets of NMD. Because NMD of hERG PTC-bearing mRNA requires splicing of a downstream intron, the effect of NMD is not observed in the context of cDNA constructs. It is noted that most of these nonsense and frameshift mutations exhibit a dominant-negative effect on WT hERG channels in these cDNA-based studies (Choe et al., 2006; Christé et al., 2008; Gong et al., 2004; Li et al., 1997; Nof et al., 2010; Paulussen et al., 2005; Trudeau et al., 2011). However, if these studies were performed using an NMD-competent construct, the PTC-containing mRNA would be degraded and the dominant-negative effect caused by truncated proteins would be minimized.
In conclusion, our results identify the positional requirements that determine the susceptibility of hERG PTC mutations to NMD. Our data strongly support an EJC-dependent mechanism of NMD and indicate that the majority of reported LQT2 nonsense and frameshift mutations are potential targets of NMD.
Supplementary Material
Highlights.
Recognition of PTC-containing hERG transcripts by NMD machinery is studied.
Positional requirements governing NMD susceptibility of LQT2 PTCs are identified.
LQT2 PTC recognition and elimination by NMD require downstream intron.
LQT2 PTCs >54-60 nt upstream of the 3'-most exon-exon junction trigger NMD.
The majority of reported LQT2 nonsense and frameshift mutations are targets of NMD.
Acknowledgments
This study was supported in part by the National Institutes of Health grant HL-68854 and the American Heart Association Established Investigator Award. MRS is supported by National Institutes of Health training grant T32HL094294.
Abbreviations
- CHX
cycloheximide
- EJC
exon junction complex
- hERG
human ether-a-go-go-related gene
- HPH
hygromycin B phosphotransferase
- LQT2
long QT syndrome type 2
- MO
morpholino oligonucleotide
- NMD
nonsense-mediated mRNA decay
- nt
nucleotide
- PTC
premature termination codon
- RPA
RNase protection assay
- shRNA
short hairpin RNA
- UPF1
up-frameshift protein 1
- WT
wild-type.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bateman JF, Wilson R, Freddi S, Lamandé SR, Savarirayan R. Mutations of COL10A1 in Schmid metaphyseal chondrodysplasia. Hum. Mutat. 2005;25:525–534. doi: 10.1002/humu.20183. [DOI] [PubMed] [Google Scholar]
- Bhuiyan ZA, et al. Recurrent intrauterine fetal loss due to near absence of HERG: clinical and functional characterization of a homozygous nonsense HERG Q1070X mutation. Heart Rhythm. 2008;5:553–561. doi: 10.1016/j.hrthm.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler M, Steiner S, Mohn F, Paillusson A, Mühlemann O. EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3' UTR length. Nat. Struct. Mol. Biol. 2006;13:462–464. doi: 10.1038/nsmb1081. [DOI] [PubMed] [Google Scholar]
- Carter MS, Li S, Wilkinson MF. A splicing-dependent regulatory mechanism that detects translation signals. Embo J. 1996;15:5965–5975. [PMC free article] [PubMed] [Google Scholar]
- Choe C-U, et al. C-terminal HERG (LQT2) mutations disrupt IKr channel regulation through 14-3-3epsilon. Hum. Mol. Genet. 2006;15:2888–2902. doi: 10.1093/hmg/ddl230. [DOI] [PubMed] [Google Scholar]
- Christé G, et al. A new C-terminal hERG mutation A915fs+47X associated with symptomatic LQT2 and auditory-trigger syncope. Heart Rhythm. 2008;5:1577–1586. doi: 10.1016/j.hrthm.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- Eberle AB, Stalder L, Mathys H, Orozco RZ, Mühlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3' untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhang P, Li X-B, Wu C-C, Guo J-H. A novel deletion-frameshift mutation in the S1 region of HERG gene in a Chinese family with long QT syndrome. Chin. Med. J. 2013;126:3093–3096. [PubMed] [Google Scholar]
- Gong Q, Keeney DR, Robinson JC, Zhou Z. Defective assembly and trafficking of mutant HERG channels with C-terminal truncations in long QT syndrome. J. Mol. Cell. Cardiol. 2004;37:1225–1233. doi: 10.1016/j.yjmcc.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Gong Q, Stump MR, Dunn AR, Deng V, Zhou Z. Alternative splicing and polyadenylation contribute to the generation of hERG1 C-terminal isoforms. J. Biol. Chem. 2010;285:32233–32241. doi: 10.1074/jbc.M109.095695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q, Stump MR, Zhou Z. Inhibition of nonsense-mediated mRNA decay by antisense morpholino oligonucleotides restores functional expression of hERG nonsense and frameshift mutations in long-QT syndrome. J. Mol. Cell. Cardiol. 2011;50:223–229. doi: 10.1016/j.yjmcc.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q, Zhang L, Vincent GM, Horne BD, Zhou Z. Nonsense mutations in hERG cause a decrease in mutant mRNA transcripts by nonsense-mediated mRNA decay in human long-QT syndrome. Circulation. 2007;116:17–24. doi: 10.1161/CIRCULATIONAHA.107.708818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapplinger JD, et al. Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION long QT syndrome genetic test. Heart Rhythm. 2009;6:1297–1303. doi: 10.1016/j.hrthm.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajavi M, Inoue K, Lupski JR. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur. J. Hum. Genet. 2006;14:1074–1081. doi: 10.1038/sj.ejhg.5201649. [DOI] [PubMed] [Google Scholar]
- Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmiak HA, Maquat LE. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol Med. 2006;12:306–316. doi: 10.1016/j.molmed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Li X, Xu J, Li M. The human delta1261 mutation of the HERG potassium channel results in a truncated protein that contains a subunit interaction domain and decreases the channel expression. J. Biol. Chem. 1997;272:705–708. doi: 10.1074/jbc.272.2.705. [DOI] [PubMed] [Google Scholar]
- Lieve KV, et al. Results of genetic testing in 855 consecutive unrelated patients referred for long QT syndrome in a clinical laboratory. Genet Test Mol Biomarkers. 2013;17:553–561. doi: 10.1089/gtmb.2012.0118. [DOI] [PubMed] [Google Scholar]
- Nagaoka I, et al. Mutation site dependent variability of cardiac events in Japanese LQT2 form of congenital long-QT syndrome. Circ. J. 2008;72:694–699. doi: 10.1253/circj.72.694. [DOI] [PubMed] [Google Scholar]
- Napolitano C, et al. Genetic testing in the long QT syndrome: development and validation of an efficient approach to genotyping in clinical practice. JAMA. 2005;294:2975–2980. doi: 10.1001/jama.294.23.2975. [DOI] [PubMed] [Google Scholar]
- Nof E, et al. A common single nucleotide polymorphism can exacerbate long-QT type 2 syndrome leading to sudden infant death. Circ Cardiovasc Genet. 2010;3:199–206. doi: 10.1161/CIRCGENETICS.109.898569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulussen ADC, et al. HERG mutation predicts short QT based on channel kinetics but causes long QT by heterotetrameric trafficking deficiency. Cardiovasc. Res. 2005;67:467–475. doi: 10.1016/j.cardiores.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Rajavel KS, Neufeld EF. Nonsense-mediated decay of human HEXA mRNA. Mol. Cell. Biol. 2001;21:5512–5519. doi: 10.1128/MCB.21.16.5512-5519.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splawski I, et al. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102:1178–1185. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- Stump MR, Gong Q, Zhou Z. Isoform-Specific Dominant-Negative Effects Associated with hERG1 G628S Mutation in Long QT Syndrome. PLoS ONE. 2012a;7:e42552. doi: 10.1371/journal.pone.0042552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump MR, Gong Q, Packer JD, Zhou Z. Early LQT2 nonsense mutation generates N-terminally truncated hERG channels with altered gating properties by the reinitiation of translation. J. Mol. Cell. Cardiol. 2012b;53:725–733. doi: 10.1016/j.yjmcc.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump MR, Gong Q, Zhou Z. LQT2 Nonsense mutations generate trafficking defective NH2-terminally truncated channels by the reinitiation of translation. Am. J. Physiol. Heart Circ. Physiol. 2013;305:H1397–H1404. doi: 10.1152/ajpheart.00304.2013. [PubMed: 23997099] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, et al. A novel nonsense mutation Y652X in the S6/pore region of human ether-go-go gene found in a long QT syndrome family. Scand. Cardiovasc. J. 2009;43:181–186. doi: 10.1080/14017430802582610. [DOI] [PubMed] [Google Scholar]
- Tester DJ, Will ML, Haglund CM, Ackerman MJ. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2005;2:507–517. doi: 10.1016/j.hrthm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- Trudeau MC, Leung LM, Roti ER, Robertson GA. hERG1a N-terminal eag domain-containing polypeptides regulate homomeric hERG1b and heteromeric hERG1a/hERG1b channels: a possible mechanism for long QT syndrome. J. Gen. Physiol. 2011;138:581–592. doi: 10.1085/jgp.201110683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarraga IG, Zhang L, Stump MR, Gong Q, Vincent GM, Zhou Z. Nonsense-mediated mRNA decay caused by a frameshift mutation in a large kindred of type 2 long QT syndrome. Heart Rhythm. 2011;8:1200–1206. doi: 10.1016/j.hrthm.2011.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.