Summary
Coreceptor CD4 and CD8αβ double negative (DN) TCRαβ+ intraepithelial T cells, although numerous, have been greatly overlooked and their contribution to the immune response is not known. Here we used T cell receptor (TCR) sequencing of single cells combined with retrogenic expression of TCRs, to study the fate and the major histocompatibility complex (MHC) restriction of DN TCRαβ+ intraepithelial T cells. The data show that commitment of thymic precursors to the DN TCRαβ+ lineage is imprinted by their TCR specificity. Moreover, the TCRs they express display a diverse and unusual pattern of MHC restriction that is non-overlapping with that of CD4+ or CD8αβ+ T cells, indicating that they sense antigens that are not recognized by the conventional T cell subsets. The new insights indicate that DN TCRαβ+ T cells form a third lineage of TCRαβ T lymphocytes expressing a variable TCR repertoire, which serve non-redundant immune functions.
Introduction
Cellular immunity mediated by the T-cell pool is essential for responses against invading pathogens and for elimination of transformed cells. Separate T cell subsets can be characterized by their T cell receptors (TCRs) (αβ and γδ), their antigen specificity and function. TCRαβ+ T cells expressing either the CD8αβ or CD4 coreceptor recognize antigens presented by major histocompatibility complex (MHC) class I or class II molecules, respectively (Davis and Bjorkman, 1988) and they represent the main T cell pools in peripheral lymphoid organs. The TCRαβ+ T cell compartment contains also other subsets that are phenotypically and functionally different from CD4+ and CD8αβ+ T cells and are often highly represented in particular tissues. For instance, the natural killer T cells (NKT) or the coreceptor CD4− and CD8αβ-double negative (DN or coreceptor negative) TCRαβ+ intraepithelial T cells can represent up to one fourth of the total T cell pool of the liver or the epithelium of the small intestine, respectively (Abadie et al., 2012; Fang et al., 2010). NKT cells have been clearly defined as a separate lineage of T cells that are able to recognize self or foreign antigens presented by CD1d molecules and elicit a protective or harmful role in microbial infections, cancers, autoimmune or allergic diseases (Brennan et al., 2013; Engel and Kronenberg, 2012). On the contrary, the lineage affiliation, the MHC specificity and function of DN TCRαβ+ intraepithelial T cells remain enigmatic (Lambolez et al., 2007).
DN TCRαβ+ intraepithelial T cells are non-circulating T lymphocytes (Guy-Grand et al., 2013) that comprise about one third of the TCRαβ+ cells in the intestinal epithelium. They exhibit unusual features compared to conventional T cells, including their phenotype, TCR repertoire, and thymic selection pathway (Abadie et al., 2012; Cheroutre et al., 2011; Lambolez et al., 2007; Pobezinsky et al., 2012). Indeed, DN TCRαβ+ intraepithelial T cells lack expression of molecules typically expressed by mature CD8αβ+ or CD4+ T cells, including CD5, CD28, and Thy1 (Lefrancois, 1991; Ohteki and MacDonald, 1993; Shires et al., 2001) whereas they express natural killer receptors such as Ly49 family members, CD314 or CD244 (Denning et al., 2007; Guy-Grand et al., 1996; Yamagata et al., 2004). In addition, like other T cell subsets in the intestine, most of the DN TCRαβ+ intraepithelial T cells acquire expression of CD69 and CD8αβ, which are hallmark features of their activated phenotype (Cheroutre and Lambolez, 2008). DN TCRαβ+ intraepithelial T cells were historically called CD8αβ+ TCRαβ+ T cells (Guy-Grand et al., 1991), however, unlike CD4 and CD8αβ, CD8αβ does not function as a TCR coreceptor on these cells (Cheroutre and Lambolez, 2008). Precursors of DN TCRαβ+ intraepithelial T cells are found in the thymus where they undergo agonist positive selection (Gangadharan et al., 2006; Pobezinsky et al., 2012; Stritesky et al., 2012; Yamagata et al., 2004), meaning that the TCR must engage self-ligands with relatively high affinity, which results in the generation of post-selected DN TCRαβ+ thymocytes (Gangadharan et al., 2006). The latter exit the thymus and reside mainly within the epithelium of the small intestine (Gangadharan et al., 2006; Pobezinsky et al., 2012). As a consequence of agonist selection, DN TCRαβ+ intraepithelial T cells have an oligoclonal TCR repertoire enriched for self-reactive clones (Regnault et al., 1994; Rocha et al., 1991). Despite a myriad of studies focused on the development of these cells, the characteristics that determine their fate and their MHC restriction remain unknown.
Previous analyses of mouse strains deficient in various major histocompatibility complex (MHC) molecules indicated that the development of these cells is either not impaired, or only moderately impaired, in the absence of MHC class II or in mice deficient for one of the MHC class I molecules, such as H-2K, and -D, CD1d, Thymic Leukemia antigen (TL) or Qa-2 (Das et al., 2000; Gapin et al., 1999; Park et al., 1999). Deficiency of the transporter associated with antigen presentation (TAP), which leads to reduced peptide loading and classical MHC class I expression on the cell surface, impairs CD8αβ+ T cell development. Tap1−/− mice, however, still contain, albeit in reduced numbers, DN TCRαβ+ intraepithelial T cells (Fujiura et al., 1996; Sydora et al., 1996). In contrast, the accumulation of DN TCRαβ+ intraepithelial T cells is completely abolished in mice deficient for β2-microglobulin (B2m−/−), suggesting that their development and/or survival might be controlled by a non-classical β2m-dependent MHC class I molecule, like CD1d-reactive iNKT cells (Engel and Kronenberg, 2012) or MR1-reactive mucosal associated invariant T (MAIT) cells (Treiner et al., 2003). Alternatively, they might express TCRs with diverse specificities restricted by different β2m-dependent classical and/or non-classical MHC class I molecules.
To determine the MHC restriction of DN TCRαβ+ intraepithelial T cells and to shed light on their antigen specificity, we sequenced and cloned TCRs from DN TCRαβ+ intraepithelial T cells. All TCRs were variable and therefore, each cloned TCR was transfected retrovirally into bone marrow (BM) cells and used to generate BM chimera on MHC-sufficient or on various MHC-deficient backgrounds. Surprisingly, all precursor cells expressing a variable cloned TCR gave rise exclusively to DN TCRαβ+ T cells in the periphery, indicating that the TCRs in part imprint commitment to this unique T cell lineage and that the TCR repertoire is non-overlapping with that of CD4+ or CD8αβ+ T cells. We further demonstrate that the TCR specificity of DN TCRαβ+ intraepithelial T cells is not only variable but also unique and that DN TCRαβ+ intraepithelial T cell-derived TCRs are not restricted by a single type of MHC class I molecules, but instead they display unusual specificities, either for β2m-dependent non-classical (non polymorphic) class I MHC molecules, or for Kb or Db classical (polymorphic) class I molecules. Interestingly, in the case of H-2Kb or -Db restriction, the function of DN TCRs did not depend on TAP, suggesting that the antigens recognized by DN TCRαβ+ intraepithelial T cells are different from those presented to MHC class I restricted CD8αβ+ T cells. Overall, the results show, for the first time, that DN TCRαβ+ intraepithelial T cells represent a unique T cell lineage with divers MHC restriction. In addition, this implies that the antigen repertoire sensed by these cells is distinct from that of the CD4+ or CD8αβ+ T cell subsets.
Results
Cloned TCRs are properly expressed in vitro and in vivo
To evaluate the functional potential of TCRs expressed by DN TCRαβ+ intraepithelial T cells, we first sequenced TCRs from naturally arising DN TCRαβ+ T cells isolated from the epithelium of the small intestine. To this end, the TCRs were either from a DN TCRαβ+ intraepithelial T cell-derived hybridoma or from freshly isolated DN TCRαβ+ intraepithelial T cells enriched for TRBV16 and TRAV9N-3 co-expression (See methods and Table S1). We cloned five individual TCRs named: TCR-Mathilde, -Lucienne, -Diego, -Ronja and -Ninon. The TCR α and β chain sequence of each TCR was diverse and did not show any unusual features with regard to CDR3 length as compared to TCRs expressed by CD4+ or CD8αβ+ TCRαβ+ cells (Rock et al., 1994) (See Supplemental Table S1).
To express the cloned TCRs in vivo, we selected a retroviral transduction system for several reasons. First, this strategy has been proven reliable (Alli et al., 2008; Arnold et al., 2004; Hofstetter et al., 2012; Lathrop et al., 2011) and it allowed us to compare several cloned TCRs in parallel. More importantly, each TCR could be analysed directly in several recipients with various genetic deficiencies in antigen presenting molecules without the need for laborious backcrossing of TCR transgenes. Individual TCRα and TCRβ pairs were linked by a 2A peptide and inserted as multicistronic constructs into an IRES-GFP-containing retroviral vector (Figure S1A) (Bettini et al., 2012; Holst et al., 2006). Expression of the cloned TCRs was tested in vitro using retroviral transfection of 58α−β− cells (Figure S1B and S1C). The 2A peptide was cleaved in all expressed TCRs as evidenced by the presence of a 37kDa product. This cleavage enabled efficient pairing of the α- and β-chains and the subsequent TCRαβ expression on the cell surface. To evaluate the accuracy of the retrogenic system in vivo, a TCR cloned from a CD8αβ+ TCRαβ+ intraepithelial T cell, was used to generate and analyze BM chimeras on a Rag1−/− background (Figure S1D and S1E). Consistent with development within the CD8αβ+ TCRαβ+ lineage, the control chimera contained normal subsets of thymocytes, as well as, naive CD8αβ+ TCRαβ+ T cells in the periphery, but no CD4+ or DN TCRαβ+ T cells. This was consistent with results from retrogenic chimera expressing well-described TCRs, such as the OT I or HY TCR in absence of cognate antigen (Holst et al., 2006).
The TCR imprints the DN TCRαβ+T cell lineage
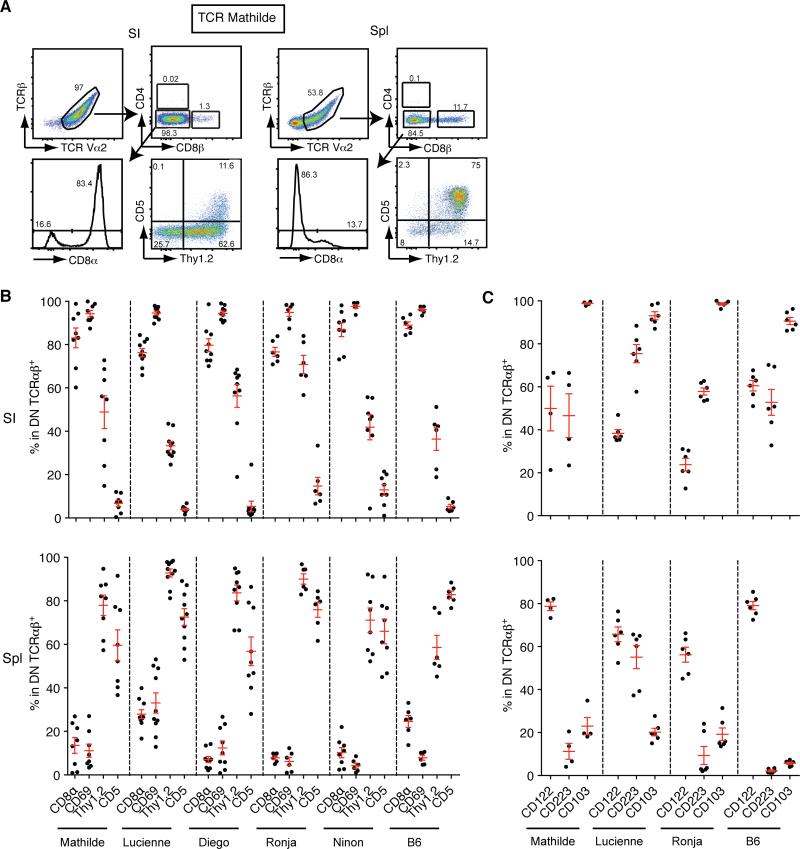
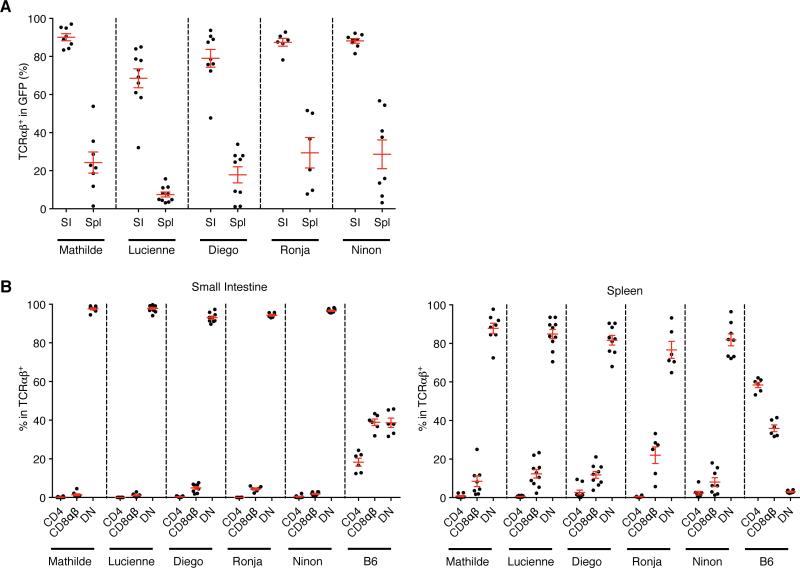
Various TCR retrogenic BM chimeras on a Rag1−/− background each expressing a single TCR isolated from an individual DN TCRαβ+ intraepithelial T cell (Figure 2A for gating strategy) were generated and GFP+ donor BM derived cells in both epithelium of the small intestine (SI) and spleen were analyzed. The presence of mature TCRαβ+ T cells within the GFP+ subset could be detected in both compartments with a lower frequency in the spleen as compared to the SI (Figure 1A). We then evaluated the lineage fate by the coreceptor expression or absence thereof. Interestingly, the GFP+ TCRαβ+ T cells lacked expression of both CD4 and CD8αβ coreceptors (Figure 1B), indicating that the GFP+ BM progenitors only gave rise to mature DN TCRαβ+ T cells. Furthermore, GFP+ DN TCRαβ+ intraepithelial T cells were predominantly CD8αβ+CD69+Thy1.2lo/intCD5− (Figure 2B) and many of them expressed CD122, Lag-3 and CD103 (Figure 2C). This is consistent with the typical phenotype of DN TCRαβ+ intraepithelial T cells found in wild type mice (Cheroutre et al., 2011; Denning et al., 2007; Kilshaw and Murant, 1990; Lambolez et al., 2007). In contrast the spleen GFP+ DN TCRαβ+ T cells expressed a phenotype typical of lymphoid resident lymphocytes, lacking expression of the activation markers, CD8αβ and CD69 (Figure 2B) and low in Lag-3 or CD103 expression although most of them did express CD122 (Figure 2C). The expression of CD122 on DN TCRαβ+ cells, which confers them the ability to respond to interleukin-2 (IL-2) or (IL-15) in vivo, agrees with the expression reported for lymph node DN TCRαβ+ T cells or those generated in TCR transgenic models (Mixter et al., 1999). The presence of mature DN TCRαβ+ T cells, arising from precursors expressing the same TCR, in both intestinal epithelium and spleen, suggests that these DN TCRαβ+ T cells share the same origin and lineage. Indeed, DN TCRαβ+ splenocytes isolated from retrogenic mice and adoptively transferred into Rag1−/− recipients were able to migrate to the small intestine where they acquired the typical DN TCRαβ+ intraepithelial T cell phenotype (Figure S2). Further investigation will be required to elucidate the developmental kinetics between these two mature DN TCRαβ T cell subsets. Finally, BM cells expressing cloned TCRs developed in a competitive setting together with WT C57BL/6 (B6) BM cells and gave rise to DN TCRαβ+ intraepithelial T cells with an identical phenotype as those that developed in the TCR retrogenic BM chimeras on non-competitive setting (Figure S3). These data not only confirm that the TCRs expressed by DN TCRαβ+ intraepithelial T cells imprint the coreceptor-negative lineage fate, but they also exclude a potential artifact induced by a monoclonal lymphopenic setting. All together the results show, for the first time, that the TCR repertoire of DN TCRαβ+ T cells is unique and non-overlapping with that of CD4+ or CD8αβ+ T cells. In addition, the data also imply that the nature of the TCRs expressed by DN TCRαβ+ T cells directs the commitment and developmental fate of their precursors into this unique lineage.
Figure 2. The DN TCRαβ+ T cell phenotype is identical for each TCR clone tested.
(A) Flow cytometry analysis of GFP+ T cells from the small intestinal epithelium (SI) (left panels) and the spleen (Spl) (right panels) from Rag1−/− chimera expressing TCR Mathilde analysed 28 days post-reconstitution. Representative plots are shown from four independent experiments (n=8). (B) Graphs show the expression of CD8α, CD69, Thy1.2 and CD5 on GFP+ DN TCRαβ+ cells isolated from the small intestinal epithelium or spleen from Rag1−/− chimeras expressing TCR Mathilde, Lucienne, Diego, Ronja and Ninon as described in a. For comparison, the percentage of expression of each marker is shown for DN TCRαβ+ αGalCer-CD1d-tetramer − T cells in B6. The number of mice analysed for each construct is provided in figure legend 1C. (C) Graphs show the expression of CD122, CD223 and CD103 on GFP+ DN TCRαβ+ T cells isolated from the small intestinal epithelium or spleen from Rag1−/− chimeras expressing TCR Mathilde, Lucienne, and Ronja 28 days post reconstitution. Data points are shown for each mouse with mean plus s.e.m. from at least two independent experiments (TCR Mathilde (n=4), Lucienne (n=6) and Ronja (n=6)). For comparison, the percentage of expression of each marker is shown for DN TCRαβ+ αGalCer-CD1d-tetramer − T cells in C57BL/6. (See also Figure S2 and S3)
Figure 1. The T cell receptor dictates commitment to the DN TCRαβ+ T cell lineage.
Flow cytometry analysis of GFP+ cells isolated from the small intestinal epithelium (SI) and the spleen (Spl) from Rag1−/− BM chimera expressing TCR Mathilde, Lucienne, Diego, Ronja or Ninon analysed 28 days post-reconstitution. (A) Graph shows the percentage of T cells among GFP+ cells isolated from the small intestinal epithelium (SI) or spleen (Spl). (B) Panels represent the percentage of CD4+, CD8αβ+ or CD4− CD8αβ− (DN, double negative) cells gated on GFP+ TCRαβ+ T cells isolated from the small intestine or spleen as in B. For comparison, the percentage of CD4+, CD8αβ+ or DN cells gated on TCRαβ+ αGalCer-CD1d-tetramer − T cells in B6 is shown. Data points are shown for each mouse, with mean plus s.e.m., from at least three independent experiments (TCR Mathilde (n=8), Lucienne (n=10), Diego (n=9), Ronja (n=6), Ninon (n=8) and C57BL/6 (n=6)). (See also Figure S1)
Thymic precursors of DN TCRαβ+ intraepithelial T cells
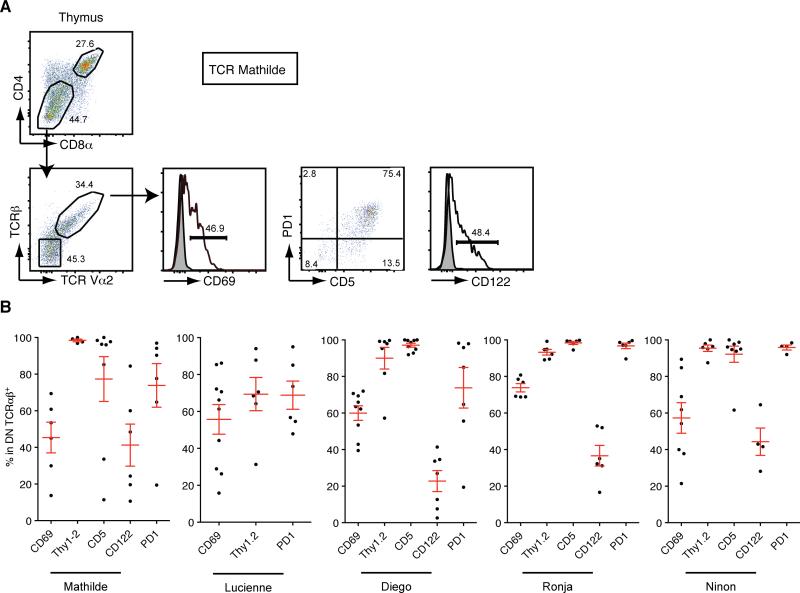
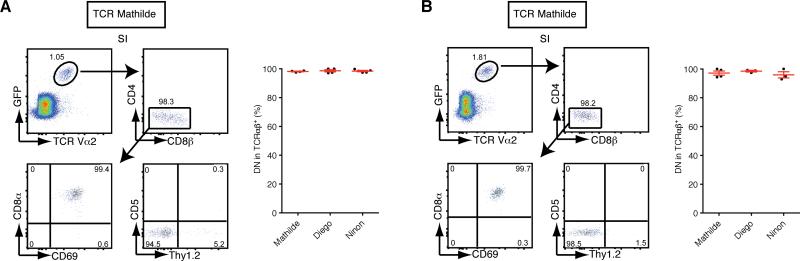
To identify the thymic precursor cells, we analyzed the thymus of the various TCR BM chimeras. In all cases, a defined population of GFPhi thymocytes that were DN or CD4loCD8lo cells expressed the cloned TCR (Figure 3A). Furthermore, these DN TCRαβ+ thymocytes were Thy1.2+CD5+CD122+PD-1+ (Figure 3A and 3B). This is consistent with the phenotype previously defined for mature thymic precursor cells of DN CD8αα+ TCRαβ+ intraepithelial T cells from transgenic mice expressing both a TCR isolated from conventional T cells and the cognate antigen or from wild type mice (Gangadharan et al., 2006; Pobezinsky et al., 2012; Yamagata et al., 2004). In addition, these DN TCRαβ+ thymocytes expressed CD69, indicating that they were post-selected cells. As previously reported, the amount of CD69 expression was modest (Pobezinsky et al., 2012 and supplemental Figure S4). To test if the thymus was the source of precursors for the DN TCRαβ+ intraepithelial T cells, we grafted the thymus tissue from retrogenic mice under the kidney capsule of Rag1−/− recipients. Five weeks after engraftment, GFP+ donor cells were found almost exclusively in the epithelium of the small intestine and they expressed the typical features of DN TCRαβ+ intraepithelial T cells (Figure 4A). These results demonstrate that, similar to the mainstream CD4+ and CD8αβ+ T cells, precursors of the DN TCRαβ+ lineage T cells are present in the thymus. We also adoptively transferred sorted DN TCRαβ+ thymocytes isolated from retrogenic mice into Rag1−/− hosts and detected GFP+ DN TCRαβ+ T cells in the intestine 5 weeks after the initial transfer (Figure 4B). These results support the notion that DN TCRαβ+ intraepithelial T cells originate from the thymus (Gangadharan et al., 2006; Pobezinsky et al., 2012).
Figure 3. DN TCRαβ+ thymocyte precursors have a mature post-selected phenotype.
(A) Representative flow cytometry analysis of GFPhi thymocytes from chimeras expressing TCR Mathilde analyzed 28 days post reconstitution. CD4 and CD8α profile of GFPhi cells (top panel); TCRβ and TCR Vα2 profile of gated DN thymocytes (bottom left). Expression of CD69, PD1, CD5 and CD122 on gated DN TCRαβ+ thymocytes is depicted (bottom middle and left panels). (B) Graphs represent percentage of GFP+ DN TCRαβ+ thymocytes expressing CD69, Thy1.2, CD5, CD122 and PD1 in chimeras expressing TCR Mathilde (n=8), TCR Lucienne (n=10), TCR Diego (n=9), TCR Ronja (n=6) or TCR Ninon (n=8). Data points are shown for each mouse, with mean plus s.e.m. from at least two independent experiments with a minimum of two mice per experiment. (See also Figure S4)
Figure 4. Precursors of DN TCRαβ+ intraepithelial T cells are present in the thymus.
(A) Thymi from retrogenic donor mice were grafted under kidney capsules of Rag1−/− mice. Representative flow cytometry plots of GFP+ intraepithelial T cells isolated from the small intestine for TCR Mathilde analysed five weeks post transplantation. The graph represents the percentage of DN TCRαβ+ cells in gated GFP+ intraepithelial T cells for each TCR clone. Data points are shown for each mouse, with mean plus s.e.m., from two independent experiments for TCR Mathilde (n=4), TCR Diego (n=4) and TCR Ninon (n=5). (B) Sorted GFP+ DN TCR+ thymocytes isolated from retrogenic donors mice were adoptively transferred to Rag1−/− recipients. Representative flow cytometry plots of GFP+ intraepithelial T cells from chimeras injected with GFP+ DN TCRαβ+ Mathilde thymocytes analysed five weeks post-transfer. The graph represents the percentage of DN TCRαβ+ cells in gated GFP+ intraepithelial T cells for each TCR clone. Data points are shown for each mouse, with mean plus s.e.m. from two independent experiments for TCR Mathilde (n=4), TCR Diego (n=3) and TCR Ninon (n=3).
DN TCRαβ+ intraepithelial T cells display variable MHC specificity
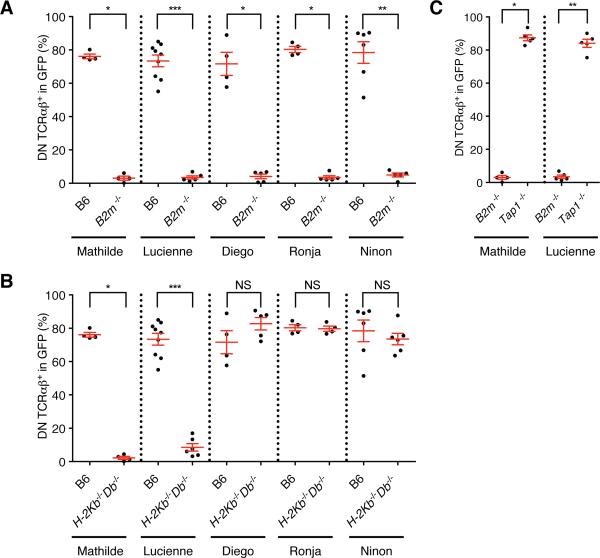
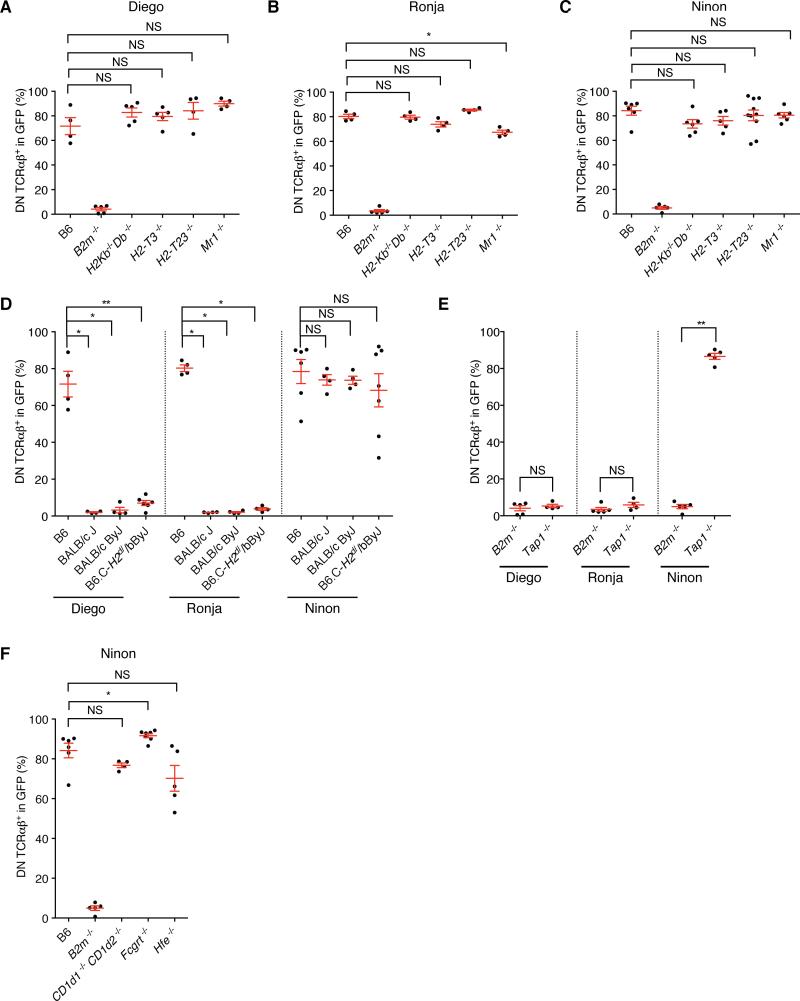
To examine if the DN TCRαβ+ thymic precursors undergo an MHC-based selection process, and to identify the nature of their MHC restriction, we generated BM chimeras on backgrounds deficient for various MHC molecules. Consistent with a requirement forβ2 m molecule, the percentage of DN TCRαβ+ intraepithelial T cells was greatly decreased in B2m−/− recipients as compared to control B6 mice (Figure 5A). In contrast, only two cloned TCRs (TCRs Mathilde and Lucienne) led to a drastic reduction in the percentage of DN TCRαβ+ intraepithelial T cells in H2-Kb−/−Db−/−mice but not in CD1d-, Qa-1- or TL- deficient mice, indicating that these TCRs required one of the polymorphic MHC-encoded class I molecules for positive selection and/or survival (Figure 5B and S5A and S5B). The percentage of DN TCRαβ+ intraepithelial T cells for the other cloned TCRs was not affected in H2-Kb−/−Db−/− mice, indicating that these TCRs could be restricted to a single or several β2m-dependent non-classical MHC I molecules (Figure 5B). Surprisingly, DN TCRαβ+ intraepithelial T cells expressing a cloned H2-KbDb restricted TCR were present in Tap1−/− recipient mice, indicating that, in contrast to conventional CD8αβ+ T cells, H-2K or-D restricted DN TCRαβ+ intraepithelial T cells can sense antigens in a TAP-independent fashion (Figure 5C).
Figure 5. A subset of DN TCRαβ intraepithelial T cells is restricted to classical MHC Class I.
(A-C) Frequency of DN TCRαβ+ cells in GFP+ gated intraepithelial T cells isolated from various MHC deficient chimeras expressing each individual TCR clone and analysed five weeks after reconstitution. Data points are shown for each mouse, with mean plus s.e.m. (A) Analysis of WT B6 BM chimera from at least two independent experiments for TCR Mathilde (n=4), TCR Lucienne (n=8), TCR Diego (n=4), TCR Ronja (n=4) and TCR Ninon (n=6). Analysis of B2m−/− BM chimera from at least two independent experiments for TCR Mathilde (n=4), TCR Lucienne (n=5), TCR Diego (n=4), TCR Ronja (n=5) and TCR Ninon (n=4). (B) Analysis of WT B6 BM chimera as in A and H2-Kb−/−Db−/− BM chimera from at least two independent experiments for TCR Mathilde (n=4), TCR Lucienne (n=6), TCR Diego (n=5), TCR Ronja (n=4) and TCR Ninon (n=6). (C) Analysis of B2m−/− BM chimera as in (A) and Tap1−/− BM chimera from at least two independent experiments for TCR Mathilde (n=5) and TCR Lucienne (n=5). A two-tailed Mann-Whitney test was performed. * P<0.05; ** P <0.01; *** P <0.001; and NS, not significant. (See also Figure S5 and S6)
The other three TCR clones (TCRs Diego, Ronja and Ninon) that gave rise to DN TCRαβ+ intraepithelial T cells in the absence of H2-KbDb, were also not dependent on TL, Qa-1 or MR1 (Figure 6A, 6B and 6C), despite expression of these MHC class I paralogs in the thymus and/or the intestinal epithelium. Although, this is consistent with TL not being an antigen-presenting molecule (Leishman et al., 2001; Weber et al., 2002), for Qa-1 and MR1 it is possible that TCRs with specificity for these molecules can be detected in a larger pool of DN TCRαβ+ intraepithelial T cell TCRs. DN TCRαβ+ intraepithelial T cells were reported to be partially dependent on the non-classical MHC class I molecule, Qa-2, which is encoded by multiple genes located in the Qa region (Q6 to Q9) (Das et al., 2000). Qa-2 MHC molecules, however, display very minor structural polymorphism and inbred mice are either Qa2+ (B6 and BALB/cJ) or Qa2null (BALB/cByJ or B6.C-H2d/bByJ congenic) (Mellor et al., 1985). Therefore, comparison of BM chimera generated on a BALB/cJ (Qa-2+) or BALB/cByJ (Qa-2null) background indicated that the TCR Ninon- was Qa-2-independent (Figure 6D). Unexpectedly, DN TCRαβ+ intraepithelial T cells expressing the TCR Diego or Ronja were unable to develop in both (Qa-2+) and (Qa-2null) strains indicated that both TCRs required an MHC molecule other than Qa-2 that differs between BALB/cJ and B6 strains. In order to define that chromosomal region better, we analyzed the B6.C-H2d/bByJ mice, which is a congenic strain on a B6 background with the D17mit198 to D17mit152 distal MHC region replaced by the BALB/cByJ sequence. The MHC region contains the MHC class II loci, H2-K and H2-D loci as well as H2-Q, H2-T and H2-M regions. If the MHC class I molecule is localized outside the MHC region on chromosome 17, then we expected that the DN TCRαβ+ intraepithelial T cells expressing the Diego or Ronja TCR would develop on a B6.C-H2d/bByJ background. Since, however no DN TCRαβ+ intraepithelial T cells developed in the B6.C-H2d/bByJ chimeric mice, we concluded that these TCRs are not restricted to an MHC molecule localized outside the MHC locus on chromosome 17 (Figure 6D). Both TCRs, Diego and Ronja depended on TAP (Figure 6E), indicating that they are restricted to non-classical, β2m-and TAP-dependent MHC class I molecules localized within a region on chromosome 17 for which B6 and BALB/cJ mice differ (therefor excluding H2-M3 (Lindahl et al., 1997) and H2-Q10 (Mellor et al., 1984)), or which are pseudogenes on the BALB/cJ background (summarized in Figure S6). In contrast, TCR Ninon was TAP-independent (Figure 6E), and except in B2m−/− hosts, this TCR gave rise to mature DN TCRαβ+ intraepithelial T cells in all other class I deficient backgrounds tested, indicating that this TCR was restricted to a β2m-dependent, TAP- independent non classical MHC class I molecule, other than TL, CD1d, Qa-1, Qa2, MR1, FcRn or HFE (Figures 6C, 6D and 6F).
Figure 6. A subset of DN TCRαβ intraepithelial T cells is restricted to non classical MHC Class I.
(A-F) Frequency of DN TCRαβ+ cells in GFP+ gated intraepithelial T cells isolated from various MHC deficient chimeras expressing each individual TCR clone and analysed five weeks after reconstitution. Data points are shown for each mouse from at least two independent experiments, with mean plus s.e.m. (A) Analysis of chimera expressing TCR Diego (B6 (n=4), B2m−/− (n=5), H2-Kb−/−Db−/− (n=5), H2-T3b−/− (n=5), H2-T23−/− (n=4) and Mr1−/− (n=4). (B) Analysis of chimera expressing TCR Ronja (B6 (n=4), B2m−/− (n=5), H2-Kb−/−Db−/− (n=4), H2-T3b−/− (n=4), H2-T23−/− (n=4), and Mr1−/− (n=4)). (C) Analysis of chimera expressing TCR Ninon (B6 (n=6), B2m−/− (n=5), H2-Kb−/−Db−/− (n=6), H2-T3b−/− (n=5), Cd1d1−/− Cd1d2−/− (n=4), H2-T23−/− (n=10) and Mr1−/− (n=6)). (D) Analysis of chimera expressing TCR Diego (B6 (n=4), BALB/cJ (n=4), BALB/cByJ (n=4), and B6.C-H2d/bByJ (n=6)), TCR Ronja (B6 (n=4), BALB/cJ (n=6), BALB/cByJ (n=6), and B6.C-H2d/bByJ (n=4)) and TCR Ninon (B6 (n=6), BALB/cJ (n=4), BALB/cByJ (n=5), and B6.C-H2d/bByJ (n=7)). (E) Analysis of B2m−/− BM chimera for TCR Diego (n=4), TCR Ronja (n=5) and TCR Ninon (n=4). Analysis of Tap1−/− BM chimera for TCR Diego (n=4), TCR Ronja (n=4) and TCR Ninon (n=5)). (F) Analysis of chimera expressing TCR Ninon from at least two independent experiments (B6 (n=6), B2m−/− (n=5), Cd1d1−/− Cd1d2−/− (n=4), Fcgrt−/− (n=7) and Hfe−/− (n=4). A two-tailed Mann-Whitney test was performed. * P<0.05; ** P <0.01; *** P <0.001; and NS, not significant. (See also Figure S6)
Selection of DN TCRαβ+precursor thymocytes is MHC-dependent
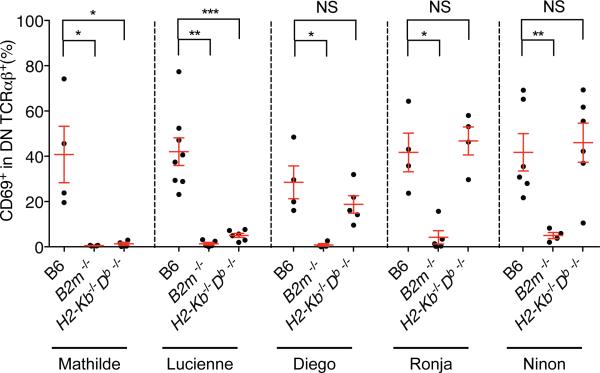
To evaluate if precursors of the DN TCRαβ+ intraepithelial T cells are positively selected in the thymus through a cognate MHC interaction, we examined the CD69 expression, which should be absent on DN TCRαβ+ thymocytes in mice deficient for the MHC molecule recognized by the TCR in question. Indeed, a significant decrease in CD69-expressing DN TCRαβ+ thymocytes was noted in B2m−/− mice for each of the cloned TCRs, as well as in the H2-Kb−/−Db−/− mice for the KbDb restricted TCRs (Figure 7). This is consistent with an MHC-dependent maturation process in the thymus that leads to the positive selection of DN TCRαβ+ lineage precursor cells.
Figure 7. DN TCRαβ+ thymocytes fail to induce CD69 on non-selective MHC background.
Frequency of CD69 expression on GFP+ DN TCRαβ+ thymocytes isolated from WT B6, B2m−/− and H2-Kb−/−Db−/− BM chimera expressing each individual TCR clone and analysed five weeks after reconstitution. Data points are shown for each mouse, with mean plus s.e.m. A two-tailed Mann-Whitney test was performed. * P<0.05; ** P <0.01; *** P <0.001; and NS, not significant.
Discussion
Although DN TCRαβ+ T cells represent a significant fraction of all peripheral T lymphocytes, very little is known about their differentiation, MHC restriction or antigen specificity. This has significantly hampered progress in understanding the function(s) of these cells and their possible contributions to disease or health. This present study is the first to examine the biology of DN TCRαβ+ T cells using endogenous TCRs directly cloned from single mature DN TCRαβ+ intraepithelial T cells. The findings led to several major conclusions. First, the TCRs expressed by DN TCRαβ+ intraepithelial T cells dictate in part the unique lineage commitment most likely through their self-antigen specificities and “TCR-instructive” selection signals in the thymus. Second, the data provide direct evidence that the precursors of these cells undergo an MHC-dependent thymic selection process. Third, the DN TCRαβ+ intraepithelial T cells express a variable TCR repertoire restricted to various MHC molecules. Finally, the specificities of these cells are non-overlapping with those of mainstream CD4+ and CD8αβ+ T cells or any known unconventional T cell subset, including NKT cells and MAIT cells.
Our conclusion that DN TCRαβ+ T cells are a unique T cell lineage, developmentally distinct from conventional and unconventional TCRαβ+ subsets, is supported by the observation that thymic precursors expressing TCRs cloned from naturally arising DN TCRαβ+ intraepithelial T cells gave rise almost exclusively to the DN TCRαβ+ T cell lineage. Furthermore, our findings support the notion that DN TCRαβ+ T cell precursors undergoing self-agonist selection in the thymus (Gangadharan et al., 2006; Yamagata et al., 2004), (Pobezinsky et al., 2012). Notably, the presence of post-selected CD69+ TCRαβ+ thymocytes that lack expression of both the CD4 and CD8αβ coreceptors, and that express PD1, is consistent with agonist selection (Pobezinsky et al., 2012). In the same line of evidence, thymic DN TCRαβ T cell precursors express CD122, which also marks thymocytes and peripheral T cells that differentiated under agonist stimulation (Mixter et al., 1999; Pobezinsky et al., 2012; Priatel et al., 2001). In contrast, unlike for Treg and iNKT cell precursors (Moran et al., 2011), Nur77 does not faithfully identify agonist selected DN TCRαβ T cell precursors (data not shown). Further study is required to identify the conditions that lead to agonist selection of the DN TCRαβ+ subset. The data here also support the notion that both, DN TCRαβ+ splenocytes and intraepithelial T cells share the same lineage, consistent with the fact that both subsets can be generated under agonist selection conditions (Gangadharan et al., 2006). The relatively high frequency of DN TCRαβ+ T cells in the spleen of the chimeric mice is in agreement with similar observations in transgenic mice that express a mainstream TCR together with the cognate antigen as transgenes in the thymus (Leishman et al., 2002; Mixter et al., 1999; Priatel et al., 2001; von Boehmer et al., 1991). This further supports that DN TCRαβ+ T cells require a self-agonist-based TCR signal for their selection and lineage differentiation. It should be emphasized that the generation of these T cells in the spleen in our model is not the result of forced expression of the TCR, since splenic DN TCRαβ+ T cells were absent in mice expressing a mainstream TCR retrogenically or as shown before as a single transgene in the absence of the cognate antigen (Leishman et al., 2002; Holst et al., 2006). This is also in agreement with the fact that the nature of the TCR in part drives the commitment to the DN TCRαβ+ T cell lineage. It remains possible that DN TCRαβ+ splenocytes and DN TCRαβ+ intraepithelial T cells are two related subpopulations of agonist-selected cells with similar MHC specificities but targeting different tissues.
The new findings further provide compelling evidence indicating that DN TCRαβ+ T cells are not restricted to a single MHC molecule, but rather express a variable TCR repertoire with diverse MHC class I specificities. It is particularly striking that none of the TCRs tested had an overlapping specificity with that of either MHC class II-restricted CD4+- or classical, TAP-dependent, MHC I-restricted CD8αβ+ T lymphocytes or any known unconventional T cell subset. At this point however, we cannot exclude the possibility that other DN T cell TCRs with those restrictions could be found if additional clones would be screened. Furthermore, the TAP-independency of some of these TCRs is consistent with earlier reports (Medina et al., 2009; Oliveira and van Hall, 2013; Rock et al., 2010; Tiwari et al., 2007) and indicates that some DN TCRαβ+ intraepithelial T cells have the capacity to sense antigens processed and presented in alternative pathways than those sensed by mainstream T cells. The capacity to recognize non-redundant antigen specificities together with their prevalence within the intestine epithelium likely reflects a unique role for the DN TCRαβ+ T cells to provide adapted immune surveillance at the mucosal borders. Indeed, intestinal epithelial cells employ unique mechanism to process antigens such as absorption, autophagy or other TAP-independent pathways (Goto and Kiyono, 2012; Patel and Stappenbeck, 2013; Rock et al., 2010). However, DN TCRαβ+ T cells are also found outside the intestine, and therefore it is likely that their particular antigen specificities have broader implications, such as immune surveillance in the face of viral immune evasion strategies that often target the MHC class I pathway or for the detection of self-antigens expressed by transformed cells that frequently lose TAP expression (Aptsiauri et al., 2007).
In conclusion, given their accumulation at strategic locations and the findings reported here, we propose that DN TCRαβ+ T cells are designed to play a non-redundant role in immune surveillance and protection, especially under conditions of immune evasion, where CD4+ or CD8αβ+ T cells or the conventional pathway of antigen-processing and presentation is inadequate or impaired (Zhu et al., 2013). Based on the presence of DN TCRαβ+ T cells in healthy humans as well as in disease conditions, such as autoimmune diseases, infections and cancers (D'Acquisto and Crompton, 2011), it is of utmost importance to finally begin to analyze and understand the biology of this significant yet greatly neglected T cell population.
Experimental procedures
Mice
C57BL/6J (B6), B6.129S7-Rag1tm1Mom/J (Rag1−/−), B6.129P2-B2mtm1Unc/J (B2m−/−), B6.129S2-Tap1tm1Arp/J (Tap1−/−), B6.C-H2d/bByJ (Qa-2null), B6.129X1-Fcgrttm1Dcr/DcrJ (Fcgrt−/− or FcRn-deficient mice), B6.129S6-Hfetm2Nca/J (Hfe−/−), B6.129S6-Cd1d1/Cd1d2tm1Spb/J (Cd1d1−/− Cd1d2−/−), BALB/cJ and BALB/cByJ mice were purchased from the Jackson Laboratory. B6.129P2-H2-Kbtm1H2-Dbtm1 N12 (H2-Kb−/−Db−/−) mice were a gift from Dr. A. Sette. H2-T3b−/− mice (TL-deficient mice) were provided by Dr. L. van Kaer (Vanderbilt University, Nashville, TN), H2-T23−/− mice (Qa-1-deficient mice) were provided by Dr. H. Cantor (Dana-Farber Cancer Institute, Boston, MA) and Mr1−/− mice were provided by Dr. T. Hansen (Washington University, St Louis, MO). All mice used in this study were bred and maintained at the La Jolla Institute for Allergy and Immunology under specific pathogen-free conditions. Animal care and experimentation protocols were consistent with National Institutes of Health guidelines, and were approved by the Institutional Animal Care and Use Committee at the La Jolla Institute for Allergy and Immunology.
Preparation of Cell Suspensions
Thymocytes, splenocytes and intraepithelial T cell suspensions are described in Supplemental Experimental Procedures.
Flow Cytometry
Cells were stained in PBS supplemented with 5% FBS, 0.16% azide and anti-CD16/CD32 (2.4G2) antibody (BD Biosciences, San Diego, CA, USA) was included to prevent nonspecific binding through Fc-receptors. Antibodies used are listed in Supplemental Experimental Procedures. Flow cytometric analysis was performed using an LSR Fortessa flow cytometer (BD Biosciences, San Jose, CA, USA), and the obtained data were analysed with FlowJo software (Tree Star, Ashland, OR, USA).
Hybridoma Generation
Hybridomas were generated according to the standard method described by Sydora et al. (Sydora et al., 1993) and described in Supplemental Experimental Procedures.
Single Cell 5’-Rapid Amplification of cDNA Ends (RACE)-PCR and RT-PCR for TCRαβChain Sequencing
Sequencing of TCR chains from the hybridoma cell line was performed using 5’-RACE-PCR at a single cell level. This method was previously developed by Ozawa et al. and was adapted in this study for mouse TCR sequences (Ozawa et al., 2008). All oligonucleotide primer sequences and usages are shown in Supplemental Experimental Procedures. PCR products were analysed using nested TCRα or TCRβ primer by either direct sequencing or sequencing after subcloning using the TOPO ® TA Cloning ® Kit (Invitrogen, Carlsbad, CA).
A preliminary screening of TCR chain expression indicates that TRAV9N-3 and TRBV16 positive cells represent the highest TCRα and TCRβ co-expressing chains among CD8αβ expressing TCRαβ intraepithelial T cells. Therefore for single-cell sequencing of TCRα and TCRβ chains, the cell suspension was enriched for the most co-expressing TRAV and TRBV chains by cell sorting. One cell was plated in PCR tubes that contained RT mix using a FACS Aria equipped with an automatic cell deposition unit. PCR was performed using specific primers for the known TCR variable region (See Supplemental Experimental Procedures). PCR products were separated on an agarose gel and purified using the Wizard Gel clean-up kit (Promega) prior to sequencing by Eton Biosciences (San Diego, CA, USA) using TCR nested primers. TCR sequences were analysed using the IMGT/V-Quest tool (http://imgt.cines.fr/) (Lefranc, 2006).
TCRαβConstructs and Retroviral Plasmids
Genes encoding TCRα and TCRβ chains were obtained from five CD8αβ expressing TCRαβ intraepithelial T cells (Table S1). Four of the TCR sequences were sequenced from single cell sorted B6 CD8αβ expressing TCRαβ intraepithelial T cells and one TCR was from an intraepithelial T cell hybridoma. Each verified TCRα and TCRβ pair was then cloned into a 2A peptide-linked multicistronic vector (Fig. S1A). An MSCV-IRES-GFP retroviral vector was used (kind gift from Dr K. Murphy, University of Washington, USA), which has an MSCV2.2 backbone with an IRES-GFP cassette to facilitate identification of cells expressing the construct. A GSG spacer was incorporated between the TCRα and the T2A peptide to ensure maximal cleavage between TCRα and TCRβ. Synthesis of the insert coding for the TCR-α and -β chains and subsequent cloning into the vector were performed by GenScript (Piscataway, NJ, USA).
Generation of Retroviral Bone Marrow Chimeras
Single-cell suspensions of BM cells were prepared from femur and tibia bones isolated from Rag-deficient, MHC-deficient or B6 mice. Cells were filtered through a 70μm nylon cell strainer (Fisher Scientific, San Diego, CA, USA) and treated with red blood cell lysis buffer (Sigma Aldrich, St Louis, MO, USA). BM cells were cultured at 37°C in 24-well cell culture plates (Costar, Corning Incorporated, Corning, NY, USA) for 44-48h at 1.5×106 cells per well in 1 ml DMEM supplemented with 15% ES FBS (Omega Scientific, Tarzana, CA, USA), 10 ηg/ml recombinant murine IL-6 (Peprotech, Rocky Hill, NJ, USA), 6 ηg/ml recombinant murine IL-3 (Peprotech), 100 ηg/ml recombinant murine SCF (Peprotech), with Penicillin-Streptomycin-Glutamine (Gibco, Life Technologies, Grand Island, NY, USA). BM cells were retrovirally infected three times (once each on days 2, 3 and 4 post-collection). Retroviral plasmids were co-transfected into the Platinum-E Retroviral Packaging cell line (Cell Biolabs, San Diego, CA, USA) using the TransIT-LT1 transfection reagent (Mirus, Madison, WI, USA) according to the manufacturer's instructions. Retrovirus-containing supernatants were collected 48 h after transfection and filtered through a 0.22 μm filter. For infection, cells were spin-infected with retroviral supernatants containing 8mg/ml polybrene (Sigma Aldrich, St Louis, MO, USA) for 90 min at room temperature at 2000rpm. During spin-infection, retroviral supernatants were supplemented with IL-3-, IL-6- and SCF. After spin-infection, cells were returned to the culture conditions described above. The next day, following a third spin-infection, BM cells were harvested, washed and suspended in PBS to a concentration of 3 to 8×106 cells/100 μl. BM cells were assessed for retroviral reporter expression by flow cytometry. For all experiments, GFP expression on total BM cells ranged from 5% to 15%. Recipient mice were irradiated at 450 rad or 900 rad for Rag1−/− or B6 and MHC-deficient mice, respectively. Cells were injected into anesthetised mice in the volume of 100ul through the retro-orbital vein. Mice were euthanized and their tissues analysed 4 to 5 weeks post reconstitution.
Kidney Capsule Transplantation
Thymus tissue used for the transplantation was collected from retrogenic mice 17 days after receiving the BM injection. Thymus tissue was grafted under the kidney capsule of an 8 week-old Rag1−/− recipient. These mice were euthanized and their tissues analysed 5 weeks post transplantation. Detailed method is described in Supplemental Experimental Procedures.
Adoptive Transfer of Thymocytes
Thymus tissue from BM chimeric mice day 17 post reconstitution were harvested in PBS supplemented with 5% FBS and single cell suspensions were prepared. To enrich for GFP+ DN TCRβ+ thymocytes, cells were stained with CD4, CD8α, TCRα, TCRβ, Thy1.2 and CD69 and purified by two rounds of cell sorting using a BD FACS Aria (BD Biosciences, San Jose, CA, USA). Cell contaminants, if present, were below the detection limit. The recipient mice were injected with 104-105 sorted GFP+ DN TCR+ thymocytes in a volume of 100 μl PBS through the retro-orbital vein. These mice were euthanized and their tissues analysed 5 weeks post reconstitution.
Statistical Analysis
A two-tailed Mann-Whitney test was performed to analyse the data using GraphPad software (Prism). Differences indicated and considered as significant are as followed: * p<0.05, ** p<0.01, *** p<0.001 and **** p<0.0001. N.S. not significant.
Supplementary Material
Highlights.
TCRs express by DN TCRαβ+ intraepithelial T cells dictate their unique lineage fate
Precursors of DN TCRαβ+ T cells undergo an MHC-dependent thymic selection process
DN TCRαβ+ intraepithelial T cells display a diverse pattern of MHC restriction
Acknowledgements
We thank C. Lena and Y. Wang-Zhu for technical help, the LIAI Imaging Core Facility for cell sorting and the LIAI Department of Laboratory Animal Care for mouse colony management. We thank S. Bécart for providing cellular biology expertise, E. Girardi for advice regarding TCR sequences analysis, B. Peters for providing statistical analysis expertise and B. Lucas for suggestions, advice and critical reading of the manuscript. We thank M. Cheroutre for her contribution. We thank K. Murphy for the retroviral vector, N. Gascoigne for the 58α−β− cell line, P. Marrack and J. Kappler for the BW5147 cell line, H. Cantor, L van Kaer, A. Sette, and T. Hansen for providing mice. This work was supported by grants from the National Institutes of Health (R21 AI090504 to F.L.; R21 AI099726 to F.L; DP1 OD006433 to H.C: PO1 DK46763 to M.K.). S.M. was supported by a fellowship from the Swedish Research Council (Dnr 623-2010-6567). B.A. was supported by an Amgen Summer Research Program Scholarship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, the National Institutes of Health, or other funding agencies. This is manuscript number 1626 from the La Jolla Institute for Allergy and Immunology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: S.M. and F.L. designed and performed experiments; D.S. generated hybridoma with help from F.L.; B.A., S.P. and F.L. developed methodologies for TCR sequencing; A.L. did western blot experiments; J. D. provided substantial technical help; R.S. provided molecular biology expertise; S.M. and F.L. analyzed data and prepared figures; M.K. provided advice and insight; S.M. and F.L. wrote the manuscript with editing help from H.C. and M.K.; all authors contributed to the manuscript editing; and F.L. conceived, supervised and obtained funding for the project with help and insight from H.C.
Competing financial interests
The authors declare no competing financial interests.
References
- Abadie V, Discepolo V, Jabri B. Intraepithelial lymphocytes in celiac disease immunopathology. Semin Immunopathol. 2012;34:551–566. doi: 10.1007/s00281-012-0316-x. [DOI] [PubMed] [Google Scholar]
- Alli R, Nguyen P, Geiger TL. Retrogenic modeling of experimental allergic encephalomyelitis associates T cell frequency but not TCR functional affinity with pathogenicity. J Immunol. 2008;181:136–145. doi: 10.4049/jimmunol.181.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aptsiauri N, Cabrera T, Garcia-Lora A, Lopez-Nevot MA, Ruiz-Cabello F, Garrido F. MHC class I antigens and immune surveillance in transformed cells. International review of cytology. 2007;256:139–189. doi: 10.1016/S0074-7696(07)56005-5. [DOI] [PubMed] [Google Scholar]
- Arnold PY, Burton AR, Vignali DA. Diabetes incidence is unaltered in glutamate decarboxylase 65-specific TCR retrogenic nonobese diabetic mice: generation by retroviral-mediated stem cell gene transfer. J Immunol. 2004;173:3103–3111. doi: 10.4049/jimmunol.173.5.3103. [DOI] [PubMed] [Google Scholar]
- Bettini ML, Bettini M, Vignali DA. T-cell receptor retrogenic mice: a rapid, flexible alternative to T-cell receptor transgenic mice. Immunology. 2012;136:265–272. doi: 10.1111/j.1365-2567.2012.03574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13:101–117. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- Cheroutre H, Lambolez F. Doubting the TCR coreceptor function of CD8alphaalpha. Immunity. 2008;28:149–159. doi: 10.1016/j.immuni.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat Rev Immunol. 2011;11:445–456. doi: 10.1038/nri3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Acquisto F, Crompton T. CD3+CD4-CD8-(double negative) T cells: saviours or villains of the immune response? Biochem Pharmacol. 2011;82:333–340. doi: 10.1016/j.bcp.2011.05.019. [DOI] [PubMed] [Google Scholar]
- Das G, Gould DS, Augustine MM, Fragoso G, Sciutto E, Stroynowski I, Van Kaer L, Schust DJ, Ploegh H, Janeway CA., Jr. Qa-2-dependent selection of CD8alpha/alpha T cell receptor alpha/beta(+) cells in murine intestinal intraepithelial lymphocytes. J Exp Med. 2000;192:1521–1528. doi: 10.1084/jem.192.10.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- Denning TL, Granger SW, Mucida D, Graddy R, Leclercq G, Zhang W, Honey K, Rasmussen JP, Cheroutre H, Rudensky AY, Kronenberg M. Mouse TCRalphabeta+CD8alphaalpha intraepithelial lymphocytes express genes that down-regulate their antigen reactivity and suppress immune responses. J Immunol. 2007;178:4230–4239. doi: 10.4049/jimmunol.178.7.4230. [DOI] [PubMed] [Google Scholar]
- Engel I, Kronenberg M. Making memory at birth: understanding the differentiation of natural killer T cells. Curr Opin Immunol. 2012;24:184–190. doi: 10.1016/j.coi.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Du P, Liu Y, Tang J. Efficient isolation of mouse liver NKT cells by perfusion. PLoS One. 2010;5:e10288. doi: 10.1371/journal.pone.0010288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiura Y, Kawaguchi M, Kondo Y, Obana S, Yamamoto H, Nanno M, Ishikawa H. Development of CD8 alpha alpha+ intestinal intraepithelial T cells in beta 2-microglobulin- and/or TAP1-deficient mice. J Immunol. 1996;156:2710–2715. [PubMed] [Google Scholar]
- Gangadharan D, Lambolez F, Attinger A, Wang-Zhu Y, Sullivan BA, Cheroutre H. Identification of pre- and postselection TCRalphabeta+ intraepithelial lymphocyte precursors in the thymus. Immunity. 2006;25:631–641. doi: 10.1016/j.immuni.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Gapin L, Cheroutre H, Kronenberg M. Cutting edge: TCR alpha beta+ CD8 alpha alpha+ T cells are found in intestinal intraepithelial lymphocytes of mice that lack classical MHC class I molecules. J Immunol. 1999;163:4100–4104. [PubMed] [Google Scholar]
- Goto Y, Kiyono H. Epithelial barrier: an interface for the cross-communication between gut flora and immune system. Immunol Rev. 2012;245:147–163. doi: 10.1111/j.1600-065X.2011.01078.x. [DOI] [PubMed] [Google Scholar]
- Guy-Grand D, Cerf-Bensussan N, Malissen B, Malassis-Seris M, Briottet C, Vassalli P. Two gut intraepithelial CD8+ lymphocyte populations with different T cell receptors: a role for the gut epithelium in T cell differentiation. J Exp Med. 1991;173:471–481. doi: 10.1084/jem.173.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy-Grand D, Cuenod-Jabri B, Malassis-Seris M, Selz F, Vassalli P. Complexity of the mouse gut T cell immune system: identification of two distinct natural killer T cell intraepithelial lineages. Eur J Immunol. 1996;26:2248–2256. doi: 10.1002/eji.1830260942. [DOI] [PubMed] [Google Scholar]
- Guy-Grand D, Vassalli P, Eberl G, Pereira P, Burlen-Defranoux O, Lemaitre F, Di Santo JP, Freitas AA, Cumano A, Bandeira A. Origin, trafficking, and intraepithelial fate of gut-tropic T cells. J Exp Med. 2013;210:1839–1854. doi: 10.1084/jem.20122588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstetter AR, Ford ML, Sullivan LC, Wilson JJ, Hadley A, Brooks AG, Lukacher AE. MHC class Ib-restricted CD8 T cells differ in dependence on CD4 T cell help and CD28 costimulation over the course of mouse polyomavirus infection. J Immunol. 2012;188:3071–3079. doi: 10.4049/jimmunol.1103554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst J, Vignali KM, Burton AR, Vignali DA. Rapid analysis of T-cell selection in vivo using T cell-receptor retrogenic mice. Nat Methods. 2006;3:191–197. doi: 10.1038/nmeth858. [DOI] [PubMed] [Google Scholar]
- Kilshaw PJ, Murant SJ. A new surface antigen on intraepithelial lymphocytes in the intestine. Eur J Immunol. 1990;20:2201–2207. doi: 10.1002/eji.1830201008. [DOI] [PubMed] [Google Scholar]
- Lambolez F, Kronenberg M, Cheroutre H. Thymic differentiation of TCR alpha beta(+) CD8 alpha alpha(+) IELs. Immunol Rev. 2007;215:178–188. doi: 10.1111/j.1600-065X.2006.00488.x. [DOI] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP. Using bioinformatics tools for the sequence analysis of immunoglobulins and T cell receptors. Curr Protoc Immunol Appendix 1, Appendix 1W. Lefrancois, L. (1991). Phenotypic complexity of intraepithelial lymphocytes of the small intestine. J Immunol. 2006;147:1746–1751. doi: 10.1002/0471142735.ima01ws71. [DOI] [PubMed] [Google Scholar]
- Leishman AJ, Gapin L, Capone M, Palmer E, MacDonald HR, Kronenberg M, Cheroutre H. Precursors of functional MHC class I- or class II-restricted CD8alphaalpha (+) T cells are positively selected in the thymus by agonist self-peptides. Immunity. 2002;16:355–364. doi: 10.1016/s1074-7613(02)00284-4. [DOI] [PubMed] [Google Scholar]
- Leishman AJ, Naidenko OV, Attinger A, Koning F, Lena CJ, Xiong Y, Chang HC, Reinherz E, Kronenberg M, Cheroutre H. T cell responses modulated through interaction between CD8alphaalpha and the nonclassical MHC class I molecule, TL. Science. 2001;294:1936–1939. doi: 10.1126/science.1063564. [DOI] [PubMed] [Google Scholar]
- Lindahl KF, Byers DE, Dabhi VM, Hovik R, Jones EP, Smith GP, Wang CR, Xiao H, Yoshino M. H2-M3, a full-service class Ib histocompatibility antigen. Annu Rev Immunol. 1997;15:851–879. doi: 10.1146/annurev.immunol.15.1.851. [DOI] [PubMed] [Google Scholar]
- Medina F, Ramos M, Iborra S, de Leon P, Rodriguez-Castro M, Del Val M. Furin-processed antigens targeted to the secretory route elicit functional TAP1−/− CD8+ T lymphocytes in vivo. J Immunol. 2009;183:4639–4647. doi: 10.4049/jimmunol.0901356. [DOI] [PubMed] [Google Scholar]
- Mellor AL, Antoniou J, Robinson PJ. Structure and expression of genes encoding murine Qa-2 class I antigens. Proc Natl Acad Sci U S A. 1985;82:5920–5924. doi: 10.1073/pnas.82.17.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor AL, Weiss EH, Kress M, Jay G, Flavell RA. A nonpolymorphic class I gene in the murine major histocompatibility complex. Cell. 1984;36:139–144. doi: 10.1016/0092-8674(84)90082-5. [DOI] [PubMed] [Google Scholar]
- Mixter PF, Russell JQ, Morrissette GJ, Charland C, Aleman-Hoey D, Budd RC. A model for the origin of TCR-alphabeta+ CD4-CD8− B220+ cells based on high affinity TCR signals. J Immunol. 1999;162:5747–5756. [PubMed] [Google Scholar]
- Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohteki T, MacDonald HR. Expression of the CD28 costimulatory molecule on subsets of murine intestinal intraepithelial lymphocytes correlates with lineage and responsiveness. Eur J Immunol. 1993;23:1251–1255. doi: 10.1002/eji.1830230609. [DOI] [PubMed] [Google Scholar]
- Oliveira CC, van Hall T. Importance of TAP-independent processing pathways. Mol Immunol. 2013;55:113–116. doi: 10.1016/j.molimm.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Ozawa T, Tajiri K, Kishi H, Muraguchi A. Comprehensive analysis of the functional TCR repertoire at the single-cell level. Biochem Biophys Res Commun. 2008;367:820–825. doi: 10.1016/j.bbrc.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Park SH, Guy-Grand D, Lemonnier FA, Wang CR, Bendelac A, Jabri B. Selection and expansion of CD8alpha/alpha(1) T cell receptor alpha/beta(1) intestinal intraepithelial lymphocytes in the absence of both classical major histocompatibility complex class I and nonclassical CD1 molecules. J Exp Med. 1999;190:885–890. doi: 10.1084/jem.190.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KK, Stappenbeck TS. Autophagy and intestinal homeostasis. Annual review of physiology. 2013;75:241–262. doi: 10.1146/annurev-physiol-030212-183658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobezinsky LA, Angelov GS, Tai X, Jeurling S, Van Laethem F, Feigenbaum L, Park JH, Singer A. Clonal deletion and the fate of autoreactive thymocytes that survive negative selection. Nat Immunol. 2012;13:569–578. doi: 10.1038/ni.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priatel JJ, Utting O, Teh HS. TCR/self-antigen interactions drive double-negative T cell peripheral expansion and differentiation into suppressor cells. J Immunol. 2001;167:6188–6194. doi: 10.4049/jimmunol.167.11.6188. [DOI] [PubMed] [Google Scholar]
- Regnault A, Cumano A, Vassalli P, Guy-Grand D, Kourilsky P. Oligoclonal repertoire of the CD8 alpha alpha and the CD8 alpha beta TCR-alpha/beta murine intestinal intraepithelial T lymphocytes: evidence for the random emergence of T cells. J Exp Med. 1994;180:1345–1358. doi: 10.1084/jem.180.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha B, Vassalli P, Guy-Grand D. The V beta repertoire of mouse gut homodimeric alpha CD8+ intraepithelial T cell receptor alpha/beta + lymphocytes reveals a major extrathymic pathway of T cell differentiation. J Exp Med. 1991;173:483–486. doi: 10.1084/jem.173.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock EP, Sibbald PR, Davis MM, Chien YH. CDR3 length in antigen-specific immune receptors. J Exp Med. 1994;179:323–328. doi: 10.1084/jem.179.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, Farfan-Arribas DJ, Shen L. Proteases in MHC class I presentation and cross-presentation. J Immunol. 2010;184:9–15. doi: 10.4049/jimmunol.0903399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shires J, Theodoridis E, Hayday AC. Biological insights into TCRgammadelta+ and TCRalphabeta+ intraepithelial lymphocytes provided by serial analysis of gene expression (SAGE). Immunity. 2001;15:419–434. doi: 10.1016/s1074-7613(01)00192-3. [DOI] [PubMed] [Google Scholar]
- Stritesky GL, Jameson SC, Hogquist KA. Selection of self-reactive T cells in the thymus. Annu Rev Immunol. 2012;30:95–114. doi: 10.1146/annurev-immunol-020711-075035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydora BC, Brossay L, Hagenbaugh A, Kronenberg M, Cheroutre H. TAP-independent selection of CD8+ intestinal intraepithelial lymphocytes. J Immunol. 1996;156:4209–4216. [PubMed] [Google Scholar]
- Sydora BC, Mixter PF, Houlden B, Hershberg R, Levy R, Comay M, Bluestone J, Kronenberg M. T-cell receptor gamma delta diversity and specificity of intestinal intraepithelial lymphocytes: analysis of IEL-derived hybridomas. Cell Immunol. 1993;152:305–322. doi: 10.1006/cimm.1993.1293. [DOI] [PubMed] [Google Scholar]
- Tiwari N, Garbi N, Reinheckel T, Moldenhauer G, Hammerling GJ, Momburg F. A transporter associated with antigen-processing independent vacuolar pathway for the MHC class I-mediated presentation of endogenous transmembrane proteins. J Immunol. 2007;178:7932–7942. doi: 10.4049/jimmunol.178.12.7932. [DOI] [PubMed] [Google Scholar]
- Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, Affaticati P, Gilfillan S, Lantz O. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–169. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- von Boehmer H, Kirberg J, Rocha B. An unusual lineage of alpha/beta T cells that contains autoreactive cells. J Exp Med. 1991;174:1001–1008. doi: 10.1084/jem.174.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber DA, Attinger A, Kemball CC, Wigal JL, Pohl J, Xiong Y, Reinherz EL, Cheroutre H, Kronenberg M, Jensen PE. Peptide-independent folding and CD8 alpha alpha binding by the nonclassical class I molecule, thymic leukemia antigen. J Immunol. 2002;169:5708–5714. doi: 10.4049/jimmunol.169.10.5708. [DOI] [PubMed] [Google Scholar]
- Yamagata T, Mathis D, Benoist C. Self-reactivity in thymic double-positive cells commits cells to a CD8 alpha alpha lineage with characteristics of innate immune cells. Nat Immunol. 2004;5:597–605. doi: 10.1038/ni1070. [DOI] [PubMed] [Google Scholar]
- Zhu J, Peng T, Johnston C, Phasouk K, Kask AS, Klock A, Jin L, Diem K, Koelle DM, Wald A, et al. Immune surveillance by CD8alphaalpha+ skin-resident T cells in human herpes virus infection. Nature. 2013;497:494–497. doi: 10.1038/nature12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.