Significance
Insulin provides a model for analysis of protein structure and evolution. Here we describe in detail a conformational switch that enables otherwise hidden nonpolar surfaces in the hormone to engage its receptor. Whereas the classical closed conformation of insulin enables its stable storage in pancreatic β cells, its active conformation is open and susceptible to nonnative aggregation. Our findings illuminate biophysical constraints underlying the evolution of an essential signaling system and provide a structural foundation for design of therapeutic insulin analogs.
Keywords: diabetes mellitus, signal transduction, receptor tyrosine kinase, metabolism, protein structure
Abstract
Insulin provides a classical model of a globular protein, yet how the hormone changes conformation to engage its receptor has long been enigmatic. Interest has focused on the C-terminal B-chain segment, critical for protective self-assembly in β cells and receptor binding at target tissues. Insight may be obtained from truncated “microreceptors” that reconstitute the primary hormone-binding site (α-subunit domains L1 and αCT). We demonstrate that, on microreceptor binding, this segment undergoes concerted hinge-like rotation at its B20-B23 β-turn, coupling reorientation of PheB24 to a 60° rotation of the B25-B28 β-strand away from the hormone core to lie antiparallel to the receptor's L1–β2 sheet. Opening of this hinge enables conserved nonpolar side chains (IleA2, ValA3, ValB12, PheB24, and PheB25) to engage the receptor. Restraining the hinge by nonstandard mutagenesis preserves native folding but blocks receptor binding, whereas its engineered opening maintains activity at the price of protein instability and nonnative aggregation. Our findings rationalize properties of clinical mutations in the insulin family and provide a previously unidentified foundation for designing therapeutic analogs. We envisage that a switch between free and receptor-bound conformations of insulin evolved as a solution to conflicting structural determinants of biosynthesis and function.
How insulin engages the insulin receptor has inspired speculation ever since the structure of the free hormone was determined by Hodgkin and colleagues in 1969 (1, 2). Over the ensuing decades, anomalies encountered in studies of analogs have suggested that the hormone undergoes a conformational change on receptor binding: in particular, that the C-terminal β-strand of the B chain (residues B24–B30) releases from the helical core to expose otherwise-buried nonpolar surfaces (the detachment model) (3–6). Interest in the B-chain β-strand was further motivated by the discovery of clinical mutations within it associated with diabetes mellitus (DM) (7). Analysis of residue-specific photo–cross-linking provided evidence that both the detached strand and underlying nonpolar surfaces engage the receptor (8).
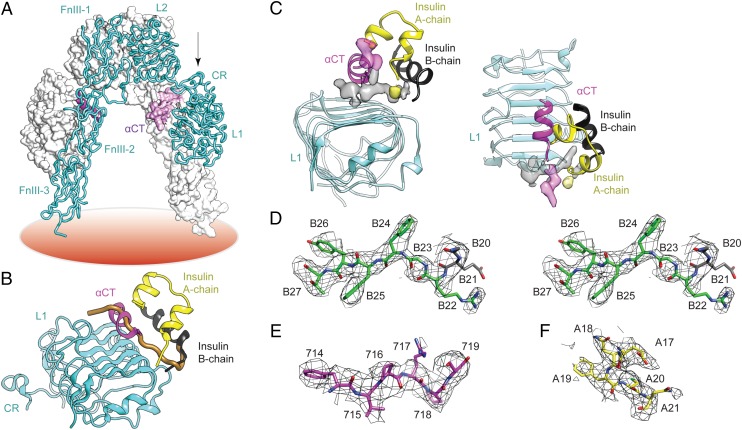
The relevant structural biology is as follows. The insulin receptor is a disulfide-linked (αβ)2 receptor tyrosine kinase (Fig. 1A), the extracellular α-subunits together binding a single insulin molecule with high affinity (9). Involvement of the two α-subunits is asymmetric: the primary insulin-binding site (site 1*) comprises the central β-sheet (L1–β2) of the first leucine-rich repeat domain (L1) of one α-subunit and the partially helical C-terminal segment (αCT) of the other α-subunit (Fig. 1A) (10). Such binding initiates conformational changes leading to transphosphorylation of the β-subunits’ intracellular tyrosine kinase (TK) domains. Structures of wild-type (WT) insulin (or analogs) bound to extracellular receptor fragments were recently described at maximum resolution of 3.9 Å (11), revealing that hormone binding is primarily mediated by αCT (receptor residues 704–719); direct interactions between insulin and L1 were sparse and restricted to certain B-chain residues. On insulin binding, αCT was repositioned on the L1–β2 surface, and its helix was C-terminally extended to include residues 711–714. None of these structures defined the positions of C-terminal B-chain residues beyond B21. Support for the detachment model was nonetheless provided by entry of αCT into a volume that would otherwise be occupied by B-chain residues B25–B30 (i.e., in classical insulin structures; Fig. 1B) (11).
Fig. 1.
Insulin B-chain C-terminal β-strand in the μIR complex. (A) Structure of apo-receptor ectodomain. One monomer is in tube representation (labeled), the second is in surface representation. L1, first leucine-rich repeat domain; CR, cysteine-rich domain; L2, second leucine-rich repeat domain; FnIII-1, -2 and -3; first, second and third fibronectin type III domains, respectively; αCT, α-subunit C-terminal segment; coral disk, plasma membrane. (B) Insulin bound to μIR; the view direction with respect to L1 in the apo-ectodomain is indicated by the arrow in A. Only B-chain residues indicated in black were originally resolved (11). The brown tube indicates classical location of residues B20-B30 in free insulin, occluded in the complex by αCT. (C) Orthogonal views of unmodeled 2Fobs-Fcalc difference electron density (SI Appendix), indicating association of map segments with the αCT C-terminal extension (transparent magenta), insulin B-chain C-terminal segment (transparent gray), and AsnA21 (transparent yellow). Difference density is sharpened (Bsharp = −160 Å2). (D–F) Refined models of respective segments insulin B20–B27, αCT 714–719, and insulin A17-A21 within postrefinement 2Fobs-Fcalc difference electron density (Bsharp = −160 Å2). D is in stereo.
We describe here the structure and interactions of the detached B-chain C-terminal segment of insulin on its binding to a “microreceptor” (μIR), an L1–CR domain-minimized version of the α-subunit (designated IR310.T) plus exogenous αCT peptide 704–719 (11). Our analysis defines a hinge in the B chain whose opening is coupled to repositioning of αCT between nonpolar surfaces of L1 and the insulin A chain. To understand the role of this hinge in holoreceptor binding and signaling, we designed three insulin analogs containing structural constraints (Table 1): [d-AlaB20, d-AlaB23]-insulin, ∆PheB25-insulin, and ∆PheB24-insulin, where ∆Phe is (α,β)-dehydrophenylalanine (Fig. 2) (12). The latter represents, to our knowledge, the first use of ∆Phe—a rigid “β-breaker” with extended electronic conjugation between its side chain and main chain (SI Appendix, Fig. S1)—as a probe of induced fit in macromolecular recognition. In addition, a fourth analog, active but with anomalous flexibility in the B chain (5, 6) (Table 1), was used to investigate the relationship between the hinge and insulin’s susceptibility to misfolding.
Table 1.
Summary of insulin analogs
| Analog | Modification | Templates* | Rationale |
| 1 | d-AlaB20, d-AlaB23 | Insulin; KP-insulin | Locked β-turn |
| 2 | ∆PheB25 | KP-insulin; DKP-insulin | β-breaker at B25 |
| 3 | ∆PheB24 | KP-insulin; DKP-insulin | β-breaker at B24 |
| 4 | GlyB24 | KP-insulin; DKP-insulin | Destabilized hinge |
All templates use the human insulin sequence, with KP-insulin (“lispro”) having substitutions ProB28Lys and LysB29Pro and DKP-insulin having the additional substitution HisB10Asp.
Fig. 2.
Structure of ∆Phe. (A and B) Respective line drawings of E and Z configurational isomers of (α,β)-dehydro-Phe. The present studies use the more stable Z isomer (23).
Despite the limitations of domain minimization, our structure of the μIR complex illuminates the properties of DM-associated mutations in insulin and rationalizes a wealth of prior biochemical data. Of broader importance, our findings demonstrate that hidden within insulin sequences lie multiple layers of structural information, encoding a complex conformational life cycle from biosynthesis to function. As such, they provide a structural foundation for design of therapeutic analogs.
Results
Insulin Binding to μIR vs. Holoreceptor.
To distinguish whether the apparent disorder of insulin segment B22–B30 in the extant μIR crystal structure (11) was a consequence of receptor truncation or a technical limitation of prior crystals, we tested whether PheB24, PheB25, and TyrB26 contribute to μIR binding in a fashion equivalent to holoreceptor binding (2, 4). Each residue was singly substituted by alanine (Ala), and assembly of the μIR complex was monitored as a function of the added concentration of insulin or analog. Relative stabilities of the variant μIR complexes correlated with analog affinities for the holoreceptor (SI Appendix, Fig. S2 A and B). Consistency was also obtained in assays of insulin variants containing single Ala substitutions at classical receptor contact sites resolved in the original μIR structure (SI Appendix, Fig. S2 C and D and Table S1) (2, 13–15). Together, these data imply that residues B24–B26 make similar contributions to the binding of insulin to the μIR and holoreceptor.
Transverse Relaxation-Optimized NMR Spectroscopy.
Given the critical role of the B24–B26 segment in insulin binding to the holoreceptor, the above findings suggested that this segment is ordered in the μIR complex. We investigated this via transverse relaxation-optimized NMR spectroscopy (TROSY) (16). Complexes were prepared containing 13C-labeled insulin and complementary labels in αCT; TROSY-defined 1H- and 13C chemical shifts provided probes of the labeled sites in the free molecules and in the μIR complex. A monomeric insulin analog [LysB28, ProB29]-porcine insulin (KP-p-insulin) (17) was prepared containing uniformly [13C, 15N]-enriched amino acids at positions B23, B24, B26, and B30 (the 15N labels were not exploited). An αCT peptide derived from receptor α-subunit sequences 703–717 (with an N-terminal Arg and C-terminal Lys added to enhance solubility) was likewise labeled at positions Tyr708, Leu709, Val713, and Phe714. [1H-13C]-TROSY spectra in D2O were obtained from a complex comprising unlabeled IR310.T, labeled KP-p-insulin, and labeled αCT peptide (complex 1; Fig. 3 and SI Appendix, Fig. S3) and from a complex comprising unlabeled IR310.T, unlabeled KP-insulin, and labeled αCT (complex 2; used to distinguish αCT cross-peaks in complex 1). Chemical shifts were assessed in relation to spectra of the isolated labeled components.
Fig. 3.
TROSY NMR analysis of unlabeled L1–CR fragment of the receptor α-subunit (construct IR310.T) complexed with labeled KP-porcine insulin and labeled αCT peptide (complex 1). [1H-13C]-TROSY spectrum of selected aromatic ring resonances in the μIR complex (black) is shown relative to [1H-13C] HSQC spectra of KP-porcine insulin (red) and free αCT (blue). Labeled aromatic sites in the insulin analog were PheB24 and TyrB26; labeled aromatic sites in αCT were Tyr708 and Phe714 as indicated. The characteristic upfield secondary chemical shifts of PheB24 and TyrB26 in free insulin are attenuated in the complex. For aliphatic [1H-13C] HSQC spectra, see SI Appendix, Fig. S3.
Each of the labeled sites in insulin (except flexible chain terminus AlaB30) and in αCT underwent significant changes in 1H and/or 13C chemical shifts on μIR assembly (SI Appendix, Table S2). In general, labeled αCT sites exhibited complexation shifts away from random-coil values (i.e., enhanced secondary shifts) in accordance with stabilization of the α-helical secondary structure on assembly. Although aliphatic 1H-13C heteronuclear single-quantum coherence (HSQC) signals from the labeled KP-p-insulin were attenuated in complex 1 (due to resonance broadening; SI Appendix, Fig. S3), the aromatic cross-peaks of PheB24 and TyrB26 remained sharp and exhibited large changes in chemical shift (Fig. 3). Depending on the site, secondary shifts were enhanced or attenuated, suggesting local differences in magnetic environments between free and bound hormone. These findings imply that, on μIR binding, PheB24 and TyrB26 adopt nonrandom conformations.
Crystallographic Resolution of the Bound B22–B27 Segment.
The above motivated further crystallographic analysis. We collected diffraction data from >40 crystals of the prior insulin μIR complex (11); although these data were mostly of lower resolution than originally described, combination of the best three datasets enabled technical improvement on merging of intensities to 3.5-Å resolution as assessed by <I/σ(I)> and CC1/2 criteria (18). Refinement against the combined diffraction dataset yielded interpretable density segments adjacent to insulin B21 and to αCT Val715, respectively (Fig. 1C). The first of these segments was successfully modeled and crystallographically refined (Rwork/Rfree = 0.264/0.284) as B22–B27 docked largely within a crevice between the C-terminal region of αCT and the L1 surface. The second of these refined segments encompassed αCT residues 716–719. Full details regarding X-ray data processing and crystallographic refinement are provided in the SI Appendix, with statistics in SI Appendix, Table S3.
Corresponding to well-defined electron density (Fig. 1D), the side chain of PheB24 is anchored in a hydrophobic pocket bounded by B-chain residues ValB12, LeuB15, TyrB16, and CysB19 and receptor residues Asn15, Leu37, Phe39, and Phe714 (Fig. 4 A and B)†. Modeling connected PheB24 through density to CysB19 via a type 1 β-turn with positive ϕ angles at GlyB20 and GlyB23—similar to those of WT insulin (2). The B20–B23 β-turn has minimal interaction with the μIR. The side chain of PheB25 (in less well-defined density than that of PheB24; Fig. 1D) projects away from L1–β2 to insert between αCT residues Val715 and Pro718 (Fig. 4C); contacts are also made to the αCT main chain at 715–718. The PheB25 main chain adjoins the side chain of Arg14, but no contacts occur between the aromatic ring and either L1 or other insulin side chains. The side chain of TyrB26 lies within a shallow depression defined by L1 side chains Asp12 and Arg14; B26 side-chain density is less well defined than either PheB24 or PheB25 (Fig. 1D). Although the B26 side chain contacts only L1, its main chain abuts both Arg14 and Val715. The orientation of ThrB27 is unclear (Fig. 1D), with little apparent μIR interaction (Fig. 4B). Electron density extending C-terminal to αCT residue 715 was readily modeled (Fig. 1E). Residues 716–719 are directed away from L1 in an irregular conformation (Fig. 4 A–C); the helical portion of αCT terminates at Phe714. Bulges in density correspond to Pro716 and Pro718 (Fig. 1E), whereas the Arg717 side chain appears disordered. Although detailed conformations are uncertain, it is clear that the displaced B24–B26 segment inserts between the first strand of the L1–β2 sheet and αCT residues 715–718 (Fig. 4C); contacts with PheB25 (above) stabilize residues 716–719.
Fig. 4.
Structural change in the B-chain C-terminal segment on μIR engagement. (A) Overlay of the B-chain of free insulin onto the μIR complex. Brown, B1–B30 conformation in free insulin; black, B7–B19 conformation of μIR-bound insulin; green, B20–B27 conformation of μIR-bound insulin; yellow, A-chain conformation of μIR-bound insulin; magenta, αCT segment. In the μIR complex, αCT side chains His710 and Phe714 fill a volume occupied by TyrB26 in free insulin; the location of the PheB24 side chain is approximately maintained by rotameric change. (B and C) Environments of PheB24 (B) and residues B25–B27 (C) in the μIR complex. Colors are as in A, with the L1 domain in cyan. (D) Dissection of the hinge-like opening of residues B20–B30 into a ∼10° rotation of B20–B30 about B20 followed by a ∼50° rotation of B24–B30 about B24. Brown, B20–B30 conformation in free insulin; black/green, B-chain conformation in μIR complex; blue, intermediate conformation illustrating ∼10° rotation of the B20–B23 β-turn. (E) Packing of residues B20–B27 between αCT and L1–β2 sheet surfaces (C, green; N, blue; O, red). Black, residues B8–B19; yellow, insulin A chain. Contact surfaces of αCT with B24–B26 are highlighted in magenta, and those of L1 with B24–B26 in cyan; surfaces not abutting B24–B26 are in lighter shades. Main- and side-chain surfaces of Val715 are labeled M and S. (F) Orthogonal view to E, showing interactions of PheB24 side chain with nonpolar surface of L1–β2 sheet and those of conserved residues A1–A3 with αCT.
Detachment of the B-chain C-terminal strand from the hormone core can be factored into two rotations that result in its repositioning between αCT and L1. The first is rotation of the B20–B27 segment by ∼10° about B20, and the second is a further rotation of B24–B27 by ∼50° about B24 (Fig. 4D). Although the first rotation effects maximal displacement at PheB24, the latter side chain undergoes a compensating rotameric change (Fig. 4B), preserving contacts with the B-chain helix. The second rotation positions B25–B27 antiparallel to the first strand of L1–β2 and nearly perpendicular to the B-chain α-helix (Fig. 4 A and B). This series of conformational changes couples detachment of the insulin B-chain C-terminal strand with insertion of the αCT helix (residues 705–714) into the volume occupied by residues B25–B30 in the free hormone (11). Such detachment motivated design of the following insulin analogs to enable interpretation in the context of the holoreceptor.
Analog 1: Stabilization of a β-Turn.
[d-AlaB20, d-AlaB23]-insulin (analog 1) contains pairwise d-Ala substitutions, designed to reinforce the native positive ϕ dihedral angles of GlyB20 and GlyB23 in the B20–B23 β-turn (19, 20). This chiral “lock” enhanced thermodynamic stability (∆∆Gu = 1.0 ± 0.2 kcal/mol; SI Appendix, Fig. S4A and Table S4). Whereas in WT insulin crystal structures the β-turn exhibits unusually high thermal B-factors with incomplete side-chain electron density (2), the crystal structure of receptor-free analog 1 (determined at 1.40-Å resolution; SI Appendix and SI Appendix, Table S5) exhibits a well-ordered β-turn with the d-methyl groups projecting into solvent (Fig. 5); the overall structure closely resembles free insulin (SI Appendix, Fig. S5A). Molecular modeling suggests that the d-methyl groups can readily be accommodated within the μIR complex (SI Appendix, Fig. S5A).
Fig. 5.
Structure of analog 1. (A) Ribbon model of zinc-free dimer showing residues d-AlaB20 and d-AlaB23 in each protomer in red (side chain methyl groups) and green (Cα atoms). Side chains of PheB24, PheB25, and TyrB26 (and their dimer-related mates) are shown in blue. B-chain ribbons are pink (residues B1–B8) or light gray (B9–B30); A chains are black. (B) Expanded view of residues B12–B26, overlaid with weighted 2Fobs-Fcalc difference electron density. The sulfur atoms of cystine B19-A20 are shown in gold; the coloring scheme is otherwise as in A.
To probe dynamics, we used 1H-NMR, using (here and below) a monomeric insulin template‡ to circumvent self-association of the WT hormone (21). Aromatic 1H-NMR resonances at sites flanking the stabilized β-turn (TyrB16, PheB24, and TyrB26) exhibited enhanced chemical-shift dispersion (SI Appendix, Fig. S6 A and B) and a denser network of interresidue nuclear Overhauser enhancements (NOEs) than were observed in the parent template (SI Appendix, Fig. S6 C and D), indicating that conformational fluctuations within the B-chain supersecondary structure are damped by the chiral modifications. These NOEs are in accordance with the analog’s crystal structure (Fig. 5). The activity of analog 1 as an engineered monomer, evaluated by receptor-binding affinity in vitro and glycemic potency in a rat model of DM (22), was at least as high as that of WT insulin (SI Appendix, Figs. S4B and 7). Together, these data suggest that the B20–B23 β-turn, as visualized in the refined μIR complex, is maintained as a distinct structural element on holoreceptor binding and activation.
Analog 2: β-Breaker at PheB25.
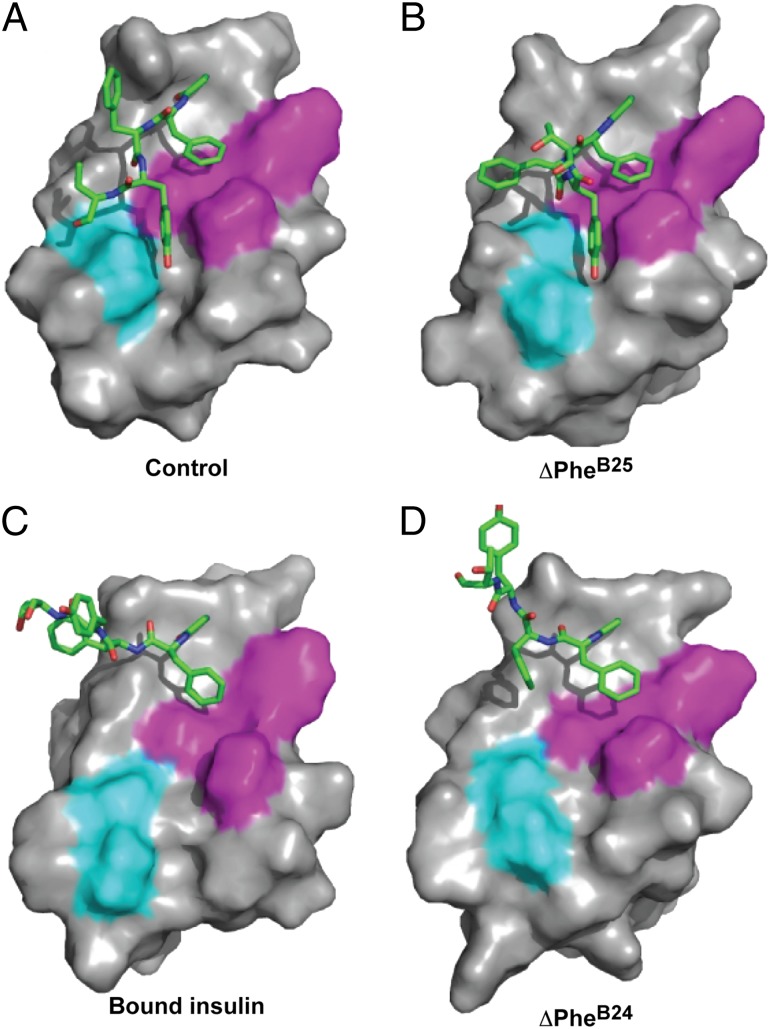
We next probed the importance of the B-chain hinge at B24–B25 via analog 2, which contains a rigid kink at B25 introduced by ∆Phe (Z isomer; Fig. 2B). Preserving side-chain size and aromaticity, ∆Phe contains trigonal Cα/Cβ configurations, leading to extended conjugation of π electrons (from aromatic ring to formal double bonds Cα=Cβ and C=O) (23). Such delocalization enforces near planarity, associated in turn with a break in β-strand–associated main-chain dihedral angles (23). Introduction of ∆Phe at position B25 is well tolerated in the free hormone as monitored by circular dichroism (CD; SI Appendix, Fig. S8A); thermodynamic stability was indistinguishable from that of its monomeric parent (SI Appendix, Fig. S8B and Table S4).
1H-NMR spectra of engineered insulin monomers containing ∆PheB25 are similar to those of the native templates (Fig. 6 A and C). The solution structure of analog 2 (Fig. 6B and D), determined by heteronuclear multidimensional NMR (24) (SI Appendix, Fig. S9 A–D; restraints and statistical parameters are given in SI Appendix, Table S6), closely resembles WT insulin. Native-like positioning of the B24–B25–B26 aromatic rings is maintained relative to the α-helical domain with a single family of main-chain dihedral angles at B25 (Fig. 6D). Control ensembles imposing alternative main-chain conformations [based on peptide structures (23); SI Appendix] exhibited similar overall features, including positioning of TyrB26. Although recapitulating a native-like fold, ∆PheB25 exhibited severe deficits in receptor binding (SI Appendix, Fig. S4B), μIR assembly (SI Appendix, Fig. S4C), downstream signaling in cell culture (SI Appendix, Fig. S10 A–C), and biological activity in vivo (SI Appendix, Fig. S10 F and G). These impairments were more severe than ordinarily encountered among insulin analogs (2). Such marked inactivity suggests that a planar B25 constraint limits the main-chain flexibility required for detachment of the B24–B27 segment and subsequent engagement with the receptor. Indeed, modeling predicts steric clashes between αCT and both ∆PheB25 and TyrB26 and also between L1 and the B27–B30 segment, despite the latter’s flexibility (Fig. 7 and SI Appendix, Fig. S5B). Resolution of these clashes presumably requires conformational distortion of either αCT or the variant B chain.
Fig. 6.
NMR studies of ∆Phe insulin analogs. (A) Baseline NOESY spectrum of KP-insulin. Cross-peak (ω1, ω2) assignments are (a) LeuB15 δ1-CH3/PheB24 Hδ, (b) LeuB15 δ1-CH3/PheB24 Hε, (c) LeuB15 δ1-CH3/PheB24 Hζ, (d) LeuB15 δ1-CH3/TyrB26 Hε, (e) LeuB15 δ1-CH3/TyrB26 Hδ, and (f) LeuB15 δ1-CH3/TyrA19 Hδ. *B15-δ1-CH3/B25 Hδ. (B) Solution structure of parent DKP-insulin (PDB ID code 2JMN). (C) NOESY spectrum of ∆PheB25-KP-insulin. Peak assignments are as labeled in A. (D) Superposition of 20 DG/RMD structures of ∆PheB25-DKP-insulin. In each case the A chain is shown in yellow, residues B1–B20 in orange, and B-chain segment B21–B30 in dark brown. Selected side chains are labeled. (E) NOESY spectrum of ∆PheB24-KP-insulin. Red box indicates upfield region of NOESY spectrum in which contacts from the native A19, B24, and B26 aromatic rings to the methyl resonances of LeuB15 δ1,2-CH3 are ordinarily observed due to ring-current effects; such upfield-shifted methyl resonances are absent in spectra of the ∆PheB24 analog. (F) Models of ∆PheB24-DKP-insulin. NOESY spectra in A–C were acquired at 25 °C with a mixing time of 200 ms.
Fig. 7.
Solution structure of analog 2 superposed on the μIR complex. Yellow, insulin A chain; black, insulin residues B1–B20; brown, B21–B24; gray, B27–B30; magenta, αCT; cyan, L1 domain. Side-chain atoms are shown for αCT residues His-710 and Phe-714 (magenta), ∆PheB25 (dark pink, space-filling), and TyrB26 (light green, space-filling). Conformational restriction by ∆PheB25 predicts steric clashes (see main text).
Analog 3: β-Breaker at PheB24.
Analog 3, containing a ∆Phe-induced kink at position B24, retained high holoreceptor affinity (SI Appendix, Fig. S10A) and TK activation (SI Appendix, Fig. S10 B and C) with substantial potency in vivo (SI Appendix, Fig. S10 D and E). Its CD spectrum by contrast exhibited attenuated α-helix content (SI Appendix, Fig. S8A), and stability was markedly reduced (SI Appendix, Fig. S8B): ∆∆Gu = −0.8 ± 0.2 kcal/mol at 25 °C with respect to the monomeric template (baseline ∆Gu = −3.0 ± 0.1 kcal/mol; SI Appendix, Table S4). Further, 1H-NMR spectra of analog 3 were broadened with reduced chemical shift dispersion (Fig. 6E and SI Appendix, Fig. S8F). Broadening worsened over hours at 25 °C, suggesting progressive aggregation despite the analog’s intended monomeric design (17). Similar trends were observed under a variety of conditions (temperature, buffer composition, and pH). Although these features precluded detailed NMR analysis, modeling suggested that the analog populates an ensemble of partial folds encompassing the μIR-bound state of insulin (Fig. 6F). Indeed, under a variety of structural assumptions (SI Appendix), the near-planar residue at B24 is found to direct the C-terminal B-chain segment away from the helical core of the hormone, rationalizing its productive engagement with the insulin receptor (SI Appendix, Fig. S5C).
Together, the contrasting properties of analogs 2 and 3 elucidate key structural requirements of the B-chain hinge, with immobilization (via ∆PheB25) preserving native structure and assembly at the price of receptor binding and forced opening (via ∆PheB24) preserving activity at the price of protein instability and nonnative aggregation.
Analog 4: GlyB24 Detachment and Fibrillation.
Insulin fibrillation, a central concern in pharmaceutical formulation (25), provides a model of aggregation-coupled misfolding (26). Whereas protective substitutions are rare among standard variants (22, 27), analog 1 was resistant to fibrillation. On gentle agitation at 37 °C (22), KP-insulin (made 60 μM in PBS at pH 7.4) exhibited a lag time of 3 ± 0.3 d (n = 20); under the same conditions, [d-AlaB20, d-AlaB23]-KP-insulin exhibited lag times of 15 and 18 d (n = 2), presumably due to its greater dynamic and thermodynamic stability. Analogs 2 and 3 also exhibited longer lag times,§ but interpretation of these findings is confounded by potential effects of the planar ∆Phe conformation on cross–β-assembly (28, 29).
To test the relationship between C-terminal B-chain detachment (as seen in WT insulin at elevated temperatures that promote insulin fibrillation) (30) and misfolding, we prepared an analog in which PheB24 was substituted by glycine. Analog 4 (GlyB24-KP-insulin) thus lacks a nonpolar side chain to anchor the B24–B28 segment to the α-helical hormone core. Consistent with past studies (4, 5, 31), this analog retains native potency (SI Appendix, Fig. S11A), despite its decreased thermodynamic stability (∆∆Gu = −0.7 ± 0.2 kcal/mol at 25 °C relative to KP-insulin) and despite its seeming lack of a conserved receptor contact (Discussion). 1H-NMR studies (at neutral pH) demonstrated marked flexibility of the B20–B30 segment without precise packing of PheB25 or TyrB26 against the central B-chain α-helix (SI Appendix, Fig. S11 B–D and Table S7). The fibrillation lag time of analog 4 is foreshortened (1.0, 1.6, and 1.6 d; n = 3). We envisage that the analog’s anomalous activity and susceptibility to fibrillation have a common origin: enhanced detachability of the B-chain hinge. Such a shared mechanism is consistent with the accelerated fibrillation of analogs containing C-terminal truncated B chains (25) and the forestalled fibrillation of single-chain insulins (SCIs) whose topology favors the closed hinge conformation (32).
Discussion
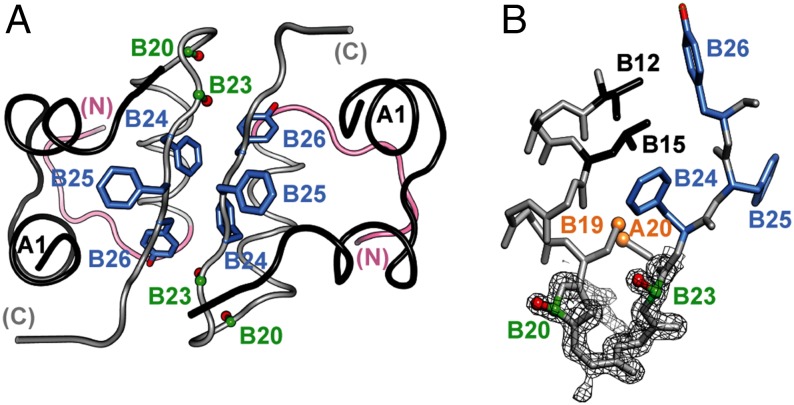
The classical structure of insulin, determined as a zinc-coordinated hexamer, is remarkable for an aromatic-rich dimerization element. This interface contains six invariant residues (PheB24-PheB25-TyrB26 and their dimer-related partners) within an antiparallel β-sheet. The present study defines how the elements of this sheet reorganize to engage conserved receptor surfaces.
Concordance with Prior Biochemical Data.
The salient feature of the B24–B27 segment in the μIR complex is its alternating pattern of side-chain contacts with L1 and αCT: PheB24 and TyrB26 are primarily directed toward L1, PheB25, and ThrB27 toward αCT. Such alternation was foreshadowed by photo–cross-linking studies of the insulin-holoreceptor complex (8), supporting the relevance of the μIR model.
Conservation of both PheB24 and its cognate binding pocket among vertebrate sequences is striking. Ala scanning mutagenesis of the pocket-lining residues Asn15, Leu37, Phe39, and Phe714 impairs high-affinity hormone binding to the holoreceptor (33) or ectodomain (34).
The B25-related surface is partly accessible to solvent. Lined by Pro716 and Pro718 in IR-A (Pro716 and Lys718 in IR-B) and multiple main-chain atoms, this open surface presumably contributes to the rigid requirement for an aromatic side chain at B25 (2, 4, 35). In the holoreceptor, substitution of Val715 by Ala eliminates detectable hormone binding (33). It is unclear, however, why substitution of PheB25 by Ala, deleterious in full-length insulin (13), is by contrast well tolerated in truncated analog des-pentapeptide[B26-B30]-insulin-amide (see below) (35). It is possible that native PheB25–receptor interactions contribute to coordinated displacement of the B26–B30 segment on receptor binding and are not required by the truncated analog.
The interaction of TyrB26 with site 1 is sparse and restricted to Asp12 and Arg14. This observation rationalizes (i) the high activity of an AlaB26 analog (36), (ii) the dispensability of TyrB26 in truncated analogs (37), and (iii) that whereas holoreceptor substitution Arg14Ala (near both TyrB26 and the B25 main chain) impairs insulin binding >103-fold, Ala substitution of Asp12 (in contact only with TyrB26) impairs hormone binding by only 6-fold (33). We suggest that the conservation of TyrB26 (2) arises not from its role in receptor binding but instead from its contribution to proinsulin folding (38) and insulin self-assembly (2, 39). Indeed, in the free hormone, TyrB26 inserts within a conserved nonpolar interchain crevice (2); on μIR binding, this crevice is occupied by key αCT side chains His710 and Phe714 (11).
The present structure rationalizes the reduced affinity of clinical variant LeuB25-insulin Chicago (7) and essential inactivity of LeuA3-insulin Wakayama (7, 40). Receptor engagement at these sites reflects mutual induced fit. LeuA3, predicted to clash with αCT, exhibits a residual receptor-binding affinity (0.1% relative to WT insulin) similar to that of ∆PheB25. The μIR structure further accounts for effects of C-terminal truncation or modification of the B chain¶ (41). Truncation at B25 (with C-terminal amide), B26, or B27 yields analogs with complete activity, in accordance with our observations that, beyond PheB24, successive C-terminal residues exhibit progressively more limited (B25–B27) or absent (B28–B30) interactions with the μIR. Modification of the latter residues can modulate pharmacokinetic properties without loss of activity as exploited in clinical formulations (41).
Analog 4 exploits the intrinsic flexibility of glycine. Its high activity (like that of related analogs) (4, 5, 31, 42) poses a seeming paradox: how can the hormone-receptor interface tolerate the absence of an invariant side chain—especially an aromatic ring anchored within a canonical binding pocket? This question is made more pointed by the enhanced receptor-binding affinity conferred by d-AlaB24 and other d substitutions at B24 (6, 42, 43). On the one hand, destabilization of the B20–B30 segment (GlyB24) or its predetachment (d-AlaB24) might mitigate the cost of induced fit and so enhance binding affinity. Such enhancement might be reinforced by an entropic advantage gained by desolvation of nonpolar surfaces—anomalously exposed due to the B24 substitutions—releasing interfacial water molecules into bulk solvent. On the other hand, the entropic cost of immobilizing a flexible B-chain segment on receptor binding would weaken affinity. Further, were such analogs to bind in the same mode as WT insulin, the variant complex would incur a cavity penalty due to an unoccupied B24-related pocket (44). It is possible that these effects cancel each other, rationalizing native activities despite complex perturbations to underlying thermodynamic drivers as a striking example of entropy-enthalpy compensation (EEC).
An alternative and simpler explanation posits a one-residue shift in register between the C-terminal B-chain β-strand and site 1 surface (45). This model envisages that PheB25 occupies the erstwhile B24-binding pocket (and likewise TyrB26 occupies the B25 pocket), leaving a noncanonical five-residue loop between this FY motif and the B-chain α-helix. This model is consistent with the structural compatibility of a register-shifted ThrB27 with the exposed B26-related depression in the μIR and lack of interactions by residues B28–B30. Although in their free states GlyB24-DKP-insulin and d-AlaB24-DKP-insulin (6) lack 1H-NMR features of a register shift (at least not as a stable structural element), we imagine that such a reorganized interface may be more favorable than a binding mode that leaves the B24-related pocket empty. The register-shifted and EEC models may be distinguished through crystallographic studies of variant μIR complexes.
Induced Fit and Biological Signaling.
The function of the B-chain hinge in cellular signaling was probed through comparative studies of ∆Phe analogs. Remarkably, although ∆PheB24-KP-insulin (analog 3) is unstable and prone to aggregation, native receptor-binding affinity was maintained. Further, analog 3 was able to direct reduction of blood glucose concentration in vivo. Modeling, based on qualitative CD and NMR features, suggests that the ∆PheB24 analog populates an ensemble of partial folds consistent with displacement of the B20–B27 segment in the μIR complex. Unlike ∆PheB24-KP-insulin, the free ∆PheB25 analog exhibited unperturbed stability with the B26–B30 segment directed toward the hormone’s helical core as in WT insulin. The contrasting conformations of the B-chain C-terminal segments in the ∆Phe analogs are shown in Fig. 8 in relation to the free structure of insulin and its μIR-bound conformation. ∆PheB25-DKP-insulin thus recapitulates classical structural relationships (1, 2). Strikingly, however, this analog has essentially no biological activity, mirroring LeuA3-insulin (31, 46) and inactive SCIs (3). Loss of activity presumably reflects steric clash between the B25–B30 segment and αCT. Thus, the μIR model—despite its intrinsic limitations as a small fragment of the holoreceptor complex—may rationalize the contrasting properties of the constrained analogs and broadly inform classical structure–activity relationships as discussed above.
Fig. 8.
Orientation of B23–B27 segments in free insulin analogs. The closed B-chain conformation in DKP-insulin (A) is recapitulated in the inactive constrained conformation of ∆PheB25-DKP-insulin (B). The open conformation observed in the μIR complex (C) is in accordance with molecular models of the active but less stable ∆PheB24 analog (D). Green, B23–B27 segment; magenta, the insulin internal contact surface of PheB24 (which includes ValB12, LeuB15, TyrB16, and CysB19; TyrB26 also packs against ValB12); cyan, additional contact surface of TyrB26 with A-chain residues IleA2 and ValA3. The latter nonpolar side chains are exposed in C and D and so poised to engage αCT on receptor binding.
Insulin-Like Growth Factor System.
Concordant with their sequence homology, insulin and the two insulin-like growth factors (IGF-I and IGF-II) are each capable of binding to both the insulin receptor (isoforms A and B) and IGF-1R (47). In structures of free IGFs (48, 49), the corresponding B22–B26 segment has an almost identical disposition (with respect to the α-helical core) to that in insulin; the L1 structure and αCT sequence are also conserved between the insulin receptor and IGF-1R (11). We thus expect that IGF B domains undergo a similar hinge-like detachment on binding to site 1 of IGF-1R. We note in particular that (i) the B20–B23 β-turn (sequence GERG in insulin) is conserved among IGFs both in sequence (GDRG) and structure (48, 49)—suggesting that the homologous turn is also maintained as IGFs engage IGF-1R; and (ii) the aromatic triplet in insulin (PheB24-PheB25-TyrB26) is conserved in IGFs (as Phe-Tyr-Phe) as are its cognate binding residues in IGF-1R (SI Appendix, Fig. S12 A and B).
Determinants of Foldability.
A monogenic form of DM is due to dominant mutations in the insulin gene leading to misfolding of proinsulin (50, 51). Age of onset reflects the severity of the folding defect and extent of interference with WT insulin biosynthesis (38). Of particular interest is GlyB23→Val (50), predicted (as an l-amino acid) to destabilize a positive ϕ dihedral angle at B23 (20). We suggest that ValB23 impairs disulfide pairing in the variant proinsulin by perturbing the orientation of PheB24 and adjoining C-terminal B-domain segment. This model is supported by observations that this segment facilitates classical insulin chain combination (37) and that PheB24 stabilizes nascent structure in peptide models of proinsulin folding intermediates (38).
Substitution of PheB24 by serine (insulin Los Angeles) leads to variable onset of DM in early adulthood (46). In both the free and μIR-bound hormone, the side chain of PheB24 abuts cystine B19–A20. Although secretion of SerB24-insulin was observed in a patient (46), SerB24 was found in a β-cell line to perturb disulfide pairing (52). SerB24-associated DM may thus have dual origins—impaired receptor binding by the mutant insulin (46) and β-cell dysfunction resulting from chronic endoplasmic reticular (ER) stress (52). The extent of impaired binding (14-fold) is less marked than that of insulins Chicago or Wakayama: PheB25→Leu (50-fold) or ValA3→Leu (1000-fold) (31, 46). Because SerB24 introduces segmental B-chain destabilization similar to that of GlyB24 (53), its partial activity may be rationalized by the above register-shifted or EEC models.
Fibrillation.
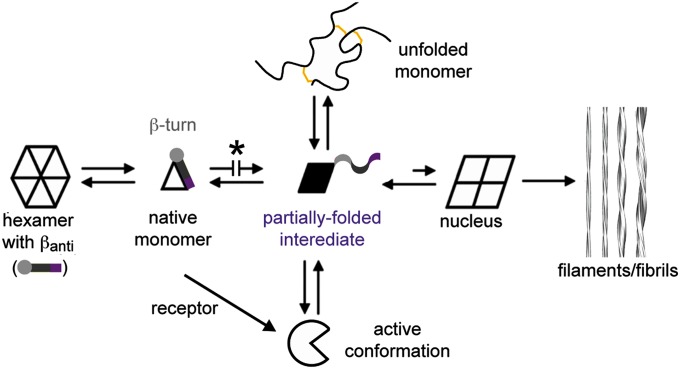
Chiral stabilization of the B20–B23 β-turn (analog 1) delays insulin fibrillation, whereas its destabilization (analog 4) promotes fibrillation. These findings may be rationalized in relation to a general scheme of insulin fibrillation (Fig. 9) based on conformational distortion of a susceptible monomer (27). Whereas the native hormone is protected within storage hexamers (Fig. 9, Left), nonnative aggregates can form amyloidogenic nuclei leading to cross–β-assembly in protofilaments and fibrils (Fig. 9, Right). We envisage hinge opening both enables receptor binding and underlies the susceptibility of the monomeric hormone to fibrillation. Like populated partial folds in amyloidogenic proteins in general (28, 29), conformational distortion of insulin represents a breakdown of the cooperativity of protein folding.
Fig. 9.
Mechanism of insulin fibrillation via partial unfolding. The native state is protected by classical self-assembly (far left), mediated in part by an anti-parallel β-sheet (βanti) at the dimer interfaces of the hexamer. C-terminal segment of B-chain is represented by light gray circle (B20–B23 β-turn), gray bar (B24–B28 β-strand), and purple bar (less ordered C-terminal residues B29 and B30) (1, 2). Disassembly leads to equilibrium between native- and partially folded monomers (open triangle and black trapezoid). Detachment of the C-terminal B-chain segment within a partial fold (5, 6) may lead to the off-pathway, active conformation (open circle) or to an aggregated nucleus en route to a protofilament assembly (far right). Asterisk highlights protective modifications stabilizing the closed conformation: d-Ala–locked β-turn (analog 1), ∆PheB25 (analog 2 relative to ∆PheB24), and SCIs (25, 32, 70, 71). The unfolded state, constrained by native disulfide bridges (gold), shown in schematic form at the top, is off-pathway. Reproduced with permission from ref. 26.
Lack of correlation between fibrillation lag times of insulin analogs and native state stability (22, 27) reflects a key aspect of fibrillation: the unfolded-state ensemble is off-pathway (Fig. 9, Top). Whereas measurements of stability ordinarily probe free-energy differences (∆Gu) relative to this ensemble, susceptibility to fibrillation may be influenced by the relative stabilities of partial folds, intervening kinetic barriers, and mechanisms of secondary nucleation (29). That hinge opening enhances susceptibility to fibrillation is in accordance with the stereospecific effects of l- and d-Ala substitutions at B24: although these diastereomers exhibit similar thermodynamic stabilities, the lag time of the (active) d-AlaB24 analog (with unstructured B20-B30 segment) is threefold shorter than that of the (inactive) l-AlaB24 analog, whose hinge is partially closed (6). It would be of future interest to dissect which individual kinetic steps in the mechanism of insulin fibrillation (54, 55) are influenced by substitutions at position B24 in relation to the dynamics of hinge opening. Such analogs provide a model of segmental disorder, a general feature of mutant proteins associated with diverse diseases of toxic protein misfolding and amyloid deposition (29).
Aggregation-coupled misfolding may have evolutionary consequences. In the caviomorph rodent Octodon degus, for example, endogenous fibrillation of a divergent insulin can lead to selective islet amyloidosis associated with β-cell loss and DM (56, 57). The degu B chain contains multiple anomalous substitutions predicted to impair dimerization (TyrB26→Arg, ThrB27→Pro, and ProB28→His) and the zinc-stabilized hexamer assembly (HisB10→Asn). The marked susceptibility of such rodents to β-cell dysfunction when fed a diet high in carbohydrates (especially free sugars, which are almost absent in its natural plant-based diet) may reflect both extracellular- and intracellular proteotoxicity, respectively, due to amyloidogenesis and impaired folding of the divergent proinsulin—the latter leading to ER stress as in human neonatal DM (52). The anomalous molecular features of degu insulin may thus honor in the breach our suggestion that the closed conformation of insulin and its stable self-assembly in secretory vesicles (2, 39) evolved to protect the β cell from proteotoxicity.
Concluding Remarks.
The present study illustrates the power of nonstandard structural constraints (as exemplified by chiral β-turn stabilization and by main-chain planarity enforced through extended π conjugation) to interrogate a functional hinge in a globular protein. Our results have extended the repertoire of insulin structures through multicrystal refinement (18) of the conformation of the hormone bound to the L1 and αCT domains of the receptor α-subunit (11). In this μIR complex, the C-terminal β-turn (residues B20–B23) and β-strand (B24–B27) rotate away from the α-helical core of insulin to enable its conserved aromatic motif (PheB24-PheB25-TyrB26) to dock between L1 and αCT. The B chain thus contains a hinge whose closed setting functions in β-cell biosynthesis—from the nascent folding of proinsulin (38) to zinc-mediated self-assembly (2, 39)—and whose open setting mediates biological activity.
The μIR model provides a simplified view of the structural complexity of the intact insulin receptor. Indeed, asymmetric binding of insulin to site 1, as visualized here, defines only the first step of what is likely to be a complex choreography of conformational change leading to activation of the receptor tyrosine kinase domain and its multiple signaling outputs. Clues regarding such subsequent steps have been provided by analysis of mutations at the edge of the present site 1-binding surface (58) or at sites in contact with site 2 of the holoreceptor (9). These analogs distinguish between the thermodynamics of receptor binding and determinants of biological signaling. The μIR surface in contact with the conserved aromatic side chains at insulin positions B24–B26 is intimately connected to putative signaling surfaces of αCT and insulin. In particular, the deep and distinctive B24-binding pocket may provide an attractive target for engineering insulin analogs with enhanced potency or selective activities as biased agonists. Such molecular engineering defines a key emerging frontier of pharmacology.
Materials and Methods
Detailed materials and methods are available in SI Appendix.
Preparation of Insulin Analogs.
[13C, 15N]-labeled peptides were prepared by solid-phase peptide synthesis, purified by HPLC, and assessed by MS. Unlabeled B23–B30 octapeptides containing Ala, Gly, or ∆Phe substitutions were likewise prepared; ∆Phe-containing octapeptides were also synthesized as described (12). Labeled or variant insulins were prepared by semisynthesis (42). Analog 1 was prepared by chain combination (15). Biosynthetic isotopic labeling of the α-helical domain of analogs 2 and 3 (residues B1–B22 and A1–A21) was accomplished in Pichia pastoris (14).
μIR Stability and Holoreceptor Binding.
Relative stabilities of variant μIR complexes were assessed in a competitive binding assay using polyethylene glycol (PEG) precipitation (59); holoreceptor binding affinities were obtained as previously described (60).
TK Activation Assays.
Relative activities of insulin analogs were evaluated in an in vitro assay of hormone-stimulated receptor autophosphorylation (33).
Biological Studies.
Insulin-stimulated receptor autophosphorylation and protein kinase B phosphorylation were assessed in an IGF-1R–deficient mouse embryo fibroblast cell line expressing the human insulin receptor (isoform B) (61). Potency of insulin analogs was tested in male Sprague–Dawley rats rendered diabetic by streptozotocin (22). Statistical significance was assessed using a Student t test.
Fibrillation Assays.
Fibrils were induced at 37 °C by gentle agitation of a solution of insulin analog at 60 μM in PBS (pH 7.4) and monitored by thioflavin T fluorescence and transmission electron microscopy (EM) (22).
Crystallographic Analysis of the μIR Complex.
Crystal growth and data collection protocols were as previously described (11). Six datasets displayed diffraction to higher than 4.0-Å resolution—two successfully comerged to 3.5 Å with that originally reported. The original structure [Protein Data Bank (PDB) ID code 3W11 (11)] was then refined against the merged set. Inspection of the 2Fobs-Fcalc difference electron density map revealed two polypeptide-like segments: one running approximately parallel to the first strand of L1–β2 and the other extending onward from IR 715 (the last modeled residue of the αCT); these segments could be successfully mapped to insulin B22–B27 and αCT 716–719, respectively. These additional residues were then included in the model, and the entire model further refined at 3.5-Å resolution. Data processing and refinement statistics are in SI Appendix, Table S3.
Crystallographic Analysis of Analog 1.
Zinc-free crystals of analog 1 were obtained during attempts to cocrystallize with the μIR, Fab 83-7, and αCT peptide; conditions were subsequently refined to 0.7 M trisodium citrate and 0.1 M imidazole-HCl (pH 8.0). Diffraction data were collected at beamline MX2 (Australian Synchrotron). The structure was refined (following molecular replacement) to a resolution of 1.40 Å; data processing and refinement statistics are in SI Appendix, Table S5.
Circular Dichroism and Stabilities.
CD spectra acquisition, assays of guanidine denaturation, and thermodynamic modeling were undertaken as previously described (22).
NMR Spectroscopy and Structure Calculations.
Spectra were acquired at 600, 700, and 800 MHz; [1H, 13C]-TROSY spectra were acquired at 900 MHz (16). Data were processed using NMRPipe (62). Structures of analogs 1 and 4 were determined as previously described (6, 15); interpretation of long-range NOEs was aided by use of multiple mixing times and model-based back-calculation of predicted NOESY spectra and aromatic ring-current shifts. Heteronuclear studies of analog 2 were performed as previously described (22). In brief, assignments were based on 3D HNCACB, CBCA(CO)NH, C(CO)NH, H(CCO)NH, and HCCH-TOCSY spectra and extended by analyses of 3D 13C- and 15N-separated NOESY-HSQC spectra. Distance restraints in the labeled domain were derived from 4D-NOESY spectra. The solution structure of analog 2 was calculated using X-PLOR-NIH. Models of analog 3 were similarly calculated based on structural assumptions.
Molecular Modeling of μIR Complexes.
Modeling was performed using MODELLER (63) with μIR fragments from PDB ID code 3W11 (11) and native insulin from PDB ID code 2G4M (64). Molecular dynamics calculations were performed with GROMACS (v4.5.5) (65) using the OPLS-aa force field (66, 67). Partial atomic charges for ∆Phe were obtained from the electrostatic potential calculated with GAUSSIAN-09 (68).
Supplementary Material
Acknowledgments
We thank Q. X. Hua and K. Huang for initial studies; M. Gupta for assistance with ∆Phe-related synthesis; P. G. Katsoyannis, S. B. Kent, and S. H. Nakagawa for advice; K. Siddle for supplying the 83-7 hybridomas; and L. Lu and the fermentation group at the Commonwealth Scientific and Industrial Research Organization (CSIRO) Materials Science and Engineering laboratory (Parkville, Australia). This work was supported by Australian National Health and Medical Research Council (NHMRC) Project Grants 1005896 and 1058233 and the Hazel and Pip Appel Fund (to M.C.L.); National Institutes of Health (NIH) Grants DK04949 and DK079233 and The Leona M. and Harry B. Helmsley Charitable Trust (to M.A.W.); Grant 7-13-IN-31 from the American Diabetes Association (to N.B.P.); and NHMRC Independent Research Institutes Infrastructure Support Scheme Grant 361646 and a Victorian State Government Operational Infrastructure support grant (to the Walter and Eliza Hall Institute). S.J.C. and D.F.S. received support from National Institutes of Health Grants DK013914 and UC DRTC DK020595. V.P. received support from Institutional NIH Medical Scientist Training Program Grant T32 GM007250 and NIH Fellowship F30 DK094685-04. Structural data were obtained at the Australian Synchrotron (beam line MX2) and Cleveland Center for Membrane and Structural Biology.
Footnotes
Conflict of interest statement: F.I.-B. and M.A.W. hold stock in Thermalin Diabetes, LLC (Cleveland, OH), for which N.B.P. and J.W. are consultants; M.A.W. is Chief Scientific Officer and a Director. Part of M.C.L.'s research is funded by Sanofi (Germany).
*Whereas site 1 contacts insulin’s classical receptor-binding surface, site 2 (proposed to reside at the junction of the α-subunit’s fibronectin homology domains 1 and 2) is defined in relation to cognate substitutions in the hormone’s trimer-forming surface that affect the lifetime of the holoreceptor complex (9).
†In the original map, PheB24 side-chain density was overlain in part by AsnA21; the latter was found here to project away from the axis of the A-chain C-terminal helix (Fig. 4C).
‡Monomeric insulin analogs contained substitutions ProB28→Lys and LysB29→Pro (i.e., parent KP-insulin; PDB ID code 2KJJ) or these with addition of HisB10→Asp (DKP-insulin; PDB ID code 2JMN) (Table 1).
§Analog 2 exhibited lag times of 9, 10, and 10 d (N = 3); analog 3 exhibited lag times of 5, 5, and 6 d (N = 3). Whereas the latter may have been influenced by both the generic properties of ∆Phe and the initial state of ∆PheB24-KP-insulin (i.e., protective noncanonical α-helical aggregates), the more marked delay characteristic of monomeric ∆PheB25-KP-insulin is likely to reflect the dynamic and thermodynamic stabilization of its closed conformation (Fig. 6D). Negative-stain transmission electron microscopic studies demonstrated fibrillar structures in all end point samples except those formed by ∆PheB24 -KP-insulin, which appeared amorphous.
¶Nonstandard analogs directing B26 (and, where present, residues beyond) away from the insulin core exhibit high affinity presumably due to general predisplacement of B26–B30 rather than specific mimicry of the receptor-bound state as originally envisioned (69).
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (insulin/μIR/Fab 83-7 complex, PDB ID code 4OGA; insulin analog 1, PDB ID code 4NIB; analog 2, PDB ID code 2MLI; and analog 4, PDB ID code 2MPI).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412897111/-/DCSupplemental.
References
- 1.Adams MJ, et al. Structure of rhombohedral 2 zinc insulin crystals. Nature. 1969;224(5218):491–495. [Google Scholar]
- 2.Baker EN, et al. The structure of 2Zn pig insulin crystals at 1.5 A resolution. Philos Trans R Soc Lond B Biol Sci. 1988;319(1195):369–456. doi: 10.1098/rstb.1988.0058. [DOI] [PubMed] [Google Scholar]
- 3.Derewenda U, et al. X-ray analysis of the single chain B29-A1 peptide-linked insulin molecule. A completely inactive analogue. J Mol Biol. 1991;220(2):425–433. doi: 10.1016/0022-2836(91)90022-x. [DOI] [PubMed] [Google Scholar]
- 4.Mirmira RG, Nakagawa SH, Tager HS. Importance of the character and configuration of residues B24, B25, and B26 in insulin-receptor interactions. J Biol Chem. 1991;266(3):1428–1436. [PubMed] [Google Scholar]
- 5.Hua QX, Shoelson SE, Kochoyan M, Weiss MA. Receptor binding redefined by a structural switch in a mutant human insulin. Nature. 1991;354(6350):238–241. doi: 10.1038/354238a0. [DOI] [PubMed] [Google Scholar]
- 6.Hua QX, et al. Enhancing the activity of a protein by stereospecific unfolding: Conformational life cycle of insulin and its evolutionary origins. J Biol Chem. 2009;284(21):14586–14596. doi: 10.1074/jbc.M900085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shoelson S, et al. Three mutant insulins in man. Nature. 1983;302(5908):540–543. doi: 10.1038/302540a0. [DOI] [PubMed] [Google Scholar]
- 8.Xu B, et al. Decoding the cryptic active conformation of a protein by synthetic photoscanning: Insulin inserts a detachable arm between receptor domains. J Biol Chem. 2009;284(21):14597–14608. doi: 10.1074/jbc.M900087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Meyts P, Whittaker J. Structural biology of insulin and IGF1 receptors: implications for drug design. Nat Rev Drug Discov. 2002;1(10):769–783. doi: 10.1038/nrd917. [DOI] [PubMed] [Google Scholar]
- 10.Smith BJ, et al. Structural resolution of a tandem hormone-binding element in the insulin receptor and its implications for design of peptide agonists. Proc Natl Acad Sci USA. 2010;107(15):6771–6776. doi: 10.1073/pnas.1001813107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menting JG, et al. How insulin engages its primary binding site on the insulin receptor. Nature. 2013;493(7431):241–245. doi: 10.1038/nature11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta M, et al. Dehydrophenylalanine (DeltaPhe) as a β breaker: Extended structure terminated by a DeltaPhe-induced turn in the pentapeptide Boc-Phe1-Ala2-Ile3-DeltaPhe4-Ala5-OMe. ChemBioChem. 2008;9(9):1375–1378. doi: 10.1002/cbic.200800053. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi M, et al. Changes in receptor binding, biological activity and immunoreactivity of insulin caused by replacing the residues B23-B26 with alanine. Biomed Res. 1984;5(3):267–272. [Google Scholar]
- 14.Huang Y, Liang Z, Feng Y. The relationship between the connecting peptide of recombined single chain insulin and its biological function. Sci China C Life Sci. 2001;44(6):593–600. doi: 10.1007/BF02879353. [DOI] [PubMed] [Google Scholar]
- 15.Huang K, et al. How insulin binds: The B-chain α-helix contacts the L1 β-helix of the insulin receptor. J Mol Biol. 2004;341(2):529–550. doi: 10.1016/j.jmb.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 16.Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94(23):12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua QX, Jia W, Weiss MA. Conformational dynamics of insulin. Front Endocrinol (Lausanne) 2011;2:48. doi: 10.3389/fendo.2011.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336(6084):1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anil B, Song B, Tang Y, Raleigh DP. Exploiting the right side of the Ramachandran plot: Substitution of glycines by D-alanine can significantly increase protein stability. J Am Chem Soc. 2004;126(41):13194–13195. doi: 10.1021/ja047119i. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa SH, et al. Chiral mutagenesis of insulin. Contribution of the B20-B23 β-turn to activity and stability. J Biol Chem. 2006;281(31):22386–22396. doi: 10.1074/jbc.M603547200. [DOI] [PubMed] [Google Scholar]
- 21.Weiss MA, Hua QX, Lynch CS, Frank BH, Shoelson SE. Heteronuclear 2D NMR studies of an engineered insulin monomer: Assignment and characterization of the receptor-binding surface by selective 2H and 13C labeling with application to protein design. Biochemistry. 1991;30(30):7373–7389. doi: 10.1021/bi00244a004. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, et al. An Achilles’ heel in an amyloidogenic protein and its repair: Insulin fibrillation and therapeutic design. J Biol Chem. 2010;285(14):10806–10821. doi: 10.1074/jbc.M109.067850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta M, Chauhan VS. De novo design of α,β-didehydrophenylalanine containing peptides: From models to applications. Biopolymers. 2011;95(3):161–173. doi: 10.1002/bip.21561. [DOI] [PubMed] [Google Scholar]
- 24.Yang Y, et al. Solution structure of proinsulin: Connecting domain flexibility and prohormone processing. J Biol Chem. 2010;285(11):7847–7851. doi: 10.1074/jbc.C109.084921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brange J, Andersen L, Laursen ED, Meyn G, Rasmussen E. Toward understanding insulin fibrillation. J Pharm Sci. 1997;86(5):517–525. doi: 10.1021/js960297s. [DOI] [PubMed] [Google Scholar]
- 26.Jiménez JL, et al. The protofilament structure of insulin amyloid fibrils. Proc Natl Acad Sci USA. 2002;99(14):9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen L, Frokjaer S, Brange J, Uversky VN, Fink AL. Probing the mechanism of insulin fibril formation with insulin mutants. Biochemistry. 2001;40(28):8397–8409. doi: 10.1021/bi0105983. [DOI] [PubMed] [Google Scholar]
- 28.Booth DR, et al. Instability, unfolding and aggregation of human lysozyme variants underlying amyloid fibrillogenesis. Nature. 1997;385(6619):787–793. doi: 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- 29.Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 30.Hua QX, Weiss MA. Mechanism of insulin fibrillation: The structure of insulin under amyloidogenic conditions resembles a protein-folding intermediate. J Biol Chem. 2004;279(20):21449–21460. doi: 10.1074/jbc.M314141200. [DOI] [PubMed] [Google Scholar]
- 31.Shoelson SE, Lu ZX, Parlautan L, Lynch CS, Weiss MA. Mutations at the dimer, hexamer, and receptor-binding surfaces of insulin independently affect insulin-insulin and insulin-receptor interactions. Biochemistry. 1992;31(6):1757–1767. doi: 10.1021/bi00121a025. [DOI] [PubMed] [Google Scholar]
- 32.Huang K, Dong J, Phillips NB, Carey PR, Weiss MA. Proinsulin is refractory to protein fibrillation: Topological protection of a precursor protein from cross-β assembly. J Biol Chem. 2005;280(51):42345–42355. doi: 10.1074/jbc.M507110200. [DOI] [PubMed] [Google Scholar]
- 33.Whittaker J, Whittaker L. Characterization of the functional insulin binding epitopes of the full-length insulin receptor. J Biol Chem. 2005;280(22):20932–20936. doi: 10.1074/jbc.M411320200. [DOI] [PubMed] [Google Scholar]
- 34.Williams PF, Mynarcik DC, Yu GQ, Whittaker J. Mapping of an NH2-terminal ligand binding site of the insulin receptor by alanine scanning mutagenesis. J Biol Chem. 1995;270(7):3012–3016. doi: 10.1074/jbc.270.7.3012. [DOI] [PubMed] [Google Scholar]
- 35.Mirmira RG, Tager HS. Disposition of the phenylalanine B25 side chain during insulin-receptor and insulin-insulin interactions. Biochemistry. 1991;30(33):8222–8229. doi: 10.1021/bi00247a019. [DOI] [PubMed] [Google Scholar]
- 36.Kristensen C, et al. Alanine scanning mutagenesis of insulin. J Biol Chem. 1997;272(20):12978–12983. doi: 10.1074/jbc.272.20.12978. [DOI] [PubMed] [Google Scholar]
- 37.Cosmatos A, Ferderigos N, Katsoyannis PG. Chemical synthesis of [des(tetrapeptide B27--30), Tyr(NH2)26-B] and [des(pentapeptide B26--30), Phe(NH2)25-B] bovine insulins. Int J Pept Protein Res. 1979;14(5):457–471. doi: 10.1111/j.1399-3011.1979.tb01957.x. [DOI] [PubMed] [Google Scholar]
- 38.Weiss MA. Diabetes mellitus due to the toxic misfolding of proinsulin variants. FEBS Lett. 2013;587(13):1942–1950. doi: 10.1016/j.febslet.2013.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dodson G, Steiner D. The role of assembly in insulin’s biosynthesis. Curr Opin Struct Biol. 1998;8(2):189–194. doi: 10.1016/s0959-440x(98)80037-7. [DOI] [PubMed] [Google Scholar]
- 40.Nanjo K, et al. Diabetes due to secretion of a structurally abnormal insulin (insulin Wakayama). Clinical and functional characteristics of [LeuA3] insulin. J Clin Invest. 1986;77(2):514–519. doi: 10.1172/JCI112331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayer JP, Zhang F, DiMarchi RD. Insulin structure and function. Biopolymers. 2007;88(5):687–713. doi: 10.1002/bip.20734. [DOI] [PubMed] [Google Scholar]
- 42.Mirmira RG, Tager HS. Role of the phenylalanine B24 side chain in directing insulin interaction with its receptor. Importance of main chain conformation. J Biol Chem. 1989;264(11):6349–6354. [PubMed] [Google Scholar]
- 43.Žáková L, et al. Structural integrity of the B24 site in human insulin is important for hormone functionality. J Biol Chem. 2013;288(15):10230–10240. doi: 10.1074/jbc.M112.448050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthews BW. Studies on protein stability with T4 lysozyme. Adv Protein Chem. 1995;46:249–278. doi: 10.1016/s0065-3233(08)60337-x. [DOI] [PubMed] [Google Scholar]
- 45.Ludvigsen S, Olsen HB, Kaarsholm NC. A structural switch in a mutant insulin exposes key residues for receptor binding. J Mol Biol. 1998;279(1):1–7. doi: 10.1006/jmbi.1998.1801. [DOI] [PubMed] [Google Scholar]
- 46.Shoelson SE, Polonsky KS, Zeidler A, Rubenstein AH, Tager HS. Human insulin B24 (Phe--Ser). Secretion and metabolic clearance of the abnormal insulin in man and in a dog model. J Clin Invest. 1984;73(5):1351–1358. doi: 10.1172/JCI111338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Meyts P, Whittaker J. Structural biology of insulin and IGF1 receptors: Implications for drug design. Nat Rev Drug Discov. 2002;1(10):769–783. doi: 10.1038/nrd917. [DOI] [PubMed] [Google Scholar]
- 48.Vajdos FF, et al. Crystal structure of human insulin-like growth factor-1: Detergent binding inhibits binding protein interactions. Biochemistry. 2001;40(37):11022–11029. doi: 10.1021/bi0109111. [DOI] [PubMed] [Google Scholar]
- 49.Brzozowski AM, et al. Structural origins of the functional divergence of human insulin-like growth factor-I and insulin. Biochemistry. 2002;41(30):9389–9397. doi: 10.1021/bi020084j. [DOI] [PubMed] [Google Scholar]
- 50.Støy J, et al. Neonatal Diabetes International Collaborative Group Insulin gene mutations as a cause of permanent neonatal diabetes. Proc Natl Acad Sci USA. 2007;104(38):15040–15044. doi: 10.1073/pnas.0707291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colombo C, et al. Early Onset Diabetes Study Group of the Italian Society of Pediatric Endocrinology and Diabetes (SIEDP) Seven mutations in the human insulin gene linked to permanent neonatal/infancy-onset diabetes mellitus. J Clin Invest. 2008;118(6):2148–2156. doi: 10.1172/JCI33777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu M, et al. Mutant INS-gene induced diabetes of youth: Proinsulin cysteine residues impose dominant-negative inhibition on wild-type proinsulin transport. PLoS ONE. 2010;5(10):e13333. doi: 10.1371/journal.pone.0013333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hua QX, Shoelson SE, Inouye K, Weiss MA. Paradoxical structure and function in a mutant human insulin associated with diabetes mellitus. Proc Natl Acad Sci USA. 1993;90(2):582–586. doi: 10.1073/pnas.90.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knowles TPJ, et al. An analytical solution to the kinetics of breakable filament assembly. Science. 2009;326(5959):1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- 55.Buell AK, et al. Electrostatic effects in filamentous protein aggregation. Biophys J. 2013;104(5):1116–1126. doi: 10.1016/j.bpj.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishi M, Steiner DF. Cloning of complementary DNAs encoding islet amyloid polypeptide, insulin, and glucagon precursors from a New World rodent, the degu, Octodon degus. Mol Endocrinol. 1990;4(8):1192–1198. doi: 10.1210/mend-4-8-1192. [DOI] [PubMed] [Google Scholar]
- 57.Hellman U, et al. Amino acid sequence from degu islet amyloid-derived insulin shows unique sequence characteristics. Biochem Biophys Res Commun. 1990;169(2):571–577. doi: 10.1016/0006-291x(90)90369-x. [DOI] [PubMed] [Google Scholar]
- 58.Whittaker J, et al. α-Helical element at the hormone-binding surface of the insulin receptor functions as a signaling element to activate its tyrosine kinase. Proc Natl Acad Sci USA. 2012;109(28):11166–11171. doi: 10.1073/pnas.1205681109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kristensen C, Andersen AS, Ostergaard S, Hansen PH, Brandt J. Functional reconstitution of insulin receptor binding site from non-binding receptor fragments. J Biol Chem. 2002;277(21):18340–18345. doi: 10.1074/jbc.M112249200. [DOI] [PubMed] [Google Scholar]
- 60.Whittaker L, Hao C, Fu W, Whittaker J. High-affinity insulin binding: Insulin interacts with two receptor ligand binding sites. Biochemistry. 2008;47(48):12900–12909. doi: 10.1021/bi801693h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Denley A, et al. Differential activation of insulin receptor substrates 1 and 2 by insulin-like growth factor-activated insulin receptors. Mol Cell Biol. 2007;27(10):3569–3577. doi: 10.1128/MCB.01447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 63.Fiser A, Sali A. Modeller: Generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 64.Mueller-Dieckmann C, et al. On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 3):366–380. doi: 10.1107/S0907444906055624. [DOI] [PubMed] [Google Scholar]
- 65.Hess B, Kutzner C, Van Der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 66.Jorgensen WL, Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 67.Damm W, Frontera A, Tirado–Rives J, Jorgensen WL. OPLS all-atom force field for carbohydrates. J Comput Chem. 1997;18(16):1955–1970. [Google Scholar]
- 68.Frisch M, et al. Gaussian 09, Revision A. 02. Wallingford, CT: Gaussian; 2009. [Google Scholar]
- 69.Jirácek J, et al. Implications for the active form of human insulin based on the structural convergence of highly active hormone analogues. Proc Natl Acad Sci USA. 2010;107(5):1966–1970. doi: 10.1073/pnas.0911785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hua QX, et al. Design of an active ultrastable single-chain insulin analog: Synthesis, structure, and therapeutic implications. J Biol Chem. 2008;283(21):14703–14716. doi: 10.1074/jbc.M800313200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Phillips NB, Whittaker J, Ismail-Beigi F, Weiss MA. Insulin fibrillation and protein design: Topological resistance of single-chain analogs to thermal degradation with application to a pump reservoir. J Diabetes Sci Tech. 2012;6(2):277–288. doi: 10.1177/193229681200600210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.