Significance
This study defines the biochemical and biological specificity of RNA 5′-end discrimination by the human innate immune response protein denoted IFN-induced protein with tetratricopeptide repeats 5 (IFIT5). Cellular and recombinant protein interaction assays revealed an adaptable RNA binding site that, considering both WT and mutant proteins, can broadly accommodate and distinguish phosphate-containing 5′ ends. Comprehensive profiling of IFIT5-associated cellular RNAs by thermostable group II intron reverse transcriptase sequencing identified a broad spectrum of IFIT5-bound precursor and processed tRNA transcripts and revealed a surprisingly flexible order of human tRNA processing reactions. Our findings for IFIT5 illuminate new structural specificities of protein–RNA recognition important for innate immunity.
Keywords: innate immunity, poly-U tailing, protein–RNA interaction, RNA post-transcriptional modifications, tRNA processing
Abstract
Interferon (IFN) responses play key roles in cellular defense against pathogens. Highly expressed IFN-induced proteins with tetratricopeptide repeats (IFITs) are proposed to function as RNA binding proteins, but the RNA binding and discrimination specificities of IFIT proteins remain unclear. Here we show that human IFIT5 has comparable affinity for RNAs with diverse phosphate-containing 5′-ends, excluding the higher eukaryotic mRNA cap. Systematic mutagenesis revealed that sequence substitutions in IFIT5 can alternatively expand or introduce bias in protein binding to RNAs with 5′ monophosphate, triphosphate, cap0 (triphosphate-bridged N7-methylguanosine), or cap1 (cap0 with RNA 2′-O-methylation). We defined the breadth of cellular ligands for IFIT5 by using a thermostable group II intron reverse transcriptase for RNA sequencing. We show that IFIT5 binds precursor and processed tRNAs, as well as other RNA polymerase III transcripts. Our findings establish the RNA recognition specificity of the human innate immune response protein IFIT5.
Innate immune responses provide a front-line defense against pathogens. Unlike adaptive immune responses, innate immunity relies on general principles of discrimination between self and pathogen epitopes to trigger pathogen suppression (1). Pathogen-specific features that can provide this discrimination come under evolutionary selection to evade host detection, and in turn, host genes adapt new recognition specificities for pathogen signatures. Among the most clearly established targets of innate immune response recognition are nucleic acid structures not typical of the host cell, such as cytoplasmic double-stranded RNA (2). Detection of a pathogen nucleic acid signature robustly induces type I IFN, which activates a cascade of pathways for producing antiviral effectors (3).
Cytoplasmic viral RNA synthesis occurs without cotranscriptional coupling to the 5′-capping machinery, which acts pervasively on host cell nuclear RNA polymerase II transcripts (4, 5). Eukaryotic mRNA 5′ ends are first modified by addition of a cap0 structure containing N7-methylated guanosine, which is joined to the first nucleotide (nt) of RNA by a 5′-5′ triphosphate linkage (7mGpppN). In higher eukaryotes including humans, cap0 is further modified by ribose 2′-O-methylation of at least 1 nt (7mGpppNm, cap1) and sometimes 2 nt (7mGpppNmpNm, cap2). Cap0 addition makes essential contributions to mRNA biogenesis and function in steps of mRNA splicing, translation, and protection from decay (4, 5). In contrast, the biological role of mRNA cap0 modification to cap1 and cap2 structures is largely enigmatic. Some viruses encode enzymes for 7mGpppN formation and, less frequently, the ribose 2′-O-methylation necessary to generate cap1 (6). Recent studies show that virally encoded cap 2′-O-methyltransferase activity can inhibit the innate immune response (7–11).
The IFN-induced protein with tetratricopeptide repeats (IFIT) family of tetratricopeptide repeat (TPR) proteins are among the most robustly accumulated proteins following type I IFN signaling (12, 13). Phylogenetic analyses reveal different copy numbers and combinations of four distinct IFIT proteins (IFIT1, 2, 3, and 5) even within mammals, generated by paralog expansions and/or gene deletions, including the loss of IFIT5 in mice and rats (14). Human IFIT1, IFIT2, and IFIT3 coassemble in cells into poorly characterized multimeric complexes that exclude IFIT5 (15, 16). Recombinant IFIT family proteins range from monomer to multimer, with crystal structures solved for a human IFIT2 homodimer (17), the human IFIT5 monomer (16, 18, 19), and an N-terminal fragment of human IFIT1 (18). Studies of IFIT1 report its preferential binding to either 5′ triphosphate (ppp) RNA (15) or cap0 RNA (11, 20) or optimally cap0 without guanosine N7-methylation (10). Reports of IFIT5 RNA binding specificity are likewise inconsistent: the protein has been described to bind RNA single-stranded 5′ ends with ppp and monophosphate (p) but not OH (16); ppp but not p, OH, or cap0 (18); ppp but not cap0 (10, 20); or single-stranded 5′-p RNA and double-stranded DNA (19).
Here we establish the structural principles of RNA ligand recognition by human IFIT5 from both the protein and RNA perspectives, using structure-guided mutagenesis coupled with quantitative binding assays of purified recombinant protein and comprehensive sequencing of IFIT5-bound cellular RNAs. Our results define an IFIT5 RNA binding cavity that discriminates 5′ ends with the monophosphate, triphosphate, cap0, and/or cap1 structure. With a new method for high-throughput sequencing (21), we characterize cellular IFIT5 ligands including precursor and processed tRNAs and polyuridine (poly-U)–tailed tRNA fragments. Overall, this work elucidates the RNA interaction specificity that underlies IFIT5 function in the innate immune response.
Results
IFIT5 Sequence Determinants of Binding to Cellular RNAs.
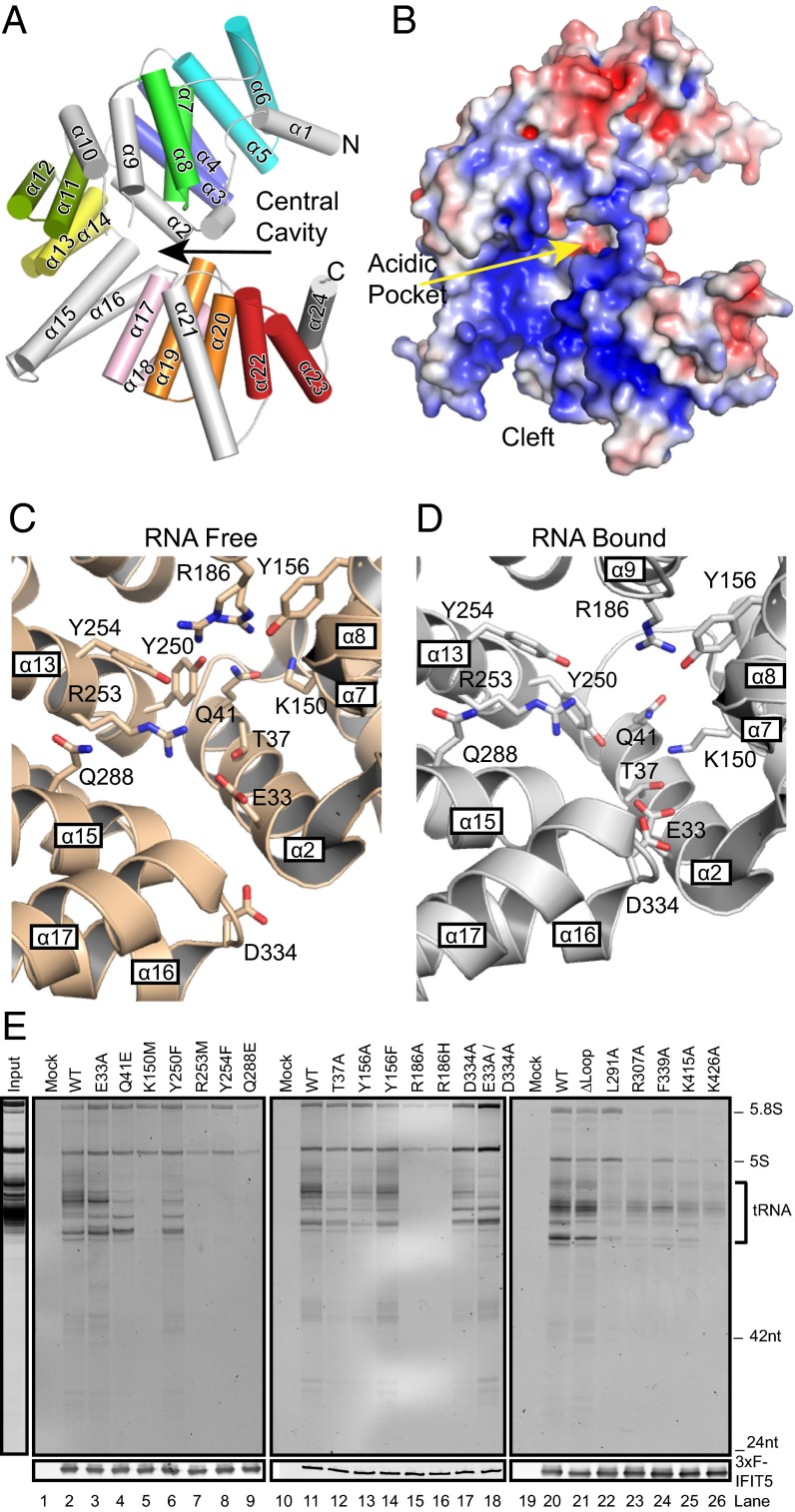
We built on recent high-resolution structures of human IFIT5 alone and with bound homopolymer 5′-ppp RNA oligonucleotides (16, 18, 19) by mutagenesis of numerous side chains brought together in a central cavity at the base of a wide cleft created by an atypical TPR Eddy topology of repeat packing (Fig. 1A, the eight tandem α helix TPRs are differentially colored and non-TPR elements are in gray). The wide cleft has a basic surface generally favorable for RNA interaction, whereas the central cavity has a contrasting acidic pocket (Fig. 1B). Comparison of central cavity residues in the RNA-free and RNA-bound IFIT5 structures (Fig. 1 C and D) suggests that side chains reorient on RNA binding (18).
Fig. 1.
Cellular RNA binding by IFIT5 variants. (A) The TPR Eddy of IFIT5 (RNA-free; PDB ID code 3ZGQ) with each TPR in color and non-TPR helices in gray. Helices are numbered consecutively. (B) Surface electrostatic potential generated with the APBS plugin (32), with the central cavity acidic pocket and wide RNA binding cleft labeled. Contours are red to blue at −4 to +4 kTe−1. (C and D) Expanded views of the central cavity side chains for RNA-free (PDB ID code 3ZGQ) and RNA-bound (PDB ID code 4HOT) IFIT5. (E) RNA bound to WT and mutant 3xF-IFIT5 proteins purified from HEK293T cell extracts, with cell extract lacking tagged protein as a background control (Mock). RNAs were analyzed by denaturing gel electrophoresis and SYBR Gold staining, with parallel FLAG antibody immunoblot of an aliquot of the samples as a recovery control. (Left) A representative input extract RNA profile.
We first expressed WT and mutant IFIT5 proteins with an N-terminal triple-FLAG tag (3xF) in HEK293T cells. IFIT5 RNPs were purified from cell extract, and bound RNAs were detected by SYBR Gold staining after denaturing gel electrophoresis (Fig. 1E and Fig. S1). Using this approach and blot hybridization, IFIT5 was shown to coenrich 5′-p tRNAs and also cellular 5′-ppp RNAs of uncharacterized identity (16). Crystal structures predict a key role for E33 on α2, which positions a sodium or magnesium ion that coordinates with 5′-ppp α and γ phosphate groups deep in the central cavity (18). In a previous study, the E33A substitution gave partial inhibition of IFIT5 association with 5′-ppp RNA oligonucleotide-coated beads (18), with incomplete rather than complete inhibition hypothesized to arise from functional substitution of E33A by the spatially adjacent D334 (Fig. 1D). However, we found that substitutions E33A and D334A, singly or together, did not dramatically affect the overall amount of cellular RNA copurified with IFIT5 but did change the RNA size profile (Fig. 1E, lanes 2–3 and 17–18). Also against previous conclusions, the α2 substitutions Q41E, T37A, and T37V adjacent to the E33/D334 acidic pocket did not preclude RNA binding (Fig. 1E, lanes 4 and 12, and Fig. S1). On the other hand, the substitution K150M on α7 greatly reduced the amount of copurifying RNA, as did K150E but not K150R (Fig. 1E, lane 5, and Fig. S1). IFIT5 with a Y250F substitution of α13 retained RNA binding activity (Fig. 1E, lane 6), inconsistent with this residue providing a critical contact with the 5′-ppp γ phosphate (18). Farther out the RNA binding channel, consistent with previous results (18), α13 substitutions R253M and Y254F and α15 substitution Q288E inhibited IFIT5 association with RNA (Fig. 1E, lanes 7–9).
In RNA-bound IFIT5 structures, Y156 on α8 and R186 on α9 contact the 5′ nt ribose and base hydroxyl, respectively (18). IFIT5 substitutions Y156F and Y156A had only modest influence on cellular RNA binding, whereas R186H and R186A imposed a large loss of copurifying RNA (Fig. 1E, lanes 13–16), consistent with previous assays of R186H and Y156F (18). We also tested the impact of replacing the loop sequence between non-TPR α9 and α10, which is disordered in IFIT5 alone but becomes ordered with bound RNA (16, 18). Deletion of this loop imposed no obvious RNA binding defect (Fig. 1E, lanes 20–21). Substitutions L291A, R307A, K415A, and K426A in the wide RNA binding cleft (16), outside the central cavity, resulted in intermediate reductions of copurifying cellular RNA as did α17 F339A (Fig. 1E, lanes 22–23 and 24–26, and Fig. S2A). Overall, this IFIT5 mutagenesis suggests that the structural determinants of RNA binding are not fully accounted for by predictions of IFIT5 binding specificity based on the cocrystal structures of protein and 5′-ppp homopolymer oligonucleotides (18).
IFIT5 Discrimination of RNA 5′ End Structures.
We next used purified, bacterially expressed WT and mutant IFIT5 proteins (Fig. S2B) in electrophoretic mobility shift assays (EMSAs) with purified radiolabeled RNAs. We used an RNA sequence corresponding to the first 30 nt of West Nile Virus (WNV) positive-strand genomic RNA (WNV30). The WNV30 sequence should be generally representative, because IFIT5 binding is not sequence specific (16, 18). We generated WNV30 RNAs with different 5′ end structures (Fig. 2A and SI Methods) and confirmed quantitative 5′-p, 5′-ppp, or cap0 modification by RNA susceptibility to nuclease degradation using the 5′-p–dependent exonuclease XRN1, with or without prior RNA 5′ polyphosphatase (5′ PPtase) conversion of 5′-ppp to 5′-p, or tobacco acid pyrophosphatase (TAP) conversion of cap0 or cap1 to 5′-p (Fig. 2B).
Fig. 2.
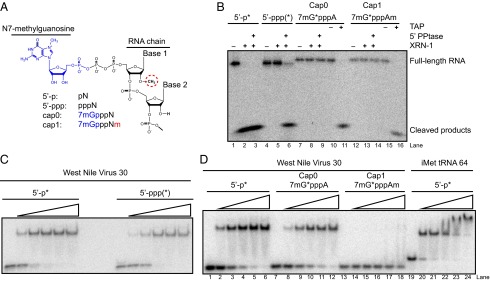
RNA structure discrimination by purified recombinant IFIT5. (A) Illustration of the RNA 5′ structures compared in this study. (B) Verification of 5′-end structure on the WNV30 RNAs used for EMSAs. The radiolabeled phosphate is indicated by an asterisk (*), except that the 5′-ppp RNA contains internal radiolabel from incorporation during transcription. Treatments included 5′ polyphosphatase (5′ PPtase), terminator exonuclease (XRN-1), tobacco acid pyrophosphatase (TAP), or the corresponding buffers without enzyme (–). Products were resolved by denaturing PAGE. (C and D) EMSAs of bacterially expressed WT IFIT5 with the panel of WNV30 RNAs. Protein concentration was titrated from 0.32 to 200 nM. The 64-nt iMet tRNA used in previous EMSA studies (16) is included for comparison.
Purified IFIT5 bound to the 5′-p and 5′-ppp WNV30 RNAs with similar, nanomolar affinity (Fig. 2C, Table 1, and Fig. S3). In addition, IFIT5 bound cap0 but not cap1 WNV30 with an affinity similar to 5′-p WNV30 (Fig. 2D, Table 1, and Fig. S3). None of the WNV30 RNAs bound to IFIT5 with as high affinity as the previously assayed 3′-truncated iMet tRNA (Fig. 2D, lanes 19–24), indicating that regions beyond an RNA 5′ end can contribute to the IFIT5 interaction. Paralleling the results for cellular RNA binding, recombinant IFIT5 E33A, D334A, and E33A/D334A retained near WT binding affinity for 5′-p and 5′-ppp WNV30 RNAs (Table 1 and Fig. S3). The substitutions Q41E, T37A, and Y250F also had little impact on RNA binding, whereas K150M and R253M decreased IFIT5 binding to both 5′-p and 5′-ppp RNAs. Curiously, although R186H and Q288E strongly compromised IFIT5 binding to 5′-p WNV30 RNA and cellular RNA, these substitutions did not substantially affect IFIT5 binding to 5′-ppp WNV30 RNA by EMSA. This differential recognition suggests that changes to IFIT5 amino acid sequence can alter the specificity as well as the affinity of RNA binding.
Table 1.
Dissociation constants
| Protein | 5′-p | 5′-ppp | Cap0 | Cap1 |
| WT* | 0.53 ± 0.13 | 1.4 ± 0.18 | 1.7 ± 0.15 | NC |
| E33A | 0.9 (1)† | 3 | 0.8 (0.6) | 30 (20) |
| E33A/D334A | 2 (1) | 2 | 1 (1) | 20 (20) |
| T37A | 1 (2) | 0.9 | 2 | NC |
| Q41E | 0.4 (0.3) | 2 | >200‡ | NC |
| K150M | 50 (50) | 100 | >200‡ | NC |
| Y156A | 1 | ND | 6 (5) | NC |
| Y156F | 2 (2) | 4 | 3 (2) | NC |
| R186A | >200 | ND | >200 | NC |
| R186H | 70 (80) | 3 (4) | >200‡ | NC |
| Δ Loop | 20 (20) | 4 | 40 (40) | NC |
| Y250F | 1 (2) | 2 | 1 (1) | NC |
| R253M | 90 (100) | 30 | >200 | NC |
| Y254F | 2 (2) | 2 | 90 (90) | NC |
| Q288E | >200‡ | 3 (2) | >200‡ | NC |
| L291A | 0.9 | 1 | 4 | NC |
| R307A | 4 (5) | 1 | 13 | NC |
| D334A | 3 (3) | 2 | 2 (2) | NC |
| F339A | 2 (2) | 3 | 3 | NC |
| K415A | 3 (3) | 3 | 13 | NC |
| K426A | 1 (2) | 2 | 2 | NC |
Values are in nanomolar. NC, not calculated (RNA binding not quantified); ND, not determined (RNA binding not tested).
Values are the mean ± SD of three independent experiments.
Values in parentheses are experimental replicates.
Replicate >200.
We next investigated the influence of IFIT5 sequence substitutions on the binding of cap0 and cap1 WNV30 RNAs. Remarkably, E33A and E33A/D334A IFIT5 proteins gained binding affinity for cap1 WNV30 RNA (Table 1 and Fig. S3). Binding remained specific for the 5′ end structure, because E33A and E33A/D334A IFIT5 proteins did not gain comparable affinity for 5′ OH RNA (Fig. S4). The substitutions Q41E, R186H, Y254F, and Q288E decreased binding to cap0 RNA while retaining 5′-ppp binding, whereas other substitutions did not differentially influence RNA 5′-end discrimination (Table 1 and Fig. S3). Overall, this quantitative protein–RNA interaction analysis establishes a surprisingly adaptable IFIT5 recognition specificity for the RNA 5′ structure.
Comprehensive Profiling of IFIT5-Bound Cellular RNAs.
The broad RNA recognition specificity of IFIT5 in vitro suggested that it could bind to many cellular RNAs. To investigate the diversity of IFIT5-bound cellular RNAs in an unbiased manner, we deep-sequenced RNAs copurified with IFIT5 from HEK293 cells. Because IFIT5 binds tRNAs, which are recalcitrant to standard methods of sequencing, we exploited a new thermostable group II intron reverse transcriptase sequencing (TGIRT-seq) method (21). TGIRTs have higher fidelity and processivity than retroviral reverse transcriptases, as well as a novel template-switching activity that eliminates the need for an RNA 3′ adaptor ligation step for RNA-seq library construction (21, 22).
TGIRT-seq was first performed for cellular RNAs copurified with IFIT5 from a HEK293 cell line with 3xF-IFIT5 expressed at a physiological level (16). To capture in vivo protein–RNA interactions, we used stringent purification after formaldehyde cross-linking, which was reversed before analyzing the bound RNAs (Fig. S5A). We also compared IFIT5-bound RNAs isolated under native affinity purification conditions from extracts of cells with or without prior IFN-β treatment (Fig. S5B). In addition, we compared WT and mutant E33A and E33A/D334A IFIT5 proteins expressed in HEK293 cells by transient transfection (Fig. S5C). In the first set of three samples, comparing WT IFIT5 with or without formaldehyde cross-linking or IFN-β treatment before cell lysis, gel-purified cDNA products were pooled and amplified together (Fig. S5D and Table 2). In the second set of three samples, because RNAs bound to WT and mutant IFIT5 had different size profiles, we amplified and sequenced discrete pools of cDNA lengths (Fig. S5E and Table 2). Finally, in a third sample, we pooled cDNAs before amplification and sequencing for a biological replicate of the WT vs. mutant IFIT5 comparison (Fig. S5C and Table S1). For each purification condition, cDNAs were sequenced on an Illumina MiSeq to a depth of 1,000,000 or more reads, which were mapped to the Ensembl GRCh37 human genome reference sequence (Methods and SI Methods).
Table 2.
TGIRT-seq read mapping
| RNA class | Cross-linked* | Native* | WT† | E33A | E33A/D334A | |||||||
| – | – | + | a | b | c | a | b | c | a | b | c | |
| tRNA | 89 | 90 | 89 | 95 | 91 | 51 | 84 | 72 | 33 | 75 | 54 | 12 |
| snaR | 7.6 | 3.9 | 3.9 | 0.2 | 0.6 | 19 | 0.2 | 1.0 | 9.9 | 0.3 | 0.7 | 4.6 |
| protein_coding‡ | 0.4 | 1.3 | 1.6 | 2.7 | 5.3 | 5.5 | 11 | 21 | 17 | 17 | 33 | 9.1 |
| rRNA‡ | 1.9 | 4.3 | 5.0 | 0.4 | 0.7 | 21 | 1.1 | 1.4 | 35 | 2.5 | 3.6 | 70 |
| snRNA‡ | 0.1 | 0.1 | 0.2 | 0.0 | 0.1 | 0.8 | 0.2 | 0.5 | 1.4 | 0.4 | 0.9 | 1.7 |
| snoRNA‡ | 0.0 | 0.1 | 0.1 | 0.0 | 0.2 | 0.2 | 0.1 | 0.8 | 0.8 | 0.2 | 1.8 | 0.7 |
| misc_RNA‡ | 0.6 | 0.4 | 0.4 | 0.3 | 0.5 | 0.4 | 1.3 | 1.5 | 1.0 | 1.8 | 1.9 | 0.5 |
| tRNA % poly-U tailed | 2.6 | 2.1 | 2.1 | 2.4 | 1.8 | 2.3 | 1.6 | 1.7 | 1.2 | 1.0 | 1.1 | 0.5 |
Table values are rounded percentages. RNA classes >1.5% of reads in any sample are shown.
Cross-linked and native extract purifications were done following induction of IFIT5 expression in cells without (−) or with (+) IFN-β.
Size categories of cDNA a, b, and c (defined in text) were analyzed for WT and mutant IFIT5 proteins expressed by transfection.
Transcript categories from Ensembl GRCh37.
RNA from IFIT5 purifications gave TGIRT-seq reads that mapped predominantly to tRNA gene loci in all samples (Table 2 and Table S1). Cross-linked and native extract purifications showed a large diversity of bound tRNAs, with reads from different samples mapping to 507–527 of the 625 annotated human tRNA and tRNA pseudogene loci (Fig. S6). For IFIT5 expressed by transient transfection with size-selected cDNA pools sequenced separately (Table 2; size categories a, b, and c correspond to cDNA of ∼55–82, 84–150, and 150–230 nt, respectively, including the 42-nt primer added by template switching; Fig. S5 and SI Methods), the largest cDNA size pool contained substantial amounts of 5S rRNA, which is less abundant in the cross-linked RNA purification (Table 2) and thus could in part reflect IFIT5 binding of a highly abundant RNA in native cell extract (16).
To further characterize IFIT5-bound tRNAs, we plotted read coverage across individual tRNA loci from 50 bp upstream to 50 bp downstream of the mature tRNA ends, with representative coverage plots shown for the cross-linked RNA sample (Fig. 3, mature RNA ends are indicated with dashed lines). Some tRNA loci were represented by reads abundant only across the mature tRNA region (iMetCAT, AspGTC, and HisGTG). Read alignments to the genome sequence revealed that many IFIT5-bound mature tRNAs were full length, including the posttranscriptionally added 3′ CCA tail (Fig. S7). In the case of HisGTG, the alignments also detected the expected posttranscriptional 5′ guanosine addition (23). Posttranscriptionally modified nucleotides within the tRNA were evident from positions of frequent read mismatch to the genome sequence (Fig. S7). Some IFIT5-bound tRNA reads had truncated 5′ and/or 3′ ends (Fig. 3 and Fig. S7), resulting from nuclease cleavage of tRNAs and, for 5′-truncated ends, potentially from premature reverse transcription stops.
Fig. 3.
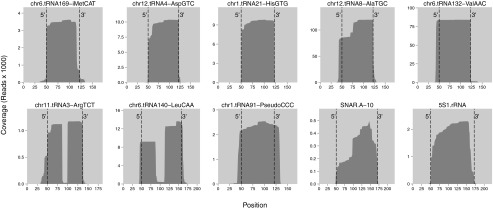
Individual gene coverage by reads from the WT IFIT5 cross-linked RNA sample. Read coverage of loci was mapped for tRNA, snaR, and 5S rRNA genes across a window from 50 bp upstream to 50 bp downstream of the mature RNA ends, which are indicated with dashed lines. The top left plot shows coverage for an iMet tRNA gene, followed by six additional tRNA genes, a tRNA pseudogene, a snaR locus (National Center for Biotechnology Information NR_024229.1), and 5S rRNA (Ensembl ENSG00000199352.1). Each tRNA gene is identified by chromosome number, chromosome position, charged amino acid, and anticodon sequence (5′–3′). Read alignments to genome sequence are shown for each RNA in Fig. S7. The apparent excess of 3′ exon fragments for LeuCAA likely reflects misalignment of truncated 5′ exon sequences by Bowtie 2 local alignment after the gap resulting from intron removal (Fig. S7).
In addition to mature tRNAs, we were surprised to find that numerous tRNA loci were represented by abundant IFIT5-bound tRNAs with the 5′ extension of a primary Pol III transcript (Fig. 3 and Fig. S7; AlaTGC, ValAAC, ArgTCT, and LeuCAA). Many of these 5′-extended tRNAs included the full-length mature tRNA sequence with a 3′ CCA tail (Fig. S7). Also, some IFIT5-bound tRNAs with a 5′ precursor extension and CCA tail had undergone splicing to remove the intron (Fig. 3; ArgTCT and LeuCAA), which is unexpected given that 5′ processing precedes splicing in known tRNA biogenesis pathways (23). Furthermore, some of the spliced tRNAs had aberrant splice junctions suggestive of missplicing (Fig. S7; ArgTCT). Of interest, some 5′ and/or 3′ extended or truncated tRNAs had posttranscriptionally appended poly-U tails (Table 2, Table S1, and Fig. S7). We also found tRNA pseudogene transcripts (Fig. 3, PseudoCCC; full list in Fig. S8), as well as a few tRNAs with atypically long 5′ or 3′ extensions (Fig. S9; AsnGTT, GlnCTG, and ThrCGT) or with sequence reads ending at an internal modified nucleotide position suggestive of a reverse transcription stop (Fig. S9; CysGCA).
Cellular IFIT5 binding to the RNAs described above is consistent with its biochemical specificity of RNA interaction in vitro: precursor tRNAs are expected to have 5′-ppp from RNA polymerase III initiation, whereas mature tRNAs are expected to have 5′-p generated by RNase P. Although biochemically consistent, some types of incompletely processed IFIT5-bound tRNAs should be nuclear, whereas IFIT5 is cytoplasmic (16). The cytoplasmic localization of IFIT5 suggests that some immature or aberrantly processed tRNA transcripts escape the nucleus to become available for IFIT5 binding, either via mis-transport or during mitosis.
IFIT5 also bound to a family of cytoplasmic, ∼120 nt, Alu-related, primate-specific RNA polymerase III small NF90-associated RNA (snaR) transcripts and 5S rRNA (Fig. 3, Table 2, Table S1, and Fig. S7). The snaRs have a single-stranded 5′ end but extensive secondary structure that impedes cDNA synthesis by a conventional reverse transcriptase (24, 25). Nonetheless TGIRT-seq gave coverage across the full snaR (Fig. 3 and Fig. S7). We confirmed snaR association with IFIT5 using blot hybridization (Fig. S5C). Notably, the poly-U tailing of IFIT5-bound tRNAs was also observed for IFIT5-bound snaRs (Fig. S7).
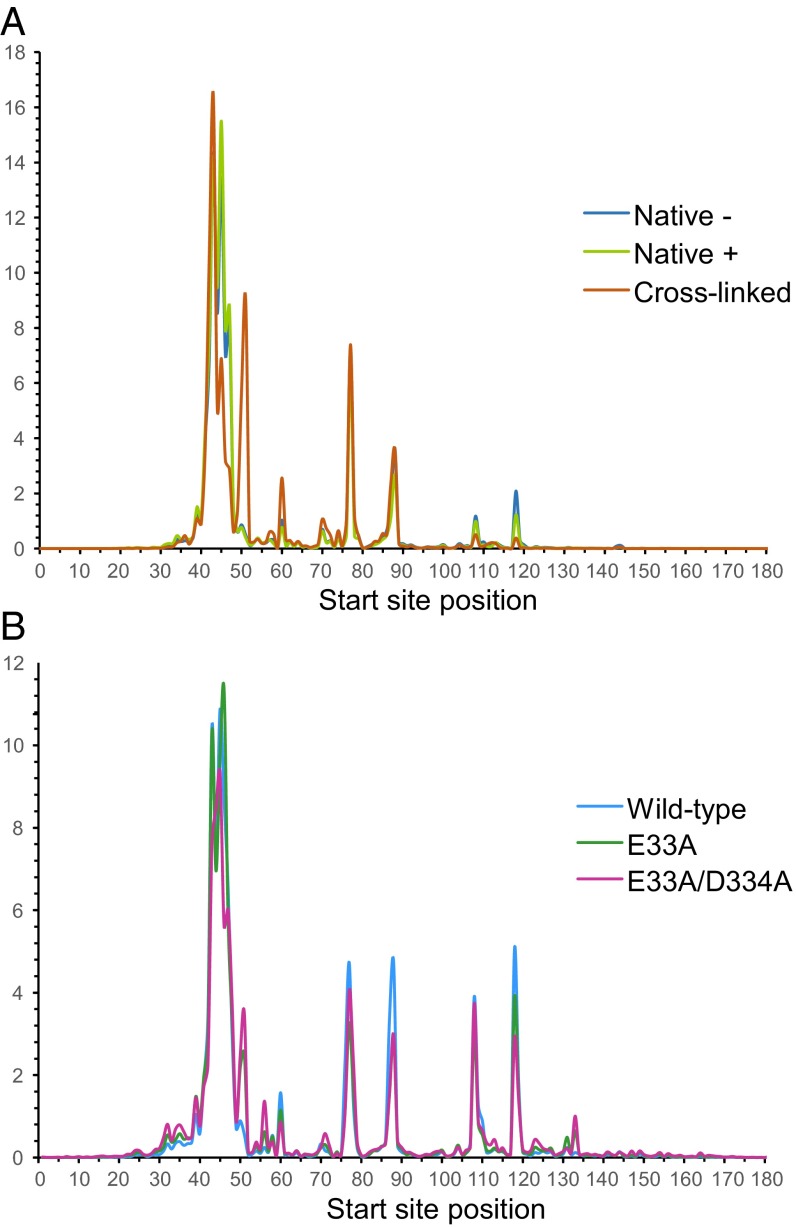
Compared with WT IFIT5 assayed in parallel, E33A or E33A/D334A IFIT5 purifications contained an increased proportion of rRNA and mRNA (Table 2 and Table S1). The mRNA reads showed no obvious bias for 5′ ends (Fig. S10) and were more abundant in native than in cross-linked samples (Table 2), suggestive of IFIT5 binding to 5′-p mRNA fragments generated in cell extract. To investigate a potential change in specificity of IFIT5 binding to tRNA 5′ ends imposed by the E33A and E33A/D334A substitutions, we determined the overall frequency of tRNA read start-site positions for all tRNA loci combined (Fig. 4). Using reads mapped against tRNA loci from 50 bp upstream to 50 bp downstream of the mature tRNA ends, most read start sites corresponded to 5′-extended precursor (positions 1–50) or the mature tRNA 5′ end (position 51). The cross-linked sample had a higher fraction of mature tRNA start sites at position 51 than the two native purifications from the same cell line (Fig. 4A). Mutant E33A or E33A/D334A IFIT5 purifications also showed an increased fraction of read start sites at the mature tRNA 5′ end (position 51) compared with the parallel purification of WT IFIT5 (Fig. 4B), possibly reflecting some shift of the mutant IFIT5 proteins toward binding of 5′-p vs. 5′-ppp RNAs.
Fig. 4.
Composite tRNA read start sites. Cross-comparison of tRNA read start sites for WT IFIT5 variously purified from extracts of a stable cell line (A) or WT and mutant IFIT5 proteins purified after expression by transient transfection (B). Native extract was from cells without (−) or with (+) IFN-β treatment. x axis positions are as in Fig. 3, and the y axis represents the percentage of reads starting at each position. Precursor tRNA ends are at positions 1–50 and the mature tRNA 5′ end is at position 51. Read start sites at positions within the tRNA correlate with positions of reverse transcription stops at or near modification sites common among eukaryotic tRNAs (Fig. S7): position 59, G9/1-methylguanosine (m1G); position 70, U20/dihydrouridine (D); position 77, G26/N2,N2-dimethylguanosine (m2,2G) or U27/pseudouridine (Ψ) depending on on the length of the tRNA d-loop; position 87, A37/N6-isopentenyladenosine (i6A), N6-threonylcarbamoyladenosine (t6A) or 1-methylinosine (m1I), and G37/m1G or wybutosine (yW); position 108, A58/1-methyladenosine (m1A).
Discussion
Here we broadly profiled the biochemical and biological specificity of RNA association with human IFIT5. We found that recombinant IFIT5 binds RNAs with cap0 and 5′-p or 5′-ppp ends. The breadth of physiological RNA 5′-end groups permissive for IFIT5 binding was not predicted from high-resolution structures of RNA-bound IFIT5, which showed triphosphate coordination by the side chains of E33 and Y250 and led to the conclusion that IFIT5 exclusively binds 5′-ppp RNAs (18). In contrast to previous biochemical evidence supporting that conclusion, we found that substitutions of E33, Q41, T37, and Y250 had relatively little impact on 5′-ppp RNA binding assayed in vitro. The disparity between previous and current findings could derive from differences in RNA and protein preparation, which using methods described here support 100-fold greater RNA binding affinity than reported in other studies (18, 20).
Although the cap0 structure did not decrease the affinity of the IFIT5–RNA interaction relative to 5′-p or 5′-ppp RNAs, the cap1 structure greatly inhibited interaction. Our studies demonstrate that this discrimination between cap0 and cap1 derives in part from an acidic pocket in the central cavity. However, even the combination of acidic pocket substitutions E33A and D334A did not permit IFIT5 binding to cap1 RNA with an affinity comparable to cap0, 5′-ppp, or 5′-p RNA. In general, despite the clear role of the IFIT5 central cavity in determining RNA binding affinity, there was surprisingly unpredictable structure/function correlation in the specificity of RNA 5′-end discrimination. Also, our binding assays suggest that the preferential interaction of IFIT5 with tRNAs derives in large part from tRNA structure beyond the 5′ end. We speculate that IFIT5 RNA binding specificity relies on changes in RNA-bound protein conformation that have yet to be understood. We note that if IFIT5 conformation varies depending on the identity of the bound RNA, different RNA ligands could induce different effector responses.
We profiled IFIT5-bound cellular RNAs using a new method of sequencing ideal for characterizing noncoding RNAs. TGIRT-seq analysis supports IFIT5 binding to both 5′-p and 5′-ppp cellular RNAs and also the poly-U tailing of IFIT5-bound RNA fragments speculated based on a tRNA fragment sequenced previously (16). Recent studies describe poly-U tailing as a commitment step for RNA degradation by the human cytoplasmic exonuclease DIS3L2, which is deficient in human Perlman syndrome (26–28). Because IFIT5-bound tRNAs include 3′-extended or truncated poly-U tailed forms that would be a minority of total cellular tRNA forms, we suggest that IFIT5 may not only sequester cellular tRNAs but also trigger their subsequent degradation by DIS3L2. Analogous modes of action have been found for RNaseL, which degrades cellular RNA to mediate its function in innate immunity (29), and human schlafen 11, which binds tRNAs to alter translation as its antiviral effector mechanism (30). We speculate that any cytoplasmic single-stranded viral RNA 5′-p or 5′-ppp end would be bound by IFIT5, potentially inhibiting viral mRNA capping and/or translation. In addition, by recruiting RNA degradation enzymes to bound RNAs, IFIT5 could target virally encoded RNAs for rapid turnover. Finally, our results suggest that IFIT5 could also play a general role, beyond its function in innate immunity, in cytoplasmic surveillance for 5′-ppp RNA polymerase III transcripts that escape the nucleus.
Methods
Recombinant IFIT5 proteins were purified using nickel-charged agarose under stringent salt wash conditions (16). RNAs were transcribed by T7 RNA polymerase and gel purified. TGIRT-seq template switching reactions were done essentially as previously described (21), with modifications for construction of Illumina RNA-seq libraries. Samples were sequenced on an Illumina MiSeq to a depth of 1,000,000 or more 100- or 250-nt paired-end reads, of which only read 1 was used for analysis. Adaptor-trimmed reads were filtered for quality and aligned to the human genome reference sequence (Ensembl GRCh37) with Bowtie 2 (31) using local alignment to identify posttranscriptional additions at RNA 5′ and 3′ ends. Read mapping distribution was tabulated using annotations accompanying the genome sequence from the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu). Posttranscriptionally added poly-U tails (two or more consecutive U after the last genome-matched read position) were identified using Samtools and quantified using a custom computer script. See SI Methods for additional information.
Supplementary Material
Acknowledgments
We thank members of the K.C. and Tjian laboratories for discussion and reagents and Elisa Zhang, Brian Farley, and Philip Kranzusch for advice and comments on the manuscript. Funding for this research was from National Institutes of Health R01 Grants HL079585 (to K.C.) and GM37949 and GM37951 (to A.M.L.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive, http://www.ncbi.nlm.nih.gov (accession nos. SRR1508385, SRR1508404, and SRR1508427).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412842111/-/DCSupplemental.
References
- 1.Gürtler C, Bowie AG. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013;21(8):413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38(5):855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1(6):519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA. 2011;2(2):277–298. doi: 10.1002/wrna.52. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh A, Lima CD. Enzymology of RNA cap synthesis. Wiley Interdiscip Rev RNA. 2010;1(1):152–172. doi: 10.1002/wrna.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decroly E, Ferron F, Lescar J, Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol. 2012;10(1):51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daffis S, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468(7322):452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Züst R, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12(2):137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szretter KJ, et al. 2′-O methylation of the viral mRNA cap by West Nile virus evades ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog. 2012;8(5):e1002698. doi: 10.1371/journal.ppat.1002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Habjan M, et al. Sequestration by IFIT1 impairs translation of 2’O-unmethylated capped RNA. PLoS Pathog. 2013;9(10):e1003663. doi: 10.1371/journal.ppat.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura T, et al. Ifit1 inhibits Japanese encephalitis virus replication through binding to 5′ capped 2′-O unmethylated RNA. J Virol. 2013;87(18):9997–10003. doi: 10.1128/JVI.00883-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou X, et al. Interferon induced IFIT family genes in host antiviral defense. Int J Biol Sci. 2013;9(2):200–208. doi: 10.7150/ijbs.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13(1):46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Zhang YB, Liu TK, Gui JF. Lineage-specific expansion of IFIT gene family: An insight into coevolution with IFN gene family. PLoS ONE. 2013;8(6):e66859. doi: 10.1371/journal.pone.0066859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pichlmair A, et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol. 2011;12(7):624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 16.Katibah GE, et al. tRNA binding, structure, and localization of the human interferon-induced protein IFIT5. Mol Cell. 2013;49(4):743–750. doi: 10.1016/j.molcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z, et al. Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms. Cell Res. 2012;22(9):1328–1338. doi: 10.1038/cr.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abbas YM, Pichlmair A, Górna MW, Superti-Furga G, Nagar B. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature. 2013;494(7435):60–64. doi: 10.1038/nature11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng F, et al. Crystal structure and nucleotide selectivity of human IFIT5/ISG58. Cell Res. 2013;23(8):1055–1058. doi: 10.1038/cr.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar P, et al. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5’ppp- mRNAs. Nucleic Acids Res. 2014;42(5):3228–3245. doi: 10.1093/nar/gkt1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohr S, et al. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA. 2013;19(7):958–970. doi: 10.1261/rna.039743.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins K, Nilsen TW. Enzyme engineering through evolution: Thermostable recombinant group II intron reverse transcriptases provide new tools for RNA research and biotechnology. RNA. 2013;19(8):1017–1018. doi: 10.1261/rna.040451.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24(17):1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parrott AM, Mathews MB. Novel rapidly evolving hominid RNAs bind nuclear factor 90 and display tissue-restricted distribution. Nucleic Acids Res. 2007;35(18):6249–6258. doi: 10.1093/nar/gkm668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parrott AM, et al. The evolution and expression of the snaR family of small non-coding RNAs. Nucleic Acids Res. 2011;39(4):1485–1500. doi: 10.1093/nar/gkq856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Astuti D, et al. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet. 2012;44(3):277–284. doi: 10.1038/ng.1071. [DOI] [PubMed] [Google Scholar]
- 27.Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497(7448):244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malecki M, et al. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013;32(13):1842–1854. doi: 10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448(7155):816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature. 2012;491(7422):125–128. doi: 10.1038/nature11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.