Abstract
Nonhost resistance (NHR) is a plant immune response to resist most pathogens. The molecular basis of NHR is poorly understood, but recognition of pathogen effectors by immune receptors, a response known as effector-triggered immunity, has been proposed as a component of NHR.
We performed transient expression of 54 Phytophthora infestansRXLR effectors in pepper (Capsicum annuum) accessions. We used optimized heterologous expression methods and analyzed the inheritance of effector-induced cell death in an F2 population derived from a cross between two pepper accessions.
Pepper showed a localized cell death response upon inoculation with P. infestans, suggesting that recognition of effectors may contribute to NHR in this system. Pepper accessions recognized as many as 36 effectors. Among the effectors, PexRD8 and Avrblb2 induced cell death in a broad range of pepper accessions. Segregation of effector-induced cell death in an F2 population derived from a cross between two pepper accessions fit 15 : 1, 9 : 7 or 3 : 1 ratios, depending on the effector.
Our genetic data suggest that a single or two independent/complementary dominant genes are involved in the recognition of RXLR effectors. Multiple loci recognizing a series of effectors may underpin NHR of pepper to P. infestans and confer resistance durability.
Keywords: multiple interactions, nonhost resistance, pepper, Phytophthora infestans, RXLR effectors
Introduction
Plants are challenged by numerous pathogens including fungi, bacteria and viruses. However, most plants are resistant to most pathogens and disease is the exception in nature (Huitema et al., 2003). This phenomenon is known as nonhost resistance (NHR), an immune response of plant species against all isolates of a microorganism that cause disease on other plant species (Sumit et al., 2012). NHR is the most durable and strong resistance form of plants (Heath, 2000; Gurr & Rushton, 2005), and may have the potential to improve crop resistance to pathogens (Fan & Doerner, 2012). However, the molecular mechanism underlying NHR remains to be elucidated, because classical genetic approaches are limited as most nonhost plants cannot be crossed with susceptible host plants (Niks & Marcel, 2009).
The mechanism of NHR comprises of several components, including pre-formed and induced immunity (Thordal-Christensen, 2003). Pre-formed immunity includes physical barriers, such as cuticular wax, and chemical, antimicrobial compounds. Induced immunity includes pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Thordal-Christensen, 2003; Zhao et al., 2005; Jones & Dangl, 2006). PAMPs are conserved components of pathogens that are recognized by pattern recognition receptors at the surface of plant cells, which activate basal defense responses referred collectively as PTI. To suppress PTI, pathogens secrete effectors into plant cells that are specifically recognized by plant resistance (R) proteins leading to ETI (Jones & Dangl, 2006). Defense responses induced by PTI and ETI involve an oxidative burst and transcriptional reprogramming, mediated by signaling machinery (Tsuda & Katagiri, 2010). However, ETI can be distinguished from PTI by the induction of localized programmed cell death in the former, known as the hypersensitive response (HR) (Hammond-Kosack & Jones, 1997; Heath, 2000; Cunnac et al., 2009). The relative contribution of ETI and PTI to NHR may relate to the evolutionary divergence time between host and nonhost plants. It was suggested that the relative contribution of ETI increases with decreasing phylogenetic divergence time between two plant species (Schulze-Lefert & Panstruga, 2011).
There is some evidence to show the involvement of ETI in NHR. HopQ1-1, a type III effector of Pseudomonas syringae pv. tomato, elicited HR in nonhost Nicotiana benthamiana and a HopQ1-1 deletion strain caused disease in the nonhost plants, suggesting a role for ETI in NHR (Wei et al., 2007). Similarly, HopAS1 broadly present in P. syringae strains triggered NHR in Arabidopsis, whereas pathogenic strains had a truncated form of HopAS1 (Sohn et al., 2012). Resistance genes may also play a role in NHR. Rxo1 identified from maize encodes R protein that interacts with avrRxo1 and confers resistance against the nonhost pathogen Xanthomonas oryzae pv. oryzicola which causes bacterial leaf streak in rice (Zhao et al., 2005). When Rxo1 was transferred to susceptible rice, multiplication of the bacteria was inhibited. Another R protein in NHR is WRR4, which was identified from Arabidopsis ecotype Col-0 infected with Albugo candida using a natural variation in immunity of Arabidopsis accessions (Borhan et al., 2008). Transfer of WRR4 to susceptible Brassica napus rendered the plant resistant to A. candida (Borhan et al., 2010). However, nonhost plants carrying a nonfunctional copy of WRR4 retained resistance to the pathogen, suggesting polygenic control of NHR (Borhan et al., 2008).
Phytophthora infestans, an oomycete pathogen, is one of the most devastating pathogens in the world. It causes potato late blight on Solanaceae crops such as potato and tomato, but not on pepper. Oomycete pathogens secrete effectors into host cells that suppress host defense responses and modify host cell processes (Dodds & Rathjen, 2010; Bozkurt et al., 2012). The effectors have conserved RXLR motifs in their N-terminal domains that play an important role in targeting the host cells, and they contain W, Y and L motifs in their C-terminal domains (Whisson et al., 2007; Jiang et al., 2008). The genomic sequence of P. infestans reveals c. 550 genes encoding possible RXLR effectors. Most of these are found in the expanded repetitive DNA-rich regions, suggesting that they have rapidly evolved (Haas et al., 2009; Win et al., 2012). Using genomic information, a collection of nonredundant RXLR effectors of P. infestans has been cloned into Potato virus X (PVX)-based transient expression vectors (Torto et al., 2003; Vleeshouwers et al., 2008; Oh et al., 2009). High-throughput screening systems, such as PVX agro-infection, have been optimized in Solanaceae plants, allowing rapid identification of cognate R genes that interact with RXLR effectors (Takken et al., 2000; Torto et al., 2003; Vleeshouwers et al., 2006). Two of these genes are Rpi-blb1, which interacts with Avrblb1 (Vleeshouwers et al., 2008), and Rpi-blb2, which recognizes Avrblb2 (Oh et al., 2009). This genomic information and the development of experimental methods that enable effector-omics analyses of plant–microbe interactions, provide a promising approach for studying nonhost interaction.
Here, we hypothesized that multiple interactions between nonhost plant factors and pathogen effectors establish durable NHR. To test this hypothesis, we optimized in planta transient expression methods to screen interactions between pepper accessions and RXLR effectors of P. infestans. Using the results from the screening, we performed genetic analyses on an F2 population to investigate how plant factor(s) control effector-induced cell death. From the inheritance study, we conclude that multiple interaction between plant factor(s) and effectors of P. infestans, could be one of the determinants of the durable nature of NHR in pepper against P. infestans.
Materials and Methods
Plant materials and growth conditions
In order to screen nonhost interactions between pepper and RXLR effectors of P. infestans, a total of 100 pepper accessions from various regions were kindly provided by Dr Byoung-Cheorl Kang (Seoul National University, Korea). Seeds of the pepper accessions were sterilized with 0.1% sodium hypochlorite (NaOCl). Seven days after germination at 30°C, all of the seedlings were transferred to controlled chambers with a 16 h : 8 h, light : dark period at room temperature until cotyledons were fully expanded. Four plants from each accession were transplanted into a 32-plug form tray filled with horticultural bed soil composed of cocopeat (65–70%), peat moss (8–12%), and vermiculite (10–14%) according to the manufacturer's information (Baroker, Seoul Bio Co., Ltd, Seoul, Korea).
Phytophthora infestans spore infection
For P. infestans inoculation assays, 4-wk-old Capsicum annuum L. cv CM334 (nonhost), Solanum lycopersicum L. cv Heinz (susceptible host) and Solanum tuberosum L. cv Daeji (susceptible host) were used. The P. infestans isolates 88069, 40707, 40718, 43071 and 43072 provided by the RDA Gene Bank of Korea were used. All isolates were grown on rye sucrose agar media as previously described at 17°C in the dark for 10 d (Kamoun et al., 1998). To release the zoospores from sporangia, the plates were flooded with autoclaved distilled cold water, gently rubbed with a sterile cell scraper, and incubated at 4°C for 1.5 h. The zoospores were counted under a hemocytometer, and the concentration was adjusted to 5 × 104 zoospores ml−1. The detached leaves of pepper, potato and tomato were placed in the rectangle plate overlaid with a wet paper towel to maintain high humidity. The 10-μl droplets of zoospores were applied onto the detached leaves and incubated at 17°C. The inoculated leaves were sampled at 0, 24, 48 and 72 h post-inoculation (hpi).
Trypan blue staining
Upon inoculation of P. infestans, leaf discs containing zoospore droplets were excised at 5 d post-inoculation (dpi) and stained with lactophenol-trypan blue solution (10 ml lactic acid, 10 ml glycerol, 9.3 ml phenol, 10 ml distilled water, and 20 mg trypan Blue) diluted 1 : 2 in ethanol under vacuum as described (Yeom et al., 2012). The leaf discs were boiled in the lactophenol-trypan solution, incubated overnight in staining solution, and cleared with chloral hydrate (2.5 g ml−1). The cleared leaf discs were mounted in 50% glycerol solution and observed using stereo microscopy (Dimis-M, Siwon Optical Technology Co., Ltd, Anyang, Korea) and differential interference contrast microscopy.
Reverse transcription-PCR
Six leaf disks were harvested using a cork borer from the detached leaves inoculated with P. infestans. Total RNA were extracted using the Trizol reagent (Invitrogen) and 2 μg of total RNA were reverse-transcribed using Superscript II (Invitrogen). To determine in planta effector expression, PCR was performed using gene-specific primers (10 pmol μl−1, listed in Supporting Information Table S1). The following cycling conditions were used: 1 cycle of 94°C for 3 min; 30 cycles (or 35 cycle for pepper) of 94°C for 20 s, 58°C for 20 s and 72°C for 45 s. The actin gene (designated ActA) for the pathogen was used as control for monitoring transcript levels (Bos et al., 2010). The PCR products were then analyzed on 2% agarose gel and visualized by ethidium bromide.
PexRD effectors of P. infestans
All effectors were cloned into a binary Potato Virus X-based pGR106 vector in A. tumefaciens strain GV3101 (Holsters et al., 1980; Jones et al., 1999) for in planta Agrobacterium-mediated transient expression. Both pGR106-dGFP and pGR106-PexRD2 were used as negative and positive controls, respectively.
PVX agro-infection
Recombinant Agrobacterium carrying pGR106-PexRD effectors was incubated at 28°C on a YEP agar plate containing kanamycin (50 mg l−1) and rifampicin (50 mg l−1) for 2 d. Using toothpicks, the Agrobacterium was inoculated by piercing 5-wk-old pepper leaves. Duplicate experiments were performed twice on different days. Cell death induced by effectors was monitored at 14 dpi.
In planta expression using recombinant PVX virions
Agrobacterium carrying PexRD effector genes in binary vector pGR106 were cultured overnight at 28°C in YEP liquid media with antibiotics as described above and resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES and 150 μM acetosyringone). The culture was diluted to a final OD600 of 0.5 and incubated with gentle shaking at room temperature for 3 h, then infiltrated into the expanded leaves of N. benthamiana with a 1-ml disposable syringe without a needle. At 7 dpi, a 1-g sample of upper leaves that emerged after systemic infection of PVX was collected and ground in 5 ml of 0.05 M potassium phosphate buffer (pH 7.4) using a pestle and mortar to make the sap inoculum carrying recombinant PVX virions. The inoculum was rubbed onto the leaves of 4-wk-old pepper plants using carborundum 400 mesh. Each virion was applied to two leaves of each plant to prevent virions from being mixed. Then, distilled water was applied onto the inoculated leaves to wash out the carborundum. Cell death on the inoculated leaves was monitored up to 7 dpi, and necrotic cell death was observed by destaining chlorophyll with 100% ethanol.
Results
Nonhost resistance of pepper against P. infestans is associated with hypersensitive cell death
In order to investigate how nonhost pepper plants respond to P. infestans and characterize its nonhost defense response, the detached leaves of pepper, tomato and potato plants were inoculated with a zoospore suspension of P. infestans isolate 88069 (5 × 104 spores ml−1). The interaction with P. infestans was examined cytologically using UV light and trypan blue staining to visualize dead cells. At 5 dpi, dead cells at the infection site were readily shown by their brown color under white light and auto-fluoresced on a red background under UV light. The germinated zoospores of P. infestans were shown and germ tubes were elongated on pepper leaves (Fig. 1a, upper right). The HR on penetrated epidermal cells was apparent on pepper leaves and remained limited to those cells (Fig. 1a, upper right). In contrast to the responses in pepper, the biotrophic growth of P. infestans was apparent 5 dpi in tomato and potato leaves and hyphae with sporangia, necrotic death of mesophyll cells spread throughout the leaves (Fig. 1a, lower right panel).
Fig. 1.
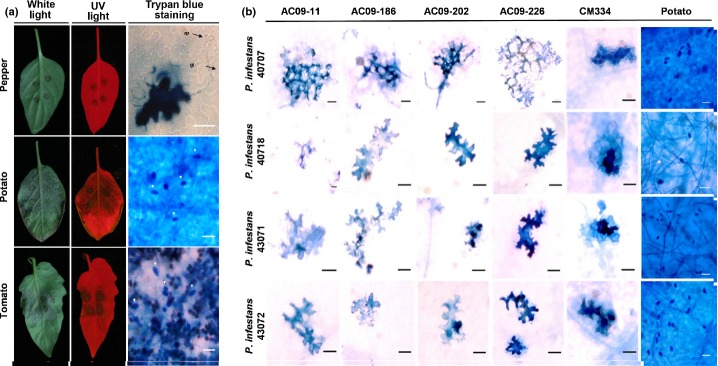
Nonhost response of pepper (Capsicum annuum) against Phytophthora infestans is associated with hypersensitive cell death. (a) Detached leaves of nonhost pepper landrace CM334 and host plants (potato (Solanum tuberosum) cv Daeji and tomato (S. lycopersicum) cv Heinz) were infected with a zoospore suspension of P. infestans 88069 (5 × 104 ml−1). Leaf pieces containing zoospore droplet were excised at 5 d post-inoculation, examined for hypersensitive cell death and sporangia development under UV illumination, and visualized by trypan blue staining. White arrows indicate sporangia formation on potato and tomato leaves. Zp, zoospore; gt, germ tube. (b) Hypersensitive cell death was confirmed by interactions between pepper accessions and four isolates of P. infestans 40707, 40718, 43071 and 43072. Bars, 50 μm.
NHR is defined as resistance displayed by all accessions of a plant species against all strains of a pathogen. To further examine the pepper–P. infestans interaction, we tested five accessions of pepper with four isolates of P. infestans (40707, 40718, 43071 and 43072) collected from different regions of Korea (Fig. 1b). All accessions displayed localized cell death typical of the HR to all four isolates. The HR was limited to penetrated epidermal cells in all accessions inoculated with three of the isolates, whereas in pepper infected with isolate 40707, the induced HR was not completely confined to the infected cells and extended to adjacent cells. The finding that NHR of pepper to P. infestans is associated with the HR suggests the possible presence of pepper R genes that interact with P. infestans effectors.
Nonhost pepper shares expression profiles of the effector genes of P. infestans with host plants
We first examined whether effector genes of P. infestans are expressed during the nonhost interaction. The 54 RXLR effectors used in this study can be grouped into 32 PexRD genes of homologs in Table 1. Expression of 24 of 32 PexRD genes was investigated in pepper (AC09-226) and tomato (cv Heinz) leaves following infection with P. infestans 43072 using gene-specific primers (Fig. 2; Table S1). ActA expression increased during the infection in tomato but decreased in pepper at 3 dpi, reflecting the susceptible and resistant responses of each plant species. The Kazal-like serine protease inhibitor EPI1 is an in planta-induced gene that may be involved in suppression of PTI (Tian et al., 2005; Attard et al., 2008). EPI1 was expressed in P. infestans-infected pepper.
Table 1.
Description of the tested PexRD genes*
| Effector names | PITG gene ID | Number of homologs tested | RXLR | dEER | Average number of accessions with cell death |
|---|---|---|---|---|---|
| PexRD1 | PITG_15287 | 1 | RQLR | EDGEER | 0 |
| PexRD2 | PITG_21422, PITG_11350, PITG_11383, PITG_11384, PITG_22935 | 1 | RLLR | ENDDDSEAR | 43 |
| PexRD3 | PITG_09160 | 1 | RFLR | EGDNEER | 13 |
| PexRD4 | NA | 1 | RFLR | DEER | 2 |
| PexRD6, ipiO, Avrblb1 | PITG_21388 | 3 | RSLR | DEER | 5 |
| PexRD7, Avr3a | PITG_14371 | 2 | RLLR | EENEETSEER | 7 |
| Pex147-2, Avr3a paralog | 1 | RLLR | EESEETSEER | 6 | |
| Pex147-3, Avr3a paralog | 1 | RFLR | EENEETSEER | 3 | |
| PexRD8 | PITG_14739 | 1 | RLLR | DDDDEEER | 22 |
| PexRD10 | PITG_23206 | 1 | RKLR | EER | 4 |
| PexRD11 | PITG_23206 | 2 | RLLR | DEGELTEER | 3 |
| PexRD12 | PITG_16233, PITG_16240 | 2 | RSLR | DSDDGEER | 7 |
| PexRD13 | PITG_08812 | 2 | RQLR | 7 | |
| PexRD14 | PITG_14961, PITG_14962, PITG_14954, PITG_14959 | 2 | RLLR | ETGNQEER | 6 |
| PexRD16 | PITG_06087 | 2 | RSLR | EER | 4 |
| PexRD17 | PITG_08599 | 2 | RVLR | EIEAETER | 1 |
| PexRD21 | PITG_13452 | 1 | RLLR | EREVQEER | 2 |
| PexRD22 | PITG_13306 | 2 | RFLR | EDASDEER | 4 |
| PexRD24 | PITG_04314 | 2 | RSLR | ETSEDEEER | 8 |
| PexRD26 | PITG_07947 | 2 | RVLR | DEER | 1 |
| PexRD27 | PITG_13628 | 1 | RLLR | DSEER | 0 |
| PexRD28 | PITG_03192 | 1 | RSLR | ETSEDEEER | 1 |
| PexRD31 | PITG_16402 | 1 | RSLR | EDQEGDEER | 2 |
| PexRD36 | PITG_23132 | 2 | RHLR | DDEER | 3 |
| PexRD39/40, Avrblb2 | PITG_20300 | 3 | RSLR | 19 | |
| PexRD41 | PITG_04089 | 3 | RSLR | 3 | |
| PexRD44 | PITG_17063 | 1 | RFLR | QEEGVFEER | 2 |
| PexRD45 | PITG_09632 | 2 | RSLR | 2 | |
| PexRD46 | PITG_18685 | 3 | RSLR | 3 | |
| PexRD49 | PITG_05750 | 1 | RLLR | EEER | 3 |
| PexRD50 | PITG_06099 | 2 | RLLR | 2 | |
| PexRD51 | 2 | RFLR | EER | 0 |
Table modified from Oh et al. (2009).
Fig. 2.
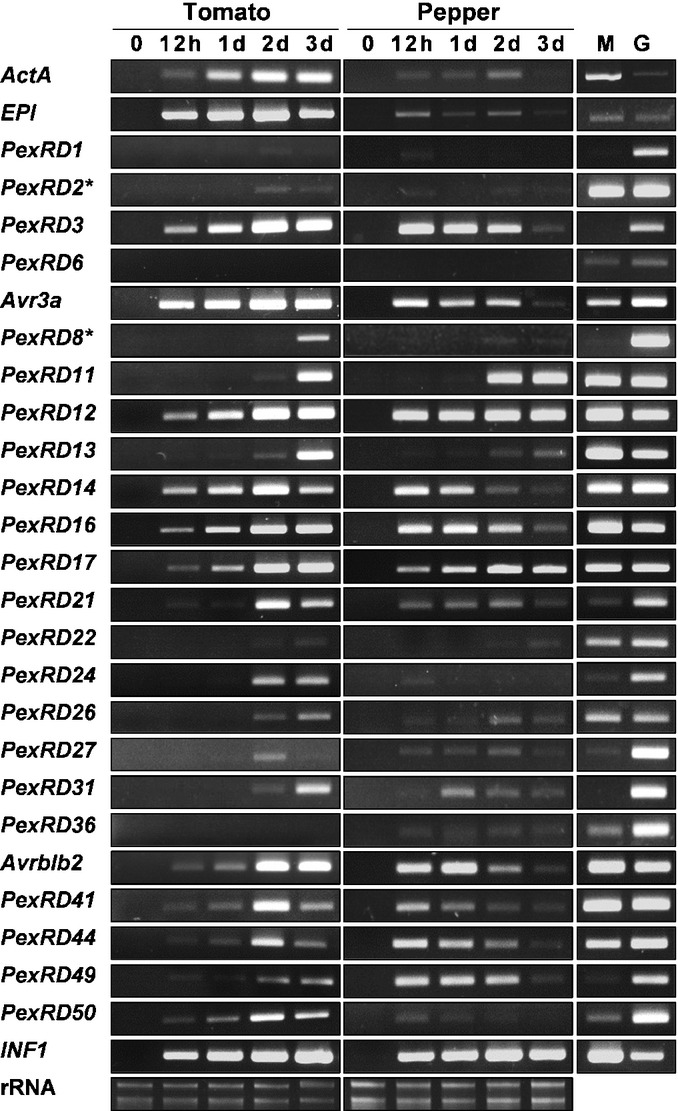
Nonhost pepper shares expression profiles of the effector genes of Phytophthora infestans with host plants. Detached leaves of pepper (Capsicum annuum) and tomato (Solanum lycopersicum) upon inoculation with P. infestans isolate 43072 were harvested at 0, 12 h, and 1, 2 and 3 days post-inoculation. Total RNA was extracted from the harvested samples. Reverse transcription-PCR was performed to observe in planta expression of PexRD effector genes. Tomato cDNAs was subjected to a 30-cycle PCR and a 35-cycle PCR was performed with pepper cDNAs. Constitutive ActA and in planta-induced EPI1 were used as controls and those genes were detected using 30-cycle PCR in both pepper and tomato. This experiment was repeated twice with similar results. M, P. infestans mycelium RNA; G, P. infestans genomic DNA. *PexRD2 and PexRD8 in tomato cDNAs: subjected to a 35-cycle PCR.
Of 24 PexRD genes, the expression of 23 PexRD genes was detected in the interaction between pepper and P. infestans. Even considering the different cycles of PCR amplification, all of the PexRD genes expressed in tomato were also detected in the pepper–P. infestans interaction. Twelve effectors of PexRD3, Avr3a, PexRD12, PexRD14, PexRD16, PexRD17, PexRD21, Avrblb2, PexRD41, PexRD44, PexRD49 and PexRD50 were expressed in pepper at 12 hpi. The expression of those effectors increased in tomato from 12 hpi. PexRD8, PexRD11, PexRD13 and PexRD22 showed detectable expression in pepper after 2 dpi, which is similar to the expression in tomato. In conclusion, the expression of all the tested P. infestans effector genes during interactions with pepper indicates that these effectors could be targeted by the pepper immune response.
Optimization of an heterologous effector expression system in pepper
Effector-omics or high-throughput approaches for the rapid assignment of activities to effector genes require effective heterologous expression systems. To establish such a system in pepper, we applied a PVX agro-infection method for the in planta expression of effectors in pepper, as depicted in Fig. 3(a). This method is well suited for high-throughput screening of effector activity in Solanaceae plants. To confirm the suitability of this approach in pepper, four elicitors known to induce the necrotic response in Solanaceae plants, specifically INF1 (Kamoun et al., 1998), PiNPP1 (Kanneganti et al., 2006), CRN2 (Torto et al., 2003) and PexRD2 (Oh et al., 2009), were wound-inoculated on leaves of 4-wk-old pepper plants. Agrobacterium strain GV3101 carrying those elicitors and negative control dGFP were inoculated onto both sides of the mid-vein of fully expanded pepper leaves using toothpicks. Among the four different elicitors, only PexRD2 caused cell death on pepper leaves at 14 dpi (Fig. 3b). Considering the induced cell death by PexRD2, the PVX agro-infection method is as applicable in pepper as it is in other Solanaceae plants.
Fig. 3.
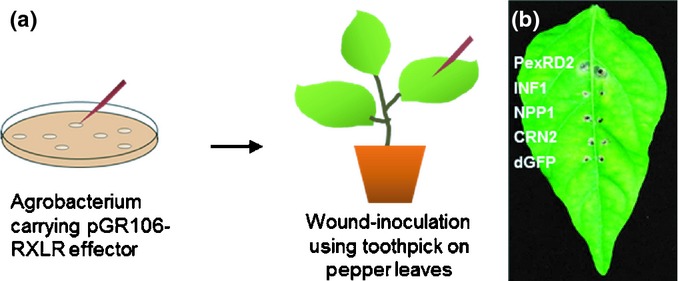
Optimization of an heterologous effector expression system in pepper (Capsicum annuum). (a) The PVX agro-infection method is depicted. Agrobacterium carrying pGR106-RXLR effectors was wound-inoculated onto pepper leaves. (b) PexRD2 and elicitins (INF1, NPP1 and CRN2) which are known to induce cell death were inoculated on both sides of mid veins in pepper leaves. PexRD2 but not elicitins induced cell death on inoculated pepper leaves at 14 d post-inoculation .
Pepper accessions respond to a diversity of P. infestans effectors
In order to perform a primary screen of the nonhost interaction, we inoculated the infection-ready collection of 54 P. infestans RXLR effectors onto the leaves of 100 pepper accessions using the PVX agro-infection method (Fig. S1). The effectors are putative secreted effectors from expressed-sequence tag data of P. infestans and have been cloned into the binary PVX vector pGR106 for high-throughput screening of effector activities (Randall et al., 2005; Oh et al., 2009). Among 54 effectors, nine effector families including PexRD6, PexRD7, PexRD16, PexRD24, Avrblb2, PexRD41, PexRD44, PexRD49 and PexRD50 were recently determined as core effectors using criteria that measure induction of expression during infection and presence in the genome of three P. infestans isolates (Cooke et al., 2012). The pepper accessions originating from various areas including the United States, China, and Japan were selected for screening of nonhost interactions. Most of them belong to Capsicum annuum with a few accessions of Capsicum chinense (Table S2). After inoculation of RXLR effectors in duplicate, effector-induced cell death was monitored at 14 dpi. Agrobacterium carrying pGR106-dGFP and pGR106-PexRD2 were used as negative and positive controls, respectively. Considering the relatively low efficiency of Agrobacterium-mediated transient expression on pepper, we judged ‘cell death’ when either one of the duplicate inoculation sites showed cell death. Cell death confirmed twice was marked as ‘+’ (Tables 2, S2).
Table 2.
Pepper (Capsicum annuum) accessions respond to a diversity of Phytophthora infestans effectors
 |
Among 100 pepper accessions, 55 showed no cell death response by inoculation of Agrobacterium-carrying the positive control (PexRD2), which may indicate incompatibility of these accessions with Agrobacterium and low growth of Agrobacterium resulting in insufficient expression of effectors. Three accessions (AC09-15, AC09-20 and R09-135) showed the necrotic cell-death response triggered by the negative control pGR106-dGFP, as evidenced by susceptible response to PVX developing systemic mosaic symptoms and leaf crinkling at 14 dpi (Table S2). Those three accessions were excluded for further screening. From the screening results of the other 42 accessions showing the HR against positive control PexRD2, each accession variously responded to at least one effector, showing a distributed cell death response among pepper accessions. The average number of effectors inducing cell death on each pepper accession was seven and many accessions reacted to multiple effectors. Each pepper accession showed cell death by in planta expression of 1–36 effectors, respectively. These results support the hypothesis that there are multiple recognitions in nonhost interactions between pepper and effectors of P. infestans (Table 2).
The RXLR effectors that interacted with pepper accessions could be divided into two groups: broadly recognized and specifically recognized effectors. The effectors recognized by a broad variety of pepper accessions included PexRD8 and PexRD39/40, known as the Avrblb2 family (Table 1; Fig. 4). Over half of the tested accessions showed the HR to those effectors. The specifically recognized effectors showed various responses, depending on the pepper accession. Generally, homologous effectors induced cell death on the same pepper accessions. For example, PexRD12-1 and PexRD12-2 exhibited 99% nucleotide sequence similarity and both triggered cell death on pepper accessions AC09-12, R09-120, AC09-202 and AC09-52. Furthermore, PexRD24-1 and PexRD24-2, sharing 94% identity, elicited cell death on pepper accessions AC09-72, AC09-73 and AC09-10. Those accessions could be postulated to have cell death-inducing factors that interact with an effector family. Taken together, these results revealed that nonhost pepper plants show cell death by interactions with multiple RXLR effectors of P. infestans.
Fig. 4.
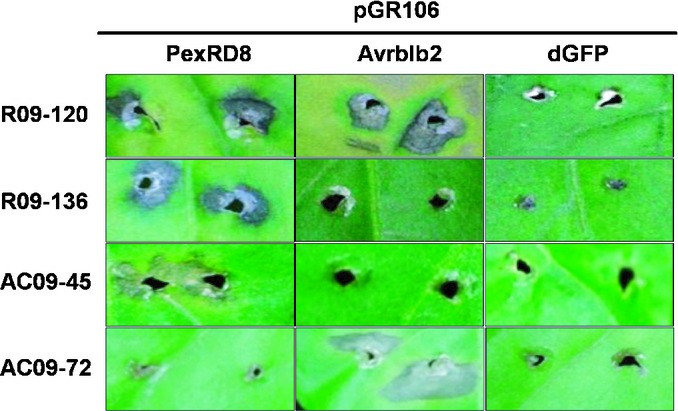
PexRD8 and Avrblb2 are recognized by a variety of pepper (Capsicum annuum) accessions. Hypersensitive cell death was observed in pepper accessions after wound inoculation with Agrobacterium carrying pGR106-PexRD8 and Avrblb2 at 14 d post-inoculation.
Validation of the pepper hypersensitive response to P. infestans effectors using recombinant PVX virions
Some of the pepper accessions that did not show any reaction with the PVX agro-infection assay may be recalcitrant to A. tumefaciens-mediated transient assays. To confirm the positive interactions of pepper to P. infestans effectors for a further inheritance study and exclude the effect of Agrobacterium, we developed a PVX-virion-mediated screening method. As depicted in Fig. 5(a), recombinant PVX virions carrying RXLR effectors propagated in N. benthamiana were harvested from upper leaves that showed PVX symptoms and were manually applied on carborundum-dusted leaves of the pepper accessions. HR cell death was solely developed by PVX-mediated in planta expression of RXLR effectors.
Fig. 5.
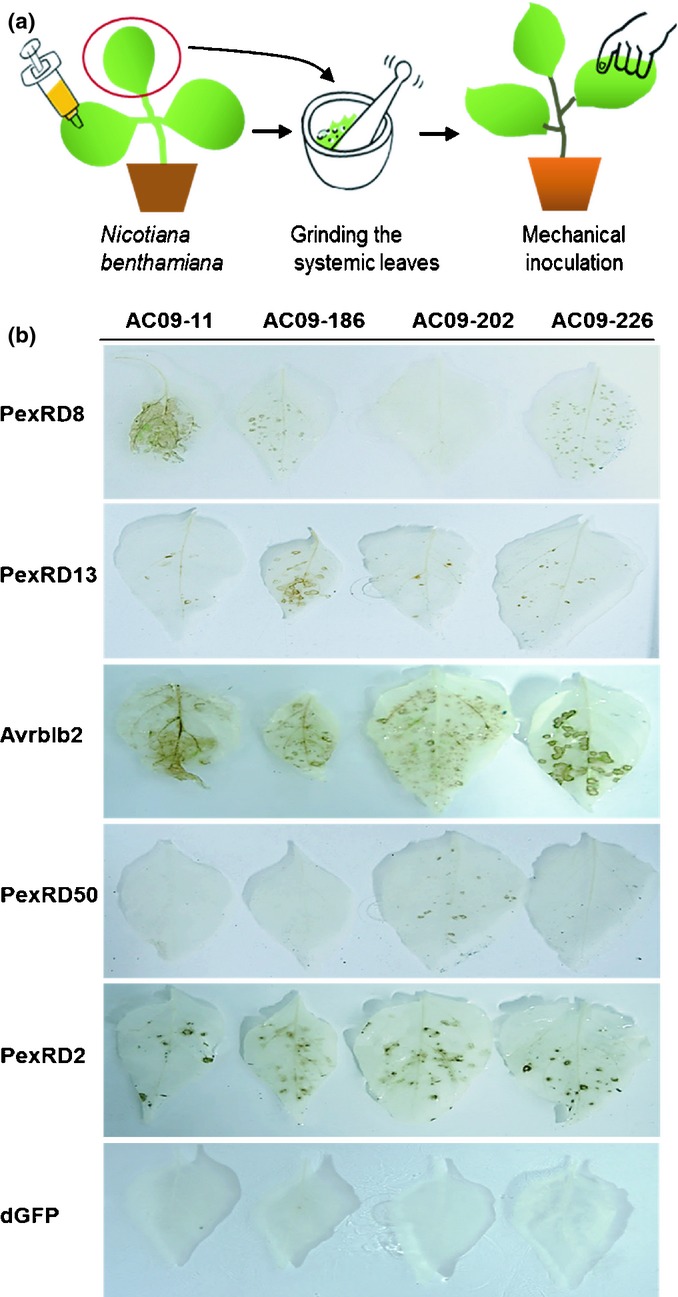
Validation of effector-induced cell death using recombinant PVX virions. (a) Overview of the PVX-mediated transient expression method is shown. Agrobacterium carrying RXLR effectors was inoculated into lower leaves of Nicotiana benthamiana. Upper leaves showing virus symptom were ground to produce a PVX-inoculum carrying RXLR effectors. The inoculum was mechanically inoculated on pepper (Capsicum annuum) leaves to validate the effector-induced cell death. (b) Four pepper accessions (AC09-11, 186, 202 and 226) were wound-inoculated with recombinant PVX virions (PexRD8, PexRD13, Avrblb2 and PexRD50). PVX virions of PexRD2 and dGFP were used as positive control and negative control, respectively. Inoculated leaves were harvested at 5 d post-inoculation and cleared in 100% ethyl alcohol to remove chlorophyll in order to visualize the cell death.
Susceptible responses of pepper against PVX, including severe necrosis, have been reported in 11 cultivars of C. annuum species (Shi et al., 2008). To select accessions for the application of the recombinant virion method, we screened 14 pepper accessions for their response to PVX following inoculation of the negative control PVX-dGFP and positive control PVX-PexRD2 (Fig. S2; Table S3). Twelve out of 14 pepper accessions were randomly selected from the pepper accessions showing cell death induced by PexRD2. The other two accessions (AC09-186 and AC09-226) were chosen from the pepper accessions showing no effector-induced cell death. Upon inoculation of the recombinant virions, some accessions, including AC09-56 and AC09-200, showed cell death against PVX-dGFP, which may indicate that these accessions contain intrinsic R genes against PVX. For further study of inheritance of effector-induced cell death, we selected four pepper accessions, AC09-11, AC09-186, AC09-202 and AC09-226 showing no cell death response induced by PVX-dGFP and a clear response against PVX-PexRD2. All accessions showed cell death against PexRD2 but no cell death against PVX-dGFP (Fig. 5b).
Of 54 RXLR effectors, 13 effectors (PexRD6-3, PexRD12-1, PexRD12-2, PexRD24-2, PexRD26-1, PexRD26-2, PexRD31, PexRD39, Pex46-2, PexRD49, PexRD50-2 and PexRD51) were excluded, due to the difficulty of obtaining recombinant PVX virions. In total, recombinant PVX virions of 41 RXLR effectors were tested on four accessions (Table 3). Effector-induced cell death by PexRD8, PexRD13, Avrblb2 and PexRD50 is shown in Fig. 5(b). PexRD8 resulted in cell death on all accessions except AC09-202, confirming the results of the primary screening. Interestingly, the PexRD13-1 and Avrblb2 families (PexRD40-1 and PexRD40-2) were recognized by all four accessions. In potato, PexRD13-1 elicited cell death on Solanum bulbocastanum and S. pinnatisectum which are both resistant to P. infestans. Avrblb2 families induced cell death in both resistant and susceptible Solanum species (Vleeshouwers et al., 2008). These similar patterns of cell death induced by the PexRD8 and Avrblb2 families suggest that pepper has common plant factors or cognate R genes interacting with those effectors. Some effectors elicited cell death on one or two accessions. For example, PexRD24 induced cell death on AC09-11 and AC09-226. PexRD50-induced cell death was only observed on AC09-202. Overall, we confirmed the multiple interactions between pepper and effectors of P. infestans.
Table 3.
Hypersensitive cell death induced by 41 RXLR effectors in pepper (Capsicum annuum) accessions using inoculation with recombinant PVX virions
| Phytophthora infestans | Pepper accessions | |||
|---|---|---|---|---|
| RXLR effectors | AC09-11 | AC09-186 | AC09-202 | AC09-226 |
| PexRD1 | − | − | − | − |
| PexRD2 | + | + | + | + |
| PexRD3 | − | − | − | − |
| PexRD4 | − | − | − | − |
| PexRD6-1 | − | − | − | − |
| PexRD6-2 | − | − | − | − |
| PexRD7-1 | − | − | − | − |
| PexRD7-2 | + | − | + | − |
| Pex147-2 | − | − | − | − |
| Pex147-3 | − | − | nd | − |
| PexRD8 | + | + | − | + |
| PexRD10 | + | − | − | − |
| PexRD11-1 | − | − | − | − |
| PexRD11-2 | − | − | − | − |
| PexRD13-1 | + | + | + | + |
| PexRD13-2 | + | − | + | − |
| PexRD14-1 | − | − | − | − |
| PexRD14-2 | − | − | − | − |
| PexRD16-1 | − | − | − | − |
| PexRD16-2 | + | − | + | − |
| PexRD17-1 | − | − | − | − |
| PexRD17-2 | − | − | − | − |
| PexRD21 | nd | nd | + | − |
| PexRD22-1 | − | − | − | − |
| PexRD22-2 | − | − | − | − |
| PexRD24-1 | + | − | − | + |
| PexRD27 | − | − | − | − |
| PexRD28 | − | − | − | − |
| PexRD36-1 | + | − | − | − |
| PexRD36-2 | − | − | − | − |
| PexRD40-1 | + | + | + | + |
| PexRD40-2 | + | + | + | + |
| PexRD41-1 | + | − | − | − |
| PexRD41-2 | nd | − | − | − |
| PexRD41-3 | − | nd | + | − |
| PexRD44 | − | − | − | − |
| PexRD45-1 | − | − | − | − |
| PexRD45-2 | − | − | − | nd |
| PexRD46-1 | + | − | + | − |
| PexRD46-3 | − | − | + | − |
| PexRD50-1 | − | − | + | − |
PexRD40-1/PexRD40-2, Avrblb2 family effectors.
−, No cell death; +, cell death; nd, not determined.
Multiple loci determine responses of pepper to P. infestans RXLR effectors
Based on the diversity of effector-induced cell death in pepper accessions, an inheritance study was performed to determine the genetic basis of effector-induced cell death in pepper. Two accessions, AC09-226 and AC09-202, were crossed to generate an F2 population. Among the effectors to which these two pepper accessions reacted differentially, PexRD8 and PexRD24 caused cell death on AC09-226, whereas PexRD41-3, PexRD46 and PexRD50 induced cell death on AC09-202.
PVX virions expressing the five effectors (PexRD8, PexRD24, PexRD41, PexRD46 and PexRD50) were manually inoculated onto two leaves per plant of the F1 and F2 populations. Each F2 plant was treated with only one recombinant virion. Six days later, all inoculated leaves were scored for effector-induced cell death. To determine whether PVX affects the cell death induced by effectors on F2 plants, we applied the virion of negative control PVX-dGFP on 65 F2 plants, resulting in no cell death responses on any of the tested F2 plants. Observations of cell death induced by five effectors in F1 progeny indicated that plant factor(s) to determine the cell death phenotype were dominant alleles. The F2 distribution of PexRD8-induced cell death showed no significant deviation from the 15 : 1 ratio of cell death at the 0.05 probability level, suggesting that two unlinked dominant genes interact with the PexRD8 effector (Table 4). The F2 segregation of cell death induced by PexRD24, PexRD46 and PexRD50 fitted the expected 9 : 7 ratio, consistent with two independent gene loci that interact in complementary dominant epistasis to trigger effector-induced cell death. These results indicate that pepper has a set of genes that recognize a P. infestans RXLR effector. The cell death phenotype induced by PexRD41-3 segregated in a 3 : 1 ratio, suggesting that cell death is conditioned by a single dominant gene. Taken together, our data support the idea that multiple interactions of plant factors with effectors underpin the NHR of pepper against P. infestans.
Table 4.
Multiple dominant genes mediate effector-induced cell death in F2 population from the cross of AC09-226 to AC09-202
| F2 (Pepper (Capsicum annuum) accessions AC09-226 × AC09-202) | ||||||
|---|---|---|---|---|---|---|
| Observed ratio | Expected ratio | |||||
| Effectors | F1 HR+/HR− | HR+ | HR− | HR+ : HR− | Chi-square | P-value |
| PexRD8 | 8 : 0 | 141 | 14 | 15 : 1 | 2.047 | 0.152 |
| PexRD24-1 | 15 : 0 | 101 | 81 | 9 : 7 | 0.042 | 0.837 |
| PexRD41-3 | 15 : 0 | 101 | 31 | 3 : 1 | 0.162 | 0.687 |
| PexRD46-1 | 15 : 0 | 83 | 57 | 9 : 7 | 0.524 | 0.469 |
| PexRD50-1 | 15 : 0 | 27 | 17 | 9 : 7 | 0.468 | 0.494 |
HR, hypersensitive response-like cell death.
Discussion
NHR is a durable resistance that includes both preformed immunity and induced immunity. Here, we report on the interaction between pepper, a Solanaceae plant, and the destructive potato late blight pathogen, the oomycete Phytophthora infestans. We performed a cytological investigation to examine the cellular responses of this interaction, and concluded that a localized HR is a major response of NHR in pepper against P. infestans. This finding is consistent with a previous study, in which NHR to P. infestans in Solanum spp. and Arabidopsis was associated with the HR (Vleeshouwers et al., 2000; Huitema et al., 2003). It was recently reported that PAMPs such as flagellin and the glycoprotein CBEL have elicitor activity and induce HR-like cell death (Khatib et al., 2004). However, the HR, a form of programmed cell death, is more frequently induced by interactions between R genes and the cognate effectors (Khatib et al., 2004; Naito et al., 2008; Coll et al., 2011). Based on our finding, a set of effectors of P. infestans were expressed during interactions with both host potato and nonhost pepper. We suggest that multiple recognition of effectors by an arsenal of R genes in pepper is an important factor in durable NHR. Pepper, tomato and potato are all in the Solanaceae family and have a relatively close evolutionary distance; thus, it seems reasonable that pepper might carry R genes that detect RXLR effectors of P. infestans and that ETI would therefore contribute to NHR (Schulze-Lefert & Panstruga, 2011).
As an initial step in the characterization of the interaction between RXLR effectors of P. infestans and pepper, we inoculated 54 PexRD effectors into 100 pepper accessions using a PVX agro-infection method. This method has been used to study the response by effectors in several Solanum species (Vleeshouwers et al., 2006, 2008) and is considered appropriate for the high-throughput screening of effector activity that induces cell death. However, pepper has a relative low efficiency of Agrobacterium-mediated transformation, and gene transfer by Agrobacterium might not be easily performed (Lee et al., 2004). Functional studies in pepper mainly have been conducted by gene silencing using viruses rather than overexpression. In this study, we optimized the method and confirmed the response to effectors of pepper with recombinant PVX virions for in planta overexpression of effectors. The method made it possible to exclude Agrobacterium and achieved high efficiency in a transient expression assay. For instance, pepper accessions AC09-186 and AC09-226 used for the inheritance study did not show any cell death induced by effectors using the PVX agro-infection method but clear cell death induced by effectors was observed after inoculation with recombinant PVX virions.
Several RXLR effectors of P. infestans are known to be recognized by R genes of Solanum species. R gene-avirulence effector pairs include Rpi-blb2 (Lokossou et al., 2009), R3a-Avr3a (Armstrong et al., 2005), Rpi-blb1-Avrblb1 (Vleeshouwers et al., 2008) and Rpi-blb2-Avrblb2 (Oh et al., 2009). Our finding that pepper recognized several RXLR effectors of P. infestans, which is a nonhost pathogen, suggests that pepper has R genes that interact with RXLR effectors from this pathogen. Based on comparative genomic analysis of Arabidopsis accessions, R genes were found to rapidly evolve, having higher polymorphism than plant gene families involved in basic biological processes such as ribosomal function and transcriptional regulation (Clark et al., 2007). Each pepper accession showed a cell death response to each RXLR effector, which could reflect a high number of polymorphisms of the R gene family among the pepper accessions. Each pepper accession exhibited cell-death responses to at least one effector, and 1–36 of the tested 54 RXLR effectors, indicating multiple interactions of the nonhost pepper with RXLR effectors. The 54 effectors used in this study account for 10% of the effectors in the P. infestans genome, indicating that pepper could have more interactions with effectors of P. infestans. As a consequence, we cannot conclude that AC09-221, showing a cell death response to only the PexRD2 effector, would be compatible to a P. infestans isolate not producing a PexRD2 effector. Furthermore, we identified several effectors including PexRD8 and PexRD39/40 – termed Avrblb2 – which trigger cell death on a broad range of pepper accessions. PexRD8 is known to suppress INF1-induced cell death and Avrblb2 is recognized by the late blight resistance gene Rpi-blb2 identified from Solanum bulbocastanum (van der Vossen et al., 2005; Oh et al., 2009). PexRD8 and Avrblb2 also induce cell death on most of P. infestans-resistant wild Solanum species (Vleeshouwers et al., 2008). However, Nicotiana benthamiana, a member of the Solanaceae family, did not respond to either PexRD8 or Avrblb2 (Oh et al., 2009). These results suggest that conserved genes exist in pepper and potato that can interact with PexRD8 and Avrblb2. We also tested interactions with Avrblb2 using recombinant PVX virion in 10 pepper accessions, which included 9 out of 14 pepper accessions used in the screening of the response against PVX in addition to the reference cultivar ‘CM334’ (Table S3). Avrblb2, which is conserved in P. infestans isolates, induced cell death in the 10 pepper accessions tested, suggesting that pepper might have target(s) or R gene(s) with conserved functions that interact with Avrblb2. The putative R gene(s) that interact with Avrblb2 could play an important role in NHR and are likely to be distinct from Rpi-blb2 as Rpi-blb2 orthologous cloned from several pepper accessions did not recognize Avrblb2 (H.A. Lee et al., unpublished).
Our genetic data suggest that several major genes are associated with the interaction with P. infestans effectors. Those genes could be R gene(s). In a previous study, the genetic analysis of the response to P. syringae effectors in nonhost lettuce showed that a linkage group, containing chromosome regions including multiple R gene analogs, determines the cell death phenotype (Wroblewski et al., 2009). In this study, PexRD8-induced cell death in an F2 population from a cross between AC09-226 and AC09-202 showed a 15 : 1 phenotypic segregation ratio, indicating that two independent dominant alleles (e.g. A-B-, aaB- and A-bb) are required to confer cell death. A dual R-gene system (RRS1-Ws and RPS4-Ws) showed resistance to Colleotrichum higginisianum (Narusaka et al., 2009). Arabidopsis RPP2A and RPP2B, members of TIR-NB-LRR, are required for resistance against Peronospora parasitica isolate Cala2 (Sinapidou et al., 2004). Furthermore, the segregation of effector-induced cell death by PexRD24, PexRD41-3 and PexRD46 in the F2 population was consistent with a 9 : 7 ratio, suggesting that two dominant complementary genes (e.g. A-B-) are associated with effector-induced cell death. The mode of action of two putative R genes in NHR could be supported by several studies in which two R genes act cooperatively in the resistance response. The tomato NRC1 gene encodes a coiled-coil (CC)-nucleotide binding (NB)-leucine-rich repeat (LRR) type resistance protein and involves an HR signaling pathway mediated by the Cf-4 resistance gene (Gabriels et al., 2007). Similarly, NRG1, encoding CC-NB-LRR, mediates resistance together with the tobacco N gene against Tobacco mosaic virus (Peart et al., 2005). In our experimental system, the inverse model, the interaction of an R gene with multiple effectors, could not be demonstrated. Arabidopsis RPM1 induces the resistance response by the interaction with two unrelated effectors of P. syringae, AvrB and AvrRpm1. Similarly, a dual interaction of an R gene with different effectors is likely to occur in nonhost interactions. We cannot rule out the possibility that the combination of the interactions between multiple R genes and effectors establishes durable NHR.
Our finding that pepper has the ability to respond to P. infestans effectors raises an interesting question. Phytophthora infestans is a specialized pathogen of Solanum spp. native to Central and South America but is known to infect neither pepper nor related Capsicum spp. Why, then, would the nonhost pepper carry genes that detect effectors of P. infestans? There are several possibilities. It is possible that pepper was formerly a host of P. infestans during its evolutionary history. Another explanation is that pepper does not directly respond to P. infestans effectors but rather to the cellular perturbations that these effectors trigger. This would be consistent with models of indirect recognition as suggested by the guard model (Innes, 2004). Recently, recognition of the P. infestans AVR2 effector by the Solanum R2 gene was shown to require an additional host protein, phosphatase BSL1 (Saunders et al., 2012). R2-mediated resistance to P. infestans was compromised specifically by the silencing of BSL1. The association of AVR2 with BSL1 and the requirement of AVR2 for interaction of R2 with BSL1 suggest the role of BSL1 as an intermediary host protein. In addition, Antonovics et al. (2013) hypothesized that the evolutionary mechanism for pathogen specialization by co-evolution with the host results in NHR that reduces the virulence on other potential hosts (Antonovics et al., 2013). These authors explained that nonhost plant species that are closely related to the host may possess the gene variants required for the resistance. However, those variants would be less exposed to selection pressure. As hypothesized, it is possible that pepper has R genes orthologous to Solanum species that recognize the effectors. On the basis of criteria such as sequence identity and chromosome location, 319 putative orthologous pairs of R gene families were identified from the comparative analysis between tomato and potato (Andolfo et al., 2013). Analysis of the pepper genome revealed that 395 NB-LRRs are clustered with those of tomato and potato. Additionally, pepper has several NB-LRRs with orthology to known Solanaceae R genes (Kim et al., 2014). Inversely, pepper is host to Phytophthora capsici which causes serious blight disease. This means that pepper R genes should have evolved to resist P. capsici in the evolutionary arms race. As a consequence of adaptation to P. capsici, pepper R genes could recognize P. infestans effectors that have homology with P. capsici effectors. Genome-wide analysis of the evolution of NB-LRRs and effectors could provide clues to elucidate the mechanism of NHR.
NHR is the most durable form of plant resistance and therefore practical and scientific interest in NHR has increased. However, it is poorly understood, partly due to interspecific genetic incompatibility. We performed functional profiling of the responses of nonhost pepper using effectors of P. infestans based on effector genomics. This experimental approach could possibly be exploited in other nonhost plant–pathogen interactions, which might lead to the identification of the corresponding active R genes and to the understanding of the molecular mechanism of NHR. The completion of the sequencing of the pepper genome combined with our results could offer a new approach for identifying nonhost R genes against late blight (Kim et al., 2014). R genes interacting with multiple effectors may confer durable resistance through functional interfamily transfer and provide a powerful source to breed broad-spectrum disease resistance.
Acknowledgments
We thank Dr Byoung-Cheorl Kang and Hee-Jin Jeong (Seoul National University) for providing the pepper germplasm. This work was supported by grants from the National Research Foundation of the Korea Ministry of Education, Science and Technology (Project number 2010-0015105), the Agricultural Genome Center Program (Project number 710001-03), Rural Development Administration of the Korean government and the National Junior Research Fellowship (Project number NRF-2011-0001763).
Supporting Information
Additional supporting information may be found in the online version of this article.
Fig. S1 Work-flow of this study.
Fig. S2 The response of pepper accessions against PVX upon inoculation of the negative control PVX-dGFP and positive control PVX-PexRD2.
Table S1 The list of primers used in this study
Table S2 Cell death induced by RXLR effectors of Phytophthora infestans in pepper accessions
Table S3 The response of pepper accessions upon inoculation of negative control PVX-dGFP
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
References
- Andolfo G, Sanseverino W, Rombauts S, Van de Peer Y, Bradeen JM, Carputo D, Frusciante L, Ercolano MR. Overview of tomato (Solanum lycopersicum) candidate pathogen recognition genes reveals important Solanum R locus dynamics. New Phytologist. 2013;197:223–237. doi: 10.1111/j.1469-8137.2012.04380.x. [DOI] [PubMed] [Google Scholar]
- Antonovics J, Boots M, Ebert D, Koskella B, Poss M, Sadd BM. The origin of specificity by means of natural selection: evolved and nonhost resistance in host-pathogen interactions. Evolution. 2013;67:1–9. doi: 10.1111/j.1558-5646.2012.01793.x. [DOI] [PubMed] [Google Scholar]
- Armstrong MR, Whisson SC, Pritchard L, Bos JI, Venter E, Avrova AO, Rehmany AP, Bohme U, Brooks K, Cherevach I, et al. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proceedings of the National Academy of Sciences, USA. 2005;102:7766–7771. doi: 10.1073/pnas.0500113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attard A, Gourgues M, Galiana E, Panabieres F, Ponchet M, Keller H. Strategies of attack and defense in plant-oomycete interactions, accentuated for Phytophthora parasitica Dastur (syn. P. Nicotianae Breda de Haan) Journal of Plant Physiology. 2008;165:83–94. doi: 10.1016/j.jplph.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Borhan MH, Gunn N, Cooper A, Gulden S, Tor M, Rimmer SR, Holub EB. WRR4 encodes a TIR-NB-LRR protein that confers broad-spectrum white rust resistance in Arabidopsis thaliana to four physiological races of Albugo candida. Molecular Plant-Microbe Interactions. 2008;21:757–768. doi: 10.1094/MPMI-21-6-0757. [DOI] [PubMed] [Google Scholar]
- Borhan MH, Holub EB, Kindrachuk C, Omidi M, Bozorgmanesh-Frad G, Rimmer SR. WRR4, a broad-spectrum TIR-NB-LRR gene from Arabidopsis thaliana that confers white rust resistance in transgenic oilseed Brassica crops. Molecular Plant Pathology. 2010;11:283–291. doi: 10.1111/j.1364-3703.2009.00599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JI, Armstrong MR, Gilroy EM, Boevink PC, Hein I, Taylor RM, Zhendong T, Engelhardt S, Vetukuri RR, Harrower B, et al. Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proceedings of the National Academy of Sciences, USA. 2010;107:9909–9914. doi: 10.1073/pnas.0914408107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt TO, Schornack S, Banfield MJ, Kamoun S. Oomycetes, effectors, and all that jazz. Current Opinion in Plant Biology. 2012;15:483–492. doi: 10.1016/j.pbi.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL. Programmed cell death in the plant immune system. Cell Death and Differentiation. 2011;18:1247–1256. doi: 10.1038/cdd.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke DE, Cano LM, Raffaele S, Bain RA, Cooke LR, Etherington GJ, Deahl KL, Farrer RA, Gilroy EM, Goss EM, et al. Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PLoS Pathogens. 2012;8:e1002940. doi: 10.1371/journal.ppat.1002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnac S, Lindeberg M, Collmer A. Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Current Opinion in Microbiology. 2009;12:53–60. doi: 10.1016/j.mib.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nature Reviews Genetics. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Fan J, Doerner P. Genetic and molecular basis of nonhost disease resistance: complex, yes; silver bullet, no. Current Opinion in Plant Biology. 2012;15:400–406. doi: 10.1016/j.pbi.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Gabriels SH, Vossen JH, Ekengren SK, van Ooijen G, Abd-El-Haliem AM, van den Berg GC, Rainey DY, Martin GB, Takken FL, de Wit PJ, et al. An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant Journal. 2007;50:14–28. doi: 10.1111/j.1365-313X.2007.03027.x. [DOI] [PubMed] [Google Scholar]
- Gurr SJ, Rushton PJ. Engineering plants with increased disease resistance: how are we going to express it? Trends in Biotechnology. 2005;23:283–290. doi: 10.1016/j.tibtech.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Kamoun S, Zody MC, Jiang RH, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JD. Plant disease resistance genes. Annual Review of Plant Physiology and Plant Molecular Biology. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- Heath MC. Nonhost resistance and nonspecific plant defenses. Current Opinion in Plant Biology. 2000;3:315–319. doi: 10.1016/s1369-5266(00)00087-x. [DOI] [PubMed] [Google Scholar]
- Holsters M, Silva B, Van Vliet F, Genetello C, De Block M, Dhaese P, Depicker A, Inzé D, Engler G, Villarroel R, et al. The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid. 1980;3:212–230. doi: 10.1016/0147-619x(80)90110-9. [DOI] [PubMed] [Google Scholar]
- Huitema E, Vleeshouwers VG, Francis DM, Kamoun S. Active defence responses associated with non-host resistance of Arabidopsis thaliana to the oomycete pathogen Phytophthora infestans. Molecular Plant Pathology. 2003;4:487–500. doi: 10.1046/j.1364-3703.2003.00195.x. [DOI] [PubMed] [Google Scholar]
- Innes RW. Guarding the goods. New insights into the central alarm system of plants. Plant Physiology. 2004;135:695–701. doi: 10.1104/pp.104.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang RH, Tripathy S, Govers F, Tyler BM. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proceedings of the National Academy of Sciences, USA. 2008;105:4874–4879. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Jones L, Hamilton AJ, Voinnet O, Thomas CL, Maule AJ, Baulcombe DC. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell. 1999;11:2291–2301. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S, van West P, Vleeshouwers VG, de Groot KE, Govers F. Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell. 1998;10:1413–1426. doi: 10.1105/tpc.10.9.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti TD, Huitema E, Cakir C, Kamoun S. Synergistic interactions of the plant cell death pathways induced by Phytophthora infestans Nepl-like protein PiNPP1.1 and INF1 elicitin. Molecular Plant-Microbe Interactions. 2006;19:854–863. doi: 10.1094/MPMI-19-0854. [DOI] [PubMed] [Google Scholar]
- Khatib M, Lafitte C, Esquerre-Tugaye MT, Bottin A, Rickauer M. The CBEL elicitor of Phytophthora parasitica var. nicotianae activates defence in Arabidopsis thaliana via three different signalling pathways. New Phytologist. 2004;162:501–510. [Google Scholar]
- Kim S, Park M, Yeom SI, Kim YM, Lee JM, Lee HA, Seo E, Choi J, Cheong K, Kim KT, et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nature Genetics. 2014;46:270–278. doi: 10.1038/ng.2877. [DOI] [PubMed] [Google Scholar]
- Lee YH, Kim HS, Kim JY, Jung M, Park YS, Lee JS, Choi SH, Her NH, Lee JH, Hyung NI, et al. A new selection method for pepper transformation: callus-mediated shoot formation. Plant Cell Reports. 2004;23:50–58. doi: 10.1007/s00299-004-0791-1. [DOI] [PubMed] [Google Scholar]
- Lokossou AA, Park TH, van Arkel G, Arens M, Ruyter-Spira C, Morales J, Whisson SC, Birch PR, Visser RG, Jacobsen E, et al. Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Molecular Plant-Microbe Interactions. 2009;22:630–641. doi: 10.1094/MPMI-22-6-0630. [DOI] [PubMed] [Google Scholar]
- Naito K, Taguchi F, Suzuki T, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y. Amino acid sequence of bacterial microbe-sssociated molecular pattern flg22 is tequired for virulence. Molecular Plant-Microbe Interactions. 2008;21:1165–1174. doi: 10.1094/MPMI-21-9-1165. [DOI] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant Journal. 2009;60:218–226. doi: 10.1111/j.1365-313X.2009.03949.x. [DOI] [PubMed] [Google Scholar]
- Niks RE, Marcel TC. Nonhost and basal resistance: how to explain specificity? New Phytologist. 2009;182:817–828. doi: 10.1111/j.1469-8137.2009.02849.x. [DOI] [PubMed] [Google Scholar]
- Oh SK, Young C, Lee M, Oliva R, Bozkurt TO, Cano LM, Win J, Bos JI, Liu HY, van Damme M, et al. In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell. 2009;21:2928–2947. doi: 10.1105/tpc.109.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart JR, Mestre P, Lu R, Malcuit I, Baulcombe DC. NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Current Biology. 2005;15:968–973. doi: 10.1016/j.cub.2005.04.053. [DOI] [PubMed] [Google Scholar]
- Randall TA, Dwyer RA, Huitema E, Beyer K, Cvitanich C, Kelkar H, Fong AM, Gates K, Roberts S, Yatzkan E, et al. Large-scale gene discovery in the oomycete Phytophthora infestans reveals likely components of phytopathogenicity shared with true fungi. Molecular Plant-Microbe Interactions. 2005;18:229–243. doi: 10.1094/MPMI-18-0229. [DOI] [PubMed] [Google Scholar]
- Saunders DG, Breen S, Win J, Schornack S, Hein I, Bozkurt TO, Champouret N, Vleeshouwers VG, Birch PR, Gilroy EM, et al. Host protein BSL1 associates with Phytophthora infestans RXLR effector AVR2 and the Solanum demissum immune receptor R2 to mediate disease resistance. Plant Cell. 2012;24:3420–3434. doi: 10.1105/tpc.112.099861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P, Panstruga R. A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends in Plant Science. 2011;16:117–125. doi: 10.1016/j.tplants.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Shi J, Choi D, Kim BD, Kang BC. Study on inheritance of Potato virus X Resistance in Capsicum annuum. Plant Pathology Journal. 2008;24:433–438. [Google Scholar]
- Sinapidou E, Williams K, Nott L, Bahkt S, Tor M, Crute I, Bittner-Eddy P, Beynon J. Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant Journal. 2004;38:898–909. doi: 10.1111/j.1365-313X.2004.02099.x. [DOI] [PubMed] [Google Scholar]
- Sohn KH, Saucet SB, Clarke CR, Vinatzer BA, O'Brien HE, Guttman DS, Jones JD. HopAS1 recognition significantly contributes to Arabidopsis nonhost resistance to Pseudomonas syringae pathogens. New Phytologist. 2012;193:58–66. doi: 10.1111/j.1469-8137.2011.03950.x. [DOI] [PubMed] [Google Scholar]
- Sumit R, Sahu BB, Xu M, Sandhu D, Bhattacharyya MK. Arabidopsis nonhost resistance gene PSS1 confers immunity against an oomycete and a fungal pathogen but not a bacterial pathogen that cause diseases in soybean. BMC Plant Biology. 2012;12:87. doi: 10.1186/1471-2229-12-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FL, Luderer R, Gabriels SH, Westerink N, Lu R, de Wit PJ, Joosten MH. A functional cloning strategy, based on a binary PVX-expression vector, to isolate HR-inducing cDNAs of plant pathogens. Plant Journal. 2000;24:275–283. doi: 10.1046/j.1365-313x.2000.00866.x. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H. Fresh insights into processes of nonhost resistance. Current Opinion in Plant Biology. 2003;6:351–357. doi: 10.1016/s1369-5266(03)00063-3. [DOI] [PubMed] [Google Scholar]
- Tian M, Benedetti B, Kamoun S. A Second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiology. 2005;138:1785–1793. doi: 10.1104/pp.105.061226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torto TA, Li S, Styer A, Huitema E, Testa A, Gow NA, van West P, Kamoun S. EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Research. 2003;13:1675–1685. doi: 10.1101/gr.910003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Katagiri F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Current Opinion in Plant Biology. 2010;13:459–465. doi: 10.1016/j.pbi.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers VG, van Dooijeweert W, Govers F, Kamoun S, Colon LT. The hypersensitive response is associated with host and nonhost resistance to Phytophthora infestans. Planta. 2000;210:853–864. doi: 10.1007/s004250050690. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers VG, Driesprong JD, Kamphuis LG, Torto-Alalibo T, Van't Slot KA, Govers F, Visser RG, Jacobsen E, Kamoun S. Agroinfection-based high-throughput screening reveals specific recognition of INF elicitins in Solanum. Molecular Plant Pathology. 2006;7:499–510. doi: 10.1111/j.1364-3703.2006.00355.x. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers VG, Rietman H, Krenek P, Champouret N, Young C, Oh SK, Wang M, Bouwmeester K, Vosman B, Visser RG, et al. Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE. 2008;3:e2875. doi: 10.1371/journal.pone.0002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vossen EA, Gros J, Sikkema A, Muskens M, Wouters D, Wolters P, Pereira A, Allefs S. The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant Journal. 2005;44:208–222. doi: 10.1111/j.1365-313X.2005.02527.x. [DOI] [PubMed] [Google Scholar]
- Wei CF, Kvitko BH, Shimizu R, Crabill E, Alfano JR, Lin NC, Martin GB, Huang HC, Collmer A. A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant Journal. 2007;51:32–46. doi: 10.1111/j.1365-313X.2007.03126.x. [DOI] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- Win J, Chaparro-Garcia A, Belhaj K, Saunders DG, Yoshida K, Dong S, Schornack S, Zipfel C, Robatzek S, Hogenhout SA, et al. Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harbor Symposia on Quantitative Biology. 2012;77:235–247. doi: 10.1101/sqb.2012.77.015933. [DOI] [PubMed] [Google Scholar]
- Wroblewski T, Caldwell KS, Piskurewicz U, Cavanaugh KA, Xu H, Kozik A, Ochoa O, McHale LK, Lahre K, Jelenska J, et al. Comparative large-scale analysis of interactions between several crop species and the effector repertoires from multiple pathovars of Pseudomonas and Ralstonia. Plant Physiology. 2009;150:1733–1749. doi: 10.1104/pp.109.140251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom SI, Seo E, Oh SK, Kim KW, Choi D. A common plant cell-wall protein HyPRP1 has dual roles as a positive regulator of cell death and a negative regulator of basal defense against pathogens. Plant Journal. 2012;69:755–768. doi: 10.1111/j.1365-313X.2011.04828.x. [DOI] [PubMed] [Google Scholar]
- Zhao B, Lin X, Poland J, Trick H, Leach J, Hulbert S. A maize resistance gene functions against bacterial streak disease in rice. Proceedings of the National Academy of Sciences, USA. 2005;102:15383–15388. doi: 10.1073/pnas.0503023102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Work-flow of this study.
Fig. S2 The response of pepper accessions against PVX upon inoculation of the negative control PVX-dGFP and positive control PVX-PexRD2.
Table S1 The list of primers used in this study
Table S2 Cell death induced by RXLR effectors of Phytophthora infestans in pepper accessions
Table S3 The response of pepper accessions upon inoculation of negative control PVX-dGFP


