Abstract
BACKGROUND & AIMS
Autophagy is an intracellular lysosomal degradation process that plays an important role in regulating normal physiological functions of the liver. The purpose of the present study was to investigate the mechanism(s) by which the loss of hepatic autophagy leads to liver inflammation, fibrosis and tumorigenesis.
METHODS
Hepatocyte-specific Atg5 knockout mice were generated by crossing Atg5 Flox/Flox mice with albumin Cre mice. These mice were also crossed with Nrf2 knockout mice to generate Atg5 Flox/Flox, Albumin Cre+/Nrf2−/− double knockout mice. These mice were housed for various time points up to 15 months, and blood and liver tissues were harvested for biochemical and histological analysis.
RESULTS
Hepatocyte-specific deletion of Atg5 resulted in increased apoptosis, inflammation and fibrosis in the liver. Increased apoptosis in hepatocyte-specific Atg5 knockout mice was likely due to accumulation of aberrant polyubiquitinated proteins (proteotoxicity) and disruption of the homeostasis of pro-and anti-apoptotic proteins. All of these pathological changes started as early as one month and persisted for 12–15 months. At 9–15 months of age, these mice also developed hepatocellular adenomas. Interestingly, deletion of Nrf2 in Atg5 liver-specific knockout mice markedly abolished these pathological changes, indicating a key role for this transcription factor in the mechanism of hepatic pathology.
CONCLUSIONS
Our results provide genetic evidence that loss of autophagy in hepatocytes causes cell death resulting in liver inflammation, fibrosis and tumorigenesis. We also demonstrate that persistent activation of Nrf2 is critical for liver inflammation, fibrosis and eventual tumorigenesis that occur in mice with defects in hepatocyte autophagy.
Keywords: Atg5, autophagy, Nrf2, inflammation, fibrosis, liver tumor
INTRODUCTION
Accumulating evidence suggests that autophagy, an intracellular lysosomal degradation pathway, acts as a tumor suppressor. Mice with heterozygous disruption of Beclin 1, an essential autophagy gene, develop spontaneous tumors in multiple tissues including hepatocellular carcinoma [1]. Moreover, hepatocyte-specific Atg7 knockout (KO) mice and mice with systemic mosaic deletion of Atg5 develop benign liver adenomas [2, 3]. Both hepatocytes-specific Atg7 KO and Atg5 mosaic mice have increased liver injury and severe hepatomegaly. However, the mechanism by which liver tumorigenesis develops in autophagy-deficient mice is not clear. Autophagy-deficient livers have increased accumulation of p62/SQSTM1, an autophagy substrate and receptor protein. p62 competes with Nrf2 (Nuclear factor (erythroid-derived 2)-like 2) for binding to Keap1 (Kelch-like ECH-associated protein 1) resulting in dissociation of Nrf2 from Keap1 and activation of Nrf2, a transcription factor which regulates expression of cytoprotective genes and hepatic detoxification enzymes [4–6]. Interestingly, accumulation of p62 and activation of Nrf2 have been found in human HCCs [3]. More importantly, it has been shown that deletion of p62 reduces the size and number of tumors in liver-specific Atg7 KO mice [2]. However, the molecular mechanism by which p62 promotes liver tumorigenesis is not clear.
Liver tumorigenesis is caused by chronic liver injury produced by agents such as alcohol, hepatitis C or B infections, exposure to hepatotoxins and obesity. These conditions often lead to hepatocyte death, which triggers a cyclical inflammatory response that further induces cell death and subsequent liver repair and compensatory proliferation. Concomitantly, the liver also develops fibrosis, cirrhosis and eventual liver tumorigenesis [7–9]. Hepatic fibrosis is a reversible wound-healing process resulting from continuous injury to the liver. Interestingly, it has been reported that autophagy of activated stellate cells promotes fibrosis by increasing the degradation of lipids [10, 11]. However, it is not known how autophagy in hepatocytes, the major parenchymal cell type in the liver, would regulate fibrosis. The aim of this study was to determine the role of autophagy on pathogenesis of hepatic fibrosis and tumorigenesis. Furthermore, we also tested the hypothesis that p62 promotes liver tumor formation in part by stimulating persistent Nrf2 activation.
MATERIALS AND METHODS
Animal experiments
Atg5 Flox/Flox (Atg5 F/F) mice (C57BL/6/129) were generated by Dr. N. Mizushima and have been backcrossed with C57BL/6 for at least 5 generations and further crossed with Albumin-Cre mice (Alb-Cre, C57BL/6) (Jackson Laboratory) as described previously [12, 13]. The generation of Nrf2−/− mice was described previously [14], and the mice were kindly provided by Dr. Curtis Klaassen (University of Kansas Medical Center). All animals received humane care. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Kansas Medical Center. Atg5 F/F, Alb Cre+ mice and Atg5 F/F, Alb Cre- matched littermates were used in this study. Mice were sacrificed at 1, 2, 4, 6, 9, 12, and 15 months. Atg5 F/F, Alb Cre+ mice were further crossed with Nrf2−/− mice to generate Atg5 F/F, Alb Cre+/Nrf2−/− double knockout (DKO) mice. All others see Supplemental Materials.
RESULTS
Atg5-deficient hepatocytes have increased apoptosis likely due to disruption of the homeostasis of pro- and anti-apoptotic proteins
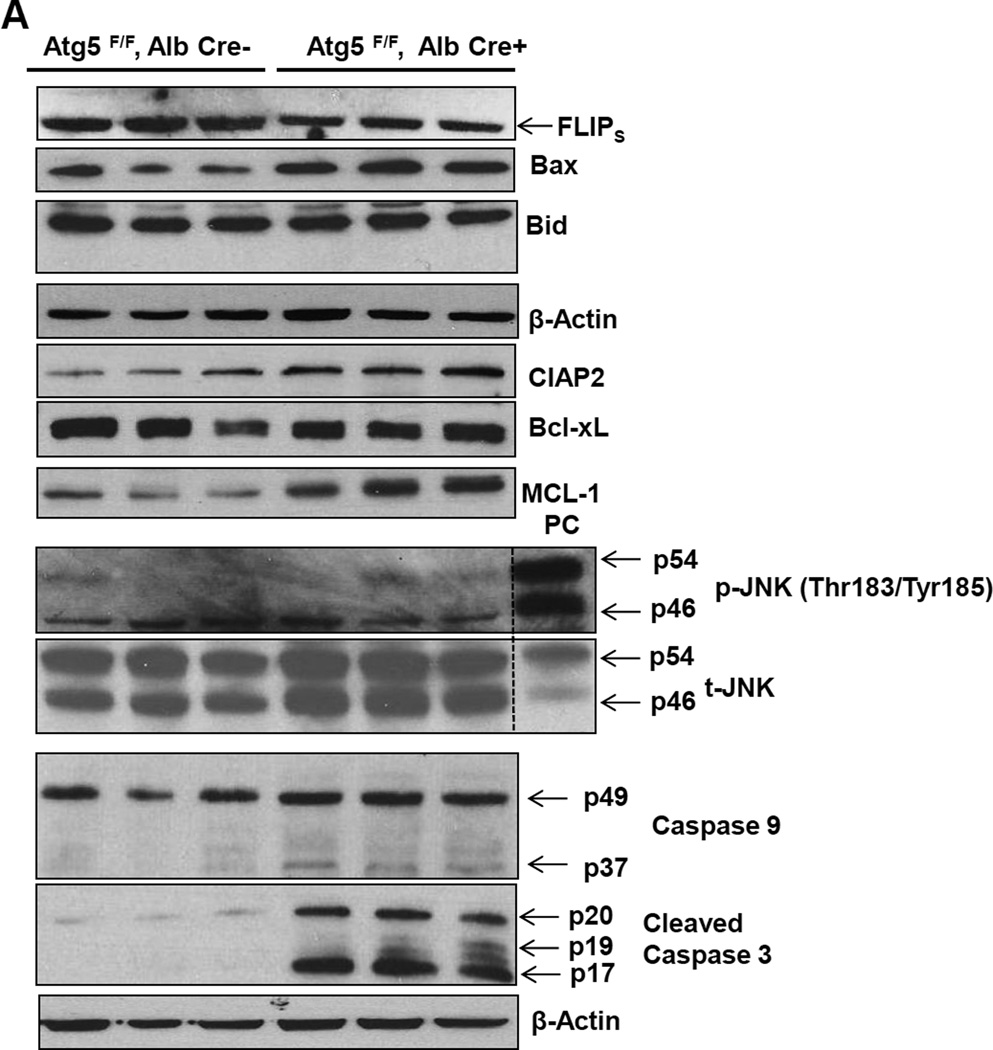
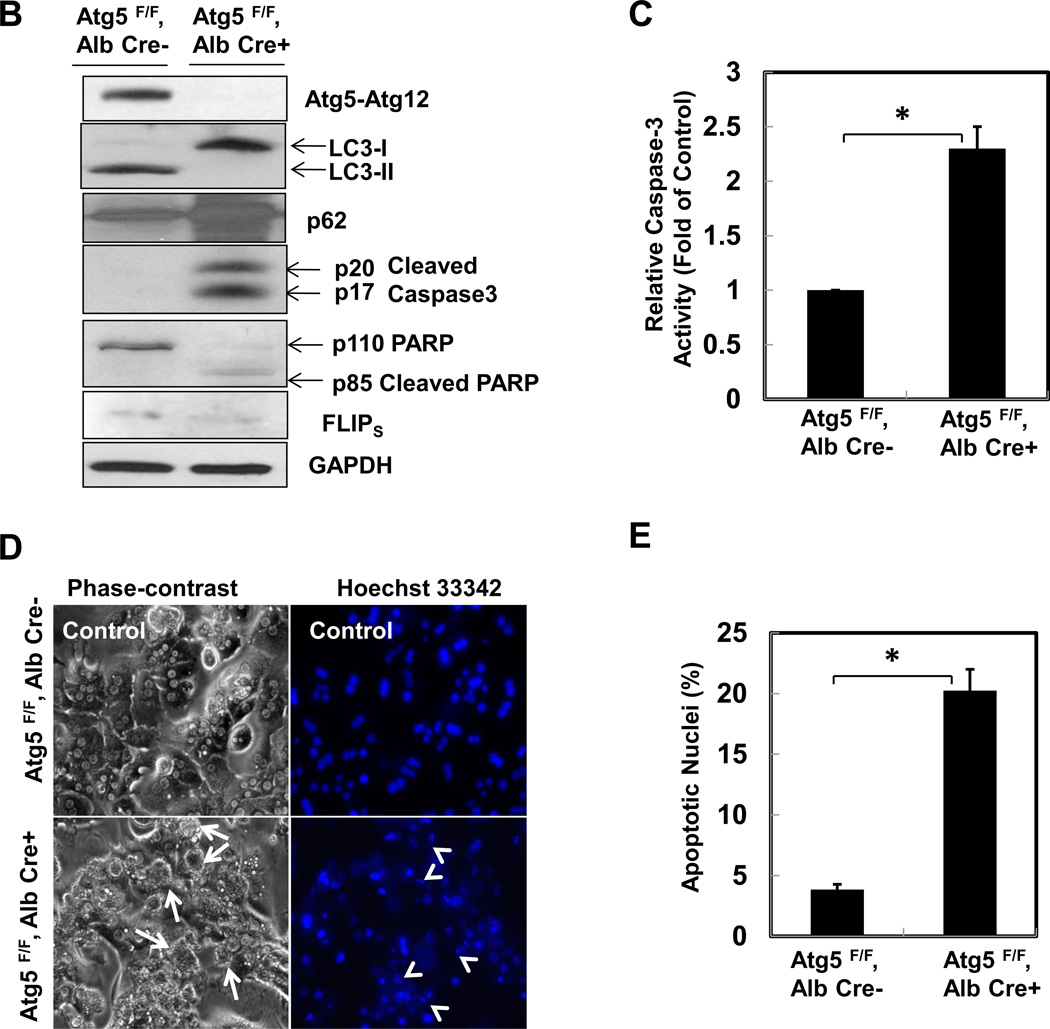
To gain detailed information on the chronologic pathological changes in the autophagy-deficient liver, we measured liver to body weight ratios and serum alanine aminotransferase (ALT) levels in 1, 2, 4, 6, 9, 12 and 15 month old hepatocyte specific Atg5-deficient mice (Hep-Atg5 KO) and matched wild type (WT) littermates. The liver to body weight ratio was increased as early as 1 month (approximately 75% increase) in Hep- Atg5 KO mice and was increased as high as four fold by 15 months (sFigure 1A). Serum ALT activities (sFigure 1B) were increased significantly at all time points assessed in Hep-Atg5 KO mice, indicating liver injury. We found that the level of FLICE-like protein (FLIP), which inhibits caspase-8 activation, was decreased but the expression of Bax (a pro-apoptotic protein) was increased in Hep-Atg5 KO mouse livers. No differences in the expression of Bcl-XL or phosphorylated JNK were found between Hep-Atg5 KO and WT mice, but the expression levels of anti-apoptotic Mcl-1 and CIAP2 were increased in Hep-Atg5 KO mice, likely due to a compensatory adaptive response to injury. As a result, the activation of caspase-8, -9 and -3 were all increased (Figure 1A & sFigure 1C-E). We did not find obvious Bid cleavage, likely due to the relatively weak activation of caspase-8 in Hep-Atg5 KO mice. Primary cultured Atg5 KO hepatocytes had no detectable Atg5-Atg12, LC3-II but increased p62 levels, which also had increased caspase-3 and PARP cleavage, caspase-3 activities and apoptosis compared to WT hepatocytes (Figure 1 B-E). Histological analysis of H & E-stained liver sections demonstrated increased inflammation (sFigure 2A, arrows) and apoptosis (sFigure 2A arrow heads) as well as focal necrosis (sFigure 2A, stars) in Hep-Atg5 KO mice. Immunostaining using specific antibodies for neutrophils (Ly6B) and macrophages (F4/80) confirmed the presence of neutrophils (sFigure 2B, upper panel, arrow heads) and macrophages (sFigure 2B lower panel, arrows) in Hep-Atg5 KO mouse livers. Consistent with the immunostaining data, mRNA levels of F4/80, CD68 and Ly6G as well as the number of neutrophils and macrophages were also significantly elevated in Hep-Atg5 KO mouse livers (sFigure 2C-E). In addition, increased expression of various inflammatory cytokines was observed at all time points assessed in Hep-Atg5 KO mouse livers (sFigure 3A-D). These data suggest that loss of autophagy in hepatocytes leads to apoptosis likely due to decreased FLIP expression, which results in caspase activation followed by compensatory activation of some anti-apoptotic proteins and subsequent inflammation.
Figure 1. Atg5-deficient hepatocytes have disrupted homeostasis of pro-and anti-apoptotic proteins and increased apoptosis.
(A) Total liver lysates of the indicated genotypes of 2-month-old mice were subjected to western blot analysis. PC (positive control): liver lysates from acetaminophen (500 mg/kg, 6hrs)-treated mice. Primary hepatocytes isolated from indicated genotypes and cultured for 6 hrs. Total cell lysates were subjected to western blot analysis (B) and caspase-3 activity assay (C). Apoptosis was determined by Hoechst 33342 staining for apoptotic nuclei (D). Apoptotic nuclei were quantified from counting more than 300 cells for each experiment and data are presented as means± SEM from 3 independent experiment (E). * p<0.05 Student t Test.
Loss of Atg5 in hepatocytes causes fibrosis
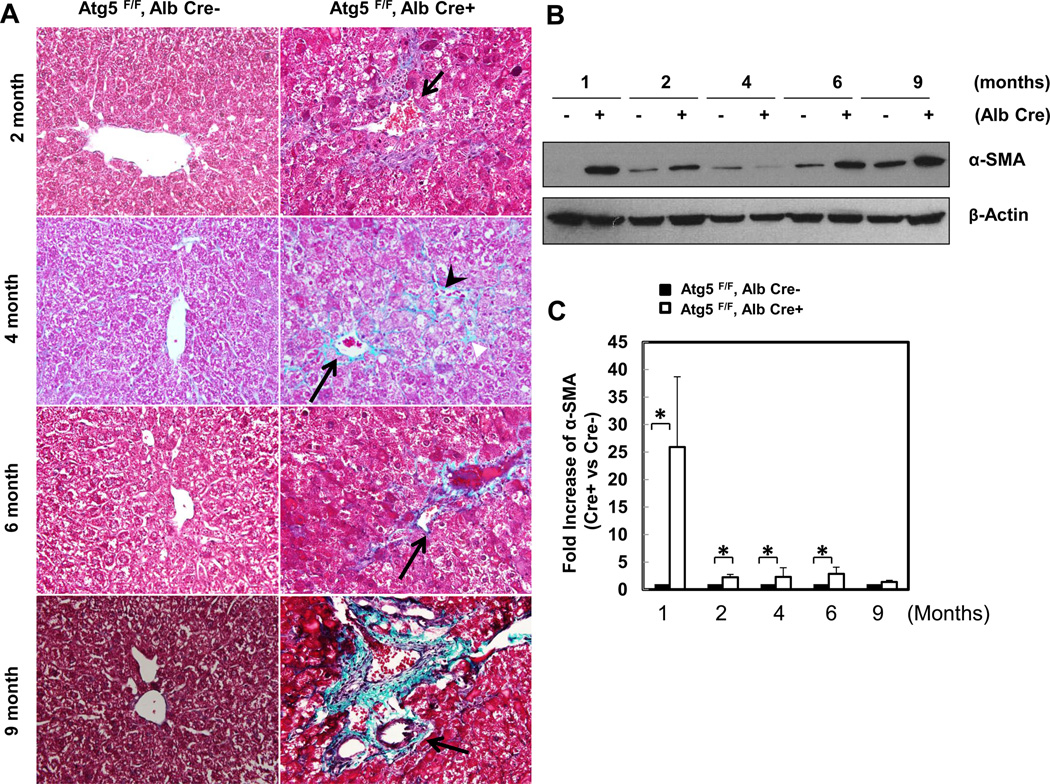
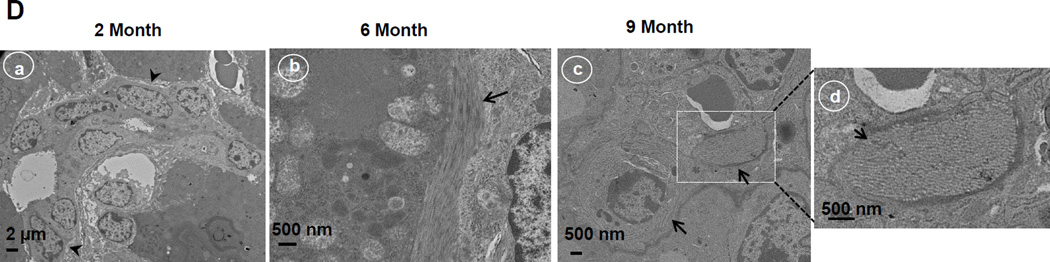
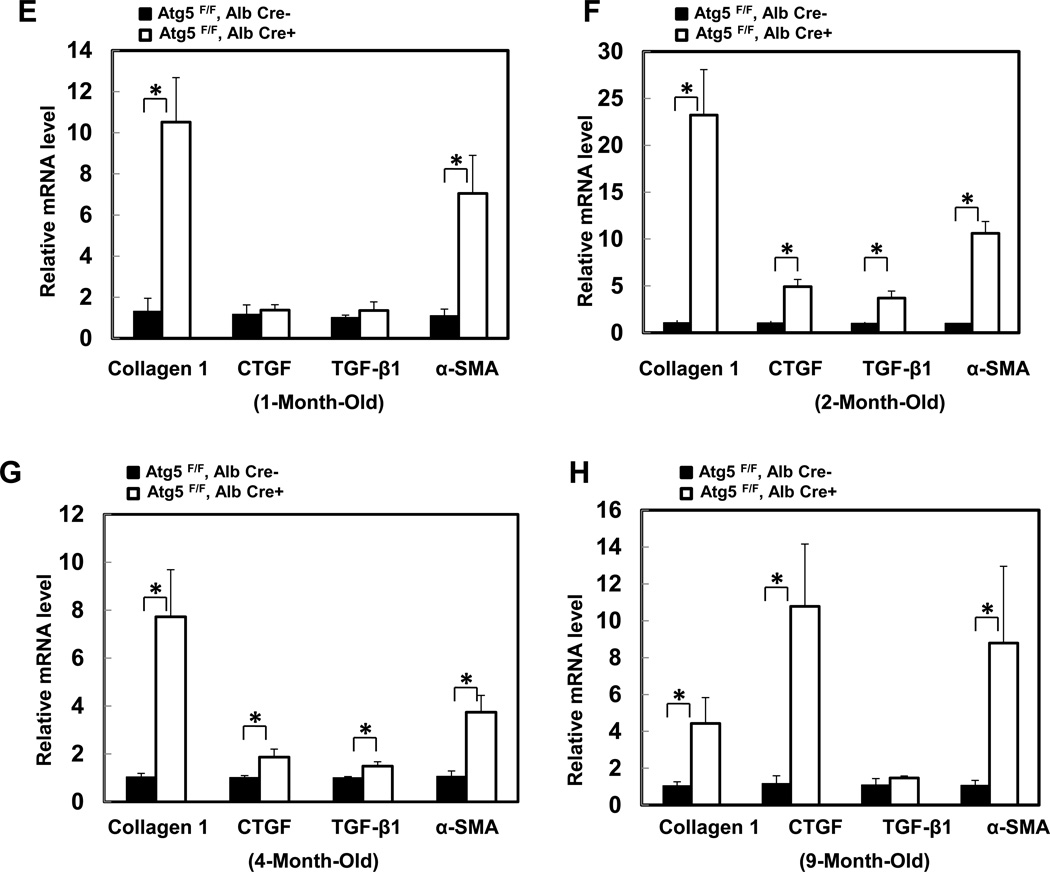
We next evaluated hepatic fibrosis in Hep-Atg5 KO mice. Extensive perivenular, portal (Figure 2A, arrows) and pericellular (Figure 2A, arrow heads) collagen deposition was evident in Hep-Atg5 KO mouse livers, as demonstrated by Gomori’s trichrome staining (Figure 2A & sFigure 4A). Western blot analysis revealed that α-smooth muscle actin (α-SMA) levels were persistently higher in Hep-Atg5 KO mouse livers indicating the presence of myofibroblasts (Figure 2B &C). Furthermore, immunostaining for cytokeratin 19 (CK19), a liver precursor cell marker, showed increased CK19 positive duct-like structures in Hep-Atg5 KO livers with barely detectable levels in WT mice (sFigure 4B, arrows). Duct-like structures (Figure 2D, panel a) and collagen fibers (Figure 2D, panels b-d) were also detected in liver tissues from Hep-Atg5 KO mice under EM analysis. In line with these fibrotic changes, the expression of profibrotic genes including collagen type 1, connective tissue growth factor (CTGF), transforming growth factor β1 (TGF-β1) and α-SMA were increased (Figure 2E-H). Since it has been reported that autophagy in HSC promotes liver fibrosis by increasing the release of free fatty acids through lipophagy [11], we next determined autophagy activity in HSC isolated from Hep-Atg5 KO mice. We found that HSC isolated from Hep-Atg5 KO mice proliferated during a 10 day culture as demonstrated by increased cell number and density at day 8 and day 10 compared to day 1 (sFigure 5A). More importantly, typical double-membrane autophagosome structures that contained lipid droplets (LD) (sFigure 5B, panel a) or other cellular contents and membrane structures (sFigure 5B, panel b), were readily detected in cultured HSC isolated from Hep-Atg5 KO mice. Western blot analysis showed that unlike the Atg5-deficient hepatocytes, which had higher unlipidated LC3-I form, there was an increased level of lipidated LC3-II form with barely detectable LC3-I form in cultured HSC from Hep-Atg5 KO mice. Interestingly, the level of p62 decreased in HSC cultured for 10 days compared to cells cultured for 2 days (sFigure 5C), suggesting increased autophagic flux during culture. These data clearly indicate that autophagy is functional in HSCs in Hep-Atg5 KO mice, suggesting the deletion of Atg5 by Alb Cre mainly affected hepatocytes but not HSC. Collectively, these data indicate that Hep-Atg5 KO mice develop hepatic fibrosis.
Figure 2. Deletion of Atg5 in the liver causes liver fibrosis.
(A) Gomori’s trichrome staining of liver tissues. Mice were sacrificed at indicated ages and liver tissues were processed for Gomori’s trichrome staining (20 ×). Arrows: peribiliary fibrosis. Arrow heads: interstitial fibrosis. (B) Total liver lysates of the indicated genotypes from different ages were subjected to western blot analysis for α-SMA, and one representative experiment from 3 independent experiments is shown. (C) Densitometry analysis of the changes of α-SMA of (B) (n=3). (D) Liver tissues from different ages of Atg5 F/F, Alb Cre+ mice were processed for EM analysis. Panel a: bile duct epithelial cells (arrow heads). Panels b-c: collagen fibers (arrows). Panel d is an enlarged micrograph from the boxed area in panel c. (E-H) Quantitative real-time PCR analysis of fibrogenic genes in mouse livers. Total RNAs were prepared from livers of the indicated genotypes of different ages. Data are presented as means± SEM (n=3–10). * p<0.05 Student t Test.
Liver injury, inflammation and fibrosis in Hep-Atg5 KO mice are suppressed by deletion of Nrf2
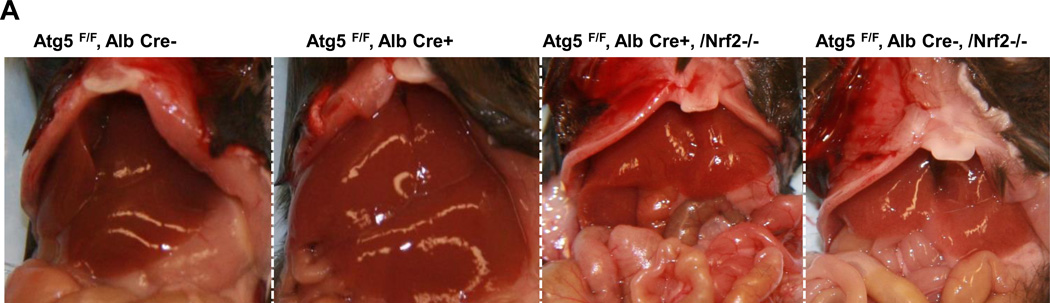
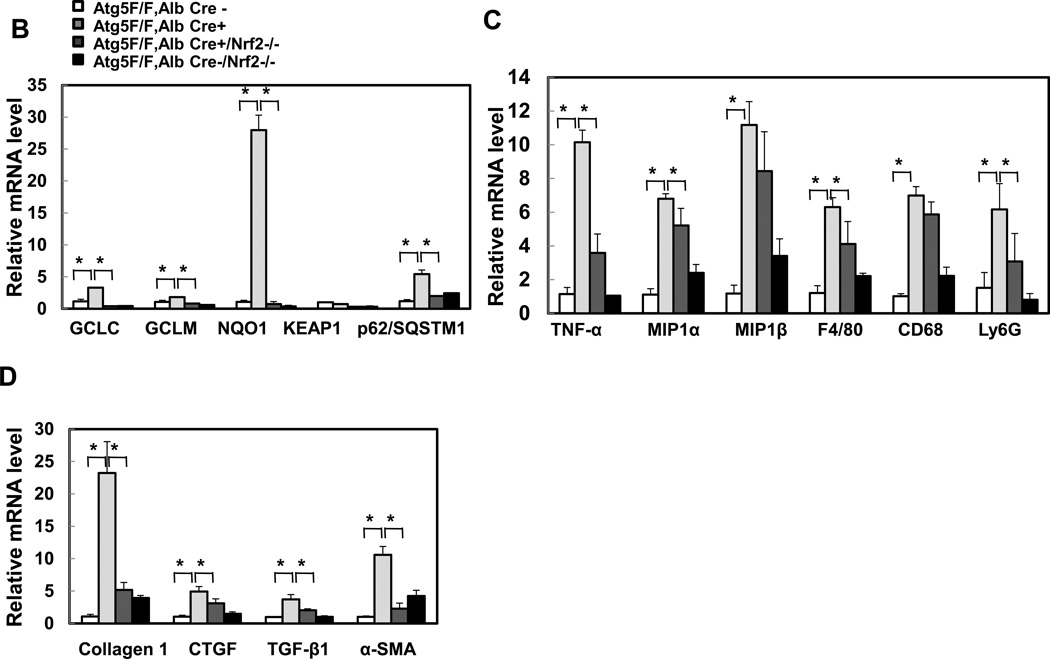
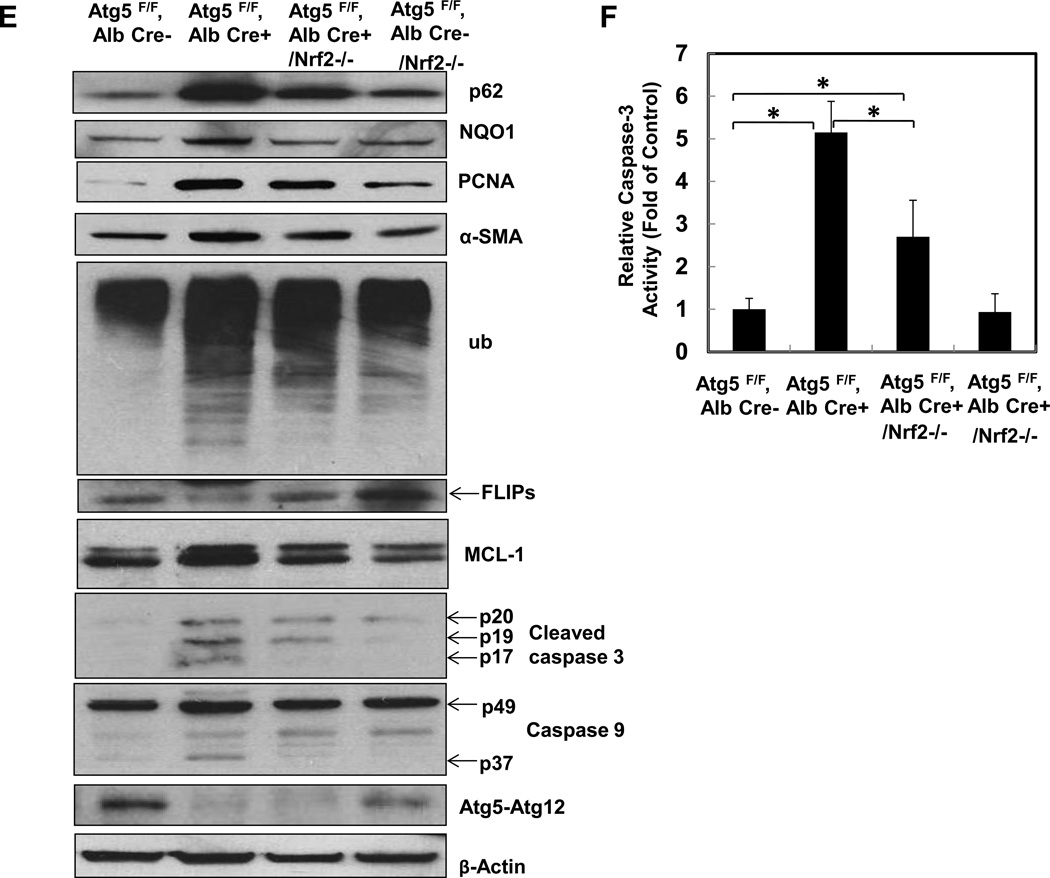
Previous studies, including ours, showed that loss of autophagy in livers caused persistent activation of Nrf2 by activating the noncanonical p62-Keap1-Nrf2 pathway [2, 13, 15]. Consistent with previous studies, p62 and Nrf2 target protein NAD(P)H:quinone oxidoreductase (NQO1), were increased in Hep-Atg5 KO mouse livers. The lack of LC3-II form and increased LC3-I form and p62 levels in Hep-Atg5 KO mouse liver tissues confirmed the lack of autophagy (sFigure 6A). In agreement with previous findings, we also found that over-expression or knockdown of p62 increased or decreased NQO1 expression, respectively (sFigure 6B-C), indicating that accumulating p62 activates Nrf-2. To further determine the role of Nrf2 in the pathogenesis of Hep-Atg5 KO mouse livers, we deleted Nrf2 in Hep-Atg5 KO mice by crossing Atg5F/F, Alb Cre+ mice with Nrf2−/− mice. We found that loss of Nrf2 completely abolished hepatomegaly and liver injury in Hep-Atg5 KO mice (Figure 3A & sFigure 7). In the absence of Nrf2, expression of glutamate-cysteine ligase catalytic subunit (GCLC), glutamate-cysteine ligase modifier subunit (GCLM) and NQO1 in Hep-Atg5 KO mouse livers was significantly blunted. The expression of Keap1 was not affected. Interestingly, we found that the mRNA level of p62 was significantly increased in Hep-Atg5 KO mouse livers, which was inhibited by the further deletion of Nrf2 (Figure 3B). These results are in agreement with previous findings that there is a positive feedback loop that regulates hepatic p62 levels in which p62 activates Nrf2 through competitive binding with Keap1, and activated-Nrf2 further upregulates p62 at the transcription level[16]. Furthermore, increased expression of inflammatory (Figure 3C) and fibrotic genes (Figure 3D) in Hep-Atg5 KO mouse livers was also significantly inhibited by deletion of Nrf2. In addition to mRNA changes, we also confirmed that increased protein levels of p62, NQO1, PCNA and α-SMA as well as activation of caspase-3 and -9 were all inhibited by deletion of Nrf2 (Figure 3E-F). Infiltration of inflammatory cells and apoptosis/necrosis (sFigure 8A), aberrant membrane structures (sFigure 8B), fibrosis (sFigure 8C) and hepatic triglyceride (sFigure 8D) in Hep-Atg5 KO livers were also dramatically decreased or absent in the Atg5 F/F, Alb Cre+/Nrf2−/− double knockout (DKO) mouse livers. Hep-Atg5 KO mice had increased phosphorylated 4EBP1, suggesting increased mTOR activity, which was also blunted in the DKO mice (sFigure 9A-B). Furthermore, increased polyubiquitinated proteins and Mcl-1 as well as decreased FLIPs were also suppressed in DKO mice (Figure 3E). Interestingly, no obvious ER stress was found in Hep-Atg5 KO mouse livers (sFigure 9C). These results indicate that persistent activation of Nrf2 contributes to Atg5-deficiency-induced liver injury by enhancing aberrant protein accumulation and disrupting the homeostasis of pro-and anti-apoptotic proteins.
Figure 3. Suppression of liver hepatomegaly and liver injury in hepatocyte specific Atg5-deficient mice by Nrf2 deletion.
(A) Representative gross anatomy of livers of indicated genotypes at 2 months. *p<0.05, one way anova analysis with Scheffé’s post hoc test. Quantitative real-time PCR analyses of (B) Nrf2 target genes, (C) inflammatory genes and (D) fibrogenic genes. Total RNAs were prepared from livers of the indicated genotypes of 2-month-old mice. Data are presented as means± SEM (n=3–10). *p<0.05, one way anova analysis with Scheffé’s post hoc test. (E) Total liver lysates of the indicated genotypes of 2-month-old mice were subjected to western blot analysis. One representative experiment from 3 independent experiments is shown. (F) Total cell lysates from (E) were used to measure the caspase-3 activities. Data are presented as fold of control (means± SEM, n=3–10). * p<0.05 Student t Test
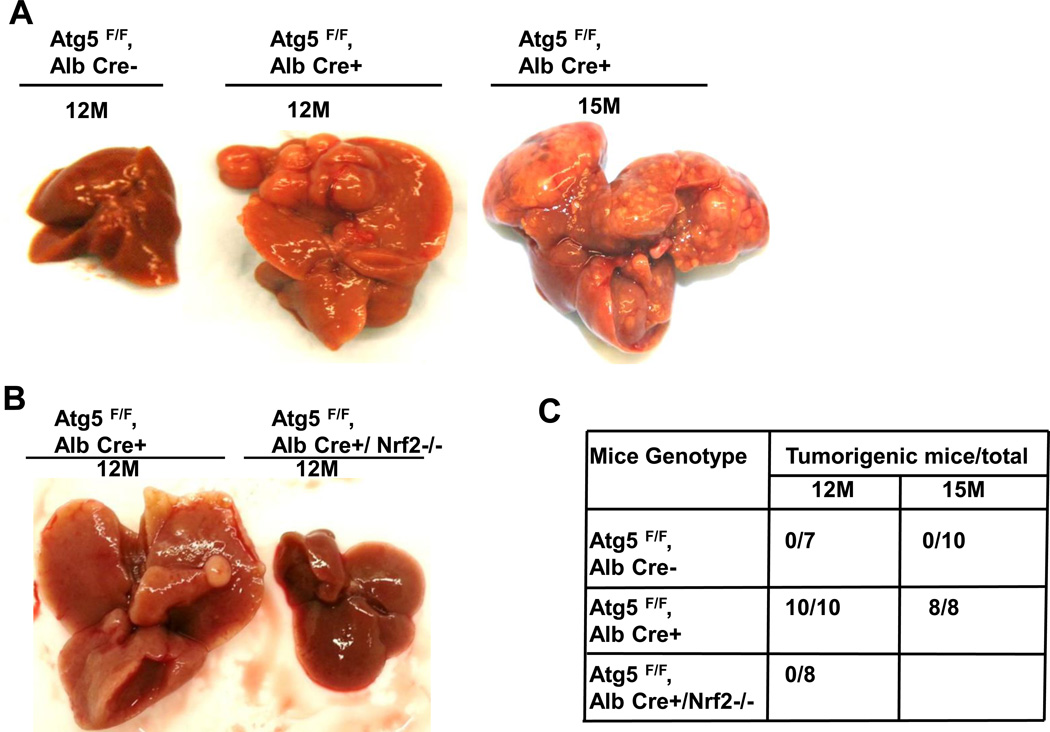
Deletion of Nrf2 suppresses Atg5-deficiency-induced spontaneous liver tumors
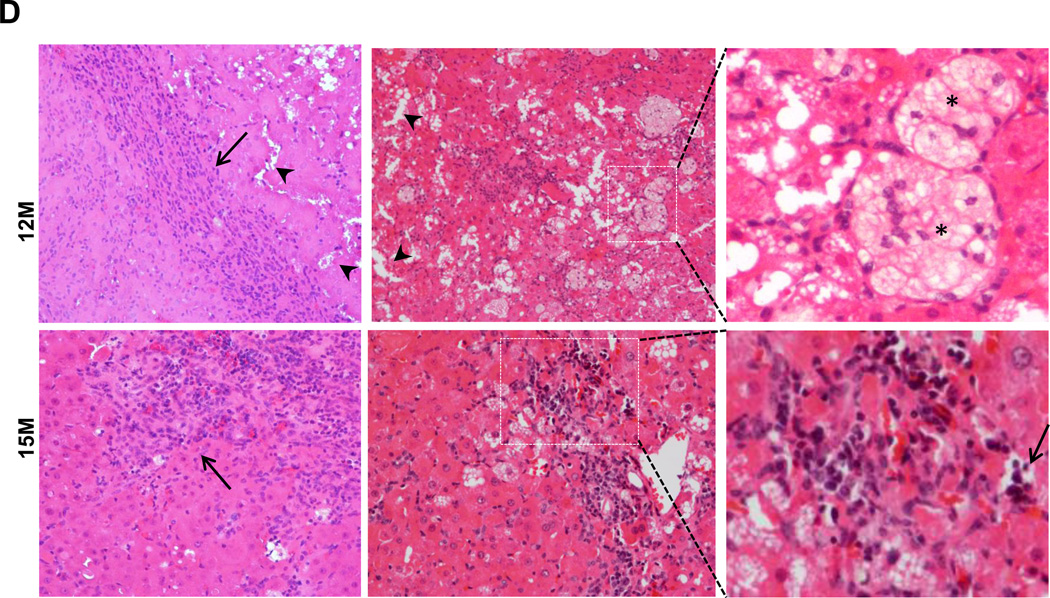
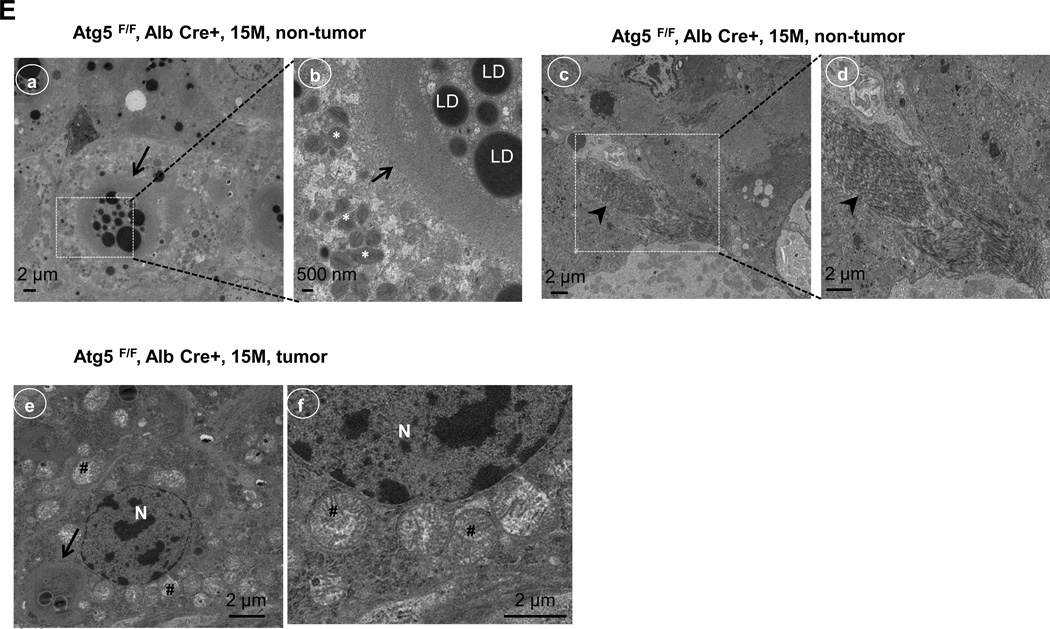
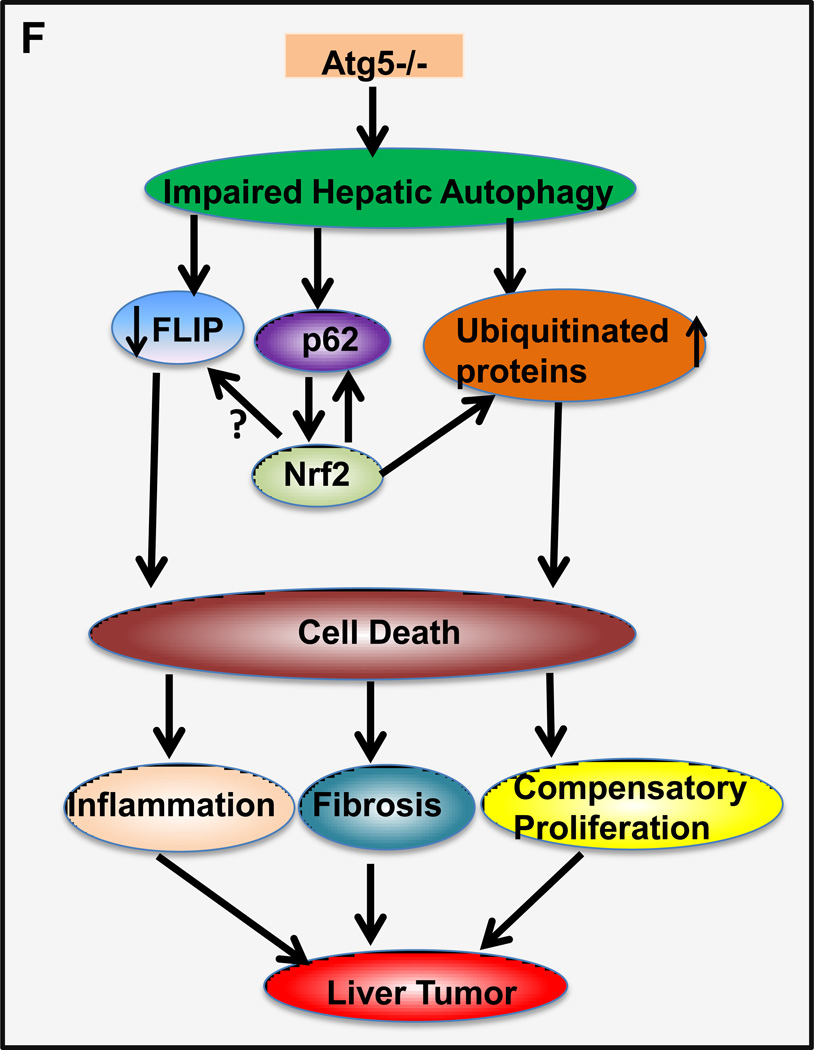
Previous studies showed that spontaneous liver tumors develop in mice with a loss of Atg7 in the liver or mosaic deletion of Atg5 [2, 17]. In agreement with these results, Hep-Atg5 KO mice also developed spontaneous liver tumors, which were evident as early as 9 months (data not shown). The number and size of tumors increased with age, occurring in all 12 and 15 month old Hep-Atg5 KO mice (Figure 4A-C). Interestingly, we did not observe any liver tumors in the Atg5 F/F, Alb Cre+/Nrf2−/− DKO mice (Figure 4B & C). Taken together, these results indicate that liver inflammation, fibrosis and tumorigenesis in Hep-Atg5 KO mice are reversed by deletion of Nrf2. Histological analysis showed that the tumors were ill-demarcated and were composed of benign hepatocytes arranged in regular plates, usually one or two cells thick. Sinusoidal dilatation, ductular reaction, and inflammation were also found in Hep-Atg5 KO mouse liver tumors (Figure 4D). Focal necrosis was also observed in some of the tumors (Figure 4D, stars). These tumors also had abundant reticulin staining, which is normally absent or decreased in hepatocellular carcinoma (sFigure 10). These phenotypes were similar to tumors reported in Atg7-deficient mouse livers and were pathologically diagnosed as inflammatory hepatocellular adenoma [17]. The tumor cells had a higher proliferation rate as demonstrated by increased protein levels of PCNA in tumor tissues compared to adjacent non-tumor tissues from the same Hep-Atg5 KO mouse liver (sFigure 11). EM analysis for non-tumor tissues from 15-month-old Hep-Atg5 KO livers showed aberrant multimembrane structures surrounded by lipid droplets (Figure 4E, panels a-b, arrows) which was similar to the 2-month-old Hep-Atg5 KO livers we reported previously[18]. Increased numbers of peroxisomes and collagen fibers were also evident in Atg5-deficient non tumor mouse liver tissue (Figure 4E, panels b-d, stars & arrow heads). Aberrant multimembrane structures were also observed in Hep-Atg5 KO liver tumors as well as abnormal swollen mitochondria (Figure 4E, panels e-f, arrow and #). Taken together, these results indicate that liver injury and tumorigenesis in Hep-Atg5 KO mice is reversed by deletion of Nrf2. The cellular and molecular events for how impaired autophagy induces liver pathogenesis are summarized in Figure 4F.
Figure 4. Further deletion of Nrf2 suppresses hepatocyte-specific deletion of Atg5-induced-hepatocellular adenoma.
Representative gross anatomy of livers of Atg5 F/F, Alb Cre- and Atg5 F/F, Alb Cre+ mice aged at 12 and 15 months (A), and Atg5 F/F, Alb Cre+ and Atg5 F/F, Cre+/Nrf2−/− mice aged at 12 months (B). (C) Summary of tumor formation in Atg5 F/F, Alb Cre-, Atg5 F/F, Alb Cre+ and Atg5 F/F, Cre+/Nrf2−/− mice. (D) Histological analysis of mouse livers with tumors of Atg5 F/F, Alb Cre+ mice. Mice were sacrificed at 12 and 15 months and liver tissues were processed for H & E staining (20 ×). The right panels are enlarged micrographs from the squared areas. Arrows: ductal-like and inflammatory cells. Arrow heads: dilated sinusoids. *: focal necrosis. (E) Non-tumor and tumor liver tissues from 15 month Atg5 F/F, Alb Cre+ mice were processed for EM analysis. Panels a-d: non-tumor. Panel e: tumor. Panels b, d & f are enlarged micrographs from the boxed region in panel a, d & e respectively. LD: lipid droplets. N: nucleus. Arrows: aberrant multimembrane structures. Arrow heads: collagen fibers. #: damaged mitochondria. (F) A proposed model of the molecular events in autophagy-deficiency-induced liver pathogenesis. Atg5-deficient liver had increased accumulation of p62 resulting in persistent activation of Nrf-2. Atg5-deficient liver also had decreased FLIP expression and increased accumulation of ubiquitinated proteins, which could be further exacerbated by Nrf2 activation resulting in cell death followed by increased inflammation, fibrosis, compensatory proliferation and hepatocellular adenoma.
DISCUSSION
In the present study, we characterized the time-course of pathologic changes occurring in the livers of Hep-Atg5 KO mice. Our results show that loss of hepatocyte basal autophagy due to the deletion of Atg5 causes apoptosis, inflammation, fibrosis, and eventual hepatocellular adenoma. More importantly, we demonstrated that persistent activation of Nrf2 contributes to the pathogenesis of cell death, inflammation, fibrosis, and liver tumorigenesis.
It is likely that accumulation of hepatocyte p62, an autophagy substrate protein that is normally degraded by autophagy, could play a role to trigger cell death in autophagy-deficient hepatocytes. Indeed, one cell culture study showed that p62 promotes apoptosis by increasing caspase-8 activation through activating cullin3 (CUL3) E3 ligase, which polyubiquitinates caspase-8 [19]. However, several later reports suggested that the LC3 positive autophagosome membrane serves as a platform for caspase-8 aggregation and activation, which seems to be more important than accumulation of p62 [20–22]. The later notion is further supported by the observation that forced overexpression of p62 in Atg7/p62-DKO hepatocytes showed no cytotoxicity [15]. In agreement with these findings, our results also show that Atg5/Nrf2 DKO mice, which have higher p62 levels compared to WT mice, do not have liver injury. The relatively lower p62 levels in DKO mice compared to Hep-Atg5 KO mice could be due to the lack of feedback loop regulation of p62 mediated by Nrf2, which increases transcription of p62 as previous suggested [23].These observations suggest that activation of Nrf2 but not the accumulation of p62 is the detrimental factor in cell death in autophagy-deficient mouse livers. However, this conclusion seems to be paradoxical to the well-known role of Nrf2 in protecting against oxidative and electrophilic stress-induced tissue injury. It is worth noting that liver-specific Keap1 KO mice only develop mild liver abnormalities which are much lower in magnitude compared to liver-specific Atg7 or Atg5 KO mice [16], suggesting that liver injury in autophagy-deficient livers is not solely due to activation of Nrf2 alone. It is possible that the absence of autophagy may create primed conditions that turn Nrf2 from a protective factor into a detrimental one. Persistent activation of Nrf2 leads to the robust synthesis of proteins including detoxification cytochrome P450 enzymes and antioxidant proteins. Moreover, we found that mTOR, a key intracellular regulator for protein synthesis, was also elevated in Hep-Atg5 KO mouse livers. With the lack of autophagic degradation caused by deletion of Atg5, it is possible that newly synthesized proteins are not cleared efficiently and that accumulation of these proteins may cause proteotoxicity resulting in hepatocyte malfunction and eventual cell death. Besides these possibilities, it was reported that deletion of Atg7 in the mouse liver decreased FLIP expression, which promotes TNF-α-induced caspase-8 activation and apoptosis [24]. We found that Hep-Atg5 KO had decreased FLIP but increased Bax expression. The increased polyubiquitinated proteins (proteotoxicity) and decreased FLIP expression, together with increased TNFα expression, could contribute to cell death in Hep-Atg5 KO mouse livers. Interestingly, we also found increased Mcl-1 expression in Hep-Atg5 KO mice, which could be the secondary adaptive response to the cellular injury triggered by the proteotoxicity and decreased FLIP expression due to the chronic lack of autophagy in hepatocytes.
Fibrosis is the result of a wound-healing response following chronic liver injury [8, 9]. Our results show increased fibrosis in Atg5-deficient mouse livers, suggesting that basal hepatocyte autophagy may protect against fibrosis, which is in agreement with several other previous studies. For instance, increased collagen deposition and fibrosis is observed in the kidney of Beclin 1 heterozygous mice, suggesting autophagy may suppress fibrosis in the kidney [25]. Pharmacological inhibition of autophagy by bafilomycin A1 or genetic knockdown of Beclin 1 by siRNA leads to increased protein levels of collagen in TGF-β-treated primary cultured cells [26]. Interestingly, collagen is found in LC3 and LAMP1 positive vesicles, suggesting that collagen might be degraded via the autophagy pathway. Rapamycin activates autophagy and reduces bile duct ligation-induced fibrosis in rat liver[27]. Moreover, induction of autophagy by carbamazepine attenuates liver fibrosis in the mouse mutant alpha-1 antitrypsin-deficiency model [28]. Intriguingly, in contrast to the role of autophagy in hepatocytes, it has been shown that autophagy in HSC is required for its activation and hepatic fibrosis by degrading lipid droplets [10, 11]. Since HSC activation is an energy consuming process, it is suggested that lipophagy in HSC cells may provide a key energy source of free fatty acids from the breakdown of lipid droplets to fuel HSC activation [10, 11]. Our results show that autophagy function in HSC in Hep-Atg5 KO mice is not disrupted and is actually activated during the primary culture process. These findings generally support the previous notion that autophagy in HSC may favor hepatic fibrogenesis. Since the fibrogenic cells only account for only a small portion of the cells in the liver, it will be very challenging to develop drugs that would specifically target fibrogenic cells without affecting other cell types. However, based on this study together with other previous studies, global autophagy inducers may be an ideal treatment for improving hepatic functions and inhibiting fibrosis by preventing early hepatocyte cell death.
Autophagy acts as a tumor suppressor likely by selectively removing damaged proteins and organelles, especially damaged and senescent mitochondria, which are major cellular sources of ROS. Accumulation of p62 due to the deficiency of autophagy can lead to cellular metabolic stress and genome-instability which may promote tumorigenesis [29]. Indeed, deletion of p62 in liver-specific Atg7 KO mice reduced tumor size and numbers in liver-specific Atg7 KO mice [2]. However, the incomplete suppression of liver tumorigenesis in liver-specific Atg7 KO mice by further deletion of p62 suggests that other factor(s) may be critical in promoting tumor progression in addition to p62 [2]. As discussed above, accumulation of p62 can lead to Nrf2 activation by competing with Keap1 binding with Nrf2, and increasing evidence has indicated that activated Nrf2 is associated with tumorigenesis [30, 31]. The protein level of p62 and the activation of Nrf2 are closely associated with human HCC development [17]. In the present study, our results provide convincing genetic evidence that persistent activation of Nrf2 plays a critical role in promoting tumorigenesis in autophagy-deficient livers. At first glance, these results seem to be paradoxical to the known functions of Nrf2, which plays a critical role in regulating induction of cellular detoxification enzymes and antioxidant genes against various cellular stresses. However, while on the one hand ROS can promote tumorigenesis by inducing DNA damage, activating inflammatory pathways and stabilizing hypoxia-inducible factor transcription factors; on the other hand, ROS per se can also be harmful to cancer cells by inducing cell death. Up-regulation of Nrf2 target genes, including heme oxygenase-1, peroxiredoxin and NQO-1, has been found in many cancers and may contribute to chemo resistance and tumor progression [32]. Therefore, targeting activation of the p62-Keap1-Nrf2 pathway may be a novel approach for chemoprevention.
In conclusion, our results suggest that impaired hepatocyte autophagy can lead to cell death resulting in inflammation, fibrosis and tumorigenesis in the liver, which is mediated by the persistent activation of Nrf2. These findings provide a molecular basis to explain how impaired autophagy promotes tumorigenesis. These findings imply that induction of hepatic autophagy could be a novel therapeutic approach to mitigate liver fibrosis and liver tumorigenesis.
Supplementary Material
ACKNOWLDGEMENTS
We thank Ms. Barbara Fegley (KUMC Electron Microscopy Research Laboratory) for her excellent technical assistance. We thank Drs. Noboru Mizushima (University of Tokyo) and Curtis Klaassen (University of Kansas Medical Center) for providing Atg5 F/F and Nrf2−/− mice. This study was supported in part by the National Institute of Health (NIH) funds R01 AA020518 (W.X.D) and AA12916 and DK070195 (to H.J.) and by grants from the National Center for Research Resources (5P20RR021940-07) and the National Institute of General Medical Sciences (8 P20 GM103549-07). B.C is supported by DK073566 and JL by R01 ES017537.
List of Abbreviations
- Alb
Albumin
- ALT
Alanine aminotransferase
- α-SMA
α-smooth muscle actin
- CTGF
Connective tissue growth factor
- CK 19
cytokeratin 19
- ECM
Extracellular matrix
- EM
Electron microscopy
- FLIP
FLICE-like protein (FLIP) protein
- GCLC
glutamate-cysteine ligase catalytic subunit
- GCLM
glutamate-cysteine ligase modifier subunit
- HCC
Hepatocellular carcinoma
- HSC
Hepatic stellate cells
- ICAM-1
Intercellular adhesion molecule 1
- IL-6
interleukin 6
- KC
C-X-C motif ligand 1
- Keap1
Kelch-like ECH-associated protein 1
- KO
Knockout
- LC3
Microtubule light chain 3
- LD
Lipid droplets
- Ly6B
lymphocyte antigen B superfamily
- MIP1α
Macrophage inflammatory protein 1α
- NQO1
NAD(P)H quinone oxidoreductase
- Nrf2
Nuclear factor (erythroid-derived 2)-like 2
- PCNA
Proliferating cell nuclear antigen
- ROS
Reactive oxygen species
- TGF-β1
transforming growth factor beta 1
- TNF-α
Tumor necrosis factor alpha
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors have declared that no conflict of interest exists.
REFERENCES
- 1.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvationinduced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 7.Zhang DY, Friedman SL. Fibrosis-dependent mechanisms of hepatocarcinogenesis. Hepatology. 2012;56:769–775. doi: 10.1002/hep.25670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425–456. doi: 10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- 9.Brenner DA. Molecular pathogenesis of liver fibrosis. Trans Am Clin Climatol Assoc. 2009;120:361–368. [PMC free article] [PubMed] [Google Scholar]
- 10.Thoen LF, Guimaraes EL, Dolle L, Mannaerts I, Najimi M, Sokal E, et al. A role for autophagy during hepatic stellate cell activation. J Hepatol. 2011;55:1353–1360. doi: 10.1016/j.jhep.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 13.Ni HM, Bockus A, Boggess N, Jaeschke H, Ding WX. Activation of autophagy protects against acetaminophen-induced hepatotoxicity. Hepatology. 2011;55:222–232. doi: 10.1002/hep.24690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A. 1996;93:13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 16.Taguchi K, Fujikawa N, Komatsu M, Ishii T, Unno M, Akaike T, et al. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc Natl Acad Sci U S A. 2012;109:13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ni HM, Boggess N, McGill MR, Lebofsky M, Borude P, Apte U, et al. Liver-specific loss of Atg5 causes persistent activation of Nrf2 and protects against acetaminophen-induced liver injury. Toxicol Sci. 2012;127:438–450. doi: 10.1093/toxsci/kfs133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 20.Pan JA, Ullman E, Dou Z, Zong WX. Inhibition of protein degradation induces apoptosis through a microtubule-associated protein 1 light chain 3-mediated activation of caspase-8 at intracellular membranes. Mol Cell Biol. 2011;31:3158–3170. doi: 10.1128/MCB.05460-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ullman E, Pan JA, Zong WX. Squamous cell carcinoma antigen 1 promotes caspase-8-mediated apoptosis in response to endoplasmic reticulum stress while inhibiting necrosis induced by lysosomal injury. Mol Cell Biol. 2011;31:2902–2919. doi: 10.1128/MCB.05452-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young MM, Takahashi Y, Khan O, Park S, Hori T, Yun J, et al. Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J Biol Chem. 2012;287:12455–12468. doi: 10.1074/jbc.M111.309104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujita K, Maeda D, Xiao Q, Srinivasula SM. Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc Natl Acad Sci U S A. 2011;108:1427–1432. doi: 10.1073/pnas.1014156108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amir M, Zhao E, Fontana L, Rosenberg H, Tanaka K, Gao G, et al. Inhibition of hepatocyte autophagy increases tumor necrosis factor-dependent liver injury by promoting caspase-8 activation. Cell Death Differ. 2013;20:878–887. doi: 10.1038/cdd.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SI, Na HJ, Ding Y, Wang Z, Lee SJ, Choi ME. Autophagy promotes intracellular degradation of type I collagen induced by Transforming Growth Factor (TGF)-beta1. J Biol Chem. 2012 doi: 10.1074/jbc.M111.308460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SI, Na HJ, Ding Y, Wang Z, Lee SJ, Choi ME. Autophagy promotes intracellular degradation of type I collagen induced by transforming growth factor (TGF)-beta1. J Biol Chem. 2012;287:11677–11688. doi: 10.1074/jbc.M111.308460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bridle KR, Popa C, Morgan ML, Sobbe AL, Clouston AD, Fletcher LM, et al. Rapamycin inhibits hepatic fibrosis in rats by attenuating multiple profibrogenic pathways. Liver Transpl. 2009;15:1315–1324. doi: 10.1002/lt.21804. [DOI] [PubMed] [Google Scholar]
- 28.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 29.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 31.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008;58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.