Analyses of toxin binding to a homology model of Nav1.4 indicate similar folding of the outer pore region in eukaryotic and prokaryotic sodium channels.
Abstract
Voltage-gated sodium channels are targets for many drugs and toxins. However, the rational design of medically relevant channel modulators is hampered by the lack of x-ray structures of eukaryotic channels. Here, we used a homology model based on the x-ray structure of the NavAb prokaryotic sodium channel together with published experimental data to analyze interactions of the μ-conotoxins GIIIA, PIIIA, and KIIIA with the Nav1.4 eukaryotic channel. Using Monte Carlo energy minimizations and published experimentally defined pairwise contacts as distance constraints, we developed a model in which specific contacts between GIIIA and Nav1.4 were readily reproduced without deformation of the channel or toxin backbones. Computed energies of specific interactions between individual residues of GIIIA and the channel correlated with experimental estimates. The predicted complexes of PIIIA and KIIIA with Nav1.4 are consistent with a large body of experimental data. In particular, a model of Nav1.4 interactions with KIIIA and tetrodotoxin (TTX) indicated that TTX can pass between Nav1.4 and channel-bound KIIIA to reach its binding site at the selectivity filter. Our models also allowed us to explain experimental data that currently lack structural interpretations. For instance, consistent with the incomplete block observed with KIIIA and some GIIIA and PIIIA mutants, our computations predict an uninterrupted pathway for sodium ions between the extracellular space and the selectivity filter if at least one of the four outer carboxylates is not bound to the toxin. We found a good correlation between computational and experimental data on complete and incomplete channel block by native and mutant toxins. Thus, our study suggests similar folding of the outer pore region in eukaryotic and prokaryotic sodium channels.
INTRODUCTION
Sodium channels, which play key roles in physiology of excitable cells, have been a subject of experimental studies during the last decades (Trimmer et al., 1989; Priestley, 2004; Wood et al., 2004; Rogers et al., 2006). The pore-forming α subunit of voltage-gated sodium channels (Nav1) is composed of four homologous repeats (I–IV), which are quasi-symmetrically arranged around the central pore. Each repeat contains six transmembrane segments (S1–S6), a membrane-diving P-loop (P), and the intracellular N and C termini. P-loops and extracellular thirds of transmembrane S5 and S6 helices contribute to the outer pore region, whereas cytoplasmic two-thirds of helices S5 and S6 contribute to the inner pore region.
Actions of naturally occurring toxins and synthetics drugs on Nav1 channels have been intensively studied. These include the hallmark sodium channel blockers tetrodotoxin (TTX), saxitoxin, and μ-conotoxins; sodium channel agonists batrachotoxin, veratridine, aconitine, and grayanotoxin; local anesthetics; and many other ligands (French et al., 1984; Moczydlowski et al., 1984; Linford et al., 1998; French and Terlau, 2004; Eijkelkamp et al., 2012; Waszkielewicz et al., 2013). However, in the absence of x-ray structures of eukaryotic sodium channels, understanding of atomistic mechanisms of action is still incomplete for some classes of ligands. X-ray structures of potassium channels in the closed and open states (Doyle et al., 1998; Li et al., 1998; Jiang et al., 2003; Long et al., 2005) have been used to build homology models of sodium channels, which provide structural explanation for numerous experimental data (Lipkind and Fozzard, 2000; Tikhonov and Zhorov, 2005; Choudhary et al., 2007; Tikhonov et al., 2014). However, potassium channels are poor templates for homology modeling of the outer pore region, which includes the selectivity filter. In potassium channels, the TVGYG motif downstream from the P-loop turn forms a narrow tunnel where five rings of backbone carbonyls catalyze the selective ion permeation. In sodium channels, the selectivity is provided by side chains of the D, E, K, and A residues from four P-loops (the DEKA ring), and the selectivity-filter region is wide enough to accommodate blockers as large as TTX. Therefore, attempts to rationalize the data on ligand action in the outer pore necessarily involved speculations about folding of the outer pore region (Lipkind and Fozzard, 2000; Tikhonov and Zhorov, 2005; Choudhary et al., 2007).
The x-ray structures of NavAb and other prokaryotic sodium channels have provided a new and possibly more reliable structural basis for understanding ligand action in the outer pore region (Payandeh et al., 2011; McCusker et al., 2012; Zhang et al., 2012). Selectivity in these channels is provided by a single EEEE ring. Unlike potassium channels, each subunit of a prokaryotic sodium channel has two helices before (P1) and after (P2) the selectivity filter glutamate. The P2 helices line the outer pore and form relatively wide extracellular vestibule. Action of TTX has been recently modeled in view of the NavAb folding (Tikhonov and Zhorov, 2012). However, TTX interacts only with a local region between the DEKA ring and the more extracellular ring of outer carboxylates. It remains unclear whether or not available structures of prokaryotic sodium channels can be used to rationalize action of much bigger toxins that target the outer pore.
Particularly interesting is the action of μ-conotoxins (Olivera and Cruz, 2001; Lewis et al., 2012). Conotoxins are biologically active peptides found in the venom of cone snails. μ-Conotoxins are rather small peptides (16–26 amino acids) with several disulfide cross-links that stabilize a typical folding pattern called framework III (Lewis et al., 2012). μ-Conotoxins do not target precisely the same binding area as TTX and saxitoxin (Zhang et al., 2010) and do not strongly interact with the selectivity filter, but interact with outer carboxylates and several residues downstream. The family of μ-conotoxins includes three toxins, which are particularly well studied: GIIIA (Cruz et al., 1989), PIIIA (Shon et al., 1998), and KIIIA (Zhang et al., 2007). GIIIA, the best-studied μ-conotoxin, binds with a high affinity to the Nav1.4 sodium channel (Sato et al., 1991; Becker et al., 1992; Dudley et al., 1995, 2000; French et al., 1996; Chang et al., 1998; Li et al., 2001, 2002; Cummins et al., 2002; Hui et al., 2002, 2003; Xue et al., 2003; Choudhary et al., 2007). PIIIA is a close homologue of GIIIA, but with some distinguishing features, which make it an interesting object for studying (Safo et al., 2000; McArthur et al., 2011a,c; Chen and Chung, 2012; Tietze et al., 2012). KIIIA, a short peptide of 16 amino acids (Zhang et al., 2007), is of special interest because of its small size and ability to bind to the channel simultaneously with TTX (Zhang et al., 2009; Van Der Haegen et al., 2011; Wilson et al., 2011; Khoo et al., 2012; Stevens et al., 2012).
Previous models of μ-conotoxin binding (Choudhary et al., 2007; McArthur et al., 2011b,c) were elaborated to rationalize solid experimental data using KcsA-based homology models. The fact that the outer pore regions in KcsA and NavAb have different folding calls for reconsidering structural aspects of μ-conotoxin–channel interactions. Here we used our NavAb-based model of Nav1.4 (Tikhonov and Zhorov, 2012) to dock μ-conotoxins GIIIA, PIIIA, and KIIIA in the outer pore region. The predicted complexes of Nav1.4 with μ-conotoxins are in agreement with a large body of experimental data. Furthermore, they allowed us to rationalize previously unexplained experimental data on residual currents in μ-conotoxin–bound channels.
MATERIALS AND METHODS
All calculations were performed by using the ZMM program (ZMM Software, Inc.). Nonbonded interactions were calculated with the AMBER force field (Weiner et al., 1984, 1986) and a cutoff distance of 8 Å. Electrostatic interactions were calculated by using the distance- and environment-dependent dielectric function (Garden and Zhorov, 2010). No specific energy terms were used for cation-π interactions, which were accounted for with partial negative charges at the aromatic carbons (Bruhova et al., 2008). Bond lengths and bond angles were kept rigid during the calculations.
The model of rat Nav1.4 was taken from our previous study (Tikhonov and Zhorov, 2012). The sequence alignment is shown in Fig. 1. Initial conformations of toxins were taken from the Protein Data Bank (PDB IDs are 1TCJ for GIIIA, 1R9I for PIIIA, and 2LXG for KIIIA). Voltage-sensing domains were not modeled because they are far from the binding site of μ-conotoxins. Big extracellular loops were truncated in our model because no experimental data on their folding, as well as interaction with μ-conotoxins, is available.
Figure 1.
Sequence alignment of P-loops in NavAb and Nav1.4 and sequences of μ-conotoxins GIIIA, PIIIA, and KIIIA. In the P-loops the selectivity filter residues and outer carboxylates are shown in bold. The polar residues, which stabilize relative folding of P1 and P2 helices in the NavAb structure (underlined), are in matching positions in Nav1.4. In the toxins, critical basic residues in matching positions are shown in bold.
The Monte Carlo minimization (MCM) method (Li and Scheraga, 1987) was used to optimize the models. All side chain torsions were randomly sampled during the MCM trajectories. Both side chains and backbones were flexible during energy minimizations. MCM of each model was performed until 6,000 consecutive energy minimizations did not decrease the energy of the apparent global minimum. To incorporate experimental data in our calculations we used a system of constraints. A distance constraint is a flat-bottom energy function that allows a particular atom–atom distance to deviate energy-free between lower and higher limits and imposes a parabolic energy penalty if the distance is beyond the limits. Two types of constraints were used. To maintain the template folding we used “pin” constraints. A pin allows an α carbon of an amino acid residue to deviate up to 1 Å from the respective template position without a penalty and imposes a parabolic energy function to penalize larger deviations. Pins were applied to all residues in the P1 and P2 helices, but not to the selectivity filter region (residues 176–181 in NavAb). Pins are not suitable for maintaining the folding of conotoxins because the latter were mobile in the docking procedure. Instead we applied atom–atom distance constraints to keep distances between α carbons of a toxin close to values obtained in the respective nuclear magnetic resonance study. To bias experimentally identified toxin–channel contacts, distance constraints between centers of charged groups in the contacting amino acids were used. For all constraints, the energy penalty was calculated using the force constant of 10 kcal/mol/Å2. To minimize possible influence of constraints on the docking results, each optimization was performed in two stages. After MCM with constraints (the first stage), the model was refined by the second MCM, in which all constraints except pins were removed (the second stage).
Online supplemental material
Figs. S1, S2, and S3 show details of GIIIA docking procedure. Tables S1, S2, and S3 show residue–residue interaction energies for Nav1.4 complexes with GIIIA, PIIIA, and KIIIA, respectively. Figs. S4, S7, and S8 show main contacts for these complexes. Fig. S5 shows interactions that impose orientation of GIIIA in the model. Fig. S6 and Video 1 demonstrate switching of contacts between GIIIA and the channel because of the flexibility of long side chains of charged residues. Fig. S9 shows distributions of favorable ion positions in the model with wild-type and mutant toxins. Online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201411226/DC1.
RESULTS
Selection of the model
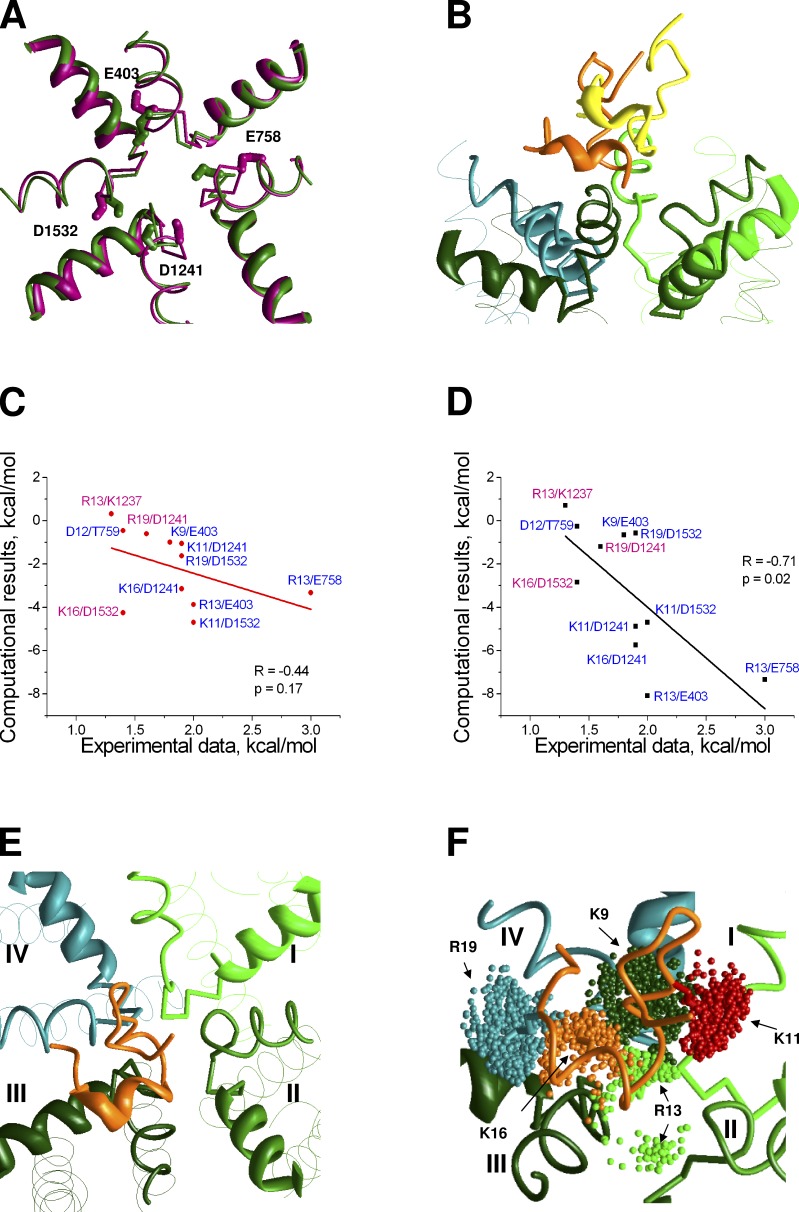
Available x-ray structures of prokaryotic sodium channels (NavAb, NavMs, and NavRh) are very similar in the outer pore region. Particularly the P1 and P2 helices are very close at the superimposed x-ray structures (Fig. S1). Therefore, selection of a particular template is not essential for the modeling. In contrast, the sequence alignment between prokaryotic and eukaryotic channels is critical, as only one position shift in the alignments results in ∼100° reorientation of a residue in the α helix. The sequence similarity between prokaryotic and eukaryotic sodium channels is rather small. For example, the sequence identity between P-loops of NavAb and repeat I of Nav1.4 is only ∼21%. Recently, we demonstrated that a straightforward sequence alignment (without insertions/deletions) does not allow us to build a homology model consistent with the experimental data on TTX binding (Tikhonov and Zhorov, 2012). We proposed an adjusted alignment with insertions/deletions in the selectivity filter region (Fig. 1). The adjusted alignment allowed us to build a 3D model in which TTX-sensing residues are oriented inside the pore, whereas long polar residues before and after the selectivity filter form interhelical H bonds that stabilize the structure. The orientations of the outer carboxylates in the models with the straightforward and adjusted sequence alignments are significantly different (Fig. 2 A).
Figure 2.
GIIIA in Nav1.4 models. (A) Different orientation of the outer carboxylates (sticks) in the NavAb-based models with the straightforward (magenta) and adjusted (green) sequence alignments. P1 helices are shown as ribbons. Selectivity-filter region and P2 helices are shown as thin rods. (B) Lowest-energy GIIIA-binding modes obtained in the models based on the straightforward (yellow) and adjusted (orange) alignments. (C and D) Correlation between energies of pairwise interactions in experiments (Table 1) and the models, which are based on the straightforward (C) and adjusted (D) sequence alignments. Contacts, which were used in docking procedure, have blue labels. Other contacts have magenta labels. (E) Asymmetric binding of GIIIA. The toxin body (orange) is shifted toward repeat III. (F) Ensemble of predicted orientations of side chains of key basic residues of GIIIA. Dark green, red, light green, orange, and cyan dots show the centers of the charged moieties in K9, K11, R13, K16, and R19, respectively.
We compared the abilities of the NavAb-based models of Nav1.4 with the straightforward and adjusted sequence alignments to reproduce available data on GIIIA binding. Particularly valuable are results of the mutant cycle analysis, which revealed pairwise interactions between the toxin and the channel residues (Table 1). Here we used experimental data (Choudhary et al., 2007) to define a set of distance constraints, which would impose H bonds or salt bridges between side chains of respective residues in the channel and toxins. In some cases, unambiguous definition of specific pairwise channel–toxin contacts was impossible. For example, according to experimental data, outer carboxylates of repeats III and IV of the channel are involved in contacts with three basic residues of the toxin (Choudhary et al., 2007). Therefore, we have chosen only the strongest pairwise contacts for these residues (Table 1).
Table 1.
Experimental data (Choudhary et al., 2007) used to dock GIIIA
| Number | Toxin residue | Channel residue | Strength of interactiona | Coupling energy |
| kcal/mol | ||||
| 1 | K9 | E403 | medium | 1.8 |
| 2 | K11 | D1241 | strong | 1.9 |
| 3 | K11 | D1532 | strong | 2 |
| 4 | D12 | T759 | medium | 1.4 |
| 5 | R13 | E403 | strong | 2 |
| 6 | R13 | E758 | strong | 3 |
| 7 | R13 | K1237 | medium | 1.3 |
| 8 | K16 | D1241 | strong | 1.9 |
| 9 | K16 | D1532 | medium | 1.4 |
| 11 | R19 | D1241 | medium | 1.6 |
| 12 | R19 | D1532 | strong | 1.9 |
Contacts, which were used to impose constraints in the docking procedure, are underlined.
According to Choudhary et al. (2007).
We placed GIIIA above the channel manually in such a way that the toxin mass center was at the pore axis and the toxin face, which contains basic residues (K9, K11, R13, K16, and R19), was oriented toward the channel. The distance between the plane of α carbons of the outer carboxylate ring and the toxin backbone atom, which is closest to the ring, was 11 Å. At this distance, toxin did not interact with the channel. Using this general orientation, we generated 36 starting toxin–channel mutual dispositions by consecutively turning the toxin around the pore axis. From each starting point, the energy was Monte Carlo–minimized with the toxin–channel distance constrains. A representative MCM trajectory is shown in Fig. S2. Next, each model was refined by an unconstrained MCM. We obtained generally the same orientation of the toxin for every starting point in models with both the straightforward and adjusted alignments. Fig. S3 shows the superimposition of all the models obtained from different GIIIA orientations for the model with the adjusted alignment. This result indicates that the system of toxin–channel distance constraints unambiguously determines orientation of the toxin.
In both models, the large toxin body did not fit into the selectivity filter. Rather it bound above the narrowing vestibule. Side chains of GIIIA basic residues reached the outer carboxylates, which are located at the extracellular entrance to the vestibule. Importantly, the toxin mass center shifted from the pore axis toward repeat III (Fig. 2 E). The C-end helix of the toxin strongly interacted with the P2 helix of repeat III. The N-end of the toxin projected to the extracellular space. The middle part, which contains arginine R13, occurred within the outer pore, near repeat II.
Despite this general similarity, results of docking showed significant advantage of the model with the adjusted alignment. For this model, the root-mean-square deviation (RMSD) of α carbons in the toxin-binding region from the NavAb structure was 1.39 Å. During the refining unconstrained MCM, the toxin slightly moved (the RMSD of α carbons between the constrained and unconstrained models was as small as 0.5 Å). In contrast, the experimental distance constraints caused significant deformation of the model, which is based on the straightforward sequence alignment. Indeed, the RMSD for α carbons in the toxin-binding region between the model and the NavAb structure was as large as 2.04 Å. The refining unconstrained MCM shifted the toxin ∼4.5 Å away from the channel (Fig. 2 B). The toxin–channel interaction energy in the straightforward alignment model was 33% higher (less preferable) than in the adjusted alignment model.
We calculated energy of interactions between individual residues of the toxin and the channel in the two refined models (Fig. 2, C and D). The toxin–channel pairwise interactions were plotted against the experimental free energies of respective contacts (Table 1). Note that not all of the experimental contacts were used as constraints during the docking procedure. For the straightforward alignment model, the correlation is weak (Pearson correlation coefficient P = −0.44 and significance of correlation P = 0.17). Contrarily, for the adjusted alignment model, P = −0.71 and the correlation appeared statistically significant (P = 0.02).
These results clearly show that the adjusted alignment model fits experimental data much better than the straightforward alignment model. Therefore, all further analysis was performed with the adjusted alignment model.
Binding of GIIIA
Although our model readily reproduced many experimentally found specific toxin–channel interactions (Table S1 and Fig. S4), the contact between hydroxyproline hP17 and M1240 (Dudley et al., 2000) was not observed. hP17 leaned toward repeat III P2-helix (DIIIP2), where M1240 is located, but no specific interactions between the two residues were found. The hydroxyl group of hP17 occurred in a close proximity to D1248 at the C-end of DIIIP2. We imposed an additional distance constraint to bias an H bond between the hydroxyl group of hP17 and carboxyl oxygen of D1248 and obtained a model (Fig. S5, top) in which the contacts obtained in the previous model were preserved. Thus, our model provides a structural explanation for the role of hP17 in the toxin action. Importantly, this new contact, which is predicted by our calculations, is not yet reported in experimental studies. D1248 does not have any matching residues in homologous positions of other repeats, and thus the new toxin–channel contact may be an important determinant of the toxin–channel mutual disposition.
Substitutions of D762 and E765 in DIIP2 affect GIIIA binding (Xue et al., 2003). In our model these residues are located far from GIIIA and cannot directly interact with it. However, according to the NavAb crystal structure, D762 stabilizes the outer pore by making a salt bridge with R395. E765 is situated on the same side of the P2-helix as D762, thus facing toward the P1-helix of repeat I. According to our model, both D762 and E765 make specific contacts with R395, contributing to stabilization of the outer pore folding (Fig. S5, bottom). We suggest that alanine substitutions of D762 and E765 (Xue et al., 2003) affect the toxin binding allosterically rather than directly.
In our model the body of GIIIA, which contains the α helix, interacts mainly with the P2 helix of repeat III. Interestingly, repeat III aspartate D1248, which is highly conserved in sodium channels, is an attractive target for electrostatic interaction with the toxin. Other repeats of the channel have either neutral or positively charged residues in the matching positions of the sequence alignment (Fig. 1). Another possible cause for the asymmetric binding of the toxin is the asymmetric ring of the outer carboxylates. Although the outer carboxylates in repeats I, II, and IV are in the matching positions of the alignment, repeat III D1241 is shifted one position toward the C-end.
The calculated energies of pairwise interactions correlate reasonably well with the experimentally determined values. However, rather poor correlation was found for some medium-strength contacts between the outer carboxylates in repeats III and IV and toxin basic residues (Fig. 2 D). A possible reason is that we considered only a single-complex structure and thus our calculations ignored the entropy. Because of the large conformational flexibility of long side chains in the toxin basic residues and the channel acidic residues, the experimentally determined pairwise energies likely integrate contributions from many micro-states of the channel–toxin complex. To explore this possibility, we performed additional calculations. We used the above described model to generate 20,000 starting points with random conformations of side chains of GIIIA basic residues. From each starting points the energy was Monte Carlo–minimized with the backbone geometry preserved by the system of distance constrains between α carbons of the toxin (see Materials and methods). The results are shown in Fig. 2 F. In the ensemble of lowest-energy complexes, the charged terminal groups in flexible side chains of basic residues fluctuated up to 5 Å. This flexibility may switch some contacts (Fig. S6 and Video 1). Key basic residues of the toxin are K11, R13, K16, and R19. According to our results, K11 mostly interacted with the channel repeats I, III, and IV, but not with repeat II because of the presence of R13 in close proximity of repeat II. R13 interacted mostly with repeats I and II, but can interact with any repeat because of its long side chain. K16 in the middle of the helical part of the toxin can interact with any channel repeat, depending on contacts of other toxin residues. R19 can only reach repeats III and IV because it is situated close to the C-end of the toxin, downstream from the helical part, and cannot approach closer repeats I and II. Thus, our model predicts that within the same binding mode and backbone geometry of the toxin, its basic residues can form different contacts with acidic residues of the channel.
Binding of PIIIA
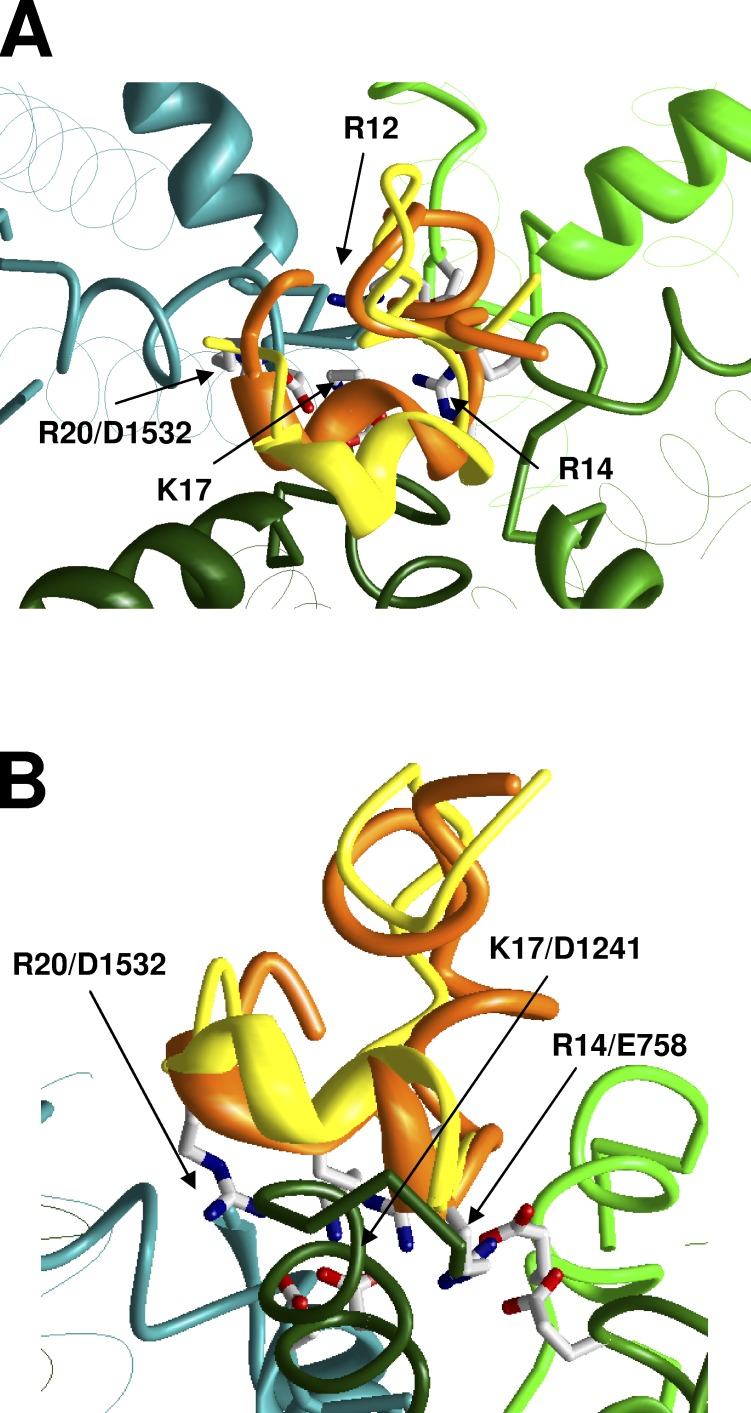
Residues R12, R14, K17, and R20 are important for PIIIA–channel interactions (McArthur et al., 2011c). However, pairwise contacts between PIIIA and Nav1.4 are not known. Therefore, the docking methodology, which we used for GIIIA, is not applicable for PIIIA. Because PIIIA folds like GIIIA, we suggested that in the complex with the channel, the α helix and key basic residues of PIIIA are oriented like those of GIIIA and generated respective starting point for our calculations. The Monte Carlo–minimized binding mode is shown in Fig. 3. In our model, the key PIIIA residues are involved in the following pairwise interactions: R12/E403, R14/E758, K17/D1241, K17/D1532, and R20/D1532 (Fig. S7 and Table S2).
Figure 3.
Predicted complex of PIIIA (orange backbone) with Nav1.4. (A and B) Side and top views. For comparison, GIIIA (yellow backbones) is shown in the same binding mode as in Fig. 2. Superposition of the toxins indicates similarity of their binding models.
Although the overall binding modes for GIIIA and PIIIA are rather similar (RMSD for α carbons of key basic residues is 1.8 Å), there are certain differences in pairwise toxin–channel interactions. For example, specific interaction between D12 and T759 (Choudhary et al., 2007) significantly contributed to the stability of the GIIIA–channel complex in our model. PIIIA lacks an acidic residue in the homologous position. As a result, in our models, specific interactions of arginine R12 in PIIIA differ from interactions of the homologous lysine K11 in GIIIA. R12 in PIIIA interacted strongly with repeat I, whereas K11 in GIIIA interacted with repeats III and IV because contacts of K11 with repeat I were sterically precluded by D12. R14 formed tight contacts with repeats I and II, but weakly interacted with repeat III, and even weaker with repeat IV. K17 interacted with repeat III and IV like its analogue in GIIIA, K16. Finally, R20 (the analogue of R19 in GIIIA) interacted only with repeat IV. Like in the case of GIIIA–channel interactions, the shown macrostate may represent many microstates that exist as a result of the possibility of switching contacts between long flexible side chains.
McArthur et al. (2011c) explored the voltage dependence of action of PIIIA and its mutants and determined the depths (δ) of residues in the membrane electric field. The δ values rank as follows: R14 > K17 > R20 > R12 > S13 > R2 > G6. In our model, the relative depth of the charged residues (determined by positions of their basic atoms) is ranked as follows: R14 > K17 > R20 > R12 > R2. S13 is located approximately at the same depth as R12, whereas G6 and R2 are located at the extracellularly projected N-end of the toxin. Thus, the toxin-binding orientation in our model is consistent with experimental data. It should be noted that no data on voltage dependence were used to obtain the toxin orientation.
Binding of KIIIA
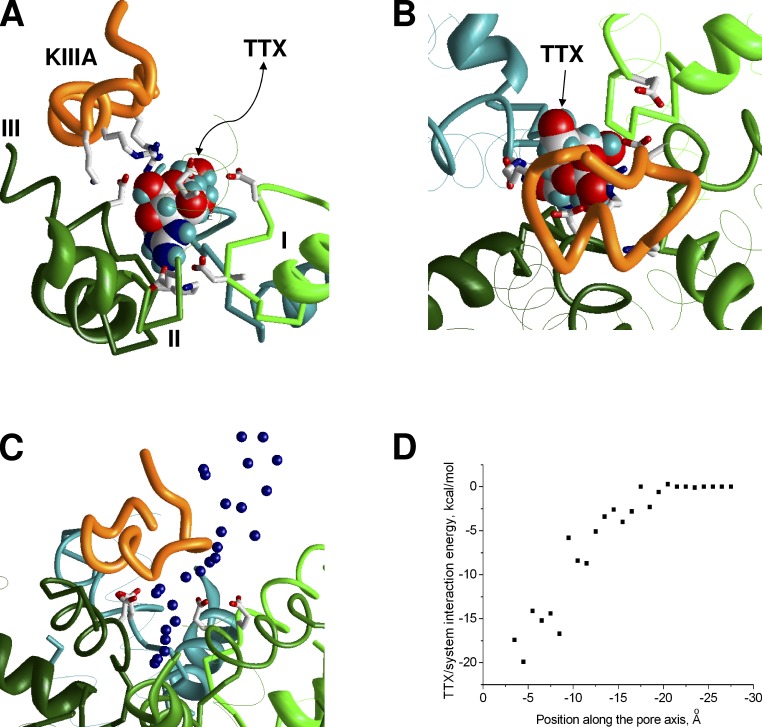
KIIIA, the smallest μ-conotoxin, lacks an analogue of the N-terminal segment, which is present in GIIIA and PIIIA, and contains only three basic residues in the α helix (Fig. 1). The data on pairwise interactions (McArthur et al., 2011b) suggest that all of these residues interact with repeat III outer carboxylate (D1241). We used these data to impose distance constraints, which clamped H bonds between the toxin basic side chains and the outer carboxylate of repeat III. MCM yielded a model (Fig. 4) in which the position and orientation of the KIIIA α helix are similar to those of PIIIA and GIIIA. The entire set of contacts of basic residues in KIIIA is shown in Fig. S8 and Table S3. Importantly, because of its small size and the lack of N-terminal segment, which would be analogous to N-terminal segments in GIIIA and PIIIA, KIIIA did not cover the pore with its body and weakly interacted with repeat I.
Figure 4.
A ternary complex of Nav1.4 with KIIIA and TTX. (A and B) Side and top views of the complex. TTX (space filled)-binding mode is taken from a previous study (Tikhonov and Zhorov, 2012). KIIIA is shifted toward repeat III, leaving the space for TTX to access its binding site in the narrow part of the outer pore. (C) MCM trajectory of forced TTX egress from its binding site in the presence of KIIIA. Blue dots show the central nitrogen atom of the TTX charged group. (D) TTX system energy along the trajectory (C). The smooth energy profile indicates absence of steric hindrances.
Intriguingly, TTX can reach its binding site deeply in the outer pore in the presence of KIIIA, although with a slower kinetics than in the absence of KIIIA (Zhang et al., 2009). To rationalize these data, we have built a ternary complex model of the channel with KIIIA and TTX. We used the TTX-binding mode from Tikhonov and Zhorov (2012) and KIIIA-binding mode described above. The ternary complex model (Fig. 4, A and B) did not reveal toxin–channel or toxin–toxin sterical clashes. The compact TTX molecule is bound deeply in the narrow portion of the pore between the DEKA ring and the ring of outer carboxylates, whereas the larger KIIIA toxin did not penetrate deep into the pore, approached the outer carboxylates from the extracellular side, and formed salt bridges with them. Each carboxylate can accept several H bonds so that simultaneous contacts with TTX and KIIIA were easily formed. The repeat III outer carboxylate, which unlike other outer carboxylates faces away from the pore axis, did not interact with TTX, but formed a salt bridge with R10 of KIIIA.
To find a possible passage for TTX between KIIIA and the channel, we pulled TTX out from the ternary complex model in the extracellular direction. To do this we imposed a distance constraint between the central carbon atom in the TTX guanidinium group and a plane drawn through four α carbons of the DEKA ring. Next we systematically increased the minimal value of the distance constraint with the step 1 Å and Monte Carlo–minimized energy in each step. The lowest-energy structure found in a given step was used as the starting point for the next step. During the calculations, the binding mode of KIIIA was preserved by distance constraints, which clamped contacts of its basic residues with the outer carboxylates. The TTX egress trajectory is rather smooth (Fig. 4 C). The corresponding energy plot (Fig. 4 D) does not have energy barriers, indicating the absence of steric obstacles in the predicted pathway for the TTX egress.
Mechanisms of incomplete block
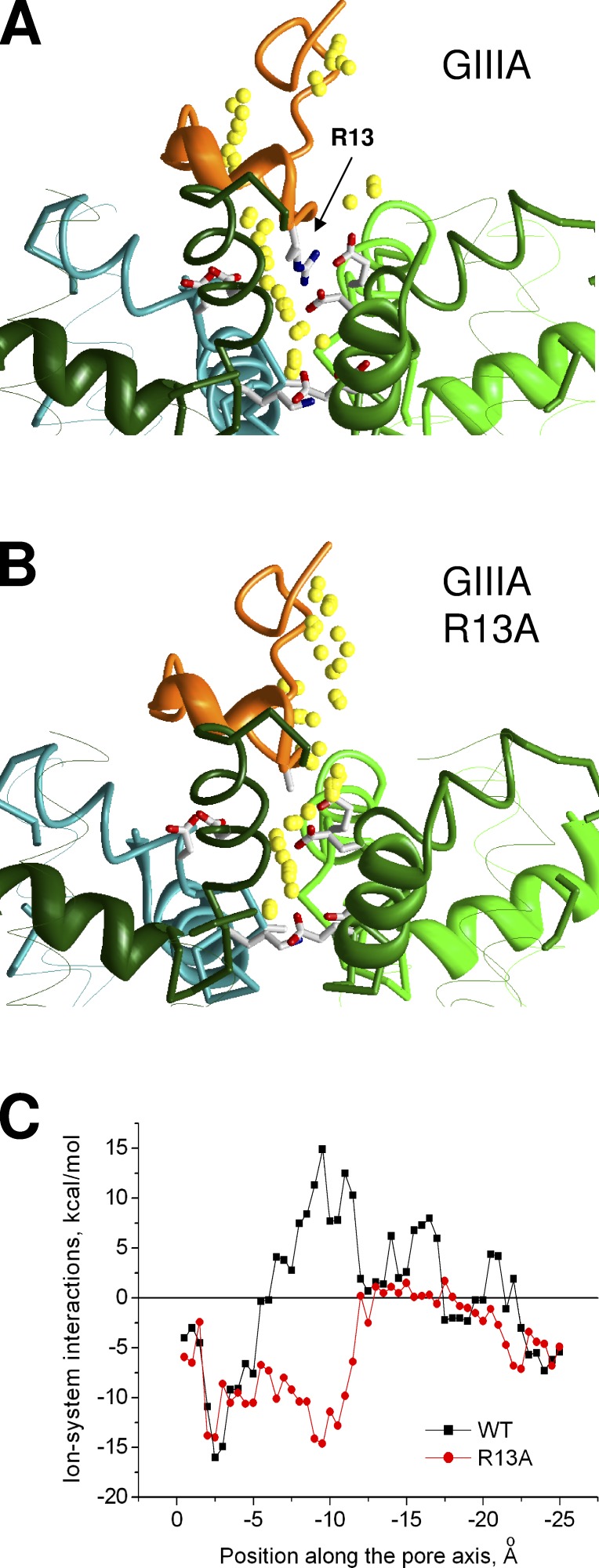
Experimental studies revealed incomplete block of Nav1 channels by KIIIA and some GIIIA and PIIIA mutants (Hui et al., 2002; McArthur et al., 2011a; Wilson et al., 2011), but structural rationale for these data is lacking. To address the problem, we used the approach that was used to predict the TTX egress pathway. A sodium ion was placed in the DEKA ring, and the egress pathway in the presence of GIIIA was calculated. The ion trajectory is interrupted near the toxin (Fig. 5 A). The energy profile contains zones of negative values, where the ion experiences favorable electrostatic interactions with the DEKA ring and the outer carboxylates, and the zone of positive values, which corresponds to repulsive interactions with the toxin (Fig. 5 C).
Figure 5.
MCM trajectory for a sodium ion (small yellow spheres) pulled from the selectivity filter in the extracellular direction. (A) Ion positions in the complex of Nav1.4 with GIIIA, which produces a complete block. The trajectory is interrupted by R13 residue. (B) Ion positions in the complex of Nav1.4 with GIIIA mutant R13A, which produces a partial block. The trajectory is smooth. (C) Energy of the ion-system interactions. For the wild-type toxin, but not its mutant, the energy plot has a region of positive (repulsive) energies.
Next we computed a similar energy profile for the R13A mutant of GIIIA bound in Nav1.4. We did not search systematically the mutant-binding mode, as we did for GIIIA, but just replaced R13 with alanine in our GIIIA–channel model and then Monte Carlo–minimized the energy of the complex. The toxin body did not move, and all of the toxin–channel contacts (except those at the mutated reside) were preserved. However, the egress pathway of the sodium ion in this model was quite different from that predicted for the wild-type toxin in terms of energy and geometry (Fig. 5 B). The trajectory became smooth. Comparison of panels A and B in Fig. 5 demonstrates a key role of mutation R13A in this difference. The zone of positive energies disappeared (Fig. 5 C), indicating that the mutant toxin does not impose obstacles for the ion movement between the selectivity filter and the extracellular space. Thus, our calculations allow us to explain the different action of GIIIA, which produces complete block, and its R13A mutant, which produces only a partial block.
To further explore possibilities of sodium movement through the toxin-bound channels, we generated 10,000 staring structures in which the ion was randomly placed in the outer pore region and briefly optimized the starting positions (10 minimizations in each of the 10,000 MCM trajectories). For the analysis, we selected only those structures in which the energy of interaction of the ion with the rest of the system was negative (attracting interactions dominate). This approach allowed us to find regions in the outer pore that can be populated by ions. The calculations were performed for the toxin-free channel model and for the complexes of the channel with the wild-type and mutant toxins. The results are shown in Fig. S9, and some representative data are shown in Fig. 6.
Figure 6.
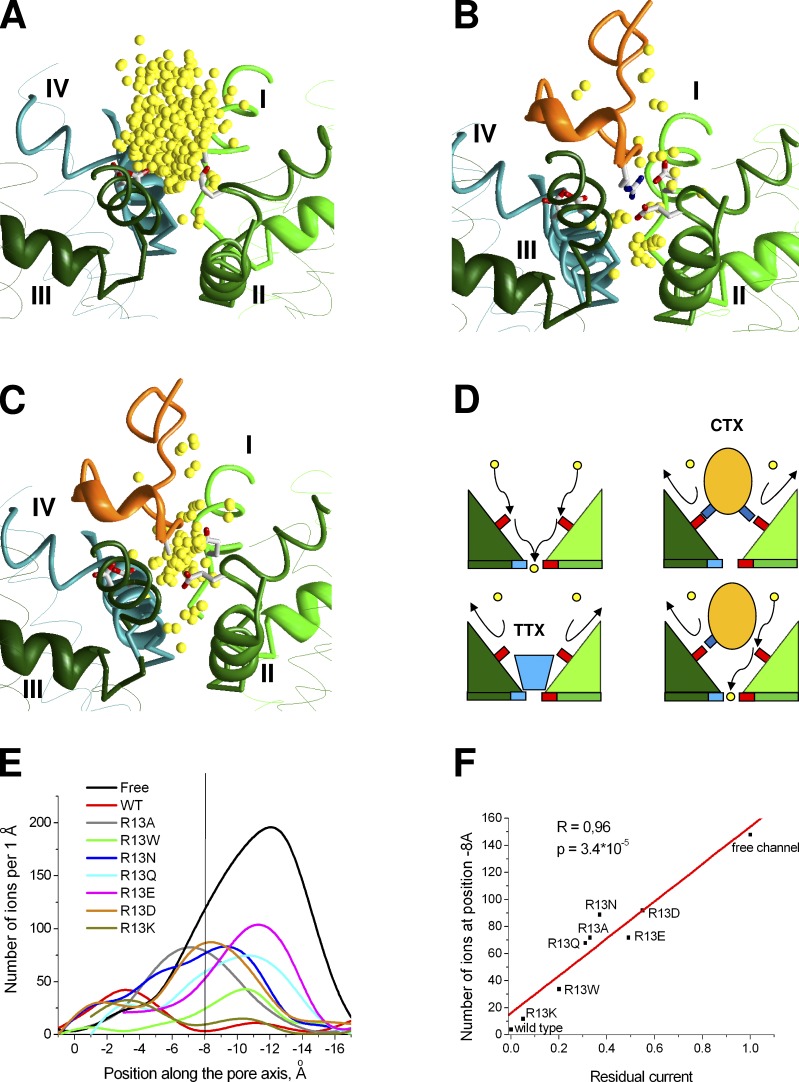
Mechanism of Nav channel block. (A–C) Examples of ion distribution in the toxin-free channel (A) and in the presence of the wild-type GIIIA (B) and GIIIA R13A mutant (C). (D) Proposed schemes of ion permeation in toxin-free channel and complete or incomplete channel block by toxins. Side chains of acidic and basic residues are shown by small red and blue rectangles, respectively. The DEKA locus is at the bottom and the outer carboxylates are at the upper level. Incoming Na+ ions first bind to the outer carboxylates and then to the DEKA locus. TTX binds between the DEKA locus and the outer carboxylates, thus sterically blocking the ion permeation. Conotoxins (CTX) bind superficially and block permeation by occupying ion-binding sites at the outer carboxylates. KIIIA and some GIIIA and PIIIA mutants leave one of the outer carboxylates free, thus producing only a partial block. (E) Plots of ion distribution along the pore axis. For the toxin-free model and for the R13A mutants of GIIIA, which produce incomplete block, there are many ions in the outer pore region with clear maxima at the levels of −12 Å and −7 Å where the ions are attracted to the outer carboxylates. In contrast, for GIIIA, which produces a complete block, the plot has two maxima in the selectivity filter region and near the extracellular entrance to the outer pore, which are separated by a nonpopulated region around −8 Å. (F) Correlation between the residual current value (fraction of single channel current relative to the current through nonblocked channel) and the number of ions at the level of outer carboxylate ring (z = −8 Å) for ion distributions shown in E.
In the toxin-free Nav1.4 model, continuous occupancy of the outer pore region is seen (Fig. 6 A). A large number of energetically possible ion positions in the extracellular half of the outer pore reflects the large free space at this region. The bound GIIIA toxin displaces most of ions from this region (Fig. 6 B), but two constellations of ions are seen at the extracellular part of the outer pore and at the selectivity filter. Importantly, between these constellations there is a zone, which practically lacks energetically favorable positions for the ion (Fig. 6, B and E). It should be noted that the absence of favorable ion positions in this zone is not caused by complete sterical occlusion of the pore. Rather, the ions cannot occupy this region because of electrostatic repulsion with positively charged residues of GIIIA. Indeed, when we disabled the electrostatic interactions for the randomly placed ion, energetically favorable ion positions appeared all along the pore axis without a break (not depicted). Thus, in our model the nature of the channel block by μ-conotoxin is predominantly electrostatic.
For the R13A mutant of GIIIA, the shape of ion distribution is more similar to that in the toxin-free channel. Although many possible ion positions in the outer pore of toxin-free channel are eliminated by the toxin, such positions are seen at all the levels of the outer pore, implying that the pathway for the ions is not interrupted (Fig. 6 C). Similar uninterrupted ion distributions were obtained for KIIIA and PIIIA mutants, which produce only incomplete block. In contrast, for the GIIIA mutants (K9A, K11A, K16A, and R19A) and for the R20A mutant of PIIIA, which produce complete block (McArthur et al., 2011a), the ion distributions were similar to that of GIIIA wild type, i.e., two regions populated by ions are separated by the zone, where ions are repelled by the toxin (Fig. S9).
To analyze the obtained results, we generated a list of specific contacts in the channel complexes with the wild-type and mutated toxins (Table 2). We found that uninterrupted ion distributions in the models are observed if and only if at least one of the outer carboxylates is not involved in the interactions with the toxin basic residues. Thus, KIIIA, which has only three basic residues, is unable to block permeation completely. GIIIA and PIIIA, which completely block the current, specifically interact with all four outer carboxylates. The models with toxin mutants, in which any of the outer carboxylates is not engaged in specific interactions with the toxin, correspond to the incomplete block. If an outer carboxylate can form salt bridges with two or more basic residues of the toxin, mutation of a single basic residue does not release the outer carboxylate, which remains neutralized and therefore unattractive for a sodium ion. The block remains complete in such cases. For example, alanine substitution of R19 in GIIIA does not produce a residual current (McArthur et al., 2011a) because K16 and K11 readily interact with D1532, which is the strongest contact for the native R19 in GIIIA.
Table 2.
Major contacts of basic residues in μ-conotoxins with outer carboxylates
| Toxin | Outer carboxylates | |||
| E (repeat I) | E (repeat II) | D (repeat III) | D (repeat IV) | |
| GIIIA | R13 | R13 | K11, K16 | K11, R19 |
| PIIIA | R12, R14 | R14 | K17 | K17, R20 |
| KIIIA | - | R10 | R10, R14 | R14 |
Thus, our study proposes a critical role of the outer carboxylates in the process of ion permeation. These residues likely serve as intermediate binding sites for ions moving from the extracellular space toward the selectivity filter (Fig. 6 D). If at least one of the outer carboxylates remains free, the ion permeation is possible, although it is reduced to some extent. This model agrees with experimental data (McArthur et al., 2011a,b) and with the previous modeling suggestion about the role of outer carboxylates (Tikhonov and Zhorov, 2007).
Effects of R13 substitutions in GIIIA were systematically studied (Hui et al., 2002). It was demonstrated that the value of residual current depends on both size and charge/polarity of a substitute. To reproduce these results, we modeled R13 substitutions by K, Q, W, Q, N, A, E, and D and calculated the ion distributions as described above. Ion distributions for all the mutants are not completely interrupted, suggesting the existence of some residual current (Fig. 6 E). To obtain quantitative correlation with experimental data (Hui et al., 2002), we plotted the number of ion at axis coordinate z = 8 Å (where the ion distribution is interrupted for the wild-type toxin) against the experimental data on residual current. Results of our calculations (Fig. 6 F) demonstrate a good correlation with experiments (R = 0.96, P = 3.4 × 10−5). Thus, the model reproduced residual currents in the channel complexed with different GIIIA mutants not just qualitatively, but quantitatively.
DISCUSSION
In the present work we have used a NavAb-based homology model of the outer pore region of the voltage-gated sodium channel Nav1.4 to dock μ-conotoxins GIIIA, PIIIA, and KIIIA and some of their mutants. A key question addressed in our work was whether or not the NavAb structure is suitable for rationalizing a large body of available experimental data on the action μ-conotoxins in eukaryotic channels. Unlike previous models of the outer pore region in Nav1 channels, which were necessarily based on potassium channel structures, we did not modify the NavAb template folding to obtain agreement between the modeling predictions and experimental data. Our calculations have demonstrated that the NavAb-based model does allow us to rationalize various experimental data. This result indicates that the outer pore in prokaryotic and eukaryotic voltage-gated sodium channels folds similarly. In other words, our results demonstrate that the NavAb structure allows us to proceed from “hand-made” models of the outer pore region in voltage-gated sodium channels to structure-based models.
This conclusion is not a trivial one. The sequences of NavAb and eukaryotic channels in the selectivity filter and outer pore regions are significantly different. NavAb contains the EEEE ring in the selectivity filter, whereas eukaryotic channels have the DEKA ring (Fig. 1). The EEEE ring is typical for calcium-selective rather than sodium-selective channels. Furthermore, NavAb lacks the ring of outer carboxylates, which has been demonstrated to participate in the ion permeation. This suggests that mechanisms of selectivity and permeation in NavAb and eukaryotic sodium channels have different structural bases.
Recently, PIIIA was predicted to block NavAb channel in the binding mode that significantly differs from that in our model of PIIIA with Nav1.4 (Chen and Chung, 2012). The large difference between sequences of prokaryotic and eukaryotic sodium channels in the outer pore region may explain different binding modes of μ-conotoxins in these channels. A model of PIIIA in Nav1.4 was proposed, which is based on the straightforward alignment of Nav1.4 and NavAb sequences (Chen et al., 2014). Using MD simulations and constraints that pulled the toxin basic side chains toward the selectivity filter, the authors obtained models in which PIIIA is located in the outer pore deeper than in our model. The proposed toxin–channel interactions also differ significantly from those in our models. In the absence of experimental data on PIIIA–channel pairwise contacts, it is hardly possible to select the most realistic PIIIA-binding model. It should be noted that we addressed diverse experimental data on the action of GIIIA, PIIIA, and KIIIA and the obtained models readily reproduced various experimental observations, which were not used during docking procedure. For instance, the binding mode elaborated for GIIIA explains simultaneous binding of KIIIA and TTX. The PIIIA-binding mode agrees with the experimental data on relative electric depths of individual residues.
Incomplete block by some μ-conotoxins and their mutants is well known (McArthur et al., 2011a,b), but structural bases of this effect were unclear. It is demonstrated that the current decreases because of decrease of the channel conductance (Wang et al., 2000, 2001). Our modeling suggests a molecular mechanism of this effect. Unlike TTX and related small-size ligands, μ-conotoxins do not penetrate into the narrow portion of the outer pore and do not completely occlude the pore lumen by their bodies. According to our models, conotoxins block the current by catching and neutralizing the outer carboxylates. If all four outer carboxylates are involved in the interactions with a toxin, the permeation is completely blocked. If some of the outer carboxylates remain free, the toxin only partially blocks the current. This proposed scheme (Fig. 6 D) agrees with the data that mutations of the outer carboxylates reduce the channel conductance. Although our models emphasize the role of electrostatic interactions in compete or incomplete channel block, the value of residual current obviously depends on the steric factors. This is particularly true for R13 substitutions in GIIIA because this residue directly faces the pore. The size of the side chain in this position largely determines the number of ions, which are displaced from the critical region of the outer carboxylates, in agreement with experimental data (Hui et al., 2002).
Based on our models, some testable predictions can be proposed. Particularly, we found that the hydroxyl group of hP17 can specifically interact with carboxyl group of D1248. Also, according to our model, D762 and E765 make specific contacts with R395 and contribute to stabilization of the outer pore folding, suggesting that alanine substitutions of D762 and E765 (Xue et al., 2003) affect the toxin binding allosterically rather than directly. From this point of view, other pairs of specifically interacting residues (Q407-E1524, Y1244-R750, N1536-Q1232), which stabilize mutual disposition of P1 and P2 helices, can also have a certain impact on conotoxin binding.
Certain limitations of our models should be discussed. First, we modeled the asymmetric channel using a symmetric template. Certainly, sequence differences between individual repeats should cause some structural asymmetry on the backbone. To take this effect into account, we did not clamp the template backbone but used pin constraints, which allow penalty-free deviation of α carbons up to 1 Å from the respective template positions. However, the asymmetry of the pore domain may be more significant, in particular because of the influence of the channel regions, which were not considered in our model. Second, we did not take into account all possible factors that affect conotoxin binding. For example, the toxin–channel affinity significantly depends on the ionic strength (Li et al., 2003). Qualitatively, it can be explained by competition between metal ions and basic residues of the toxin for interactions with acidic residues of the channel. However, precise quantification of this effect is difficult. Third, we did not perform hands-free docking of the toxins by systematically exploring all possible binding modes. Inherent limitations of the homology modeling, some of which are mentioned above, make hands-free calculations not completely reliable. That is why we used available experimental data as constraints for the docking procedure. Fourth, we assumed that point mutations do not change the overall binding mode. For some mutants this is not evident. For example, the R13A mutant of GIIIA subtly, but clearly stands apart from other mutants (Hui et al., 2002), suggesting that the binding mode is changed. However, at the present level of model precision, it is hardly possible to correctly predict the difference between the binding modes of wild-type and mutant toxins.
Given these limitations, we did not attempt to predict binding affinities of the native and mutant toxins. However, the limitations do not affect the main conclusions of our study. Our results suggest that NavAb-based models of the outer pore region of eukaryotic voltage-gated sodium channels provide significant progress in understanding the structural organization of these important proteins. Until the x-ray structure of a eukaryotic sodium channel becomes available, the NavAb-based model can be used to rationalize experimental data and design new experiments.
Supplementary Material
Acknowledgments
Computations were made possible by the facilities of the Shared Hierarchical Academic Research Computing Network (SHARCNET). We thank an anonymous reviewer of the manuscript for bringing our attention to results of systematic analysis of GIIIA_R13 mutations and to the problem of steric versus electrostatic nature of the sodium channel block by μ-conotoxins.
This work was supported by grants to D.B. Tikhonov (the program of Russian Academy of Sciences “Molecular and Cellular Biology” and RFBR-13-04-00724) and to B.S. Zhorov (GRPIN-2014-04894 from the Natural Sciences and Engineering Research Council of Canada and RFBR-14-04-378).
The authors declare no competing financial interests.
Kenton J. Swartz served as editor.
Footnotes
Abbreviations used in this paper:
- MCM
- Monte Carlo minimization
- RMSD
- root-mean-square deviation
- TTX
- tetrodotoxin
References
- Becker, S., Prusak-Sochaczewski E., Zamponi G., Beck-Sickinger A.G., Gordon R.D., and French R.J.. 1992. Action of derivatives of μ-conotoxin GIIIA on sodium channels. Single amino acid substitutions in the toxin separately affect association and dissociation rates. Biochemistry. 31:8229–8238 10.1021/bi00150a016 [DOI] [PubMed] [Google Scholar]
- Bruhova, I., Tikhonov D.B., and Zhorov B.S.. 2008. Access and binding of local anesthetics in the closed sodium channel. Mol. Pharmacol. 74:1033–1045 10.1124/mol.108.049759 [DOI] [PubMed] [Google Scholar]
- Chang, N.S., French R.J., Lipkind G.M., Fozzard H.A., and Dudley S. Jr. 1998. Predominant interactions between μ-conotoxin Arg-13 and the skeletal muscle Na+ channel localized by mutant cycle analysis. Biochemistry. 37:4407–4419 10.1021/bi9724927 [DOI] [PubMed] [Google Scholar]
- Chen, R., and Chung S.H.. 2012. Binding modes of μ-conotoxin to the bacterial sodium channel (NaVAb). Biophys. J. 102:483–488 10.1016/j.bpj.2011.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R., Robinson A., and Chung S.H.. 2014. Mechanism of μ-conotoxin PIIIA binding to the voltage-gated Na+ channel NaV1.4. PLoS ONE. 9:e93267 10.1371/journal.pone.0093267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary, G., Aliste M.P., Tieleman D.P., French R.J., and Dudley S.C. Jr. 2007. Docking of μ-conotoxin GIIIA in the sodium channel outer vestibule. Channels (Austin). 1:344–352 10.4161/chan.5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz, L.J., Kupryszewski G., LeCheminant G.W., Gray W.R., Olivera B.M., and Rivier J.. 1989. μ-Conotoxin GIIIA, a peptide ligand for muscle sodium channels: chemical synthesis, radiolabeling, and receptor characterization. Biochemistry. 28:3437–3442 10.1021/bi00434a043 [DOI] [PubMed] [Google Scholar]
- Cummins, T.R., Aglieco F., and Dib-Hajj S.D.. 2002. Critical molecular determinants of voltage-gated sodium channel sensitivity to μ-conotoxins GIIIA/B. Mol. Pharmacol. 61:1192–1201 10.1124/mol.61.5.1192 [DOI] [PubMed] [Google Scholar]
- Doyle, D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., and MacKinnon R.. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77 10.1126/science.280.5360.69 [DOI] [PubMed] [Google Scholar]
- Dudley, S.C. Jr, Todt H., Lipkind G., and Fozzard H.A.. 1995. A mu-conotoxin-insensitive Na+ channel mutant: possible localization of a binding site at the outer vestibule. Biophys. J. 69:1657–1665 10.1016/S0006-3495(95)80045-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley, S.C. Jr, Chang N., Hall J., Lipkind G., Fozzard H.A., and French R.J.. 2000. μ-Conotoxin GIIIA interactions with the voltage-gated Na+ channel predict a clockwise arrangement of the domains. J. Gen. Physiol. 116:679–690 10.1085/jgp.116.5.679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelkamp, N., Linley J.E., Baker M.D., Minett M.S., Cregg R., Werdehausen R., Rugiero F., and Wood J.N.. 2012. Neurological perspectives on voltage-gated sodium channels. Brain. 135:2585–2612 10.1093/brain/aws225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French, R.J., and Terlau H.. 2004. Sodium channel toxins—receptor targeting and therapeutic potential. Curr. Med. Chem. 11:3053–3064 10.2174/0929867043363866 [DOI] [PubMed] [Google Scholar]
- French, R.J., Worley J.F. III, and Krueger B.K.. 1984. Voltage-dependent block by saxitoxin of sodium channels incorporated into planar lipid bilayers. Biophys. J. 45:301–310 10.1016/S0006-3495(84)84156-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French, R.J., Prusak-Sochaczewski E., Zamponi G.W., Becker S., Kularatna A.S., and Horn R.. 1996. Interactions between a pore-blocking peptide and the voltage sensor of the sodium channel: an electrostatic approach to channel geometry. Neuron. 16:407–413 10.1016/S0896-6273(00)80058-6 [DOI] [PubMed] [Google Scholar]
- Garden, D.P., and Zhorov B.S.. 2010. Docking flexible ligands in proteins with a solvent exposure- and distance-dependent dielectric function. J. Comput. Aided Mol. Des. 24:91–105 10.1007/s10822-009-9317-9 [DOI] [PubMed] [Google Scholar]
- Hui, K., Lipkind G., Fozzard H.A., and French R.J.. 2002. Electrostatic and steric contributions to block of the skeletal muscle sodium channel by μ-conotoxin. J. Gen. Physiol. 119:45–54 10.1085/jgp.119.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, K., McIntyre D., and French R.J.. 2003. Conotoxins as sensors of local pH and electrostatic potential in the outer vestibule of the sodium channel. J. Gen. Physiol. 122:63–79 10.1085/jgp.200308842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y., Lee A., Chen J., Ruta V., Cadene M., Chait B.T., and MacKinnon R.. 2003. X-ray structure of a voltage-dependent K+ channel. Nature. 423:33–41 10.1038/nature01580 [DOI] [PubMed] [Google Scholar]
- Khoo, K.K., Gupta K., Green B.R., Zhang M.M., Watkins M., Olivera B.M., Balaram P., Yoshikami D., Bulaj G., and Norton R.S.. 2012. Distinct disulfide isomers of μ-conotoxins KIIIA and KIIIB block voltage-gated sodium channels. Biochemistry. 51:9826–9835 10.1021/bi301256s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, R.J., Dutertre S., Vetter I., and Christie M.J.. 2012. Conus venom peptide pharmacology. Pharmacol. Rev. 64:259–298 10.1124/pr.111.005322 [DOI] [PubMed] [Google Scholar]
- Li, H.L., Sui H.X., Ghanshani S., Lee S., Walian P.J., Wu C.L., Chandy K.G., and Jap B.K.. 1998. Two-dimensional crystallization and projection structure of KcsA potassium channel. J. Mol. Biol. 282:211–216 10.1006/jmbi.1998.2016 [DOI] [PubMed] [Google Scholar]
- Li, R.A., Ennis I.L., French R.J., Dudley S.C. Jr, Tomaselli G.F., and Marbán E.. 2001. Clockwise domain arrangement of the sodium channel revealed by μ-conotoxin (GIIIA) docking orientation. J. Biol. Chem. 276:11072–11077 10.1074/jbc.M010862200 [DOI] [PubMed] [Google Scholar]
- Li, R.A., Sato K., Kodama K., Kohno T., Xue T., Tomaselli G.F., and Marbán E.. 2002. Charge conversion enables quantification of the proximity between a normally-neutral μ-conotoxin (GIIIA) site and the Na+ channel pore. FEBS Lett. 511:159–164 10.1016/S0014-5793(01)03316-6 [DOI] [PubMed] [Google Scholar]
- Li, R.A., Hui K., French R.J., Sato K., Henrikson C.A., Tomaselli G.F., and Marbán E.. 2003. Dependence of μ-conotoxin block of sodium channels on ionic strength but not on the permeating [Na+]: implications for the distinctive mechanistic interactions between Na+ and K+ channel pore-blocking toxins and their molecular targets. J. Biol. Chem. 278:30912–30919 10.1074/jbc.M301039200 [DOI] [PubMed] [Google Scholar]
- Li, Z., and Scheraga H.A.. 1987. Monte Carlo-minimization approach to the multiple-minima problem in protein folding. Proc. Natl. Acad. Sci. USA. 84:6611–6615 10.1073/pnas.84.19.6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linford, N.J., Cantrell A.R., Qu Y., Scheuer T., and Catterall W.A.. 1998. Interaction of batrachotoxin with the local anesthetic receptor site in transmembrane segment IVS6 of the voltage-gated sodium channel. Proc. Natl. Acad. Sci. USA. 95:13947–13952 10.1073/pnas.95.23.13947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkind, G.M., and Fozzard H.A.. 2000. KcsA crystal structure as framework for a molecular model of the Na+ channel pore. Biochemistry. 39:8161–8170 10.1021/bi000486w [DOI] [PubMed] [Google Scholar]
- Long, S.B., Campbell E.B., and Mackinnon R.. 2005. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903 10.1126/science.1116269 [DOI] [PubMed] [Google Scholar]
- McArthur, J.R., Ostroumov V., Al-Sabi A., McMaster D., and French R.J.. 2011a. Multiple, distributed interactions of μ-conotoxin PIIIA associated with broad targeting among voltage-gated sodium channels. Biochemistry. 50:116–124 10.1021/bi101316y [DOI] [PubMed] [Google Scholar]
- McArthur, J.R., Singh G., McMaster D., Winkfein R., Tieleman D.P., and French R.J.. 2011b. Interactions of key charged residues contributing to selective block of neuronal sodium channels by μ-conotoxin KIIIA. Mol. Pharmacol. 80:573–584 10.1124/mol.111.073460 [DOI] [PubMed] [Google Scholar]
- McArthur, J.R., Singh G., O’Mara M.L., McMaster D., Ostroumov V., Tieleman D.P., and French R.J.. 2011c. Orientation of μ-conotoxin PIIIA in a sodium channel vestibule, based on voltage dependence of its binding. Mol. Pharmacol. 80:219–227 10.1124/mol.111.071779 [DOI] [PubMed] [Google Scholar]
- McCusker, E.C., Bagnéris C., Naylor C.E., Cole A.R., D’Avanzo N., Nichols C.G., and Wallace B.A.. 2012. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat. Commun. 3:1102 10.1038/ncomms2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczydlowski, E., Hall S., Garber S.S., Strichartz G.S., and Miller C.. 1984. Voltage-dependent blockade of muscle Na+ channels by guanidinium toxins. J. Gen. Physiol. 84:687–704 10.1085/jgp.84.5.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera, B.M., and Cruz L.J.. 2001. Conotoxins, in retrospect. Toxicon. 39:7–14 10.1016/S0041-0101(00)00157-4 [DOI] [PubMed] [Google Scholar]
- Payandeh, J., Scheuer T., Zheng N., and Catterall W.A.. 2011. The crystal structure of a voltage-gated sodium channel. Nature. 475:353–358 10.1038/nature10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestley, T.2004. Voltage-gated sodium channels and pain. Curr. Drug Targets CNS Neurol. Disord. 3:441–456 10.2174/1568007043336888 [DOI] [PubMed] [Google Scholar]
- Rogers, M., Tang L., Madge D.J., and Stevens E.B.. 2006. The role of sodium channels in neuropathic pain. Semin. Cell Dev. Biol. 17:571–581 10.1016/j.semcdb.2006.10.009 [DOI] [PubMed] [Google Scholar]
- Safo, P., Rosenbaum T., Shcherbatko A., Choi D.Y., Han E., Toledo-Aral J.J., Olivera B.M., Brehm P., and Mandel G.. 2000. Distinction among neuronal subtypes of voltage-activated sodium channels by mu-conotoxin PIIIA. J. Neurosci. 20:76–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, K., Ishida Y., Wakamatsu K., Kato R., Honda H., Ohizumi Y., Nakamura H., Ohya M., Lancelin J.M., Kohda D., et al. . 1991. Active site of mu-conotoxin GIIIA, a peptide blocker of muscle sodium channels. J. Biol. Chem. 266:16989–16991 [PubMed] [Google Scholar]
- Shon, K.J., Olivera B.M., Watkins M., Jacobsen R.B., Gray W.R., Floresca C.Z., Cruz L.J., Hillyard D.R., Brink A., Terlau H., and Yoshikami D.. 1998. mu-Conotoxin PIIIA, a new peptide for discriminating among tetrodotoxin-sensitive Na channel subtypes. J. Neurosci. 18:4473–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, M., Peigneur S., Dyubankova N., Lescrinier E., Herdewijn P., and Tytgat J.. 2012. Design of bioactive peptides from naturally occurring μ-conotoxin structures. J. Biol. Chem. 287:31382–31392 10.1074/jbc.M112.375733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietze, A.A., Tietze D., Ohlenschläger O., Leipold E., Ullrich F., Kühl T., Mischo A., Buntkowsky G., Görlach M., Heinemann S.H., and Imhof D.. 2012. Structurally diverse μ-conotoxin PIIIA isomers block sodium channel NaV1.4. Angew. Chem. Int. Ed. Engl. 51:4058–4061 10.1002/anie.201107011 [DOI] [PubMed] [Google Scholar]
- Tikhonov, D.B., and Zhorov B.S.. 2005. Modeling P-loops domain of sodium channel: homology with potassium channels and interaction with ligands. Biophys. J. 88:184–197 10.1529/biophysj.104.048173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov, D.B., and Zhorov B.S.. 2007. Sodium channels: ionic model of slow inactivation and state-dependent drug binding. Biophys. J. 93:1557–1570 10.1529/biophysj.106.100248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov, D.B., and Zhorov B.S.. 2012. Architecture and pore block of eukaryotic voltage-gated sodium channels in view of NavAb bacterial sodium channel structure. Mol. Pharmacol. 82:97–104 10.1124/mol.112.078212 [DOI] [PubMed] [Google Scholar]
- Tikhonov, D.B., Bruhova I., Garden D.P., and Zhorov B.S.. 2014. State-dependent inter-repeat contacts of exceptionally conserved asparagines in the inner helices of sodium and calcium channels. Pflugers Arch. In press 10.1007/s00424-014-1508-0 [DOI] [PubMed] [Google Scholar]
- Trimmer, J.S., Cooperman S.S., Tomiko S.A., Zhou J.Y., Crean S.M., Boyle M.B., Kallen R.G., Sheng Z.H., Barchi R.L., Sigworth F.J., et al. . 1989. Primary structure and functional expression of a mammalian skeletal muscle sodium channel. Neuron. 3:33–49 10.1016/0896-6273(89)90113-X [DOI] [PubMed] [Google Scholar]
- Van Der Haegen, A., Peigneur S., and Tytgat J.. 2011. Importance of position 8 in μ-conotoxin KIIIA for voltage-gated sodium channel selectivity. FEBS J. 278:3408–3418 10.1111/j.1742-4658.2011.08264.x [DOI] [PubMed] [Google Scholar]
- Wang, S.Y., Nau C., and Wang G.K.. 2000. Residues in Na+ channel D3-S6 segment modulate both batrachotoxin and local anesthetic affinities. Biophys. J. 79:1379–1387 10.1016/S0006-3495(00)76390-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S.Y., Barile M., and Wang G.K.. 2001. Disparate role of Na+ channel D2-S6 residues in batrachotoxin and local anesthetic action. Mol. Pharmacol. 59:1100–1107 [DOI] [PubMed] [Google Scholar]
- Waszkielewicz, A.M., Gunia A., Szkaradek N., Słoczyńska K., Krupińska S., and Marona H.. 2013. Ion channels as drug targets in central nervous system disorders. Curr. Med. Chem. 20:1241–1285 10.2174/0929867311320100005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner, S.J., Kollman P.A., Case D.A., Singh U.C., Ghio C., Alagona G., Profeta S., and Weiner P.. 1984. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 106:765–784 10.1021/ja00315a051 [DOI] [Google Scholar]
- Weiner, S.J., Kollman P.A., Nguyen D.T., and Case D.A.. 1986. An all atom force field for simulations of proteins and nucleic acids. J. Comput. Chem. 7:230–252 10.1002/jcc.540070216 [DOI] [PubMed] [Google Scholar]
- Wilson, M.J., Yoshikami D., Azam L., Gajewiak J., Olivera B.M., Bulaj G., and Zhang M.M.. 2011. μ-Conotoxins that differentially block sodium channels NaV1.1 through 1.8 identify those responsible for action potentials in sciatic nerve. Proc. Natl. Acad. Sci. USA. 108:10302–10307 10.1073/pnas.1107027108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, J.N., Boorman J.P., Okuse K., and Baker M.D.. 2004. Voltage-gated sodium channels and pain pathways. J. Neurobiol. 61:55–71 10.1002/neu.20094 [DOI] [PubMed] [Google Scholar]
- Xue, T., Ennis I.L., Sato K., French R.J., and Li R.A.. 2003. Novel interactions identified between μ-conotoxin and the Na+ channel domain I P-loop: implications for toxin-pore binding geometry. Biophys. J. 85:2299–2310 10.1016/S0006-3495(03)74654-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M.M., Green B.R., Catlin P., Fiedler B., Azam L., Chadwick A., Terlau H., McArthur J.R., French R.J., Gulyas J., et al. . 2007. Structure/function characterization of micro-conotoxin KIIIA, an analgesic, nearly irreversible blocker of mammalian neuronal sodium channels. J. Biol. Chem. 282:30699–30706 10.1074/jbc.M704616200 [DOI] [PubMed] [Google Scholar]
- Zhang, M.M., McArthur J.R., Azam L., Bulaj G., Olivera B.M., French R.J., and Yoshikami D.. 2009. Synergistic and antagonistic interactions between tetrodotoxin and μ-conotoxin in blocking voltage-gated sodium channels. Channels (Austin). 3:32–38 10.4161/chan.3.1.7500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M.M., Gruszczynski P., Walewska A., Bulaj G., Olivera B.M., and Yoshikami D.. 2010. Cooccupancy of the outer vestibule of voltage-gated sodium channels by μ-conotoxin KIIIA and saxitoxin or tetrodotoxin. J. Neurophysiol. 104:88–97 10.1152/jn.00145.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Ren W., DeCaen P., Yan C., Tao X., Tang L., Wang J., Hasegawa K., Kumasaka T., He J., et al. . 2012. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 486:130–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.