The authors used comparative sequence analysis to delineate, using 12 flowering plants, conserved noncoding sequences in Arabidopsis thaliana and demonstrated a high enrichment for functional cis-regulatory elements in these conserved regions. Based on known binding sites, a gene regulatory network was generated, revealing new and condition-specific functional gene regulatory interactions.
Abstract
Transcriptional regulation plays an important role in establishing gene expression profiles during development or in response to (a)biotic stimuli. Transcription factor binding sites (TFBSs) are the functional elements that determine transcriptional activity, and the identification of individual TFBS in genome sequences is a major goal to inferring regulatory networks. We have developed a phylogenetic footprinting approach for the identification of conserved noncoding sequences (CNSs) across 12 dicot plants. Whereas both alignment and non-alignment-based techniques were applied to identify functional motifs in a multispecies context, our method accounts for incomplete motif conservation as well as high sequence divergence between related species. We identified 69,361 footprints associated with 17,895 genes. Through the integration of known TFBS obtained from the literature and experimental studies, we used the CNSs to compile a gene regulatory network in Arabidopsis thaliana containing 40,758 interactions, of which two-thirds act through binding events located in DNase I hypersensitive sites. This network shows significant enrichment toward in vivo targets of known regulators, and its overall quality was confirmed using five different biological validation metrics. Finally, through the integration of detailed expression and function information, we demonstrate how static CNSs can be converted into condition-dependent regulatory networks, offering opportunities for regulatory gene annotation.
INTRODUCTION
Transcriptional regulation is a complex and dynamic process in which transcription factors (TFs) play a fundamental role. Although being subject to many potentially overlapping regulatory mechanisms, such as microRNA (miRNA) regulation and chromatin accessibility coordinated by histone modifications and DNA methylation, the binding of TFs on specific genomic locations modulating gene expression levels is pivotal for the proper regulation of different biological processes. TF binding events can have a direct or indirect effect on the activation or repression of gene transcription. More complex regulation of gene expression is achieved through cooperative binding of different TFs, adding an extra combinatorial level of regulation (Riechmann and Ratcliffe, 2000). These regulatory mechanisms allow organisms to process different endogenous signals related to growth and development and to respond to changing environmental conditions including different types of (a)biotic stresses.
Despite the functional importance of transcriptional regulation and the fact that 1500 to 1700 TFs have been identified in Arabidopsis thaliana (Riechmann et al., 2000; Jin et al., 2014), knowledge about the genes regulated by different TFs is still very limited. AtRegNet, which is a part of the AGRIS database (Yilmaz et al., 2011), summarizes regulatory interactions collected from small- and large-scale experiments and contains 728 interactions when filtering on direct and confirmed targets. This paucity of experimentally validated regulatory interactions can be partially explained by the fact that previously used methods like electrophoretic mobility shift assay (Garner and Revzin, 1981), systematic evolution of ligands by exponential enrichment (Roulet et al., 2002), and yeast one-hybrid analysis (Meng et al., 2005) are labor-intensive and only yield a small number of interactions (Mejia-Guerra et al., 2012). More recent techniques such as protein binding microarrays, chromatin immunoprecipitation (ChIP) with readout through microarray (ChIP-chip), or next-generation sequencing (ChIP-Seq) allow TF protein-DNA binding to be analyzed in a high-throughput manner. However, published binding results using these methods have revealed a weak correlation between the binding of a TF and transcriptional regulation of the potential target genes (Ferrier et al., 2011).
Dozens of software tools have been developed to delineate regulatory regions based on experimental features, such as coregulation, or using advanced computational methods (MacIsaac and Fraenkel, 2006). Although the naïve mapping of known DNA sequence motifs to promoter regions is frequently used to explore cis-regulatory elements, this approach yields many false positives because TF binding sites are often short and typically contain some level of degeneracy in the binding motif (Tompa et al., 2005). Although experimentally characterized open chromatin regions, profiled through DNase I hypersensitive (DH) sites, offer a global picture of accessible regions throughout the genome and can aid in reducing the motif search space (Zhang et al., 2012), determining individual TF binding events remains a major challenge. A promising solution for the computational detection of functional elements is phylogenetic footprinting, which identifies conservation in orthologous genomic sequences (Tagle et al., 1988; Håndstad et al., 2011). Orthologs are homologous genes derived from a speciation event in the last common ancestor of the compared species. Regions of noncoding DNA in the genome that are conserved across related species are likely to be under purifying selection and this signature can be seen as evidence for functionality (Blanchette and Tompa, 2002; Kaplinsky et al., 2002; Guo and Moose, 2003; Inada et al., 2003; Vandepoele et al., 2006, 2009; Thomas et al., 2007; Baxter et al., 2012). Overall, it is not trivial to make the distinction between conserved noncoding sequences (CNSs) that have arisen due to neutral sequence carryover and functionally constrained CNSs in closely related species. With the advent of methods such as PhastCons (Siepel et al., 2005), which make use of aligned genomes and statistical models of sequence evolution, it has become possible to determine CNSs in closely related species. These methods have shown greater power in the detection of functional elements and lineage-specific conservation than detection methods based on comparing more distantly related genomes in vertebrates, insects, worms, and yeast (Siepel et al., 2005). However, these approaches require aligned genomes and the fraction of the genome that can be aligned drops drastically (<40%) when comparing species from different genera in flowering plants (Hupalo and Kern, 2013). This is due to large-scale genome rearrangements and high sequence divergence. Furthermore, taxon sampling is still limited for flowering plants with the exception of the Brassicaceae lineage. These factors make global alignment strategies for the detection of CNSs impractical for many of the currently available plant genomes (Reineke et al., 2011). An additional difficulty for phylogenetic footprinting in plants lays in the fact that it is not trivial to identify one-to-one orthology in plants, due to a wealth of paralogs (homologous genes created through a duplication event) in almost all plant lineages (Van Bel et al., 2012). Besides continuous duplication events, for instance, via tandem duplication, many plant paralogs are remnants of whole-genome duplications. In flowering plants, the frequent whole-genome duplications in several lineages result in the establishment of one-to-many and many-to-many orthologs (or co-orthologs). As a consequence, methods for identifying CNSs that were successfully applied in yeast or vertebrates don’t work well in plants, as these methods cannot cope with complex orthology relationships (De Bodt et al., 2006; Vandepoele et al., 2006).
Recently, three approaches to identify genome-wide CNSs using multiple plant genomes have been published. Baxter and coworkers used a local pairwise alignment approach, implemented in the Seaweeds alignment plot tool (Picot et al., 2010), to search for CNSs in the 2 kb upstream of the transcription start site in Arabidopsis (Baxter et al., 2012). Pairwise alignments were generated between orthologous genes of Arabidopsis and three highly diverged dicots: papaya (Carica papaya), poplar (Populus trichocarpa), and grapevine (Vitis vinifera). The conservation scores associated with each pairwise alignment were aggregated, while orthologs were delineated using a combination of synteny and reciprocal best BLAST hits. Haudry et al. (2013) generated a whole-genome alignment approach using a combination of the LASTZ (Harris, 2007) and MULTIZ (Blanchette et al., 2004) tools across nine closely related Brassicaceae species. In this study, a genomic region was aligned with one or multiple regions in another species as a means to cope with polyploidy. Conservation in the aligned regions was determined using PhyloP (Pollard et al., 2010), yielding a set of 95,142 Arabidopsis CNSs. Similarly, Hupalo and Kern (2013) created a whole-genome alignment between 20 closely and distantly related angiosperm genomes by making use of the LASTZ tool and used PhastCons (Siepel et al., 2005) to identify sequence constraint.
To generate a comprehensive overview of cis-regulatory elements in the Arabidopsis genome, we developed a phylogenetic footprinting framework that identifies CNSs between 12 distantly related genomes. Through the integration of information about known transcription factor binding sites (TFBSs), gene expression profiles, open chromatin states, and different gene function annotations, the static CNSs were annotated and translated into a gene regulatory network capturing known and condition-specific regulatory interactions. In addition, we confirm the quality of the inferred network using different experimental data sets and biological validation metrics.
RESULTS
Detection of CNSs Using a Multispecies Footprinting Approach
We used a comparative genomics approach across 12 dicot plants to discover CNSs in Arabidopsis. A computational framework was developed that uses the mapping of known motifs as well as de novo local alignments to identify regulatory motifs conserved in multiple species. A local alignment-based approach between orthologous regions was applied because global alignment strategies are impractical for many of the currently available plant genomes due to massive loss of synteny conservation (Supplemental Figure 1). The selected comparator dicot species used in this study are reported in Supplemental Figure 1. The first method, called Comparative Motif Mapping (CMM), requires a candidate motif (e.g., a transcription factor binding site represented as a consensus sequence or position count matrix) as input and assesses the motif conservation on, for example, the 2-kb promoter of an Arabidopsis gene. Conservation is scored based on the occurrence of the motif in the promoter regions of the orthologs from the query gene in 11 other species, allowing for incomplete motif conservation. The statistical significance of a motif conserved in a set of orthologous genes is determined by comparing the observed conservation score to a background model that is built from conservation scores generated by processing the same motif on a large number of randomly assembled nonorthologous families, containing the same species composition and having the same sequence length distribution as in the real set of orthologs (see Methods). Based on the phylogenetic footprinting principle, the assumption behind this statistical model is that conservation of functional motifs will be higher between orthologous genes than between randomly chosen nonorthologous genes. As orthologous genes between Arabidopsis and all other comparator species show saturated substitution patterns (the fraction of synonymous substitutions per synonymous site, Ks > 1; see Methods), the identified CNSs show selective constraint, indicating biological functionality.
The second method is alignment based and uses a multispecies scoring approach to detect CNSs, without requiring prior motif information. All footprints extracted from pairwise local alignments between the query gene and its orthologs are collapsed onto the corresponding region of the query gene. As such, the number of species that supports each nucleotide through a pairwise alignment is determined. In the next step, conserved footprints are extracted and scored based on the number of species in which they are conserved. Significant footprints are determined using a precomputed background model built with scores of footprints derived from nonorthologous families to which each real footprint is compared. The same assumption regarding higher functional sequence conservation between orthologous genes than between randomly chosen genes is made. For the alignment-based approach, four alignment tools were implemented in the framework and their performance was compared. These tools were DIALIGN-TX (Subramanian et al., 2008), Sigma (Siddharthan, 2006), ACANA (Huang et al., 2006), and the Seaweeds alignment plot tool (Picot et al., 2010). The proposed methods are able to cope with high sequence divergence when aligning noncoding sequences between related species. As many motif and alignment comparisons are being made for thousands of genes, the false discovery rate (FDR) was estimated by comparing the significant results of the real runs with those of control runs. The FDR is defined as the ratio between the number of false positives estimated by the control run and the number of rejected null hypotheses in the real run and provides a better measure for controlling false positives compared with the false positive rate, as the latter does not correct for the multiple tests performed per query gene. Control runs are identical to real runs with the exception that the orthologous families are randomly generated, maintaining the species constitution and gene size as observed in the real families (see Methods). Unless mentioned otherwise, all presented results have an FDR below 10%.
After updating the TAIR10 genome annotation with 791 new miRNA loci obtained from the plant microRNA database (Zhang et al., 2010), three different genomic sequence types were defined to identify CNSs (2 kb upstream, 1 kb downstream, and intron). In this analysis, upstream and downstream are used relative to the translation start site and translation stop site, respectively, because it has been shown, both through promoter deletion experiments as well as using genome-wide ChIP analyses, that regulatory elements can be found in 5′ and 3′ untranslated region (UTR) (Chabouté et al., 2002; Liu et al., 2010; Wang and Xu, 2010). Another reason to include UTRs is that not all genes have information about their UTR available. In total, the different genomic sequences cover 83% of the noncoding Arabidopsis genome and 84% of all complete intergenics. Gene orthology information was retrieved from the PLAZA 2.5 integrative orthology method (Van Bel et al., 2012), which uses a combination of different detection methods to infer consensus orthology predictions, both for simple one-to-one as well as for more complex many-to-many gene relationships. Here, two different orthology definitions were used to delineate orthologs. The first definition uses a simple best BLAST hit-derived method that includes inparalogs, called best-hit and in-paralogous families (BHIF), while the second definition, called consensus orthology, requires that at least two PLAZA detection methods confirm an orthologous gene relationship (see Methods). Orthologs could be obtained for 24,241 Arabidopsis genes using BHIF and for 21,300 genes using the consensus definition. For Arabidopsis genes with orthology information, 70 and 90% have orthologs in at least 10 species for the consensus and BHIF definition, respectively (Supplemental Figure 2).
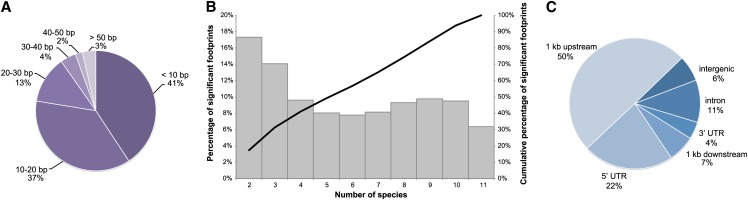
Combining phylogenetic footprinting experiments from the alignment-based and CMM runs, we identified 69,361 significant CNSs associated with 17,895 genes. These conserved regions cover 1070 kb of the Arabidopsis genome, and all CNSs are available through a genome browser (see Methods). The median length of a CNS was 11bp, while the largest and smallest CNS were 514 and 5 bp, respectively (Figure 1A). All of the significant CNSs were conserved in at least two comparator species, while the median number of supporting species was six (Figure 1B). This result illustrates the strong multispecies nature and potential functionality of the identified CNSs. Analyzing the contribution of comparator species to footprints conserved in only two species showed no bias toward the most closely related comparator species. Half of the CNSs are located in the 1-kb promoter region of annotated genes and a large number of conserved regions were associated with introns (10,872) and downstream sequences (6953) (Figure 1C). The alignment-based and CMM detection methods detect 30 and 60% of all CNSs uniquely, respectively, while 10% is shared by both methods. CMM covers 473 kb and the alignment-based approach covers 686 kb. The complementarity of the two different orthology definitions was evaluated by determining the uniquely detected CNSs and revealed that 70% of detected CNSs were found using both definitions. The consensus and BHIF definition detected 19 and 11% unique CNSs, respectively.
Figure 1.
Overview of CNS Properties.
(A) Length distribution of significantly conserved footprints. All footprints are grouped in bins of size 10 bp.
(B) Overview of significantly conserved footprints in relation to the number of species in which the footprint was conserved. For all conservation scores, the relative percentage of significant footprints is shown (gray boxes) as well as a cumulative distribution (black line).
(C) Breakdown of CNS over different genomic regions.
[See online article for color version of this figure.]
Besides regulatory elements, other structural features such as incorrectly annotated exons or missing genes may show significant conservation across related genomes. To determine whether any of the identified footprints represent coding features, we performed a sequence similarity search of all CNSs against a large set of known plant proteins (see Methods). Only 499 CNSs (0.01% of all footprints) showed a significant hit against the plant protein database and were discarded for downstream analysis.
Evaluation of Different Phylogenetic Footprinting Approaches Using an Experimental Gold Standard
In order to evaluate whether our footprints correspond with known regulatory sequences, we compared our CNSs against the AtProbe data set, which contains 144 experimentally determined cis-regulatory elements (see Methods; Supplemental Data Set 1). Overall, our CNSs recovered 26% of the experimental binding sites. This global true positive rate (TPR) was analyzed in more detail per detection method (Supplemental Figures 3A and 3B). Sigma, the best performing alignment tool, scores equally well compared with CMM as both methods have a TPR of 19%. This result indicates that Sigma, which finds conserved regions without any prior information, has sensitivity comparable to CMM, for which prior motif information is required. Additionally, these methods are complementary as they uniquely detected 22 and 16% of the recovered AtProbe elements, respectively. Whereas ACANA and Seaweeds-60 recovered experimental instances (TPR of 5 and 3%, respectively), DIALIGN-TX and Seaweeds-30 did not, which is due to the generation of spurious alignments yielding many false positives in the control runs.
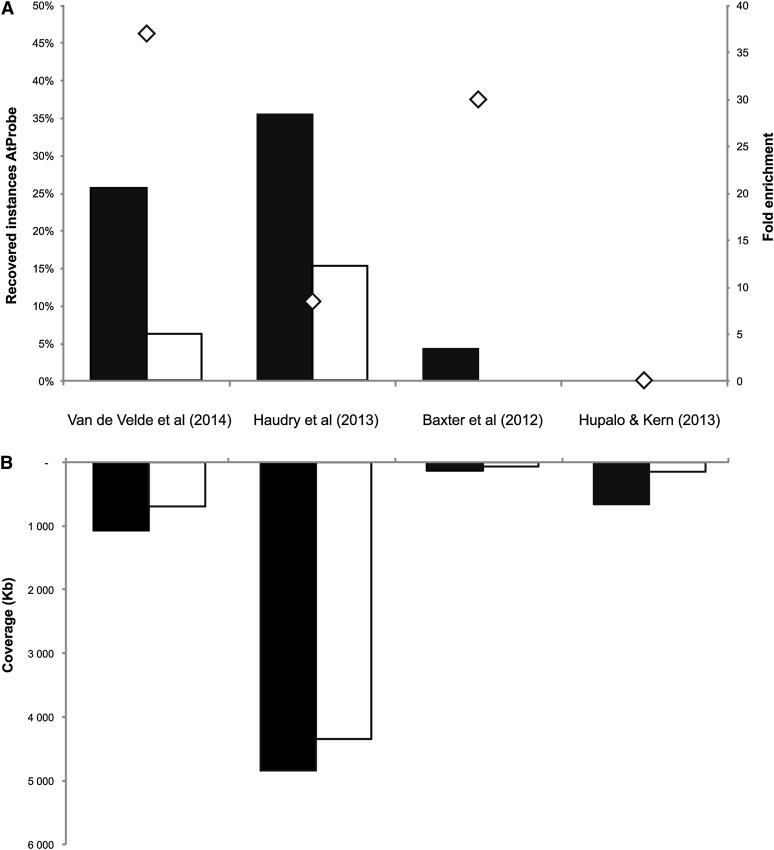
To further validate our set of CNSs, we compared our results with three other CNS data sets from published genome-wide phylogenetic footprinting approaches (Figure 2) (Baxter et al., 2012; Haudry et al., 2013; Hupalo and Kern, 2013). Apart from evaluating the sensitivity of the different studies, which relates to finding true positive AtProbe results, we also assessed the specificity, which relates to identifying negative results. The latter is important, as a method that would assign each noncoding nucleotide to a CNS would yield a high sensitivity but a low specificity, due to many false positives. Although it is not trivial to assemble a negative data set of genomic regions free from any regulatory sequence, we estimated false positives by reshuffling the AtProbe genomic locations 1000 times and determining the overlap with CNSs detected per footprinting study. The estimated number of false positives was used to determine enrichment for known regulatory elements (observed number of elements over expected number of elements; see Methods). This approach does not guarantee that the reshuffled data set, which covers in essence randomly selected noncoding genomic regions that have no overlap with real AtProbe instances, contains only true negatives. However, the reshuffled data set can be used as a proxy to estimate the specificity of different footprinting studies, as the same biases are present in the negative data set for all methods.
Figure 2.
Recovery of AtProbe Elements and Comparison of CNSs from Different Phylogenetic Footprinting Studies.
(A) Overview of the recovery of experimental AtProbe elements in four different CNS studies. Black boxes show the percentage of recovered elements, and white boxes shows the percentage of uniquely recovered elements. Diamonds depict fold enrichments, which are defined as the ratio of the observed overlap over the expected overlap by chance.
(B) Genome-wide coverage of CNSs. Black boxes show the total number of nucleotides assigned to CNSs per study, while white boxes show the number of nucleotides in CNSs that are unique to a single study.
Comparing the CNSs from the different studies showed that Haudry et al. (2013) has the highest recovery of experimental binding sites (35% TPR), followed by our results (26% TPR) and Baxter et al. (2012) (4% TPR). An overview of retrieved CNSs for the AtProbe genes for this study and Haudry et al. (2013) can be found in Supplemental Figure 4. However, comparing the specificity using the shuffled AtProbe data sets reveals that Haudry et al. (2013) has a lower enrichment toward experimentally determined elements (8.5-fold enriched) than our approach (37-fold enriched) (Figure 2). Determining the genome-wide coverage for the different CNS data sets revealed that Haudry et al. (2013) identified constraint for 4834 kb of noncoding DNA. This coverage is substantially larger than our data set (1070 kb) and those of Baxter et al. (2012) and Hupalo and Kern (2013), which cover 137 and 658 kb, respectively (Figure 2). Overall, our method, which we have shown to be accurate based on the analysis of known regulatory sites, identifies 64% of the nucleotides covered by our CNSs as evolutionary constrained that were not identified by the other methods, indicating that our phylogenetic footprinting approach covers a large fraction of unique CNSs.
Conserved Motif Instances Identify in Vivo Functional Regions
To evaluate the functionality of the identified CNSs and to verify whether these conserved footprints can provide a template to computationally map TF-target interactions, detailed comparisons of the CNSs were made against different experimentally determined data sets. DH sites are associated with regions of open chromatin where the DNA is accessible and as such provide a global perspective on possible protein binding to the genome. Overall, 48 and 47% of our CNSs overlapped with a recently published set of DH sites in flower and leaf tissue, respectively (Zhang et al., 2012). This overlap is significant (P value < 0.001) and shows high fold enrichment (4.0 for both DH sets; see Methods), revealing that a large part of the CNSs can be accessed by TFs and as such can act as a functional TFBS. Our set of CNSs also exhibited a significant overlap with H3K4me3, H3K9ac, and H3K4me2 marks (2.6-, 2.2-, and 1.7-fold enriched, respectively; Supplemental Figure 5). These histone modifications are indicative of active promoters and enhancer elements (Roudier et al., 2009; He et al., 2011). Interestingly, our regions showed an even higher enrichment for regions where DH sites, H3K4me3, H3K9ac, and H3K4me2 coincide (6.3-fold enriched, P value < 0.001), corroborating that several of the conserved regions are associated with actively transcribed genes.
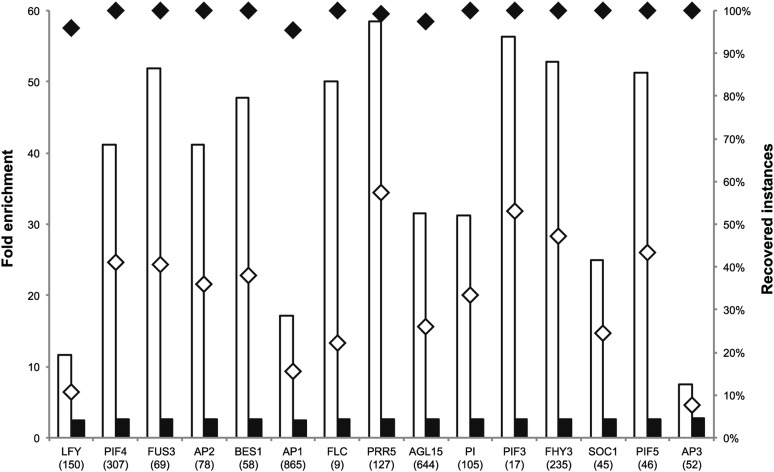
Whereas the experimental data sets profiling different chromatin states act as a proxy for functionality, more detailed regulatory information can be obtained by comparing the CNSs with experimental data sets comprising functional TFBS. To delineate a high-quality data set of in vivo functional TF targets covering directly regulated genes, publicly available ChIP-Seq data were combined with enriched motifs in ChIP-Seq peaks and TF perturbation expression profiles (see Methods). This was done for 15 TFs (AGAMOUS-LIKE15 [AGL15], APETALA1 [AP1], AP2, AP3, SUPPRESSOR OF OVEREXPRESSION OF CO1 [SOC1], PISTILLATA [PI], LEAFY [LFY], FLOWERING LOCUS C [FLC], PSEUDO RESPONSE REGULATOR5 [PRR5], PHYTOCHROME INTERACTING FACTOR3 [PIF3], PIF4, PIF5, FAR-RED ELONGATED HYPOCOTYLS3 [FHY3], BRI1-EMS-SUPPRESSOR1 [BES1], and FUSCA3 [FUS3]), yielding a data set of 2807 regulatory interactions (Supplemental Data Set 2). Importantly, these in vivo functional targets were determined independently of any comparative information and thus provide an independent data set to evaluate our footprints. Overlap analysis revealed that in total 787 functional binding sites (28%) were successfully recovered by our CNSs. Although the recovery rate for individual TF varies from 8% for AP3 to 57% for PRR5 (median recovery 36%), the number of recovered genes for all 15 TFs was significantly higher compared with the number of recovered target genes expected by chance (P < 0.001; Supplemental Data Set 2 and Supplemental Figure 3).
To compare the specificity by which our CNSs identified functional TFBS with other computational methods, two other protocols were evaluated. Whereas the first approach is based on the simple mapping of all positional count matrices of all 15 TFs on the noncoding genomic DNA, the second approach comprises motif mapping in open noncoding chromatin regions that were identified through DH sites (Zhang et al., 2012). Enrichment analysis using shuffled data sets of the in vivo functional regions (see Methods) revealed that our CNSs yielded higher specificity for functional regulatory elements than either of these alternative protocols (median fold enrichment of 41.2 for CNSs versus 2.6- and 12.8-fold enrichment for the simple and DH site-based mapping methods, respectively) (Figure 3; Supplemental Data Set 3 and Supplemental Figure 6).
Figure 3.
Recovery of in Vivo Functional Targets Using CNS Information.
White and black boxes show fold enrichments for CNSs and naïve motif mapping, respectively. White and black diamonds show the fraction of recovered elements for CNSs and a simple motif mapping approach, respectively.
Construction and Biological Evaluation of an Arabidopsis Gene Regulatory Network
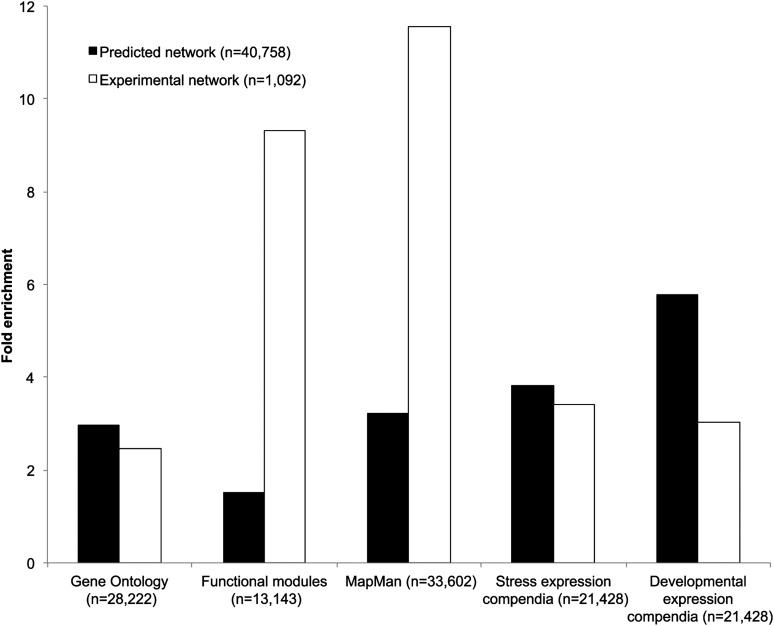
To get an overview of how transcriptional regulation is organized on a genome-wide level, motif information was combined with our CNSs to construct a gene regulatory network (GRN) containing 40,758 interactions (see Methods). This GRN includes 157 TFs that, based on conserved binding sites, have one or more target genes and covers 11,354 genes in total (Supplemental Data Set 4). On average, a TF in the predicted network has 259 target genes while each target gene is regulated by four TFs. The number of target genes per TF and their associated Gene Ontology (GO) enrichment can be seen in Supplemental Figure 7. For these interactions, 64.6% of the conserved binding sites are overlapping with a leaf or flower DH site. To evaluate our network, we used an experimental GRN of 1092 confirmed interactions derived from AtRegNet (Davuluri et al., 2003) and a collection of regulatory interactions obtained from small-scale studies concerning secondary cell wall metabolism (Hussey et al., 2013). Overlap analysis between the predicted network and the experimental network revealed that edges present in the predicted network are significantly more likely to also be present in the experimental network than would be expected by chance (4.65-fold enrichment, P value < 0.001; see Methods). Apart from comparing the global overlap between both networks, we also assessed the overlap between the predicted and experimental TF-target interactions for individual TFs for which motif information was available. For a subset of TFs with 10 or more known target genes, a significant overlap was found for nine out of 13 TFs (P value < 0.001), which covers 99 out of 385 (26%) experimentally determined gene regulatory interactions.
To evaluate which role intronic regions have in transcriptional gene regulation through TF binding, an intron-specific GRN was generated. This network consists of 2821 interactions between 123 TFs and 1552 target genes. Six out of the 99 experimentally confirmed interactions that were retrieved were unique to this network (Supplemental Data Set 5). Examples of correctly inferred intron interactions are binding events of AP2 and LFY to the intron of AGAMOUS (AG) (Hong et al., 2003). Similarly, TF-miRNA regulation was studied by constructing a small subnetwork containing 24 TF-miRNA targets for 14 TFs and 10 target miRNAs (Supplemental Data Set 6). One of the retrieved interactions is the known binding of the ABRE BINDING FACTOR1 (ABF1) to the promoter of mir168a (Li et al., 2012). Another interesting, however unconfirmed, interaction is that between AP2 and mir167a, the latter which is known to play a role in flowering maturation (Rubio-Somoza and Weigel, 2013).
In addition to the recovery of known regulatory interactions, the biological relevance of the predicted target genes was studied using five independent biological data sets. GO (Ashburner et al., 2000), MapMan (Thimm et al., 2004), and functional gene modules (Heyndrickx and Vandepoele, 2012) describe functional annotations and were used to assess if target genes of the same TF participate in similar biological processes or have similar functions. The functional modules comprise a set of 13,142 genes (1562 modules) annotated with specific functional descriptions based on experimental GO information, protein-protein interaction data, protein-DNA interactions, or AraNet gene function predictions. The evaluation of our GRN is made based on the assumption that a set of true target genes of a TF will have a higher enrichment for functional annotations than randomized networks (Marbach et al., 2012). For each TF, the enriched functional annotations were determined and compared against that of randomized networks (see Methods). Next to the three functional data sets, two general gene expression compendia were used, stress and development (De Bodt et al., 2012), to investigate if genes targeted by the same TFs (called coregulated targets) are more likely to be expressed at similar developmental stages or under similar stress conditions. Following Marbach et al. (2012), coregulated gene pairs are defined as genes having 50% or more shared regulators. The average level of coexpression was calculated using correlation analysis for all coregulated gene pairs and compared with that of randomized networks (see Methods). All five biological metrics were performed on the CNS-based GRN as well as on the experimental GRN, and we observed that both networks were significantly enriched for all five biological data sets (P value < 0.05; Figure 4). A detailed comparison revealed that GO fold enrichment was higher in the predicted network. Although the opposite is true for both MapMan and the functional modules, there is still a significant enrichment in our predicted GRN, illustrating the functional coherence of the predicted target genes. The discrepancy between different functional annotation data sets can largely be explained by the fact that for GO annotations a filtering step using GO slim terms was performed in order to have sufficient annotations for all genes in the network. These terms are very broad and as such enrichment will be lower compared with the two other functional classification data sets. Based on the stress and development expression data sets, a higher level of coexpression was observed for coregulated genes in the predicted and experimental GRN, compared with random GRNs (Figure 4). The CNS-based network outperformed the experimental network, as the fold enrichments were higher for the predicted GRN in both expression data sets. A similar evaluation was performed on two subsets of the predicted network, which were defined based on the number of species in which a regulatory interaction is conserved. The predicted network was divided into a highly (conservation CNS more than six species) and a moderately conserved (conservation CNS two to six species) subnetwork. Both the highly and the moderately conserved subnetworks showed significant enrichment for coexpression and functional coherence, indicating that CNSs with support from a lower number of species are also biologically meaningful (Supplemental Figure 8).
Figure 4.
Evaluation of the Biological Relevance of the Predicted Network Using Different Biological Metrics Assessing Functional and Expression Coherence.
GO annotations, MapMan annotations, and functional modules together with a stress and developmental expression compendium were used to evaluate the biological relevance of the predicted GRN. A comparison of fold enrichment is depicted between the predicted network (black bars) and the experimental network (white bars). All reported fold enrichments are significant (P value < 0.05). Numbers in parentheses report the number of regulatory interactions in the two networks and the number of genes having functional or expression information, respectively.
Combining the CNS-Based Network with Expression Information to Identify Condition-Specific Gene Regulatory Interactions
To investigate the biological role of the predicted GRN, the static gene regulatory interactions were converted into condition-specific interactions through the integration of expression information. Coexpression was determined between a TF and each predicted target gene based on 11 expression compendia from the CORNET database (De Bodt et al., 2012), comprising gene expression profiles from microarray experiments performed for different organs (flower, leaf, root, and seed), during development, under different treatments and stresses (hormone, biotic, and abiotic stress) (see Methods). Coexpression between a TF and a predicted target gene can act as a proxy for regulation as both are frequently expressed in the same conditions (Ma and Wang, 2012). A total of 6957 interactions between a TF and its predicted target genes showed significant coexpression in one or a maximum of three expression compendia (Supplemental Data Set 7). Examples of specific coexpression patterns of predicted TF-target interactions that are confirmed by experimentally confirmed target genes include interactions for MYB DOMAIN PROTEIN58 (MYB58) under biotic stress, MYB83 in leaf and for AP2 and ELONGATED HYPOCOTYL5 (HY5) under abiotic and biotic stress. MYB63 shows coexpression of target genes in five different compendia, including (a)biotic stress and hormone (Supplemental Figure 9). The following paragraphs highlight examples of condition-dependent GRNs.
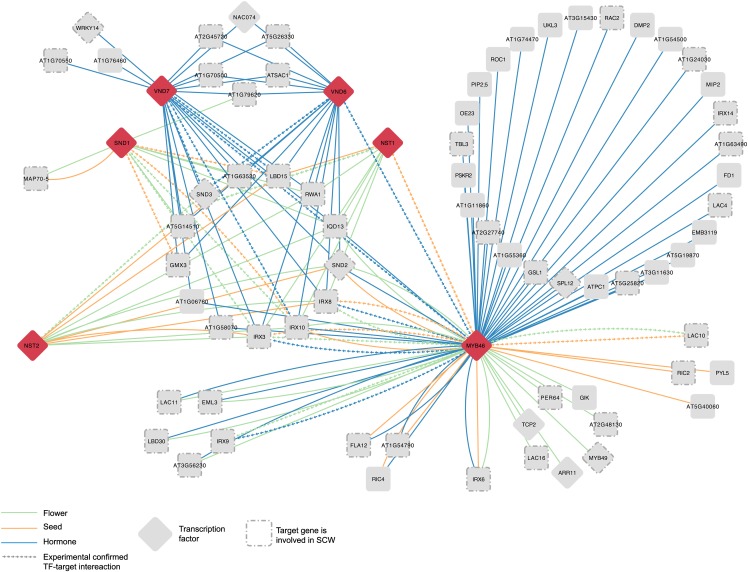
Five secondary wall NAM-ATAF1/2-CUC2 (NAC) TFs were selected to illustrate how integrating coexpression information into the predicted GRN can be used for modeling of the transcriptional network in different conditions and plant organs. SECONDARY WALL-ASSOCIATED NAC DOMAIN1 (SND1) is a master transcriptional regulator activating the developmental program of secondary cell wall (SCW) biosynthesis. SND1 and its functionally related homologs NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1), NST2, VASCULAR-RELATED NAC-DOMAIN6 (VND6), and VND7 regulate the same downstream targets in different cell types (Zhong et al., 2008). While SND1 and NST1 activate the SCW biosynthetic program in fibers, VND6 and VND7 specifically regulate SCW biosynthesis in vessels, and NST1 and NST2 act together in regulating SCW biosynthesis in endothecium of anthers (Mitsuda and Ohme-Takagi, 2008; Zhong et al., 2008). These five TFs bind to an imperfect palindromic 19-bp consensus sequence designated as secondary cell wall NAC binding element, (T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T), in the promoters of their direct targets (Zhong et al., 2010). For VND6, an additional binding site has been described (CTTNAAAGCNA) (Ohashi-Ito et al., 2010). Based on the predicted targets of these five TFs, we used the coexpression information to introduce specificity through condition-dependent regulation. For SND1, NST1, and NST2, we studied target genes coexpressed in a flower and a seed expression compendium because of their role in SCW biosynthesis in flower and reproductive organs (Mitsuda and Ohme-Takagi, 2008; Zhong et al., 2008) (Figure 5). Auxin, cytokinin, and brassinosteroids play pivotal roles in xylem vessel formation (Fukuda, 2004), and VND6 and VND7 show elevated expression levels in presence of these three hormones (Kubo et al., 2005). Both TFs reside in the same functional module, which is annotated with the GO term “response to brassinosteroid stimulus” (Heyndrickx and Vandepoele, 2012). Therefore, VND6 and VND7 targets coexpressing in a hormone compendium were selected. For all TFs, predicted target genes were only selected if they were part of a functional module grouping two or more predicted target genes. This network groups five TFs showing 69 condition-specific interactions with 24 target genes (Figure 5). The SCW network contains a large number of experimentally confirmed interactions (14/69) and nearly all genes in the network are involved in SCW metabolism based on GO annotations (21/24). In this network, two TFs, namely, MYB46 and SND3, which are known direct targets involved in the SCW pathway, are present. Interestingly, these genes do not have a coexpression link with SND1 in flower or seed, but a coexpression link is present with NST1, a TF that cooperates with SND1 in SCW biosynthesis in fibers (Zhong et al., 2008). Overexpression of MYB46 leads to activation of the entire SCW biosynthetic program and its coexpressing targets in seed, flower, and hormone expression compendia show a large number of shared targets with the five master regulators as well as a large set of genes involved in SCW biosynthesis (Zhong et al., 2008).
Figure 5.
A Condition-Specific Secondary Cell Wall Gene Regulatory Network.
Nodes and edges depict genes and regulatory interactions, while condition-specific seed, flower, and hormone coexpression edges are shown using orange, green, and blue lines, respectively. Experimentally confirmed interactions are shown using an arrow line. Red diamonds are the source TFs, gray diamonds are target genes that are TFs, and rounded rectangles are other target genes. Target genes with a gray border are known to be involved in secondary cell wall biosynthesis based on GO.
A similar approach was applied to delineate condition-specific targets for AP3 and PI, two TFs that have been shown to act as bifunctional transcription factors in flower development (Wuest et al., 2012). AP3 and PI are necessary for the proper development of the petals and stamens (Jack et al., 1992; Goto and Meyerowitz, 1994). Plant hormones such as jasmonic acid have been shown to play a role in both stamen and petal development (Brioudes et al., 2009; Song et al., 2013). The expression data for these two TFs shows induction in jasmonic acid treatment conditions. Therefore, coexpressed target genes in the hormone expression compendium were selected. This approach resulted in a hormone-specific GRN with 223 target genes and 237 interactions. The network shows a strong enrichment for genes involved in flower development (53/223) (Supplemental Figure 10). Additional evidence for the relevance of this network was generated through integrating ChIP-Seq and differential gene expression data. The ChIP and differential expression experiments were performed at the early-intermediate floral stage (stage 4 to 5 flowers) (Wuest et al., 2012). In this network, we observe 11 interactions that are confirmed through binding of the TF in the ChIP-Seq data and six interactions that are confirmed through differential expression of the gene after TF perturbation. Interestingly, AG is a predicted coexpressed target gene of AP3 in the hormone-specific network and AG has been shown to be involved in stamen development through regulation of jasmonic acid biosynthesis genes (Ito et al., 2007).
DISCUSSION
In this study, we developed a phylogenetic footprinting approach to identify conserved noncoding sequences in Arabidopsis through the comparison with 11 dicot genomes. Distantly related species were used based on the premise that, in comparison to one another, all noncoding regions that are not under functional constrained will have undergone one or more mutations. A set of 69,361 CNSs associated with 17,895 genes was delineated through the combination of an alignment-based and a non-alignment-based approach. Twenty-eight percent of the CNSs were found downstream of genes, in introns or more than 1 kb upstream of a gene, indicating that regulatory elements are not restricted to the first hundreds of base pairs upstream of a gene (Reineke et al., 2011; Korkuc et al., 2014).
A previous evaluation study reported that phylogenetic footprinting in plants works best by comparing genomes that have diverged less than 100 million years ago or have nonsaturated substitution patterns (Reineke et al., 2011). Phylogenetic footprinting methods that use genome synteny inferred through genome alignments as primary source of orthology information indeed have difficulties integrating distantly related genomes (Hupalo and Kern, 2013). This is due to the frequent nature of polyploidy and genome rearrangements in dicot plants (Supplemental Figure 1) causing problems for global genome alignment methods. Here, a combination of different gene orthology prediction methods was used that do not rely on synteny information. As such, our approach is well suited to incorporate more distantly related species including many-to-many gene orthology relationships. Our alignment-based approach is best summarized as a multiple local alignment strategy, since first local pairwise alignments are identified which are subsequently aggregated on the Arabidopsis reference genome in order to obtain multispecies footprints. We demonstrated that this approach is very suitable for detecting CNSs over large phylogenetic distances, as half of our CNS are conserved in six or more species, spanning >100 million years of evolution (Figure 1B). Furthermore, approaches based exclusively on pairwise alignments lack the power to detect a large set of our CNSs over a similar evolutionary distance (Reineke et al., 2011; Baxter et al., 2012).
Comparing our CNSs with the experimental AtProbe benchmark data set showed that both alignment and non-alignment-based approaches have a similar performance, recovering 19% of the experimental regulatory elements. Both approaches are complementary as they together recovered 26% of the AtProbe elements. This is largely explained by the fact that the alignment-based approach identifies large conserved regions, typically covering clusters of individual TFBS, whereas the non-alignment-based approach will also identify short conserved motifs. Based on a comparison of our footprints with three recently published studies (Baxter et al., 2012; Haudry et al., 2013; Hupalo and Kern, 2013), 64% of our CNSs represent newly discovered constrained sequences. This finding is in agreement with Haudry et al. (2013) who found that their CNSs show limited conservation outside the Brassicaceae lineage. Compared with Baxter et al. (2012) and Hupalo and Kern (2013), both the number of comparator species as well as the different alignment strategy contribute to the difference in identified CNSs. Comparison with the three previously published CNS data sets revealed that our CNSs have the highest enrichment for experimentally determined regulatory elements. Haudry et al. (2013) recovered a larger number of bases covered by CNSs with a lower enrichment toward the AtProbe elements. Although these results could indicate that their higher coverage is associated with a reduced specificity, additional explanations can be formulated. As demonstrated by Haudry et al. (2013), their CNSs also contain other types of functional noncoding sequences, such as RNA genes, which are not accounted for in our benchmark. CNSs could also cover long-range enhancers. Also, the conservation of functional noncoding sequences is likely greater within the Brassicaceae lineage due to more specialized developmental processes and adaptation to environmental conditions, whereas our set of CNSs covers the regulation of processes that are highly conserved across a wide range of dicot plants. A subset of the AtProbe regulatory elements recovered was unique to this analysis, corroborating the complementarity of our CNSs with these previous studies.
The biological relevance of our CNSs was further evaluated by overlap analysis with a number of different chromatin modification marks. Enrichment analysis showed that our CNSs are highly enriched for DH sites as well as for histone marks promoting transcription, indicating that our CNSs are located within open chromatin regions or nearby actively transcribed regions. Processing of 15 TF ChIP-chip/seq experiments together with the corresponding transcriptome profiling studies after TF perturbation generated a high-quality data set of 2807 in vivo functional binding sites. In total, 28% of these regions were successfully recovered. Mapping the position count matrices for all 15 TFs genome-wide and retaining only instances overlapping with a CNS proved to be more specific for recovering functional binding sites compared with filtering using DH sites. In contrast to simple motif mapping approaches that are associated with high false positive rates, computationally identified CNSs as well as experimental DH sites offer two complementary data sources to start performing systematic regulatory genome annotation in plants. The largest bottleneck for identifying all functional regions through conservation analysis is caused by the highly degenerative nature of certain binding sites, such as CArG boxes for AP1 and AP3 [CC(A/T)6GG] (Riechmann et al., 1996). The algorithm developed in the current study will not detect these binding sites as significantly conserved because these sites will have high conservation scores in both the real and control run. Another explanation for the low recovery of functional binding sites for some TFs is the fact that the position count matrices that are used to evaluate conservation in the orthologous regions of distantly related organisms might be too specific for Arabidopsis, making it more difficult to identify conserved instances. Finally, in some cases, a regulatory interaction might be species or clade specific, making comparative methods impractical. Overlap analysis of the recovered in vivo binding sites elements with CNSs from the three other studies showed that 52.3% of the 787 recovered functional regions were uniquely discovered by our approach.
Whereas several studies reporting plant CNSs have suggested different lines of evidence to indicate that sequence conservation implies functional conservation and a role for CNSs in transcriptional regulation (Kaplinsky et al., 2002; Guo and Moose, 2003; Inada et al., 2003; Baxter et al., 2012; Haudry et al., 2013; Hupalo and Kern, 2013), their success in inferring regulatory networks has been hampered by the difficulty of converting CNSs into TF-target interactions. Based on different publicly available databases and ChIP studies, TFs for which motif information was available were integrated with the CNSs to generate a gene regulatory network containing 40,758 TF-target interactions. Overlap analysis with an experimental GRN containing 1092 confirmed regulatory interactions showed that the predicted network is highly enriched for experimental edges. In addition, the functional and expression coherence of the target genes in the different GRNs was evaluated by integrating five different biological data sets. Application of these different validation metrics to the experimental and predicted network was used to assess the functional and coregulatory properties of the different TF-target interactions. Whereas both GRNs showed significant enrichment for all biological data sets, the predicted network outperformed the experimental network for the stress and developmental expression compendia and also for GO functional annotations. Application of the coexpression metric on two subnetworks with edges supported by CNSs showing conservation in a different number of species revealed that regulatory interactions with lower species support are also biologically relevant. Although the predicted GRN, like the experimental network, lacks many true regulatory relationships, comparison with experimentally validated targets as well as validation through the different biological data sets showed that the predicted network is of high overall quality. Compared with the experimental network, where each TF regulates on average 12 target genes, our GRN predicts on average 20 times more target genes for 157 TFs. As our GRN likely identifies many true interactions, which have not been detected and validated experimentally, it provides an important step forward toward the systematic regulatory annotation of individual genes.
A subnetwork containing unique regulatory interactions based on intronic CNSs recovered a small subset of experimental interactions, confirming that intronic regions also play an important role in transcriptional regulation in plants. The TF-miRNA network contained only 24 TF-miRNA interactions, for which one previously described interaction between ABF1 and mir168a could be confirmed. A major challenge for phylogenetic footprinting of miRNA genes and the construction of miRNA GRNs is the lack of miRNA orthology information across a number of related species, which is a prerequisite for most phylogenetic footprinting methods.
Although the predicted GRN offers additional information on the transcriptional regulators of individual target genes, the static nature of these CNS-based interactions offers few insights about the biological context of these regulatory events. We demonstrated how integrating expression data for different organs and conditions with the predicted interactions through coexpression analysis provides an effective approach to obtain condition-specific networks. Based on 11 compendia containing gene expression profiles in different biological contexts, we identified 6597 regulatory interactions where a TF specifically coexpressed with its target gene in one or a few conditions. As shown for the secondary cell wall and AP3/PI networks, this coexpression information can be used to filter the set of predicted interactions and to identify previously unknown target genes as well as new regulators acting downstream of the TF under investigation. Furthermore, for different TFs and signaling cascades, it also becomes possible to investigate how the transcriptional regulation of some direct target genes changes in different conditions while other targets show constitutive coexpression.
Apart from integrating sequence conservation and expression information, other approaches combining complementary functional data sets may improve the power to correctly identify regulatory interactions. For example, the incorporation of additional regulatory information such as differentially expressed genes from TF perturbation experiments or genomic regions marked with transcription-promoting chromatin modifications can offer new ways to identify functional target genes. With the advent of TF binding data from protein binding microarray experiments for an increasing number of TFs (Franco-Zorrilla et al., 2014; Lindemose et al., 2014) our CMM approach combined with coexpression analysis offers a practical means to convert in vitro TF binding information from protein binding microarrays into functional and condition-specific GRNs.
METHODS
Sequence and Orthology Information
The 12 dicotyledonous genomes used in this study were Arabidopsis thaliana (TAIR10), Carica papaya (Hawaii Agriculture Research Center), Glycine max (JGI 1.0), Malus domestica (IASMA), Populus trichocarpa (JGI 2.0), Fragaria vesca (Strawberry Genome 1.0), Medicago truncatula (Mt 3.5), Lotus japonicus (Kazusa 1.0), Theobroma cacao (CocoaGen v1.0), Ricinus communis (JCVI 1.0), Manihot esculenta (Cassava4), and Vitis vinifera (Genoscope_v1) and were obtained from the PLAZA 2.5 database (Van Bel et al., 2012). The structural annotation of the genomes in PLAZA 2.5 was updated by adding all known miRNAs obtained from the plant microRNA database (Zhang et al., 2010). miRNA sequences were downloaded from plant microRNA database and mapped to the genomes using BLASTN (Altschul et al., 1990) and GenomeThreader (-mincoverage 0.89 -minalignmentscore 0.95) (Gremme et al., 2005) and only unique mappings were retained. The overlap with existing RNA gene annotations in PLAZA 2.5 and the database was determined by using BLASTN (e-value < 1e-10) against all transcripts, and only RNA genes lacking overlap with already annotated loci were added. In total, 791 new miRNA loci were added in Arabidopsis and 20% of all miRNAs have orthologs in one or more related dicot genome.
Three sequence types, upstream, downstream, and intronic, were used to identify CNSs. Upstream sequences were restricted to the first 1000/2000 bp upstream from the translation start site or to a shorter region if the adjacent upstream gene is located within a distance smaller than 1000/2000 bp (n = 33,703). The 1000- and 2000-bp upstream sequences were processed as two independent runs. Downstream sequences were restricted to the first 1000 bp downstream from the stop codon or to a shorter region if the adjacent downstream gene was within 1000bp (n = 33,809). The intronic sequence type is defined as the complete gene locus with exons masked (n = 20,608).
Orthologs for each Arabidopsis gene were determined in 11 comparator dicot species using the PLAZA Integrative Orthology method (Van Bel et al., 2012). The included orthology detection methods are OrthoMCL (Li et al., 2003), phylogenetic tree-based orthologs, and BHIF. Through Ks graphs in the PLAZA 2.5 platform, we confirmed that all included dicot species have saturated substitution patterns (mean Ks > 1) when comparing orthologous gene pairs with Arabidopsis (Van Bel et al., 2012).
Synteny Conservation
Orthologs were determined for each Arabidopsis protein-coding gene using the PLAZA Integrative Orthology method requiring that the orthology prediction is supported by at least two detection methods. The conservation of the orthologous relationship for the flanking gene upstream and downstream of each ortholog was determined for each of the comparator species.
Comparative Motif Mapping
Known motifs were mapped on the regions covered for the three sequence types for all included species using DNA-pattern allowing no mismatches (Thomas-Chollier et al., 2008). A total of 692 cis-regulatory elements were obtained from AGRIS (Davuluri et al., 2003), PLACE (Higo et al., 1999), and Athamap (Steffens et al., 2004). In addition, 44 positional count matrices were obtained from Athamap and for 15 TFs positional count matrices were obtained from ChIP-Seq data (see section “ChIP-Seq in Vivo Targets”). Positional count matrices were mapped genome-wide using MatrixScan using a P value cutoff <1e-05 (Thomas-Chollier et al., 2008).
For each Arabidopsis gene and per sequence type, a conservation score SCMM is determined per motif. The SCMM is calculated as the number of species in which this motif was conserved in an orthologous family context. The statistical significance of each motif with SCMM was tested through a comparison with the SCMM derived from 1000 random gene families that have the same number of orthologs and species but are lacking an orthologous relationship to the query gene. Evaluation of the statistical significance using larger sets of random families (1000 to 100,000) confirmed that the P values obtained using 1000 nonorthologous families are robust.
The FDR was calculated through a control experiment in which the entire analysis, including all Arabidopsis genes, was performed using nonorthologous genes. For each query gene, a family was randomly assembled sampling nonorthologous genes, but maintaining the number of genes and the species composition of the real orthologous family. The real and control run were compared, and footprints in the real run with a P value that corresponds to a FDR ≤ 10% were retained.
Alignment-Based Phylogenetic Footprinting
Pairwise alignments were generated between all Arabidopsis query genes and their orthologous genes for all three sequence types and two orthology definitions. ACANA and DIALIGN-TX were run with standard parameters. Seaweeds was run with the step size parameter set to 1 and window size to 60 and 30 bp (referred to as Seaweeds-60 and Seaweeds-30, respectively), and only alignments with an alignment score higher than 40 and 20, respectively, were retained. Sigma was run with the -x parameter set to 0.5.
All pairwise alignments were aggregated on the query sequence generating a multispecies conservation plot that shows for each position of the investigated region how many species support this nucleotide through pairwise footprints. All footprints for each level of conservation are extracted from the multispecies conservation plot and each footprint is defined by its length and a multispecies level conservation score SMSP, which denotes the number of comparator species supporting that footprint.
For each alignment tool and sequence type, a precomputed pairwise background library, including >25 million alignments, was used to determine significant conservation of footprints. The background model was created by binning all investigated regions of all species on length, selecting 150 genes from each bin and making pairwise alignments for all possible length bin combinations. The reasoning behind this binning approach is that we wanted to compare the investigated region of the query gene with a background model consisting of genes that have regions of similar size. For each Arabidopsis gene, 1000 nonorthologous (random) gene families with the same species and ortholog composition as the query gene were generated and their pairwise alignments were obtained from the background library. Multispecies conservation is calculated for each family and the footprints obtained from all random families are binned on length. Each bin needs to contain at least 1000 multispecies footprints together with their associated scores; otherwise, one or more subsequent bins (with greater lengths) were added. Finally, the statistical significance of each real footprint was then evaluated by counting the number of footprints in random families that have an equally good or better SMSP in the associated background length bin. Comparison of results between using a background library and generating these random families on-the-fly for each gene has pointed out that the results are not altered but processing time is greatly improved. Again, the real and control run were compared and footprints in the real run with a P value that corresponds to a FDR ≤ 10% were retained.
Browsing Results in GenomeView
The complete set of CNSs, overlapping known motifs and DH sites can be browsed through the link http://bioinformatics.psb.ugent.be/cig_data/Ath_CNS/Ath_CNS.php. While loading, when asked, the file format needs to be specified to BED format.
Protein-Coding Potential of CNSs
The coding potential of a CNS was determined using BLASTX (Altschul et al., 1990) against the PLAZA 2.5 protein database (780,667 proteins from 25 Viridiplantae species), and all significant hits were removed. To establish an appropriate e-value cutoff for a significant hit, we randomly permuted each sequence in our CNS data set and performed the BLASTX search using this set of sequences to obtain the distribution of e-values for random sequences with the same length distribution (Baxter et al., 2012). We then performed the same BLASTX search on the real sequences, using the minimum e-value from the random set (e-value < 0.001) as the cutoff for a significant hit.
Overlap of CNSs with Benchmarks
Our CNS data set was compared with different functional data sets. The first one was the Arabidopsis promoter binding element database (AtProbe) (http://exon.cshl.org/cgi-bin/atprobe/instance.pl), which contains 172 experimentally determined regulatory sequences in 76 Arabidopsis genes. This data set was curated by removing results from promoter deletion experiments and CREs for which mapping data was not correct with the coordinates in the data set, resulting in a data set of 144 CREs present in 63 genes (Supplemental Data Set 1). The benchmark data set was formatted as a BED file and the overlap (recovery of elements) was determined using the BEDTools function intersectBed with –u parameter and the –f parameter on 0.5 (Quinlan and Hall, 2010). This means that an experimental CRE was considered “correctly identified” if more than half of the region was overlapping with a CNS. CNS data sets from three recent studies were obtained through the UCSC genome browser at http://genome.genetics.rutgers.edu/ (table top10conserved) from Hupalo and Kern (2013), the authors of the CNS data of Arabidopsis from Haudry et al. (2013), or were assembled from supplementary data (Baxter et al., 2012). These files were also formatted as BED files and compared with the AtProbe benchmark. False positives were determined by shuffling the AtProbe data set 1000 times using shuffleBed, excluding coding sequences and the actual AtProbe instances. The overlap with CNS files was determined for each shuffled file and the median number of recovered elements over 1000 shuffled files was used as a measure for false positives. This estimation of false positives was used to calculate the fold enrichment, defined as the ratio between observed overlap and expected overlap by chance.
A list of 2807 in vivo functional targets was assembled from genes that were annotated to a TF ChIP-Seq peak in noncoding DNA in which a DNA motif was significantly enriched and that show regulatory response in the corresponding TF perturbation experiment (Supplemental Data Set 2). Overlap and enrichment for in vivo functional targets was determined in the same way as for the AtProbe benchmark. For DH site and histone modification data sets, the number of overlapping CNSs was also determined using BEDTools. Enrichment of our CNS data set for these marked chromatin regions was determined as described above.
Detection of DNase I Hypersensitive Sites and Histone Modifications
The BED files with the flower and leaf DH sites and histone modification data sets (H3K4me3, H3K4me2, and H3K9ac) were downloaded from the SRA database (Luo et al., 2012; Zhang et al., 2012). For the histone modification data sets, the reads were mapped to the unmasked TAIR10 reference genome of Arabidopsis (TAIR10_chr_all.fas; ftp.arabidopsis.org) using CLC assembly cell 4.2.0 with -c parameter for colorspace reads and -r to ignore redundant reads. Peak calling was performed using DFilter 1.0 with -std 2 (Kumar et al., 2013).
ChIP-Seq in Vivo Targets
For the ChIP-Seq data sets (PIF4, PIF5, AP1, AP2, FLC, FHY3, PRR5, AP3, PI, and PIF3), raw reads were downloaded from the SRA database (Kaufmann et al., 2010; Yant et al., 2010; Deng et al., 2011; Ouyang et al., 2011; Hornitschek et al., 2012; Nakamichi et al., 2012; Oh et al., 2012; Wuest et al., 2012; Zhang et al., 2013)). The quality of the raw data was checked with FASTQC (v0.10.0; http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/). Adaptors and other overrepresented sequences were removed using the fastx-toolkit (v0.0.13; http://hannonlab.cshl.edu/fastx_toolkit/). The reads were mapped to the unmasked TAIR10 reference genome of Arabidopsis (TAIR10_chr_all.fas; ftp.arabidopsis.org) using BWA with default settings for all parameters (v0.5.9; Li and Durbin, 2009). Reads that could not be assigned to a unique position in the genome were removed using samtools (v0.1.18; Li et al., 2009) by setting the mapping quality threshold (-q) at 1. Redundant reads were removed, retaining only one read per start position, using Picard tools (v1.56; http://picard.sourceforge.net). Peak calling was performed using MACS (v2.0.10; Zhang et al., 2008). The genome size (-g) was set at 1.0e8, and the FDR cutoff was set at 0.05. Other parameters were set at their default values.
For the ChIP-chip data (BES1, SOC1, AGL15, LFY, and FUS3) (Zheng et al., 2009; Winter et al., 2011; Yu et al., 2011; Tao et al., 2012; Wang and Perry, 2013), raw CEL files were downloaded from GEO. The Affymetrix Tiling array bpmap files were updated to the current TAIR10 annotation with Starr (Zacher et al., 2010). Peak calling was performed with rMAT (Droit et al., 2010). The PairBinned method was used to normalize the arrays. Peaks were called using a FDR cutoff of 0.05 except for the data sets BES1 and FUS3 in which the P value was set at 10−3 (in analogy to the original study and necessary to obtain peak calling results). The minimum requirement of consecutive enriched probes was set at of eight. Other parameters were left at their default setting.
Peak regions were annotated based on the location of their summits as determined by MACS. A peak was assigned to the closest gene as annotated in the TAIR10 release present in the PLAZA2.5 database (Van Bel et al., 2012). Both upstream, intron, and downstream regions of the peak were taken into account. The complete (exon-masked) peak regions were submitted to the Peak-Motifs algorithm using default settings (Thomas-Chollier et al., 2012). The P value for motif enrichment in the peak set compared with the genomic background was calculated by mapping the motifs using matrix-scan (Turatsinze et al., 2008) (using the same default parameters of Peak-Motifs) in 1000 random sets of peaks of the same size and length distribution sampled without replacement from the complete intergenic genome space. Only motifs with significant enrichment (P value < 0.05) toward peak regions for a specific TF were retained. Lists of differentially expressed genes following perturbation of the TF were gathered from their respective publications (for SOC1, the original study describing the data was Seo et al., 2009).
Construction and Analysis of a CNS-Based Gene Regulatory Network
Based on the known motifs compiled from the different databases and literature (see section “Comparative Motif Mapping”), we retained 157 TFs for which specific motif information was available. A conserved gene regulatory network was created with intersectBed (-f parameter was set to 1 demanding complete motif presence in the conserved region, -u parameter was also used), which determined the overlap between a BED file containing all CNSs, together with their associated genes, and BED files with genome-wide occurrences of the motifs of all 157 TFs. Although in most cases experiments have confirmed the specificity of the association between a TF and its binding site, we cannot exclude that predicted target genes identified through a CNS are regulated by a member of the same TF family. Overlap between the predicted GRN and the experimental network (n = 1092) was evaluated by counting how may TF-target interactions from the experimental network were present in the predicted network and enrichment between two networks was defined as the number of interactions that are present in both networks divided by the number of interactions expected by chance. The number of common interactions expected by chance is given by the mean of the hypergeometric distribution: N1*N2/T, where N1 and N2 are the number of interactions in the two networks, and T is the total number of possible interactions. Statistical significance of the observed number of overlapping edges was evaluated using the hypergeometric distribution (Marbach et al., 2012). Overlap was also determined per TF, demanding that a TF had at least 10 target genes.
Functional enrichment was determined for each network by using five biological data sets, including three functional data sets, Gene Ontologies (Ashburner et al., 2000), MapMan (Thimm et al., 2004), functional modules (Heyndrickx and Vandepoele, 2012) as well as two expression data sets, a stress expression compendia (336 microarray experiments) and a developmental expression compendia (135 microarray experiments) (De Bodt et al., 2012).
For the functional annotation data sets, the enrichment of functional terms was determined within the set of target genes for each TF through the hypergeometric distribution with Bonferroni correction. A enrichment score (-log(p-value)*fold enrichment) was created for each significantly enriched term and the average of all enrichment scores within the network was determined. For GO, only GO slim terms were taken into account. For the expression data sets, a gene pair was considered to be coregulated in the given network if the two genes had >50% of their regulators in common. These gene pairs were identified by computing the Jaccard similarity coefficient between the set of regulators of the first gene and the second gene. For each coregulated gene pair, we then measured the similarity of the expression profile between both genes using the Pearson correlation coefficient. Finally, the biological similarity was summarized by taking the average over all coregulated gene pairs. For both functional annotation and expression data sets, the same procedure was repeated for 100 randomized versions of the network, and fold enrichment was computed as the ratio of the average functional enrichment score, or average Pearson correlation coefficient, of the original network to the average of the randomized networks. Network randomization was done by permuting the labels of all TFs and permuting the labels of all genes, which preserves the network structure. This assures that the observed enrichment is not due to potential biases arising from structural properties of the network. Statistical significance was assessed at a level of 0.05 using a one-sided Wilcoxon rank-sum test to compare the functional enrichment scores or Pearson correlation coefficient from the original network with a random sample from the randomized networks that has the same size as the real set of scores (Marbach et al., 2012). P values obtained using 100 randomizations were identical to those from obtained through 1000 randomizations.
Construction and Analysis of Condition-Specific GRNs
Coexpression was determined between all TFs and target genes using the Pearson correlation coefficient based on 11 CORNET expression compendia: abiotic stress TAIR10 (256 exp), biotic stress TAIR10 (69 exp), microarray compendium 2 TAIR10 (111 exp), development TAIR10 (135 exp), flower TAIR10 (72 exp), hormone treatment TAIR10 (140 exp), leaf TAIR10 (212 exp), root TAIR10 (258 exp), seed TAIR10 (83 exp), stress (abiotic+biotic) TAIR10 (336 exp), and whole plant TAIR10 (85 exp) from De Bodt et al. (2012). A Z-score transformation of correlation coefficients was performed in order to determine significant coexpression. A TF-target interaction was deemed significantly coexpressing if the Z-score was greater or less than 2. Only TF-target interactions that showed significant coexpression in less than four compendia were used as an additional filter to obtain specificity. This threshold was selected due to the presence of three stress-related compendia.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource database under the following accession numbers: AGL15 (AT5G13790), AP1 (AT1G69120), AP2 (AT4G36920), AP3 (AT3G54340), SOC1 (AT2G45660), PI (AT5G20240), LFY (AT5G61850), FLC (AT5G10140), PRR5 (AT5G24470), PIF3 (AT1G09530), PIF4 (AT2G43010), PIF5 (AT3G59060), FHY3 (AT3G22170), BES1 (AT1G19350), FUS3 (AT3G26790), AG (AT4G18960), ABF1 (AT1G49720), mir168a (AT4G19395), mir167a (AT3G22886), MYB58 (AT1G16490), MYB83 (AT3G08500), HY5 (AT5G11260), MYB63 (AT1G79180), SND1 (AT1G32770), NST1 (AT2G46770), NST2 (AT3G61910), VND6 (AT5G62380), VND7 (AT1G71930), MYB46 (AT5G12870), and SND3 (AT1G28470). Next-generation sequence data used in this article can be found in the GenBank Sequence Read Archive (SRA)/Gene Expression Omnibus (GEO) database under the following accession numbers: DH sites (SRP009678), H3K4me3, H3K4me2, and H3K9ac (GSE28398), PIF4 (SRP010570), PIF5 (SRP010315), AP1 (SRP002174), AP2 (SRP002328), FLC (SRP005412), FHY3 (SRP007485), PRR5 (SRP011389), AP3 and PI (SRP013458), PIF3 (SRP014179), BES1 (GSE24684), SOC1 (GSE33297), AGL15 (GSE17717), LFY (GSE28063), and FUS3 (GSE43291).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Overview of Synteny Conservation between Arabidopsis thaliana and Other Dicot Species.
Supplemental Figure 2. Distribution of Genes That Have Orthologs in the Dicot Comparator Species for Each Orthology Detection Method.
Supplemental Figure 3. Recovery of Experimental AtProbe Elements Using Different Phylogenetic Footprinting Approaches.
Supplemental Figure 4. Recovery of AtProbe Elements for the CNSs Described in This Paper and by Haudry et al. (2013).
Supplemental Figure 5. Enrichment and Overlap of in Vivo Functional Regions with CNSs.
Supplemental Figure 6. Comparison of Fold Enrichment for in Vivo Functional Binding Site Regions.
Supplemental Figure 7. GO Enrichment for All TF Targets in the Predicted GRN.
Supplemental Figure 8. Evaluation of the Biological Relevance of Highly and Moderately Conserved Interactions Using the Biological Validation Metrics.
Supplemental Figure 9. Comparison between Gold Standard and Predicted GRN of Coexpressed Target Genes in Different Conditions.
Supplemental Figure 10. A Condition-Specific GRN for PI and AP3 Based on Hormone-Specific TF Target Coexpression Edges.
Supplemental Data Set 1. AtProbe Experimentally Determined cis-Regulatory Elements.
Supplemental Data Set 2. High-Quality Benchmark Data Set of ChIP Bound and Motif Enriched Regions Associated to Differentially Expressed Genes.
Supplemental Data Set 3. Overview of the Enrichment Analysis Results for the Conserved versus Simple Motif Approaches.
Supplemental Data Set 4. Gene Regulatory Network with Conservation Scores.
Supplemental Data Set 5. Intron-Specific Gene Regulatory Network with Conservation Scores.
Supplemental Data Set 6. miRNA-Specific Gene Regulatory Network with Conservation Scores.
Supplemental Data Set 7. Condition-Specific Gene Regulatory Network with Pearson Correlation Coefficients.
Acknowledgments
We thank Bram Verhelst for technical assistance during the processing of the miRNA annotations, M. Blanchette for sending us the Arabidopsis CNS data set, and Edward Himelblau for proofreading. This work was supported by the Multidisciplinary Research Partnership “Bioinformatics: from nucleotides to networks” Project (01MR0310W) of Ghent University. K.S.H. and J.V.d.V. are indebted to the Agency for Innovation by Science and Technology in Flanders for a predoctoral fellowship.
AUTHOR CONTRIBUTIONS
J.V.d.V., K.S.H., and K.V.d.V. designed the research methodology, performed data analysis, and wrote the article.
Glossary
- TF
transcription factor
- miRNA
microRNA
- ChIP
chromatin immunoprecipitation
- DH
DNase I hypersensitive
- CNS
conserved noncoding sequence
- TFBS
transcription factor binding site
- CMM
Comparative Motif Mapping
- FDR
false discovery rate
- UTR
untranslated region
- BHIF
best-hit and in-paralogous families
- TPR
true positive rate
- GRN
gene regulatory network
- GO
Gene Ontology
- SCW
secondary cell wall
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Ashburner M., et al. The Gene Ontology Consortium (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter L., Jironkin A., Hickman R., Moore J., Barrington C., Krusche P., Dyer N.P., Buchanan-Wollaston V., Tiskin A., Beynon J., Denby K., Ott S. (2012). Conserved noncoding sequences highlight shared components of regulatory networks in dicotyledonous plants. Plant Cell 24: 3949–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Tompa M. (2002). Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Res. 12: 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Haussler D., Miller W. (2004). Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14: 708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brioudes F., Joly C., Szécsi J., Varaud E., Leroux J., Bellvert F., Bertrand C., Bendahmane M. (2009). Jasmonate controls late development stages of petal growth in Arabidopsis thaliana. Plant J. 60: 1070–1080. [DOI] [PubMed] [Google Scholar]
- Chabouté M.E., Clément B., Philipps G. (2002). S phase and meristem-specific expression of the tobacco RNR1b gene is mediated by an E2F element located in the 5′ leader sequence. J. Biol. Chem. 277: 17845–17851. [DOI] [PubMed] [Google Scholar]
- Davuluri R.V., Sun H., Palaniswamy S.K., Matthews N., Molina C., Kurtz M., Grotewold E. (2003). AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S., Theissen G., Van de Peer Y. (2006). Promoter analysis of MADS-box genes in eudicots through phylogenetic footprinting. Mol. Biol. Evol. 23: 1293–1303. [DOI] [PubMed] [Google Scholar]
- De Bodt S., Hollunder J., Nelissen H., Meulemeester N., Inzé D. (2012). CORNET 2.0: integrating plant coexpression, protein-protein interactions, regulatory interactions, gene associations and functional annotations. New Phytol. 195: 707–720. [DOI] [PubMed] [Google Scholar]
- Deng W., Ying H., Helliwell C.A., Taylor J.M., Peacock W.J., Dennis E.S. (2011). FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 6680–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droit A., Cheung C., Gottardo R. (2010). rMAT—an R/Bioconductor package for analyzing ChIP-chip experiments. Bioinformatics 26: 678–679. [DOI] [PubMed] [Google Scholar]
- Ferrier T., Matus J.T., Jin J., Riechmann J.L. (2011). Arabidopsis paves the way: genomic and network analyses in crops. Curr. Opin. Biotechnol. 22: 260–270. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., López-Vidriero I., Carrasco J.L., Godoy M., Vera P., Solano R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 111: 2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H. (2004). Signals that control plant vascular cell differentiation. Nat. Rev. Mol. Cell Biol. 5: 379–391. [DOI] [PubMed] [Google Scholar]
- Garner M.M., Revzin A. (1981). A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 9: 3047–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K., Meyerowitz E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8: 1548–1560. [DOI] [PubMed] [Google Scholar]
- Gremme G., Brendel V., Sparks M.E., Kurtz S. (2005). Engineering a software tool for gene structure prediction in higher organisms. Inf. Softw. Technol. 47: 965–978. [Google Scholar]
- Guo H., Moose S.P. (2003). Conserved noncoding sequences among cultivated cereal genomes identify candidate regulatory sequence elements and patterns of promoter evolution. Plant Cell 15: 1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håndstad T., Rye M.B., Drabløs F., Sætrom P. (2011). A ChIP-Seq benchmark shows that sequence conservation mainly improves detection of strong transcription factor binding sites. PLoS ONE 6: e18430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, R.S. (2007). Improved Pairwise Alignment of Genomic DNA. PhD dissertation (Pennsylvania State University). [Google Scholar]
- Haudry A., et al. (2013). An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat. Genet. 45: 891–898. [DOI] [PubMed] [Google Scholar]
- He G., Elling A.A., Deng X.W. (2011). The epigenome and plant development. Annu. Rev. Plant Biol. 62: 411–435. [DOI] [PubMed] [Google Scholar]
- Heyndrickx K.S., Vandepoele K. (2012). Systematic identification of functional plant modules through the integration of complementary data sources. Plant Physiol. 159: 884–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong R.L., Hamaguchi L., Busch M.A., Weigel D. (2003). Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15: 1296–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711. [DOI] [PubMed] [Google Scholar]
- Huang W., Umbach D.M., Li L. (2006). Accurate anchoring alignment of divergent sequences. Bioinformatics 22: 29–34. [DOI] [PubMed] [Google Scholar]
- Hupalo D., Kern A.D. (2013). Conservation and functional element discovery in 20 angiosperm plant genomes. Mol. Biol. Evol. 30: 1729–1744. [DOI] [PubMed] [Google Scholar]
- Hussey S.G., Mizrachi E., Creux N.M., Myburg A.A. (2013). Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front. Plant Sci. 4: 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada D.C., Bashir A., Lee C., Thomas B.C., Ko C., Goff S.A., Freeling M. (2003). Conserved noncoding sequences in the grasses. Genome Res. 13: 2030–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Ng K.H., Lim T.S., Yu H., Meyerowitz E.M. (2007). The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell 19: 3516–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T., Brockman L.L., Meyerowitz E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68: 683–697. [DOI] [PubMed] [Google Scholar]
- Jin J., Zhang H., Kong L., Gao G., Luo J. (2014). PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 42: D1182–D1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplinsky N.J., Braun D.M., Penterman J., Goff S.A., Freeling M. (2002). Utility and distribution of conserved noncoding sequences in the grasses. Proc. Natl. Acad. Sci. USA 99: 6147–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muiño J.M., Ferrier T., Wuest S.E., Kumar V., Serrano-Mislata A., Madueño F., Krajewski P., Meyerowitz E.M., Angenent G.C., Riechmann J.L. (2010). Orchestration of floral initiation by APETALA1. Science 328: 85–89. [DOI] [PubMed] [Google Scholar]
- Korkuc P., Schippers J.H., Walther D. (2014). Characterization and identification of cis-regulatory elements in arabidopsis based on single-nucleotide polymorphism information. Plant Physiol. 164: 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H., Demura T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19: 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Muratani M., Rayan N.A., Kraus P., Lufkin T., Ng H.H., Prabhakar S. (2013). Uniform, optimal signal processing of mapped deep-sequencing data. Nat. Biotechnol. 31: 615–622. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C.J., Jr, Roos D.S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Cui X., Meng Z., Huang X., Xie Q., Wu H., Jin H., Zhang D., Liang W. (2012). Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 158: 1279–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemose S., Jensen M.K., de Velde J.V., O’Shea C., Heyndrickx K.S., Workman C.T., Vandepoele K., Skriver K., Masi F.D. (2014). A DNA-binding-site landscape and regulatory network analysis for NAC transcription factors in Arabidopsis thaliana. Nucleic Acids Res., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.X., Liu H.L., Chai Z.J., Xu X.P., Song Y.R., Qu Q. (2010). Evaluation of seed storage-protein gene 5′ untranslated regions in enhancing gene expression in transgenic rice seed. Theor. Appl. Genet. 121: 1267–1274. [DOI] [PubMed] [Google Scholar]
- Luo C., Sidote D.J., Zhang Y., Kerstetter R.A., Michael T.P., Lam E. (2012). Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J. [DOI] [PubMed] [Google Scholar]
- Ma C., Wang X. (2012). Application of the Gini correlation coefficient to infer regulatory relationships in transcriptome analysis. Plant Physiol. 160: 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIsaac K.D., Fraenkel E. (2006). Practical strategies for discovering regulatory DNA sequence motifs. PLOS Comput. Biol. 2: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbach D., Roy S., Ay F., Meyer P.E., Candeias R., Kahveci T., Bristow C.A., Kellis M. (2012). Predictive regulatory models in Drosophila melanogaster by integrative inference of transcriptional networks. Genome Res. 22: 1334–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia-Guerra M.K., Pomeranz M., Morohashi K., Grotewold E. (2012). From plant gene regulatory grids to network dynamics. Biochim. Biophys. Acta 1819: 454–465. [DOI] [PubMed] [Google Scholar]
- Meng X., Brodsky M.H., Wolfe S.A. (2005). A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat. Biotechnol. 23: 988–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Ohme-Takagi M. (2008). NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J. 56: 768–778. [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Kamioka M., Suzuki T., Yamashino T., Higashiyama T., Sakakibara H., Mizuno T. (2012). Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. USA 109: 17123–17128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K., Oda Y., Fukuda H. (2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22: 3461–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X., et al. (2011). Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23: 2514–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot E., Krusche P., Tiskin A., Carré I., Ott S. (2010). Evolutionary analysis of regulatory sequences (EARS) in plants. Plant J. 64: 165–176. [DOI] [PubMed] [Google Scholar]
- Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. (2010). Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke A.R., Bornberg-Bauer E., Gu J. (2011). Evolutionary divergence and limits of conserved non-coding sequence detection in plant genomes. Nucleic Acids Res. 39: 6029–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., Ratcliffe O.J. (2000). A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 3: 423–434. [DOI] [PubMed] [Google Scholar]
- Riechmann J.L., Wang M., Meyerowitz E.M. (1996). DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24: 3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110. [DOI] [PubMed] [Google Scholar]
- Roudier F., Teixeira F.K., Colot V. (2009). Chromatin indexing in Arabidopsis: an epigenomic tale of tails and more. Trends Genet. 25: 511–517. [DOI] [PubMed] [Google Scholar]
- Roulet E., Busso S., Camargo A.A., Simpson A.J., Mermod N., Bucher P. (2002). High-throughput SELEX SAGE method for quantitative modeling of transcription-factor binding sites. Nat. Biotechnol. 20: 831–835. [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I., Weigel D. (2013). Coordination of flower maturation by a regulatory circuit of three microRNAs. PLoS Genet. 9: e1003374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo E., Lee H., Jeon J., Park H., Kim J., Noh Y.S., Lee I. (2009). Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21: 3185–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddharthan R. (2006). Sigma: multiple alignment of weakly-conserved non-coding DNA sequence. BMC Bioinformatics 7: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15: 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Xie D. (2013). Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Mol. Plant 6: 1065–1073. [DOI] [PubMed] [Google Scholar]
- Steffens N.O., Galuschka C., Schindler M., Bülow L., Hehl R. (2004). AthaMap: an online resource for in silico transcription factor binding sites in the Arabidopsis thaliana genome. Nucleic Acids Res. 32: D368–D372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A.R., Kaufmann M., Morgenstern B. (2008). DIALIGN-TX: greedy and progressive approaches for segment-based multiple sequence alignment. Algorithms Mol. Biol. 3: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagle D.A., Koop B.F., Goodman M., Slightom J.L., Hess D.L., Jones R.T. (1988). Embryonic epsilon and gamma globin genes of a prosimian primate (Galago crassicaudatus). Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J. Mol. Biol. 203: 439–455. [DOI] [PubMed] [Google Scholar]
- Tao Z., Shen L., Liu C., Liu L., Yan Y., Yu H. (2012). Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J. 70: 549–561. [DOI] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939. [DOI] [PubMed] [Google Scholar]
- Thomas B.C., Rapaka L., Lyons E., Pedersen B., Freeling M. (2007). Arabidopsis intragenomic conserved noncoding sequence. Proc. Natl. Acad. Sci. USA 104: 3348–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M., Herrmann C., Defrance M., Sand O., Thieffry D., van Helden J. (2012). RSAT peak-motifs: motif analysis in full-size ChIP-seq datasets. Nucleic Acids Res. 40: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M., Sand O., Turatsinze J.V., Janky R., Defrance M., Vervisch E., Brohée S., van Helden J. (2008). RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 36: W119–W127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa M., et al. (2005). Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 23: 137–144. [DOI] [PubMed] [Google Scholar]
- Turatsinze J.V., Thomas-Chollier M., Defrance M., van Helden J. (2008). Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat. Protoc. 3: 1578–1588. [DOI] [PubMed] [Google Scholar]
- Van Bel M., Proost S., Wischnitzki E., Movahedi S., Scheerlinck C., Van de Peer Y., Vandepoele K. (2012). Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 158: 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Casneuf T., Van de Peer Y. (2006). Identification of novel regulatory modules in dicotyledonous plants using expression data and comparative genomics. Genome Biol. 7: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Quimbaya M., Casneuf T., De Veylder L., Van de Peer Y. (2009). Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol. 150: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.T., Xu Y.N. (2010). The 5 ' untranslated region of the FAD3 mRNA is required for its translational enhancement at low temperature in Arabidopsis roots. Plant Sci. 179: 234–240. [Google Scholar]
- Wang F., Perry S.E. (2013). Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 161: 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter C.M., et al. (2011). LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell 20: 430–443. [DOI] [PubMed] [Google Scholar]
- Wuest S.E., O’Maoileidigh D.S., Rae L., Kwasniewska K., Raganelli A., Hanczaryk K., Lohan A.J., Loftus B., Graciet E., Wellmer F. (2012). Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl. Acad. Sci. USA 109: 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T.T., Ott F., Lanz C., Wollmann H., Chen X., Schmid M. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A., Mejia-Guerra M.K., Kurz K., Liang X., Welch L., Grotewold E. (2011). AGRIS: the Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res. 39: D1118–D1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P., Rodermel S., Yin Y. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65: 634–646. [DOI] [PubMed] [Google Scholar]
- Zacher B., Torkler P., Tresch A. (2011). Analysis of Affymetrix ChIP-chip data using starr and R/Bioconductor. Cold Spring Harb Protoc 2011: top110. [DOI] [PubMed] [Google Scholar]
- Zhang W., Zhang T., Wu Y., Jiang J. (2012). Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis. Plant Cell 24: 2719–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Yu J., Li D., Zhang Z., Liu F., Zhou X., Wang T., Ling Y., Su Z. (2010). PMRD: plant microRNA database. Nucleic Acids Res. 38: D806–D813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Ren N., Wang H., Stromberg A.J., Perry S.E. (2009). Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Lee C., Ye Z.H. (2010). Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant 3: 1087–1103. [DOI] [PubMed] [Google Scholar]
- Zhong R., Lee C., Zhou J., McCarthy R.L., Ye Z.H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]