Arabidopsis thaliana papain-like cysteine protease, CEP1, is a crucial executor that participates directly in tapetal cell wall hydrolysis and secretory structure formation during tapetal cell programmed cell death. Correct expression of CEP1 in the tapetum is necessary for timely tapetal cell degeneration and functional pollen formation.
Abstract
Tapetal programmed cell death (PCD) is a prerequisite for pollen grain development in angiosperms, and cysteine proteases are the most ubiquitous hydrolases involved in plant PCD. We identified a papain-like cysteine protease, CEP1, which is involved in tapetal PCD and pollen development in Arabidopsis thaliana. CEP1 is expressed specifically in the tapetum from stages 5 to 11 of anther development. The CEP1 protein first appears as a proenzyme in precursor protease vesicles and is then transported to the vacuole and transformed into the mature enzyme before rupture of the vacuole. cep1 mutants exhibited aborted tapetal PCD and decreased pollen fertility with abnormal pollen exine. A transcriptomic analysis revealed that 872 genes showed significantly altered expression in the cep1 mutants, and most of them are important for tapetal cell wall organization, tapetal secretory structure formation, and pollen development. CEP1 overexpression caused premature tapetal PCD and pollen infertility. ELISA and quantitative RT-PCR analyses confirmed that the CEP1 expression level showed a strong relationship to the degree of tapetal PCD and pollen fertility. Our results reveal that CEP1 is a crucial executor during tapetal PCD and that proper CEP1 expression is necessary for timely degeneration of tapetal cells and functional pollen formation.
INTRODUCTION
Proper development and release of pollen from the anther are essential for successful reproduction in the vast majority of angiosperms. The formation of fertile pollen in anther locules depends on nutritive contributions from the surrounding sporophytic tissues, which consist of four somatic layers from exterior to interior: the epidermis, endothecium, middle layer, and tapetum. In particular, the tapetum plays a crucial secretory role in the development of microspores by contributing to nutrients, enzymes, and sporopollenin precursors for pollen wall synthesis and pollen coat deposition via programmed cell death (PCD). Either premature or abrogated PCD of tapetal cells disrupts the supply of these nutrients to microspores resulting in sterile pollen (Ku et al., 2003; Varnier et al., 2005; Kawanabe et al., 2006).
Many genes encoding putative transcription factors are associated with tapetal PCD, including Arabidopsis thaliana DYSFUNCTIONAL TAPETUM1 (DYT1) (Feng et al., 2012), MYB33/MYB65 (Millar and Gubler, 2005), DEFECTIVE IN TAPETAL DEVELOPMENT AND FUNCTION1 (TDF1) (Zhu et al., 2008), ABORTED MICROSPORES (AMS) (Sorensen et al., 2003; Xu et al., 2010), MALE STERILITY1 (MS1) (Vizcay-Barrena and Wilson, 2006; Yang et al., 2007; Ito et al., 2007), MYB80 (formerly MYB103) (Zhu et al., 2010; Phan et al., 2011, 2012), rice (Oryza sativa) TAPETUM DEGENERATION RETARDATION (TDR) (Li et al., 2006), and PERSISTENT TAPETAL CELL1 (PTC1) (H. Li et al., 2011). The absence of these genes leads to abnormal tapetal PCD and largely sterile pollen grains. In addition, the Arabidopsis SPOROCYTELESS/NOZZLE (SPL/NZZ) and EXCESS MICROSPOROCYTES 1/EXTRA SPOROGENOUS CELLS (EMS1/EXS) genes are key regulators of anther cell differentiation and required for specifying tapetal identity (Canales et al., 2002; Zhao et al., 2002; Liu et al., 2009). Although these transcription factors regulate tapetal degeneration and pollen development through various pathways, several papain-like cysteine proteases have been consistently identified as acting downstream of these transcription factors and are speculated to play important roles in tapetal degeneration. For example, rice CP1 (a papain-like cysteine protease gene) is regulated by PTC1 and TDR in rice (Li et al., 2006; H. Li et al., 2011). In Arabidopsis, the expression levels of several papain-like cysteine proteases (RD19C, THI1, and RDL1) are reduced in the ams mutant (Xu et al., 2010), and the expression levels of RD19A, THI1, RD19C, and RD21A are altered in the ms1 mutant (Yang et al., 2007).
Cysteine protease enzymes are ubiquitously involved in cell degeneration and belong to a large family of enzymes found in animals, plants, and microorganisms that play important roles in intracellular protein degradation and organism PCD (Solomon et al., 1999). In Arabidopsis, SENESCENCE-ASSOCIATED GENE12 (SAG12), SAG2, XYLEM CYSTEINE PROTEASE1 (XCP1), and XCP2 show a tight connection to cell senescence (Ahmed et al., 2000; Funk et al., 2002; Otegui et al., 2005). However, the exact status of cysteine proteases in PCD remains unconfirmed. One reason is that other proteases can complement the functions of many of these cysteine proteases. This redundancy has made it difficult to determine the functional importance of any given enzyme during plant PCD, as many are often expressed simultaneously in a given tissue undergoing PCD (Trobacher et al., 2006; Richau et al., 2012). For example, the sag12 Arabidopsis mutant does not develop a phenotype during senescence under normal growth conditions, suggesting that SAG12 is functionally redundant with other proteases (Otegui et al., 2005).
Most plant cysteine proteases belong to the papain-like, metacaspase, and legumain families, but other cysteine proteases such as the calpain family have also been found. The papain-like cysteine proteases are the most thoroughly investigated family among cysteine proteases. Many of these are typically active in acid pH environments including the apoplast, vacuole, and lysosomes (Trobacher et al., 2006), and many are implicated in a variety of PCD events in various tissues, including xylogenesis (Han et al., 2012), leaf and flower senescence (Chen et al., 2002; Eason et al., 2002; Rabiza-Świder et al., 2003; Senatore et al., 2009), and seed development and germination (Wan et al., 2002; Greenwood et al., 2005).
A unique group of papain-like cysteine proteases for which no homologous genes have been found in mammals or yeast (Helm et al., 2008) are characterized by a C-terminal KDEL endoplasmic reticulum (ER) retention signal. A phylogenetic tree of KDEL-tailed papain-like cysteine proteases contains a distinct group among dicots, monocots, and gymnosperms (Hierl et al., 2012), and the KDEL-tailed cysteine proteases are found extensively in tissues undergoing PCD, particularly in cells that finally collapse. Examples are proteinase A in the hypogeous cotyledons of Vicia sativa (Becker et al., 1997), EP-C1 in the maturing pods of Phaseolus vulgaris (Tanaka et al., 1991), Cys-EP-related protein in the megagametophyte cells after germination of spruce (Picea glauca seeds) (He and Kermode, 2003), CysEP in the PCD of nuclear and endospermic cells in Ricinus seed (Schmid et al., 1999), SEN10 in the senescing flower of daylily (Hemerocallis hybrid cv Cradle Song) (Valpuesta et al., 1995), and CysEP in the ricinosomes of the tomato (Solanum lycopersicum) anther middle layer, endothecium, and epidermal cells (but not tapetal cells) (Senatore et al., 2009). Previous studies in our lab identified one KDEL-tailed papain-like cysteine protease gene from tobacco (Nicotiana tabacum) named CP56 and revealed that suppression of this gene results in pollen grain sterility (Zhang et al., 2009). Three KDEL-tailed papain-like cysteine proteases, CEP1, CEP2, and CEP3, are expressed in Arabidopsis roots, flowers, and green siliques where the cells are about to collapse(Helm et al., 2008). In particular, CEP1 expression is detected in the stigma, anther, sepals, petals, and young siliques after stage 15 of flower development (Helm et al., 2008). For these reasons, CEP1 has been speculated to play an essential role in flower development, but its exact function and the corresponding mechanism remain uncertain.
To investigate the role of CEP1 in anther development, we characterized the phenotype of the cep1 mutant, CEP1 expression in Arabidopsis, enzymatic characteristics of CEP1 in vitro, and the phenotype of overexpressing transgenic lines. We also performed transcriptomic analyses to compare gene expression profiles between the wild type and cep1 mutant. Our results indicate that CEP1 acts as an irreplaceable executor during tapetal cell PCD, which affects pollen development.
RESULTS
Identification of the cep1 Mutant
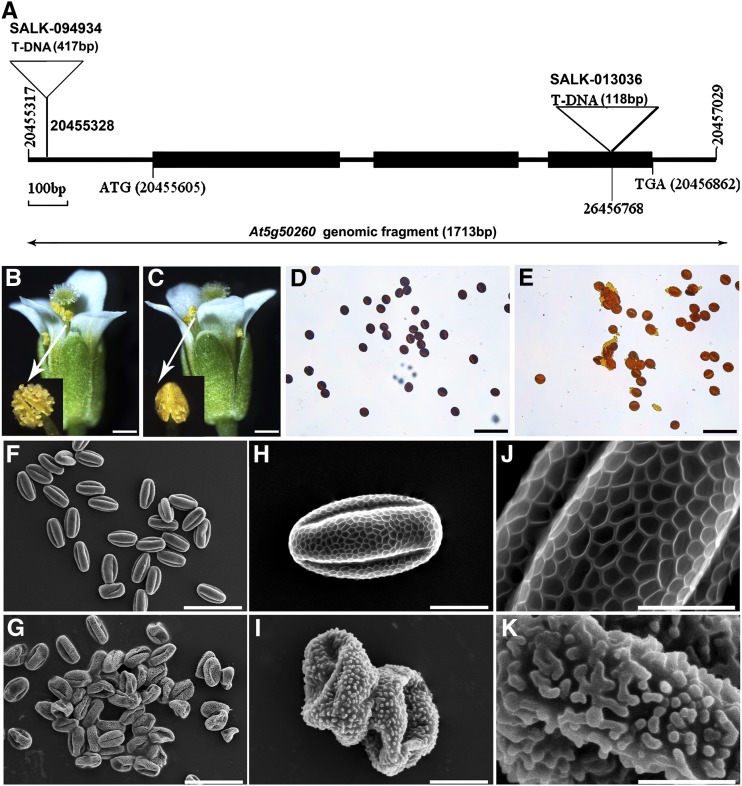
To identify the function of CEP1 during Arabidopsis anther development, we obtained two SALK T-DNA insertion mutant lines, named SALK_094934 and SALK_013036, from the ABRC. The T-DNA is inserted into the 5′ untranslated region and the third exon of CEP1 in the two mutant lines (Figure 1A). In both lines, plants displayed reduced male fertility, and normal female development was confirmed by reciprocal cross analysis. Given that both mutants had similar phenotypes, SALK_013036, designated cep1-1, was used for further analysis and is referred to as cep1 throughout. Defective cep1 mutants were crossed with the wild type, and all F1 progeny were fertile with an approximate 3:1 phenotypic segregation ratio in F2 plants (fertility:sterility = 208: 81; χ2 =1.256; P < 0.05), suggesting that this mutation is controlled by a single recessive locus. Compared with wild-type plants, cep1 mutant plants displayed a normal phenotype during vegetative and early generative development stages. However, fewer pollen grains were released from cep1 dehisced anthers (<40% of the wild type) (Figures 1B and 1C). Wild-type mature pollen grains were blue-black upon staining with iodium-potassium iodide solution (Figure 1D), whereas cep1 pollen grains were brown (Figure 1E). The germination rate of the pollen grains in vitro decreased significantly in cep1 (10.50%, 23 of 219) compared with that in the wild type (86.76%, 249 of 287). A scanning electron microscopy examination revealed that wild-type mature pollen grains were uniformly spheroid and had finely reticulate ornamentation on their surface (Figures 1F, 1H, and 1J), whereas surviving pollen grains in the cep1 mutant adhered to one another, formed irregular clumps, and exhibited collapsed and gemmate-baculate sculpture without regularly reticulate ornamentation (Figures 1G, 1I, and 1K). These results show that the absence of CEP1 markedly impaired pollen development and resulted in sterile pollen grains with abnormal pollen morphology.
Figure 1.
Comparison of the Wild Type and the cep1 Mutant.
(A) SALK_094934 and SALK_013036 T-DNA insertion positions in At5g50260. Filled boxes represent exons.
(B) Wild-type flower and dehiscing anthers.
(C) cep1 flower and dehiscing anthers.
(D) Wild-type pollen grains stained with iodium-potassium iodide solution.
(E) cep1 pollen grains stained with iodium-potassium iodide solution.
(F) to (K) Mature wild-type pollen grains ([F], [H], and [J]) and cep1 pollen grains ([G]), [I], and [K]).
Bars = 200 μm in (B) and (C), 50 μm in (D) to (G), 10 μm in (H) and (I), and 5 μm in (J) and (K).
Anther Development in the cep1 Mutant
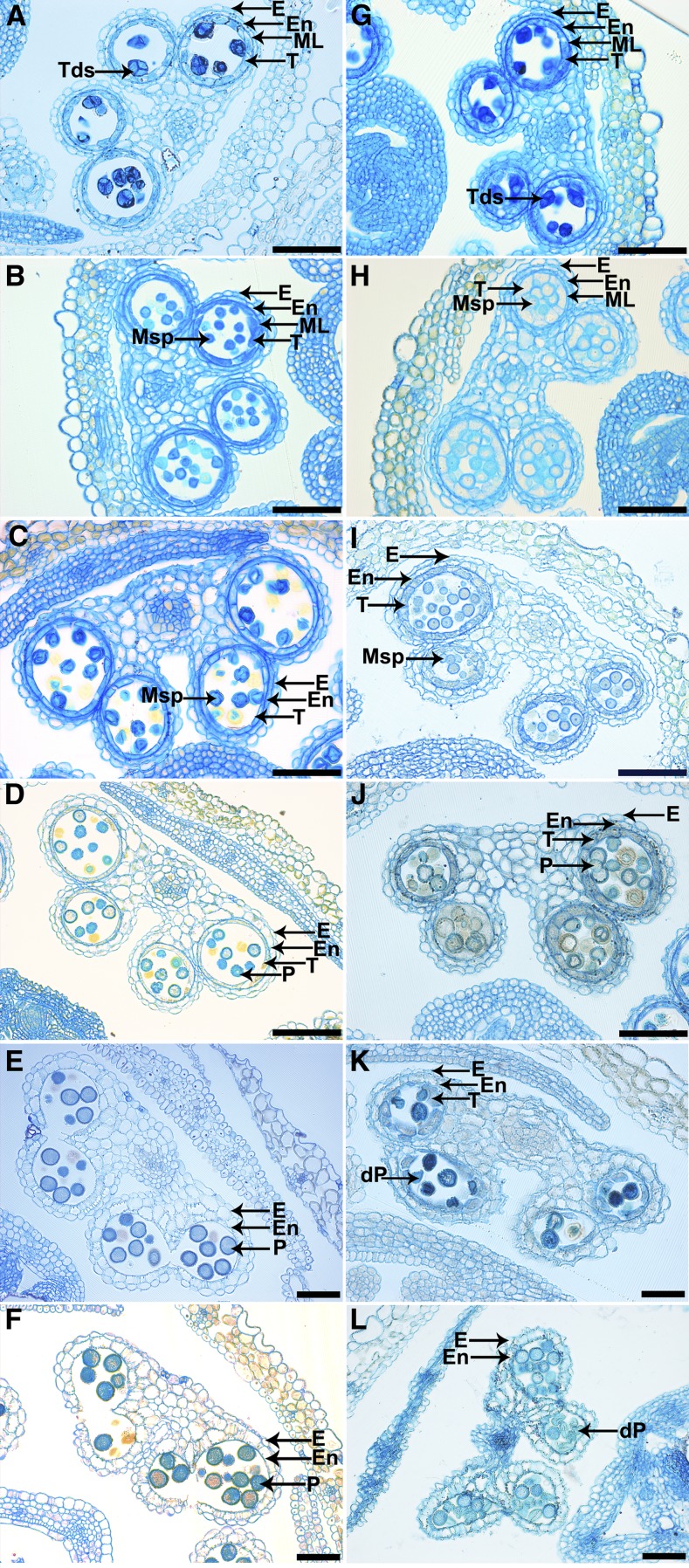
Both wild-type and cep1 anthers were examined by microscopy of anther semithin sections to further clarify the pollen development process in the cep1 mutant. The development of Arabidopsis anthers is divided into 14 stages based on morphological landmarks that correspond to cellular events visible under a microscope (Sanders et al., 1999). During stage 7, no obvious differences were observed between the wild type and the cep1 mutant (Figures 2A and 2G). At stage 8, young angular-shaped microspores were released from tetrads in the wild type, whereas the tapetal cells shrank and their cytoplasm became deeply stained (Figure 2B). However, in cep1 anthers, the microspore shape was slightly round without showing a clearly angular shape, the tapetal cells did not shrink, and the cytoplasm appeared less stained (Figure 2H). The size of the microspores was evidently different from stage 9 to stage 10 in cep1 anthers, and many of them crowded together; the tapetal cells were slightly turgid and less stained (Figure 2I). During the same stages, vacuolated uninucleate microspores of uniform size were distributed in the anther locules of the wild type, and a more condensed tapetum was visible (Figure 2C). At stage 11, the wild-type anther was filled with well-developed pollen entering mitosis, and the tapetum was clearly degenerated (Figure 2D). Nevertheless, in the cep1 mutant, the pollen size was obviously different, and many pollen grains were crowded together. The tapetal cells did not degenerate and adhered closely to other sporophytic tissues (Figure 2J). The anther became bilocular at stage 12 in the wild type, the tapetum had already degenerated completely, and mature pollen grains were densely stained (Figure 2E). However, the anther shape was shriveled in the cep1 mutant, and the anther was not dehiscent; the remnants of undegenerated tapetal cells adhered to other sporophytic tissues; the number of pollen grains had obviously decreased, and the few surviving defective pollen grains had shrunk (Figure 2K). Mature pollen grains were released during anther dehiscence at stage 13 (Figure 2F). However, the anthers failed to dehisce completely, and most of the remaining pollen grains were atrophied in the cep1 mutant (Figure 2L). These results show that anther maturation was abnormal in the cep1 mutant, particularly tapetal degeneration and pollen development.
Figure 2.
Transverse Section Comparison of Anther Development in the Wild Type and cep1 Mutant.
Semithin sections of anthers from the wild type ([A] to [F]) and cep1 ([G] to [L]) at anther development stage 7 [[A] and [G]), stage 8 ([B] and [H]), stage 10 ([C] and [I]), stage 11 ([D] and [J]), stage 12 ([E] and [K]), and stage 13 ([F] and [L]). dP, degenerated pollen; E, epidermis; En, endothecium; ML, middle layer; Msp, microspore; P, pollen; T, tapetum; Tds, tetrads. Bars = 50 μm.
Tapetal PCD Is Retarded in the cep1 Mutant
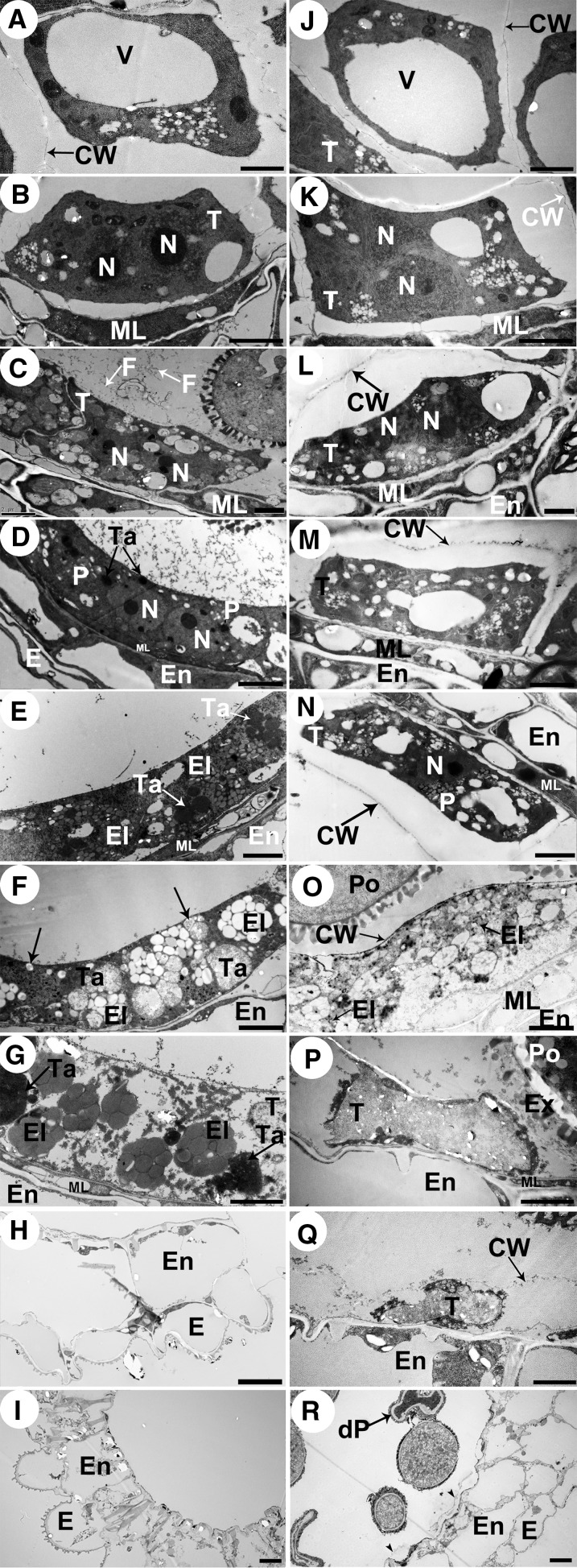
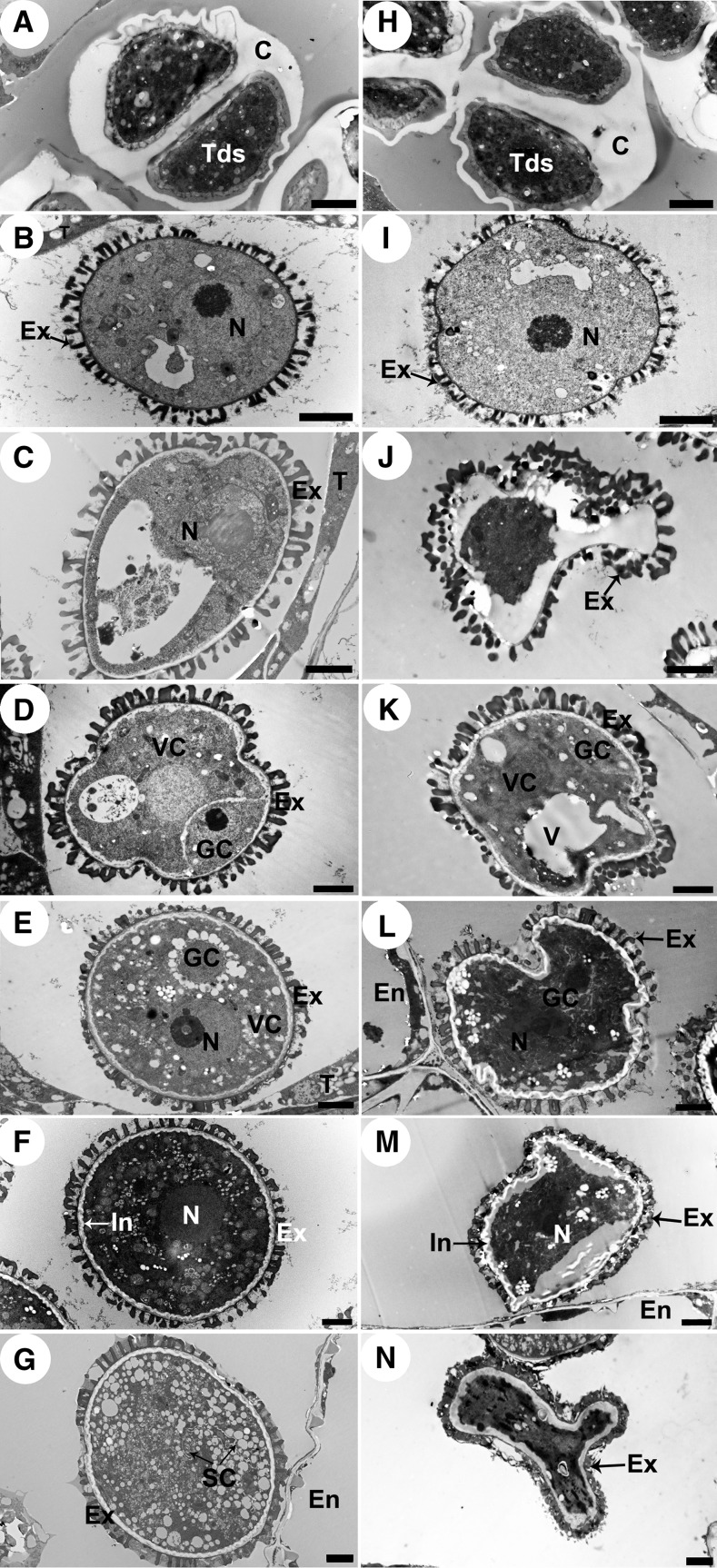
We used transmission electron microscopy (TEM) to investigate the differences in tapetal cell development between the wild type and cep1. The tapetal cell wall had visibly degraded at late stage 7 in the wild type (Figure 3B), but the tapetal cell wall remained intact in the cep1 mutant, indicating that the tapetal cells had failed to transform properly into the polar secretory type (Figure 3K). At stage 8, the tapetal cells were spongy in appearance due to the presence of numerous secretory vacuoles and vesicles in the tapetal cells, and the anther locules were filled with fine fibrillar materials that appeared to have been released from tapetal cells (Figure 3C). In the cep1 mutant, the tapetal cells were also slightly spongy with fewer secretory vacuoles and reduced vesicle formation than in the wild type, and the existing cell wall and sparse secretory vacuole formation resulted in few fibrillar materials released to the anther locules (Figure 3L). At stage 9, plastids containing numerous electron-transparent deposits and the tapetosome containing many lipid materials appeared in the wild-type binucleate tapetal cell (Figure 3D). However, the tapetal cells did not form typical plastids and tapetosomes in the cep1 mutant but only formed a few secretory vacuoles and vesicles, which was different from the wild type (Figure 3M). At stage 10, the nucleus had already degraded in wild-type tapetal cells, the numbers of tapetosomes and elaioplasts (transformed from plastids) evidently increased, and the tapetal cells continuously released fibrillar materials (lipids) into the anther locules (Figure 3E). In contrast, the tapetal cell nucleus of the cep1 mutant failed to degenerate in a timely way, and few elaioplasts and almost no tapetosomes formed, resulting in few fibrillar materials being released from tapetal cells (Figure 3N). At early stage 11, a large amount of osmiophilic material released from the tapetosomes fused with the cell membrane (arrows) and continuously deposited sporopollenin onto the pollen surface of the wild type (Figure 3F). However, tapetal cells in the cep1 mutant adhered tightly to the anther middle layer, the cell wall was still surviving, and the cytoplasm had undergone degeneration but showed no obvious osmiophilic inclusions and few elaioplasts (Figure 3O). At late stage 11, wild-type tapetal cells reached the end of their PCD and were filled with tapetosomes and elaioplasts (Figure 3G). However, tapetal cells experienced abnormal degeneration in the cep1 mutant, and no obvious tapetosomes or elaioplasts were present in tapetal cells (Figure 3P). By stage 12, tapetal cell degeneration had finished in the wild type (Figure 3H), whereas cep1 tapetal cell remnants with undegenerated cell walls remained (Figure 3Q). By stage 13, a few tapetal cell wall remnants still remained in the cep1 anthers (Figures 3I and 3R, arrowheads). These observations indicate that degeneration of the tapetal cell wall and nucleus was delayed and that the formation of secretory organelles largely decreased in the cep1 mutant.
Figure 3.
Transmission Electron Micrographs of the Anthers from the Wild Type and the cep1 Mutant.
(A) to (I) are the wild type, and (J) to (R) are the cep1 mutant. (A) and (J), stage 6; (B) and (K), stage 7; (C) and (L), stage 8; (D) and (M), stage 9; (E) and (N), stage 10; (F) and (O), early stage 11; (G) and (P), late stage 11; (H) and (Q), stage 12; (I) and (R), stage 13. CW, cell wall; dP, degenerated pollen; E, epidermis; El, elaioplast; En, endothecium; Ex, exine; F, fibrillar materials; ML, middle layer; N, nucleus; P, plastid; Po, pollen; T, tapetal cell; Ta, tapetosome; V, vacuole. Bars = 2 μm in (A) to (H) and (J) to (Q) and 5 μm in (I) and (R).
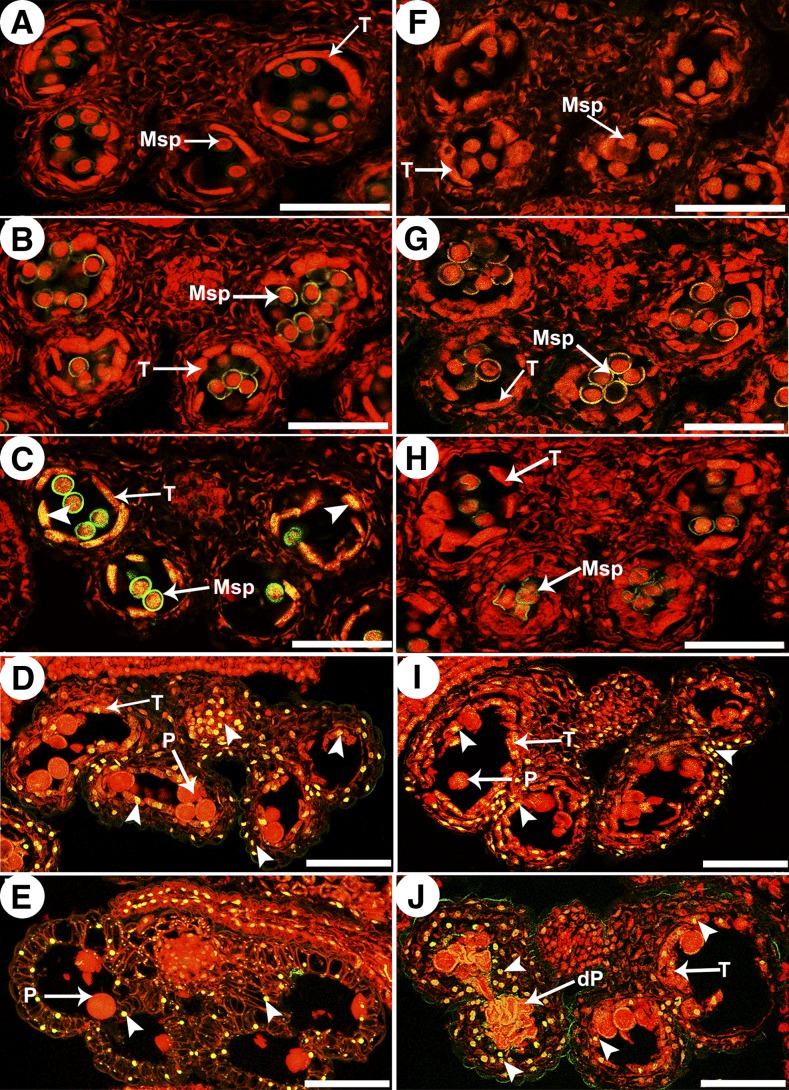
In plants, the degeneration of the tapetum is considered to be the result of PCD, which is characterized by the cleavage of nuclear DNA. To further confirm the abnormalities in cep1 tapetal PCD, the terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay was performed in wild-type and cep1 anthers at different developmental stages. TUNEL-positive signals were not detected at stages 8 or 9 in either wild-type or cep1 anthers (Figures 4A, 4B, 4F, and 4G). At stage 10, yellow TUNEL-positive signals appeared in the wild-type tapetum, indicating that the tapetal cell nucleus had degenerated during this stage (Figure 4C). Compared with the wild type, no TUNEL-positive signal was observed in the cep1 mutant at this stage (Figure 4H). At stage 11, intensely TUNEL-positive signals were present in the degenerating tapetum of the wild type (Figure 4D), whereas TUNEL-positive signals started to appear in the cep1 tapetal cells at this stage (Figures 4I). At stage 12, no TUNEL-positive signals were observed in the wild-type anther tapetum due to complete degeneration of the tapetal cells (Figure 4E). However, TUNEL-positive signals were still present in the tapetal cell remnants of the cep1 mutant (Figure 4J). From stage 11 to stage 12, intense TUNEL-positive signals were also observed in the outer cell layers of the anther (endothecium and middle layer) and the vascular bundle cells in both the wild type and cep1 mutant (Figures 4D, 4E, 4I, and 4J). These observations further confirmed that the starting time of tapetal cell DNA fragmentation was retarded and that the degree of DNA fragmentation also decreased in the cep1 mutant, suggesting that cep1 tapetal PCD was abnormal.
Figure 4.
DNA Fragmentation in Wild-Type and cep1 Anthers.
Anthers of the five developmental stages in the wild type ([A] to [E]) and the cep1 mutant ([F] to [J]) were compared for nuclear DNA fragmentation using the TUNEL assay. Nuclei were stained with propidium iodide indicated by the red fluorescence, and the yellow fluorescence is TUNEL-positive nuclei staining (arrowheads). (A) and (F), stage 8; (B) and (G), stage 9; (C) and (H), stage 10; (D) and (I), stage 11; (E) and (J), stage 12. dP, degenerated pollen; P, pollen; Msp, microspore; T, tapetum. Bars = 50 μm.
Abnormal Pollen Development in the cep1 Mutant
We also used TEM to observe pollen development to further determine whether the abnormal pollen development was synchronized with tapetal PCD defects in the cep1 mutant. No obvious differences in cep1 microspore development were observed compared with that in the wild type at stage 7 (Figures 5A and 5H). At late stage 8, the primary structure of the exine, composed of the nexine, baculum, and tectum, was observed on the surface of wild-type microspores (Figure 5B). In contrast, a reduced and irregular primary exine structure appeared on the cep1 microspore surface, and the contents of sporopollenin precursors decreased greatly in the anther locules of the cep1 mutant (Figure 5I). The nucleus in wild-type microspores at stage 9 was displaced to the side by a single large vacuole, and development of the orderly microspore exine structure proceeded (Figure 5C). However, the shape of the microspore was irregular in the cep1 mutant and showed no obvious nucleus or single large vacuole, and the exine was coarse with a disorganized baculum and tectum (Figure 5J). At stage 10, many oil bodies were present in the microspore, and one nucleus was present in the lenticularly shaped generative cell (Figure 5D). In contrast, the sparse and disordered exine structure appeared on the surface of cep1 pollen grains. Pollen cytoplasm development was incomplete, with few oil bodies and an indistinct generative cell (Figure 5K). Subsequently, the immature pollen underwent cytoplasmic enlargement to fill the pollen grain and the generative cell had moved away from the edge of the pollen grain at early stage 11. Sporopollenin materials continued to accumulate on the surface of the pollen to form the exine in the wild type (Figure 5E). Unlike the wild type, the partially degenerated cep1 pollen appeared shrunken with no obvious generative cell, and formation of the exine was not evident on the pollen surface (Figure 5L). At late stage 11, the remaining lipid content in the wild type was released from the tapetum into the locules and eventually accumulated among the bacula of the exine; a spherical vegetative nucleus with a prominent nucleolus was present in each pollen grain and numerous small vacuoles were distributed throughout the cytoplasm of the pollen grains (Figure 5F). However, the surviving cep1 pollen grains were abnormal in shape at the same stage, indicating a defective exine structure, and the pollen cytoplasm had fewer vacuoles compared with those of the wild type (Figure 5M). At stage 12, the typical pollen wall in the wild type was completely established with distinct intine and exine layers (including tectum, bacula, and nexine) (Figure 5G). In the cep1 mutant, the pollen was shriveled and collapsed with an abnormal pollen wall (Figure 5N).
Figure 5.
Transmission Electron Micrographs of Microspores from the Wild Type and cep1 Mutant.
Microspores of the six developmental stages in the wild type ([A] to [G]) and cep1 mutant ([H] to [N]). (A) and (H), stage 7; (B) and (I), stage 8; (C) and (J), stage 9; (D) and (K), stage 10; (E) and (L), early stage 11; (F) and (M), late stage 11; (G) and (N), stage 12. C, callose; En, endothecium; Ex, exine; GC, generative cell; In, intine; N, nucleus; SC, sperm cell; T, tapetal cell; Tds, tetrads; V, vacuole; VC, vegetative cell. Bars = 2 μm.
These results indicated that cep1 pollen development was impaired, resulting in defective pollen exine, abnormal pollen shape, and collapsed pollen grains.
The ProCEP1:CEP1 Translational Fusion Complements the cep1 Mutation
A complementation experiment was performed to confirm that the cep1 mutant phenotype was attributable to the loss of CEP1 function. A 3.06-kb genomic DNA fragment including the 1353-bp promoter of CEP1 (ProCEP1) and the 1713-bp CEP1 genomic sequence was cloned into the pCAMBIA 1300 vector and introduced into cep1 homozygous plants. Among 55 independent transgenic plants that grew to maturity, 37 plants showed restored normal fertility, 10 were partially fertile, and the remaining eight plants were sterile, similar to the cep1 mutant (Supplemental Figure 1A). Thirteen plants in negative control lines transformed with the empty pCAMBIA 1300 vector showed the sterile phenotype similar to that of the cep1 mutant. The 37 restored transgenic lines displayed normal tapetal degeneration, pollen development, and dehisced with fertile pollen grains (Supplemental Figures 1C, 1E, and 1G to 1L). Quantitative RT-PCR (qRT-PCR) analysis of CEP1 transcript levels in the transgenic plants showed that the restored-fertility transgenic lines contained similar numbers of transcripts as the wild type (Supplemental Figure 1B, transgenic lines 3, 11, and 36), whereas the sterile transgenic lines contained extremely low numbers of transcripts (Supplemental Figure 1B, transgenic lines 17 and 41). The partially fertile lines contained an intermediate amount of transcripts compared with the wild type and sterile lines (Supplemental Figure 1B, transgenic lines 5 and 53).
We thus confirmed that the loss of function of CEP1 was the cause of the retarded tapetal degeneration and defective pollen development in cep1.
CEP1 Is a KDEL-Tailed Cysteine Protease and Belongs to the Papain-Like Family
A 1092-bp cDNA fragment, including the 1086-bp open reading frame (ORF) of CEP1, was cloned from the Arabidopsis inflorescence and sequenced. The corresponding CEP1 genomic sequence (1713-bp fragment) was also amplified. CEP1 contained three exons and two introns. The ORF encoded a peptide of 362 residues with a predicted molecular mass of 38 kD. Similar to other papain-like cysteine proteases, the mature CEP1 peptide had highly conserved Cys-150, His-286, and Asn-307 residues that constituted the catalytic triad (Supplemental Figure 2). The ERFNIN (EX3-RX3FX3NX3I/VX3N) motif was conserved (Supplemental Figure 2) and supported membership of the CEP1 cysteine proteinase in the cathepsin L-like, but not the B-like, proteinases of the papain family (Wiederanders, 2003). Moreover, the GCNGG motif and the KDEL tail present at the C terminus (Supplemental Figure 2) showed that CEP1 belonged to the KDEL-tailed cysteine proteases, a distinct subfamily in the plant papain-like cysteine proteases. The phylogenetic tree including all Arabidopsis cysteine proteases (papain family 31, metacaspase family 9, legumain family 4, and calpain family 1) further confirmed that the CEP1 protein belongs to the papain family and was located in the group containing KDEL-tailed subfamily members (Supplemental Figure 3 and Supplemental Data Set 1).
Plant papain-like enzymes are usually synthesized as small preproproteins (40 to 50 kD) and undergo proteolytic processing to yield mature, fully active enzymes (22 to 35 kD). To further characterize the properties of CEP1, induction of the expression construct pET30a-CEP1, which encodes the full-length CEP1 cDNA minus the signal peptide, was expressed in Escherichia coli, resulting in overexpression of the recombinant protein. The molecular weight of the purified recombinant CEP1 was ∼38 kD according to SDS-PAGE analysis (Supplemental Figure 4, lanes 1 to 3). This recombinant protein did not have determinable activity. A 26-kD mature protein, which self-cleaved from the recombinant protein at pH 3.0 (Supplemental Figure 4, lane 4), was sequenced, and the self-cleavage site was between A121 and N122 (Supplemental Figure 2). The kinetics of the mature enzyme was measured using Z-Phe-Arg-7-amido-4-methylcoumarin (Z-FR-AMC) or l-Arg -7-amido-4-methylcoumarin (R-AMC) as the substrate. The Km and Vmax were (13.92 ± 0.18) μmol·L–1 and (235.14 ± 2.19) pmol·s–1·mg–1 for Z-Phe-Arg-AMC and (159.62 ± 2.34) μmol·L–1 and (537.93 ± 4.98) pmol·s–1·mg–1 for l-arginine-AMC, respectively (Table 1). An inhibition assay was performed using several inhibitors, including the papain-like cysteine protease inhibitor E-64, the cysteine protease inhibitor leupeptin, the caspase inhibitors Z-VAD-fmk and Ac-YVAD-cmk, the aspartic protease inhibitor pepstatin A, and the serine proteases inhibitors PMSF and EDTA, respectively. CEP1 was inhibited significantly by the papain-like cysteine protease inhibitor E-64 and the cysteine protease inhibitor leupeptin and was inhibited only weakly by inhibitors (Z-VAD-fmk and Ac-YVAD-cmk) belonging to the other cysteine protease classes, but not by inhibitors (pepstatin A, PMSF, and EDTA) of other protease classes (Table 2). Assays including different pH and temperature conditions showed that the optimum pH and temperature for CEP1 were pH 3.0 and 40°C, respectively. These findings confirm that CEP1 is a KDEL-tailed cysteine protease belonging to the papain-like family and that CEP1 was synthesized as a prepropeptide (38 kD) without activity, then underwent proteolytic processing of the pre- and propeptides to yield a mature, fully active 26-kD enzyme.
Table 1. Kinetic Properties of Recombinant CEP1 Protein.
| Z-FR-AMC | R-AMC | |
|---|---|---|
| Km (μmol·L−1) | 13.92 ± 0.18 | 159.62 ± 2.34 |
| Vmax (pmol·s−1·mg−1) | 235.14 ± 2.19 | 537.93 ± 4.98 |
| Kcat (10−3 s−1) | 6.11 | 13.99 |
| kcat/Km (M−1·s−1) | 439.08 | 87.62 |
Values represent the means and sd of three replicates.
Table 2. Inhibition of Recombinant CEP1 Protein.
| Inhibitor | Activity (%) |
|
|---|---|---|
| Z-FR-AMC | R-AMC | |
| None | 100 | 100 |
| E-64 1 μM | 0 | 0 |
| E-64 10 μM | 0 | 0 |
| Leupeptin 1 μM | 1 | 2 |
| Leupeptin 10 μM | 0 | 0 |
| Z-VAD-fmk 10 μM | 85 | 61 |
| Z-VAD-fmk 100 μM | 48 | 31 |
| Ac-YVAD-cmk 10 μM | 99 | 90 |
| Ac-YVAD-cmk 100 μM | 62 | 71 |
| Pepstatin A 1 μM | 100 | 100 |
| Pepstatin A 10 μM | 100 | 100 |
| PMSF 100 μM | 100 | 100 |
| PMSF 1 mM | 97 | 100 |
| EDTA 1 mM | 100 | 100 |
Activity is expressed as percentage of activity relative to a control reaction.
Expression Characterization of CEP1 in Arabidopsis
According to the morphological analysis, the timely degeneration of the tapetum and the development of microspores were greatly affected in the cep1 mutant, but no obvious effect was observed on Arabidopsis vegetative growth. CEP1 expression characteristics were investigated to further determine the relationship between the function and expression of CEP1.
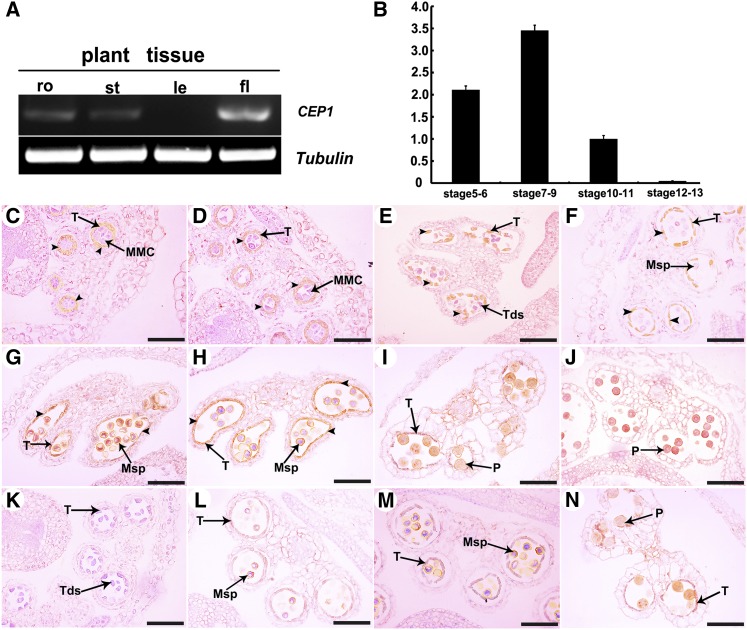
We performed RT-PCR analysis with total RNA extracted from various organs, including roots, stems, leaves, and buds. CEP1 was highly expressed in buds but relatively low expression was observed in the roots and stems, and almost no expression was detected in leaves (Figure 6A). The CEP1 expression level in buds was also evaluated by qRT-PCR. The Arabidopsis ACTIN1 gene (AT2G37620) was used as the reference for normalization. CEP1 expression in buds appeared at stages 5-6, reached the maximum level at stages 7-9, and then declined sharply to a barely detectable level at stage 10-11 (Figure 6B). Previously research reported CEP1:GUS (β-glucuronidase) expression was visible in anthers near the end of the filament (Helm et al., 2008). RNA in situ hybridization was performed at different anther development stages to further determine the spatial and temporal expression patterns of CEP1 during anther development. The CEP1-specific RNA fragment was designed as an antisense probe, and a sense probe was used as the control. Compared with the negative control (purple), CEP1 was highly expressed in tapetal cells (brown, arrowheads) during anther development. At stage 5, CEP1 transcripts began to be detected in anther tapetal cells (Figure 6C). CEP1 transcripts increased continuously in the tapetum from stages 6 to 10 (Figures 6D to 6H). At stage 11, the CEP1 expression signals in the tapetum decreased visibly along with tapetal degeneration and almost no signal was detected in the tapetum (Figure 6I). At stage 12, CEP1 transcripts were undetectable in the tapetum due to tapetal degeneration (Figure 6J). A sense probe was used for hybridization in the control, and only background signals were detected (Figures 6K to 6N). Thus, specific CEP1 expression was consistent with its inferred function during tapetal degeneration and anther development.
Figure 6.
CEP1 Expression Pattern.
(A) CEP1 spatial and temporal expression analyses by RT-PCR. fl, flower; le, leaf; ro, root; st, stem.
(B) qRT-PCR of CEP1 expression in wild-type bud tissues at different developmental stages. Bars represent standard deviations. The expression of CEP1 in stage 10-11 was selected as 1.
(C) to (N) In situ analyses of CEP1 in wild-type anthers. (C), stage 5; (D), stage 6; (E), stage 7; (F), stage 8; (G), stage 9; (H), stage 10; (I), stage 11; (J), stage 12; (K), negative controls at stage 7; (L), negative controls at stage 8; (M), negative controls at stage 9; (N), negative controls at stage 11.
MMC, microspore mother cell; Msp, microspore; P, pollen; T, tapetum; Tds, tetrads. Bars = 50 μm.
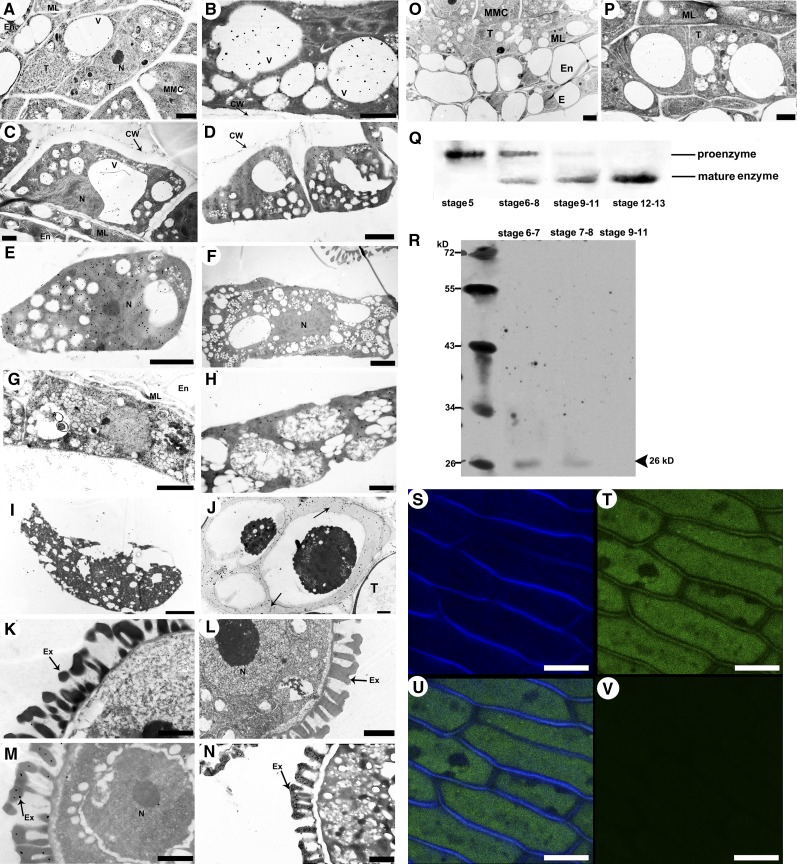
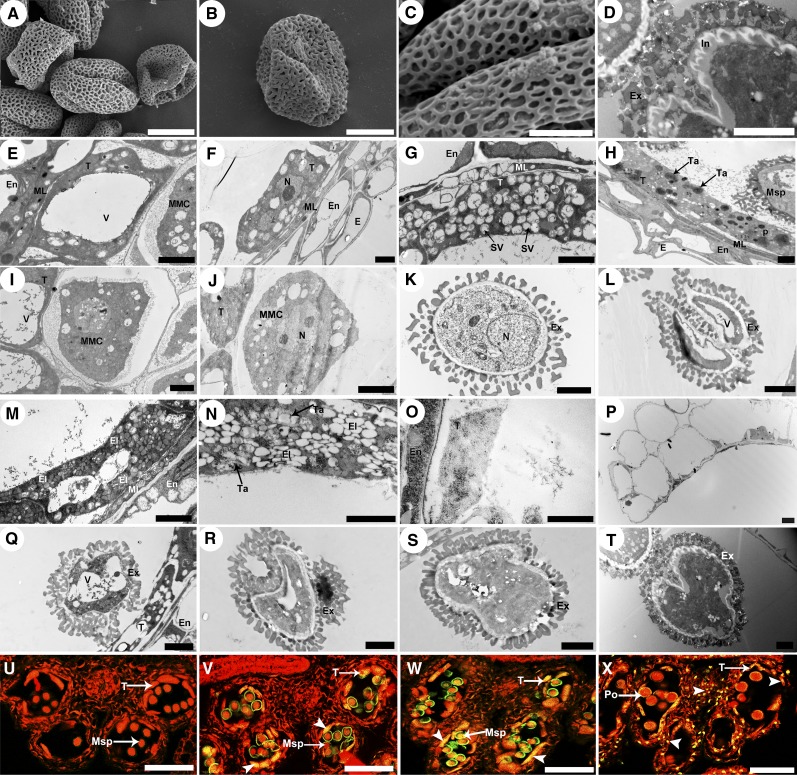
Immunoelectron microscopy was used to determine the subcellular location of CEP1 in tapetal cells. Immunogold labeling using rabbit antimature-CEP1 antibody showed that CEP1 was located in the tapetal vacuole, cytosol, cell wall, and in the mature pollen exine. At stage 5, the CEP1 protein was concentrated in the precursor protease vesicles of the tapetal cytoplasm (Figure 7A). At early stage 6, the CEP1 protein appeared in tapetal cell vacuoles with fusion of these protease vesicles (Figure 7B). Subsequently, the CEP1 protein was released from the vacuoles into the tapetal cytosol. The CEP1 protein was also released to the cell wall by small vesicles fusing with the cell membrane during late stage 6 and stage 7 (Figures 7C and 7D). At late stage 7, the CEP1 protein entered the callose wall and probably participated in callose wall degradation and the release of microspores from the tetrad (Figure 7J). CEP1 protein content reached a maximum in the tapetal cytosol at stages 8-9 (Figures 7E and 7F). At stage 10, the tapetal cell began to degenerate and the CEP1 protein content decreased slightly in the tapetal cytosol (Figure 7G). By early stage 11, with the degeneration of the tapetal cells, the content of CEP1 protein in the cytosol decreased as it was released into the anther locule and became distributed on the pollen exine (Figures 7H and 7M), whereas no CEP1 protein signals in pollen or on pollen exine were observed before this stage (Figures 7K and 7L). At late stage 11, the CEP1 protein disappeared from the tapetal cells and was abundantly distributed on the pollen exine (Figures 7I and 7N). In addition, analysis of the other layers of the wild-type anther showed that no distribution of CEP1 protease during stages 5 and 6 (Figures 7O and 7P). Given that the functioning of CEP1 requires a transformation from the proenzyme to the mature enzyme, we performed immunoblotting for CEP1 in the anthers from stages 5 to 13 to evaluate CEP1 maturation time using an antimature-CEP1 antibody. The analysis detected only the 38-kD proenzyme in stage 5 (Figure 7Q, line 1), but the mature 26-kD mature enzyme appeared during stages 6-8 (Figure 7Q, line 2), and the mature 26-kD enzyme was found abundantly in the anther during stages 9-11 (Figure 7Q, line 3). Only the mature 26-kD enzyme was found in the anther after stage 11 (Figure 7Q, line 4).
Figure 7.
Subcellular Distribution Analysis of the CEP1.
(A) to (I) Immunoelectron microscopy analysis of CEP1 in the tapetum.
(J) to (N) Immunoelectron microscopy analysis of CEP1 in microspores.
(A), stage 5; (B), early stage 6; (C), late stage 6; (D), stage 7; (E), stage 8; (F), stage 9; (G), stage 10; (H), early stage 11; (I), late stage 11; (J), late stage 7, labeling signals in the callose wall (arrows); (K), stage 9; (L), stage 10; (M), early stage 11; (N), late stage 11; (O), the anther layers at stage 5; (P), the tapetum and middle layer of the anther at stage 6. CW, cell wall; E, epidermis; En, endothecium; Ex, exine; ML, middle layer; MMC, microspore mother cell; N, nucleus; T, tapetal cell; V, vacuole. Bars = 1 μm in (A) to (C), (H), and (K) to (N) and 2 μm in (D) to (F), (G), (I), (J), (O), and (P).
(Q) Immunoblot analysis of total anther protein extracts from stages 5 to 13 with antimature-CEP1 antibody.
(R) Immunoblotting analysis of total buds cell wall dehiscing protein extracts from stages 6 to 11 using anti-mature-CEP1 antibodies.
(S) to (V) Expression of the CEP1-GFP fusion protein. Bars= 10 μm
(S) Calcofluor white stain for the detection of cell wall.
(T) Expression of the CEP1-GFP fusion protein in transgenic Arabidopsis anther filaments.
(U) Merged image of both signals of (S) and (T).
(V) the green-channel signal of the wild-type Arabidopsis anther filament.
To further investigate the distribution of mature CEP1 protein in the tapetal cell wall, intact cell walls from the buds of wild-type Arabidopsis were purified using Percoll density gradients to provide organelles devoid of contamination from the other subcellular compartments. Soluble proteins from the purified tapetal cell walls were then analyzed by immunoblotting using antimature-CEP1 antibodies. The mature 26-kD enzyme was detected in the purified tapetal cell wall during stages 6-8 (Figure 7R). As a complementary approach, the subcellular localization of CEP1 was also investigated by fusing its full-length coding sequence upstream to the green fluorescent protein (GFP). CEP1-GFP was then introduced into wild-type Arabidopsis under the control of the CEP1 promoter. As shown in Figures 7T and 7U, GFP fluorescence was detected in the cytoplasm and cell wall (Figure 7T), and the cell wall localization was confirmed by staining with the cell wall fluorescence marker calcofluor white stain (Figure 7U). These results strongly suggest that CEP1 protein was localized to the tapetal cell wall during stage 7 in Arabidopsis.
Taken together, these results indicate that the CEP1 gene is expressed abundantly in tapetal cells during Arabidopsis anther development from stages 5 to 11. First, the premature protease was transported to the protease vesicles presumably via the ER at stage 5 and was then transported into the vacuole by fusion of the precursor protease vesicles. Then, the proenzyme was transformed to the mature form at stage 6 with acidification of the vacuole but before vacuole rupture. Following rupture of the vacuole at late stage 6, the mature CEP1 proteases were released into the cytosol. CEP1 was also released to the tapetal cell wall during late stage 6 and stage 7. After the tapetal cell wall degenerated, the CEP1 enzyme entered the callose wall from the degenerated tapetal cell wall and was probably involved in degeneration of the callose wall. At early stage 11, the mature enzyme began to be released into anther locules from the tapetal cell remnants and localized to the pollen exine. The mature enzyme was present only on the pollen exine after stage 11.
Changes in the Expression of Genes Involved in Tapetal Cell Degradation and Pollen Development in cep1 Anthers
To further characterize the effect of CEP1 on Arabidopsis tapetal PCD and pollen development, we employed transcriptomic analysis using RNA from the buds of the wild type and cep1 mutants at three developmental stages: (1) stages 5-6; (2) stages 7-9; and (3) stages 10-11.
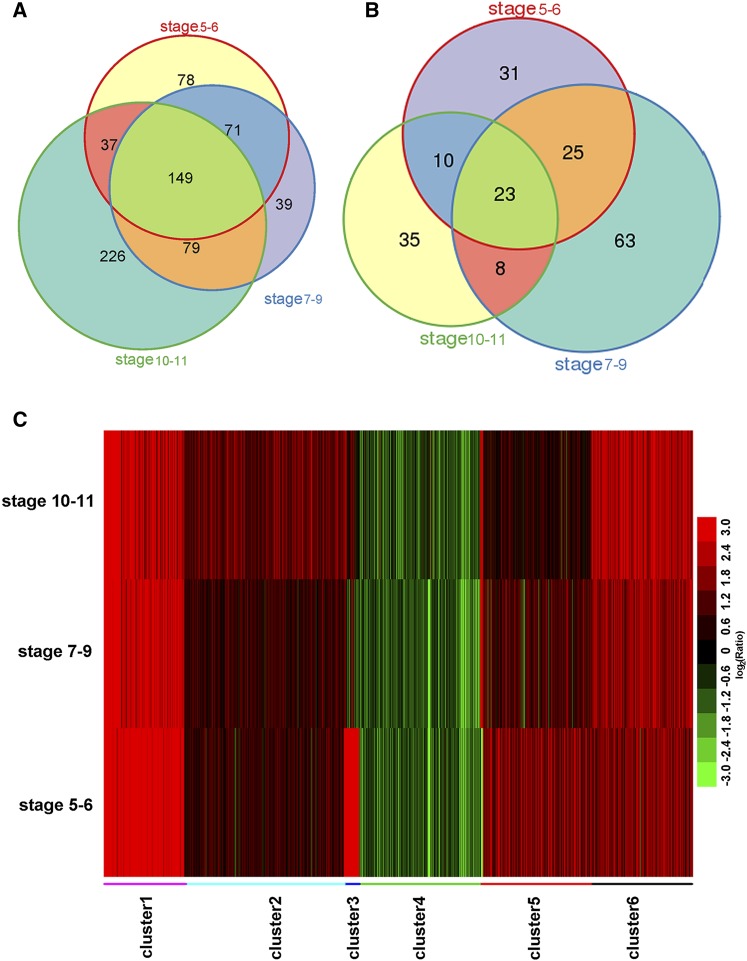
We identified 872 genes with significantly different expression levels (fold change >2 or <–2, and corrected P value < 0.005) in at least one of the three time points (Supplemental Data Set 2). The upregulated genes in the cep1 mutant significantly outnumbered the downregulated genes at the three time points. The numbers of upregulated genes were 335, 338, and 491 (Figure 8A), and those of the downregulated genes were 89, 119, and 76 (Figure 8B) at stages 5-6, stages 7-9, and stages 10-11, respectively. Moreover, many more shared upregulated genes were found than downregulated genes during the three development stages (149 shared up versus 23 shared down).
Figure 8.
Genome-Wide Transcription Analysis of Three Stages of Anther Development in cep1 Buds.
(A) and (B) Venn diagrams of differentially expressed genes that were significantly upregulated (A) or downregulated (B) (corrected P value < 0.005, fold change >2 or <–2) in stages 5-6, stages 7-9, and stages 10-11.
(C) Clustering displays of expression ratios (cep1 versus the wild type) for genes that were differentially expressed in at least one of the three stages (stages 5-6, stages 7-9, and stages 10-11). Red color indicates upregulation, and green denotes downregulation.
Expression data obtained from the transcriptomic analysis were verified by qRT-PCR to confirm the reliability of differentially expressed genes identified by transcriptomic analysis. Sixteen genes were selected based on high fold change and/or functional relevance (Supplemental Table 1). The close agreement of the fold changes between the qRT-PCR and transcriptomic data indicates that the transcriptomic data are reliable (Pearson’s correlation coefficient of 0.93).
We clustered genes with similar expression patterns across the three time points to further investigate the dynamic trend of these RNAs in the cep1 mutant compared with that in the wild type. As shown in Figure 8C, the 872 genes were grouped into six clusters (Supplemental Figures 5A to 5F). Most genes in clusters 1, 2, and 6 were upregulated, and the genes in cluster 4 were downregulated over the three time points in the cep1 mutant, indicating that these genes were probably associated directly with abnormal tapetal PCD and pollen infertility throughout tapetum development. The genes in clusters 3 and 5 were significantly upregulated at stages 5-6 and were then downregulated or returned to normal expression levels at stages 10-11, indicating that the expression of these genes was significantly affected by CEP1 during meiosis.
We performed a Gene Ontology (GO) term enrichment analysis for each cluster to further characterize the function of the differentially expressed genes in the cep1 mutant. In cluster 2, the GO term “cell wall” (P < 10−6) showed clear enrichment (Supplemental Figure 5B). In particular, three extensin family genes (AT1G21310, AT5G25550, and AT2G43150) all showed increased expression in the cep1 mutant (Supplemental Table 2). Extensins, which are among the most important components in the plant cell wall, form positively charged scaffolds in the cell plate and subsequently provide a template for deposition of the other cell wall components (Cannon et al., 2008). This scaffold may uphold the cells three-dimensional structure even after breakdown of cell turgor. The increased expression of the extensin family genes in cep1 during the three time points hints that tapetal cell wall biogenesis was ongoing, which was consistent with the phenomenon that the tapetal cell wall failed to degenerate in cep1 but disappeared in the wild type. Other cell wall biogenesis genes such as proline-rich protein 4 (PRP4), a leucine-rich repeat (LRR) family protein (AT1G33590), xyloglucan endotransglucosylase/hydrolase 3 (XTH3), laccase-like multicopper oxidase 4 (LAC4), and pectin methylesterase 44 (PME44) also showed increased expression in the cep1 mutant (Supplemental Table 2).
The GO terms “chloroplast parts” (P < 10−7) and “plastid part” (P < 10−6) were significantly enriched in cluster 4 (Supplemental Figure 5D). These genes were downregulated in the cep1 mutant. For example, lists of well-known groups of genes encoding chloroplast development associated protein, including the photosystems I and II reaction center proteins and chloroplast ribosomal protein families, were all downregulated during anther development, particularly at stages 7-9 (Supplemental Table 2), suggesting that formation of chloroplasts or plastids was repressed in the cep1 mutant. Plastids develop into relatively translucent elaioplasts during stage 10 in wild-type tapetum (Ariizumi et al., 2004). Correspondingly, the marked decrease in the formation of two mainly lipid storage organelles in the tapetal cell, the tapetosome, and the elaioplast was detected in the cep1 mutant (Figure 3). Formation of the mature pollen exine requires steryl esters secreted from elaioplasts as well as alkanes, oleosins, and flavonoids secreted from tapetosomes during tapetal PCD. The decreased formation of tapetosomes and elaioplasts suggests that pollen development, particularly pollen exine formation, would be seriously impaired.
In cluster 6, the GO term “pollen development” (P < 10−5) was also enriched, particularly the GO term “pollen wall assembly” (P < 10−6) (Supplemental Figure 5F). Genes associated with pollen wall development, particularly formation of the exine structure, increased expression, suggesting abnormal pollen well formation in the cep1 mutant. Callose deposition is critical for pollen exine formation, as it acts as a mold wherein the primexine provides a blueprint for formation of the exine pattern on the mature pollen soon after completion of meiosis during microsporogenesis (Zhang et al., 2002; Nishikawa et al., 2005). In Arabidopsis, the callose separates in developing microspores, preventing their underlying walls (exine) from fusing during meiosis. Callose synthase 5 (CalS5) is responsible for the synthesis of callose deposited in the primary cell wall of meiocytes, tetrads, and microspores, and expression of this gene is essential for exine formation in the pollen wall (Dong et al., 2005). Absence or overexpression of CalS5 causes aberrations in callose accumulation, exine patterning, and pollen wall permeability alterations (Chang et al., 2012; Xie et al., 2012). The expression of CalS5 increased 2.24- and 2.62-fold at stages 7-9 and stages 10-11 in the cep1 mutant, respectively, suggesting aberrant callose deposition and abnormal exine development. Correspondingly, obvious pollen grain aggregation and abnormal pollen wall development were observed in the cep1 mutant by a TEM analysis.
The exine consists mostly of sporopollenin, which contains mainly phenols and fatty acid derivatives (Morant et al., 2007; Ariizumi and Toriyama, 2011). The expression of several sporopollenin biosynthetic genes, including two cytochrome P450 family genes (CYP703A2 and CYP704B1) (Morant et al., 2007; Dobritsa et al., 2009), acyl-CoA synthetase 5 (ACOS5) (de Azevedo Souza et al., 2009; Lallemand et al., 2013), polyketide synthase A (PKSA) and polyketide synthase B (PKSB) (Kim et al., 2010), tetraketide α-pyrone reductase 1 (TKPR1) (Grienenberger et al., 2010), and a sporopollenin transport gene (ABCG26/WBC27) (Quilichini et al., 2010; Choi et al., 2011) all increased (Supplemental Table 2). The steryl esters, alkanes, and oleosins secreted from the elaioplasts and tapetosomes of the tapetum are needed for mature pollen wall formation (Hsieh and Huang, 2007). Steryl esters and alkanes are lipids that waterproof the pollen; oleosins on the pollen surface assist with water uptake from the stigma to the pollen for germination (Hsieh and Huang, 2004; Li et al., 2012). Therefore, normal lipid and fatty acid metabolism is crucial for fertile pollen formation. In cluster 2, the expression of genes belonging to the tapetal “fatty acid biosynthetic process (P < 10−8)” and “lipid biosynthetic processes (P < 10−5)” GO terms all increased in the cep1 mutant (Supplemental Table 2). The increased expression of these genes hints at the abnormal pollen exine wall development in the cep1 mutant.
Besides the above genes, expression of other hydrolases, such as three papain-like cysteine proteases (AT5G45890, AT1G06260, and AT1G20850) and three aspartic proteases (AT4G30040, AT4G04460, and AT3G52500), clearly increased in the cep1 mutant, suggesting that these genes participate in tapetal cell PCD.
The transcriptomic data show that the expression of genes participating in tapetal PCD and pollen development, including tapetal secretory structure formation, pollen wall development, and tapetal cell wall generation, clearly changed, corresponding to abnormal tapetum degeneration and pollen development in the cep1 mutant.
Overexpression of CEP1 Results in Premature Tapetal PCD and Pollen Infertility
The cDNA sequence of CEP1 was overexpressed under the control of the cauliflower mosaic virus 35S promoter to further confirm CEP1 function in Arabidopsis. Pollen grain fertility decreased significantly in the 22 35S:CEP1 transgenic lines. Compared with wild-type plants, some pollen grains in the 35S:CEP1 transgenic lines adhered to each other, the deposition of pollen exine was chaotic and excessive, and the intine was thicker (Figures 9A to 9D). Correspondingly, tapetal cell degeneration was clearly accelerated in the transgenic lines. At early stage 7, tapetal cell wall degeneration had already concluded in the 35S:CEP1 transgenic lines (Figures 9F and 9J). At stage 8, tapetal cells formed more secretory vacuoles, remnants of primexine were disorganized, and the baculum was particularly more serried (Figures 9G and 9K). At stages 9 and 10, the numbers of tapetosomes and plastids were excessively enriched in tapetal cells and secreted a large amount of sporopollenin into the anther locules. The nucleus degenerated at stage 9 (Figures 9H and 9M). The vacuolated and shriveled microspore showed a disordered and thick exine (Figures 9L and 9Q). At early stage 11, tapetal cells were filled with more and larger tapetosomes and elaioplasts (Figure 9N), and the partially malformed pollen grains showed a chaotic and thick exine (Figure 9R). At late stage 11, only a few tapetal remnants remained in the anther locules, and the pollen showed an aberrant and overly thick exine and intine (Figures 9O and 9S). The anther locule dehisced at stage 12, releasing aberrant pollen grains with an abnormally thick exine and intine (Figures 9P and 9T).
Figure 9.
Pollen and Tapetum Development in the 35S:CEP1 Overexpressing Transgenic Lines
(A) to (C) Scanning electron micrographs of mature overexpressing transgenic line pollen grains. Bars = 50 μm in (A), 10 μm in (B), and 5 μm in (C).
(D) Transmission electron micrographs of mature overexpressing transgenic line pollen exine. Bars = 2 μm.
(E) to (T) Transmission electron micrographs of the tapetum and microspore development in overexpressing transgenic lines at stage 6 ([E] and [I]), early stage 7 ([F] and [J]), stage 8 ([G] and [K]), stage 9 ([H] and [L]), stage 10 ([M] and [Q]), early stage 11 ([N] and [R]), late stage 11 ([O] and [S]), and stage12 ([P] and [T]). Bars = 2 μm.
(U) to (X) DNA fragmentation in overexpressing transgenic line anthers (arrowheads) at stage 8 (U), stage 9 (V), stage 10 (W), and stage 11 (X). Bars = 50 μm.
E, epidermis; El, elaioplast; En, endothecium; Ex, exine; In, intine; ML, middle layer; MMC, microspore mother cell; Msp, microspore; N, nucleus; P, plastid; Po, pollen; SV, secretory vacuole; T, tapetal cell; Ta, tapetosome; V, vacuole.
Tapetal TUNEL-positive signals appeared during stage 9, suggesting that tapetal degeneration had accelerated (Figure 9V) in the 35S:CEP1 overexpressing lines. At stage 10, intense TUNEL-positive signals appeared in tapetal cells with accelerated degeneration (Figure 9W). At stage 11, most of the tapetal cells had degenerated, and TUNEL-positive signals appeared in the residual tapetal cells in the anther locules (Figure 9X).
These observations indicate that overexpressing CEP1 under the control of the 35S promoter causes premature tapetal PCD, including advanced tapetal cell wall degeneration and increased secretory structure formation, resulting in partial pollen infertility with a disorganized and excessively thick exine and intine. In addition, the finding of no visible abnormal phenotype in the other three anther layers (epidermis, endothecium, and middle layer) or in the other vegetative tissues (roots, stems, and leaves) between 35S:CEP1 transgenic lines and the wild-type lines (Supplemental Figures 6A to 6G) indicates that the primary function of the CEP1 was confined to the tapetum rather than the other three anther layers or the other vegetative tissues.
The cDNA sequence of CEP1 was overexpressed under the control of the ProCEP1 promoter to further characterize the influence of CEP1 expression on pollen development. The developmental morphology of the ProCEP1:CEP1 transgenic line was broadly similar to that of the 35S:CEP1 transgenic lines. Pollen development was slightly abnormal and partial pollen grains were infertile in the 19 transgenic lines. Compared with wild-type plants, some pollen grains adhered to each other and the deposition of pollen exine was disordered (Supplemental Figures 6H and 6I). We accordingly speculate that the main function of CEP1 is to participate in tapetal degeneration and pollen development and that CEP1 overexpression causes premature tapetal cell PCD.
CEP1 Is Confined within a Specific Expression Level during Pollen Development
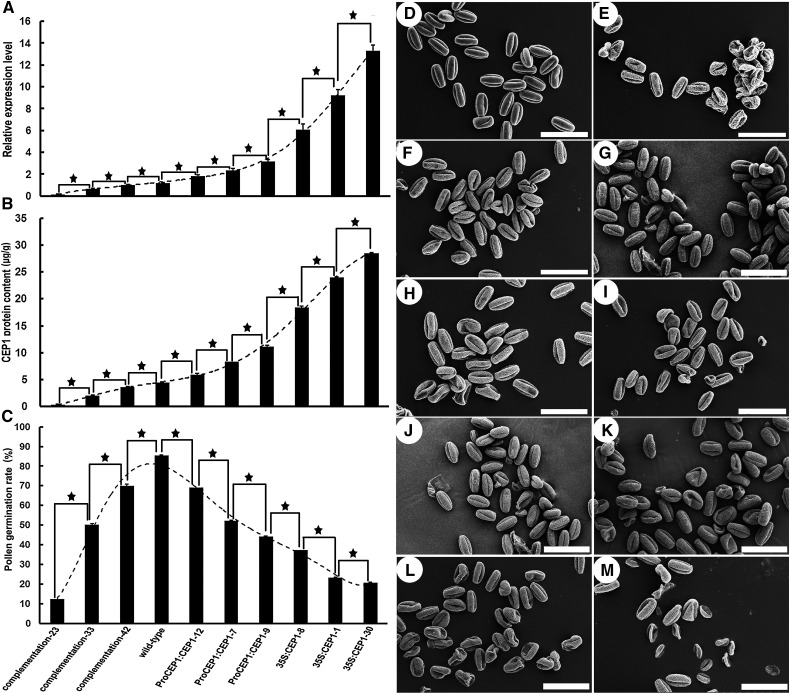
The CEP1 transcription level was detected by qRT-PCR, and the CEP1 protein content was analyzed by an ELISA to further characterize the relationship between the CEP1 expression level and pollen development. A strong relationship was observed between the CEP1 expression level and the degree of pollen fertility in transgenic plants (Figure 10). Wherever the CEP1 transcript level (Figure 10 A) and CEP1 protein content (Figure 10B) in the transgenic lines were lower or higher than in the wild type, the degree of pollen fertility (Figures 10C to 10K) in transgenic lines was lower than in the wild type. Compared with the wild type, a greater decrease in the CEP1 transcription level and CEP1 protein content accompanied a greater decrease in the degree of pollen fertility in the transgenic complementation lines, and the greater the increase in the CEP1 transcription level and CEP1 protein content, the greater the increase in pollen fertility in ProCEP1:CEP1 or 35S:CEP transgenic lines. In general, when the expression level of CEP1 was >200% or <50% of the wild type, the pollen germination rate in vitro decreased to nearly half that of normal fertile wild-type plants. These results show that the CEP1 expression level is closely associated with pollen development and that an accurately regulated CEP1 content is essential for tapetal cell PCD and normal development of pollen grains in Arabidopsis.
Figure 10.
The Relationship between the CEP1 Expression Level and Pollen Development.
(A) qRT-PCR for CEP1 expression in different lines of buds at stages 7-9.
(B) ELISA analysis of CEP1 protein content in different lines of buds at stages 7-9.
(C) Pollen germination rates of the different lines. Bars represent standard deviations.
(D) to (K) Scanning electron micrographs of pollen grains in the wild type (D), complementation transgenic line 23 (E), complementation transgenic line 33 (F), complementation transgenic line 42 (G), ProCEP1:CEP1-12 (H), ProCEP1:CEP1-7 (I), ProCEP1:CEP1-9 (J), 35S:CEP1-8 (K), 35S:CEP1-1 (L), and 35S:CEP1-30 (M). Bars = 50 μm. Columns labeled with a star are significantly different at P < 0.01.
DISCUSSION
CEP1 Acts as a Crucial and Early Executor during Arabidopsis Tapetal PCD
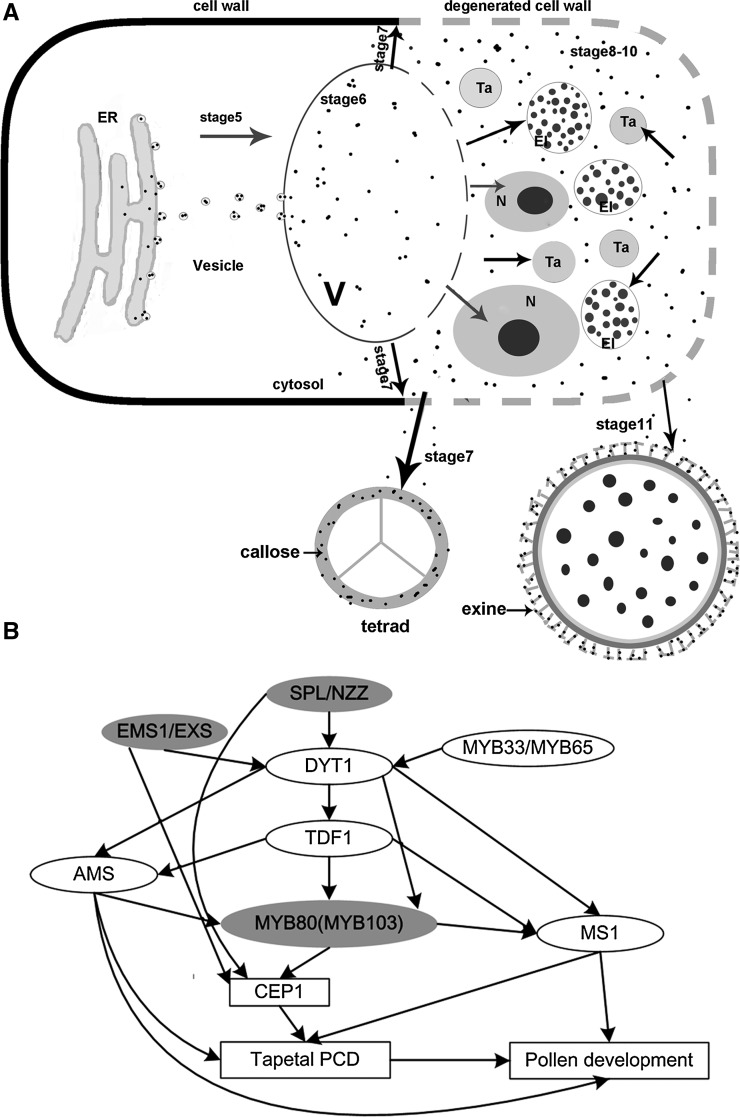
The tapetum plays a vital secretory role during the development of microspores into pollen grains, as it provides enzymes for the release of microspores from tetrads, nutrients for pollen development, and pollen wall components (Varnier et al., 2005). The premature or aborted death of tapetal cells disrupts the nutrient supply to the microspores, leading to male sterility (Ku et al., 2003; Kawanabe et al., 2006; H. Li et al., 2011; X. Li et al., 2011). We identified a KDEL-tailed cysteine protease, CEP1, which is expressed particularly in anthers from stages 5 to 11. CEP1 affected cell nucleus degeneration and the formation of the tapetal secretory structures and participated in pollen development (Figure 11A). Further analysis showed a strong relationship between the CEP1 expression level and the degree of pollen fertility. Taken together, these results suggest that CEP1 acts as a crucial hydrolase during Arabidopsis tapetal PCD and pollen maturation.
Figure 11.
Diagram of the Model of the CEP1 Reactions during Tapetum and Pollen Development.
(A) Model depicting the synthesis and transportation of CEP1 during tapetum degeneration and pollen development. Solid arrows represent transport and function. Broken lines represent ruptured vacuoles and degenerated cell walls. El, elaioplast; N, nucleus; Ta, tapetosome; V, vacuole.
(B) A model for CEP1 regulation during anther and pollen development. Solid arrows represent gene regulation and gray ovals represent direct gene regulation.
Because the C-terminal KDEL motif targets proteins by acting as an ER retrieval signal for recycling from the Golgi apparatus (Møgelsvang and Simpson, 1998), many KDEL-tailed papain-like cysteine proteases are targeted to the ricinosome, which is derived from the ER and eventually separates from it. Ricinosomes that do not fuse with the central vacuole lyse upon collapse of the central vacuole at death, releasing cysteine proteases or hydrolases into the cytoplasm (Greenwood et al., 2005). Thus, the functions of the active enzymes released from the ricinosomes require collapse of the central vacuole and acidification of the cytoplasm. Previous reports indicate that the castor bean (Ricinus communis) KDEL-tailed cysteine protease, CysEP, targets the ricinosomes and is involved in the final stages of corpse processing during the PCD of endospermic cells and subsequent germination. The tomato KDEL-tailed cysteine protease CysEP is also expressed specifically in ricinosomes of the middle layer, the endothecium, and the epidermal cells, but not the tapetal cells, and is speculated to be a late player involved in corpse processing (Senatore et al., 2009). Different from the castor bean CysEP and tomato CysEP located in the ricinosomes, the KDEL-tailed cysteine protease CEP1, which is not a vacuolar protease (as are αVPE, βVPE, γVPE, and δVPE) in Arabidopsis, first appeared in the precursor protease vesicles as a proenzyme, then aggregated in the tapetal vacuole with fusion of these vesicles and was transformed into the mature form before rupture of the vacuole. In addition, the recombinant pro-CEP1 underwent self-hydrolysis in vitro at pH 3.0, which is different from pH 4.8 of the tomato pro-CysEP enzyme in vitro self-hydrolysis and that of the castor pro-CysEP at pH 5.5. Thus, CEP1 should function immediately after release from the collapsed central vacuole and earlier than the other proteases, which are released from ricinosomes. Furthermore, the CEP1 transcript and protein accumulation also corresponded with the process of tapetal degeneration in Arabidopsis and occurred earlier than that of several known cysteine proteases, including rice CP1, tomato CysEP, and maize (Zea mays) PCP. CP1 in the rice tapetum is expressed mainly during stages 8 and 9, during which the microspores undergo meiosis and release from the tetrad (Li et al., 2006). Tomato CysEP transcript and protein accumulation begins mainly at stage 13 and is complete by stage 20 during tomato flower development (Senatore et al., 2009). The maize cysteine protease PCP is synthesized at a very late stage of anther development, during which tapetal degeneration is nearly finished (Li et al., 2012). Thus, we infer that CEP1 acts as an early executor, functions throughout the process of tapetal degeneration (from stage 6 to 11), and plays a novel role in tapetal PCD.
Further transcriptomic analysis of the gene expression between the wild type and cep1 mutant at three developmental stages revealed that a total of 872 genes had significantly different expression levels in at least one of the three time points. At stages 5-6, 7-9, and 10-11, 335, 338, and 491 genes, respectively, were upregulated (Figure 8A), and 89, 119, and 76 genes, respectively, were downregulated (Figure 8B). We then used GO term enrichment analysis to further characterize the differentially expressed genes in the cep1 mutant lines. Most of the upregulated genes encode proteins that were associated with the cell wall, and cell component organization and biosynthesis (Supplemental Figure 5G), whereas most of the downregulated genes encode proteins that were associated with tapetosome development (e.g., oleosins; Supplemental Figure 5H). This is consistent with the phenotype of cep1 mutants, such as delayed tapetal cell PCD and reduced tapetosome (e.g., oleosins) formation. These differentially expressed genes might not be regulated directly by CEP1, but rather by the delayed tapetal cell development in the cep1 mutant.
CEP1 Directly Participates in Tapetal Cell PCD
Normal tangential tapetal cell wall breakdown is necessary for materials to be secreted into the locules. The existing tapetal cell wall prevents transformation of the secretory-type tapetum (Zhang et al., 2007; H. Li et al., 2011). Degeneration of the plant cell wall requires the KDEL-tailed proteases, which possess the rare ability to accept proline or highly glycosylated hydroxyprolines, allowing them to hydrolyze the extensins, which are key plant cell wall components (Than et al., 2004; Helm et al., 2008). In our study, the immunoelectron microscopy observations confirmed that the mature CEP1 enzyme was transported specifically to the tapetal cell wall after tonoplast breakdown at stage 7. Correspondingly, loss of CEP1 function in the cep1 mutant caused failure of tapetal cell wall degeneration during this stage. Overexpression of CEP1 in the 35S:CEP1 transgenic lines advanced tapetal cell wall degeneration to early stage 7. These observations indicate that the CEP1 protease participates directly in tapetal cell wall hydrolysis. The transcriptomic analysis supported this conclusion. Compared with the wild type, the expression of genes associated with cell wall organization or biogenesis obviously increased during the three development stages in the cep1 mutant, suggesting that the tapetal cell wall continues to be synthesized after stage 7. Therefore, we infer that CEP1 participates directly in tapetal cell wall hydrolysis and affects transformation of the secretory-type tapetum.
Tapetosomes and elaioplasts are the two specific and predominant storage organelles that store and transfer major pollen wall materials in Brassicaceae species (Li et al., 2012). Elaioplasts in the tapetum are specialized plastids derived from proplastids for temporary storage of steryl esters, and tapetosomes are storage organelles, each having numerous oleosin-coated alkane droplets linked ionically with flavonoid-containing vesicles (Wu et al., 1997). Our results show that formation of the elaioplasts and tapetosomes correlated closely with CEP1 expression. In the cep1 mutant, the reduced expression of genes participating in the plastid formation at stages 7-9, including the PSB and RPS families, suggests that the formation of elaioplasts and tapetosomes had decreased. The TEM analysis confirmed that the formation of elaioplasts and tapetosomes largely decreased in the cep1 mutant, but markedly increased in the 35S:CEP1 overexpressing transgenic lines. Therefore, we infer that CEP1 could directly or indirectly participate in formation of the secretory structure during tapetal cell PCD, although the exact mechanism remains unknown.
CEP1 Affects Pollen Development
The pollen exine, as the outer sculptured part of the pollen wall, not only provides a protective barrier against pathogen attack, dehydration, and UV irradiation, but also facilitates pollen recognition and adhesion to the stigma (Edlund et al., 2004; Ariizumi and Toriyama, 2011). The exine consists mostly of sporopollenin, which is secreted from the tapetum during PCD. In the cep1 mutant, few fibrillar materials, which consist mainly of sporopollenin and other lipid/flavonoid derivatives, were released from the tapetum to the anther locule, resulting in markedly abnormal pollen and pollen exine development. According to these observations, the synthesis of lipids, flavanoids, and sporopollenin would be downregulated in the cep1 mutant. Unexpectedly, many genes involved in lipid, flavanoid, and sporopollenin biosynthesis were upregulated in the cep1 mutant, suggesting that the lower content of fibrillar materials in the anther locule is not caused by decreased synthesis of lipids/flavanoids and sporopellenin in the tapetum (Supplemental Table 2). However, the presence of the tapetal cell wall and the reduced formation of lipid secretory structures in the cep1 mutant suggest that the transport of these materials from the tapetum to the anther locule is severely obstructed. Thus, the main causes of abnormal pollen development in the cep1 mutant are expected to be the obstructed transport of the lipids, flavonoids, and sporopollenin from the tapetum to the anther locule and not their decreased synthesis in the tapetum. Consistently, increased lipid, flavanoid, and sporopollenin release from the tapetum to the anther locule was detected by TEM analysis in the 35S:CEP1 overexpressing transgenic lines, accompanied by accelerated tapetal cell wall degeneration and increased secretory structure formation.
Microspores are enclosed by a callose wall near the end of meiosis, which serves as a mold for primexine patterning and also protects the meiocytes and tetrads (Worrall et al., 1992; Dong et al., 2005). The immunoelectron microscopy observations showed that the CEP1 protein was delivered into the callose wall after degeneration of the tapetal cell wall. Although no obvious abnormalities in callose wall deposition were observed in the cep1 mutant by TEM analysis (Figure 5), marked pollen grain aggregation was detected during cep1 microspore development (Figure 2). In addition, CALS5 expression in the cep1 mutant increased sharply during stages 7-9 and 10-11. CALS5 is required for the production of temporary callose walls that separate developing microspores, so the callose layer is critical for pollen exine development (Dong et al., 2005). The aberrant CALS5 expression suggests abnormal exine formation in cep1 mutants. Therefore, CEP1 should play an important role in callose deposition, and the loss of CEP1 function probably accounts for abnormal callose wall deposition and impaired pollen exine development, resulting in pollen aggregation.
The tapetum not only contributes to nutrition for pollen development but provides materials responsible for sculpting the pollen surface, which is important during pollen recognition (Vizcay-Barrena and Wilson, 2006). In maize, the cysteine protease PCP, which becomes abundant on the mature pollen surface, is speculated to act on the stigma surface, where it hydrolyzes and modifies proteins to generate a hole for pollen tube entry (Li et al., 2012). Similar to the PCP cysteine protease, CEP1 was also located specifically on the mature pollen coat after tapetal degeneration. Thus, CEP1 likely also plays an important role in pollen recognition and pollen tube germination.
CEP1 Is Likely Involved in the SPL/NZZ, EMS1/EXS, and MYB80 Pathways Regulating Tapetal Development
Tapetal development is controlled by a complex transcriptional regulatory network. Many transcription factors are involved in anther cell differentiation and tapetal development. Studies have suggested that EMS1/EXS is differentially regulated by SPL/NZZ and that DYT1 acts genetically downstream of SPL/NZZ and EMS1/EXS but upstream of TDF1, AMS, MS1, and MYB80 (Zhu et al., 2011; Feng et al., 2012). The MYB33 and MYB65 proteins are speculated to form heterodimers with DYT1 to regulate tapetum-preferential gene expression and are not regulated by DYT1 (Millar and Gubler, 2005; Zhang et al., 2006). These regulatory genes coordinate the expression of downstream genes associated with late tapetal development, tapetal PCD, and pollen wall formation, which are essential for anther development and maturation. Although microarray data for many mutants, including spl/nzz, ems1/exs, dyt1, tdf1, ams, and myb80, have been analyzed, the possible CEP1 regulatory pathway has been ignored. An analysis of the known microarray data showed that CEP1 expression was downregulated 5.7-fold and 3.2-fold in the spl/nzz and ems1/exs mutants, respectively, and upregulated 4.8-fold in myb80 mutants (Wijeratne et al., 2007; Phan et al., 2011). However, CEP1 expression was not significantly altered in the dyt1, tdf1, ams, and ms1 mutants, whereas other papain-like cysteine proteases such as RD19C, RDL1, RD19A, THI1, and RD21A show different degrees of expression change. The cep1 mutant transcriptomic data also show that the expression of these transcription factors (SPL/NZZ, EMS1/EXS, DYT1, TDF1, MS1, and MYB80) did not change obviously. These results suggest that the CEP1 cysteine protease is involved in the SPL/NZZ, EMS1/EXS, and MYB80 pathways regulating tapetal development and degeneration and that the above-mentioned other papain-like cysteine proteases are involved in the DYT1, TDF1, AMS, and MS1 pathways (Figure 11B). SPL/NZZ and EMS1/EXS are expressed during the anther differentiation stages and probably act as early regulators of CEP1 expression around stage 5. MYB80, which is expressed mainly during stages 7-10 in the tapetum (Higginson et al., 2003; Li et al., 2007), probably acts as a later regulator of CEP1 expression. During these stages, tapetal cell vacuolation and breakdown are premature in the myb80 mutant, in which CEP1 expression increased 4.8-fold; this result is similar to that for the 35S:CEP1 overexpressing transgenic lines in our study. Furthermore, a microarray analysis of the Arabidopsis MYB80:GR inducible lines showed that 79 genes were most likely directly regulated by MYB80 (Phan et al., 2011), but CEP1 was not included. This result suggests that CEP1 probably acts downstream of SPL/NZZ, EMS1/EXS, and MYB80 to participate in tapetal development and degeneration but is not directly regulated by MYB80.
In conclusion, the CEP1 cysteine protease is a key executor in tapetal cell PCD and pollen development. Correct expression of CEP1 in the tapetum is necessary for tapetal PCD and pollen development. The characterization of CEP1 expression and regulatory metabolism during tapetal development in this study should provide valuable insight into the process of tapetal PCD and pollen development and accelerate the understanding of the exact function of the cysteine proteases during anther maturation and in identifying the cysteine proteases involved in plant cell PCD.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana accession Columbia-0 was used for all gene transfer experiments and as the wild-type control. Plants were grown on a soil mixture (3:1:1 mixture of peat moss-enriched soil:vermiculite:perlite) with a 14-h-light/10-h-dark photoperiod at 23°C. Homozygous T-DNA insertion mutants were identified by PCR using the CEP1M3-F/R/B and CEP1M4-F/R/B primers, respectively.
Characterization of the Mutant Phenotype
Arabidopsis plants were photographed with a digital camera (Coolpix 9100; Nikon). Arabidopsis flower and anther images were acquired with an M165 C microscope (Leica). To evaluate the viability of mature pollen grains, dehiscent anthers from mature flowers were dipped in a drop of iodium–potassium iodide solution (1 g iodine and 3 g potassium iodide in 100 mL water) (Chang et al., 2009), and images were captured with a Leica DM 6000 B microscope. Germination was assessed by culturing fresh pollen grains in germination medium containing 3 mM CaCl2, 1 mM H3BO3, 56 mM inositol, 1% agar (w/v), and 15% sucrose (w/v) (pH 5.8) at 25°C for 3 h.
Semithin Sections and TEM
For semithin sections and TEM, anthers at various developmental stages were prefixed and embedded as described by Li et al. (2006). Semithin sections of 2 μm were cut using a UC6 ultramicrotome (Leica) stained with 1% toluidine blue O (Sigma-Aldrich) and photographed using a Leica DM 6000 B microscope. Ultrathin sections (70 nm) were obtained with a UC6 ultramicrotome (Leica) and then double-stained with 2% (w/v) uranyl acetate and 2.6% (w/v) lead citrate aqueous solution. Observations and image capture were performed with an H-7650 transmission electron microscope (Hitachi) at 80 kV and an 832 charge-coupled device camera (Gatan).
Scanning Electron Microscopy
Pollen grains collected from freshly dehisced anthers were mounted on scanning electron microscopy stubs. The mounted samples were coated with palladium-gold in a sputter coater (E-1010; Hitachi) and examined by scanning electron microscopy (S-3400N; Hitachi) at an acceleration voltage of 10 kV.
TUNEL
Buds at different developmental stages were fixed in a PBS solution (1.3 M NaCl, 70 mM Na2HPO4, and 30 mM NaH2PO4, pH 7.0) containing 4% paraformaldehyde for 24 h at 4°C. The 8-μm Parafilm sections of treated buds were assessed with a TUNEL apoptosis detection kit (DeadEnd Fluorometric TUNEL system; Promega) according to the supplier’s instructions. Samples were observed using a Leica DMI6000 CS confocal laser scanning microscope.
Molecular Cloning and Plasmid Construction
A 3.06-kb genomic DNA fragment containing the CEP1 genomic sequence and the ProCEP1 promoter was amplified with the Pro-CEP1-F/R primers and inserted into the pCAMBIA 1300 binary vector for the functional complementation test. Full-length cDNA was amplified with the primer pair 35SOX -F/35SOX -R and cloned into pBI121 for the 35S:CEP1 overexpression test. Full-length cDNA was amplified with the CEP1-C-F/R primer pair, and the ProCEP1 promoter was amplified with the ProCEP1-F/R primer pair and cloned into pCAMBIA1300 for the ProCEP1:CEP1 overexpression test.
The ORF minus the first 60 bp of CEP1 cDNA was amplified by PCR with the two CEP1-CM-F/R primers and inserted downstream of the pET30a plasmid T7 promoter (Novagen). The expression, extraction, purification, and renaturation of the CEP1 protein were performed according to the procedure described by Zhang et al. (2009).
Enzyme Assays
The enzyme activity of purified recombinant CEP1 was routinely assayed with Z-FR-AMC and R-AMC as the substrates (Rozman-Pungercar et al., 2003). An inhibition assay was performed using several inhibitors, including the papain-like cysteine protease inhibitor E-64, the cysteine protease inhibitors leupeptin, the caspase inhibitors Z-VAD-fmk and Ac-YVAD-cmk, the aspartic protease inhibitor pepstatin A, and the serine proteases inhibitors PMSF and EDTA, respectively. The dependence of enzyme activity on pH and temperature was measured in a pH gradient of 1.0 to 6.0 and a temperature gradient of 20 to 80°C.
In Situ Hybridization
Tissues were fixed and hybridized according to Brewer et al. (2006) with the following modifications. A 150-bp CEP1 cDNA segment was amplified by PCR with CEP1-SH-F/R and inserted into the pSPT19 plasmid to construct a plasmid that was used to synthesize antisense and sense probes. The probes were generated using the DIG RNA labeling kit (Roche Diagnostics) according to the manufacturer’s instructions. The RNA was hybridized, and the hybridized probes were detected according to the protocol of Darby and Hewitson (2006). Slides were observed and photographed using a Leica DM 6000 B microscope.
CEP1 Immunolocalization
A 459-bp fragment encoding the CEP1 peptide was amplified from cDNA by PCR using the An-CEP1-F/R primer to make the anti-CEP1 antibody. The expression, extraction, purification, and renaturation of the CEP1 peptide were performed according to the procedures described by Zhang et al. (2009).The purified recombinant peptide (5 mg) was used to immunize rabbits at 3-week intervals. The specificity of the CEP1 antibody was confirmed by hybridization with a membrane blotted with protein extracts from the buds of Arabidopsis (Supplemental Figure 7).
Wild-type Arabidopsis anthers were fixed and embedded as described by Schmid et al. (1999). The sections were then labeled with affinity-purified CEP1 antibody (1:500) in PBGT (150 mM NaCl, 2 mM KCl, 8 mM Na2HPO4, 0.2% gelatin [w/v], 0.2% Tween 20 [v/v], and 1.0% BSA [w/v]). The negative control did not label with affinity-purified CEP1 antibody. After washing in PBGT, the sections were labeled with colloidal gold particles (10 nm) coupled to goat anti-rabbit immunoglobulin G (1:20) in PBGT. The sections were then photoelectrically sensitized using silver lactate solution after labeled with the gold particles. The grids were examined and photographed with an H-7650 transmission electron microscope (Hitachi) at 80 kV and an 832 charge-coupled device camera (Gatan).
Immunoblotting
The anti-SlCysEP antibody was diluted 1:200 followed by affinity-purified goat anti-rabbit IgG horseradish peroxidase (HRP)-conjugated antibody (CW Bio) diluted 1:4000 for immunoblotting. ECL Plus protein gel blotting detection reagents (CW Bio) were used as the HRP substrate and were exposed in the Fusion X7 chemiluminescence imaging system.
Cell Wall Protein Isolation
Buds from wild-type Arabidopsis (stages 5-6, 7-8, and 9-11) were gathered and fixed in liquid nitrogen. Two grams of fixed buds from each developmental stage were ground in liquid nitrogen. The cell wall preparations and protein extracts of Arabidopsis bud cell walls were characterized as described above (Feiz et al., 2006).
ProCEP1-GFP Fusion Targeting Analyses
A GFP gene was amplified by PCR with the two primers (GFP-F/GFP-R). The product was inserted to pBI101 (Clontech), which was minus the GUS coding region previously. Full-length cDNA were amplified with the CEP1-C-F/R primer pair, and the ProCEP1 promoter was amplified with the ProCEP1-F/R primer pair and cloned to create the ProCEP1-GFP fusion construct. The cell wall fluorescence stain calcofluor white stain was also used. To avoid the interference of GFP fluorescence originating from the plasma membrane, which is always close to the cell wall, partial plasmolysis was performed. The transgenic Arabidopsis flowers filament were analyzed using the Leica DMI6000 CS confocal laser scanning microscope.
RT-PCR and qRT-PCR Analyses
CEP1 expression in different Arabidopsis tissues was assessed by RT-PCR using the RT-CEP1-F/R primer. Tubulin was used as a control with the tubulin-F/R primer. The conditions for PCR amplification were as follows: 94°C for 5 min; 40 cycles at 94°C for 30 s, 56°C for 35 s, and 72°C for 60 s; and 72°C for 10 min.
The qRT-PCR analyses of the CEP1 in buds at different developmental stages were performed using the 2× SYBR Green qPCR mix (LabAid; Thermo Scientific) on an iQ5 Multicolor Real-Time PCR detection system (Bio-Rad) using the qRT-CEP1-F/R primer. The PCR conditions were as follows: 94°C for 3 min; 40 cycles at 94°C for 10 s, 55°C for 20 s, 72°C for 20 s, and 60°C for 30 s; and 72°C for 1 min. qRT-actin-F/R was used as the internal control to normalize the expression data. Data were analyzed using the iQ5 (Bio-Rad) software, and differences in gene expression were calculated using the 2–ΔΔCt analysis method.
ELISA Analysis
Anti-CEP1 antibody in PBS was added to each well of a microtiter plate and left overnight at room temperature. The plate was washed with washing buffer (0.05% Tween 20 in PBS [v/v], pH 7.2) and blocked with blocking buffer (SinoGene). The reagent diluents (5% BSA [w/v] and 0.05% Tween 20 in PBS, pH 7.2) including the bud protein extract to be tested were added to each well. The CEP1 antibody (1:5000 in PBS) was added to each well again, and goat anti-rabbit IgG HRP-conjugated antibody (1:5000 in PBS) was added to each well. The HRP substrate was added, and stop solution was added to stop the reaction after the color reaction. The optical density values were recorded using a Bio-Rad 680 microplate reader.
RNA-Seq Technology: Sample Collection, Illumina Sequencing, and Data Processing
Three stages of buds from the wild type and the cep1 SALK_013036 mutant were collected, and total RNA was isolated using TRIzol reagent according to the manufacturer’s protocol (Invitrogen). The samples were sequenced using the Illumina Genome Analyzer (HiSeq 2000; Illumina). The raw reads were data-filtered to obtain high-quality clean reads. The clean reads were mapped to the Arabidopsis reference genome and reference genes using SOAPaligner/SOAP2 (Li et al., 2009). No more than two mismatches were allowed in the alignment. The gene expression level was calculated using RPKM (reads per kb per million reads) (Mortazavi et al., 2008). The data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50956) under accession number GSE50956.
Identification and Analysis of Differentially Expressed Genes
Differential expression analysis between the wild type and cep1 mutant was performed using the DEGseq R package (1.12.0) (Wang et al., 2009) based on the normalized read counts. P values were adjusted using the Benjamini and Hochberg (1995) method. A corrected P value of <0.005 and |log2Ratio| > 1 were set as the threshold for significantly different expression. A cluster analysis of the gene expression patterns was performed based on the K-means method using Cluster 3.0 (Sturn et al., 2002). A GO enrichment analysis of the differentially expressed genes was performed using the GOseq R package, in which gene length bias was corrected (Young et al., 2010). GO terms with a corrected P value < 0.01 were considered significantly enriched among differentially expressed genes.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under accession number At5G50260 (CEP1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Complementation of the cep1 Mutant by CEP1 Genomic DNA.
Supplemental Figure 2. Nucleotide and the Deduced Amino Acid Sequence of At5g50260.
Supplemental Figure 3. A Phylogenetic Tree of Papain-Like Cysteine Proteases in Arabidopsis.
Supplemental Figure 4. Purified Recombinant CEP1.
Supplemental Figure 5. Gene Ontology (GO) Term Enrichment of Differentially Expressed Genes in the cep1 Mutant.
Supplemental Figure 6. CEP1 Overexpression Phenotype Using the CaMV35S and ProCEP1 Promoters.
Supplemental Figure 7. Specificity Analysis of the Antimature CEP1 Antibodies by immunoblot.
Supplemental Table 1. Quantitative RT-PCR Corroboration of the Differentially Expressed Genes.
Supplemental Table 2. Partial List of Genes Differentially Expressed in the cep1 Mutant.
Supplemental Table 3. Primer Information Used for Molecular Cloning and Plasmid Construction.
Supplemental Table 4. Primer Information Used for RT-PCR/Quantitative RT-PCR.
Supplemental Data Set 1. Alignment Corresponding to the Phylogenetic Analysis in Supplemental Figure 3.
Supplemental Data Set 2. Differentially Expressed Genes That Were Significantly Upregulated or Downregulated (P Value < 0.005, Fold Change >2 or <–2) in at Least One of the Three Stages (stages 5-6, stages 7-9, stages 10-11) in cep1 Buds.
Supplementary Material
Acknowledgments
We thank Danlong Jing (Research Institute of Forestry, Chinese Academy of Forestry) for support during data analysis. This research was supported by grants from the Natural Science Foundation of China (31170574, 31370590, and 30671697).
AUTHOR CONTRIBUTIONS
D.Z., D.L., X.L., Y.W., Z.X., and Z.L. performed all of the experiments and analyzed the results. H.L. and F.L. designed all of the experiments and wrote and revised the article.
Glossary
- PCD
programmed cell death
- ER
endoplasmic reticulum
- TEM
transmission electron microscopy
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling
- qRT-PCR
quantitative RT-PCR
- ORF
open reading frame
- Z-FR-AMC
Z-Phe-Arg-7-amido-4-methylcoumarin
- R-AMC
l-Arg -7-amido-4-methylcoumarin
- GO
Gene Ontology
- HRP
horseradish peroxidase
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Ahmed S.U., Rojo E., Kovaleva V., Venkataraman S., Dombrowski J.E., Matsuoka K., Raikhel N.V. (2000). The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J. Cell Biol. 149: 1335–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T., Toriyama K. (2011). Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 62: 437–460. [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Hatakeyama K., Hinata K., Inatsugi R., Nishida I., Sato S., Kato T., Tabata S., Toriyama K. (2004). Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39: 170–181. [DOI] [PubMed] [Google Scholar]
- Becker C., Senyuk V.I., Shutov A.D., Nong V.H., Fischer J., Horstmann C., Müntz K. (1997). Proteinase A, a storage-globulin-degrading endopeptidase of vetch (Vicia sativa L.) seeds, is not involved in early steps of storage-protein mobilization. Eur. J. Biochem. 248: 304–312. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Brewer P.B., Heisler M.G., Hejatko J., Friml J., Benkova E.(2006). In situ hybridization for mRNA detection in Arabidopsis tissue sections. Nat. Protoc. 1: 1462–1467. [DOI] [PubMed] [Google Scholar]
- Canales C., Bhatt A.M., Scott R., Dickinson H. (2002). EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 12: 1718–1727. [DOI] [PubMed] [Google Scholar]
- Cannon M.C., Terneus K., Hall Q., Tan L., Wang Y., Wegenhart B.L., Chen L., Lamport D.T., Chen Y., Kieliszewski M.J. (2008). Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. USA 105: 2226–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y., Gong L., Yuan W., Li X., Chen G., Li X., Zhang Q., Wu C. (2009). Replication protein A (RPA1a) is required for meiotic and somatic DNA repair but is dispensable for DNA replication and homologous recombination in rice. Plant Physiol. 151: 2162–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.S., Zhang C., Chang Y.-H., Zhu J., Xu X.-F., Shi Z.-H., Zhang X.-L., Xu L., Huang H., Zhang S., Yang Z.N. (2012). No primexine and plasma membrane undulation is essential for primexine deposition and plasma membrane undulation during microsporogenesis in Arabidopsis. Plant Physiol. 158: 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.H., Huang L.T., Yap M.N., Lee R.H., Huang Y.J., Cheng M.C., Chen S.C. (2002). Molecular characterization of a senescence-associated gene encoding cysteine proteinase and its gene expression during leaf senescence in sweet potato. Plant Cell Physiol. 43: 984–991. [DOI] [PubMed] [Google Scholar]
- Choi H., Jin J.Y., Choi S., Hwang J.U., Kim Y.Y., Suh M.C., Lee Y. (2011). An ABCG/WBC-type ABC transporter is essential for transport of sporopollenin precursors for exine formation in developing pollen. Plant J. 65: 181–193. [DOI] [PubMed] [Google Scholar]
- Darby, I.A., and Hewitson, T.D. (2006). In Situ Hybridization Protocols. (Totowa, NJ: Humana Press). [Google Scholar]
- de Azevedo Souza C., Kim S.S., Koch S., Kienow L., Schneider K., McKim S.M., Haughn G.W., Kombrink E., Douglas C.J. (2009). A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A.A., Shrestha J., Morant M., Pinot F., Matsuno M., Swanson R., Møller B.L., Preuss D. (2009). CYP704B1 is a long-chain fatty acid ω-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 151: 574–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Hong Z., Sivaramakrishnan M., Mahfouz M., Verma D.P. (2005). Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 42: 315–328. [DOI] [PubMed] [Google Scholar]
- Eason J.R., Ryan D.J., Pinkney T.T., O'Donoghue E.M. (2002). Programmed cell death during flower senescence: isolation and characterization of cysteine proteinases from Sandersonia aurantiaca. Funct. Plant Biol. 29: 1055–1064. [DOI] [PubMed] [Google Scholar]
- Edlund A.F., Swanson R., Preuss D. (2004). Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell 16 (suppl.): S84–S97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiz L., Irshad M., Pont-Lezica R.F., Canut H., Jamet E. (2006). Evaluation of cell wall preparations for proteomics: a new procedure for purifying cell walls from Arabidopsis hypocotyls. Plant Methods 2: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B., Lu D., Ma X., Peng Y., Sun Y., Ning G., Ma H. (2012). Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development. Plant J. 72: 612–624. [DOI] [PubMed] [Google Scholar]
- Funk V., Kositsup B., Zhao C., Beers E.P. (2002). The Arabidopsis xylem peptidase XCP1 is a tracheary element vacuolar protein that may be a papain ortholog. Plant Physiol. 128: 84–94. [PMC free article] [PubMed] [Google Scholar]
- Greenwood J.S., Helm M., Gietl C. (2005). Ricinosomes and endosperm transfer cell structure in programmed cell death of the nucellus during Ricinus seed development. Proc. Natl. Acad. Sci. USA 102: 2238–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E., Kim S.S., Lallemand B., Geoffroy P., Heintz D., Souza Cde.A., Heitz T., Douglas C.J., Legrand M. (2010). Analysis of TETRAKETIDE α-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 22: 4067–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.J., Lin W., Oda Y., Cui K.M., Fukuda H., He X.Q. (2012). The proteasome is responsible for caspase-3-like activity during xylem development. Plant J. 72: 129–141. [DOI] [PubMed] [Google Scholar]
- He X., Kermode A.R. (2003). Proteases associated with programmed cell death of megagametophyte cells after germination of white spruce (Picea glauca) seeds. Plant Mol. Biol. 52: 729–744. [DOI] [PubMed] [Google Scholar]
- Helm M., Schmid M., Hierl G., Terneus K., Tan L., Lottspeich F., Kieliszewski M.J., Gietl C. (2008). KDEL-tailed cysteine endopeptidases involved in programmed cell death, intercalation of new cells, and dismantling of extensin scaffolds. Am. J. Bot. 95: 1049–1062. [DOI] [PubMed] [Google Scholar]
- Hierl G., Vothknecht U., Gietl C. (2012). Programmed cell death in Ricinus and Arabidopsis: the function of KDEL cysteine peptidases in development. Physiol. Plant. 145: 103–113. [DOI] [PubMed] [Google Scholar]
- Higginson T., Li S.F., Parish R.W. (2003). AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J. 35: 177–192. [DOI] [PubMed] [Google Scholar]
- Hsieh K., Huang A.H. (2004). Endoplasmic reticulum, oleosins, and oils in seeds and tapetum cells. Plant Physiol. 136: 3427–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K., Huang A.H. (2007). Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell 19: 582–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Nagata N., Yoshiba Y., Ohme-Takagi M., Ma H., Shinozaki K. (2007). Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19: 3549–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanabe T., Ariizumi T., Kawai-Yamada M., Uchimiya H., Toriyama K. (2006). Abolition of the tapetum suicide program ruins microsporogenesis. Plant Cell Physiol. 47: 784–787. [DOI] [PubMed] [Google Scholar]
- Kim S.S., et al. (2010). LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22: 4045–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku S., Yoon H., Suh H.S., Chung Y.Y. (2003). Male-sterility of thermosensitive genic male-sterile rice is associated with premature programmed cell death of the tapetum. Planta 217: 559–565. [DOI] [PubMed] [Google Scholar]
- Lallemand B., Erhardt M., Heitz T., Legrand M. (2013). Sporopollenin biosynthetic enzymes interact and constitute a metabolon localized to the endoplasmic reticulum of tapetum cells. Plant Physiol. 162: 616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Yuan Z., Vizcay-Barrena G., Yang C., Liang W., Zong J., Wilson Z.A., Zhang D. (2011). PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 156: 615–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Yu C., Li Y., Lam T.-W., Yiu S.-M., Kristiansen K., Wang J. (2009). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967. [DOI] [PubMed] [Google Scholar]
- Li S.F., Iacuone S., Parish R.W. (2007). Suppression and restoration of male fertility using a transcription factor. Plant Biotechnol. J. 5: 297–312. [DOI] [PubMed] [Google Scholar]
- Li X., Gao X., Wei Y., Deng L., Ouyang Y., Chen G., Li X., Zhang Q., Wu C. (2011). Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-box adenosine 5′-triphosphate-dependent RNA helicases regulates tapetum degeneration. Plant Cell 23: 1416–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Suen D.F., Huang C.Y., Kung S.Y., Huang A.H. (2012). The maize tapetum employs diverse mechanisms to synthesize and store proteins and flavonoids and transfer them to the pollen surface. Plant Physiol. 158: 1548–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Huang J., Parameswaran S., Ito T., Seubert B., Auer M., Rymaszewski A., Jia G., Owen H.A., Zhao D. (2009). The SPOROCYTELESS/NOZZLE gene is involved in controlling stamen identity in Arabidopsis. Plant Physiol. 151: 1401–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møgelsvang S., Simpson D.J. (1998). Changes in the levels of seven proteins involved in polypeptide folding and transport during endosperm development of two barley genotypes differing in storage protein localisation. Plant Mol. Biol. 36: 541–552. [DOI] [PubMed] [Google Scholar]
- Millar A.A., Gubler F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17: 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant M., Jørgensen K., Schaller H., Pinot F., Møller B.L., Werck-Reichhart D., Bak S. (2007). CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19: 1473–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Nishikawa S., Zinkl G.M., Swanson R.J., Maruyama D., Preuss D. (2005). Callose (β-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biol. 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M.S., Noh Y.S., Martínez D.E., Vila Petroff M.G., Staehelin L.A., Amasino R.M., Guiamet J.J. (2005). Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 41: 831–844. [DOI] [PubMed] [Google Scholar]
- Phan H.A., Li S.F., Parish R.W. (2012). MYB80, a regulator of tapetal and pollen development, is functionally conserved in crops. Plant Mol. Biol. 78: 171–183. [DOI] [PubMed] [Google Scholar]
- Phan H.A., Iacuone S., Li S.F., Parish R.W. (2011). The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23: 2209–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini T.D., Friedmann M.C., Samuels A.L., Douglas C.J. (2010). ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol. 154: 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiza-Świder J., Rybka Z., Skutnik E., Łukaszewska A. (2003). Proteolysis and expression of the cysteine protease gene in senescing cut leaves of Hosta ‘Undulata Erromena’ and Zantedeschia aethiopica Spr. treated with BA or GA3. Acta Physiol. Plant. 25: 319–324. [Google Scholar]
- Richau K.H., Kaschani F., Verdoes M., Pansuriya T.C., Niessen S., Stüber K., Colby T., Overkleeft H.S., Bogyo M., Van der Hoorn R.A. (2012). Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol. 158: 1583–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozman-Pungercar J., et al. (2003). Inhibition of papain-like cysteine proteases and legumain by caspase-specific inhibitors: when reaction mechanism is more important than specificity. Cell Death Differ. 10: 881–888. [DOI] [PubMed] [Google Scholar]
- Sanders P.M., Bui A.Q., Weterings K., McIntire K., Hsu Y.-C., Lee P.Y., Truong M.T., Beals T., Goldberg R. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11: 297–322. [Google Scholar]
- Schmid M., Simpson D., Gietl C. (1999). Programmed cell death in castor bean endosperm is associated with the accumulation and release of a cysteine endopeptidase from ricinosomes. Proc. Natl. Acad. Sci. USA 96: 14159–14164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senatore A., Trobacher C.P., Greenwood J.S. (2009). Ricinosomes predict programmed cell death leading to anther dehiscence in tomato. Plant Physiol. 149: 775–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon M., Belenghi B., Delledonne M., Menachem E., Levine A. (1999). The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 11: 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen A.M., Kröber S., Unte U.S., Huijser P., Dekker K., Saedler H. (2003). The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33: 413–423. [DOI] [PubMed] [Google Scholar]
- Sturn A., Quackenbush J., Trajanoski Z. (2002). Genesis: cluster analysis of microarray data. Bioinformatics 18: 207–208. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Yamauchi D., Minamikawa T. (1991). Nucleotide sequence of cDNA for an endopeptidase (EP-C1) from pods of maturing Phaseolus vulgaris fruits. Plant Mol. Biol. 16: 1083–1084. [DOI] [PubMed] [Google Scholar]
- Than M.E., Helm M., Simpson D.J., Lottspeich F., Huber R., Gietl C. (2004). The 2.0 A crystal structure and substrate specificity of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm. J. Mol. Biol. 336: 1103–1116. [DOI] [PubMed] [Google Scholar]
- Trobacher C.P., Senatore A., Greenwood J.S. (2006). Masterminds or minions? Cysteine proteinases in plant programmed cell death. Can. J. Bot. 84: 651–667. [Google Scholar]
- Valpuesta V., Lange N.E., Guerrero C., Reid M.S. (1995). Up-regulation of a cysteine protease accompanies the ethylene-insensitive senescence of daylily (Hemerocallis) flowers. Plant Mol. Biol. 28: 575–582. [DOI] [PubMed] [Google Scholar]
- Varnier A.L., Mazeyrat-Gourbeyre F., Sangwan R.S., Clément C. (2005). Programmed cell death progressively models the development of anther sporophytic tissues from the tapetum and is triggered in pollen grains during maturation. J. Struct. Biol. 152: 118–128. [DOI] [PubMed] [Google Scholar]
- Vizcay-Barrena G., Wilson Z.A. (2006). Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J. Exp. Bot. 57: 2709–2717. [DOI] [PubMed] [Google Scholar]
- Wan L., Xia Q., Qiu X., Selvaraj G. (2002). Early stages of seed development in Brassica napus: a seed coat-specific cysteine proteinase associated with programmed cell death of the inner integument. Plant J. 30: 1–10. [DOI] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederanders B. (2003). Structure-function relationships in class CA1 cysteine peptidase propeptides. Acta Biochim. Pol. 50: 691–713. [PubMed] [Google Scholar]
- Wijeratne A.J., Zhang W., Sun Y., Liu W., Albert R., Zheng Z., Oppenheimer D.G., Zhao D., Ma H. (2007). Differential gene expression in Arabidopsis wild-type and mutant anthers: insights into anther cell differentiation and regulatory networks. Plant J. 52: 14–29. [DOI] [PubMed] [Google Scholar]
- Worrall D., Hird D.L., Hodge R., Paul W., Draper J., Scott R. (1992). Premature dissolution of the microsporocyte callose wall causes male sterility in transgenic tobacco. Plant Cell 4: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.S., Platt K.A., Ratnayake C., Wang T.W., Ting J.T., Huang A.H. (1997). Isolation and characterization of neutral-lipid-containing organelles and globuli-filled plastids from Brassica napus tapetum. Proc. Natl. Acad. Sci. USA 94: 12711–12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B., Deng Y., Kanaoka M.M., Okada K., Hong Z. (2012). Expression of Arabidopsis callose synthase 5 results in callose accumulation and cell wall permeability alteration. Plant Sci. 183: 1–8. [DOI] [PubMed] [Google Scholar]
- Xu J., Yang C., Yuan Z., Zhang D., Gondwe M.Y., Ding Z., Liang W., Zhang D., Wilson Z.A. (2010). The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22: 91–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Vizcay-Barrena G., Conner K., Wilson Z.A. (2007). MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19: 3530–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Guinel F.C., Moffatt B.A. (2002). A comparative ultrastructural study of pollen development in Arabidopsis thaliana ecotype Columbia and male-sterile mutant apt1-3. Protoplasma 219: 59–71. [DOI] [PubMed] [Google Scholar]
- Zhang W., Sun Y., Timofejeva L., Chen C., Grossniklaus U., Ma H. (2006). Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095. [DOI] [PubMed] [Google Scholar]
- Zhang X.M., Wang Y., Lv X.M., Li H., Sun P., Lu H., Li F.L. (2009). NtCP56, a new cysteine protease in Nicotiana tabacum L., involved in pollen grain development. J. Exp. Bot. 60: 1569–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.B., et al. (2007). Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 52: 528–538. [DOI] [PubMed] [Google Scholar]
- Zhao D.-Z., Wang G.-F., Speal B., Ma H. (2002). The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16: 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Chen H., Li H., Gao J.F., Jiang H., Wang C., Guan Y.F., Yang Z.N. (2008). Defective in tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 55: 266–277. [DOI] [PubMed] [Google Scholar]
- Zhu J., Lou Y., Xu X., Yang Z.N. (2011). A genetic pathway for tapetum development and function in Arabidopsis. J. Integr. Plant Biol. 53: 892–900. [DOI] [PubMed] [Google Scholar]
- Zhu J., Zhang G., Chang Y., Li X., Yang J., Huang X., Yu Q., Chen H., Wu T., Yang Z. (2010). AtMYB103 is a crucial regulator of several pathways affecting Arabidopsis anther development. Sci. China Life Sci. 53: 1112–1122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.