This study reveals a novel mechanism that regulates the mitogen-activated protein kinase (MAPK) cascade by a calcium-dependent protein kinase (CDPK) in land plants. The finding provides insight into the crosstalk mechanism between the CDPK and MAPK pathways and identifies a role for this mechanism in stress signal transduction and plant immunity.
Abstract
The mitogen-activated protein kinase (MAPK) is a pivotal point of convergence for many signaling pathways in eukaryotes. In the classical MAPK cascade, a signal is transmitted via sequential phosphorylation and activation of MAPK kinase kinase, MAPK kinase (MKK), and MAPK. The activation of MAPK is dependent on dual phosphorylation of a TXY motif by an MKK, which is considered the sole kinase to phosphorylate and activate MAPK. Here, we report a novel regulatory mechanism of MAPK phosphorylation and activation besides the canonical MAPK cascade. A rice (Oryza sativa) calcium-dependent protein kinase (CDPK), CPK18, was identified as an upstream kinase of MAPK (MPK5) in vitro and in vivo. Curiously, CPK18 was shown to phosphorylate and activate MPK5 without affecting the phosphorylation of its TXY motif. Instead, CPK18 was found to predominantly phosphorylate two Thr residues (Thr-14 and Thr-32) that are widely conserved in MAPKs from land plants. Further analyses reveal that the newly identified CPK18-MPK5 pathway represses defense gene expression and negatively regulates rice blast resistance. Our results suggest that land plants have evolved an MKK-independent phosphorylation pathway that directly connects calcium signaling to the MAPK machinery.
INTRODUCTION
Plants have evolved sophisticated signaling pathways to transduce biotic or abiotic stimuli into proper cellular responses in order to adapt to ever-changing environmental challenges. These signaling pathways often require the activation of protein kinases to orchestrate diverse cellular processes through the phosphorylation of various substrates. The eukaryotically conserved mitogen-activated protein kinase (MAPK) cascade is a pivotal phosphorylation pathway to convey external or internal signals to downstream effectors such as transcription factors. The prototypic MAPK cascade consists of three sequentially phosphorylated and activated protein kinases: MAPK kinase kinase (MAPKKK or MKKK), MAPK kinase (MAPKK or MKK), and MAPK (Seger and Krebs, 1995). Following the dual phosphorylation at the threonine (T) and tyrosine (Y) residues in its TXY motif (where X is Asp or Glu in plants), a MAPK is activated and becomes capable of phosphorylating downstream transcription factors and other effectors to reprogram the transcriptome and metabolome (Seger and Krebs, 1995; Chang and Karin, 2001; Pitzschke et al., 2009; Rodriguez et al., 2010; Meng and Zhang, 2013). As a result, MKK is generally considered the sole kinase to phosphorylate and activate MAPKs (Andreasson and Ellis, 2010; Rodriguez et al., 2010; Meng and Zhang, 2013).
Land plants typically encode more MKKKs, MKKs, and MAPKs than yeasts or mammals. Arabidopsis thaliana and rice (Oryza sativa) contain 20 and 17 MAPKs (MAPK Group, 2002; Reyna and Yang, 2006), respectively, which can be divided into four phylogenetic groups (Supplemental Figure 1). Among them, Arabidopsis MPK3/4/6 and their orthologs from group A and B MAPKs play a key role in stress signal transduction associated with reactive oxygen species homeostasis, stomata development, hormone signaling, disease resistance, and abiotic stress tolerance (Pitzschke et al., 2009; Rodriguez et al., 2010; Meng and Zhang, 2013). For example, rice MPK5, the ortholog of Arabidopsis MPK3 (Supplemental Figure 1 and Supplemental Data Set 1), was previously shown to positively regulate abiotic stress tolerance but negatively modulate rice disease resistance (Xiong and Yang, 2003). During the past decade, many studies have focused on deciphering the components of the classical MAPK cascade involved in developmental or stress signal transduction, such as plant immunity signaling. Two extensively studied canonical MAPK cascades in Arabidopsis, MKK4/5-MPK3/6 and MEKK1-MKK1/2-MPK4, were activated by pathogen-associated molecular patterns or effectors to regulate downstream signaling components. Activated MPK3/6 was shown to phosphorylate transcription factors like WRKY33, ETHYLENE-RESPONSE FACTOR104 (ERF104), and VIRE2-INTERACTING PROTEIN1 to activate defense gene expression and induce ethylene (ET)/camalexin biosynthesis (Djamei et al., 2007; Bethke et al., 2009; Mao et al., 2011). However, how receptors/sensors transmit different signals to MAPKs remains a major gap in our understanding of plant MAPK cascade signaling (Meng and Zhang, 2013).
Besides the MAPK cascade, calcium (Ca2+) signaling is another critical pathway triggered by environmental stimuli and developmental cues. Ca2+ is a ubiquitous secondary messenger in cellular signal transduction and is decoded by various Ca2+ binding proteins and kinases such as calcium-dependent protein kinase (CDPK) (Harper et al., 2004). CDPK is an ancient family of Ser/Thr kinases that has evolved independently in green algae and land plants (Hamel et al., 2014). There are 34 and 29 CDPKs in Arabidopsis and rice (Cheng et al., 2002; Asano et al., 2005), respectively. Many CDPKs contain a myristoylation motif (Hrabak et al., 2003), and a few of them were confirmed to be associated with the plasma membrane (Dammann et al., 2003). Biochemical and genetic studies have shown that CDPKs play important roles in numerous signaling pathways and biological processes such as hormone signaling, oxidative burst, innate immunity, and abiotic stress response (Cheng et al., 2002; Harper et al., 2004; Lecourieux et al., 2006; Reddy et al., 2011).
Since the CDPK and MAPK pathways are often activated in response to the same environmental stimuli, it has been speculated that a potential crosstalk may occur between the two important families of protein kinases (Ludwig et al., 2005; Wurzinger et al., 2011). However, recent studies in Arabidopsis indicated that stress-responsive CDPKs and MAPKs acted differentially in innate immunity and salt signaling without direct crosstalk (Boudsocq et al., 2010; Mehlmer et al., 2010). Therefore, whether and how Ca2+/CDPK signaling may regulate the MAPK cascade in plants is largely unknown. In this study, a plasma membrane–associated rice CDPK, CPK18, was found to directly phosphorylate and activate MPK5 in vitro and in vivo. Genetic and functional analyses demonstrate that the CPK18-MPK5 pathway negatively regulates rice immunity and represents a novel regulatory mechanism for the MKK-independent activation of MAPKs in land plants.
RESULTS
CPK18 Is Both a Kinase and a Substrate of MPK5 in Vitro
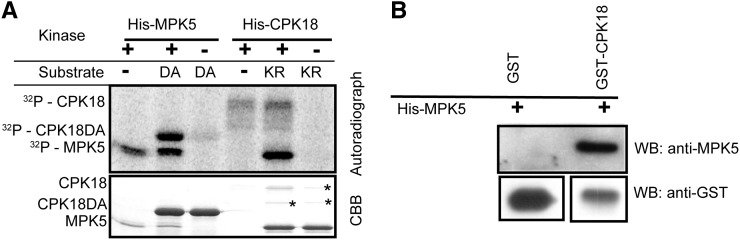
In a screening assay of MPK5 substrates by in situ solid-phase phosphorylation (Fukunaga and Hunter, 1997), CPK18 was initially identified as a potential MPK5 substrate. These two kinases were then found to phosphorylate each other when in vitro kinase assays were performed using the polyhistidine (His)-tagged recombinant proteins (His-CPK18 and His-MPK5). To confirm their phosphorylation, kinase-defective recombinant proteins, in which CPK18 Asp-178 was substituted with Ala (His-CPK18DA) and MPK5 Lys-65 was substituted with Arg (His-MPK5KR), were used as substrates. His-CPK18DA retained barely detectable autophosphorylation activity and His-MPK5KR had no detectable autophosphorylation (Figure 1A). An in vitro kinase assay showed that His-CPK18 phosphorylated His-MPK5KR, whereas His-MPK5 phosphorylated His-CPK18DA (Figure 1A). The in vitro interaction between MPK5 and CPK18 was also confirmed by the glutathione S-transferase (GST) pull-down assay (Figure 1B). These results demonstrated that direct interaction and phosphorylation occurred between CPK18 and MPK5 in vitro.
Figure 1.
Direct Interaction and Phosphorylation between CPK18 and MPK5 in Vitro.
(A) Recombinant His-CPK18 and His-MPK5 phosphorylated kinase-defective His-MPK5KR (K65R; labeled as KR) and His-CPK18DA (D178A; labeled as DA), respectively. The size difference between CPK18 and CPK18DA was due to autophosphorylation. Recombinant His-MPK5 purified from E. coli was capable of autophosphorylation and possessed the catalytic activity. Asterisks indicate bacterial protein contaminants with His-MPK5KR. CBB, Coomassie Brilliant Blue staining.
(B) MPK5 interacted with CPK18 in the GST pull-down assay. WB, immunoblotting.
The existence of a CDPK interacting with and phosphorylating MAPK (Figure 1) is highly intriguing, because the eukaryotically conserved MAPK is typically known to be only phosphorylated by MKK. Based on the protein kinase phylogeny, CDPK is a plant- and protist-specific group in the CAMK kinase superfamily and is distinct from MKK, which belongs to the STE kinase superfamily (Dardick et al., 2007). CPK18 is a typical group IV CDPK (Supplemental Figure 2) with four domains: the N-terminal variable domain (amino acids 1 to 40) with a myristoylation site for membrane association, the protein kinase domain (amino acids 40 to 312), the autoinhibitory junction domain (amino acids 313 to 350), and the calmodulin-like domain containing four EF-hand motifs (amino acids 351 to 512). Consistent with the CDPK and MKK phylogeny, the in vitro kinase assay showed that CPK18 did not affect the phosphorylation level of the MPK5 TEY motif, which was phosphorylated by the rice MKK MKK4/6 (Asano et al., 2005) or by itself (autophosphoryaltion) in the canonical MAPK cascade (Supplemental Figure 3).
CPK18 Interacts with and Phosphorylates MPK5 in Vivo
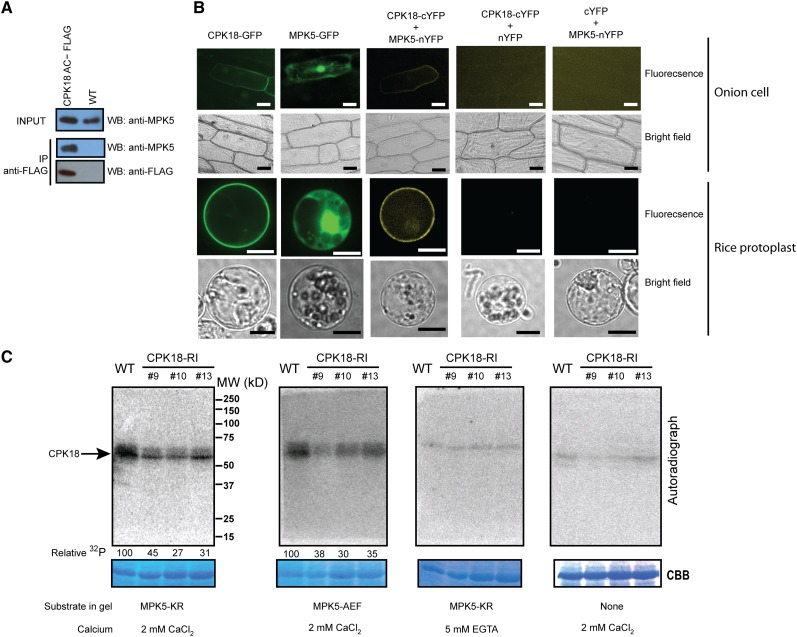
We then sought evidence that CPK18 interacts with MPK5 in vivo. Their interaction was first tested by coimmunoprecipitation. To this end, a constitutively activated CPK18 (in which the autoinhibitory and calmodulin domains were removed) with a FLAG tag at its C terminus (CPK18AC-FLAG) was expressed in rice plants. Immunoblotting, using the MPK5-specific antibody (Xiong and Yang, 2003), showed that MPK5 was specifically bound to CPK18AC-FLAG (Figure 2A). A bimolecular fluorescence complementation (BiFC) assay was also performed to investigate the subcellular localization of CPK18, MPK5, and their complex. Transient expression of green fluorescent protein (GFP)–tagged MPK5 and CPK18 in onion (Allium cepa) epidermal cells and rice protoplasts showed that MPK5 was localized in the nucleus and cytoplasm, whereas CPK18 was predominantly associated with the plasma membrane (Figure 2B). These observations are in agreement with previously reported locations of CPK18 (Campos-Soriano et al., 2011) and MPK5 (Singh et al., 2012). When MPK5-nYFP (the N-terminal half of YFP [yellow fluorescent protein]) and CPK18-cYFP (the C-terminal half of YFP) were coexpressed in onion cells or rice protoplasts, the complemented YFP signal was observed along the plasma membrane, as was CPK18-GFP (Figure 2B). These results confirmed that CPK18 interacted with MPK5 in vivo.
Figure 2.
CPK18 Interacted with and Phosphorylated MPK5 in Vivo.
(A) MPK5 was coimmunoprecipitated (IP) with FLAG-tagged CPK18 in vivo. MPK5 and CPK18 were detected in input control or coimmunoprecipitated samples by immunoblotting (WB) with anti-MPK5 and anti-FLAG antibodies, respectively.
(B) Subcellular localization of CPK18-GFP, MPK5-GFP, and their complex (BiFC) in onion epidermal cells or rice protoplasts. Bars = 100 µm (onion cells) and 10 µm (rice protoplast).
(C) In-gel kinase assay shows that CPK18 phosphorylated MPK5KR and MPK5AEF in a calcium-dependent manner. Total rice proteins from wild-type and CPK18-RI plants were resolved by SDS-PAGE containing His-MPK5KR or His-MPK5AEF. The in-gel kinase reaction was performed in the presence of Ca2+ (2 mM) or EGTA (5 mM) as indicated. A gel without a substrate was used as a control to monitor autophosphorylation (right panel). The kinase was visualized by autoradiography, and the relative activity toward phosphorylating His-MPK5KR or His-MPK5AEF was quantified with normalization to protein loading and shown at the bottom of the gel. Protein loadings are indicated by Coomassie Brilliant Blue (CBB) staining. MW, molecular mass marker in kD.
[See online article for color version of this figure.]
To confirm that native CPK18 phosphorylates MPK5, we generated transgenic plants (CPK18-RI lines) in which CPK18 expression was specifically suppressed by RNA interference (Supplemental Figure 4). In three independent CPK18-RI lines (lines 9, 10, and 13), CPK18 mRNA was reduced by 70 to 80% in comparison with the wild type, but the expression of CPK4, which is the closest homolog of CPK18 in rice (Supplemental Figure 2), was not affected (Supplemental Figure 4). By contrast, the mRNA levels of MKK4 and MKK6, two MKKs upstream of MPK5 in the classical MAPK cascade, were increased slightly (∼1.2- to 1.5-fold) in CPK18-RI lines (Supplemental Figure 4). To examine native kinases that phosphorylate MPK5 in CPK18-RI and wild-type plants, in-gel kinase assays were performed using the highly purified His-MPK5KR or His-MPK5AEF (where Thr-194, Glu-195, and Tyr-196 were replaced by Ala, Asp, and Phe) as a substrate. A protein kinase, with a molecular mass (∼58 to 60 kD) comparable to rice CDPKs but distinct from two 40-kD MKKs (MKK4 and MKK6), was the only kinase we identified to phosphorylate His-MPK5KR and His-MPK5AEF in the presence of Ca2+ (Figure 2C). We barely detected any kinases phosphorylating His-MPK5KR in the presence of EGTA (a Ca2+ chelator), nor did we detect any autophosphorylation without substrates in the gel (Figure 2C). We reasoned that the detected CDPKs included CPK18, because their ability to phosphorylate His-MPK5KR or His-MPK5AEF was specifically reduced in CPK18-RI lines (Figure 2C). In comparison with the wild type, the overall activity of CDPKs toward phosphorylating His-MPK5KR was reduced approximately 70% in the strongest CPK18-RI line (line 13), indicating that CPK18 is likely a predominant CDPK that phosphorylates MPK5 in rice. Taken together, these results suggest that CPK18 phosphorylates MPK5 at a site(s) out of its TEY motif in vivo.
Substrate Specificity of CDPK-MAPK Phosphorylation
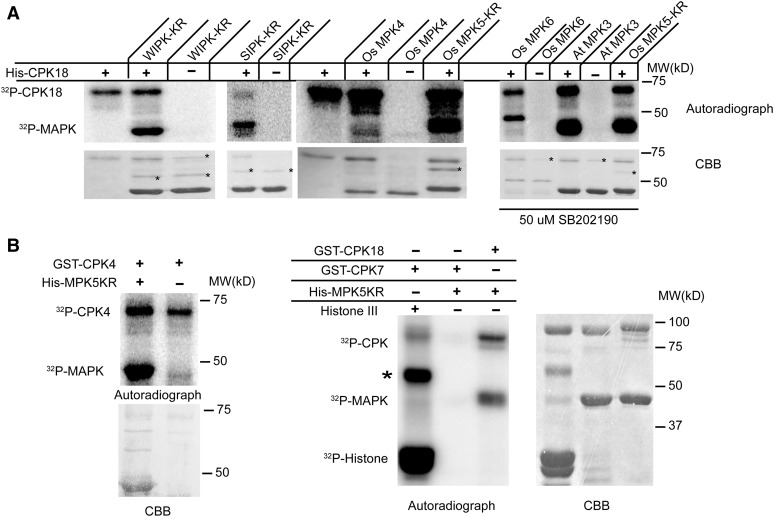
We further examined the specificity of CPK18 toward phosphorylating different MAPKs. Five additional MAPKs, rice MPK4/6, Arabidopsis MPK3, tobacco (Nicotiana tabacum) salicylic acid-induced protein kinase (SIPK) and wounding-induced protein kinase (WIPK) (Zhang and Klessig, 1998), and two rice CDPKs (CPK4 and CPK7), were fused with His or GST tags and purified from Escherichia coli. An in vitro kinase assay showed that rice CPK18 phosphorylated four MAPKs (Os MPK6, At MPK3, Nt WIPK, and Nt SIPK) from groups A and B in addition to MPK5 (Figure 3A) but hardly phosphorylated rice MPK4, which is a group C MAPK (Figure 3A). On the other hand, both rice group IV CDPKs, CPK4 and CPK18, phosphorylated MPK5KR (Figure 3B), whereas the group I rice CDPK, CPK7, did not (Figure 3B). We speculate that phosphorylation between CDPKs and MAPKs likely exists between specific subgroups of CDPK and MAPK families.
Figure 3.
Phosphorylation Specificity between CDPKs and MAPKs.
(A) CPK18 phosphorylated tobacco WIPK and SIPK, rice MPK6, and Arabidopsis MPK3 but hardly phosphorylated rice MPK4. The autophosphoryaltion of MAPKs was blocked by the mutation of essential residues (SIPK-KR and WIPK-KR) or by adding the MAPK inhibitor SB202190 (right panel; Os MPK6 and At MPK3), except for rice MPK4, which had barely detectable autophosphorylation. At, Arabidopsis; MW, molecular mass marker; Os, rice. Bacterial protein contaminants copurified with MAPKs are indicated with asterisks on the Coomassie Brilliant Blue (CBB)–stained gels.
(B) Rice CPK4 phosphorylated MPK5KR but CPK7 did not. The common CDPK substrate Histone III (Histone type III-S) was used as a positive control to show GST-CPK7 activity. The asterisk indicates a protein contaminant in Histone III.
CPK18 Predominantly Phosphorylates Thr-14 and Thr-34 in the N Terminus of MPK5
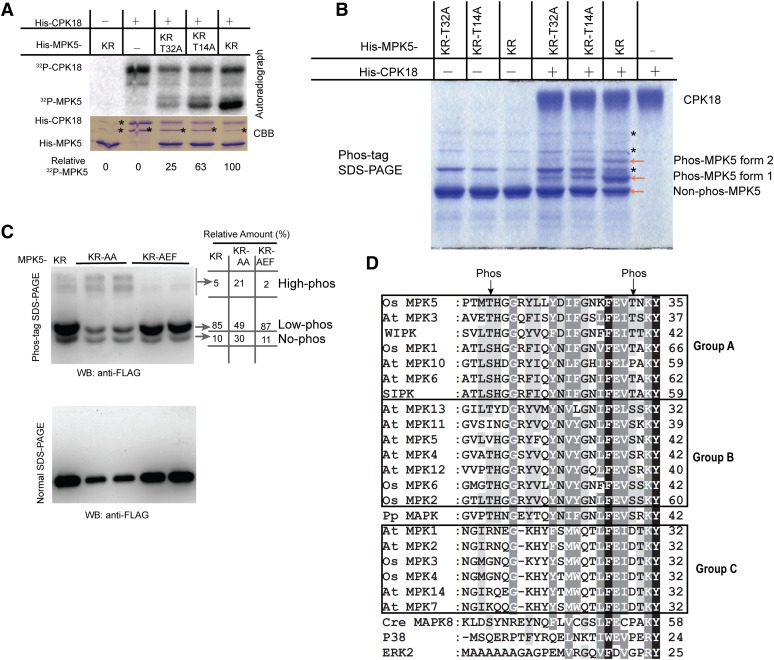
Next we determined which Thr/Ser on MPK5 was phosphorylated by CPK18. A total of 16 Ser/Thr residues (Supplemental Figure 5) were predicted as potential CPK18 phosphorylation sites according to the substrates’ specificity (Figure 3) or common CDPK phosphorylated motifs (Hegeman et al., 2006). Using site-directed mutagenesis, these candidate sites were substituted by Ala in the kinase defect MPK5KR, and 13 mutated recombinant proteins were made to cover all 16 substitutions (Supplemental Figure 6). By comparing the phosphorylation levels of these mutated proteins with MPK5KR, two Thr residues (Thr-14 and Thr-32) emerged as CPK18 phosphorylated sites (Supplemental Figure 6). In comparison with His-MPK5KR, T14A and T32A substitutions removed ∼37% and 75% of CPK18 phosphorylation, respectively (Figure 4A). However, mutated MPK5 proteins, which had both Thr-14 and Thr-32 replaced by other amino acids, were completely insoluble in E. coli. As a result, we used Phos-tag SDS-PAGE (Phos-tag gel) to confirm the phosphorylation sites. A Phos-tag gel could separate the phosphorylated proteins according to the number of phosphate groups attached (Kinoshita-Kikuta et al., 2007), and a protein containing more phosphorylated residues would have a lower migration rate in the Phos-tag gel. After phosphorylation by His-CPK18, two distinct forms of phosphorylated His-MPK5KR (phos-MPK5) were detected according to their migration rate in a Phos-tag gel (Figure 4B), suggesting that CPK18 phosphorylated at least two sites on His-MPK5KR. In agreement with the initial phosphorylated site mapping data, both T14A and T32A mutations drastically reduced the phos-MPK5 level by CPK18 (Figure 4B). Because we still observed two faint phosphorylated forms of His-MPK5KR-T14A, it is possible that CPK18 may weakly phosphorylate another site on MPK5 besides Thr-14 and Thr-32. These results suggest that MPK5 Thr-14 and Thr-32 are two major sites, and Thr-32 is the predominant site, for CPK18 phosphorylation in vitro.
Figure 4.
CPK18 Phosphorylated MPK5 at Thr-14 and Thr-32.
(A) In vitro kinase assay shows that the T14A and T32A mutations drastically impaired MPK5KR phosphorylation by CPK18. The relative phosphorylation level of MPK5 mutants is indicated at the bottom. The asterisks indicate bacterial protein contaminants.
(B) Analysis of MPK5 phosphorylation using a Phos-tag gel. Two phosphorylated forms of MPK5KR (forms 1 and 2) were detected on the Coomassie Brilliant Blue (CBB)–stained Phos-tag gel. The asterisks indicate bacterial protein contaminants.
(C) In vivo phosphorylation profile of MPK5 mutants. FLAG-tagged MPK5KR, MPK5KR-AA (T14A-T32A-K65R), and MPK5KR-AEF (K65R-T194A-Y196F) were expressed in rice protoplasts, and their phosphorylation patterns were analyzed using a Phos-tag gel and immunoblotting. The percentage of differentially phosphorylated forms (no-, low-, and high-phos) was quantified and indicated. WB, immunoblotting.
(D) CPK18 phosphorylation sites (indicated with arrows) are highly conserved in plant group A and B MAPKs but exist neither in group C plant MAPKs nor in MAPKs of humans and C. reinhardtii. N-terminal sequences of all group A/B/C MAPKs from Arabidopsis (At MPK) and rice (Os MPK) were aligned. Two tobacco MAPKs (WIPK and SIPK), human ERK2 and P38, P. patens (Pp) MAPK, and C. reinhardtii (Cre) MAPK8 were also included for comparison. The number at the end of each line indicates the coordinate of the last residue. The high- and medium-level conserved residues among MAPKs are indicated in the alignment with black and gray shading, respectively.
[See online article for color version of this figure.]
We further sought evidence that Thr-14 and Thr-32 were phosphorylated in vivo using the Phos-tag gel. We compared the in vivo phosphorylation pattern of MPK5KR, MPK5KR-AEF (K65R-T194A-Y196F), and MPK5KR-AA (K65R-T14A-T32A), which were expressed in rice protoplasts with a FLAG tag. MPK5 with these mutations showed identical migrating rates in normal SDS-PAGE, although MPK5KR-AA showed much lower protein expression than the others (Figure 4C). Consistent with multiple phosphorylation sites, multiple phosphorylation forms of MPK5KR were detected in the Phos-tag gel and are referred to as no-, low-, and high-phosphorylated forms according to their migration rates (Figure 4C). The percentage of high-phosphorylated MPK5KR, which had the lowest migrating rate, was substantially reduced when the TEY motif was replaced by AEF (Figure 4C). Although MPK5KR-AA had a similar phosphorylation pattern to that of MPK5KR, ∼30% of MPK5KR-AA remained nonphosphorylated, while only ∼10% of MPK5KR or MPK5KR-AEF remained nonphosphorylated (Figure 4C). The T14A-T32A mutations mainly impaired the low-phosphorylated form, which was likely attached with one phosphate group. We noticed that the T14A-T32A mutations appeared to increase the percentage of the high-phosphorylated form compared with MPK5KR (Figure 4C). This is probably due to compensatory phosphorylation in a protein with multiple phosphorylation sites, as observed previously (Bauer et al., 2003). Taken together, both in vitro and in vivo data have shown that CPK18 phosphorylated MPK5 mainly at the Thr-14 and Thr-32 sites.
Thr-14 and Thr-32 Residues Are Conserved in Group A and B MAPKs of Land Plants
Sequence alignment of Arabidopsis and rice MAPKs indicates that all group A/B MAPKs have Ser/Thr aligned with MPK5 Thr-14 and Thr-32, with highly conserved context sequences except Arabidopsis MPK5/10/11 (Figure 4D). Although the context sequence–flanked MPK5 Thr-32 is conserved in all MAPKs, MAPKs in other groups do not have Ser/Thr at the position aligned with the Thr-32 site (Figure 4D). We also examined the sequences of putative MPK5 orthologs in tobacco, moss (Physcomitrella patens), green algae (Chlamydomonas reinhardtii), and human (Homo sapiens). As is the case for Arabidopsis and rice, both Thr-32 and Thr-14 were also conserved in tobacco and P. patens MAPKs but absent in C. reinhardtii and human MAPKs (Figure 4D). These results suggest that CPK18 phosphorylated sites are conserved in MAPKs from both lower and higher plants and might have emerged during the evolution of land plants.
CPK18 Positively Regulates MPK5 Activity
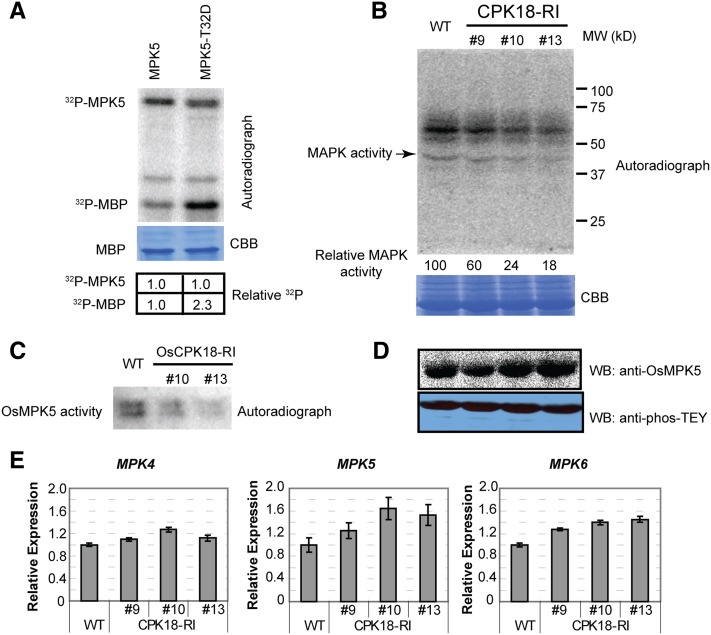
To examine the effect on MPK5 activity of CPK18 phosphorylation, MPK5-T32D was made to mimic phosphorylated Thr-32, which is a predominant phosphorylation site. Upon expression in E. coli, His-MPK5T32D showed comparable protein yield and solubility to His-MPK5. An in vitro kinase assay showed that recombinant MPK5-T32D exhibited twice as much catalytic activity toward phosphorylating the common MAPK substrate myelin basic protein (MBP) than wild-type MPK5, but their autophosphorylation levels were comparable (Figure 5A).
Figure 5.
CPK18 Positively Regulates MPK5 Activity.
(A) MPK5-T32D, containing the T32A mutation to mimic phosphorylation, exhibited increased kinase activity toward MBP. The relative radioactivity is shown at the bottom.
(B) and (C) Detection of overall MAPK activity and MPK5 activity in CPK18-RI and wild-type plants using an in-gel kinase assay. Total protein (B) and immunoprecipitated MPK5 (C) were resolved on 10% SDS-PAGE gels containing MBP. The in-gel kinase assay was performed to analyze MAPK activity. The relative MAPK activity is shown at the bottom of (B). Protein loading is shown by Coomassie Brilliant Blue (CBB) staining.
(D) MPK5 protein level and overall MAPK TEY phosphorylation level in wild type and CPK18-RI lines. MPK5 and phos-TEY levels were examined by immunoblotting (WB) with anti-MPK5 and anti-phos-TEY antibodies, respectively.
(E) RT-qPCR examination of MPK4/5/6 expression in the wild type and CPK18-RI lines. Data are presented as means ± sd (n = 3).
[See online article for color version of this figure.]
Then, we further investigated whether CPK18 regulates MPK5 activity in rice plants. We compared overall MAPK activity in CPK18-RI and wild-type plants by in-gel kinase assays using MBP as a substrate. The activity of ∼43-kD MAPKs was reduced 40 to 80% in three CPK18-RI lines compared with the wild type (Figure 5B). The result also showed that the activity of another kinase(s) with a size of ∼55 to 65 kD was reduced in CPK18-RI plants, but its identity was undefined (Figure 5B). Because most rice group A/B/C MAPKs have similar molecular masses (43 to 45 kD), the activity of native MPK5 was further measured after immunoprecipitation. As shown by the in-gel kinase assay, immunoprecipitated MPK5 from CPK18-RI plants also exhibited a lower kinase activity than that from wild-type plants (Figure 5C). By contrast, the MPK5 protein level was increased slightly in CPK18-RI plants than in the wild type, and the overall phos-TEY level of MAPKs was comparable between CPK18-RI and the wild type (Figure 5D). Further expression profiling of three rice MAPKs reveals that the transcripts of MPK5 and MPK6, which encode CPK18 substrates (Figure 3A), were increased, but MPK4 expression was not significantly changed in CPK18-RI plants compared with the wild type (Figure 5E). Thus, we conclude that CPK18 positively regulates MPK5 activity through phosphorylating Thr-14/Thr-32 without affecting MPK5 TEY phosphorylation.
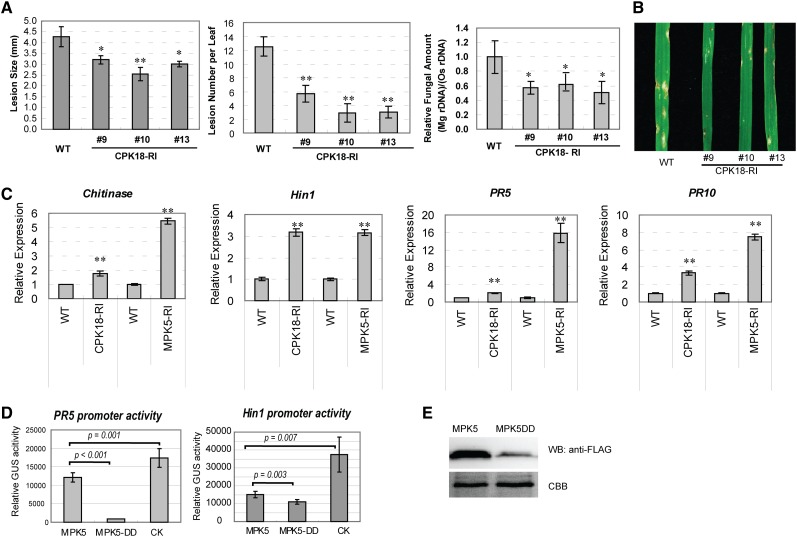
The CPK18-MPK5 Pathway Negatively Regulates Rice Blast Resistance
To explore the physiological function of the CPK18-MPK5 pathway, the phenotype of CPK18-RI lines was compared with that of MPK5-RI lines, in which MPK5 was suppressed by RNA interference (Xiong and Yang, 2003). Similar to MPK5-RI plants, three CPK18-RI lines exhibited enhanced resistance to the rice blast fungus (Magnaporthe oryzae) (Figures 6A and 6B). After fungal infection, the size and number of disease lesions or relative fungal amounts in leaves of CPK18-RI lines were significantly lower than those in wild-type plants. The enhanced resistance phenotype is consistent with the observations that the expression of rice defense-related genes (pathogenesis-related genes PR5 and PR10, Chitinase, and Harpin-induced protein1 [Hin1]) was increased in CPK18-RI and MPK5-RI lines (Figure 6C). Although MPK5 also phosphorylated CPK18 in vitro (Figure 1A), an in-gel kinase assay suggested that the CDPK activity (including CPK18) was comparable between wild-type and MPK5-RI plants (Supplemental Figure 7). To confirm that phosphorylation at MPK5 Thr-14/Thr-32 contributes to the repression of defense genes, we used the β-glucuronidase (GUS) reporter assay to examine the PR5 and Hin1 promoter activities after expressing FLAG-tagged MPK5 or MPK5-DD (T14D-T32D) in rice protoplasts. Consistent with the quantitative RT-PCR (RT-qPCR) results (Figure 6C), PR5 and Hin1 promoter activities were repressed in rice protoplasts expressing MPK5 in comparison with the vector control (Figure 6D). MPK5DD that mimicked the Thr-14 and Thr-32 phosphorylation exhibited stronger repression; even its protein level was much less than that of MPK5 (Figures 6D and 6E). These results suggest that the CPK18-MPK5 pathway negatively regulates rice immunity by repressing defense gene expression.
Figure 6.
The CPK18-MPK5 Pathway Negatively Regulates Plant Immunity.
(A) CPK18-RI plants exhibited enhanced resistance to M. oryzae. Lesion size, lesion number, and relative fungal amount were compared between the wild type and CPK18-RI lines 6 d after fungal infection. Asterisks indicate statistically significant differences (*P < 0.05, **P < 0.01, Student’s t test). The results are shown as means ± se (n = 3 biological repeats).
(B) Rice disease symptoms caused by M. oryzae strain 70-15 in the wild type and CPK18-RI lines. The photograph was taken 6 d after inoculation.
(C) RT-qPCR analysis shows that the expression of defense-related genes (Chitinase, Hin1, PR5, and PR10) was increased in CPK18-RI and MPK5-RI plants in comparison with the wild type (**P < 0.01, Student’s t test). The data are shown as means ± sd (n = 3).
(D) and (E) The phosphorylation-mimic mutant MPK5DD (T14D-T32D) exhibited stronger ability to repress PR5 and Hin1 promoter activity than did MPK5 in rice protoplast reporter assays. A plasmid containing PR5pro:GUS or Hin1pro:GUS was cotransfected with FLAG-tagged MPK5, MPK5DD, or empty vector (CK) (D). Promoter activities were determined based on the relative GUS activity. The P values from Student’s t test are shown in the plot. The results are presented as means ± sd (n = 5). Protein levels of MPK5 and MPK5DD in rice protoplasts were examined by immunoblotting (WB) using anti-FLAG antibody (E). Equal protein loading is indicated by Coomassie Brilliant Blue (CBB) staining.
[See online article for color version of this figure.]
DISCUSSION
In the classical MAPK cascade, MKK has been recognized as the sole kinase to transmit signals from upstream MAPKKKs to MAPKs in plant, animal, and yeast systems. Three exceptions have been reported for the activation of the mammalian MAPK P38α (Ge et al., 2002; Salvador et al., 2005) or the Arabidopsis MAPK MPK8 (Takahashi et al., 2011) via an MKK-independent pathway. P38α could be activated after TAK1 Binding Protein1 binding (Ge et al., 2002) or through Tyr-323 phosphorylation by T cell receptor–proximal Tyr kinases (Salvador et al., 2005), and both pathways activated P38α by triggering autophosphorylation of the TXY motif. The full activation of Arabidopsis MPK8 was found to require a calmodulin protein binding to its TDY motif in addition to MKK phosphorylation (Takahashi et al., 2011). The CPK18-MPK5 pathway described in this study is distinct from these reported examples of MKK-independent activation of MAPKs (Ge et al., 2002; Salvador et al., 2005; Takahashi et al., 2011). CPK18 triggered MPK5 activation by phosphorylating its N-terminal Thr residues (Thr-14 and Thr-32) without affecting the phos-TEY level (Figure 5; Supplemental Figure 3). Hence, the CPK18-MPK5 pathway represents a novel mechanism to directly modulate MAPK activity in addition to the canonical MAPK cascade.
It is intriguing that the N-terminal Thr-14/Thr-32 residues and their context sequence in MPK5 are conserved in almost all Arabidopsis and rice group A and B MAPKs (Figure 4D). We have demonstrated that at least five MAPKs from three plant species could be directly phosphorylated by CPK18 (Figure 3A). In addition, both group IV rice CDPKs, CPK4 and CPK18, are capable of phosphorylating MPK5 (Figure 3B). Thus, the CPK18-MPK5 pathway may not be an exception but likely represents a conserved regulatory mechanism for a specific subset of CDPKs to modulate group A and B MAPKs in plants. Interestingly, group IV CDPKs appear to have evolved only in land plants (Hamel et al., 2014), whereas Thr-14/Thr-32 residues are conserved in the MAPKs of land plants but are absent in green algae (Figure 4D). This evolutionary coincidence suggests that the CDPK-MAPK pathway might have emerged during the evolution of land plants as an adaptation to environmental changes.
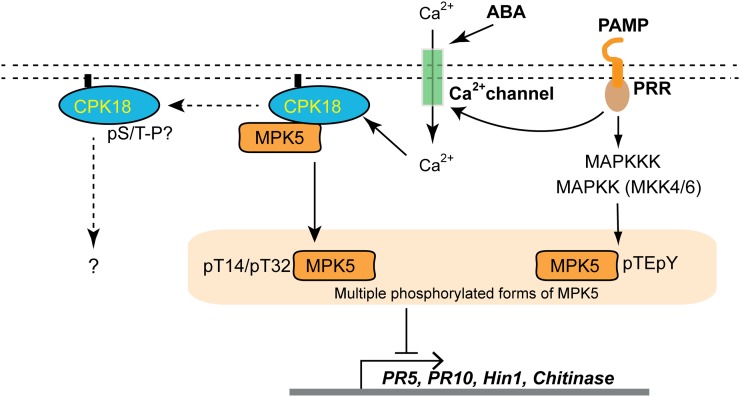
A major effort has been made to study MAPK cascades and Ca2+/CDPK signaling in plants, but little is known about the crosstalk between these two important kinase pathways. Although both pathways may share downstream components, previous studies in Arabidopsis and tobacco suggest that CDPKs and MAPKs work in parallel without direct interaction or crosstalk (Boudsocq et al., 2010). The phosphorylation of MPK5 and other MAPKs by CPK18 shown in this study indicates that the Ca2+ signaling pathway can directly activate MAPKs through specific CDPKs. Ca2+ is one of the earliest secondary signals to sense microbial infection, abiotic stresses, or phytohormones such as abscisic acid (ABA) (McAinsh et al., 1992; Kurusu et al., 2005). The CPK18-MAPK pathway potentially provides a shortcut or an alternative route to activate MAPKs without the multiple phosphorylation steps involved in the canonical MAPK cascade. As shown in our proposed working model (Figure 7), the Ca2+ signal activates the membrane-anchored CPK18, which in turn phosphorylates MPK5 to repress defense gene expression. Ca2+ signals triggered by environmental stimuli (e.g., pathogen infection, drought, or ABA) could be rapidly decoded by CPK18 and transmitted to transcriptional machinery through the CPK18-MAPK pathway by taking advantage of the fact that MAPKs target numerous transcription factors (Popescu et al., 2009). Since CPK18 does not affect MPK5 TEY phosphorylation (Figures 5A and 5D; Supplemental Figure 3), multiple phosphorylated forms of MPK5 likely coexist (Figure 4C), and the CPK18-MAPK pathway may work independently or cooperatively with the classical MAPK cascade.
Figure 7.
A Working Model for the CPK18-MPK5 Pathway.
Plasma membrane–associated CPK18 is regulated by Ca2+ signals to activate MPK5 through Thr-14/Thr-32 phosphorylation (pT14/pT32). Activated MPK5 then phosphorylates different transcription factors to activate or repress downstream gene expression. The CPK18-MPK5 pathway may work independently or cooperatively with the canonical MAPK cascade, which is activated by pathogen-associated molecular pattern (PAMP) and pattern recognition receptor (PRR) interaction. MPK5 likely exists in multiple phosphorylated forms with variable activities. Arrows indicate positive regulation; the blunt-ended line indicates negative regulation. Dashed arrows indicate undefined pathways of MPK5 feedback phosphorylation of CPK18.
[See online article for color version of this figure.]
Our observations suggest that the newly identified CPK18-MPK5 pathway represses defense gene expression, thereby increasing host susceptibility to rice blast fungus (Figure 6). Recent advances indicate that MPK5 likely mediates the interplay of plant hormones to fine-tune immunity and growth (Yang et al., 2013). Among the phytohormones, ET appears to play a prominent role in rice blast resistance (Helliwell et al., 2013). In Arabidopsis, MPK3 and MPK6 positively regulate the ET pathway by phosphorylating and stabilizing 1-aminocyclopropane-1-carboxylic acid synthase, which catalyzes ET production (Liu and Zhang, 2004), or transcription factors (ETHYLENE-INSENSITIVE3 and ERF104) that are essential components of ET signaling (Yoo et al., 2008; Bethke et al., 2009). A similar phosphorylation event likely exists in rice, but it may lead to a different biochemical and physiological consequence. Indeed, elevated ET production was observed in MPK5-RI plants (Bailey et al., 2009), suggesting the repression of the ET pathway by MPK5. By mediating antagonistic crosstalk between ET and ABA, MPK5 is a positive regulator of the ABA pathway and abiotic stress tolerance (Xiong and Yang, 2003). Interestingly, MPK5 is required for ABA-mediated brown spot resistance in rice (De Vleesschauwer et al., 2010). Taking into account the fact that abiotic stresses and ABA rapidly trigger cellular Ca2+ signals that could be decoded by CDPK, the CPK18-MPK5 pathway likely mediates the crosstalk among abiotic stress, ABA, and ET pathways that fine-tune rice growth, immunity, and abiotic stress tolerance.
Moreover, in vitro experiments suggest that phosphorylation between CPK18 and MPK5 may occur in both directions (i.e., CPK18 phosphorylates MPK5 while MPK5 feedback phosphorylates CPK18) (Figure 1A). However, the molecular and physiological consequences of the MPK5-CPK18 phosphorylation were not demonstrated or elucidated in this study (Figure 7). Bidirectional phosphorylation between two kinases is rarely reported but has been observed between specific MKKs and MAPKs in human, Xenopus, and yeast (Matsuda et al., 1993; Mansour et al., 1994; Jiménez-Sánchez et al., 2007). Further studies are needed to reveal if such bidirectional phosphorylation exists in plants to provide a simple mechanism to mediate crosstalk between the CDPK and MAPK pathways. In conclusion, our results demonstrate that plants have evolved a CDPK-mediated pathway for MAPK activation in addition to the linear model of the classical MAPK cascade.
METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa) cv Kitaake was used to generate CPK18-RI lines in this study, whereas cv Nipponbare was used previously to generate MPK5-RI lines (Xiong and Yang, 2003). The rice plants were grown in a greenhouse or a growth chamber at 28°C day/23°C night with 12 h of light.
In Situ Solid-Phase Phosphorylation Screen
To screen putative MPK5 substrate, a rice cDNA library was constructed from Magnaporthe oryzae–infected rice leaves using the ZAP Express cDNA Synthesis Kit and the ZAP Express cDNA Gigapack III Gold Cloning Kit (Stratagene). The in situ solid-phase screen was performed according to the published method that was used to identify mammalian MAPK substrates (Fukunaga and Hunter, 1997).
Gene Constructs for Recombinant Protein Expression
The full-length cDNA of CPK18 in the pBK-CMV phagemid vector (Stratagene) was cut with EcoRI and XhoI and inserted into pGEX-5x-3 and pET28c, which were used to express GST-CPK18 and His-CPK18, respectively, in Escherichia coli. MPK4/5/6, MKK4, CPK4, and CPK7 full-length cDNAs were obtained from the Rice Genome Resource Center (http://www.rgrc.dna.affrc.go.jp). They were first cloned into pENTR/D-TOPO by TOPO cloning (primers are listed in Supplemental Table 1) and then moved into pDEST17 or pDEST15 by the LR reaction for recombinant protein expression with a His or GST tag (Life Technologies). Arabidopsis thaliana MPK3 in a pENTR vector was obtained from the ABRC (http://abrc.osu.edu/) and cloned into pDEST17 to express His-MPK3.
Gene Constructs for Rice Transformation
To express CPK18 and MPK5 in plants, the complete open reading frames without a stop codon of MPK5 and CPK18 were amplified (primers are listed in Supplemental Table 1) and cloned into pENTR/D-TOPO, and then LR reactions were performed to move MPK5 and CPK18 into Gateway destination pUGW vectors for plant transient expression (Nakagawa et al., 2007). For GFP-tagged constructs, MPK5 and CPK18 were cloned into pUGW5 with GFP fused to their C termini. For the BiFC assay, MPK5 and CPK18 were cloned into the nEYFP/pUGW2 and cEYFP/pUGW2 vectors, respectively. For FLAG-tagged constructs, a CPK18 or MPK5 fragment without a stop codon was cloned into pUGW11 by the LR reaction (Life Technologies). The aforementioned gene constructs were introduced into rice protoplasts by polyethylene glycol–mediated transfection or into onion epidermal cells by particle bombardment.
To make double-stranded RNA interference constructs, a cDNA fragment of CPK18 (nucleotides 1364 to 1755) was cloned into pENTR/D-TOPO by TOPO cloning (Life Technologies) and then subcloned into the binary vector pANDA (Miki and Shimamoto, 2004). To express a FLAG-tagged constitutively activated CPK18 (CPK18AC), a cDNA fragment containing base pairs 157 to 1121 of CPK18 was amplified and cloned into pENTR/D-TOPO and then subcloned into binary vector pGWB11 with the FLAG tag fused to the C terminus (Nakagawa et al., 2007). The aforementioned gene constructs were introduced into rice cv Kitaake by Agrobacterium tumefaciens–mediated transformation.
Site-Directed Mutagenesis
To mutate CPK18 or MPK5, the Multi Site-Directed Mutagenesis Kit (Agilent Technologies) was used according to the manufacturer’s instructions. The DNA oligonucleotides used in mutagenesis were synthesized (Integrated DNA Technologies), and their sequences are listed in Supplemental Table 1. The oligonucleotide was phosphorylated using T4 PNK (New England Biolabs). After mutagenesis, all of the mutated plasmids were confirmed by Sanger sequencing.
Recombinant Protein Induction and Purification
His-CPK18 and GST-CPK18 were expressed in E. coli BL21(DE3) Codon RIL (Stratagene) upon induction with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 37°C for 4 h. His-MPK5 or its derived mutants were expressed in E. coli BL21(DE3) pLysS upon induction with 0.01 mM IPTG at 12°C for 12 h. GST-CPK18 was purified using Glutathione Sepharose 4B (GE) according to the manufacturer’s instructions, except that all buffer contained 0.1 mM EGTA. His-fused proteins (His-CPK18 and His-MPK5) were purified with the His GraviTrap Column (GE) according to the manufacturer’s instructions. All purified proteins were desalted by dialysis against the buffer containing 20 mM Tris, pH 7.6, 0.1 mM DTT, and 10% glycerol and then stored at −80°C in aliquots. The concentration of protein was determined by Protein Reagent (Bio-Rad). Other MAPKs and CDPKs were purified as MPK5 and CPK18, respectively.
To express and purify substrates for an in-gel kinase assay, His-MPK5KR or His-MPK5AEF was induced with 1 mM IPTG at 37°C for 4 h. After induction, bacteria were harvested and resuspended in lysis buffer (25 mM Tris, pH 7.4, 10 mM EDTA, and 1% Triton X-100) and then disrupted by sonication in ice (Branson Sonifier). Insoluble inclusion bodies were collected by centrifugation at 10,000g for 15 min. After washing three times with lysis buffer, inclusion bodies were dissolved in solubilized buffer (50 mM Tris, pH 9.5, and 0.3% sarkosyl) at 10 mg/mL. The insoluble cell debris was removed by centrifugation. The dissolved His-MPK5KR or His-MPK5AEF was then dialyzed against 20 mM Tris, pH 8.8, with three changes of buffer. The purity of His-MPK5KR and His-MPK5AEF was higher than 95% based on estimation by SDS-PAGE.
In Vitro Kinase Assay
For the in vitro kinase assay, 0.5 μg of recombinant kinase (His-CPK18 or His-MPK5) and 2 μg of substrate were added to a 20-μL reaction containing 25 mM Tris, pH 7.5, 10 mM MgCl2, 1 mM DTT, 0.2 mM ATP, and 0.1 μCi of [γ-32P]ATP (MP Biomedicals) supplemented with 5 mM CaCl2 (for the CDPK assay), 5 mM MnCl2 (for the MAPK assay), or 50 μM SB202190 (LC Laboratories). The reaction was incubated at room temperature (25°C) for 1 h and terminated with SDS sample buffer. After separation with SDS-PAGE and staining, gels were washed thoroughly with gel washing buffer (5% trichloroacetic acid and 1% sodium pyrophosphate) and vacuum dried on 3MM filter paper. Protein phosphorylation was analyzed by autoradiography with the Storage Phosphor Screen and Storm 820 scanner (GE). The relative phosphorylation level was quantified using ImageJ (http://rsb.info.nih.gov/ij/).
In-Gel Kinase Assay
For the in-gel kinase assay to detect CDPK, total protein was extracted from rice plants using extraction buffer (25 mM Tris, pH 7.6, 150 mM NaCl, 50 mM glycerol phosphate, 5 mM NaF, 5 mM Na3VO4, 1 mM DTT, 1% Triton X-100, 10% glycerol, 0.1 mM phenylmethylsulfonyl fluoride [PMSF], and 1% Protease Inhibitor Cocktail [Sigmal-Aldrich]). Total protein extracts (50 μg) were separated by 10% SDS-PAGE supplemented with 1 mg/mL substrate (His-MPK5KR or His-MPK5AEF) in a resolving gel. After electrophoresis, SDS was removed from the gel by thorough washing in SDS removal buffer (25 mM Tris, pH 7.6, 0.5 mM DTT, 0.1 mM Na3VO4, 5 mM NaF, 0.5 mg/mL BSA, and 0.1% Triton X-100). Then, the native kinases were renatured in gel by incubation at 4°C for 20 h with five changes of renaturation buffer (25 mM Tris, pH 7.6, 1 mM DTT, 0.1 mM Na3VO4, and 5 mM NaF). After renaturation, gels were soaked in cold CDPK kinase buffer (25 mM Tris, pH 7.6, 12 mM MgCl2, 1 mM DTT, and 0.1 mM Na3VO4) for 10 min, and then the cold CDPK kinase buffer was replaced with fresh CDPK kinase buffer containing 200 nM ATP and 2 μCi/mL [γ-32P]ATP (MP Biomedicals) to start the kinase reaction. The CDPK kinase buffer contained 2 mM CaCl2 or 5 mM EGTA, as indicated in Figure 2C. The reaction was performed at room temperature for 1 h and stopped by replacing kinase buffer with gel washing buffer (5% trichloroacetic acid and 1% sodium pyrophosphate) and then washed thoroughly to remove free radioactive ATP. The gel was stained with Coomassie Brilliant Blue R 250 and vacuum dried to 3MM filter paper. The autoradiographic detection was performed using the Storage Phosphor screen and Storm 820 scanner (GE).
For the in-gel kinase assay to detect MAPK activity, total protein (30 µg) or immunoprecipitated MPK5 was separated on 10% SDS-PAGE gels containing 0.25 mg/mL MBP (Life Technologies). The immunoprecipitation and MAPK in-gel kinase assay were performed as described previously by Xiong and Yang (2003).
In Vitro GST Pull Down
GST-CPK18 and GST (5 μg) were mixed with 10 μg of His-OsMAPK5 in 0.5 mL of binding buffer (25 mM Tris, pH 7.5, 150 mM NaCl, and 1% Triton X-100). Then, 20 μL of equilibrated Glutathione Sepharose 4B (GE) was added. After incubation at 25°C for 2 h, the Glutathione Sepharose was washed five times with binding buffer and resuspended in 0.2 mL of SDS sample buffer. The pull-down product was analyzed by immunoblotting using anti-MPK5 and anti-GST (Sigma-Aldrich) antibodies.
RT-qPCR
Total RNAs were extracted from rice tissues and treated with DNase I (New England Biolabs) before reverse transcription. The cDNA was synthesized using the Multiscript Reverse Transcription Kit (Life Technologies). Real-time PCR was performed in StepOne Plus (Life Technologies) using the GoTaq qPCR Master Mix (Promega). The relative quantification of gene expression was calculated as described previously (Xie et al., 2006), and rice UBQ10 was used as the reference gene. All primers are listed in Supplemental Table 1.
Coimmunoprecipitation and Immunoblotting
To detect proteins by immunoblotting, rice protein extracts were separated on 10% SDS-PAGE gels, blotted to polyvinylidene difluoride membranes, and then detected using anti-MPK5 (Xiong and Yang, 2003), anti-phos-TEY (anti-phos-ERK1/2, 4G12; Millipore), or anti-FLAG (Sigma-Aldrich) antibody. For coimmunoprecipitation, total proteins were extracted from wild-type and transgenic rice plants expressing FLAG-tagged CPK18AC with lysis buffer containing 50 mM Tris, pH 7.6, 150 mM NaCl, 1% Triton X-100, and 1 mM PMSF. About 1 mg of total protein was incubated with FLAG M2 agarose (Sigma-Aldrich) at 4°C overnight. Then, the FLAG M2 agarose was collected in a spin column and washed three times with lysis buffer without Triton X-100. The protein complex was eluted using 0.1 M Gly, pH 2.7, and one-fifth of the elution was analyzed by immunoblotting.
Subcellular Localization and BiFC
To transiently express tagged genes in onion, the plasmid constructs were delivered into onion epidermal cells by biolistic bombardment using the PDS-1000/He Biolistic Particle Delivery System (Bio-Rad). Briefly, 5 to 10 μg of plasmid DNA per construct was coated on 1-µm golden particles according to the manufacturer’s instructions. Then, coated particles were bombarded into onion epidermal cells using 1100 p.s.i. pressure. The YFP/GFP fluorescence was observed after 16 h using an epifluorescence microscope (Zeiss) with appropriate filters. To examine protein localization and BiFC in rice protoplasts, the plasmid constructs were used to transfect rice protoplasts as described previously (Xie and Yang, 2013). The fluorescence was then observed with a confocal microscope (Zeiss) at 16 h after transfection.
Rice Protoplast Reporter Assay
To make reporter constructs, PR5 and Hin1 promoters were amplified (see Supplemental Table 1 for primer sequences) from rice genomic DNA and cloned into pUGW3 (Nakagawa et al., 2007). For reporter assays, the reporter plasmid (PR5:GUS or Hin1:GUS) was cotransfected with the effector plasmid (pUGW11-MPK5 or pUGW11-MPK5DD) into rice protoplasts as described previously (Xie and Yang, 2013). Twenty hours later, the GUS reporter assay was performed as described previously (Yoo et al., 2007).
Phos-Tag Gel Electrophoresis and Blotting
The Phos-tag gel was prepared according to the previously reported method (Kinoshita-Kikuta et al., 2007). Acrylamide-pendant Phos-tag (WAKO Chemicals) and MnCl2 were added to the resolving SDS-PAGE gel. To analyze the phosphorylation of His-MPK5KR or its derived mutant proteins, phosphorylated proteins from in vitro kinase assays were desalted by chloroform/methanol precipitation and then dissolved in 2× SDS sample buffer. After electrophoresis in a Phos-tag gel (25 μM Mn2+-Phos-tag), proteins were visualized with Coomassie Brilliant Blue R 250 staining.
To analyze protein phosphorylation in vivo, FLAG-tagged MPK5 (pUGW11-MPK5) or its mutants were transiently expressed in rice protoplasts. Total protein was extracted from protoplasts with modified radioimmunoprecipitation assay buffer containing 50 mM Tris, pH 7.6, 150 mM NaCl, 1 mM NaF, 5 mM Na3VO4, 1% Triton X-100, 0.1% SDS, 10% glycerol, 0.1 mM PMSF, and 1 μg/mL Protease Inhibitor Cocktail (Sigma-Aldrich). Then, the protein extract was mixed with SDS sample buffer plus 10 mM MnCl2 and loaded onto a Phos-tag gel (50 μM Mn2+-Phos-tag). The protein was blotted to a polyvinylidene difluoride membrane (Millipore) using blotting buffer containing 25 mM Tris, 192 mM Gly, 10% methanol, 1 mM EDTA, and 0.025% SDS.
Rice Blast Fungus Inoculation
Rice blast fungus inoculation was performed as described previously (Qi and Yang, 2002). M. oryzae isolates 70-15 and IE1K, which are virulent on cv Nipponbare and cv Kitaake, were used in this study. Rice seedlings at the three-leaf stage (20 d after germination) were spray-inoculated with 1 × 105 spores/mL. The disease symptoms were scored 6 d after inoculation by measuring lesion size and number in the third leaf. The relative growth of the fungus was measured by quantifying the fungal rDNA as described previously (Qi and Yang, 2002). The fungal infection was repeated three times with consistent results.
Phylogenetic Analysis
Alignments of MAPKs or CDPKs (Supplemental Data Sets 1 and 2) were generated by ClustalX 1.83 (Larkin et al., 2007). Maximal likelihood phylogenetic trees were constructed using MEGA 5.0 (Tamura et al., 2011) with bootstrap values of 1000 trials.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Supplemental Table 1, except Arabidopsis MPK3 (AT3G45640), tobacco WIPK (BAA09600.1) and SIPK (BAC53772.1), P. patens MAPK (XP_001763232.1), C. reinhardtii MAPK8 (XP_001700291.1), and human P38 (NP_001306.1) and ERK2 (NP_620407.1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Phylogenetic Tree of Rice and Arabidopsis Group A/B/C MAPKs That Contain the TEY Motif in Their Activation Loops.
Supplemental Figure 2. Phylogenesis of Rice and Arabidopsis CDPKs.
Supplemental Figure 3. MPK5 TEY Motif Is Phoshorylated by MKK4 and Autophosphorylation, but Not by CPK18.
Supplemental Figure 4. Expression of CPK18, CPK4, MKK4, and MKK6 in Three CPK18-RI Lines (9, 10, and 13).
Supplemental Figure 5. Prediction of CPK18 Phosphorylated Sites on MPK5.
Supplemental Figure 6. Mapping CPK18 Phosphorylated Sites on MPK5.
Supplemental Figure 7. Native CPK18 Activities in Wild-Type and MPK5-RI Lines.
Supplemental Table 1. List of Genes and DNA Oligonucleotides Used in This Study.
Supplemental Data Set 1. Sequence Alignment of MAPKs to Construct the Phylogenetic Tree in Supplemental Figure 1.
Supplemental Data Set 2. Sequence Alignment of CDPKs to Construct the Phylogenetic Tree in Supplemental Figure 2.
Supplementary Material
Acknowledgments
We thank the late Ko Shimamoto at the Nara Institute of Science and Technology and Tsuyoshi Nakagawa at Shimane University for providing the pANDA and pUGW vectors, respectively, and Shuqun Zhang at the University of Missouri for providing the pET28-SIPK-KR and pET28-WIPK-KR plasmids. We also thank Kun-Liang Guan at the University of California San Diego for carefully reviewing the article and the Genomics Core Facility at Pennsylvania State University for providing the DNA sequencing service. This work was supported by the USDA/NIFA (Grant 2008-35301-19028 to Y.Y.) and the National Science Foundation Plant Genome Research Program (Grant DBI-0922747 to Y.Y.).
AUTHOR CONTRIBUTIONS
K.X., J.C., and Y.Y. designed the research. K.X., J.C., and Q.W. performed the experiments. K.X., J.C., and Y.Y. analyzed the data. K.X. and Y.Y. wrote the article.
Glossary
- BiFC
bimolecular fluorescence complementation
- RT-qPCR
quantitative RT-PCR
- ABA
abscisic acid
- ET
ethylene
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- PMSF
phenylmethylsulfonyl fluoride
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Andreasson E., Ellis B. (2010). Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 15: 106–113. [DOI] [PubMed] [Google Scholar]
- Asano T., Tanaka N., Yang G., Hayashi N., Komatsu S. (2005). Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: Comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 46: 356–366. [DOI] [PubMed] [Google Scholar]
- Bailey, T., Zhou, X., Chen, J., and Yang, Y. (2009). Role of ethylene, abscisic acid and MAP kinase pathways in rice blast resistance. In Advances in Genetics, Genomics and Control of Rice Blast Disease, G.-L. Wang and B. Valent, eds (Springer Science+Business Media), pp. 185–190. [Google Scholar]
- Bauer P.M., Fulton D., Boo Y.C., Sorescu G.P., Kemp B.E., Jo H., Sessa W.C. (2003). Compensatory phosphorylation and protein-protein interactions revealed by loss of function and gain of function mutants of multiple serine phosphorylation sites in endothelial nitric-oxide synthase. J. Biol. Chem. 278: 14841–14849. [DOI] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M.R., McCormack M., Lee H., Shan L., He P., Bush J., Cheng S.H., Sheen J. (2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464: 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Soriano L., Gómez-Ariza J., Bonfante P., San Segundo B. (2011). A rice calcium-dependent protein kinase is expressed in cortical root cells during the presymbiotic phase of the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 11: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Karin M. (2001). Mammalian MAP kinase signalling cascades. Nature 410: 37–40. [DOI] [PubMed] [Google Scholar]
- Cheng S.H., Willmann M.R., Chen H.C., Sheen J. (2002). Calcium signaling through protein kinases: The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 129: 469–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann C., Ichida A., Hong B., Romanowsky S.M., Hrabak E.M., Harmon A.C., Pickard B.G., Harper J.F. (2003). Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol. 132: 1840–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardick C., Chen J., Richter T., Ouyang S., Ronald P. (2007). The rice kinase database: A phylogenomic database for the rice kinome. Plant Physiol. 143: 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D., Yang Y., Cruz C.V., Höfte M. (2010). Abscisic acid-induced resistance against the brown spot pathogen Cochliobolus miyabeanus in rice involves MAP kinase-mediated repression of ethylene signaling. Plant Physiol. 152: 2036–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamei A., Pitzschke A., Nakagami H., Rajh I., Hirt H. (2007). Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science 318: 453–456. [DOI] [PubMed] [Google Scholar]
- Fukunaga R., Hunter T. (1997). MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 16: 1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge B., Gram H., Di Padova F., Huang B., New L., Ulevitch R.J., Luo Y., Han J. (2002). MAPKK-independent activation of p38alpha mediated by TAB1-dependent autophosphorylation of p38alpha. Science 295: 1291–1294. [DOI] [PubMed] [Google Scholar]
- Hamel L.P., Sheen J., Séguin A. (2014). Ancient signals: Comparative genomics of green plant CDPKs. Trends Plant Sci. 19: 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.F., Breton G., Harmon A. (2004). Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 55: 263–288. [DOI] [PubMed] [Google Scholar]
- Hegeman A.D., Rodriguez M., Han B.W., Uno Y., Phillips G.N., Jr, Hrabak E.M., Cushman J.C., Harper J.F., Harmon A.C., Sussman M.R. (2006). A phyloproteomic characterization of in vitro autophosphorylation in calcium-dependent protein kinases. Proteomics 6: 3649–3664. [DOI] [PubMed] [Google Scholar]
- Helliwell E.E., Wang Q., Yang Y. (2013). Transgenic rice with inducible ethylene production exhibits broad-spectrum disease resistance to the fungal pathogens Magnaporthe oryzae and Rhizoctonia solani. Plant Biotechnol. J. 11: 33–42. [DOI] [PubMed] [Google Scholar]
- Hrabak E.M., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132: 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Sánchez M., Cid V.J., Molina M. (2007). Retrophosphorylation of Mkk1 and Mkk2 MAPKKs by the Slt2 MAPK in the yeast cell integrity pathway. J. Biol. Chem. 282: 31174–31185. [DOI] [PubMed] [Google Scholar]
- Kinoshita-Kikuta E., Aoki Y., Kinoshita E., Koike T. (2007). Label-free kinase profiling using phosphate affinity polyacrylamide gel electrophoresis. Mol. Cell. Proteomics 6: 356–366. [DOI] [PubMed] [Google Scholar]
- Kurusu T., Yagala T., Miyao A., Hirochika H., Kuchitsu K. (2005). Identification of a putative voltage-gated Ca2+ channel as a key regulator of elicitor-induced hypersensitive cell death and mitogen-activated protein kinase activation in rice. Plant J. 42: 798–809. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lecourieux D., Ranjeva R., Pugin A. (2006). Calcium in plant defence-signalling pathways. New Phytol. 171: 249–269. [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A.A., Saitoh H., Felix G., Freymark G., Miersch O., Wasternack C., Boller T., Jones J.D., Romeis T. (2005). Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc. Natl. Acad. Sci. USA 102: 10736–10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S.J., Resing K.A., Candi J.M., Hermann A.S., Gloor J.W., Herskind K.R., Wartmann M., Davis R.J., Ahn N.G. (1994). Mitogen-activated protein (MAP) kinase phosphorylation of MAP kinase kinase: Determination of phosphorylation sites by mass spectrometry and site-directed mutagenesis. J. Biochem. 116: 304–314. [DOI] [PubMed] [Google Scholar]
- Mao G., Meng X., Liu Y., Zheng Z., Chen Z., Zhang S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAPK Group (2002). Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 7: 301–308. [DOI] [PubMed] [Google Scholar]
- Matsuda S., Gotoh Y., Nishida E. (1993). Phosphorylation of Xenopus mitogen-activated protein (MAP) kinase kinase by MAP kinase kinase kinase and MAP kinase. J. Biol. Chem. 268: 3277–3281. [PubMed] [Google Scholar]
- McAinsh M.R., Brownlee C., Hetherington A.M. (1992). Visualizing changes in cytosolic-free Ca2+ during the response of stomatal guard cells to abscisic acid. Plant Cell 4: 1113–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlmer N., Wurzinger B., Stael S., Hofmann-Rodrigues D., Csaszar E., Pfister B., Bayer R., Teige M. (2010). The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 63: 484–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Zhang S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51: 245–266. [DOI] [PubMed] [Google Scholar]
- Miki D., Shimamoto K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45: 490–495. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Pitzschke A., Schikora A., Hirt H. (2009). MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12: 421–426. [DOI] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M., Yang Y. (2002). Quantification of Magnaporthe grisea during infection of rice plants using real-time polymerase chain reaction and northern blot/phosphoimaging analyses. Phytopathology 92: 870–876. [DOI] [PubMed] [Google Scholar]
- Reddy A.S., Ali G.S., Celesnik H., Day I.S. (2011). Coping with stresses: Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23: 2010–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyna N.S., Yang Y. (2006). Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol. Plant Microbe Interact. 19: 530–540. [DOI] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649. [DOI] [PubMed] [Google Scholar]
- Salvador J.M., Mittelstadt P.R., Guszczynski T., Copeland T.D., Yamaguchi H., Appella E., Fornace A.J., Jr, Ashwell J.D. (2005). Alternative p38 activation pathway mediated by T cell receptor-proximal tyrosine kinases. Nat. Immunol. 6: 390–395. [DOI] [PubMed] [Google Scholar]
- Seger R., Krebs E.G. (1995). The MAPK signaling cascade. FASEB J. 9: 726–735. [PubMed] [Google Scholar]
- Singh R., et al. (2012). Rice mitogen-activated protein kinase interactome analysis using the yeast two-hybrid system. Plant Physiol. 160: 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F., Mizoguchi T., Yoshida R., Ichimura K., Shinozaki K. (2011). Calmodulin-dependent activation of MAP kinase for ROS homeostasis in Arabidopsis. Mol. Cell 41: 649–660. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzinger B., Mair A., Pfister B., Teige M. (2011). Cross-talk of calcium-dependent protein kinase and MAP kinase signaling. Plant Signal. Behav. 6: 8–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Yang Y. (2013). RNA-guided genome editing in plants using a CRISPR-Cas system. Mol. Plant 6: 1975–1983. [DOI] [PubMed] [Google Scholar]
- Xie K., Wu C., Xiong L. (2006). Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 142: 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Yang Y. (2003). Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15: 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.L., Yang Y., He Z. (2013). Roles of plant hormones and their interplay in rice immunity. Mol. Plant 6: 675–685. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Tena G., Xiong Y., Sheen J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. (1998). Resistance gene N-mediated de novo synthesis and activation of a tobacco mitogen-activated protein kinase by tobacco mosaic virus infection. Proc. Natl. Acad. Sci. USA 95: 7433–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.