Abstract
The regulation of cholesterol metabolism is one of the most studied biological processes since its first isolation from gallstones in 1784. High levels of plasma low-density lipoprotein (LDL) cholesterol and reduced levels of plasma high-density lipoprotein (HDL) cholesterol are widely recognized as major risk factors of cardiovascular disease. An imbalance in the production of reactive oxygen species (ROS) can oxidize LDL particles increasing the levels of the highly pro-atherogenic oxidized LDLs (ox-LDLs). Furthermore, under pathological scenarios, numerous molecules can function as pro-oxidants, such as iron or high-glucose levels. In addition to the classical mechanisms regulating lipid homeostasis, recent studies have demonstrated the important role of microRNAs (miRNAs) as regulators of lipoprotein metabolism, its oxidative derivatives and redox balance. Here, we summarize the recent findings in the field, highlighting the contribution of some miRNAs in lipid and oxidative-associated pathologies. We also discuss how therapeutic intervention of miRNAs may be a promising strategy to decrease LDL, increase HDL and ameliorate lipid and oxidative related disorders, including atherosclerosis, non-alcoholic fatty liver disease (NAFLD) and metabolic syndrome.
INTRODUCTION
MiRNAs have emerged as crucial and widely distributed post-transcriptional regulators of gene expression in the majority of biological processes, ranging from housekeeping functions to environmental stress responses [1–4]. In contrast with the high conservation of protein-coding genes between species, miRNAs appear to be an important factor in increasing the complexity of organisms, as mammalian genomes transcribe over an order of magnitude more non-coding RNAs than worms or flies [5]. MiRNAs are transcribed in the nucleus mainly by RNA polymerase II as long primary miRNAs (pri-miRNA) of 500–3000 bp that show a stem-loop hairpin structure. The pri-miRNA undergoes maturation by the sequential action of the Drosha/Pasha complex in the nucleus [6] and Dicer in the cytoplasm [7, 8]. After the pri-miRNA is processed, the mature miRNA (25–21 nt) is incorporated into the RNA-induced silencing complex (RISC) and binds preferentially to the 3´ untranslated region (3´UTR) of the mRNA target genes. Of note, a single miRNA modulates multiple genes often within the same biochemical pathway or interconnected nodes in regulatory networks and can help confer the robustness of biological processes by reinforcing transcriptional programs and attenuating dysregulated transcripts.
Regulation of miRNA function is involved in the pathogenesis of human diseases including cancer, metabolic disorders, cardiovascular diseases and neurological dysfunctions [9]. Here we review the role of miRNAs in regulating lipid metabolism, oxidative stress and cardiovascular diseases, including atherosclerosis. We will also discuss how modulating miRNA expression might be a promising therapy to combat atherosclerotic vascular disease and related dyslipidemias.
1. MiRNA regulation of Cholesterol metabolism
Cholesterol is an essential component of cell membranes and is required for vital processes [10, 11]. An excess of plasma cholesterol leads to its accumulation in the artery wall promoting atherosclerosis, the main cause of death in the Western and developing countries [12]. Cholesterol levels are maintained through a tightly regulated and complex mechanism that includes de novo biosynthesis, internalization of exogenous cholesterol and efflux of its excessive levels. All of these processes are controlled by miRNAs.
MiR-122
MiR-122 was one of the first miRNAs described in humans due to its abundance in the liver. This conserved liver-specific miRNA constitutes 70% of the total miRNA pool in this organ [13, 14], while it is absent in other tissues. Several observations underline the importance of miR-122 in liver biology and disease. First, antisense-mediated inhibition of miR-122 in mice leads to the induction of genes that are normally repressed in adult liver [15], suggesting that this miRNA is important for hepatocyte differentiation. Second, anti-miR-122 therapy in mice and non-human primates results in a significant reduction of plasma cholesterol and triglyceride levels. These effects on lipid metabolism have been associated with the modulation of genes involved in cholesterol synthesis including 3-hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS1), 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), 7-dehydrocholesterol reductase (DHCR7), and squalene epoxidase (SQLE) [15]. However, none of these genes are direct miR-122 targets and thus the mechanism underlying these effects remains unclear. Some of these observations have been recently confirmed in miR-122 deficient mouse models [16]. Specifically, miR-122 liver-specific knockout and miR-122 germline knockout mice have a significant reduction (~30%) of total serum cholesterol and triglyceride levels [17, 18]. Mechanistically, Tsai and colleagues found that the absence of miR-122 results in a significant reduction of the microsomal transfer protein (MTTP) expression, thereby decreasing very low-density lipoprotein (VLDL) secretion from the liver [17] (Figure 1). Together, these observations suggest that absence of miR-122 results in hypolipidemia through reduction of hepatic cholesterol synthesis and VLDL secretion.
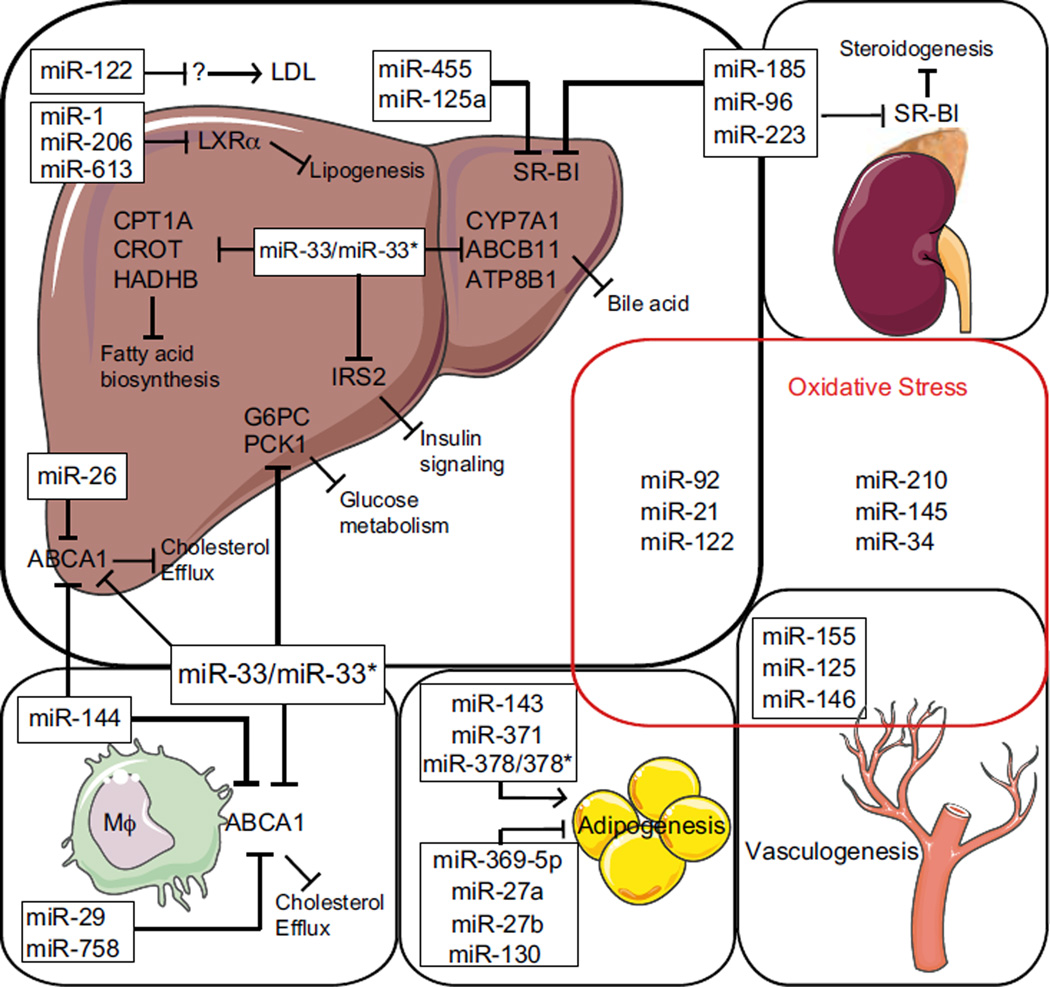
Figure 1. Schematic overview of miRNAs involved in lipid metabolism and ROS.
Different miRNAs are grouped in boxes and /or representative organs in which they target lipid metabolism regulators. Biological processes in which they are involved are also shown. ↑ indicates activation, ⊤ indicates inhibition.
miR-33/miR-33*
Different groups among us have recently identified miR-33a and miR-33b as intronic miRNAs located within the sterol regulatory element binding factor 2 (Srebf2) and Srebf1 genes [19–21]. In humans, miR-33a and miR-33b are co-transcribed with their host genes and regulate HDL biogenesis and cholesterol efflux by targeting ATP-binding cassette A1 (ABCA1) and ABCG1, and the endolysosomal transport protein, Niemann-Pick C1 (NPC1) [19–21]. These results were later confirmed genetically in miR-33 deficient mice [22]. In addition to ABCA1 and ABCG1, miR-33 also regulates the expression of two canalicular transporters, ATP-binding cassette, sub-family B (MDR/TAP), member 11 (ABCB11) and aminophospholipid transporter class I type 8B member 1 (ATP8B1), which regulate bile acid secretion [23]. Moreover, Li et al. have recently discovered that miR-33a also regulates the cytochrome P450, family 7, subfamily A, polypeptide 1 (CYP7A1), the rate-limiting enzyme in the synthesis of bile acid from cholesterol [24]. Altogether, these findings suggest that miR-33 is a key player in regulating several steps of the reverse cholesterol transport (RCT), including HDL biogenesis, macrophage cholesterol efflux and bile acid secretion (Figure 1 and Table 1). MiR-33a and miR-33b also contribute to the regulation of fatty acid metabolism, modulating the expression of carnitine palmitoyltransferase 1A (CPT1A), carnitine O-octanyl transferase (CROT), and hydroxyacyl-CoA dehydrogenase-3-ketoacyl-CoA thiolase-enoyl-CoA hydratase (trifunctional protein) β-subunit (HADHB) [25, 26]. CPT1a, CROT and HADHB regulate the transport and degradation of fatty acids in the mitochondria and the overexpression of miR-33 in human hepatic cells results in a significant decrease of fatty acid oxidation [25, 26]. In addition to lipid metabolism, miR-33 also regulates insulin sensitivity and hepatic glucose production by targeting the insulin receptor substrate 2 (IRS2) and glucose-6-phosphatase (G6PC) and phosphoenolpyruvate carboxykinase (PKC1), respectively [25, 27].
Table 1.
Summary of the prototypical functions of miRNAs in lipid metabolism and oxidative stress.
| miRNA | Target mRNA | Regulatory effect | References |
|---|---|---|---|
| Lipid metabolism | |||
| miR-122 | ?, MTTP | ↑ Cholesterol synthesis (LDL) ↑ VLDL secretion |
[17] |
| miR-33 | ABCA1 | ↓ Cholesterol efflux | [19] |
| CROT CPT1 HADHB |
↓ Fatty acid biosynthesis | [25, 26] | |
| CYP7A1 ABCB11 ATP8B1 |
↓ Bile acid synthesis and secretion | [23] | |
| IRS2 G6PC |
↓ Insulin signaling | [25, 27] | |
| PCK1 | ↓ Glucose metabolism | [27] | |
| miR-758, miR-106b, miR144 | ABCA1 | ↓ Cholesterol Efflux | [32–35] |
| miR-125a, miR-455 | SR-BI | ↓ Cholesterol metabolism and Steroidogenesis | [39] |
| miR-185, miR-96, miR-223 | SR-BI | ↓ Cholesterol metabolism and Steroidogenesis | [40] |
| miR-1, miR-206, miR-613 | LXRα | ↓ Lipogenesis | [43, 44] |
| miR-143, miR-371 miR-378/378* |
CRAT, MED13 | ↑ Adipogenesis | [45–47] |
| miR-27a/b, miR-369-5p, miR-130 | ANGPTL3 GPAM, FABP4 |
↓ Adipogenesis | [46, 48, 49] |
| Oxidative stress | |||
| miR-125a/b-5p | ECE-1 ORP-9 |
Anti-inflammatory | [56, 57] |
| miR-365 | BCL-2 | Pro-inflammatory | [58] |
| Let-7 | BCL-XL | Pro-inflammatory | [59] |
| miR-222/221 | eNOS | Endothelial homeostasis | [60] |
| miR-92a | eNOS KLF2 KLF4 |
Endothelial homeostasis | [61] |
| miR-21 | eNOS PTEN PPAR-a |
Anti-apoptotic Pro-inflammatory | [62] |
| miR-34, miR-217 | SIRT-1 | Pro-apoptotic Endothelial senescence | [63–65] |
| miR-633 | PPAR-a | Monocyte adhesion | [69] |
| miR-146a | TLR4 KLF-4 |
Anti-inflammatory VSMC proliferation Neointimal hyperplasia | [73, 79] |
| miR-9 | PPAR-γ | Anti-inflammatory | [74] |
| miR-27* a/b | NF-κB | Anti-inflammatory | [75–80] |
| Let-7g | LOX-1 | Monocyte adhesion | [84] |
| miR-29b | DNMT3b | Plaque instability | [86] |
| miR-210 | ISCU | Oxidative response to hypoxia | [87–88] |
| miR-122 | HFE HJV |
Oxidative stress | [89] |
During miRNA biosynthesis, the passenger strand (also designated as 3p or *) is often rapidly degraded and has no effect on gene expression, but in certain miRNAs, such as miR-33 [28], the Dicer generated duplex (miRNA/miRNA*) gives rise to both “5p” and “3p” mature active miRNAs. Interestingly, we have recently demonstrated that miR-33* shares a similar target network to that of miR-33 and directly represses target genes in human hepatic and macrophage cell lines. Importantly, miR-33* overexpression reduces fatty acid oxidation in hepatocytes. Altogether, these data suggest that miR-33 regulates lipid metabolism through both arms of the miR-33/miR-33* duplex [28].
Anti-miR-33 therapy: a promising approach for treating cardiometabolic diseases
Human epidemiological studies show a strong inverse association between HDL plasma levels and coronary heart disease (CHD), which led several groups to study the potential benefit of anti-miR-33 therapy in preventing atherosclerosis progression and promoting plaque regression in murine models of atherosclerosis. In the first study, Rayner and colleagues demonstrated that 2’-O-methoxyethyl (2´F/MOE) modified anti-miR33 therapy enhances the regression of atherosclerosis in Ldlr−/− mice by increasing circulating HDL-C levels and RCT. Antagonism of miR-33 in mice also reduces lipid and monocyte/macrophage accumulation in atherosclerotic plaques, thereby reducing inflammation. Another interesting finding of this study is the preferential localization of 2’F/MOE anti-miR-33 oligonucleotides in foam cells, which results in a significant derepression of miR-33 target genes, including Abca1. In addition to reduced macrophage accumulation in atherosclerotic plaques from Ldlr−/− mice treated with anti-miR-33 oligonucleotides, the inhibition of miR-33 also decreases the expression of proinflammatory and pro-oxidant genes, including inducible nitric oxide synthase (Nos2) and tumor necrosis factor alpha (Tnf). Even though these results suggest that miR-33 may increase inflammation, other studies have found the opposite results [29]. Indeed, overexpression of miR-33 reduces the expression of the receptor interacting protein 140 (RIP-140), which is a transcriptional co-activator of NF-κB-responsive genes [29].
In addition to the atherosclerosis regression studies, other groups have examined the efficacy of anti-miR-33 therapy during the progression of atherosclerosis. In our study, we found that 2’F/MOE modified anti-miR-33 therapy was effective in an atherosclerosis progression model. We observed a significant reduction in atherosclerotic plaque size in mice treated with miR-33 ASOs. Importantly, the circulating HDL levels were very similar but the cholesterol efflux capacity of HDL isolated from mice treated with miR-33 ASOs was significantly higher compared to HDL isolated from Ctrl-ASO treated mice [30]. We also observed that mice treated with anti-miR-33 oligonucleotides have a significantly higher expression of ABCA1, matrix metallopeptidase (MMP-2), collagen type I alpha 1 (COL1A-1) and COL3A-1 in the aorta. These results suggest that the anti-atherogenic effect of long-term anti-miR-33 therapy is independent of plasma HDL levels. In contrast, Marquart and colleagues have recently reported that anti-miR-33 therapy using locked nucleic acid (LNA) modified oligonucleotides fails to increase circulating HDL levels and to slow the progression of atherosclerosis in Ldlr−/− mice fed a Western diet (WD). These unexpected results might be explained by the different chemistry employed in the oligonucleotide modification, as well as by the reduced hepatic miR-33 levels observed in mice fed a WD. Finally, Horie and colleagues assessed the progression of atherosclerosis in miR-33/ApoE−/− double mutant mice [31]. Similar to the results observed in our study, miR-33 genetic deficiency results in a significant reduction in atherosclerotic plaque formation. However, the authors also found increased levels of circulating HDL in miR-33 null mice, suggesting that the anti-atherogenic effect of miR-33 deficiency might be mediated by increasing plasma HDL and RCT.
Other miRNAs that regulate ABCA1 expression
In addition to miR-33 and miR-122, other miRNAs have also been described to participate in lipid metabolism. MiR-758 and miR-106 have been shown to regulate ABCA1 expression at the post-transcriptional level in hepatocytes, macrophages and neuronal cells [32, 33]. MiR-758 is an intergenic miRNA which is down-regulated after cholesterol loading in macrophages and in the liver of mice fed a high-fat diet [32]. MiR-106 also regulates ABCA1 levels and miR-106 overexpression significantly decreases ABCA1 levels and impairs cellular cholesterol efflux in neuronal cells [33]. In addition to these miRNAs, miR-144 has also been recently reported to regulate lipid metabolism by modulating ABCA1 expression [34, 35]. Ramirez et al showed that ABCA1 is post-transcriptionally regulated by miR-144 in vitro and in vivo (Figure 1 and Table 1). MiR-144 over-expression inhibits ABCA1 expression in human and mouse cell lines, thereby attenuating cholesterol efflux to APOA1. Most importantly, delivery of miR-144 mimics to mice inhibits hepatic ABCA1 expression and reduces circulating HDL levels. Conversely, miR-144 silencing in mice increases hepatic ABCA1 expression and plasma HDL levels. Thus, miR-144 appears to regulate both macrophage cholesterol efflux and HDL biogenesis in the liver. We also reported that liver X receptor (LXR) activation increases miR-144 in macrophages, human hepatic cells and mouse livers and that ABCA1 is a target of LXR-induced miR-144. These data reveal how an inducible miRNA controls a negative feedback loop to ensure a tight regulation of cholesterol homeostasis [34]. In a second report, Vallim and colleagues described a novel pathway that links the activation of nuclear farnesoid X receptor (FXR) to the induction of hepatic miR-144 expression. They use both gain-of-function and loss-of-function experiments to demonstrate that changes in hepatic miR-144 levels are sufficient to regulate hepatic ABCA1 expression and plasma HDL levels. They also identify functional FXR response elements (FXREs) in the miR-144 promoter, consistent with direct FXR regulation [35].
SR-BI regulation by miRNAs
In addition to promoting cellular cholesterol efflux, HDL particles deliver cholesterol esters to the liver through the scavenger receptor class B type I (SR-BI) for excretion [36]. SR-BI is expressed mostly in liver and other steroidogenic cells such as adrenal glands, ovary and testis, where SR-BI delivers the bulk of the cholesterol substrate needed for steroidogenesis, the biological process that synthesizes steroid hormones [37, 38]. MiR-125a-5p and miR-455 have been recently described to regulate SR-BI expression by direct targeting [39] (Figure 1). Importantly, miR-125a and miR-455 overexpression inhibits SR-BI-mediated selective HDL uptake and SR-BI-supported steroid hormone synthesis and accordingly, anti-miR-125a and anti-miR-455 treatment stimulate both processes. In this study, in vitro treatment of primary rat granulosa cells with cyclic AMP (cAMP) or in vivo treatment of rat adrenals with adrenocorticotropic hormone (ACTH) decreased the expression of miR-125a, miR-125b, and miR-455 and reciprocally increased SR-BI expression. Most recently, Wang and colleagues have identified miR-185, miR-96 and miR-223 as important regulators of SR-BI expression and HDL uptake [40]. MiR-185, miR-96 and miR-223 overexpression in Hep-G2 cells inhibits SR-BI expression and decreases HDL uptake. Interestingly, miR-96 and miR-185 levels correlate inversely with SR-BI expression in the liver of ApoE knockout mice fed a high-fat diet [40]. These data suggest that miR-185, miR-96 and miR-223 may be involved in a novel type of regulation of hepatic SR-BI and cholesterol metabolism.
Other miRNAs that regulate lipid metabolism
Several recent reports have identified novel miRNAs that regulate lipid metabolism including miR-1, miR-206, miR-378/378*, miR-27a/b. Two independent studies have addressed the role of several miRNAs inhibiting LXRα expression post-transcriptionally. LXR is a ligand-activated nuclear receptor playing an important role in the transcriptional regulation of lipid metabolism [41]. LXR activation induces the expression of lipogenic genes such as SREBP1c, fatty acid synthase (FAS), carbohydrate responsive element-binding protein (ChREBP) and acetyl-CoA carboxylase (ACC) [42]. In the first study, Zhong and colleagues demonstrate that miR-1 and miR-206 regulate lipogenesis by repressing LXRα expression, thereby decreasing lipogenic gene expression and lipid droplet accumulation in human hepatic cell lines [43] (Figure 1). Using a similar approach, Zhong et al. found that miR-613 also inhibits lipogenesis by suppressing genes involved in lipid synthesis, including SREBP1c, FAS, ChREBP and ACC [44]. Altogether, these observations suggest that the regulation of lipid metabolism by LXRs is also controlled by LXR-induced miRNAs.
There are many other more miRNAs involved in lipid metabolism such as miR-143, miR-371 and miR-378/miR-378* that have been described to promote adipogenesis [45–47] (Figure 1). In contrast, miR-27a, miR-27b, miR-369-5p and miR-130 negatively regulate adipocyte differentiation [46, 48, 49] (Figure 2). Interestingly, miR-27b is highly expressed in the liver of mice fed a high-fat diet [48] and regulates the expression of several key lipid-metabolism genes, including angiopoietin-like 3 (ANGPTL3) and glycerol-3-phosphate acyltransferase (GPAM) [48].
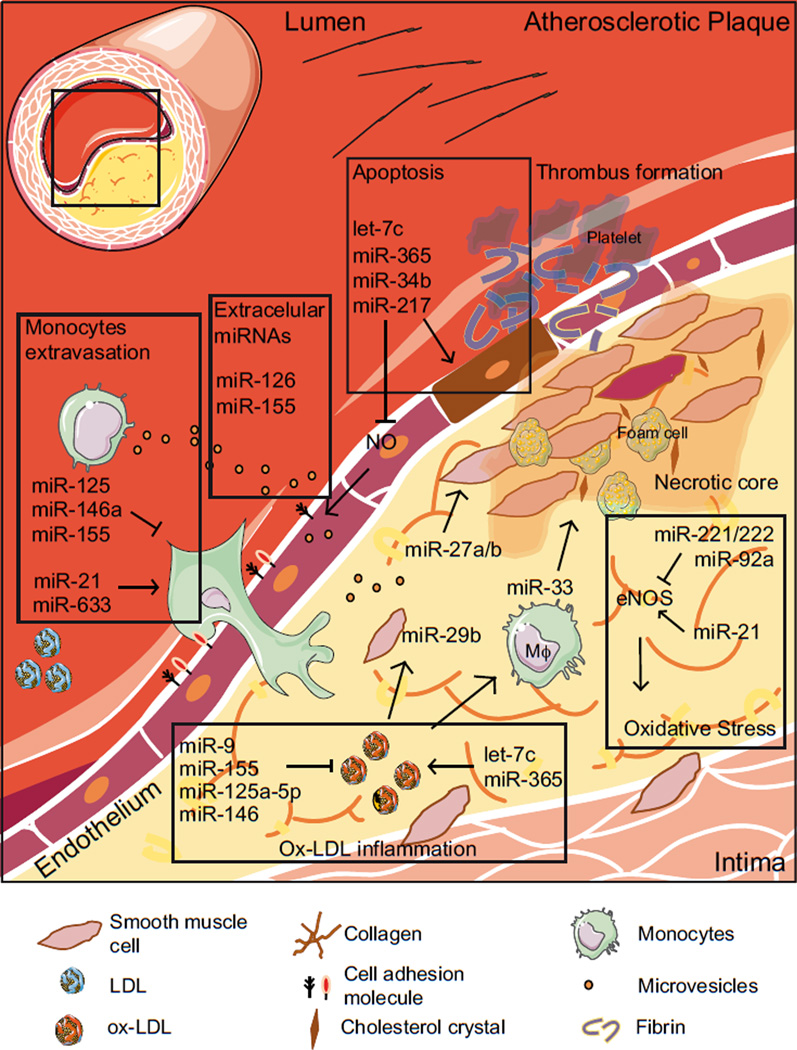
Figure 2. Overview of the potential roles of miRNAs in atherosclerosis.
A schematic picture of the pathological scenario in the atherosclerotic plaque. Boxes represent different biological processes implicated in atherosclerotic disease, such as inflammation, monocyte extravasation, apoptosis, oxidative stress or extracellular miRNAs. MiRNAs can modulate either positively or negatively several of these processes. ↑ indicates activation, ⊤ indicates inhibition.
In summary, miRNAs seem to be modulating post-transcriptionally several processes involved in cholesterol metabolism and, in general, lipid metabolism. It is clear that deeper knowledge of miRNA mechanistic functions could open new opportunities for the therapeutic treatment of lipid-related-diseases.
2. Atherosclerosis, cholesterol and reactive oxygen species (ROS)
There are a growing number of studies that demonstrate the importance of miRNAs in regulating the biology of the human atherosclerotic plaque (Figure 2 and Table 1). Furthermore, miRNA expression profiles comparing atherosclerotic plaques versus healthy arteries or plaques from symptomatic and asymptomatic subjects have been performed [50–52]. Even though these studies provide useful information about the genetic changes of human atherosclerotic plaques, they usually fail to decipher the specific source (cellular and/or area of the plaque) and the molecular mechanisms that underlie the role of these miRNAs. New techniques, such as laser capture microdissection, could help to elucidate the specific commitment of a single miRNA in oxidative stress or lipid metabolism inside a specific cell type and/or in the particular communication established by the plaque milieu [53].
ROS, endothelial cell activation and atherosclerosis
The initial stage of atherosclerotic plaque development is characterized by the infiltration and retention of LDL in the artery wall [54]. LDL particles are subsequently modified and oxidized leading to the accumulation of oxidized LDL particles (ox-LDLs) in the arterial intima. Ox-LDLs generated by mediation of ROS are a major trigger for atheroma plaque formation and development because these modified LDLs are significantly more potent than native LDLs in promoting endothelial dysfunction, one of the first steps leading to plaque formation [55]. In this regard, ox-LDL-regulated expression of miR-125a/b-5p has been shown to control endothelin-1 levels, which has an important role as one of the most potent vasoconstrictive peptides in endothelial cells (ECs) [56, 57]. Moreover, miR-365 and let-7c also regulates EC functions in response to ox-LDLs by targeting the pro-survival molecules B–Cell lymphome 2 (BCL-2) and B–Cell lymphome extra-large (BCL-XL), respectively [58, 59].
Nitric oxide (NO) bioavailabilty is another factor that regulates ROS levels in the artery wall. An imbalance in NO levels potentiates a dysregulation in ROS metabolism resulting in oxidative stress. NO levels are regulated by the nitric oxide synthases (NOS) and among them, endothelial nitric oxide synthase (eNOS) is a key player in cardiovascular diseases. The expression of this enzyme is downregulated at the post-transcriptional level by miRNAs, including miR-222/221and miR-92a [60, 61]. In contrast, miR-21 upregulates eNOS activity and protects against apoptosis through targeting of its antagonist phosphatase and tensin homologue (PTEN) [62]. Other miRNAs have also been shown to regulate EC functions in response to oxidative stress, including miR-34a and miR-217. Both miRNAs regulate SIRT1 signaling [63–65]. SIRT-1 controls EC function against oxidative stress through different mechanisms, such as the increase in NO bioavailability and promotion of mitochondrial biogenesis, thereby augmenting anti-oxidant defenses and preventing senescence [66].
In the atherosclerotic process, endothelial dysfunction and excessive ROS boost the expression of adhesion molecules, inflammatory cytokines and chemotactic factors promoting leucocyte adhesion and transmigration [67]. As a consequence, migrated leukocytes inside the neointima engulf ox-LDLs and enhance macrophage and foam cell differentiation [68]. In this scenario, miR-633 and miR-21 stimulate monocyte adhesion to the endothelium during oscillatory flow by targeting peroxisome proliferator-activated receptor α (PPARα) [69, 70], whereas miR-92a is downregulated during laminar shear flow and upregulates the atheroprotective KLF2 [61] and the pro-inflammatory KLF4 [71]. On the other hand, miR-125a-5p and miR-155 inhibit lipid uptake and the inflammatory responses in ox-LDL-stimulated monocytes/macrophages [72]. Similar effects occur through the action of miR-146a, which hampers ox-LDL uptake and the inflammatory response through targeting toll-like receptor 4 (TLR4) and as a consequence, the inhibition of its downstream signaling cascade [73]. Other miRNAs, such as miR-9, regulates the expression of peroxisome proliferator-activated receptor δ (PPARδ) in human monocytes, negatively affecting the inflammatory response [74].
ROS can also modulate miRNA levels in monocytes/macrophages. In this regard, oxidative stress downregulates miR-27a* and miR-27b* expression in RAW264.7 cells [75]. Both miRNAs inhibit the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) response induced by lipopolysacharide (LPS). Altogether these results demonstrate that miRNAs regulates ROS production and that ROS production also regulates miRNA expression.
ROS, Vascular Smooth Muscle Cells and atherogenesis
Vascular smooth muscle cells (VSMCs) are mainly involved in the regulation of blood pressure due to contractile tonus. Nonetheless, during atheroma formation both biochemical and mechanical signals promote reprogramming of these stromal cells, switching them from a contractile to a phagocyte-like phenotype [76]. These responses also include changes in migration, proliferation, secretion of proteases and finally apoptosis [77]. VSMCs, apoptosis and secretion of proteolityc enzymes are determinants of plaque instability, rupture and thrombosis when the haemo/necro/lipidic core is exposed to the bloodstream. Indeed, the lipid rich core is six times more thombogenic than any other component from the plaque [78]. The phenotypic changes suffered by VSMCs in the plaque can be modulated by miRNAs, such as miR-27 or miR-146a, among others [79]. MiR-27 has been shown to participate in mostly all known processes that take place in atherosclerosis including inflammation, lipid metabolism, oxidative stress, insulin resistance and type 2 diabetes [80]. MiR-146a controls VSMC functions by regulating the expression of the anti-proliferative KLF4, promoting VSMC proliferation in vitro and neointimal hyperplasia in vivo [79]. Similar to miR-210 and miR-21, miR-146 has also been reported to be dysregulated in human atherosclerotic plaques, while miR-21 is responsible for protecting VSMCs against oxidative stress-induced apoptosis [51, 81].
Cholesterol can promote VSMC transdifferentiation to macrophage-like cells, thereby enhancing the lipid pool in the plaque [76]. One of the mechanisms that allows the ox-LDLs to be engulfed is through the lectin-like oxidized LDL receptor 1 (LOX-1) in VSMCs. As a consequence of this uptake, the production of reactive species is increased in the vasculature [82, 83]. Let-7g has been shown to modulate the uptake of ox-LDL through LOX-1, as well as VSMC apoptosis and ROS production, which is dependent on NADPH activity [84]. Furthermore, ox-LDLs can upregulate miR-29b levels which results in an increased synthesis of metallopeptidase 2/9 (MMP2/9), thus favoring plaque instability through reduction of collagen content [85]. Accordingly, it was recently shown that miR-29 mediates the down-regulation of extracellular matrix proteins, thereby promoting plaque rupture or aneurysm formation [86].
Iron metabolism, ROS and atherogenesis
Other molecules present at high levels in the thrombogenic plaque, such as iron, are also involved in oxidative processes in atherosclerosis and are modulated by miRNAs. Iron metabolism is essential for cell survival since it is needed for the action of several enzyme complexes (i.e. ribonucleotide reductase and cytochrome P450). Nevertheless, when there is an imbalance in iron metabolism homeostasis and its levels are increased, it can promote ROS production through the FENTON reaction. Several miRNAs have been shown to regulate iron homeostasis, such as miR-210. MiR-210 has recently been described to regulate the activity of prototypical iron-sulfur scaffold protein (ISCU) controlling mitochondrial metabolism and free radical response in response to hypoxia [87, 88]. Recently, it has also been shown that miR-122 regulates systemic iron homeostasis by repressing the target genes hemochromatosis (HFE) and hemojuvelin (HJV) [89]. These mRNAs encode activators of the hormone hepcidin, which regulates iron availability and mice with reduced miR-122 levels suffer from iron deficiency [89]. MiR-122 inhibition also decreases the basic leucine zipper transcription factor 1 (BACH1) and increases heme oxygenase-1 (HO-1), a key cytoprotective enzyme with antioxidant properties repressed by BACH1 [90]. These data suggest that therapeutic targeting of miR-122 and upregulation of HO-1 may represent a new strategy for cytoprotection.
Atherosclerosis is a complex disease in which different interactions between various cell types and molecules in the vessel wall have been described. New players, such as red blood cells (RBCs) and iron content, are being highlighted to regulate lipid oxidation/accumulation and the redox status in the atheroma, besides the well-known participants such as ECs, VSMCs or monocytes/macrophages. Elevated levels of LDLs and their oxidation are believed to trigger atherosclerosis and the accumulation of modified species of lipids inside the intima is a hallmark of plaque development and instability. For example, oxidized lipids can be stored inside the cell as ceroids, or aggregates of protein and lipids typical of human plaques, which colocalize with intra- and extracellular iron content [91]. In atherosclerotic plaques, heme/iron groups are increased up to 17 times and have been reported to alter cholesterol efflux and oxidative stress in macrophages [91]. Thus, modulators of these processes, in which lipids and ROS are involved, are of high interest in research and potential targets in vascular therapeutics. While we have already revised numerous scientific findings regarding miRNA modulation of lipid metabolism and redox homeostasis, most of these recent and exciting findings still must be clearly elucidated as a single miRNA (e.g. miR-146a) can exert different functions depending on the cell type in which it is expressed. Furthermore, a single mRNA target can be regulated by several miRNAs. In addition, the complex network of communications established by the cellular milieu in the atherosclerotic plaque requires further studies to reach a therapeutic decision in atherosclerosis.
3. Circulating miRNAs
While the idea of intercellular communication through small vesicles containing mixtures of proteins and nucleic acids was proposed over 30 years ago [92], the presence of cell-type specific miRNAs in these particles has only recently been reported. Cells can selectively package certain miRNAs and are actively secreted into the blood in membrane-derived vesicles [93, 94], lipoproteins, such as HDL [95], and AGO2 ribonucleoprotein complexes [96]. Specifically, circulating miRNAs have recently been shown to be very stable in the plasma and have been implicated in vascular diseases [97, 98]. Despite this, the diagonostic and prognostic properties of these miRNAs are still highly controversial [99]. These extracellular miRNAs have become a focus of attention because of their potential as novel disease biomarkers. Among others, cardiovascular diseases, such as atherosclerosis are characterized by differential plasma miRNA profiles compared with healthy populations [97, 100, 101]. Circulating miRNAs are biologically active since recipient cells show altered gene expression [102, 103]. HDL particles are also able to deliver miRNAs to recipient cells. Interestingly, the miRNA profiles of HDL particles are different in healthy people than in subjects with familial hypercholesterolemia [95]. These interactions can be performed in several different fashions and miRNAs can serve as bridges of communication between cells in the vasculature. In recent studies, it has been shown that extracellular vesicles secreted by shear-stressed ECs enriched in miR-126 or miR-143/145 are engulfed by VSMCs thereby regulating target-gene expression in host cells [104, 105]. Conversely, increased proliferation of ECs can be achieved by exogenous miRNAs secreted by other cells from the vasculature, such as monocytes [103].
Therefore, a new potential role for miRNAs as a possible form of intercellular communication like hormones can be considered. There is increasing interest in the knowledge of the mechanisms that control miRNA extracellular export, cell-specific targeting and recognition machinery. In this regard, the physiological “in vivo” roles have to be definitively established; therefore, it is likely that future research on secreted miRNAs will open new therapeutic approaches for cardiovascular disease treatment.
CONCLUSIONS
There is a tremendous interest in the therapeutic application of miRNA inhibitors and miRNA mimics to treat lipid-related disorders. The high conservation of miRNA sequences between species suggests that the biological pathways modulated by miRNAs might also be conserved, making them attractive research targets with potential therapeutic applications. One of the better-studied miRNAs that illustrates this phenomenon is miR-33. Nonetheless, caution should be taken before translating data from murine models to human therapies. A good example derives from miR-33b, which is absent in the mouse genome. Additionally, a given miRNA can be predicted to regulate several hundred genes, in the same way that a gene can be regulated by more than one miRNA. Thus, the contribution of each miRNA in modulating its target gene expression levels will be determined by the relative abundance of each miRNA in different tissues and the biological stimuli that regulates their expression. Therefore, it seems likely that a combination therapy might improve the prognosis for patients with cardiovascular disease.
HIGHLIGHTS.
-
→
MicroRNAs are emerging regulators of gene expression, controlling specific biochemical pathways or cellular processes.
-
→
Atherosclerosis is a multifactorial disease in which lipid species, ROS, endothelial and circulating cells play important roles.
-
→
Different microRNAs can exert both pro-atherogenic and anti-atherogenic roles.
-
→
Antisense microRNA therapies represent a new promising intervention for cardiometabolic diseases.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health R01HL107953 and R01HL106063 (to C.F.-H.). N.R. is supported by the Ministerio de Educación (Programa Nacional de Movilidad de Recursos Humanos del Plan Nacional de I-D+i 2008-2011). Figures were produced using Servier Medical Art (www.servier.com).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
C.F.-H has patents on the use of miRNA-33 inhibitors.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 3.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 4.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci U S A. 2008;105:2946–2950. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 7.Davis N, Mor E, Ashery-Padan R. Roles for Dicer1 in the patterning and differentiation of the optic cup neuroepithelium. Development. 2011;138:127–138. doi: 10.1242/dev.053637. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez C, Lobo MMV, Gomez-Coronado D, Lasuncion MA. Cholesterol is essential for mitosis progression and its deficiency induces polyploid cell formation. Exp Cell Res. 2004;300:109–120. doi: 10.1016/j.yexcr.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez C, Martin M, Gomez-Coronado D, Lasuncion MA. Effects of distal cholesterol biosynthesis inhibitors on cell proliferation and cell cycle progression. J Lipid Res. 2005;46:920–929. doi: 10.1194/jlr.M400407-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006;367:1747–1757. doi: 10.1016/S0140-6736(06)68770-9. [DOI] [PubMed] [Google Scholar]
- 13.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 14.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 15.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 16.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen RM, Marquart TJ, Albert CJ, Suchy FJ, Wang DQ, Ananthanarayanan M, et al. miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol Med. 2012 doi: 10.1002/emmm.201201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li T, Francl JM, Boehme S, Chiang JY. Regulation of cholesterol and bile acid homeostasis by the CYP7A1/SREBP2/miR-33a axis. Hepatology. 2013 doi: 10.1002/hep.26427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, et al. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez CM, Goedeke L, Rotllan N, Yoon JH, Cirera-Salinas D, Mattison JA, et al. MiR-33 regulates glucose metabolism. Mol Cell Biol. 2013 doi: 10.1128/MCB.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goedeke L, Vales-Lara FM, Fenstermaker M, Cirera-Salinas D, Chamorro-Jorganes A, Ramirez CM, et al. A regulatory role for miRNA-33* in controlling lipid metabolism gene expression. Mol Cell Biol. 2013 doi: 10.1128/MCB.01714-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho PC, Chang KC, Chuang YS, Wei LN. Cholesterol regulation of receptor-interacting protein 140 via microRNA-33 in inflammatory cytokine production. FASEB J. 2011;25:1758–1766. doi: 10.1096/fj.10-179267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rotllan N, Ramirez CM, Aryal B, Esau CC, Fernandez-Hernando C. Therapeutic Silencing of MicroRNA-33 Inhibits the Progression of Atherosclerosis in Ldlr−/− Mice. Arterioscler Thromb Vasc Biol. 2013 doi: 10.1161/ATVBAHA.113.301732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horie T, Baba O, Kuwabara Y, Chujo Y, Watanabe S, Kinoshita M, et al. MicroRNA-33 deficiency reduces the progression of atherosclerotic plaque in ApoE−/− mice. J Am Heart Assoc. 2012;1:e003376. doi: 10.1161/JAHA.112.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramirez CM, Davalos A, Goedeke L, Salerno AG, Warrier N, Cirera-Salinas D, et al. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler Thromb Vasc Biol. 2011;31:2707–2714. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Yoon H, Ramirez CM, Lee SM, Hoe HS, Fernandez-Hernando C, et al. MiR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Exp Neurol. 2012;235:476–483. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramirez CM, Rotllan N, Vlassov AV, Davalos A, Li M, Goedeke L, et al. Control of Cholesterol Metabolism and Plasma HDL Levels by miRNA-144. Circ Res. 2013 doi: 10.1161/CIRCRESAHA.112.300626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de, A V, Tarling E, Kim T, Civelek M, Baldan A, Esau C, et al. MicroRNA-144 Regulates Hepatic ABCA1 and Plasma HDL Following Activation of the Nuclear Receptor FXR. Circ Res. 2013 doi: 10.1161/CIRCRESAHA.112.300648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 1996;271:518–520. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 37.Azhar S, Nomoto A, Leers-Sucheta S, Reaven E. Simultaneous induction of an HDL receptor protein (SR-BI) and the selective uptake of HDL-cholesteryl esters in a physiologically relevant steroidogenic cell model. J Lipid Res. 1998;39:1616–1628. [PubMed] [Google Scholar]
- 38.Landschulz KT, Pathak RK, Rigotti A, Krieger M, Hobbs HH. Regulation of scavenger receptor, class B, type I, a high density lipoprotein receptor, in liver and steroidogenic tissues of the rat. J Clin Invest. 1996;98:984–995. doi: 10.1172/JCI118883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu Z, Shen WJ, Kraemer FB, Azhar S. MicroRNAs 125a and 455 repress lipoprotein-supported steroidogenesis by targeting scavenger receptor class B type I in steroidogenic cells. Mol Cell Biol. 2012;32:5035–5045. doi: 10.1128/MCB.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, Jia XJ, Jiang HJ, Du Y, Yang F, Si SY, et al. MiRNA-185, MiRNA-96 and MiRNA-223 Repress Selective HDL-Cholesterol Uptake through Posttranscriptional Inhibition of Scavenger Receptor Class BI in Hepatic Cells. Mol Cell Biol. 2013 doi: 10.1128/MCB.01580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zelcer N, Tontonoz P. Liver X receptors as integrators of metabolic and inflammatory signaling. J Clin Invest. 2006;116:607–614. doi: 10.1172/JCI27883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cha JY, Repa JJ. The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. J Biol Chem. 2007;282:743–751. doi: 10.1074/jbc.M605023200. [DOI] [PubMed] [Google Scholar]
- 43.Zhong D, Huang G, Zhang Y, Zeng Y, Xu Z, Zhao Y, et al. MicroRNA-1 and microRNA-206 suppress LXRalpha-induced lipogenesis in hepatocytes. Cell Signal. 2013;25:1429–1437. doi: 10.1016/j.cellsig.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 44.Zhong D, Zhang Y, Zeng YJ, Gao M, Wu GZ, Hu CJ, et al. MicroRNA-613 represses lipogenesis in HepG2 cells by downregulating LXRalpha. Lipids Health Dis. 2013;12:32. doi: 10.1186/1476-511X-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 46.Bork S, Horn P, Castoldi M, Hellwig I, Ho AD, Wagner W. Adipogenic differentiation of human mesenchymal stromal cells is down-regulated by microRNA-369-5p and up-regulated by microRNA-371. J Cell Physiol. 2011;226:2226–2234. doi: 10.1002/jcp.22557. [DOI] [PubMed] [Google Scholar]
- 47.Gerin I, Bommer GT, McCoin CS, Sousa KM, Krishnan V, MacDougald OA. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab. 2010;299:E198–E206. doi: 10.1152/ajpendo.00179.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vickers KC, Shoucri BM, Levin MG, Wu H, Pearson DS, Osei-Hwedieh D, et al. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57:533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim MM, Srikantan S, et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol. 2011;31:626–638. doi: 10.1128/MCB.00894-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bidzhekov K, Gan L, Denecke B, Rostalsky A, Hristov M, Koeppel TA, et al. microRNA expression signatures and parallels between monocyte subsets and atherosclerotic plaque in humans. Thromb Haemost. 2012;107:619–625. doi: 10.1160/TH11-09-0607. [DOI] [PubMed] [Google Scholar]
- 51.Raitoharju E, Lyytikainen LP, Levula M, Oksala N, Mennander A, Tarkka M, et al. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis. 2011;219:211–217. doi: 10.1016/j.atherosclerosis.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 52.Cipollone F, Felicioni L, Sarzani R, Ucchino S, Spigonardo F, Mandolini C, et al. A unique microRNA signature associated with plaque instability in humans. Stroke. 2011;42:2556–2563. doi: 10.1161/STROKEAHA.110.597575. [DOI] [PubMed] [Google Scholar]
- 53.Wang S, Wang L, Zhu T, Gao X, Li J, Wu Y, et al. Improvement of tissue preparation for laser capture microdissection: application for cell type-specific miRNA expression profiling in colorectal tumors. BMC Genomics. 2010;11:163. doi: 10.1186/1471-2164-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams KJ, Tabas I. The response-to-retention hypothesis of early atherogenesis. Arterioscler Thromb Vasc Biol. 1995;15:551–561. doi: 10.1161/01.atv.15.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heinecke JW. Oxidants and antioxidants in the pathogenesis of atherosclerosis: implications for the oxidized low density lipoprotein hypothesis. Atherosclerosis. 1998;141:1–15. doi: 10.1016/s0021-9150(98)00173-7. [DOI] [PubMed] [Google Scholar]
- 56.Li D, Yang P, Xiong Q, Song X, Yang X, Liu L, et al. MicroRNA-125a/b-5p inhibits endothelin-1 expression in vascular endothelial cells. J Hypertens. 2010;28:1646–1654. doi: 10.1097/HJH.0b013e32833a4922. [DOI] [PubMed] [Google Scholar]
- 57.Yanagisawa M, Kurihara H, Kimura S, Tomobe Y, Kobayashi M, Mitsui Y, et al. A novel potent vasoconstrictor peptide produced by vascular endothelial cells. Nature. 1988;332:411–415. doi: 10.1038/332411a0. [DOI] [PubMed] [Google Scholar]
- 58.Qin B, Xiao B, Liang D, Xia J, Li Y, Yang H. MicroRNAs expression in ox-LDL treated HUVECs: MiR-365 modulates apoptosis and Bcl-2 expression. Biochem Biophys Res Commun. 2011;410:127–133. doi: 10.1016/j.bbrc.2011.05.118. [DOI] [PubMed] [Google Scholar]
- 59.Qin B, Xiao B, Liang D, Li Y, Jiang T, Yang H. MicroRNA let-7c inhibits Bcl-xl expression and regulates ox-LDL-induced endothelial apoptosis. BMB Rep. 2012;45:464–469. doi: 10.5483/BMBRep.2012.45.8.033. [DOI] [PubMed] [Google Scholar]
- 60.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 61.Wu W, Xiao H, Laguna-Fernandez A, Villarreal G, Jr., Wang KC, Geary GG, et al. Flow-Dependent Regulation of Kruppel-Like Factor 2 Is Mediated by MicroRNA-92a. Circulation. 2011;124:633–641. doi: 10.1161/CIRCULATIONAHA.110.005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weber M, Baker MB, Moore JP, Searles CD. MiR-21 is induced in endothelial cells by shear stress and modulates apoptosis and eNOS activity. Biochem Biophys Res Commun. 2010;393:643–648. doi: 10.1016/j.bbrc.2010.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ito T, Yagi S, Yamakuchi M. MicroRNA-34a regulation of endothelial senescence. Biochem Biophys Res Commun. 2010;398:735–740. doi: 10.1016/j.bbrc.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 64.Zhao T, Li J, Chen AF. MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. Am J Physiol Endocrinol Metab. 2010;299:E110–E116. doi: 10.1152/ajpendo.00192.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Menghini R, Casagrande V, Cardellini M, Martelli E, Terrinoni A, Amati F, et al. MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation. 2009;120:1524–1532. doi: 10.1161/CIRCULATIONAHA.109.864629. [DOI] [PubMed] [Google Scholar]
- 66.Chen Z, Shentu TP, Wen L, Johnson DA, Shyy JY. Regulation of SIRT1 by Oxidative Stress-Responsive miRNAs and A Systematic Approach to Identify Its Role in the Endothelium. Antioxid Redox Signal. 2013 doi: 10.1089/ars.2012.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vidal F, Colome C, Martinez-Gonzalez J, Badimon L. Atherogenic concentrations of native low-density lipoproteins down-regulate nitric-oxide-synthase mRNA and protein levels in endothelial cells. Eur J Biochem. 1998;252:378–384. doi: 10.1046/j.1432-1327.1998.2520378.x. [DOI] [PubMed] [Google Scholar]
- 68.Tabas I, Williams KJ, Boren J. Subendothelial lipoprotein retention as the initiating process in atherosclerosis: update and therapeutic implications. Circulation. 2007;116:1832–1844. doi: 10.1161/CIRCULATIONAHA.106.676890. [DOI] [PubMed] [Google Scholar]
- 69.Ni CW, Qiu H, Jo H. MicroRNA-663 upregulated by oscillatory shear stress plays a role in inflammatory response of endothelial cells. Am J Physiol Heart Circ Physiol. 2011;300:H1762–H1769. doi: 10.1152/ajpheart.00829.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou J, Wang KC, Wu W, Subramaniam S, Shyy JY, Chiu JJ, et al. MicroRNA-21 targets peroxisome proliferators-activated receptor-alpha in an autoregulatory loop to modulate flow-induced endothelial inflammation. Proc Natl Acad Sci U S A. 2011;108:10355–10360. doi: 10.1073/pnas.1107052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang Y, Davies PF. Site-specific microRNA-92a regulation of Kruppel-like factors 4 and 2 in atherosusceptible endothelium. Arterioscler Thromb Vasc Biol. 2012;32:979–987. doi: 10.1161/ATVBAHA.111.244053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen T, Huang Z, Wang L, Wang Y, Wu F, Meng S, et al. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc Res. 2009;83:131–139. doi: 10.1093/cvr/cvp121. [DOI] [PubMed] [Google Scholar]
- 73.Yang K, He YS, Wang XQ, Lu L, Chen QJ, Liu J, et al. MiR-146a inhibits oxidized low-density lipoprotein-induced lipid accumulation and inflammatory response via targeting toll-like receptor 4. FEBS Lett. 2011;585:854–860. doi: 10.1016/j.febslet.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 74.Thulin P, Wei T, Werngren O, Cheung L, Fisher RM, Grander D, et al. MicroRNA-9 regulates the expression of peroxisome proliferator-activated receptor delta in human monocytes during the inflammatory response. Int J Mol Med. 2013;31:1003–1010. doi: 10.3892/ijmm.2013.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thulasingam S, Massilamany C, Gangaplara A, Dai H, Yarbaeva S, Subramaniam S, et al. miR-27b*, an oxidative stress-responsive microRNA modulates nuclear factor-kB pathway in RAW 264.7 cells. Mol Cell Biochem. 2011;352:181–188. doi: 10.1007/s11010-011-0752-2. [DOI] [PubMed] [Google Scholar]
- 76.Rong JX, Shapiro M, Trogan E, Fisher EA. Transdifferentiation of mouse aortic smooth muscle cells to a macrophage-like state after cholesterol loading. Proc Natl Acad Sci U S A. 2003;100:13531–13536. doi: 10.1073/pnas.1735526100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lacolley P, Regnault V, Nicoletti A, Li Z, Michel JB. The vascular smooth muscle cell in arterial pathology: a cell that can take on multiple roles. Cardiovasc Res. 2012;95:194–204. doi: 10.1093/cvr/cvs135. [DOI] [PubMed] [Google Scholar]
- 78.Fernandez-Ortiz A, Badimon JJ, Falk E, Fuster V, Meyer B, Mailhac A, et al. Characterization of the relative thrombogenicity of atherosclerotic plaque components: implications for consequences of plaque rupture. J Am Coll Cardiol. 1994;23:1562–1569. doi: 10.1016/0735-1097(94)90657-2. [DOI] [PubMed] [Google Scholar]
- 79.Sun SG, Zheng B, Han M, Fang XM, Li HX, Miao SB, et al. miR-146a and Kruppel-like factor 4 form a feedback loop to participate in vascular smooth muscle cell proliferation. EMBO Rep. 2011;12:56–62. doi: 10.1038/embor.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen WJ, Yin K, Zhao GJ, Fu YC, Tang CK. The magic and mystery of microRNA-27 in atherosclerosis. Atherosclerosis. 2012;222:314–323. doi: 10.1016/j.atherosclerosis.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 81.Lin Y, Liu X, Cheng Y, Yang J, Huo Y, Zhang C. Involvement of MicroRNAs in hydrogen peroxide-mediated gene regulation and cellular injury response in vascular smooth muscle cells. J Biol Chem. 2009;284:7903–7913. doi: 10.1074/jbc.M806920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sawamura T, Kume N, Aoyama T, Moriwaki H, Hoshikawa H, Aiba Y, et al. An endothelial receptor for oxidized low-density lipoprotein. Nature. 1997;386:73–77. doi: 10.1038/386073a0. [DOI] [PubMed] [Google Scholar]
- 83.Sun Y, Chen X. Ox-LDL-induced LOX-1 expression in vascular smooth muscle cells: role of reactive oxygen species. Fundam Clin Pharmacol. 2011;25:572–579. doi: 10.1111/j.1472-8206.2010.00885.x. [DOI] [PubMed] [Google Scholar]
- 84.Ding Z, Wang X, Khaidakov M, Liu S, Mehta JL. MicroRNA hsa-let-7g targets lectin-like oxidized low-density lipoprotein receptor-1 expression and inhibits apoptosis in human smooth muscle cells. Exp Biol Med (Maywood) 2012;237:1093–1100. doi: 10.1258/ebm.2012.012082. [DOI] [PubMed] [Google Scholar]
- 85.Chen KC, Wang YS, Hu CY, Chang WC, Liao YC, Dai CY, et al. OxLDL up-regulates microRNA-29b, leading to epigenetic modifications of MMP-2/MMP-9 genes: a novel mechanism for cardiovascular diseases. FASEB J. 2011;25:1718–1728. doi: 10.1096/fj.10-174904. [DOI] [PubMed] [Google Scholar]
- 86.Boon RA, Seeger T, Heydt S, Fischer A, Hergenreider E, Horrevoets AJ, et al. MicroRNA-29 in aortic dilation: implications for aneurysm formation. Circ Res. 2011;109:1115–1119. doi: 10.1161/CIRCRESAHA.111.255737. [DOI] [PubMed] [Google Scholar]
- 87.Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C, Crosby M, et al. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One. 2010;5:e10345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Castoldi M, Vujic SM, Altamura S, Elmen J, Lindow M, Kiss J, et al. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J Clin Invest. 2011;121:1386–1396. doi: 10.1172/JCI44883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shan Y, Zheng J, Lambrecht RW, Bonkovsky HL. Reciprocal effects of micro-RNA-122 on expression of heme oxygenase-1 and hepatitis C virus genes in human hepatocytes. Gastroenterology. 2007;133:1166–1174. doi: 10.1053/j.gastro.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martin-Ventura JL, Madrigal-Matute J, Martinez-Pinna R, Ramos-Mozo P, Blanco-Colio LM, Moreno JA, et al. Erythrocytes, leukocytes and platelets as a source of oxidative stress in chronic vascular diseases: detoxifying mechanisms and potential therapeutic options. Thromb Haemost. 2012;108:435–442. doi: 10.1160/TH12-04-0248. [DOI] [PubMed] [Google Scholar]
- 92.Bastida E, Ordinas A, Escolar G, Jamieson GA. Tissue factor in microvesicles shed from U87MG human glioblastoma cells induces coagulation, platelet aggregation, and thrombogenesis. Blood. 1984;64:177–184. [PubMed] [Google Scholar]
- 93.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 94.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 95.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fichtlscherer S, De RS, Fox H, Schwietz T, Fischer A, Liebetrau C, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 98.Tan KS, Armugam A, Sepramaniam S, Lim KY, Setyowati KD, Wang CW, et al. Expression profile of MicroRNAs in young stroke patients. PLoS One. 2009;4:e7689. doi: 10.1371/journal.pone.0007689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fichtlscherer S, Zeiher AM, Dimmeler S. Circulating microRNAs: biomarkers or mediators of cardiovascular diseases? Arterioscler Thromb Vasc Biol. 2011;31:2383–2390. doi: 10.1161/ATVBAHA.111.226696. [DOI] [PubMed] [Google Scholar]
- 100.McManus DD, Ambros V. Circulating MicroRNAs in cardiovascular disease. Circulation. 2011;124:1908–1910. doi: 10.1161/CIRCULATIONAHA.111.062117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun X, Zhang M, Sanagawa A, Mori C, Ito S, Iwaki S, et al. Circulating microRNA-126 in patients with coronary artery disease: correlation with LDL cholesterol. Thromb J. 2012;10:16. doi: 10.1186/1477-9560-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 104.Zhou J, Li JY, Nguyen P, Wang KC, Weiss A, Kuo YC, et al. Regulation of Vascular Smooth Muscle Cell Turnover by Endothelial Cell-secreted MicroRNA-126: Role of Shear Stress. Circ Res. 2013 doi: 10.1161/CIRCRESAHA.113.280883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJ, Zeiher AM, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249–256. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]