SUMMARY
Sequencing efforts led to the identification of somatic mutations that could affect self-renewal and differentiation of cancer-initiating cells. One such recurrent mutation targets the binding pocket of the ubiquitin ligase FBXW7. Missense FBXW7 mutations are prevalent in various tumors, including T-cell acute lymphoblastic leukemia (T-ALL). To study the effects of such lesions, we generated animals carrying regulatable Fbxw7 mutant alleles. We show here that these mutations specifically bolster cancer-initiating cell activity in collaboration with Notch1 oncogenes, but spare normal hematopoietic stem cell function. We were also able to show that FBXW7 mutations specifically affect the ubiquitylation and half-life of c-Myc protein, a key T-ALL oncogene. Using animals carrying c-Myc fusion alleles, we connected Fbxw7 function to c-Myc abundance and correlated c-Myc expression to leukemia-initiating activity. Finally, we demonstrated that small molecule-mediated suppression of MYC activity leads to T-ALL remission, suggesting a novel effective therapeutic strategy.
INTRODUCTION
As next generation sequencing studies identify novel genetic lesions in cancer, it becomes evident that mutations affecting key regulators of diverse cellular processes ranging from metabolism to protein stability are somatically selected in cancer cells (Downing et al., 2012; Hodis et al., 2012; Zhang et al., 2012). The most obvious explanation for these paradoxical events is that such mutations are somehow able to bestow cells with tumorigenic properties while sparing normal cell functions. Heterozygosity of several such mutations further complicates the understanding of such mechanisms as it suggests that either small protein expression differences can have profound outcomes or that missense mutants could have neomorphic and/or dominant negative functions. Finally, it is conceivable that similar mutations do not act in isolation but in combination with additional oncogenic lesions. It is thus imperative to study the impact of somatic missense mutations using both genetic models closely mimicking the corresponding human cancer genotypes and studying in vivo effects of mutational cooperation.
The study of leukemia offers a large number of somatic missense mutations that target key components of cellular function. One of the most prominent examples is the large number of recurrent mutations targeting Fbxw7, a constituent of the SCF (Skp1-Cul1-Fbox) ubiquitin ligase complex that controls the degradation and half-life of key cellular regulators including Cyclin E, Notch1, c-Myc and Mcl1 (Crusio et al., 2010; Inuzuka et al., 2011; Popov et al., 2010; Wertz et al., 2011). FBXW7 is mutated in a significant fraction of human tumors, including approximately 20% of patients with pediatric T cell acute lymphoblastic leukemia (T-ALL) (O’Neil et al., 2007; Thompson et al., 2007). These mutations are predominantly heterozygous and cluster within the WD40 substrate-binding domain, and specifically affect three highly conserved arginine residues (Nash et al., 2001). Although the outcome of expressing these particular mutations in somatic tissues remains unknown, monoallelic deletion of Fbxw7 in the hematopoietic system fails to induce leukemia. Complete deletion can lead to T-ALL establishment, albeit with low penetrance (Matsuoka et al., 2008). However, the prevailing phenotype of Fbxw7 loss is progressive bone marrow failure, eventually leading to fatal anemia, suggesting that complete Fbxw7 inactivation is incompatible with physiological stem and progenitor cell differentiation. In agreement with this finding, nonsense FBXW7 mutations are relatively rare in T-ALL (O’Neil et al., 2007; Thompson et al., 2007). These studies suggest that FBXW7 missense mutants are not simply “dead” alleles and could behave differently in normal and malignant cells. Although the biochemical mechanisms behind FBXW7 mutations in T-ALL remains unclear, we and others have suggested that these lesions could affect the stability of NOTCH1, the main T-ALL oncogene, itself mutated in approximately half of T cell leukemia patients (Weng et al., 2004). In agreement with this notion, approximately 25% of NOTCH1 mutations in T-ALL truncate the protein deleting the conserved degron sequence recognized by Fbxw7. Similar mutations in either NOTCH1 or FBXW7 genes are also found in a larger number of additional cancer types, including marginal B cell lymphoma, melanoma, and squamous cell carcinoma (Akhoondi et al., 2007; Hodis et al., 2012; Rossi et al., 2012; Stransky et al., 2011), making the thorough understanding of their function critical for future therapies.
To study in vivo the transforming effects of such missense mutations, we have generated mice that carry Cre-inducible Fbxw7 heterozygote mutants, mimicking the most common substitution found in human T-ALL. Interestingly, in contrast to previous knockout models, such missense Fbxw7 mutations did not compromise normal hematopoietic stem cell and progenitor function, suggesting distinct thresholds of Fbxw7 activity in normal versus malignant hematopoiesis. Consistent with this notion, further studies demonstrated that Fbxw7 mutations lead to a marked increase in the number of leukemia-initiating cells (LIC) due to stabilization of the Fbxw7 substrate c-Myc. Using animals expressing fluorescent c-Myc fusion proteins (MycGFP) we were able to show a perfect correlation between c-Myc stabilization and leukemia-initiating activity. Moreover, we were able to demonstrate that c-Myc deletion in established T-ALL specifically ablates LIC and that inhibition of c-Myc induction using small molecule BET/BRD inhibitors (Filippakopoulos et al., 2010; Zuber et al., 2011) can suppress growth of mouse and human T-ALL cells. Finally, genomic studies identified direct c-Myc and Brd4 gene targets in T-ALL and suggested an intriguing transcriptional co-operation between Notch1 and Myc in this type of leukemia. Taken together, these studies identify FBXW7R465C as a novel class of cancer somatic mutations, as it has the ability to specifically alter cancer-initiating cell activity without consequence to normal stem cell differentiation.
RESULTS
Generation of inducible knock-in models of FBXW7 missense mutations
To test the function of FBXW7 mutations in vivo we targeted the most common recurrent mutation, an arginine to cysteine change at position 465 (468 in the mouse) (Aifantis et al., 2008). As mice that harbor a similar heterozygous germline mutation in Fbxw7 die perinatally, due to defects in lung development (Davis et al., 2011), we generated mutant alleles that could be conditionally activated using the Cre-lox system. Initially, using homologous recombination we generated an R468C mutation in the endogenous Fbxw7 gene introducing a lox-STOP-lox cassette in the upstream intron, thereby acting as a functionally null allele prior to recombination and a mutant allele in all lineages where Cre is activated (Figure S1A). Fbxw7R468Neo/+ pups were born and developed normally, yet Fbxw7R468Neo/R468Neo mice were never represented, consistent with reports that Fbxw7-null mice die in utero (Tsunematsu et al., 2004). Mice were crossed to the pI:pC-inducible Mx1-cre allele (Kuhn et al., 1995). Recombination was observed in genomic DNA of bone marrow cells and the mutation could be detected in mRNA (Figure S1B–C) after pI:pC treatment. Alternatively, to study the effect of the exact human R465C mutation, we introduced an exogenous human FBXW7R465C cDNA (with an N-terminal Flag fusion) in the ubiquitously expressed eukaryotic elongation factor 1 alpha 1 (Eef1a1) locus (Klinakis et al., 2009). As with the endogenous mouse Fbxw7 mutant, this targeting positioned a lox-STOP-lox cassette upstream of the mutant cDNA for conditional activation (Figure S1D). Upon Cre induction, we could detect robust expression of a 110-kDa protein by Flag immunoblot in lysates from primary hematopoietic tissues (Figure S1E). Furthermore, the mutant human FBXW7 was functional and incorporated into the SCF complex, as other components (Skp1 and Cul1) could be co-immunoprecipitated (not shown). Interestingly, and as it will become evident from the following studies, both alleles (Fbxw7R468 and Eef1a1mutW7) yield identical phenotypes. Most of the presented studies will focus on the Fbxw7R468 endogenous allele (referred as Fbxw7mut).
FBXW7 mutations do not affect normal hematopoietic stem and progenitor differentiation
Previous studies have shown that conditional deletion of Fbxw7 in the adult hematopoietic system leads to a rapid depletion of hematopoietic stem cells (HSC) and T cell progenitors (Matsuoka et al., 2008; Thompson et al., 2008). Given that HSC provide a long-lived reservoir of cells with an inherent capacity for self-renewal, it has been hypothesized that many of the oncogenic lesions found in hematopoietic malignancies might arise in HSC or multipotent progenitors (Francis and Richardson, 2007; Fukushima et al., 2012; Mohrin et al., 2010; Onoyama et al., 2007). However, this presents a paradox for FBXW7 mutations in particular, as these mutations could be deleterious for normal hematopoiesis and selected against prior to transformation. To test the possibility that heterozygous Fbxw7 mutations might be tolerated differently than deletion of both alleles, HSC function was assessed in Fbxw7F/+Mx1-Cre+ (Fbxw7Δ/+)/Fbxw7mut/+Mx1-Cre+ and Fbxw7F/FMx1-Cre+mice (Fbxw7Δ/Δ). As expected, the frequency and absolute number of HSC (Lin−Sca1+c-kit+CD150+CD4−) was significantly lower in Fbxw7Δ/Δ bone marrow compared to Fbxw7Δ. However, there was no significant decrease in phenotypic HSC observed in Fbxw7mut/+ mice (Figure 1A–B). Fbxw7mut/+ HSC were also more functionally competent than Fbxw7Δ/Δ as they generated a greater number of colonies in serial colony forming assays (Figure 1C) and reconstituted recipient mice to the same extent as Fbxw7Δ+ donors upon competitive bone marrow transplantation (Figure 1D). Moreover, the depletion of early T cell progenitors typically seen in Fbxw7Δ/Δ thymii was also absent in Fbxw7mut/+ mice (Figure 1E), demonstrating that the mutant progenitors could colonize the Fbxw7-deficient thymus and differentiate normally.
Figure 1. Mutant Fbxw7 does not impair hematopoietic stem cell function or T cell differentiation in vivo.
(a–b) Representative FACS plots showing frequency of c-kit+Sca-1+ mononuclear cells in Lineage-bone marrow (LSK) from Fbxw7F/+Mx1Cre+, Fbxw7mut/+Mx1Cre+, or Fbxw7F/FMx1Cre+ mice harvested 8 weeks after pI:pC injection, and number of phenotypic LT-HSC (CD150+CD48−LSK) recovered (b). (c) Primary and secondary colonies derived from LSK cells sorted from bone marrow of the indicated genotype. Mean +/− SD of 3 replicates shown. *p <0.05 **p<0.01. (d) Frequency of CD45.2+ donor-derived peripheral blood mononuclear cells (PBMC) in lethally-irradiated recipient mice transplanted with 5×105 total BMMC from either Fbxw7F/+Mx1Cre+, Fbxw7mut/+Mx1Cre+, or Fbxw7F/FMx1Cre+ mice (CD45.2+) mixed at 1:1 ratio with wild-type BMMC (CD45.1+), at 4 and 8 weeks post-transplant. (d) FACS profiling of thymocytes from Fbxw7 wild type, mutant or knock-out mice. Mice were analyzed four weeks after pI:pC injections. (e–f) Histogram and calculated mean fluorescence intensity (MFI) (f) of GFP measured by FACS in LT-HSC from mice in D–F expressing a c-Myc:GFP fusion (MycGFP/+) allele.
Fbxw7-deficient HSC fail to self-renewprimarily due to an aberrant accumulation of c-Myc protein (Reavie et al., 2010). To test whether differential c-Myc abundance could account for the inability of the mutant allele to affect hematopoiesis, the level of c-Myc protein in HSC of each genotype was determined by crossing the Fbxw7mut and Fbxw7F/F Mx1-Cre strains to a mouse expressing a c-Myc:GFP fusion from the endogenous Myc locus (MycGFP) (Huang et al., 2008). Intriguingly, the level of c-Myc stabilization in mutant HSC was significantly greater than Fbxw7Δ/+but still lower than that observed in Fbxw7-deficient HSC (Figure 1F–G). Altogether, these data imply that the intermediate levels of c-Myc stabilization resulting from a heterozygous Fbxw7 mutation is tolerated in HSC, whereas the level of c-Myc in Fbxw7-deficient HSC surpasses a threshold that is incompatible self-renewal.
FBXW7 and NOTCH1 mutations co-operate for the induction of aggressive leukemia
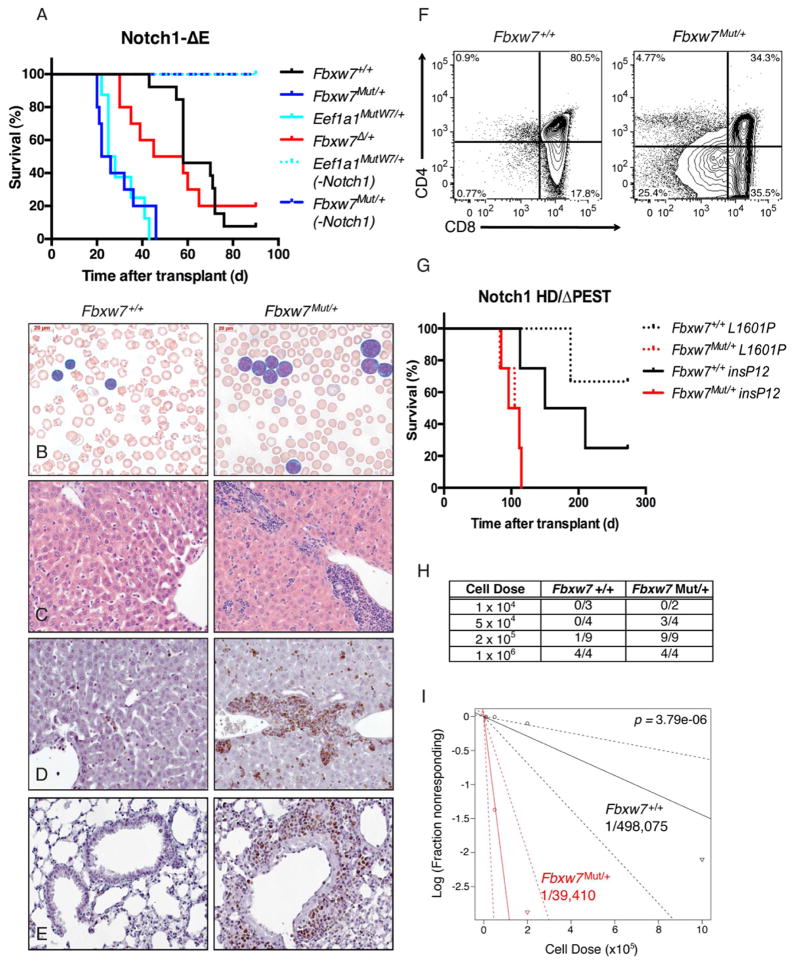
Although mutations in Fbxw7 are highly prevalent and implicated in the pathogenesis of T-ALL (O’Neil et al., 2007; Thompson et al., 2007), it is unknown whether these mutations are sufficient for leukemia induction in vivo. To address this question, a cohort of Fbxw7mut/+ or Eef1a1mutW7/+ mice were monitored periodically by peripheral blood analysis for up to 18 months. Surprisingly, none of these mutant mice ever exhibited elevated white blood cell (WBC) counts compared to their wild-type littermates or developed leukemia spontaneously (Figure 2A). Moreover, no other tumors or developmental defects were noted when a single mutant allele was activated in early stage embryos with EIIa-Cre (not shown). Next, we tested whether Fbxw7 mutations could act synergistically with other known T-ALL oncogenes. FBXW7 mutations frequently co-occur with NOTCH1 hetero-dimerization domain (HD) mutations in human T-ALL (Grabher et al., 2006; O’Neil et al., 2007; Thompson et al., 2007). This scenario was modeled in vivo by retroviral expression of Notch1ΔE, a truncated form of Notch1 that is constitutively cleaved in a ligand-independent manner (Aster et al., 1997) on an Fbxw7 wild type, Fbxw7Δ/+, Fbxw7mut/+ or Eef1a1mutW7/+ background. While all three cohorts eventually succumbed to T-ALL, the cohorts transplanted with either Fbxw7 mutant developed leukemia at a much shorter latency compared to the Fbxw7 wild-type and the Fbxw7Δ/+ groups (Figure 2A). The median survival of the Fbxw7mut/+ and Eef1a1mutW7/+ cohorts was identical. Mice transplanted with either Fbxw7 mutant or wild type Notch1-transduced cells were analyzed at 21 days post-transplant to evaluate leukemia progression. The mutant cohorts presented with a higher proportion of leukemic blasts, notably larger in size compared to their wild-type counterparts, in the peripheral blood and heavier infiltration of peripheral tissues (Figure 2B, C). In addition, a greater proportion of these blasts were actively cycling, as exhibited by more prominent Ki67 staining (Figure 2D, E). Since the extent of maturation of T-ALL often correlates with a poorer prognosis (Ferrando and Look, 2003; Zhang et al., 2012), expression of surface markers corresponding to different stages of physiological T cell development was compared between genotypes. A noticeable trend in decreased CD4 and, to a lesser extent, CD8 surface expression was observed in the Fbxw7 mutant leukemias (Figure 2F), suggesting a more immature (and potentially aggressive) phenotype. Given that Notch1 is an Fbxw7 substrate, we repeated the presented leukemia-induction experiments using human Notch1-ΔPEST mutants (Chiang et al., 2008), which are lacking the C-terminal region bound by Fbxw7. As seen in Figure 2G, Fbxw7mut/+ Notch1ΔPEST tumors also developed with significantly shorter latency, suggesting that further Notch stabilization alone could not explain the aggressiveness of the disease.
Figure 2. Fbxw7 mutations accelerate NOTCH1-induced T-ALL and expand the number of leukemia initiating cells.
(a) Kaplan-Meier curve representing morbidity in recipient mice transplanted with bone marrow from Fbxw7mut/+, Eef1a1mutW7/+, Fbxw7Δ/+ or Fbxw7+/+ transduced with a Notch1ΔE ires GFP retrovirus. As a control, mice transplanted with Fbxw7mut/+ or Eef1a1mutW7/+ cells not expressing a Notch mutation (-Notch1) are shown (Mantel-Cox log-rank test: Fbxw7mut/+ vs. Fbxw7+/+ p<0.0001, Fbxw7mut/+ vs. Fbxw7Delta;/+ p=0.0056, Fbxw7Δ/+ vs. Fbxw7+/+ p=0.5617). Peripheral blood and organs were harvested from either Fbxw7 mutant and wild-type cohorts 3 weeks post transplant and prepared for histology: (b) Wright-Giemsa stain of peripheral blood (c) H&E staining of liver and (d–e) Ki67 staining of liver and lung. (f) Representative FACS plots depicting CD4 and CD8 expression on either Fbxw7mut/+ or Fbxw7+/+ GFP+ cells in peripheral blood of recipients. (g) Kaplan-Meier curve representing morbidity in recipient mice transplanted with bone marrow from Fbxw7mut/+ or Fbxw7+/+ mice transduced with retroviruses expressing mutant, truncated forms of Notch1 lacking a C-terminal PEST domain and bearing NOTCH1 HD (L1601P and insP12) mutations (L1601P: p=0.0177, insP12: p=0.0266). (h) Fraction of secondary recipients that developed leukemia when transplanted with limiting dilutions of GFP+ splenocytes sorted from either Fbxw7 wild type or mutant T-ALL mice. (i) Log-log plot and leukemia initiating cell frequency as calculated by extreme limiting dilution analysis (ELDA). Red: Fbxw7mut/+ (1/39,410), Black: Fbxw7+/+ (1/498,075).
Initial attempts to serially transplant the disease revealed that the Fbxw7mut leukemias were more efficient at generating secondary tumors (Figure 2H). We thus hypothesized that the frequency of leukemia initiating cells (LIC) might be higher within the Fbxw7mut leukemia. To quantify the LIC frequencies in Fbxw7 mutant or wild-type T-ALL, equivalent numbers of GFP+ cells sorted from leukemias arising from either genotype were transplanted into secondary recipients at a range of doses from 104–106 cells per recipient. All of the mice transplanted with 106 cells, regardless of Fbxw7 status, eventually developed leukemia. However, the frequency of leukemia in mice that received 2×105 or 5×104 cells was significantly higher among the Fbxw7 mutant cohorts (100% vs. 11% and 75% vs. 0%, respectively) (Figure 2H). Using an algorithm adapted from studies of hematopoietic stem cells (Buchstaller et al., 2012; Quintana et al., 2008), we calculated that the LIC frequency within the Fbxw7 mutant T-ALL was more than 10-fold higher than in Fbxw7 wild-type T-ALL (Figure 2I).
FBXW7 mutations do not lead to genomic instability or cooperate with p53 loss
Previous studies observed genomic instability in human cancer cells when FBXW7 is deleted (Grim et al., 2012; Rajagopalan et al., 2004). To test whether chromosomal abnormalities were contributing to leukemia development in vivo, metaphase fluorescent in-situ hybridization (mFISH) was utilized to karyotype Fbxw7 mutant leukemias. However, the vast majority of tumor cells had a normal karyotype (Figure S2A). To test for smaller genomic events, DNA from leukemic cells was analyzed by array-comparative genomic hybridization (array CGH). Several Fbxw7 mutant or wild type leukemias were analyzed, using normal tissue from donor mice as reference. Among the Fbxw7 mutant samples, the only significant DNA gain or loss that was consistently observed was within the Tcra locus (Figure S2B), in agreement with the identity and developmental stage of the leukemia. While neither of these results rule out the possibility of collaborating point mutations, genetic instability is unlikely to be responsible for the more aggressive nature of Fbxw7 mutant leukemias. In addition to activating mutations in Notch1, Fbxw7 has been genetically associated with inactivation of p53 in a number of tumor models (Grim et al., 2012; Onoyama et al., 2007). To determine whether loss of p53 could collaborate with our Fbxw7 mutant allele, we transduced wild type, Fbxw7mut/+ or Fbxw7-deficient bone marrow with a retroviral shRNA targeting Trp53. The wild type or Fbxw7Δ/Δ donors generated tumors in recipient animals with a time of onset and disease phenotype that coincided with the previous genetic models (Figure S2C). However, the latency of leukemia development was not decreased by mutation of Fbxw7, despite efficient silencing of Trp53 (Figure S2D). Therefore, there is a clear disparity between Fbxw7-null and Fbxw7R465C mutants in the requirement for p53 inactivation. Moreover, unlike NOTCH1, TP53 mutations are not enriched in FBXW7 mutant human T-ALL (Zhang et al., 2012), supporting our findings using the Fbxw7mut animal models.
FBXW7 mutations affect c-Myc protein half-life and ubiquitylation
Having ruled out genomic instability, we next sought to determine the mechanism by which Fbxw7 mutations confer a greater leukemia initiating capacity to cells uniformly expressing a Notch1 oncogene. The protein half-life of a panel of known substrates (Inuzuka et al., 2011; Wertz et al., 2011) was measured in either Fbxw7 wild type or mutant Notch1 T-ALL cells ex vivo (Figure S3A). Of the known Fbxw7 substrates tested, we noted moderate stabilization of Notch1 and Srebp1 and a very consistent and significant stabilization of c-Myc (Figure S3A). Myc has an essential role in cell growth and self-renewal (Eilers and Eisenman, 2008; Murphy et al., 2005) and has previously been implicated in the pathogenesis of T-ALL as a transcriptional target of NOTCH1 (Palomero et al., 2006; Sharma et al., 2006). We thus focused on putative mechanisms of c-Myc stabilization by the mutant Fbxw7 allele. In an in vitro ubiquitylation assay, the R465C mutation renders Fbxw7 incapable of ubiquitylating c-Myc (Figure S3B). As SCF-Fbxw7 complexes have been shown to dimerize in certain conditions (Welcker and Clurman, 2007), we hypothesized that Fbxw7 mutations could affect the ability of Fbxw7mut:Fbxw7WT heterodimers to bind and ubiquitylate c-Myc. To test this hypothesis, we expressed either Fbxw7WT or Fbxw7R465C bearing unique epitope tags, isolated dimers and quantified binding to endogenous c-Myc. While a robust interaction with c-Myc was detected in the WT:WT dimers, binding to MUT:WT dimers was significantly decreased (Figure S3C). MUT:MUT FBXW7 homodimers failed to bind detectable levels of c-Myc.
FBW7-regulated c-Myc protein expression marks specifically leukemia initiating cells in vivo
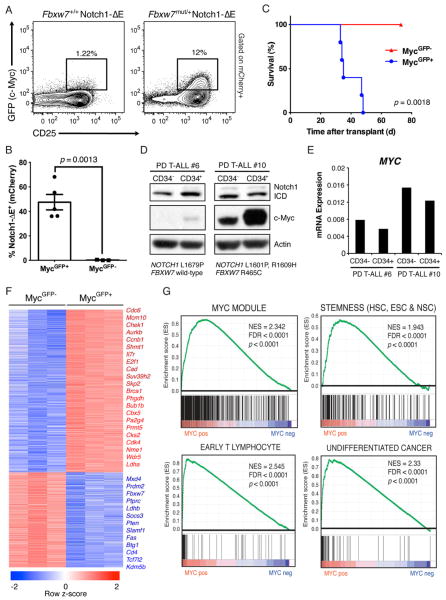
To track c-Myc protein levels in Notch1-induced T-ALL over the course of leukemia progression we once more utilized the MycGFP fusion allele. A Notch1ΔE-ires-mCherry retroviral vector was used to transduce either Fbxw7+/+MycGFP/+ or Fbxw7mut/+MycGFP/+ bone marrow cells and transplanted them into irradiated recipients, thereby allowing us to assess c-Myc protein expression within the tumor population. Since Myc is a transcriptional target of Notch1 (Palomero et al., 2006), we expected that the Notch1ΔE-transduced population would uniformly express high levels of MycGFP. However, at earlier time-points, Notch1+ cells that had detectable levels of c-Myc were virtually absent from circulation (Figure S4A) and rare in the spleen (Figure 3A). Strikingly, this population was typically 10-fold larger in the Fbxw7 mutant leukemia (Figure 3A). Upon co-staining for the T cell surface markers described earlier, the MycGFP expression was highest in the CD−CD8−CD25hi (DN3) and ISP8 (CD−CD8+TCRblow) fractions, but was virtually absent in CD4+CD8+ (DP) cells. Notably, the MycGFP+ leukemic cells also had elevated IL-7Rα expression (Figure S4B). As the leukemia progressed, the MycGFP+ population became more abundant, but still never achieved majority (Figure S4C). Interestingly, among hematopoietic and lymphoid organs, the highest frequency of MycGFP+ cells was consistently observed in the spleen (Figure S4D).
Figure 3. Myc protein stability and expression defines leukemia-initiating potential in T-ALL.
Fbxw7mut/+MycGFP/+ or Fbxw7+/+MycGFP/+ bone marrow was transduced with Notch1ΔE ires mCherry and transplanted into lethally-irradiated recipients. (a) c-MycGFP in mCherry+ splenocytes derived from either donor measured by FACS 4 weeks post-transplant (frequency of mCherry+ cells expressing both c-MycGFP and CD25 is shown). Fbxw7+/+ mCherry+ splenocytes from leukemic mice were sorted on the basis of c-MycGFP expression and transplanted into sublethally-irradiated recipients. (b) Frequency of mCherry+ PBMC 3 weeks post transplant and survival (c) of secondary recipients. (d) Immunoblot for c-Myc and intracellular Notch1 (ICD) and (e) quantitative RT-PCR (qRT-PCR) analysis for MYC transcript (normalized to GAPDH) in two independent primary human T-ALL xenografts sorted on the basis of CD34 expression. Mutations in NOTCH1 and FBXW7 in each patient sample as determined by Sanger sequencing are shown. (f) Heat-map depicting differentially expressed genes (fold change >2, p<0.05) in Notch1ΔE+c-MycGFP+ versus Notch1ΔE+c-MycGFP− T-ALL cells sorted from the spleens of 3 individual mice. (g) Selected expression signatures found to be upregulated in c-MycGFP+ population as determined by gene set enrichment analysis (GSEA).
To determine whether the MycGFP+ population was enriched in LIC activity, 105 Notch1ΔE+MycGFP+ and Notch1ΔE+MycGF− cells were sorted from the spleen of leukemic mice (Figure S4E–F) and transplanted into secondary recipients. Two weeks post-transplant, the cohort transplanted with MycGFP+ cells had significantly greater numbers of leukemic cells in peripheral blood (Figure 3B). More importantly, the cohort transplanted with MycGF− cells exhibited 100%-leukemia free survival, whereas the MycGFP+ cohort all succumbed to T-ALL by 6 weeks post-transplant (Figure 3C). Importantly, secondary leukemias arising from the MycGFP+ cohort re-established the heterogeneity of the primary tumor, including both MycGFP+ and MycGF− cells (Figure S4G), demonstrating that MycGFP+ cells can self-renew and differentiate, the defining properties of a cancer stem cell. Altogether, these data define c-Myc protein abundance as the first bona fide LIC marker in a T-ALL mouse model and suggest that this population is heavily dependent on Fbxw7 activity. To test whether this observation translates to human T-ALL, CD34+ populations enriched for T-ALL LIC (Armstrong et al., 2009) were purified from human patient samples that carry NOTCH1 and FBXW7 mutations. In agreement with the mouse data, non-LIC subsets expressed significantly lower levels of c-Myc protein compared to the CD34+ fraction (Figure 3D), despite no increase in MYC mRNA (Figure 3E) or cleaved Notch1. c-Myc levels were higher in samples carrying FBXW7 mutations, further supporting the notion that missense FBXW7 mutations augment c-Myc protein stability in T-ALL.
T-ALL leukemia initiating cells are defined by a Myc gene expression signature
To further study this subpopulation enriched in T-ALL LIC activity, we sought to determine its underlying molecular signature using gene expression analysis. Total RNA was isolated from either MycGFP+ or MycGF− leukemic cells and gene expression was determined by microarray analysis (Figure 3F). The most represented group among the top upregulated genes included those involved in the mitotic phase of the cell cycle (Cdc6, Ccnb1, Chek1, Aurkb, Brca1) and, as expected, transcriptional targets of c-Myc (Cad, Suv39h2, Bcat1, Pa2g4) (Kim et al., 2008; Margolin et al., 2009). Also, many genes encoding cell surface markers (Fas, Slamf1, Il7r, Cd4), which may be helpful for further thorough dissection of the LIC population, differed significantly at the mRNA level between the two populations. Gene-set enrichment analysis (GSEA) was performed to identify gene modules that are enriched within the MycGFP+ population (Figure 3G). Interestingly, gene-sets pertaining to stem cell identity, including common genes upregulated in embryonic (ESC) and adult stem cell populations, as well as a set of c-Myc-regulated genes in both ESC and human cancers (Kim et al., 2008), were highly enriched (Figure 3F). Additionally, gene sets associated with early T cell progenitors (Lee et al., 2004) and undifferentiated tumors (Rhodes et al., 2004) were also enriched. Fbxw7, on the other hand, was significantly downregulated in the MycGFP+ population (Figure 3F), suggesting that in the absence of mutations there is a need to transcriptionally down-regulate its activity during leukemic progression (Mavrakis et al., 2011).
Myc deletion specifically targets leukemia-initiating cells in an animal model of T-ALL
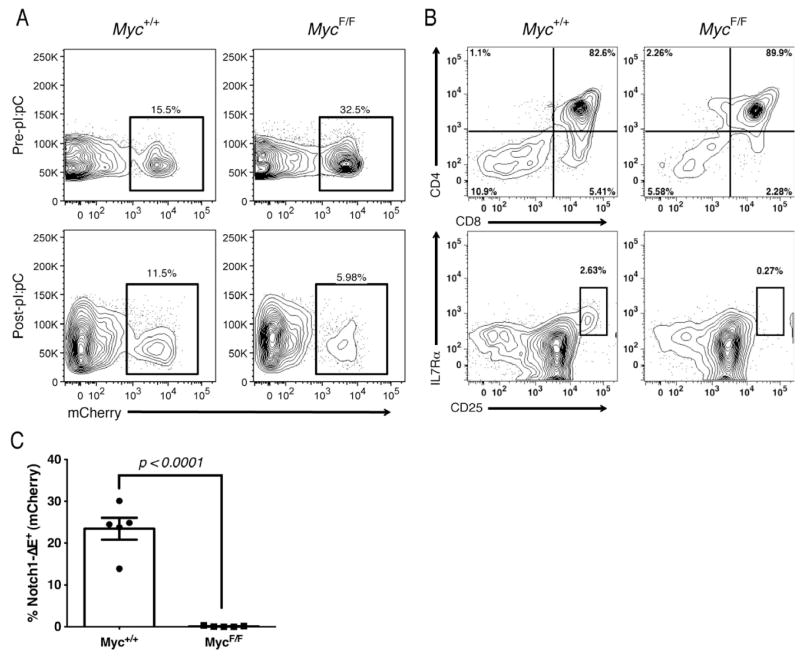
Since leukemia-initiating capacity was found to be restricted to the c-MycGFP+ population, we next tested whether depletion of Myc could specifically ablate LIC activity in T-ALL. Primary T-ALL were generated from MycF/FMx1-Cre+ or Myc+/+Mx1-Cre+ donors and recipients were injected with pI:pC 2 weeks later, after leukemic cells were detected in the periphery. Our initial observation was that leukemic burden was decreased in recipients in which c-Myc expression was deleted (Figure 4A). Strikingly, the LIC-enriched CD−CD−CD25hiIL-7R+ subset was almost completely absent in the Myc-deficient leukemias (Figure 4B), suggesting that the LIC population absolutely depends on c-Myc function in vivo. To further test this hypothesis, we transplanted equal numbers of Notch1ΔE+ cells from either the pI-pC-treated MycF/F or Myc+/+ primary T-ALL and assessed leukemia development in secondary hosts. We found that, while the Myc wild type cells effectively generated secondary tumors, no leukemic cells were detected in the Myc-deficient group (Figure 4C).
Figure 4. Genetic depletion of Myc expression abolishes leukemia-initiating activity in T-ALL.
Myc+/+Mx1Cre+ or Myc+/+Mx1Cre+ bone marrow was transduced with Notch1ΔE and transplanted into lethally-irradiated recipients. Notch1ΔE+ T-ALL cells were measured in peripheral blood 2 weeks post-transplant (a) (top panel) and 7 days following pI:pC treatment (bottom panel). (b) FACS analysis showing frequency of Notch1ΔE+ cells expressing CD4/CD8 (top panels) and CD4−CD8−CD25hiIL-7Rα+ (bottom panels) immunophenotype within spleen following Cre induction. (c) Frequency of Notch1ΔE+ PBMC in secondary recipients of Myc wild type and deficient T-ALL cells 2 weeks post transplant.
BET inhibitors efficiently suppress growth of mouse and human T-ALL
These studies suggested that LIC activity in T-ALL could feasibly be targeted via inhibition of c-Myc. Recently, a selective BET bromodomain inhibitors were identified to specifically target BRD4, a transcriptional activator of MYC (Delmore et al., 2011; Devaiah et al., 2012; Filippakopoulos et al., 2010; Mertz et al., 2011; Zuber et al., 2011). One of these molecules, JQ1, was shown to effectively inhibit expression and function of MYC and inhibited cell growth in multiple myeloma and acute myeloid leukemia (Filippakopoulos et al., 2010; Mertz et al., 2011). To determine whether inhibition of BRD4 could similarly lead to growth inhibition in T-ALL, we initially compared retroviral shRNA knockdown of Myc and Brd4 in Notch1+ mouse T-ALL cell lines. A significant loss of representation over time was observed in populations expressing shRNA targeting either gene (Figure S5A). The growth effect induced by the MYC hairpins was on-target, as complementation with non-RNAi targeted MYC cDNA restored their growth potential (Figure S5B).
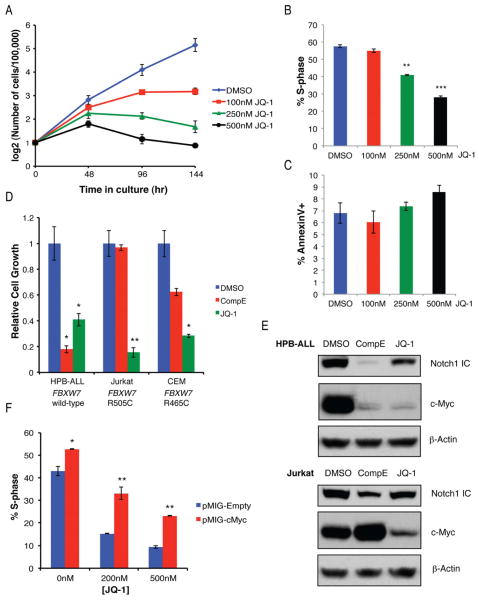
To start addressing the putative leukemia-targeting properties of BET inhibitors, growth of human TALL lines was tested in the presence of varying concentrations of JQ-1 in vitro (Figure 5A). A significant decrease in cell growth was observed after 4 days of JQ-1 treatment in dose dependent manner. BrdU incorporation assays and Annexin-V staining suggested that JQ-1 treatment resulted primarily in growth arrest rather than apoptosis (Figure 5B–C). As previous work has shown that FBXW7 mutations conferred resistance to gamma-secretase inhibitors (GSI) (Real et al., 2009; Thompson et al., 2007), response to JQ-1 or GSI (Compound E) was compared in FBXW7 mutant or wild-type T-ALL lines (Figure 5D). As expected, GSI treatment inhibited growth of HPB-ALL cells (FBXW7 wild-type), but were significantly less effective in CEM or Jurkat cells (both FBXW7 mutant). However, all T-ALL lines were unable to grow in the presence of JQ-1. The observed growth arrest was due, at least in part, to a loss of c-Myc, as JQ-1 treatment resulted in a significant reduction in c-Myc protein expression in all TALL lines tested (Figure 5E) and overexpression of c-Myc partially restored proliferation of the treated cells (Figure 5F). Together, these results show that BET bromodomain inhibitors are an appealing alternative to GSI treatment as it is able to target T-ALL cells irrespective of FBXW7 mutations.
Figure 5. BET bromodomain inhibition broadly impacts growth of human T-ALL.
(a) Dose response curve showing growth of CUTLL1 human T-ALL line (FBXW7 wild-type) treated with increasing concentrations of JQ-1 or vehicle over time. Frequency of CUTLL1 cells that incorporated BrdU during a 1hr pulse (b) or stained positive for Annexin V (c) was determined after treatment with JQ-1 for 48h. (d) Growth of human T-ALL lines treated with γ-secretase inhibitor (Compound E) or JQ-1 in vitro, after 4 days, relative to vehicle treated controls. (e) Immuno-blot for Notch1 ICD and c-Myc in T-ALL lines treated with inhibitors. (f) 720 cells transduced with pMIG, either empty or bearing a c-Myc cDNA were treated for 72 hours with JQ-1 at the indicated concentration, pulsed with BrdU for 1hr and the frequency of GFP+ cells in S-phase was determined by flow cytometry.
To determine the in vivo efficacy of BET inhibitors using mouse T-ALL models it was first necessary to establish an in vitro system to maintain and expand c-MycGFP+ LIC. Primary Notch1ΔE+c-MycGFP+ splenocytes lose expression of c-MycGFP and fail to expand in liquid culture (Figure S5C). However, when cocultured on OP9 stromal line in the presence of IL-7, these cells maintain their expression of c-MycGFP (Figure S5C) and can be expanded indefinitely. Upon treatment with JQ-1, expression of c-MycGFP rapidly returns to basal levels (Figure 6A) and the growth of the T-ALL cells is significantly inhibited (Figure 6B). To study the effect of BET bromodomain inhibitors on the progression of T-ALL in vivo we utilized a derivative of JQ1, CPI203 (Devaiah et al., 2012), which has shown superior bioavailability with oral or intraperitoneal administration. The EC50 of CPI203 was nearly 3 fold lower than JQ-1 (91.2 vs. 263nM) when tested on a primary mouse T-ALL in vitro (Figure 6B) and this corresponded to a decrease in Myc mRNA (Figure 6C). To measure the effect of CPI203 in vivo, two independently derived primary mouse T-ALL samples, either Fbxw7+/+ or Fbxw7mut/+, were transduced with a lentiviral luciferase reporter and each transplanted into two recipient animal groups, one treated with CPI203 (5 mg kg−1, BID) and the other with vehicle, and disease progression was monitored by in vivo luciferase imaging (IVIS Lumina) (Figure 6D). A significant and rapid reduction in leukemia burden was observed in both the recipient groups treated with CPI203 (Figure 6D–E), demonstrating the therapeutic potential of this compound for in vivo use.
Figure 6. BET bromodomain inhibitors deplete LIC and inhibits progression of T-ALL both in vitro and in vivo.
(a) Histogram depicting c-MycGFP levels in primary Notch1ΔE ires mCherry+c-MycGFP+ splenocytes co-cultured on OP9 stromal cells in the presence of 5 ng/ml IL-7 and either 200 nM JQ-1 or DMSO. (b) Growth of primary mouse T-ALL treated with increasing concentrations of JQ-1 or CPI203 for 72 hours. Calculated EC50 for each compound is shown. (c) Myc mRNA measured by quantitative RT-PCR, in primary mouse T-ALL cells treated with CPI203 for 4 hours at the indicated concentration. (d) Bioluminescent imaging of recipient mice transplanted either Fbxw7+/+ (top) or Fbxw7mut/+ (bottom) Notch1ΔE leukemias and treated with either CPI203 (5 mg kg−1) or vehicle BID for 7 days. (d) Quantification of total bioluminescent counts, each data point representing an individual mouse, before and following 7 days of the indicated treatment.
Finally, we tested the response of primary human T-ALL patient samples (all with activating NOTCH1 mutations +/− FBXW7 missense mutations) to both JQ1 and CPI203. Typically, a 2–3 fold reduction in cell growth occured upon treatment with either compound across all genotypes (Figure S6A). Similarly to what was observed in the cell lines, this effect could be attributed mainly to cell cycle arrest (Figure S6B), although a modest induction of apoptosis was also observed in two of the samples (Figure S6C). Taken together, these data indicate significant potential for BET inhibitors for clinical use in the treatment of T-ALL.
BET inhibitors target the Notch/Myc oncogenic transcriptional program
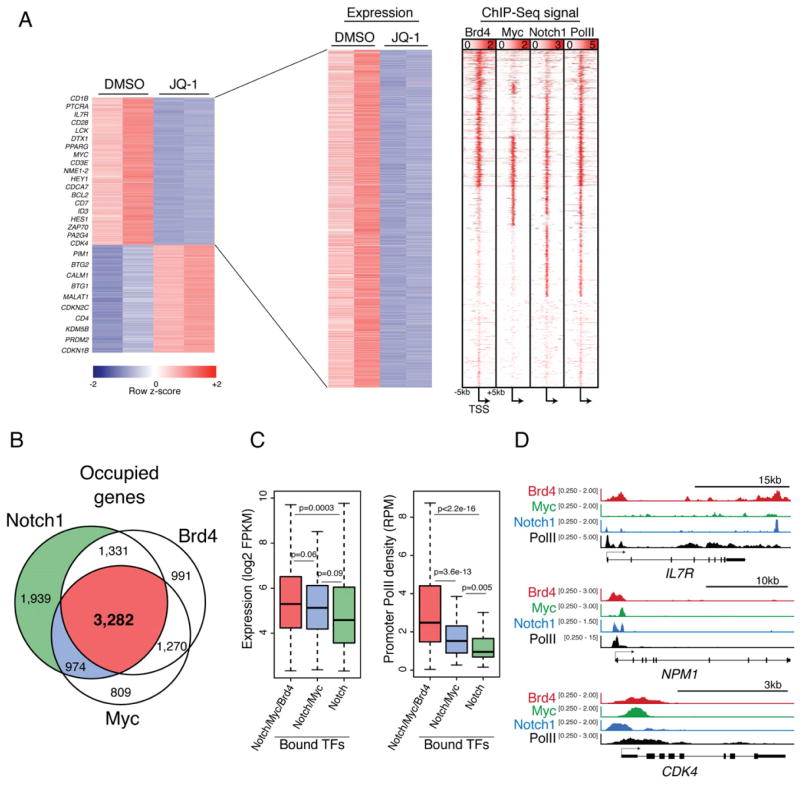
Brd4 functions as a chromatin reader by binding acetylated histones and recruiting effectors of transcriptional elongation, thereby promoting gene activation (Rahman et al., 2011). Thus, we hypothesized that Brd4 inhibition might have profound effects on T-ALL gene expression. High-throughput RNA sequencing (RNA-seq) was performed in CUTLL1 cells after treatment with inhibitor to assess immediate consequences on transcription genome-wide. A total of 1,696 genes were down-regulated and 1,287 upregulated upon JQ-1 treatment for 12 hours (400nM), compared to vehicle treated control (Max FPKM>5, Fold change > 1.3, q<0.05) (Figure 7A). Interestingly, 28% of genes down-regulated upon JQ-1 treatment were also overexpressed in the c-MycGFP+ LIC population in our mouse T-ALL model (Figure 3F). Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq) for c-Myc and Brd4 was performed in CUTLL1 cells to assess their genome-wide occupancy. A total of 6,335 genes had significant c-Myc binding and 6,874 genes showed Brd4 occupancy. Using available Notch1 and Rbpj ChIP-Seq data (Wang et al., 2011) we compared the 7,526 genes also bound by both of these factors. A significant fraction of all occupied genes (3,282/9,453) showed binding for all three of these factors suggesting an overlapping regulatory network (Figure 7B). As expected, a large portion of genes that were down-regulated upon JQ-1 treatment also showed high promoter read density for Brd4, c-Myc or Notch (Figure 7C). Accordingly, a specific loss of Brd4 binding is observed at target gene promoters and enhancers (Figure S7), and to a greater extent than Brd2, demonstrating some specificity of BET inhibition. In order to investigate the impact of c-Myc and Brd4 binding on direct Notch/Rbpj targets, we categorized expressed genes (FPKM>5) based on presence of all three factors, c-Myc and Notch only or Notch alone. We found that genes with Brd4/c-Myc/Notch binding have significantly higher expression than those with Notch binding alone. Genes with Brd4/c-Myc/Notch binding or c-Myc/Notch binding also showed significantly higher RNA PolII density at their promoters, implying that the presence of c-Myc and Brd4 in addition to Notch1 at promoters may enhance gene activity. Altogether, these data suggest that, especially in the context of FBXW7 mutations, higher thresholds of c-Myc work together with Notch1 activation to amplify a subset of genes critical for leukemic transformation.
Figure 7. BRD4 inhibition reduces expression of genes expressed in leukemia initiating cells directly co-regulated by MYC and NOTCH1.
(a) Heat maps showing genes significantly downregulated upon treatment of CUTLL1 cells with JQ-1 (400nM) for 12 hr compared to vehicle treated as determined by high throughput RNA sequencing (RNA-seq). Each column represents a biological replicate. ChIP-Seq signal density heat maps (right) show Brd4, c-Myc, Notch1, and total RNA PolII at gene loci depicted in gene expression heatmap. Genes are clustered based on Brd4, cMyc and Notch1 signal density. Scale represents reads per million (RPM) (b) Ven diagram showing overlap of total genes bound by Brd4, c-Myc and Notch1 in CUTLL1 cells. (c) Box plots showing Log2FPKM values (left) or promoter PolII density (right) for genes bound by combinations of Brd4, Myc and Notch. Whiskers represent upper and lower limits of the range. Boxes represent the first and third quartile with the line representing the median. (d) Representative ChIP-seq tracks for three gene loci: IL7R, NPM1 and CDK4. Scale corresponds to RPM.
DISCUSSION
In this study we used animal modeling of recurrent somatic FBXW7 mutations to study their function in leukemia initiation and progression. We found that such missense mutations specifically augment the function of leukemia-initiating cells (LIC) and at the same time are dispensable for normal hematopoiesis, explaining their selection during evolution of leukemia genomes and the significant abundance of FBXW7 missense (but fewer nonsense, insertion or deletion) mutations in human T-ALL. The differential function of FBXW7 mutations could be explained by the existence of distinct thresholds of c-Myc expression between physiological and malignant stem cells. This is an important finding as it suggests that genetic or pharmacological modulation of the FBXW7:MYC interaction could specifically target leukemia stem cells. Indeed, we were able to show that Myc deletion suppresses established TALL by eliminating leukemia initiating cells and pharmacologic targeting of c-Myc induction suppresses the growth of human and mouse T-ALL, including those that carry FBXW7 mutations and are resistant to NOTCH inhibition.
Another important finding illustrated here is that for the first time we are able to visualize cancer (leukemia) initiating cells using in vivo genetic fluorescent labeling. Indeed, using the c-MycGFP reporter strain, we were able to correlate c-Myc protein abundance to leukemia initiating cell (LIC) activity. The correlation was complete, as we were able to show total absence of LIC activity in the c-MycGFP negative tumor fraction. This is an exciting finding as it gives us the ability to highly purify LIC and study their biology. For example, we were able to demonstrate that T-ALL LIC have a characteristic surface phenotype and their gene expression patterns correlate to gene signatures characteristic of earlier stages of differentiation, including hematopoietic and embryonic stem cells. Interestingly, a large proportion of these LIC-specific genes are bound by both c-Myc and Notch1, both of which are key oncogenes in T-ALL. Brd4 is also present at most of these loci, a finding that can explain the therapeutic efficacy of BET inhibitors. Moreover, this is the first time that c-Myc expression directly correlated in vivo to LIC populations. Accordingly, human T-ALL LIC-enriched populations also express high levels of c-Myc protein and FBXW7 mutations lead to significant MYC stabilization, suggesting that this mechanism is evolutionarily conserved. However, this is not a universal characteristic of tumor initiating cell populations. Indeed, we have recently performed studies using animal models of BCR-ABL-driven chronic myelogenous leukemia (CML), a prototypic LIC-driven disease. In this leukemia model, Myc expression does not define the cell population that has the ability to initiate and propagate disease (Reavie et al., 2013), suggesting that different oncogenes and cellular settings could have distinct requirements for Myc expression and function.
As we have identified c-Myc as marker of LIC in animal models of T-ALL, we hypothesized that pharmacologic inhibition of MYC induction would be a feasible approach in the treatment of human TALL. Initially, inhibition of the Notch pathway using gamma-secretase inhibitors (γSI) was a promising therapeutic approach, but it has presented considerable challenges (GI toxicity, acquisition of resistance) for its adaptation in the clinic. While c-Myc is critical in the development of a wide range of tissues, recent studies have suggested that c-Myc inhibition is surprisingly well tolerated at least in pre-clinical animal studies (Soucek et al., 2008). We show here that genetic inactivation of Myc in established T-ALL specifically targeted the LIC fraction, eventually leading to tumor regression and loss of LIC self-renewal. Similarly, treatment of human T-ALL lines and primary leukemic cells with BET inhibitors (JQ1, CPI203) completely suppressed c-Myc response, leading to rapid growth arrest. Unlike γSI treatment, this effect is irrespective of FBXW7 status, suggesting such compounds could have broader applications across tumor genotypes. It remains to be seen whether BET inhibition will be as efficient for the treatment of either relapsed T-ALL or highly aggressive T-ALL subtypes, including early T cell progenitor (ETP) T-ALL that is also resistant to NOTCH pathway inhibition (Zhang et al., 2012).
The combination of disease modeling to whole genome and transcriptome studies lead to findings that could have major implications for the understanding of oncogenic (NOTCH, MYC) interactions and their role in gene expression regulation and cancer initiation. Indeed our studies suggest that Notch1 is not sufficient to induce or maintain transplantable T-ALL in the absence of c-Myc function. Interestingly, we demonstrate that the majority of LIC-specific genes are bound by both transcription factors but deletion of Myc can abrogate LIC activity. In agreement to this notion, it has been shown in vitro that MYC overexpression can induce resistance to Notch pathway inhibition (Weng et al., 2006). We thus hypothesize that Notch1 acts as a pioneering factor, influencing lineage commitment and the activity of epigenetic regulators (Ntziachristos et al., 2012), altering the chromatin structure of its transcriptional targets to a more permissive state. When c-Myc becomes expressed and stabilized (due to mutation or downregulation of FBXW7), it can bind to newly accessible E-box motifs, thereby amplifying established gene expression programs (Figure 7C). These studies support, in an in vivo tumor model, the “MYC transcriptional amplification” hypothesis recently postulated (Lin et al., 2012; Nie et al., 2012). They also prove that this transcriptional amplification mode has in vivo consequences in cancer initiation and can be pharmacologically targeted. Finally, our studies show that such key transcriptional responses can be post-translationally regulated and that oncogenic events invented ways to hijack them, as shown using in vivo modeling of the FBXW7 missense mutations.
METHODS
Animals
c-Myc GFP knock in and Myc conditional knockout mice were described previously (de Alboran et al., 2001; Huang et al., 2008). Fbxw7 knock-in mutant mice were generated by insertion of a loxP flanked splice acceptor PGK-NEO cassette with three polyA sites in the intronic sequence between exons 10 and 11, and a CCG to GCA point mutation was introduced by PCR mutagenesis in the opposite strand at coding sequence corresponding to R468. Eef1a1 human FBXW7 R465C mice were generated by introducing and R465C mutation into a N-terminally FLAG M2 tagged human FBXW7 alpha cDNA and subcloning this sequence into a the Eef1a1 lox-stop-lox targeting vector, described previously (Buonamici et al., 2009). Inducible Cre recombinase expression was achieved by injecting 4–6 week old Mx1-Cre mice intra-peritoneally three times with 20mg/kg polyI:polyC. CPI203 (Constellation Phramaceuticals) was dissolved in 5%DMSO/10% hydroxypropyl-beta cyclodextrin and administered by intraperitoneal injection twice daily at 5 mg kg−1. All animal experiments were done in accordance to the guidelines of the NYU School of Medicine or Columbia University Institutional Animal Care and Use Committee.
Bone marrow transduction and transplantation
Bone marrow was enriched for hematopoietic stem and progenitor cells by magnetic selection of cells expressing c-kit (StemCell Technologies), cultured in the presence of 50 ng/ml SCF, 50 ng/ml Flt3 ligand, 10 ng/ml IL-3 and 10 ng/ml IL-6 and infected with concentrated retroviral supernatants after 24 and 48hr. Transduction efficiency was determined by reporter fluorescence at 96h and total or sorted populations were transferred via retroorbital injection into irradiated (1100 rad) congenic recipients together with 2×105 unfractionated BMMC for hemogenic support. Sub-lethally irradiated (450 rad) mice were used for secondary transplants. For Notch1 T-ALL induction, 5×104 Notch1ΔE GFP+ bone marrow cells were transferred per recipient, unless otherwise stated.
Flow Cytometric analysis and cell sorting
Single cell suspensions were derived from bone marrow (femur and tibia), spleen and thymus from adult (>6wk) mice and red blood cells were lysed with ACK buffer. Nonspecific antibody binding was blocked by incubation with 20 μg/ml Rat IgG (Sigma) for 15 min. Cells were incubated with primary and secondary antibodies for 30 min on ice. Human CD34+ cells were isolated directly from xenografts using the EasySep Human CD34 Positive Selection Kit (StemCell Technologies). Stained cells were quantified using a BD Fortessa analyzer or isolated with a MoFlo cell sorter (Beckman Coulter) or BD ARIA II. FlowJo software (Treestar) was used to generate FACS plots, histograms and calculate mean fluorescence intensities.
Microarray and Gene set enrichment analysis
Total RNA was extracted using the RNeasy Plus Micro kit (Qiagen). RNA quantification and quality was determined using an Agilent 2100 Bioanalyzer. The Ovation RNA Amplification System V2 (NuGEN) kits were used for amplification. Amplified RNA was labeled and hybridized to the Mouse 430.2 microarrays (Affymetrix). The Affymetrix gene expression profiling data were normalized using the GC-RMA algorithm. The gene expression intensity presentations were generated with Multi Experiment Viewer software. Gene set enrichment analysis was performed using gene set as permutation type, 1,000 permutations and log2 ratio of classes as metric for ranking genes. Gene sets used in this study were identified from the Molecular Signatures Database (MSigDB Curated v3.0) or have been previously published (Kim et al., 2008). Raw expression data was deposited in the Gene Expression Omnibus (GSE46797).
In vitro drug treatments
Human and mouse T-ALL lines were grown in complete RPMI media (supplemented with 10% fetal bovine serum, penicillin/streptomycin, glutamine and 55μM β-mercaptoethanol). Primary mouse T-ALL were cocultured with OP9 stromal cells in Opti-MEM supplemented with 10% fetal bovine serum, 5 ng/ml IL-7, penicillin/streptomycin, glutamine and 55μM β-mercaptoethanol and passaged every 3–4 days onto a fresh feeder layer. JQ1 (Cayman Chemical) or Compound E (Alexis Biochemicals) prepared in DMSO was added to the cultures and the media was replaced every 24 hours. Viable cells were counted every 48 hours. BrdU (10μM) was added to the cultures for a 1hr pulse and incorporation into DNA was determined by using the APC BrdU Flow Kit (BD Pharmingen).
Chromatin Immunoprecipitation
ChIP-sequencing was performed in CUTLL1 cells as described previously (Ntziachristos et al, 2012) using rabbit polyclonal antibodies against c-Myc (N-262, Santa Cruz), Brd2 (Bethyl A302-583A) or Brd4 (Bethyl A301-985A) (5μg per IP). Peak calling was performed using MACS1.4 allowing only one duplicate read. For Notch1 we considered only peaks with p<10e-7. For c-Myc and Brd4, we considered peaks with p<10e-5. Peak annotation was performed using CEAS. Genes were considered bound if a peak was present within 2kb of the TSS.
RNAseq library preparation and analysis
Whole RNA was extracted from 1 million CUTLL1 cells per replicate using TriZol reagent (Invitrogen) according to the manufacturers protocol. Whole RNA was treated with DNase for 30 minutes (Invitrogen) and purified with RNA clean & concentrator columns (Zymo). Ribosomal RNA was depleted using RiboZero magnetic kit (Epicentre) according to manufacturers protocol. cDNA preparation and strand-specific library construction was performed using the dUTP method as described by Zhong and colleagues (doi:10.1101/pdb.prot5652). Libraries were sequenced on the Illumina HiSeq 2000 using 50bp single end reads. Base calling and quality filtering was performed as in ChIP-Seq data. Fastq files were aligned to Hg19 using TopHat allowing 2 mismatches. Differential expression tests were done using the Cuffdiff module of Cufflinks against the RefSeq annotation. We used FDR (0.05) corrected p-value of 0.05 as a cutoff for significance.
Statistical analysis
The means of each data set were analyzed using Student’s t-test, with a two-tailed distribution and assuming equal sample variance.
Supplementary Material
Highlights.
Mutant Fbxw7 alleles cooperate with Notch1 to drive aggressive T-ALL
Stabilization of c-Myc protein defines leukemia-initiating cells
BET inhibitors or c-Myc depletion impedes leukemic progression
Notch, Myc and Brd4 bind cooperatively to induce LIC gene expression
Acknowledgments
We would like to thank Constellation Pharmaceuticals for the CPI203 compound and helpful discussions. Drs. W. Pear, P. Premsrirut, J. Aster, J. Silva, M. Kelliher and A. Klinakis for sharing valuable materials. Dr. J. Zavadil and the NYU Genome Technology Center (supported in part by NIH/NCI P30 CA016087-30 grant) for assistance with microarray experiments. Drs. L. Deriano and J. Chaumeil for assistance with mFISH analysis. The NYU Flow Cytometry facility (supported in part by NIH/NCI 5 P30CA16087-31) for cell sorting, the NYU Histology Core (5P30CA16087-31), and the Transgenic Mouse Core (NYU Cancer Institute Center Grant (5P30CA16087-31). I.A. was supported by the National Institutes of Health (1RO1CA133379, 1RO1CA105129, 1RO1CA149655, 5RO1CA173636, and 1RO1GM088847). I.A. was also supported by the William Lawrence and Blanche Hughes Foundation, The Leukemia & Lymphoma Society (TRP#6340-11, LLS#6373-13), The Chemotherapy Foundation, The Irma T. Hirschl Trust, The V Foundation for Cancer Research and the St. Baldrick’s Foundation. We are particularly grateful for the support by a Feinberg Lymphoma Grant. B.K. was supported by the NYU Cell and Molecular Biology Training Program. L.R. is supported by a Ruth L. Kirschstein National Research Service F31 Award and a Marie Curie Actions International Mobility Fellowship. J.M. is supported by the Netherlands Organization for Scientific Research (NWO Rubicon) and by the Dutch Cancer Society (KWF Fellowship). I.A. is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
Author Contributions: I.A. and B.K. designed the experiments and wrote the manuscript. B.K. performed most of the experiments. L.R. generated the Eef1a1-FBW7 ES cells. L.X., A.P.G. and A.F. assisted with the in vivo and primary human T-ALL drug studies. P.S assisted with inhibitor production and provided advice for its use. C.V. J.S. and J.M. performed the BRD4 knock-down experiments. B.A.O. performed the in vitro ubiquitylation assays. P.N., T.T and S.S.S. performed and analyzed the RNA-Seq and ChIP-Seq experiments.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nature reviews. 2008;8:380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dofou D, Marth C, et al. FBXW7/hCDC4 is a generaltumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- Armstrong F, de la Grange PB, Gerby B, Rouyez MC, Calvo J, Fontenay M, Boissel N, Dombret H, Baruchel A, Landman-Parker J, et al. NOTCH is a key regulator of human T-cell acute leukemia initiating cell activity. Blood. 2009;113:1730–1740. doi: 10.1182/blood-2008-02-138172. [DOI] [PubMed] [Google Scholar]
- Aster JC, Robertson ES, Hasserjian RP, Turner JR, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J Biol Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- Buchstaller J, McKeever PE, Morrison SJ. Tumorigenic cells are common in mouse MPNSTs but their frequency depends upon tumor genotype and assay conditions. Cancer Cell. 2012;21:240–252. doi: 10.1016/j.ccr.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonamici S, Trimarchi T, Ruocco MG, Reavie L, Cathelin S, Mar BG, Klinakis A, Lukyanov Y, Tseng JC, Sen F, et al. CCR7 signalling as an essential regulator of CNS infiltration in T-cell leukaemia. Nature. 2009;459:1000–1004. doi: 10.1038/nature08020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MY, Xu L, Shestova O, Histen G, L’Heureux S, Romany C, Childs ME, Gimotty PA, Aster JC, Pear WS. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118:3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crusio KM, King B, Reavie LB, Aifantis I. The ubiquitous nature of cancer: the role of the SCF(Fbw7) complex in development and transformation. Oncogene. 2010;29:4865–4873. doi: 10.1038/onc.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis H, Lewis A, Spencer-Dene B, Tateossian H, Stamp G, Behrens A, Tomlinson I. FBXW7 mutations typically found in human cancers are distinct from null alleles and disrupt lung development. The Journal of pathology. 2011;224:180–189. doi: 10.1002/path.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alboran IM, O’Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ, 3rd, Singer DS. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci U S A. 2012;109:6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing JR, Wilson RK, Zhang J, Mardis ER, Pui CH, Ding L, Ley TJ, Evans WE. The Pediatric Cancer Genome Project. Nat Genet. 2012;44:619–622. doi: 10.1038/ng.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando AA, Look AT. Gene expression profiling in T-cell acute lymphoblastic leukemia. Semin Hematol. 2003;40:274–280. doi: 10.1016/s0037-1963(03)00195-1. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, Richardson C. Multipotent hematopoietic cells susceptible to alternative double-strand break repair pathways that promote genome rearrangements. Genes Dev. 2007;21:1064–1074. doi: 10.1101/gad.1522807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima H, Matsumoto A, Inuzuka H, Zhai B, Lau AW, Wan L, Gao D, Shaik S, Yuan M, Gygi SP, et al. SCF(Fbw7) Modulates the NFkappaB Signaling Pathway by Targeting NFkappaB2 for Ubiquitination and Destruction. Cell Rep. 2012;1:434–443. doi: 10.1016/j.celrep.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006:1–13. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- Grim JE, Knoblaugh SE, Guthrie KA, Hagar A, Swanger J, Hespelt J, Delrow JJ, Small T, Grady WM, Nakayama KI, et al. Fbw7 and p53 cooperatively suppress advanced and chromosomally unstable intestinal cancer. Mol Cell Biol. 2012;32:2160–2167. doi: 10.1128/MCB.00305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur J Immunol. 2008;38:342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]
- Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinakis A, Szabolcs M, Chen G, Xuan S, Hibshoosh H, Efstratiadis A. Igf1r as a therapeutic target in a mouse model of basal-like breast cancer. Proc Natl Acad Sci U S A. 2009;106:2359–2364. doi: 10.1073/pnas.0810221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting inmice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Lee MS, Hanspers K, Barker CS, Korn AP, McCune JM. Gene expression profiles during human CD4+ T cell differentiation. Int Immunol. 2004;16:1109–1124. doi: 10.1093/intimm/dxh112. [DOI] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional Amplification in Tumor Cells with Elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Palomero T, Sumazin P, Califano A, Ferrando AA, Stolovitzky G. ChIP-on-chip significance analysis reveals large-scale binding and regulation by human transcription factor oncogenes. Proc Natl Acad Sci U S A. 2009;106:244–249. doi: 10.1073/pnas.0806445106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Oike Y, Onoyama I, Iwama A, Arai F, Takubo K, Mashimo Y, Oguro H, Nitta E, Ito K, et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 2008 doi: 10.1101/gad.1621808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis KJ, Van Der Meulen J, Wolfe AL, Liu X, Mets E, Taghon T, Khan AA, Setty M, Rondou P, Vandenberghe P, et al. A cooperative microRNAtumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL) Nat Genet. 2011;43:673–678. doi: 10.1038/ng.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ., 3rd Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM, Morrison CG, Passegue E. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MJ, Wilson A, Trumpp A. More than just proliferation: Myc function in stem cells. Trends Cell Biol. 2005;15:128–137. doi: 10.1016/j.tcb.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc Is a Universal Amplifier of Expressed Genes in Lymphocytes and Embryonic Stem Cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoyama I, Tsunematsu R, Matsumoto A, Kimura T, de Alboran IM, Nakayama K, Nakayama KI. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J Exp Med. 2007 doi: 10.1084/jem.20062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O’Neil J, Neuberg D, Weng AP, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov N, Schulein C, Jaenicke LA, Eilers M. Ubiquitylation of the amino terminus of Myc by SCF(beta-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat Cell Biol. 2010;12:973–981. doi: 10.1038/ncb2104. [DOI] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, Sulis ML, Barnes K, Sawai C, Homminga I, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15:50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reavie L, Buckley SM, Loizou E, Takeishi S, Aranda-Orgilles B, Ndiaye-Lobry D, Abdel-Wahab O, Ibrahim S, Nakayama KI, Aifantis I. Regulation of c-Myc Ubiquitination Controls Chronic Myelogenous Leukemia Initiation and Progression. Cancer Cell. 2013;23:362–375. doi: 10.1016/j.ccr.2013.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reavie L, Della Gatta G, Crusio K, Aranda-Orgilles B, Buckley SM, Thompson B, Lee E, Gao J, Bredemeyer AL, Helmink BA, et al. Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase-substrate complex. Nat Immunol. 2010;11:207–215. doi: 10.1038/ni.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Trifonov V, Fangazio M, Bruscaggin A, Rasi S, Spina V, Monti S, Vaisitti T, Arruga F, Fama R, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012;209:1537–1551. doi: 10.1084/jem.20120904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VM, Calvo JA, Draheim KM, Cunningham LA, Hermance N, Beverly L, Krishnamoorthy V, Bhasin M, Capobianco AJ, Kelliher MA. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26:8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ, Jankovic V, Gao J, Buonamici S, Vest A, Lee JM, Zavadil J, Nimer SD, Aifantis I. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J Exp Med. 2008;205:1395–1408. doi: 10.1084/jem.20080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunematsu R, Nakayama K, Oike Y, Nishiyama M, Ishida N, Hatakeyama S, Bessho Y, Kageyama R, Suda T, Nakayama KI. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J BiolChem. 2004;279:9417–9423. doi: 10.1074/jbc.M312337200. [DOI] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell division. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.