Abstract
The use of Bacillus thuringiensis (Bt) strains with high insecticidal activity is essential for the preparation of bioinsecticide. In this study, for 60 Bt strains isolated in Taiwan, their genotypes and the correlation of some cry genes as well as the expression levels of cry1 genes, with their insecticidal activities against Plutella xylostella, were investigated. Pulsed field gel electrophoresis (PFGE) and random amplified polymorphic DNA (RAPD) results revealed that the genotypes of these Bt strains are highly diversified. Also, a considerable number of the Bt strains isolated in Taiwan were found to have high insecticidal activities. Since strains that showed individual combined patterns of PFGE and RAPD exhibited distinct insecticidal activities against P. xylostella, thus, these genotypes may be useful for the identification of the new Bt strains and those which have been used in bioinsecticides. In addition, although the presence of cry2Aa1 may have a greater effect on the insecticidal activity of Bt strains in bioassay than other cry genes, only high expression level of cry1 genes plays a key role to determine the insecticidal activity of Bt strains. In conclusion, both RAPD and PFGE are effective in the differentiation of Bt strains. The presence of cry2Aa1 and, especially, the expression level of cry1 genes are useful for the prediction of the insecticidal activities of Bt strains against P. xylostella.
Keywords: Bacillus thuringiensis, insecticidal activity, PFGE, RAPD, cry genes
1. Introduction
Bacillus thuringiensis (Bt) is a gram positive, endospore forming bacterium of the B. cereus group that forms a parasporal crystal (δ-endotoxins or Cry toxins) during the stationary phase of its growth cycle [1]. The crystal consists of proteins exhibiting a highly specific insecticidal activity to a limited number of insect species [2,3]. Due to the insecticidal properties of Bt, products based on Bt toxins have been well recognized for the biological control of insect pests in forestry and agriculture for decades [4]. For Bt, the production of insecticidal crystal inclusion inside the cell during sporulation, is controlled by cry genes, which are usually located on the plasmid [5]. Many Cry toxins have been characterized and classified according to the nomenclature of Crickmore et al. [6]. Currently, more than 70 groups of Cry toxins have been identified [7]. Major classes of Bt toxins include Lepidoptera-active Cry1, Lepidoptera and Diptera-active Cry2, Coleoptera-active Cry3, and Diptera-active Cry4 [8].
Since Bt strains have been commonly used for the preparation of bioinsecticide, the isolation of novel Bt strains has become a routine activity [9]. The discrimination between new Bt isolates with insecticidal activity and those which have been used as bioinsecticides is also important from both ecological and economical views [10]. In this regard, the survey of the genotypes, the cry genes and the insecticidal activities of Bt strains may allow us to find novel Bt strains useful for industrial product preparation. Although the distribution of some cry genes in Bt isolates in Taiwan has been reported [11], little is known about their genotypes and the insect toxicity of these strains.
The diamondback moth, Plutella xylostella (Linnaeus) (Lepidoptera: Plutellidae), is the most destructive insect pest of cruciferous crops throughout the world. The characterization of toxicity of the Bt strains to the diamondback moth is important for agricultural purposes [12]. In this study, 60 Bt strains isolated from different origins including soil and granaries in Taiwan were characterized for their toxicity against P. xylostella. The genotypes of these strains were investigated by pulsed field gel electrophoresis (PFGE) and random amplified polymorphic DNA (RAPD), which have been proven to provide a high degree of discrimination for Bt strains [10,13]. Since the information on insecticidal activities of individual crystal proteins against selected arthropod has been reviewed by Frankenhuyzen [14], a collection of cry genes, including those coding for proteins biologically active to P. xylostella, were assayed based on polymerase chain reaction (PCR) for these strains. Furthermore, the expression level of cry1 genes was determined; the concordance among insecticidal activity, PFGE and RAPD patterns, and the cry gene content as well as the expression level of cry1 genes, was assessed.
2. Materials and Methods
2.1. Bacterial Strains and Cell Cultivation
Bt strains (TT1–TT62) used in this study were obtained from Taiwan Agriculture Chemical and Toxic Substances Research Institute (TACTRI), Taichung, Taiwan. These strains were mainly isolated from soil and granaries in Taiwan. Two strains, i.e., TT12 and TT13, which were isolated from imported biopesticide products, were used as reference strains. Bacterial cells were cultivated in Brain Heart Infusion (BHI) broth (Difco™, Becton, Dickinson and Company, Sparks, MD, USA) overnight at 37 °C with rotary shaking (150 rev/min).
2.2. Bioassay of the Insecticidal Activity
The insecticidal activities of the Bt strains (TT1–TT62 except TT14) were assayed according to the methods described by Pang et al. [15]. Bt cells were cultivated, and the spores as well as crystal toxins were collected, mixed with Tris buffer (10 mM Tris-HCl, 10 mM EDTA, pH 7.4, Sigma-Aldrich, St. Louis, MO, USA), and assayed for protein concentration and insecticidal activity against P. xylostella. Two Bt strains, i.e., TT12 and TT13, isolated from two commercial insecticide products, DiPel™ and XenTari™ (Abbott, Chicago, IL, USA), were used as positive control. Tris buffer was used as negative control. Protein concentration was assayed using bicinchoninic acid (BCA) protein assay kit (Pierce, Rockford, IL, USA). For each Bt strain, two protein doses were repeated 3 times using 10 larvae per assay. The spores and crystal mixtures at protein concentrations of 25 mg/L and 250 mg/L, were sprayed onto a 15 cm2 leaf piece of cabbage by a potter spray tower (Burkard Manufacturing Co. Ltd., Hertfordshire, UK) and the leaf were then fed to the third instar larvae at 25 °C under 60% humidity and incubated for 72 h. The insecticidal activities of the Bt strains were defined as mortality rates after incubation for 24, 48 and 72 h, which were calculated according to Abbott’s formula, i.e., corrected mortality (%) = [(test mortality − blank control mortality)/(1 − blank control mortality)] × 100% [16]. All data were expressed as mean ± standard deviation (n = 3).
2.3. Genotyping by PFGE and RAPD
The isolation of chromosomal DNA of Bt strains (TT1–TT62 except TT23 and TT41) and NotI digestion were according to the methods described by Kolstø et al. [17]. Generally, bacterial cells harvested by centrifugation were washed, resuspended, and then mixed with low melting point agarose (NuSieve™ GTG™ agarose, FMC BioProducts, Rockland, ME, USA) to obtain the agarose plugs. Following the cell lysis and proteolysis, the restriction digestion was performed by placing a 2 mm slice of each plug into 100 μL of restriction buffer containing 20 U of NotI (New England Biolabs, Beverly, MA, USA). After incubation at 37 °C for 12–16 h, the plugs were placed into the slots of a 1.2% agarose gel in 0.5× TBE buffer (89 mmol/L Tris-borate, pH 8.3, 2 mmol/L EDTA, Sigma-Aldrich, St. Louis, MO, USA). Electrophoresis was performed by using CHEF-DR®II System (Bio-Rad, Hercules, CA, USA). The conditions used were 180 V for 28 h at 15 °C (pulse time: 7 to 90 s). Bacteriophage λ DNA concatemers (Bio-Rad, Hercules, CA, USA) were used as molecular weight markers.
For RAPD, bacterial genomic DNA was prepared by phenol-chloroform (Sigma-Aldrich, St. Louis, MO, USA) extraction [18], and then was subjected to RAPD reactions using a single 10-mer primer OPF-06 (5'-GGGAATTCGG-3') (MDBio, Inc., Taipei, Taiwan). RAPD reactions were performed in 25 µL reaction mixtures containing 1× PCR buffer (PROtech Technology Ent. Co., Ltd., Taipei, Taiwan), 300 µM of each deoxynucleoside triphosphate, 4 µM primer, 1.5 mM MgCl2 (PROtech Technology Ent. Co., Ltd., Taipei, Taiwan), 0.4 U of Prozyme (PROtech Technology Ent. Co., Taipei, Taiwan), and 50 ng of genomic DNA. RAPD conditions in a thermal cycler (Gene Amp PCR system 9600, Perkin Elmer, Norwalk, CT, USA) were as follows: 2 initial cycles consisted of 94 °C for 4 min, 35 °C for 2 min, and 72 °C for 2 min, followed by 40 cycles consisted of 94 °C for 20 s, 36 °C for 30 s, and 72 °C for 30 s, and a final extension at 72 °C for 7 min. The amplified products were analyzed by electrophoresis (Mupid-2 mini gel electrophoresis system, Cosmo Bio. Co. Ltd, Tokyo, Japan).
PFGE and RAPD patterns were analyzed by the NTSYSpc software (Numerical taxonomy and multivariate analysis system, version 2.10e, State University of New York, Stony Brook, NY, USA). Strains were clustered by using the Dice coefficient of similarity, and cluster analysis by unweighted pair group method with arithmetic averages (UPGMA). The final judgment of whether the patterns were identical was done by visual comparison.
2.4. Detection of cry Genes
Chromosomal DNA of each of the Bt strains was prepared by phenol-chloroform extraction [18]. All isolates were screened by PCR analysis for the presence or absence of 13 selected cry genes, including those coding for Cry proteins with insecticidal activity against P. xylostella [14]. The sequences of the primers were shown in Table 1. PCR amplifications were performed as previously described [19,20,21,22,23,24,25].
Table 1.
Sequences of primers for cry genes and the sizes of amplified products.
| Target genes | Primer sequences | Product size (bp) | Reference |
|---|---|---|---|
| cry1 | F: 5'-CTGGATTTACAGGTGGGGATAT-3' | 558 | [19] |
| R: 5'-TGAGTCGCTTCGCATATTTGACT-3' | |||
| cry1B | F: 5'-CTTCATCACGATGGAGTAA-3' | 367 | [20] |
| R: 5'-CATAATTTGGTCGTTCTGTT-3' | |||
| cry1E | F: 5'-TAGGGATAAATGTAGTACAG-3' | 1137 | [21] |
| R: 5'-MDATYTCTAKRTCTTGACTA-3' | |||
| cry2Aa1 | F: 5'-GTTATTCTTAATGCAGATGAATGGG-3' | 498 | [22] |
| R: 5'-GAGATTAGTCGCCCCTATGAG-3' | |||
| cry3A | F: 5'-CGTTATCGCAGAGAGATGACATTAAC-3' | 951 | [22] |
| R: 5'-TGGTGCCCCGTCTAAACTGAGTGT-3' | |||
| cry4A2 | F: 5'-GGGTATGGCACTCAACCCCACTT-3' | 1529 | [22] |
| R: 5'-GCGTGACATACCCATTTCCAGGTCC-3' | |||
| cry7 | F: 5'-CAACCAGACCTATTTTATTGGAGT-3' | 476 | [23] |
| R: 5'-ATTTTTACAGCTGGAATTTTGTG-3' | |||
| cry8D | F: 5'-AGAAACACAAGATAAAATACTCC-3' | 401 | [23] |
| R: 5'-ATACAGCATCCCCTTCTACAATCT-3' | |||
| cry9A | F: 5'-GGTTCACTTACATTGCCGGTTAGC-3' | 1547 | [24] |
| R: 5'-GTTTGAGCCGCTTCACAGCAATCC-3' | |||
| cry9C | F: 5'-CCACCAGATGAAAGTACCGGAAG-3' | 1232 | [24] |
| R: 5'-GTTTGAGCCGCTTCACAGCAATCC-3' | |||
| cry9Ea | F: 5'-GCGGCTGGCTTTACTTTACCGAG-3' | 824 | [24] |
| R: 5'-GTTTGAGCCGCTTCACAGCAATCC-3' | |||
| cry22 | F: 5'-CAGATGAGATAGATGGGGATTTGA-3' | 413 | [23] |
| R: 5'-ATTCGCTTCTATACTTGGCTGTC-3' | |||
| cry32Aa | F: 5'-TGGTCGGGAGAGAATGGATGGA-3' | 676–677 | [25] |
| R: 5'-ATGTTTGCGACACCATTTTC-3' |
2.5. Expression Levels of cry1 Genes
The expression levels of cry1 genes in Bt strains were determined by a two-step reverse transcription real-time PCR. For each Bt strain, total RNA was prepared from 108 CFU of Bt using PureLink™ RNA Mini Kit (Invitrogen Life Technologies, Carlsbad, CA, USA). Then, cDNA was synthesized in reactions containing 2 μL of total RNA from Bt isolates using a reverse transcription kit (SuperScript First-Strand Synthesis System, Invitrogen Life Technologies, Carlsbad, CA, USA). The cDNA was used as a template for real-time PCR to determine the expression level of cry1 genes in Bt strains. Real-time PCR was performed in 20 µL reaction mixtures containing 1× KAPA FAST SYBR green I master mix (KAPA Biosystems, Woburn, MA, USA), 2 μL of the cDNA, and 0.25 mM of each primer to amplify all cry1 genes. Real-time PCR was performed using the ABI 7500 system (Applied Biosystems, Foster, CA, USA). The amplification conditions in a thermocycler were according to the methods of Gaviria Rivera and Priest [26]. The expression levels of cry1 genes were shown by threshold cycle (Ct) values.
2.6. Statistical Analysis
Statistical tests were performed using SPSS 17.0 (SPSS Inc., Chicago, IL, USA), p < 0.05 was taken as statistically significant. A paired samples t-test was applied to compare the mortality rates at different time points. Since multiple cry genes may be involved in the insecticidal activity of a Bt strain, a multiple regression analysis was applied to determine the relationship between the presence or absence of cry genes and the insecticidal activities of Bt strains. A Mann-Whitney U test was used to determine if the Bt strains with high expression levels of cry1 genes showed higher insecticidal activities against P. xylostella than those with low expression levels of cry1 genes.
3. Results
3.1. Insecticidal Activity of Bt Strains
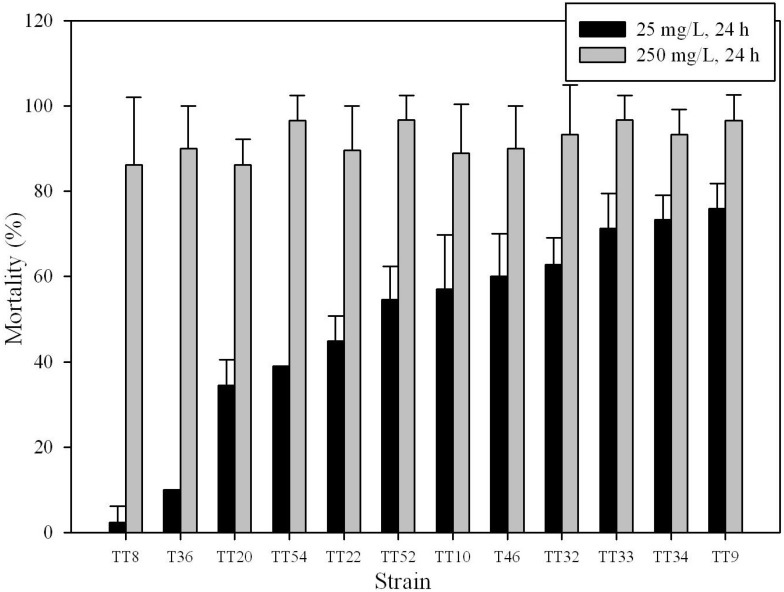
To evaluate the insecticidal activity of Bt strains, and the effect of the dose of Cry proteins and the incubation time of larvae with these proteins, two concentrations of global Cry protein, i.e., 25 mg/L and 250 mg/L, and three incubation time periods, i.e., 24, 48, and 72 h, respectively, were used. Results were shown in Table 2. At the protein concentration of 250 mg/L, it was found that 35, 46, and 46 Bt strains showed mortality rates ≥90% after incubation for 24, 48, and 72 h, respectively. On the other hand, at the protein concentration of 25 mg/L, 11, 28, and 29 strains showed mortality rates ≥90% after incubation for 24, 48, and 72 h, respectively. At least, 19 strains showed higher insecticidal activities than those of the reference strains, i.e., strains TT12 and TT13, at the toxin concentration of 25 mg/L, after incubation for 24 h (Table 1). A paired samples t-test was applied to compare the mortality rates at different time points. At the protein concentration of 25 mg/L, mortality rates after 72, and 48 h incubation were significantly higher than those after 24 as well as 48 h, and 24 h incubation, respectively (p < 0.05, degree of freedom (df) = 182). At the protein concentration of 250 mg/L, mortality rates after 48 h incubation were significantly higher than those after 24 h incubation (p < 0.05, df = 182); mortality rates after 72 h incubation were higher than those after 48 h incubation but not statistically significant. The above results indicate that the mortality of larvae gradually increases with the length of incubation time, and an assay at a protein concentration of 250 mg/L may not effectively differentiate the insecticidal activities of these Bt strains. Part of the results assayed at the global Cry protein concentration of 25 mg/L and 250 mg/L, respectively, with incubation time of 24 h, was shown in Figure 1. Thus, the insecticidal activity against P. xylostella at the global Cry protein concentration of 25 mg/L was used for further analysis.
Table 2.
Comparison of the random amplified polymorphic DNA (RAPD) and pulsed field gel electrophoresis (PFGE) patterns, and insecticidal activity of B. thuringiensis strains.
| Strain | RAPD pattern | PFGE pattern | Mortality (%) 1 | |||||
|---|---|---|---|---|---|---|---|---|
| 250 mg/L | 25 mg/L | |||||||
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | |||
| TT1 | D11 | PT11 | 6.83 ± 0.00 | 12.57 ± 10.93 | 19.00 ± 12.00 | 4.24 ± 3.71 | 11.91 ± 1.14 | 10.19 ± 4.81 |
| TT2 | D12 | PT12 | 6.90 ± 1.04 | 20.07 ± 7.28 | 22.72 ± 3.70 | 4.55 ± 3.94 | 5.28 ± 6.31 | 4.94 ± 4.28 |
| TT3 | D13 | PT13 | 2.66 ± 4.60 | 1.49 ± 1.43 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| TT4 | D14 | PT14 | 0.00 ± 0.00 | 0.00 ± 0.00 | 12.67 ± 21.94 | 3.33 ± 5.77 | 2.33 ± 4.04 | 4.62 ± 4.00 |
| TT5 | D15 | PT15 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 96.55 ± 5.98 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| TT6 | D16 | PT16 | 93.10 ± 5.98 | 96.36 ± 6.31 | 96.00 ± 6.93 | 86.20 ± 15.81 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| TT7 | D17 | PT17 | 96.86 ± 5.44 | 96.68 ± 5.74 | 96.33 ± 6.35 | 86.20 ± 5.98 | 96.33 ± 6.35 | 100.00 ± 0.00 |
| TT8 | D18 | PT18 | 86.20 ± 15.81 | 100.00 ± 0.00 | 100.00 ± 0.00 | 2.28 ± 3.94 | 16.33 ± 5.77 | 15.12 ± 6.68 |
| TT9 | D19 | PT19 | 96.55 ± 5.98 | 100.00 ± 0.00 | 100.00 ± 0.00 | 75.85 ± 5.98 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| TT10 | D20 | PT20 | 88.89 ± 11.51 | 100.00 ± 0.00 | 100.00 ± 0.00 | 57.04 ± 12.75 | 89.33 ± 0.58 | 92.63 ± 6.40 |
| TT11 | D21 | PT21 | 4.55 ± 3.95 | 30.78 ± 6.31 | 26.70 ± 6.68 | 2.28 ± 3.94 | 2.00 ± 0.00 | 0.00 ± 0.00 |
| TT14 | D24 | PT23 | ND 2 | ND | ND | ND | ND | ND |
| TT15 | D25 | PT24 | 93.10 ± 11.95 | 96.36 ± 6.31 | 96.00 ± 6.93 | 14.91 ± 20.19 | 38.00 ± 45.40 | 42.13 ± 40.09 |
| TT16 | D26 | PT25 | 30.99 ± 5.98 | 74.50 ± 12.62 | 76.85 ± 11.57 | 4.55 ± 3.94 | 5.67 ± 6.35 | 2.47 ± 4.28 |
| TT17 | D27 | PT26 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 79.30 ± 10.35 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| TT18 | D28 | PT27 | 96.17 ± 6.63 | 100.00 ± 0.00 | 100.00 ± 0.00 | 65.49 ± 33.28 | 96.33 ± 6.35 | 96.14 ± 6.68 |
| TT19 | D29 | PT28 | 4.55 ± 3.94 | 1.64 ± 0.00 | 2.47 ± 4.28 | 4.93 ± 4.31 | 17.01 ± 5.74 | 24.54 ± 9.62 |
| TT20 | D30 | PT29 | 86.20 ± 5.98 | 100.00 ± 0.00 | 100.00 ± 0.00 | 34.44 ± 5.98 | 74.33 ± 12.70 | 72.99 ± 13.36 |
| TT21 | D31 | PT30 | 68.94 ± 0.00 | 92.71 ± 6.31 | 92.00 ± 6.93 | 17.18 ± 0.00 | 38.07 ± 6.31 | 45.99 ± 6.68 |
| TT22 | D32 | PT31 | 89.65 ± 10.35 | 100.00 ± 0.00 | 100.00 ± 0.00 | 44.79 ± 5.98 | 92.67 ± 12.70 | 92.28 ± 13.36 |
| TT23 | ND | ND | 3.33 ± 5.57 | 11.51 ± 14.37 | 20.67 ± 6.35 | 3.33 ± 5.77 | 10.07 ± 6.26 | 20.10 ± 6.57 |
| TT24 | D33 | PT32 | 48.13 ± 3.23 | 89.66 ± 10.34 | 89.66 ± 10.34 | 3.33 ± 5.77 | 10.38 ± 5.97 | 13.82 ± 5.97 |
| TT25 | D34 | PT33 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 93.10 ± 11.95 | 96.33 ± 6.35 | 96.14 ± 6.68 |
| TT26 | D35 | PT34 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 93.33 ± 5.77 | 96.67 ± 5.77 | 96.49 ± 6.07 |
| TT27 | D36 | PT35 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 93.33 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| TT28 | D22 | PT36 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 89.27 ± 11.12 | 96.33 ± 6.35 | 96.11 ± 6.74 |
| TT29 | D34 | PT37 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 93.33 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| TT30 | D37 | PT38 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 93.33 ± 11.55 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| TT31 | D38 | PT39 | 96.67 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 | 90.00 ± 17.32 | 96.67 ± 5.77 | 96.49 ± 6.07 |
| TT32 | D22 | PT40 | 93.33 ± 11.55 | 100.00 ± 0.00 | 100.00 ± 0.00 | 62.77 ± 6.32 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| TT33 | D39 | PT41 | 96.67 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 | 71.20 ± 8.27 | 90.33 ± 0.58 | 89.80 ± 0.55 |
| TT34 | D40 | PT41 | 93.33 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 | 73.33 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| TT35 | D38 | PT42 | 63.33 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 | 21.67 ± 2.89 | 77.67 ± 10.97 | 87.94 ± 13.01 |
| TT36 | D41 | PT43 | 90.00 ± 10.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 10.00 ± 0.00 | 44.85 ± 5.97 | 58.63 ± 10.34 |
| TT37 | D42 | PT44 | 80.00 ± 34.64 | 100.00 ± 0.00 | 100.00 ± 0.00 | 46.67 ± 5.77 | 96.67 ± 5.77 | 96.55 ± 5.97 |
| TT38 | D43 | PT45 | 80.00 ± 26.46 | 100.00 ± 0.00 | 100.00 ± 0.00 | 33.33 ± 11.55 | 72.33 ± 11.55 | 72.42 ± 11.94 |
| TT39 | D44 | PT46 | 96.67 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 | 33.75 ± 5.30 | 76.73 ± 3.66 | 76.73 ± 3.66 |
| TT40 | D36 | PT47 | 90.00 ± 17.32 | 100.00 ± 0.00 | 100.00 ± 0.00 | 50.00 ± 43.59 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| TT41 | ND | ND | 10.00 ± 10.00 | 17.00 ± 0.00 | 8.00 ± 6.93 | 6.67 ± 5.77 | 6.93 ± 0.00 | 0.00 ± 0.00 |
| TT42 | D34 | PT48 | 96.67 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 | 43.33 ± 5.77 | 86.33 ± 15.82 | 92.03 ± 6.91 |
| TT43 | D45 | PT49 | 13.33 ± 5.77 | 37.95 ± 10.34 | 40.19 ± 11.96 | 0.00 ± 0.00 | 4.62 ± 4.00 | 2.87 ± 2.49 |
| TT44 | D46 | PT50 | 40.00 ± 0.00 | 58.63 ± 10.34 | 52.00 ± 12.00 | 3.33 ± 5.77 | 20.72 ± 5.97 | 12.28 ± 6.91 |
| TT45 | D47 | PT50 | 96.67 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 | 83.33 ± 5.77 | 96.67 ± 5.77 | 96.01 ± 6.91 |
| TT46 | D33 | PT51 | 90.00 ± 10.00 | 96.55 ± 5.87 | 96.00 ± 6.93 | 60.00 ± 10.00 | 79.33 ± 10.50 | 80.06 ± 13.81 |
| TT47 | D43 | PT52 | 96.67 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 | 74.53 ± 9.47 | 84.00 ± 10.39 | 83.42 ± 10.50 |
| TT48 | D43 | PT52 | 93.33 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 | 60.00 ± 10.00 | 82.67 ± 6.35 | 88.04 ± 11.96 |
| TT49 | D43 | PT52 | 88.57 ± 2.48 | 100.00 ± 0.00 | 100.00 ± 0.00 | 80.00 ± 10.00 | 96.67 ± 5.77 | 96.01 ± 6.91 |
| TT50 | D43 | PT52 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 83.33 ± 15.28 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| TT51 | D48 | PT1 | 70.00 ± 10.00 | 96.55 ± 5.97 | 96.00 ± 6.93 | 3.33 ± 5.77 | 24.33 ± 6.35 | 24.24 ± 6.91 |
| TT52 | D38 | PT53 | 96.67 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 | 54.53 ± 7.85 | 84.33 ± 13.80 | 82.06 ± 15.82 |
| TT53 | D41 | PT54 | 96.61 ± 5.87 | 100.00 ± 0.00 | 100.00 ± 0.00 | 86.45 ± 5.87 | 89.33 ± 10.50 | 88.91 ± 11.09 |
| TT54 | D2 | PT2 | 96.61 ± 5.87 | 100.00 ± 0.00 | 100.00 ± 0.00 | 39.02 ± 0.00 | 47.42 ± 10.52 | 51.96 ± 6.40 |
| TT55 | D49 | PT55 | 10.00 ± 10.00 | 17.27 ± 10.34 | 20.72 ± 5.97 | 3.33 ± 5.77 | 7.00 ± 0.00 | 13.82 ± 5.97 |
| TT56 | D50 | PT56 | 0.00 ± 0.00 | 3.58 ± 3.10 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.07 ± 0.13 |
| TT57 | D51 | PT57 | 6.54 ± 5.80 | 4.45 ± 4.07 | 1.00 ± 1.73 | 2.85 ± 4.93 | 3.58 ± 3.10 | 0.15 ± 0.13 |
| TT58 | D22 | PT58 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 89.84 ± 10.16 | 96.33 ± 6.35 | 96.30 ± 6.40 |
| TT59 | D38 | PT59 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| TT60 | D52 | PT60 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 96.61 ± 5.87 | 96.33 ± 6.35 | 96.30 ± 6.40 |
| TT61 | D34 | PT61 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 96.61 ± 5.87 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| TT62 | D53 | PT62 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| TT12 (positive control) | D22 | PT17 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 79.30 ± 10.35 | 96.33 ± 6.35 | 96.14 ± 6.68 |
| TT13 (positive control) | D23 | PT22 | 96.55 ± 5.98 | 100.00 ± 0.00 | 100.00 ± 0.00 | 50.17 ± 3.35 | 81.33 ± 12.42 | 80.29 ± 13.01 |
| Tris buffer (negative control) | - | - | 1.00 ± 3.05 | 4.67 ± 5.71 | 9.27 ± 7.31 | 1.00 ± 3.05 | 4.67 ± 5.71 | 9.27 ± 7.31 |
1 All data were expressed as mean ± standard deviation (n = 3); 2 ND: Not determined.
Figure 1.
Part of the insecticidal activity of Bt strains assayed at two global Cry protein concentrations after 24 h incubation. Each vertical bar represents the mean ± standard deviation (n = 3).
3.2. PFGE and RAPD Patterns
When chromosomal DNA of the 60 Bt strains (TT1–TT62 except TT23 and TT41) were cut with NotI and subjected to PFGE analysis, a total of 54 PFGE patterns were found. Only 4 patterns were shared by two or more strains. For RAPD using primer OPF-06, 41 patterns were found. Only 7 patterns were shared by two or more strains (Table 2). Strains with the same RAPD pattern could be further discriminated by PFGE typing, and vice versa. For example, strains of TT25, TT29, TT42, and TT61, within RAPD pattern of D34, could be further differentiated by PFGE typing; strains in PFGE pattern PT50, i.e., strains TT44 and TT45, could be further discriminated by RAPD typing (Table 2). For the combined use of PFGE and RAPD methods, only four strains, i.e., strains TT47–TT50, were found in a combined pattern, i.e., D43-PT52. Thus, both PFGE and RAPD results demonstrated that there is considerable genetic diversity among the Bt strains isolated in Taiwan.
Strains with individual combined patterns showed distinct insecticidal activities; strains that showed the same combined patterns of PFGE and RAPD exhibited similar levels of insecticidal activity. For example, the mortality rates of four strains, i.e., TT47–TT50, within the combined pattern of D43-PT52, were all belonging to strains of high insecticidal activities. The genotypes of Bt strains, if determined by highly discriminatory methods, may allow us to identify the newly isolated strains with potential for pesticide use and those strains which have been used in bioinsecticides.
3.3. Correlation among Insecticidal Activity, cry Genes, and the Expression Levels of cry1 Genes
A collection of randomly selected 29 Bt strains, which exhibited various insecticidal activities, was screened by PCR for the presence or absence of 13 selected cry genes, including those associated with the insecticidal activity against P. xylostella and those reported by Gaviria Rivera and Priest [26] and Ben-Dov et al. [22]. These cry genes are widely distributed among strains. All the 29 strains were positive for cry1 genes; none of the 29 strains carried cry1E, cry3A, cry4A2, cry8, cry9A, cry9C and cry32Aa genes. Variation of the presence of cry1B, cry2Aa1, cry7, cry9Ea, and cry22 genes was found among strains (Table 3). Thus, these 5 cry genes of Bt strains were used to find the correlation with the insecticidal activity against P. xylostella.
Table 3.
Comparison of cry gene profiles and insecticidal activity of B. thuringiensis strains.
| Strain | Mortality (%) 1 | cry genes 2 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25 mg/L (global Cry protein) | ||||||||||||||||
| 24 h | 48 h | 72 h | 1 | 1B | 1E | 2Aa1 | 3A | 4A2 | 7 | 8 | 9A | 9C | 9Ea | 22 | 32Aa | |
| TT59 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | + | - | - | + | - | - | - | - | - | - | - | - | - |
| TT62 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | + | - | - | + | - | - | - | - | - | - | - | - | - |
| TT61 | 96.61 ± 5.87 | 100.00 ± 0.00 | 100.00 ± 0.00 | + | - | - | + | - | - | - | - | - | - | - | - | - |
| TT5 | 96.55 ± 5.98 | 100.00 ± 0.00 | 100.00 ± 0.00 | + | - | - | - | - | - | - | - | - | - | + | - | - |
| TT27 | 93.33 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 | + | - | - | + | - | - | - | - | - | - | - | - | - |
| TT29 | 93.33 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 | + | - | - | + | - | - | - | - | - | - | - | - | - |
| TT30 | 93.33 ± 11.55 | 100.00 ± 0.00 | 100.00 ± 0.00 | + | - | - | - | - | - | - | - | - | - | + | - | - |
| TT25 | 93.10 ± 11.95 | 96.33 ± 6.35 | 96.14 ± 6.68 | + | - | - | + | - | - | - | - | - | - | - | - | - |
| TT50 | 83.33 ± 15.28 | 100.00 ± 0.00 | 100.00 ± 0.00 | + | + | - | + | - | - | - | - | - | - | - | - | - |
| TT49 | 80.00 ± 10.00 | 96.67 ± 5.77 | 96.01 ± 6.91 | + | + | - | + | - | - | - | - | - | - | - | - | - |
| TT17 | 79.30 ± 10.35 | 100.00 ± 0.00 | 100.00 ± 0.00 | + | - | - | + | - | - | - | - | - | - | - | - | - |
| TT12 | 79.30 ± 10.35 | 96.33 ± 6.35 | 96.14 ± 6.68 | + | - | - | + | - | - | - | - | - | - | - | - | - |
| TT47 | 74.53 ± 9.47 | 84.00 ± 10.39 | 83.42 ± 10.50 | + | + | - | + | - | - | - | - | - | - | - | - | - |
| TT34 | 73.33 ± 5.77 | 100.00 ± 0.00 | 100.00 ± 0.00 | + | - | - | + | - | - | - | - | - | - | - | - | - |
| TT48 | 60.00 ± 10.00 | 82.67 ± 6.35 | 88.04 ± 11.96 | + | + | - | + | - | - | - | - | - | - | - | - | - |
| TT41 | 6.67 ± 5.77 | 6.93 ± 0.00 | 0.00 ± 0.00 | + | - | - | + | - | - | - | - | - | - | - | - | - |
| TT19 | 4.93 ± 4.31 | 17.01 ± 5.74 | 24.54 ± 9.62 | + | - | - | - | - | - | - | - | - | - | - | - | - |
| TT2 | 4.55 ± 3.94 | 5.28 ± 6.31 | 4.94 ± 4.28 | + | - | - | - | - | - | - | - | - | - | - | - | - |
| TT16 | 4.55 ± 3.94 | 5.67 ± 6.35 | 2.47 ± 4.28 | + | - | - | - | - | - | - | - | - | - | - | - | - |
| TT1 | 4.24 ± 3.71 | 11.91 ± 1.14 | 10.19 ± 4.81 | + | - | - | + | - | - | - | - | - | - | - | - | - |
| TT44 | 3.33 ± 5.77 | 20.72 ± 5.97 | 12.28 ± 6.91 | + | - | - | - | - | - | - | - | - | - | + | - | - |
| TT24 | 3.33 ± 5.77 | 10.38 ± 5.97 | 13.82 ± 5.97 | + | - | - | + | - | - | - | - | - | - | - | - | - |
| TT4 | 3.33 ± 5.77 | 2.33 ± 4.04 | 4.62 ± 4.00 | + | - | - | - | - | - | - | - | - | - | - | - | - |
| TT23 | 3.33 ± 5.77 | 10.07 ± 6.26 | 20.10 ± 6.57 | + | - | - | - | - | - | - | - | - | - | - | + | - |
| TT57 | 2.85 ± 4.93 | 3.58 ± 3.10 | 0.15 ± 0.13 | + | - | - | - | - | - | + | - | - | - | - | - | - |
| TT11 | 2.28 ± 3.94 | 2.00 ± 0.00 | 0.00 ± 0.00 | + | - | - | - | - | - | - | - | - | - | - | - | - |
| TT43 | 0.00 ± 0.00 | 4.62 ± 4.00 | 2.87 ± 2.49 | + | - | - | - | - | - | - | - | - | - | - | - | - |
| TT56 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.07 ± 0.13 | + | - | - | - | - | - | + | - | - | - | - | - | - |
| TT3 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | + | - | - | - | - | - | - | - | - | - | - | - | - |
| Total | 29+ 3 | 4+ | 0+ | 16+ | 0+ | 0+ | 2+ | 0+ | 0+ | 0+ | 3+ | 1+ | 0+ | |||
1 All data were expressed as mean ± standard deviation (n = 3); 2 The presence (+) or the absence (-) of the specific cry gene by PCR was shown; 3 Number of strains positive for specific cry gene.
The correlation between the presence or the absence of these 5 cry genes and insecticidal activity of Bt strains at the global Cry protein concentration of 25 mg/L was determined using a multiple regression analysis. The presence (1) or absence (0) of these 5 cry genes were used as the explanatory variables. Statistical analysis revealed that the presence of cry2Aa1 and cry9Ea genes was positively correlated with the insecticidal activity of Bt strains (p < 0.05, Table 4). Based on the standardized regression coefficients, the presence of cry2Aa1 has a greater effect on insecticidal activity of Bt strains than other cry genes, i.e., cry1B, cry7, cry9Ea, and cry22 genes. Thus, the presence of cry2Aa1 might be useful as a reference marker to predict the insecticidal activities of Bt strains.
Table 4.
Multiple regression analysis of cry genes on insecticidal activity of B. thuringiensis strains. df, degree of freedom; Sig. F change, significance for the change in R-square for F-test.
| Variables | Parameters | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | |||||||
| Standardized coefficient | t | p | Standardized coefficient | t | p | Standardized coefficient | t | p | |
| cry1B | 0.048 | 0.324 | 0.749 | 0.107 | 0.773 | 0.447 | 0.117 | 0.822 | 0.419 |
| cry2Aa1 | 0.770 | 4.369 | 0.000 | 0.786 | 4.794 | 0.000 | 0.770 | 4.551 | 0.000 |
| cry7 | −0.008 | −0.054 | 0.957 | −0.019 | −0.138 | 0.891 | −0.031 | −0.211 | 0.835 |
| cry9Ea | 0.442 | 2.822 | 0.010 | 0.459 | 3.154 | 0.004 | 0.434 | 2.888 | 0.008 |
| cry22 | 0.002 | 0.016 | 0.988 | 0.019 | 0.143 | 0.888 | 0.058 | 0.414 | 0.683 |
| R square | 0.559 | 0.619 | 0.594 | ||||||
| df1 | 5 | 5 | 5 | ||||||
| df2 | 23 | 23 | 23 | ||||||
| Sig. F Change | 0.001 | 0.000 | 0.001 | ||||||
Since all these strains were positive for cry1 genes, the expression levels of cry1 genes were assayed (Table 5). There were 5 strains which showed high expression levels of cry1 genes (Ct values 17.83–22.76); all these 5 strains exhibited high levels of insecticidal activity (mortality rates >60% or 80%, 25 mg/L global Cry protein for 24 or 72 h). Nine strains which showed low expression levels of cry1 genes (Ct values 38.25–39.98) exhibited low levels of insecticidal activity. However, two strains, i.e., TT47 and TT49, which belonged to the strains with low expression level of cry1 genes, were found to have high insecticidal activities (mortality rates > 80%). A Mann-Whitney U test was used to determine if the Bt strains with high expression levels, i.e., low Ct values, of cry1 genes showed higher insecticidal activities than those with low expression levels, i.e., high Ct values, of cry1 genes. For such a test, Ct values <25 were recorded as high expression levels, while those >25 were recorded as low expression levels. Statistical analysis revealed that strains with high expression levels of cry1 genes exhibited significantly higher insecticidal activities against P. xylostella than those with low expression levels of cry1 genes (exact significance < 0.05 at all time points, df = 14). The above data implicate that only high expression level of cry1 genes plays a key role to determine the insecticidal activity of Bt strains against P. xylostella larvae.
Table 5.
Comparison of expression level of cry1 and insecticidal activity of B. thuringiensis strains. All data were expressed as mean ± standard deviation (n = 3); Ct, cycle threshold.
| Strain | Mortality (%) | Expression level of cry1 genes (Ct value) | ||
|---|---|---|---|---|
| 25 mg/L (global Cry protein) | ||||
| 24 h | 48 h | 72 h | ||
| TT30 | 93.33 ± 11.55 | 100.00 ± 0.00 | 100.00 ± 0.00 | 17.83 |
| TT59 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 18.92 |
| TT48 | 60.00 ± 10.00 | 82.67 ± 6.35 | 88.04 ± 11.96 | 19.00 |
| TT50 | 83.33 ± 15.28 | 100.00 ± 0.00 | 100.00 ± 0.00 | 22.73 |
| TT12 | 79.30 ± 10.35 | 96.33 ± 6.35 | 96.14 ± 6.68 | 22.76 |
| TT57 | 2.85 ± 4.93 | 3.58 ± 3.10 | 0.15 ± 0.13 | 38.25 |
| TT23 | 3.33 ± 5.77 | 10.07 ± 6.26 | 20.10 ± 6.57 | 39.59 |
| TT4 | 3.33 ± 5.77 | 2.33 ± 4.04 | 4.62 ± 4.00 | 39.79 |
| TT56 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.07 ± 0.13 | 39.80 |
| TT47 | 74.53 ± 9.47 | 84.00 ± 10.39 | 83.42 ± 10.50 | 39.86 |
| TT3 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 39.87 |
| TT11 | 2.28 ± 3.94 | 2.00 ± 0.00 | 0.00 ± 0.00 | 39.88 |
| TT19 | 4.93 ± 4.31 | 17.01 ± 5.74 | 24.54 ± 9.62 | 39.91 |
| TT49 | 80.00 ± 10.00 | 96.67 ± 5.77 | 96.01 ± 6.91 | 39.98 |
4. Discussion
For decades, worldwide screening followed by isolation and characterization of new Bt strains have been undertaken to find out strains with high insecticidal activities [27]. Screening the environment for novel Bt strains with high insecticidal activity has become one of the strategies for insect resistance management [28]. On the other hand, considerable genetic diversity among Bt strains has been reported. The diversity of Bt strains also facilitates the isolation of new types of insecticidal genes [29]. In this regard, a number of techniques have been used to discriminate Bt isolates. PFGE is generally considered as an accurate and reproducible method for typing clinically relevant bacteria, and has been used for typing Bt strains [26,30]. Another typing method, RAPD, which is fast and simple, also has been applied for the discrimination of Bt strains [13]. Although RAPD has been successfully used for the differentiation of Bt isolates based on their location of origin, further investigation is still needed to understand their insecticidal spectrum [13]. For PFGE, it has been reported that the clonal structure of Bt isolates determined by PFGE may not correlate with their cry gene content [10], however, different conclusions have been made by Gaviria Rivera and Priest [26].
In this study, both PFGE and RAPD methods were effective and informative in differentiation of Bt strains collected in Taiwan. This is evidenced by the fact that these two methods are capable to discriminate most of the Bt strains. When these two methods were combined for the analysis of 60 Bt strains isolated in Taiwan, only four strains could not be discriminated. Thus, Bt strains in Taiwan are highly diversified in their genome patterns. Such highly genetic diversity may be due to high level of heterogeneity within the chromosomal gene organization for strains. Nevertheless, since strains with individual genotypes showed distinct insecticidal activity against P. xylostella, the genotyping data may allow us to identify the Bt strains, either newly isolated strains or strains which have been used for bioinsecticide production.
In general, early instar larvae were more susceptible to Cry proteins in many insects than later instar larvae. In this regard, the larval instar used in conducting the bioassays should be considered by not only the most susceptible stage, but also with respect to other factors, such as low mortality in negative controls [31]. Even though older larvae are less susceptible to Cry proteins, to assay the insecticidal activity of Bt strains, we used the third instar larvae since such larvae may allow us to select the Bt strains with high insecticidal activity. In addition, the assay conditions at 25 mg/L of global Cry protein with 24 h incubation time were used to find the strains with high insecticidal activity since under such conditions, the insecticidal activity of different Bt strains could be discriminated effectively (Figure 1). Furthermore, since general stress of larvae, such as nutritively unbalanced food for larvae due to unsuitable host plants, may affect larval vulnerability to Bt treatment, and leads to high mortality rates in larval populations [32], the factors of general stress were also considered. P. xylostella is considered as one of the most destructive insect pests of cabbage, and effects of insecticides on P. xylostella have been evaluated with the use of cabbage [33,34]. In this study, the insecticidal activity of Bt strains was also evaluated using cabbage, one of the host plants of P. xylostella. Thus, the possibility that general stress of larvae, such as nutritively unbalanced food, affects the mortality, is little. Under such conditions, 19 of the 60 Bt strains isolated in Taiwan were found to have higher insecticidal activities than those of the reference strains.
The cry gene content of Bt strain may be useful for the prediction of its insecticidal potential, and PCR-based identifications of cry genes have been developed to help the screening process [4,8,27]. A previous study revealed that cry1 and cry2 were the most abundant genes in the Bt strains isolated in Taiwan [11]. In this study, all strains were found to be positive for cry1 genes. Also, most of the strains with high insecticidal activity were positive for cry2Aa1. Our results were in agreement with those described by Chen et al. [11]. Moreover, we found that cry2Aa1 gene was positively correlated with the insecticidal activity of Bt strains (Table 4). Regarding cry1 genes, although all Bt strains were positive for cry1 genes, their insecticidal activities varied. Ferrandis et al. [8] reported that the insecticidal toxicity could be related to gene content in most cases, however, one strain without cry1 genes, showed high toxicity against P. xylostella. A cry gene, detected by PCR, can be interrupted, mutated, or under control by a defective promoter; the corresponding Cry protein may not be present or present at reduced levels, therefore, contributing minimally to the toxicity [8]. The variety in the expression levels of individual cry genes also weakens the correlation between cry gene content and the toxicity of Bt strains [4].
Thus, the expression levels of cry1 genes in our Bt isolates collected in Taiwan were assayed. All Bt strains, which showed high expression levels of the cry1 genes, exhibited high levels of insecticidal activity. However, two strains, i.e., TT47 and TT49, belonging to the same genotypes, as strains TT48 and TT50, also exhibited high insecticidal activities despite of their low expression levels of the cry1 genes (Table 5). Thus, there might be some other cry-type genes active to P. xylostella existing in these Bt isolates. Furthermore, other factors, such as β-exotoxins, phospholipases, proteases, chitinases and the secreted VIPs (vegetative insecticidal proteins), are possibly involved in the complete pathogenic effect of a strain [4]. As for the insecticidal activity of Cyt toxins, a review from Frankenhuyzen [14] revealed that results of the activity of Cyt1Aa against P. xylostella are conflicting, while for Cyt2Aa, it was not tested against P. xylostella. The synergistic interactions of Cry toxins also contribute to the toxicity to a specific insect [35]. In this regard, Porcar and Juárez-Pérez [4] have suggested that for different Cry toxins, the expected dose needed to kill 50% of the insects need to be calculated if the relative proportions of these toxins and the individual toxicity of toxins were known. Nevertheless, although Bt strains used in this study were not checked for the presence of VIP and Cyt proteins, as well as the protein concentrations of the five Cry toxins shown in Table 4, based on the results of this study, it is possible that Bt strains with high expression levels of cry1 genes should be the strains with high insecticidal activity against P. xylostella.
5. Conclusions
In conclusion, for the isolation and characterization of novel Bt strains with high insecticidal activity, both the RAPD and PFGE methods are effective and informative in differentiation of Bt strains. Concerning the insecticidal activity against P. xylostella, a considerable number of the Bt strains isolated in Taiwan were found to have high insecticidal activity, as compared to those of the reference strains isolated from imported bioinsecticides. Also, the presence of cry2Aa1 gene determined by PCR may be used as a reference marker to predict the insecticidal activity, and only high expression level of the cry1 genes plays a key role to determine the insecticidal activity of Bt strains against P. xylostella.
Acknowledgments
The authors would like to thank the National Science Council, Taipei, Taiwan, for the support of this work. The project numbers are NSC91-2313-B005-054 and NSC101-2313-B241-004-MY2. Bacterial strains used in this study were generously provided by Ching-Chou Tzeng and Suey-Sheng Kao, Biopesticide Department, Taiwan Agriculture Chemical and Toxic Substances Research Institute (TACTRI), Taichung, Taiwan.
Author Contributions
Ming-Lun Chen, Pin-Hsin Chen, and Jen-Chieh Pang designed and performed the experiments; Jen-Chieh Pang, Chia-Wei Lin, Chin-Fa Hwang, and Hau-Yang Tsen analyzed and interpreted the data; Chia-Wei Lin drafted the manuscript; Hau-Yang Tsen was the project leader and revised it. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Schnepf E., Crickmore N., van Rie J., Lereclus D., Baum J., Feitelson J., Zeigler D.R., Dean D.H. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Höfte H., Whiteley H.R. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol. Rev. 1989;53:242–255. doi: 10.1128/mr.53.2.242-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bravo A., Gómez I., Porta H., García-Gómez B.I., Rodriguez-Almazan C., Pardo L., Soberón M. Evolution of Bacillus thuringiensis Cry toxins insecticidal activity. Microb. Biotechnol. 2013;6:17–26. doi: 10.1111/j.1751-7915.2012.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porcar M., Juárez-Pérez V. PCR-based identification of Bacillus thuringiensis pesticidal crystal genes. FEMS Microbiol. Rev. 2003;26:419–432. doi: 10.1111/j.1574-6976.2003.tb00624.x. [DOI] [PubMed] [Google Scholar]
- 5.Henderson I., Yu D., Turnbull P.C. Differentiation of Bacillus anthracis and other “Bacillus cereus group” bacteria using IS231-derived sequences. FEMS Microbiol. Lett. 1995;128:113–118. doi: 10.1111/j.1574-6968.1995.tb07509.x. [DOI] [PubMed] [Google Scholar]
- 6.Crickmore N., Zeigler D.R., Feitelson J., Schnepf E., van Rie J., Lereclus D., Baum J., Dean D.H. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 1998;62:807–813. doi: 10.1128/mmbr.62.3.807-813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crickmore N., Baum J., Bravo A., Lereclus D., Narva K., Sampson K., Schnepf E., Sun M., Zeigler D.R. Bacillus thuringiensis toxin nomenclature. [(accessed on 15 April 2014)]. Available online: http://www.btnomenclature.info/
- 8.Ferrandis M.D., Juárez-Pérez V.M., Frutos R., Bel Y., Ferré J. Distribution of cryl, cryll and cryV genes within Bacillus thuringiensis isolates from Spain. Syst. Appl. Microbiol. 1999;22:179–185. doi: 10.1016/S0723-2020(99)80064-2. [DOI] [Google Scholar]
- 9.Kuo W.S., Chak K.F. Identification of novel cry-type genes from Bacillus thuringiensis strains on the basis of restriction fragment length polymorphism of the PCR-amplified DNA. Appl. Environ. Microbiol. 1996;62:1369–1377. doi: 10.1128/aem.62.4.1369-1377.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Świȩcicka I., Mahillon J. The clonal structure of Bacillus thuringiensis isolates from north-east Poland does not correlate with their cry gene diversity. Environ. Microbiol. 2005;7:34–39. doi: 10.1111/j.1462-2920.2004.00662.x. [DOI] [PubMed] [Google Scholar]
- 11.Chen F.C., Tsai M.C., Peng C.H., Chak K.F. Dissection of cry gene profiles of Bacillus thuringiensis isolates in Taiwan. Curr. Microbiol. 2004;48:270–275. doi: 10.1007/s00284-003-4195-1. [DOI] [PubMed] [Google Scholar]
- 12.Mohan M., Gujar G.T. Toxicity of Bacillus thuringiensis strains and commercial formulations to the diamondback moth, Plutella xylostella (L.) Crop Prot. 2001;20:311–316. doi: 10.1016/S0261-2194(00)00157-5. [DOI] [Google Scholar]
- 13.Kumar D., Chaudhary K., Boora K.S. Characterization of native Bacillus thuringiensis strains by PCR-RAPD based fingerprinting. Indian J. Microbiol. 2010;50:27–32. doi: 10.1007/s12088-009-0011-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frankenhuyzen K.V. Insecticidal activity of Bacillus thuringiensis crystal proteins. J. Invertebr. Pathol. 2009;101:1–16. doi: 10.1016/j.jip.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Pang J.C., Chen M.L., Ho Y.C., Yang C.Y., Tzeng C.C., Kao S.S., Tsen H.Y. Effect of fermentation conditions on the enterotoxigenicity, cytotoxicity and pesticidal activity of Bacillus thuringiensis strains isolated in Taiwan. Bioresour. Technol. 2010;101:1871–1876. doi: 10.1016/j.biortech.2009.09.090. [DOI] [PubMed] [Google Scholar]
- 16.Abbott W. A method for computing the effectiveness of an insecticide. J. Econ. Entomol. 1925;18:265–267. [Google Scholar]
- 17.Kolstø A.B., Gronstad A., Oppegaard H. Physical map of the Bacillus cereus chromosome. J. Bacteriol. 1990;172:3821–3825. doi: 10.1128/jb.172.7.3821-3825.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ausubel F.M., Kingstone R.E., More D.D., Seidman J.G., Smith J.A., Struhl K. Current Protocols in Molecular Biology. John Wiley & Sons; New York, NY, USA: 1987. [Google Scholar]
- 19.Bravo A., Sarabia S., Lopez L., Ontiveros H., Abarca C., Ortiz A., Ortiz M., Lina L., Villalobos F.J., Pena G., et al. Characterization of cry genes in a Mexican Bacillus thuringiensis strain collection. Appl. Environ. Microbiol. 1998;64:4965–4972. doi: 10.1128/aem.64.12.4965-4972.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Céron J., Covarrubis L., Quintero R., Ortiz A., Ortiz M., Aranda E., Lina L., Bravo A. PCR analysis of the cryI insecticidal crystal family genes from Bacillus thuringiensis. Appl. Environ. Microbiol. 1994;60:353–356. doi: 10.1128/aem.60.1.353-356.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juárez-Pérez V.M., Ferrandis M.D., Frutos R. PCR-based approach for detection of novel Bacillus thuringiensis cry genes. Appl. Environ. Microbiol. 1997;63:2997–3002. doi: 10.1128/aem.63.8.2997-3002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-Dov E., Zaritsky A., Dahan E., Barak Z., Sinai R., Manasherob R., Khamraev A., Troitskaya E., Dubitsky A., Berezina N., et al. Extended screening by PCR for seven cry-group genes from field-collected strains of Bacillus thuringiensis. Appl. Environ. Microbiol. 1997;63:4883–4890. doi: 10.1128/aem.63.12.4883-4890.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ejiofor A.O., Johnson T. Physiological and molecular detection of crystalliferous Bacillus thuringiensis strains from habitats in the South Central United States. J. Ind. Microbiol. Biotechnol. 2002;28:284–290. doi: 10.1038/sj.jim.7000244. [DOI] [PubMed] [Google Scholar]
- 24.Ben-Dov E., Wang Q., Zaritsky A., Manasherob R., Barak Z., Schneider B., Khamraev A., Baizhanov M., Glupov V., Margalith Y. Multiplex PCR screening to detect cry9 genes in Bacillus thuringiensis strains. Appl. Environ. Microbiol. 1999;65:3714–3716. doi: 10.1128/aem.65.8.3714-3716.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ibarra J.E., del Rincón M.C., Ordúz S., Noriega D., Benintende G., Monnerat R., Regis L., de Oliveira C.M., Lanz H., Rodriguez M.H., et al. Diversity of Bacillus thuringiensis strains from Latin America with insecticidal activity against different mosquito species. Appl. Environ. Microbiol. 2003;69:5269–5274. doi: 10.1128/AEM.69.9.5269-5274.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaviria Rivera A.M., Priest F.G. Pulsed field gel electrophoresis of chromosomal DNA reveals a clonal population structure to Bacillus thuringiensis that relates in general to crystal protein gene content. FEMS Microbiol. Lett. 2003;223:61–66. doi: 10.1016/S0378-1097(03)00347-1. [DOI] [PubMed] [Google Scholar]
- 27.Martínez C., Ibarra J.E., Caballero P. Association analysis between serotype, cry gene content, and toxicity to Helicoverpa armiger larvae among Bacillus thuringiensis isolates native to Spain. J. Invertebr. Pathol. 2005;90:91–97. doi: 10.1016/j.jip.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Ammouneh H., Harba M., Idris E., Makee H. Isolation and characterization of native Bacillus thuringiensis isolates from Syrian soil and testing of their insecticidal activities against some insect pests. Turk. J. Agric. For. 2011;35:421–431. [Google Scholar]
- 29.Kaur S. Molecular approaches for identification and construction of novel insecticidal genes for crop protection. World J. Microbiol. Biotechnol. 2006;22:233–253. doi: 10.1007/s11274-005-9027-y. [DOI] [Google Scholar]
- 30.Carlson C.R., Caugant D.A., Kolstø A.B. Genotypic diversity among Bacillus cereus and Bacillus thuringiensis strains. Appl. Environ. Microbiol. 1994;60:1719–1725. doi: 10.1128/aem.60.6.1719-1725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asano S., Maruyama T., Iwasa T., Seki A., Takahashi M., Soares G.G., Jr. Evaluation of biological activity of Bacillus thuringiensis test samples using a diet incorporation method with diamondback moth, Plutella xylostella (Linnaeus) (Lepidoptera: Yponomeutidae) Appl. Entomol. Zoolog. 1993;28:513–524. [Google Scholar]
- 32.Bauce É., Bidon Y., Berthiaume R. Effects of food nutritive quality and Bacillus thuringiensis on feeding behaviour, food utilization and larval growth of spruce budworm Choristoneura fumiferana (Clem.) when exposed as fourth-and sixth-instar larvae. Agric. For. Entomol. 2002;4:57–70. doi: 10.1046/j.1461-9563.2002.00123.x. [DOI] [Google Scholar]
- 33.Hill T.A., Foster R.E. Effect of insecticides on the diamondback moth (Lepidoptera: Plutellidae) and its parasitoid Diadegma insulare (Hymenoptera: Ichneumonidae) J. Econ. Entomol. 2000;93:763–768. doi: 10.1603/0022-0493-93.3.763. [DOI] [PubMed] [Google Scholar]
- 34.Ayalew G. Comparison of yield loss on cabbage from Diamondback moth, Plutella xylostella L. (Lepidoptera: Plutellidae) using two insecticides. Crop Prot. 2006;25:915–919. doi: 10.1016/j.cropro.2005.12.001. [DOI] [Google Scholar]
- 35.Poncet S., Delécluse A., Klier A., Rapoport G. Evaluation of synergistic interactions among the CryIVA, CryIVB, and CryIVD toxic components of B. thuringiensis subsp. israelensis crystals. J. Invertebr. Pathol. 1995;66:131–135. doi: 10.1006/jipa.1995.1075. [DOI] [Google Scholar]