Abstract
Objective
To address the problem of mapping local laboratory terminologies to Logical Observation Identifiers Names and Codes (LOINC). To study different ontology matching algorithms and investigate how the probability of term combinations in LOINC helps to increase match quality and reduce manual effort.
Materials and methods
We proposed two matching strategies: full name and multi-part. The multi-part approach also considers the occurrence probability of combined concept parts. It can further recommend possible combinations of concept parts to allow more local terms to be mapped. Three real-world laboratory databases from Taiwanese hospitals were used to validate the proposed strategies with respect to different quality measures and execution run time. A comparison with the commonly used tool, Regenstrief LOINC Mapping Assistant (RELMA) Lab Auto Mapper (LAM), was also carried out.
Results
The new multi-part strategy yields the best match quality, with F-measure values between 89% and 96%. It can automatically match 70–85% of the laboratory terminologies to LOINC. The recommendation step can further propose mapping to (proposed) LOINC concepts for 9–20% of the local terminology concepts. On average, 91% of the local terminology concepts can be correctly mapped to existing or newly proposed LOINC concepts.
Conclusions
The mapping quality of the multi-part strategy is significantly better than that of LAM. It enables domain experts to perform LOINC matching with little manual work. The probability of term combinations proved to be a valuable strategy for increasing the quality of match results, providing recommendations for proposed LOINC conepts, and decreasing the run time for match processing.
Keywords: LOINC, Pre-coordination, Probability, Ontology Matching, Translation
Background and significance
There is increasing use of standard terminologies (ontologies1) for electronic health records2–4 and public health surveillance.5–10 One of these standard terminologies for laboratory and clinical observations is Logical Observation Identifiers Names and Codes (LOINC),11 12 which is used to facilitate information exchange —for example, for reporting infectious diseases to the Centers for Disease Control and Prevention (CDC). LOINC is a national standard in several countries such as the USA, Canada, Germany and Taiwan.13 However, hospitals usually have their own local laboratory terminologies,14 often focusing on certain aspects such as laboratory results,5 9 10 15–18 clinical reports,19 and radiology reports.20 21 This situation has led to a growing need to determine mapping between local and standard terminologies22 in order to correctly share data. Several semiautomatic LOINC matching algorithms and tools have been proposed to assist domain experts,4 7 9 10 16 19 23 but matching from local terminologies to LOINC is still a resource-intensive and time-consuming problem.9
Each LOINC concept (term) comprises a fully specified name (ie, the formal LOINC name11) and an identity code.24 Each fully specified name consists of six parts —for example, Component/Analyte, Kind of Property, etc (see table 1 for several examples)—separated by colons (eg, ‘Rotavirus Ag:ACnc:Pt:Stool:Ord:Aggl’). LOINC is primarily a pre-coordinated terminology14 25 that a priori defines which combinations of atomic concepts are allowed. For instance, the six-part combination of ‘Rotavirus Ag’, ‘ACnc’, ‘Pt’, ‘Stool’, ‘Ord’ and ‘Aggl’ is meaningful for disease surveillance and has thus been included in LOINC with a unique code (5879-2). Its long common name is ‘Rotavirus Ag [Presence] in Stool by Agglutination’. For improved coverage and completeness, there is also the possibility to include additional concepts (post-coordination) in LOINC. In particular, users or developers can suggest new combinations of existing atomic concepts (proposed LOINC concepts) to the development organization to be considered for inclusion in LOINC.4 23 24 26 27
Table 1.
An example LOINC table including five concepts (terms)
| LOINC code | Component/Analyte (COM) | Kind of property (PRO) | Time aspect (TIM) | Sample type (SAM) | Scale type (SCA) | Method type (MET) |
|---|---|---|---|---|---|---|
| 5879-2 | Rotavirus Ag | ACnc | Pt | Stool | Ord | Aggl |
| 25754-3 | Rotavirus Ab.IgG | ACnc | Pt | Ser | Qn | EIA |
| 25593-5 | Rotavirus Ab.IgG | ACnc | Pt | Ser | Ord | |
| 71701-7 | Rotavirus dsRNA | Prid | Pt | Stool | Nom | PAGE |
| 8012-7 | Rotavirus dsRNA | ACnc | Pt | Stool | Ord | Probe.amp.tar |
LOINC, Logical Observation Identifiers Names and Codes.
Existing approaches to mapping local terminologies with LOINC rely on extensive preprocessing of the local terminologies to facilitate automated match processing. Common preprocessing steps include normalization of local terminologies,10 18 adoption of synonyms,16 23 28 resolution of abbreviations,4 10 and augmenting local terms with definitions and annotations.9 29 Existing tools for matching the preprocessed local terminologies with LOINC usually calculate concept similarities by using the fully specified names —for example, ‘Rotavirus Ag:ACnc:Pt:Stool:Ord:Aggl’ (5879-2). For example, most previous studies used the Regenstrief LOINC Mapping Assistant (RELMA),4 5 9 16 which applies such a full-name strategy based on restrictive exact name matching. Only a few studies applied linguistic matching,10 19 where a match can also be identified for similar but slightly different names. Fidahussein and Vreeman investigated corpus-based matching to leverage a collection of previously matched terminologies.18 Lau et al23 considered multi-part matching with separate name matching for each part; such an approach can avoid false matches due to a name similarity in unrelated LOINC parts (eg, ‘SP’ is the abbreviation of ‘species’ in Component/Analyte as well as for ‘sputum’ in Sample Type). Moreover, Bodenreider proposed a multi-part matching technique for comparison of laboratory tests between LOINC and SNOMED CT (Systematized Nomenclature of Medicine Clinical Terms).30
Although previous approaches have already proved quite useful, they still require a substantial amount of human effort, with 11–31% of the local terms requiring manual assignment to corresponding LOINC concepts.7 16 18 Moreover, 6–37% of the local concepts could not be mapped at all, since they do not exist in LOINC (ie, unmappable/uncodable concepts).5 7 16 31 A further problem is internationalization—that is, support of local terminologies in different languages. While RELMA supports translations between some languages such as English, German and Simplified Chinese,32 there are more than 26 000 LOINC users from 157 countries,33 and therefore many languages are not yet supported.
Objective
We aimed to improve mapping of local terminologies to LOINC with respect to the amount of the remaining manual work. We built on recent advances in automated ontology matching (alignment), an area that has seen a large amount of research and development effort. 34, 35 Proposed match approaches and prototypes combine a variety of similarity measures considering the name, description and structural neighborhood of concepts, or further background information.36–38 In particular, we used the state-of-the-art match tool GOMMA (Generic Ontology Matching and Mapping Management).39 We also devised and evaluated a new multi-part match strategy. Specifically, we made the following contributions.
We studied full-name matching using the fully specified name in GOMMA to find the most applicable match approach for laboratory data. We considered different preprocessing, matching and selection methods as well as language translations.
We proposed a novel multi-part matching strategy that uses the occurrence probability of combinations of atomic LOINC concepts as a filter. The approach can also provide recommendations for possible (new) combinations of atomic concepts that could match unmapped local terms. To our knowledge this is the first work to automatically provide such recommendations, thereby supporting post-coordination.
We evaluated both strategies by comparing them with the RELMA Lab Auto Mapper (LAM) using datasets from three Taiwanese hospitals. The results show that multi-part matching with probability filtering can significantly outperform other approaches with respect to quality and execution time.
Materials and methods
We first describe the local laboratory terminologies used. We then define the assumed data model and present the full name and multi-part matching strategies. Finally, we outline our evaluation methods.
Materials
Because of a request from the Taiwanese Center of Diseases Control (Taiwan CDC), in the pilot project, we focused on their 49 most important pathogen-related tests for notifiable disease surveillance; the 49 pathogens are shown in box 1. Hence, only the tests related to these pathogens are considered in the study. To validate our algorithms, we used three local laboratory terminologies from Taiwanese hospitals. Two terminologies come from medical centers: Shin Kong Wu Ho-Su Memorial Hospital (SKH) with 919 beds and Mackay Memorial Hospital Taipei branch (Mackay) with 1118 beds. The third comes from a regional hospital, the National Taiwan University hospital Hsin-Chu branch (NTU) with 818 beds. The hospitals use different commercial laboratory information systems, TrakCare Lab, Amesdata and Tatung, which are also used in several other Taiwanese hospitals. Since the three systems do not have unified codes corresponding to LOINC codes, we collected their laboratory terminologies, including ‘test order names’, ‘sample type names’, ‘abnormal results and definitions’, ‘normal results and definitions’, ‘measurement units’ and ‘test device or reagent names’. In April 2013, the SKH, Mackay and NTU contained 452, 245 and 116 unique concepts, respectively, related to these pathogens. All the laboratory terminologies were converted into the LOINC data model.23 40 The pilot study started in May 2012. We used LOINC 2.38 (extracted through UMLS41 MetamorphoSys 2012AA for additional synonyms from Metathesaurus) released in December 2011 and took into account all LOINC concepts with class type ‘lab’ (45 896 LOINC codes).
Reference mappings (gold standard) between laboratory terminologies and LOINC were created manually by three independent experts familiar with these local laboratory concepts. We manually examined the accuracy and consistency of the preliminary reference mappings of each expert. In the case of any mapping inconsistency, these three experts and two more external experts were invited to discuss it conjointly and agree to reasonable matches and consistent correspondences. Note that the mappings also include unmappable concepts.
Data model
We assumed that an ontology (terminology)  consists of
consists of 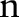 sub-ontologies (parts)
sub-ontologies (parts)  — for example, LOINC has six parts
— for example, LOINC has six parts  . Each part contains a number of atomic concepts,
. Each part contains a number of atomic concepts, 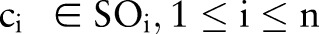 , which can be used to build combined concepts,
, which can be used to build combined concepts, 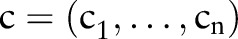 — for example, ‘Rotavirus Ag’ from COM part, ‘Aggl’ from MET part, etc form the combined concept LOINC 5879-2. Thus, it is possible to build up to
— for example, ‘Rotavirus Ag’ from COM part, ‘Aggl’ from MET part, etc form the combined concept LOINC 5879-2. Thus, it is possible to build up to  combined concepts. For instance, the multiplication of the number of each LOINC part size
combined concepts. For instance, the multiplication of the number of each LOINC part size  results in more than 37 million combined concepts (for LOINC 2.38). However, only commonly used combinations
results in more than 37 million combined concepts (for LOINC 2.38). However, only commonly used combinations 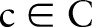 are defined as concepts in
are defined as concepts in  (pre-coordinated concepts)—that is, only 68 350 combined concepts are allowed in LOINC 2.38. Thus an ontology can be described as follows:
(pre-coordinated concepts)—that is, only 68 350 combined concepts are allowed in LOINC 2.38. Thus an ontology can be described as follows: 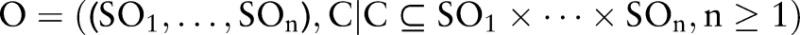 . For instance, LOINC is described as LOINC=((COM, PRO, TIM, SAM, SCA, MET), C), where
. For instance, LOINC is described as LOINC=((COM, PRO, TIM, SAM, SCA, MET), C), where  contains the 68 350 combined concepts. Other combinations
contains the 68 350 combined concepts. Other combinations 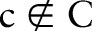 are named as proposed (post-coordinated) concepts. Such combinations present practical events and can be suggested as new concepts to the ontology's maintenance organization. According to table 1,
are named as proposed (post-coordinated) concepts. Such combinations present practical events and can be suggested as new concepts to the ontology's maintenance organization. According to table 1, 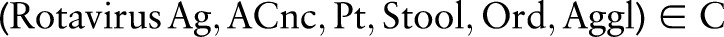 is a pre-coordinated concept in LOINC, composed of the atomic concepts Rotavirus Ag ∈ COM,
is a pre-coordinated concept in LOINC, composed of the atomic concepts Rotavirus Ag ∈ COM, 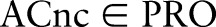 , etc. In contrast, the practically useful combination
, etc. In contrast, the practically useful combination 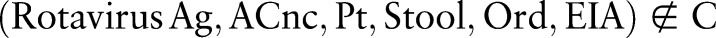 is a proposed concept and could be submitted to the Regenstrief Institute for future inclusion in LOINC.
is a proposed concept and could be submitted to the Regenstrief Institute for future inclusion in LOINC.
Ontology matching
In this section, we present our two matching strategies: full name and multi-part. Figure 1 illustrates the general situation. Full name matching (blue line) directly compares names of combined concepts from source and target ontology ( and
and  ). For example, the local concept ‘ROTAVIRUS:ACNC:PT:St:Ord:AGGL’ matches the LOINC concept ‘Rotavirus Ag:ACnc:Pt:Stool:Ord:Aggl’. The multi-part matching (red dash line) aligns names of atomic concepts from source and target sub-ontologies to finally find correspondences between combined concepts of
). For example, the local concept ‘ROTAVIRUS:ACNC:PT:St:Ord:AGGL’ matches the LOINC concept ‘Rotavirus Ag:ACnc:Pt:Stool:Ord:Aggl’. The multi-part matching (red dash line) aligns names of atomic concepts from source and target sub-ontologies to finally find correspondences between combined concepts of  and
and  . For example, the local COM concept ‘ROTAVIRUS’ matches the LOINC COM concept ‘ROTAVIRUS Ag’, the local SAM concept ‘St’ matches the LOINC SAM concept ‘Stool’, etc. In general, ontology matching is the process of determining a set of semantic correspondences (mapping) between concepts of two related ontologies
. For example, the local COM concept ‘ROTAVIRUS’ matches the LOINC COM concept ‘ROTAVIRUS Ag’, the local SAM concept ‘St’ matches the LOINC SAM concept ‘Stool’, etc. In general, ontology matching is the process of determining a set of semantic correspondences (mapping) between concepts of two related ontologies  and
and  (eg, the local terminology and LOINC):
(eg, the local terminology and LOINC): 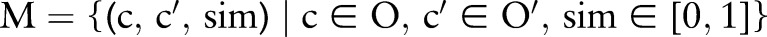 . A correspondence is a connection between two combined concepts
. A correspondence is a connection between two combined concepts  and
and 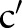 , whereby the strength of the connection is represented by a similarity value
, whereby the strength of the connection is represented by a similarity value 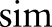 ranging from 0 to 1.
ranging from 0 to 1.
Figure 1.
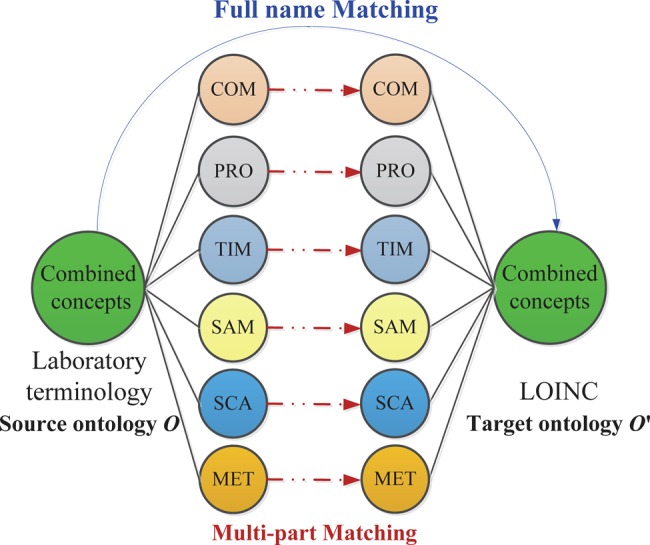
Full name and multi-part matching for Logical Observation Identifiers Names and Codes (LOINC).
Full name matching
GOMMA is freely available for research purposes and can be accessed via http://dbs.uni-leipzig.de/de/gomma. It has achieved top results in the 2012 benchmarking competition OAEI (Ontology Alignment Evaluation Initiative).42
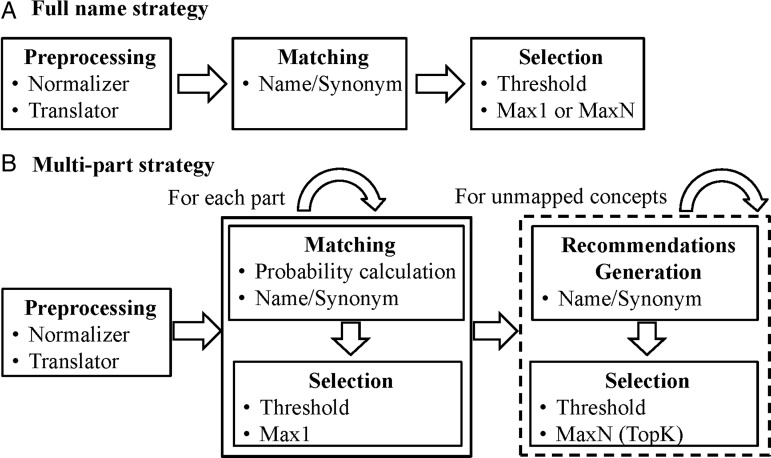
43 Our full name matching is a linguistic matching approach using GOMMA based on comparing names and synonyms of combined concepts. Figure 2A shows the overall matching process. Before matching, names and synonyms of concepts are preprocessed, including normalization to lowercase and replacement of common delimiters, abbreviations and synonyms. (The same preprocessed local concepts’ names were also imported to RELMA LAM to ensure comparability.) Some local concepts contain parts in Traditional Chinese. We translated these parts to English using MyMemory API44 and replaced them with translated English names. During the actual matching, we applied a name/synonym matcher determining the maximal string similarity for the names and synonyms per (combined) concept pair. We used trigram as the string similarity measure. Trigram tokenizes strings into tokens of length three and considers the overlap of trigram sets to compute a similarity value between two strings using the Dice metric: 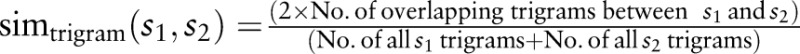 .45
46 For instance, the number of overlapping trigram tokens between ‘rotavirus acnc pt st ord aggl’ (including 31 tokens, eg, ‘rot’, ‘ota’, ‘tav’,…) and ‘rotavirus ag acnc pt stool ord aggl’ (including 37 tokens, eg, ‘rot’, ‘ota’, …) is 29. Hence, the trigram similarity value is
.45
46 For instance, the number of overlapping trigram tokens between ‘rotavirus acnc pt st ord aggl’ (including 31 tokens, eg, ‘rot’, ‘ota’, ‘tav’,…) and ‘rotavirus ag acnc pt stool ord aggl’ (including 37 tokens, eg, ‘rot’, ‘ota’, …) is 29. Hence, the trigram similarity value is 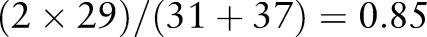 .
.
Figure 2.
The workflows of full name and multi-part strategies.
Note that an automatic match strategy may produce a set of correspondences for each (local) source concept. To provide domain experts with a manageable set of correspondences, we first applied a simple threshold filter to discard all correspondences below a minimal threshold (eg, 0.60). We then applied a second, more sophisticated selection strategy, such as MaxN.47 For each local combined concept, MaxN selects the top N correspondences. Max1 only selects the best correspondence, while 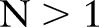 presents a larger set of match candidates, allowing expert users to select the correct match. These selection filters were applied from the source to the target ontology, since we aimed to find one or more LOINC candidate correspondences for each local concept.
presents a larger set of match candidates, allowing expert users to select the correct match. These selection filters were applied from the source to the target ontology, since we aimed to find one or more LOINC candidate correspondences for each local concept.
Multi-part matching
In contrast with full name strategy, the multi-part strategy (figure 2B) considers the atomic concepts of combined parts for matching. As depicted in figure 1 for LOINC, we matched atomic concepts from the parts COM, PRO, TIM, SAM, SCA and MET individually—that is, we applied different match approaches for the different parts and chose the best matches for each part. However, treating the atomic concepts independently from each other in the match process can cause problems. For instance, when one considers the best matches in each part for a combined concept, it is likely that the combination of these atomic concepts may not be a pre-coordinated concept in the target ontology. However, our aim was to match to pre-coordinated concepts whenever possible. For this purpose, we also considered the probability of possible combinations of target atomic concepts. We first describe how we determined the probability of combinations and then outline the overall algorithm for multi-part matching.
Each atomic concept of an ontology part has an inherent certainty, which can be derived from its occurrence probability with respect to all (pre-coordinated) combined concepts. For our example in table 1, this probability is 0.60 (three of five concepts) for the SAM concept ‘Stool’ and even 1.0 for the TIM concept ‘Pt’. We can also determine the certainty for combinations of atomic concepts from two or more ontology parts by determining their conditional occurrence probabilities. For example, the conditional probability for the COM concept ‘Rotavirus Ag’ given SAM concept ‘Stool’, P(COM=‘Rotavirus Ag’|SAM=‘Stool’) is 0.33 (one of three occurrences). In contrast, for the COM concept ‘Rotavirus Ab.IgG’, the conditional probability for the SAM concept ‘Stool’ is 0, since both atomic concepts do not occur together in any combined concept. In our multi-part match strategy described next, we used such predetermined conditional properties to focus on finding matches only for combinations of atomic concepts with non-zero conditional occurrence probabilities.
Algorithm
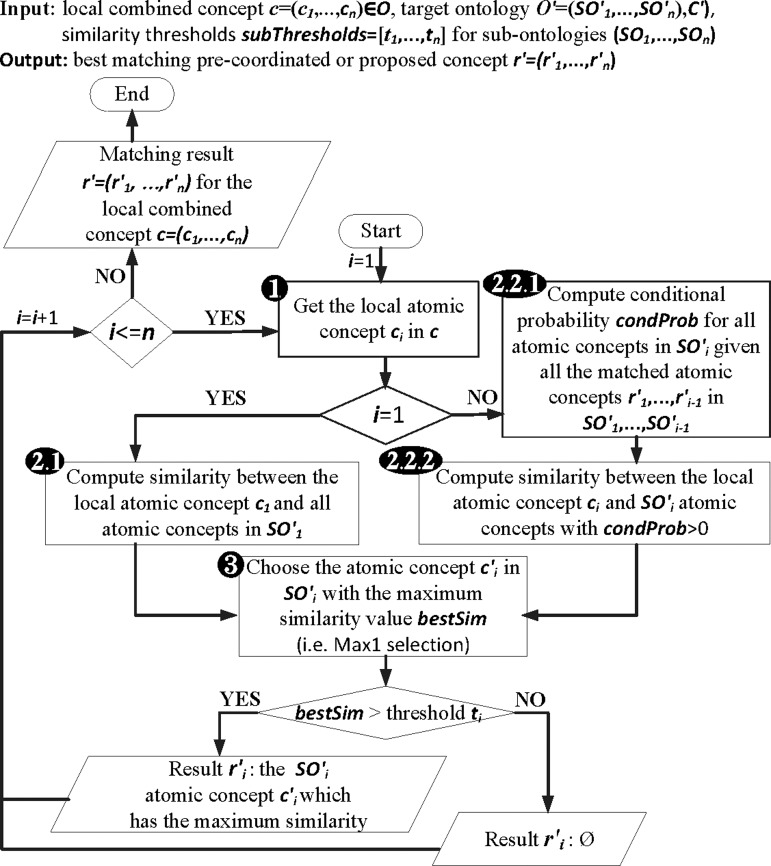
The detailed workflow of the multi-part match strategy (figure 2B) is displayed in figure 3. The pseudo code appears in online supplementary algorithm 1. The workflow (algorithm) shows how we matched the input source ontology,  , with the target ontology,
, with the target ontology,  . We computed the best matching concept in the target ontology
. We computed the best matching concept in the target ontology 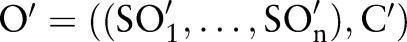 (eg, LOINC=((COM,…,MET), 68 350 combined concepts) for a given combined concept
(eg, LOINC=((COM,…,MET), 68 350 combined concepts) for a given combined concept 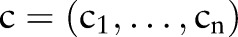 of source ontology
of source ontology  — for example, (‘ROTAVIRUS’, ‘ACNC’, ‘PT’, ‘St’, ‘Ord’, ‘AGGL’). We further used different thresholds per sub-ontology
— for example, (‘ROTAVIRUS’, ‘ACNC’, ‘PT’, ‘St’, ‘Ord’, ‘AGGL’). We further used different thresholds per sub-ontology 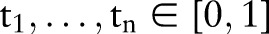 to select matches for the atomic concepts
to select matches for the atomic concepts  . The output is the best matching concept
. The output is the best matching concept 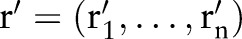 in
in  —that is,
—that is, 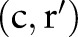 forms a correspondence in the final mapping. For example, ((‘ROTAVIRUS’, ‘ACNC’, ‘PT’, ‘St’, ‘Ord’, ‘AGGL’),(‘Rotavirus Ag’, ‘ACnc’, ‘Pt’, ‘Stool’, ‘Ord’, ‘Aggl’)) is one of the correspondences in the final mapping.
forms a correspondence in the final mapping. For example, ((‘ROTAVIRUS’, ‘ACNC’, ‘PT’, ‘St’, ‘Ord’, ‘AGGL’),(‘Rotavirus Ag’, ‘ACnc’, ‘Pt’, ‘Stool’, ‘Ord’, ‘Aggl’)) is one of the correspondences in the final mapping.
Figure 3.
Algorithm for multi-part strategy.
Multi-part matching requires the specification of an order in which the different parts are handled. In our case for 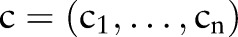 of
of  , we first matched
, we first matched 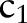 to
to 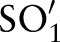 , then
, then 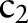 to
to 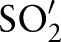 , etc. The order is provided as an input parameter. Users can manually provide (manual order (MO)) or use the following scheme (automatic order (AO)) to automatically derive an ordering. For AO, we considered the sub-ontology with the fewest concepts as
, etc. The order is provided as an input parameter. Users can manually provide (manual order (MO)) or use the following scheme (automatic order (AO)) to automatically derive an ordering. For AO, we considered the sub-ontology with the fewest concepts as 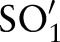 (SCA in the case of LOINC). We then iteratively selected the sub-ontology,
(SCA in the case of LOINC). We then iteratively selected the sub-ontology, 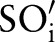 , with the minimal number of distinct concept pairs with respect to the previously selected sub-ontology,
, with the minimal number of distinct concept pairs with respect to the previously selected sub-ontology, 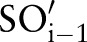 . For LOINC, we thus generated the following order—SCA-TIM-PRO-MET-SAM-COM—since the number of atomic concept pairs for SCA-TIM is smaller than for SCA-PRO, SCA-MET, etc; the number of atomic concept pairs for TIM-PRO is smaller than for TIM-MET, TIM-SAM, etc. The scheme aims at maximal filtering to reduce the search space for finding correct matches. This is because we only considered (LOINC) atomic concepts
. For LOINC, we thus generated the following order—SCA-TIM-PRO-MET-SAM-COM—since the number of atomic concept pairs for SCA-TIM is smaller than for SCA-PRO, SCA-MET, etc; the number of atomic concept pairs for TIM-PRO is smaller than for TIM-MET, TIM-SAM, etc. The scheme aims at maximal filtering to reduce the search space for finding correct matches. This is because we only considered (LOINC) atomic concepts 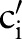 of the ith sub-ontology as a match candidate if there existed a combined concept for
of the ith sub-ontology as a match candidate if there existed a combined concept for 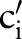 with the previously determined matches,
with the previously determined matches,  . The proposed AO ordering thus minimizes the number of match candidates,
. The proposed AO ordering thus minimizes the number of match candidates, 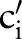 , to consider. In the evaluation, we also studied alternative orders including a manually optimized order.
, to consider. In the evaluation, we also studied alternative orders including a manually optimized order.
For each atomic concept, 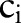 , of source concept,
, of source concept,  , the algorithm computes the best matching atomic concept,
, the algorithm computes the best matching atomic concept, 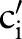 , in
, in 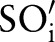 (figure 3 steps 1–3 or see online supplementary algorithm 1 lines 2–14). For the first sub-ontology (eg, SCA), we computed similarity between the local atomic concept (eg, ‘Ord’) with all the LOINC (eg, SCA) atomic concepts (eg, ‘Qn’, ‘Ord’, ‘OrdQn’, ‘Nar’, ‘Doc’, and ‘-’) using GOMMA matchers as described in the full name approach (step 2.1). However, for all further sub-ontologies, we first determined the conditional probability of the current atomic concept,
(figure 3 steps 1–3 or see online supplementary algorithm 1 lines 2–14). For the first sub-ontology (eg, SCA), we computed similarity between the local atomic concept (eg, ‘Ord’) with all the LOINC (eg, SCA) atomic concepts (eg, ‘Qn’, ‘Ord’, ‘OrdQn’, ‘Nar’, ‘Doc’, and ‘-’) using GOMMA matchers as described in the full name approach (step 2.1). However, for all further sub-ontologies, we first determined the conditional probability of the current atomic concept, 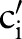 (eg,
(eg, 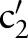 ), based on the occurrence of already matched parts in the result (eg, (‘Ord’,
), based on the occurrence of already matched parts in the result (eg, (‘Ord’, 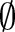 )) (step 2.2.1 or see online supplementary algorithm 1 line 5)—that is, we calculated
)) (step 2.2.1 or see online supplementary algorithm 1 line 5)—that is, we calculated 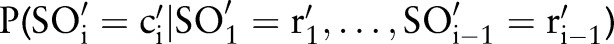 (eg, P(TIM=c2′|SCA=‘Ord’)). Only if the conditional probability is greater than zero, we consider
(eg, P(TIM=c2′|SCA=‘Ord’)). Only if the conditional probability is greater than zero, we consider 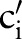 (eg,
(eg, 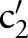 =‘Pt’ or
=‘Pt’ or 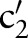 =‘24H’) and computed the similarity between
=‘24H’) and computed the similarity between 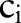 (eg,
(eg, 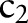 =‘Pt’) and
=‘Pt’) and 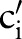 (eg,
(eg, 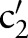 =‘Pt’ or
=‘Pt’ or 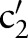 =‘24H’) (step 2.2.2 or see online supplementary algorithm 1 line 7) using GOMMA matchers as described in the full name approach. The resulting similarity was evaluated—that is, we looked for the atomic concept,
=‘24H’) (step 2.2.2 or see online supplementary algorithm 1 line 7) using GOMMA matchers as described in the full name approach. The resulting similarity was evaluated—that is, we looked for the atomic concept, 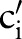 (eg,
(eg, 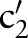 =‘Pt’), with the maximum similarity (Max1 selection). This atomic concept is regarded as the best matching concept for the particular part,
=‘Pt’), with the maximum similarity (Max1 selection). This atomic concept is regarded as the best matching concept for the particular part, 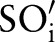 (eg, LOINC TIM) (step 3 or see online supplementary algorithm 1 line 12). If we did not find any best matching concept (ie, the similarity was too low or the probability was zero for all atomic concepts of
(eg, LOINC TIM) (step 3 or see online supplementary algorithm 1 line 12). If we did not find any best matching concept (ie, the similarity was too low or the probability was zero for all atomic concepts of 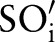 ), we left this part empty (Ø) in the result. To assess the impact of probability filtering, we also performed multi-part matching without this step (skipping steps 2.2.1 and 2.2.2 or online supplementary algorithm 1 lines 5–6).
), we left this part empty (Ø) in the result. To assess the impact of probability filtering, we also performed multi-part matching without this step (skipping steps 2.2.1 and 2.2.2 or online supplementary algorithm 1 lines 5–6).
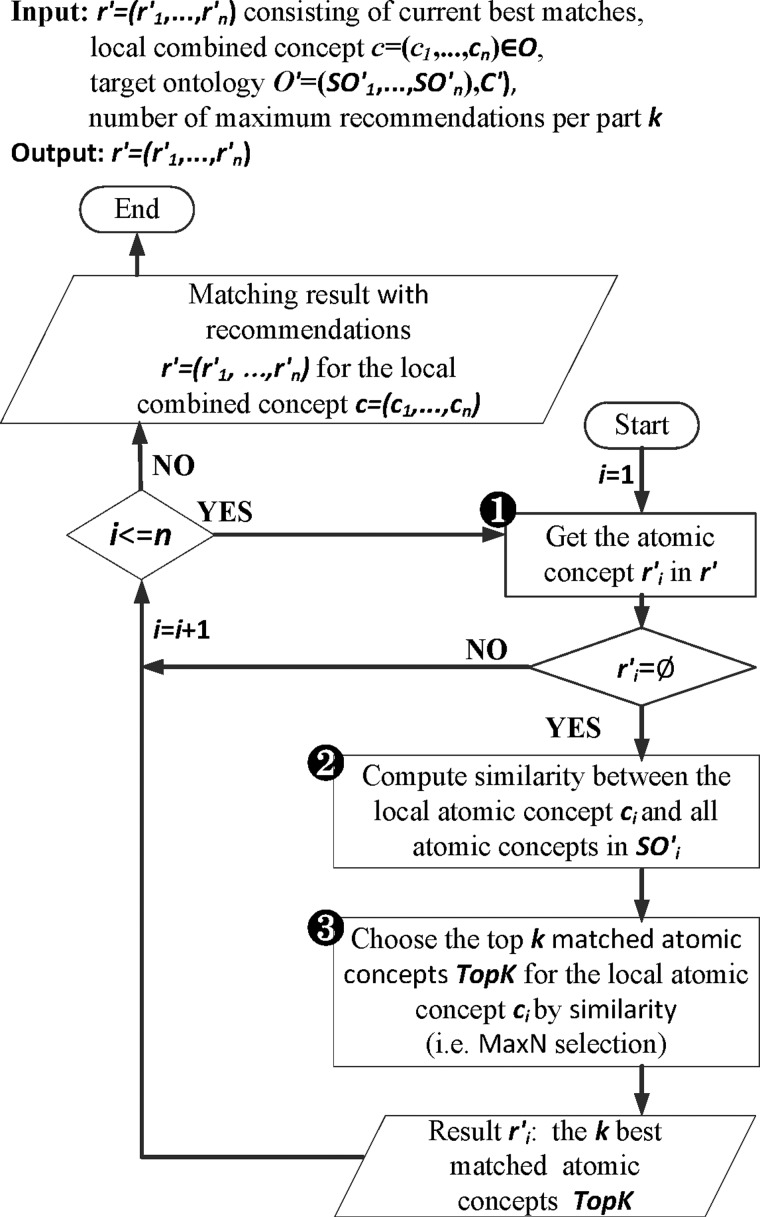
For unmapped concepts, one can generate possible recommendations (dashed line rectangle in figure 2) for such parts by applying computeRecommendations (its detailed flow is shown in figure 4; the pseudo code appears in online supplementary algorithm 2), which creates correspondences to proposed concepts. In particular, for each empty part we computed the k best matching atomic concepts (using GOMMA matchers and MaxN selection) as recommendations (steps 2 and 3 or see online supplementary algorithm 2 line 3). Users can afterwards judge these recommendations and accept them or not for their final mapping. For instance, if there were a missing SCA concept in result r′=‘Rotavirus Ag:ACnc: Pt:Stool: 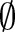 :Aggl’ , the algorithm could recommend the SCA atomic concepts, ‘Ord’, ‘Nom’ and ‘Qn’, based on the concepts from table 1.
:Aggl’ , the algorithm could recommend the SCA atomic concepts, ‘Ord’, ‘Nom’ and ‘Qn’, based on the concepts from table 1.
Figure 4.
Algorithm for computeRecommendations.
We executed a multi-part strategy algorithm (figure 3 or see online supplementary algorithm 1) for each combined concept in  to create a mapping
to create a mapping  between
between  and
and  . The result contains correspondences to pre-coordinated concepts. In contrast with the full name strategy, one can optionally use computeRecommendations to create correspondences to proposed concepts for unmapped parts and unmappable concepts as well.
. The result contains correspondences to pre-coordinated concepts. In contrast with the full name strategy, one can optionally use computeRecommendations to create correspondences to proposed concepts for unmapped parts and unmappable concepts as well.
Evaluation methods
We first present the basic information of the source and target (sub) ontologies by calculating the number of concepts and atomic concepts as well as the percentage of mappable (covmap)5 31 and unmappable (covunmap) local concepts with respect to all local concepts. We then evaluate the match algorithms RELMA v5.6 LAM (using LOINC 2.38 and enabling the option ‘Prefer Common Tests Results’), GOMMA and the multi-part strategy for the following six configurations:
F-Max10-RELMALAM: the full name strategy with Max10 selection by RELMA LAM
F-Max10-GOMMA: the full name strategy with Max10 selection by GOMMA
F-Max1-RELMALAM: the full name strategy with Max1 selection by RELMA LAM
F-Max1-GOMMA: the full name strategy with Max1 selection by GOMMA
MP-Max1-GOMMA-AO: the multi-part strategy with probability filtering and Max1 selection by GOMMA using automatic ordering (SCA-TIM-PRO-MET-SAM-COM)
MP-Max1-GOMMA-MO: the multi-part strategy with probability filtering and Max1 selection by GOMMA using optimized manual ordering (SCA-TIM-PRO-MET-COM-SAM).
We compare each approach with the gold standard established manually with respect to precision,5 19 recall and F-measure (harmonic mean of precision and recall) for mappable local concepts as well as execution run time. For the unmapped local concepts, the multi-part strategy can additionally produce recommendations. We show how the percentage of correctly mapped local concepts (number of true positives divided by total number of local concepts) changes for a different number of recommended atomic concepts. The experiments were performed on an Intel W3520 machine (4×2.67 GHz, 4GB RAM).
Results
Table 2 shows that a significant number of combined concepts can be created by atomic concepts. For example, the 45 896 LOINC codes comprise about 20 000 distinct atomic concepts. The local terminologies have 116–452 concepts, whereas there are only 1–37 atomic concepts in six parts. Some (8–25%) of the local concepts are unmappable (covunmap).
Box 1.
Pathogen names
| Haemophilus influenzae type b (Hib) | Vibrio cholera |
| Hepatitis A virus | Chikungunya virus |
| Listeria monocytogenes | West Nile virus |
| Neisseria meningitides | Rickettsia prowazekii |
| Respiratory syncytial virus | Bordetella pertussis |
| Rotavirus | Japanese encephalitis virus |
| Salmonella species | Legionella pneumophila |
| Shigella species | Leptospira interrogans |
| Streptococcus agalactiae (Group B Strep) | Burkholderia pseudomallei |
| Streptococcus pneumonia | Clostridium botulinum |
| Streptococcus pyogenes (Group A Beta Hemolytic Strep) | Coxiella burnetii |
| Yersinia enterocolitica | Rickettsia typhi |
| Campylobacter species | Borrelia burgdorferi |
| Influenza virus | Francisella tularensis |
| Parainfluenza virus | Orientia tsutsugamushi |
| Measles virus | Varicella-Zoster virus |
| Mumps virus | Bartonella henselae |
| Rubella virus | Toxoplasma gondii |
| Enterovirus | Neisseria gonorrhoeae |
| Corynebacterium diphtheriae | Hepatitis B virus |
| Dengue virus | Hepatitis C virus |
| Plasmodium vivax, P malariae, P falciparum, P ovale | Mycobacterium tuberculosis complex, multi-drug resistant (MDR-TB) |
| Entamoeba histolytica | Mycobacterium tuberculosis complex (TB) |
| STEC (E. coli O157:H7, E. coli O157:NM) | Treponema pallidum |
| Hantavirus |
Table 2.
Statistics about laboratory terminologies and LOINC 2.38
| Number of combined concepts or atomic concepts | Source (sub) ontologies | Target (sub) ontologies | ||
|---|---|---|---|---|
| SKH | Mackay | NTU | LOINC | |
| Related 49 pathogens | 37 | 28 | 37 | Class type=‘lab’ |
| Six-part combinations | 452 | 245 | 116 | 45 896 |
| COM | 36 | 32 | 37 | 19 240 |
| PRO | 6 | 4 | 7 | 101 |
| TIM | 1 | 1 | 1 | 20 |
| SAM | 12 | 25 | 23 | 322 |
| SCA | 3 | 3 | 3 | 13 |
| MET | 12 | 12 | 11 | 482 |
| Mappable combinations (covmap) | 403 (89%) | 184 (75%) | 107 (92%) | |
| Unmappable combinations (covunmap) | 49 (11%) | 61 (25%) | 9 (8%) | |
LOINC, Logical Observation Identifiers Names and Codes.
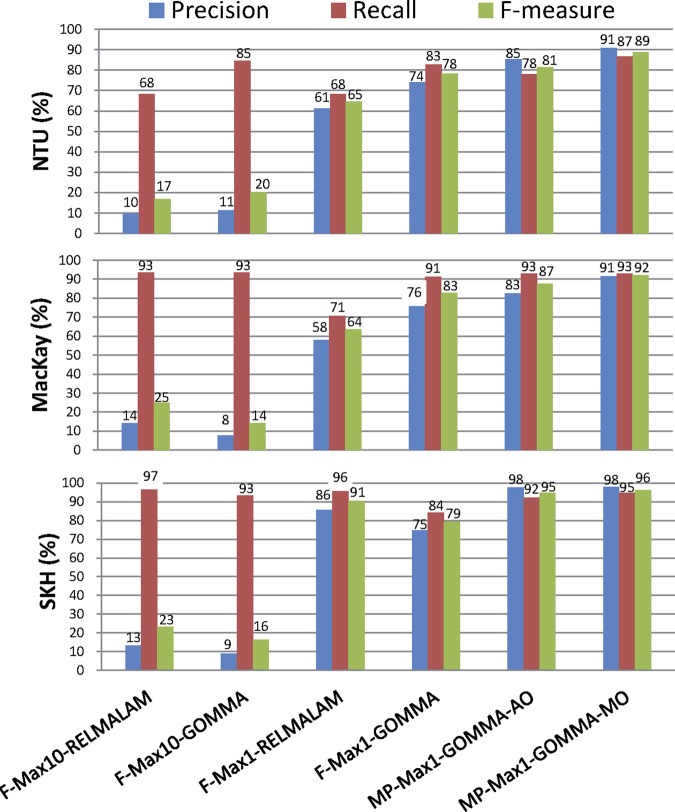
The achieved match quality of the six match configurations considered is shown in figure 5. The two Max10 configurations aim at high recall by offering up to 10 match candidates per local term. Here, GOMMA has a better recall (84% vs 68%) than RELMA LAM for the NTU terminology but performs similarly (∼ 94%) for the other two cases. Precision and thus F-measure are very low, since only 1 of the 10 proposed candidates is probably correct. Using the Max1 configuration, precision (F-measure) improves significantly by 57% (52%) in RELMA LAM and 66% (63%) in GOMMA, while recall is lower than for Max10 (average values for three datasets). For all three mapping problems, the multi-part strategies MP-Max1-GOMMA-AO and MP-Max1-GOMMA-MO achieve significantly better results than the best strategy so far, GOMMA (Max1). Using MO (AO), precision increases by 18% (14%), while recall improves slightly by 5% (2%), leading to an F-measure improvement of 12% (8%) on average. They also performed in the shortest execution time of 29–97 s, while RELMA LAM required 2–5 min—that is, about four times longer.
Figure 5.
Comparison of match algorithms and configurations. SKH, Shin Kong Wu Ho-Su Memorial Hospital; Mackay Memorial hospital Taipei branch; NTU, National Taiwan University hospital Hsin-Chu branch.
We now turn our attention to which shares of the local terminologies could be correctly mapped to LOINC. These shares are restricted not only because of less-than-perfect recall, but also because of unmappable terms (according to the reference mapping). Overall, the following percentages of local concepts (averaged over the three datasets) could be correctly mapped to pre-coordinated LOINC concepts: 72% (F-Max10-RELMALAM), 76% (F-Max10-GOMMA), 67% (F-Max1-RELMALAM), 73% (F-Max1-GOMMA), 74% (MP-Max1-GOMMA-AO) and 77% (MP-Max1-GOMMA-MO).
Figure 6 shows these percentages for MP-Max1-GOMMA-MO for the three local terminologies as green bars. The figure also shows to what degree the recommendation algorithm of MP-Max1-GOMMA-MO could find additional correspondences when varying the number k of recommended atomic concepts per part from 0 to 10 (horizontal axis). The red bars indicate the improved percentage of local terms, with a correct recommendation to an existing LOINC concept. In contrast, the blue bars show which additional improvements are feasible by mapping to recommended, new combinations of LOINC atomic concepts. The figure shows that the recommendation approach achieves an improved mapping coverage of 0%, 2% and 5% in the first case (red bars). The inclusion of proposed LOINC concepts further increases the number of correctly mapped local concepts by 10%, 18% and 4% (blue bars) to 95%, 90% and 87%, respectively—that is, 91% on average.
Figure 6.
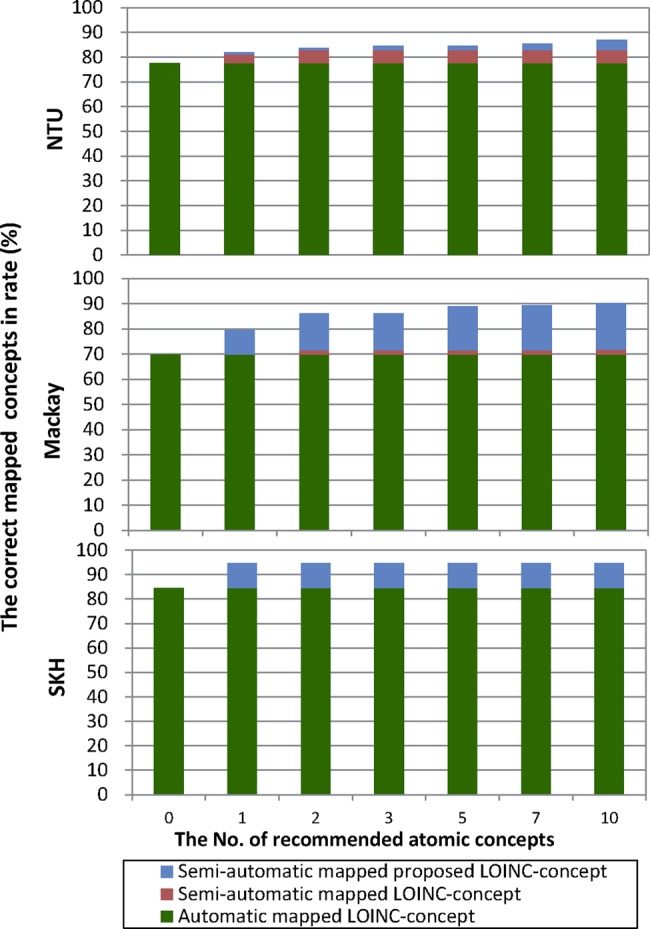
Percentage of correctly mapped concepts when varying the number of recommended atomic concepts in MP-Max1-GOMMA-MO. LOINC, Logical Observation Identifiers Names and Codes; SKH, Shin Kong Wu Ho-Su Memorial Hospital; Mackay Memorial hospital Taipei branch; NTU, National Taiwan University hospital Hsin-Chu branch.
Discussion
Full name strategy
Full name matching results are different in RELMA LAM and GOMMA, since the former adopts exact name matching, whereas the latter uses a trigram similarity function. This approximate similarity measure allows more robust and flexible matching—that is, names/synonyms of concepts with small differences can still be matched within a certain threshold. For the Max10 selection, the focus is on high recall, and GOMMA performs slightly better than RELMA LAM in this respect (90% vs 86% recall averaged over the three matching tasks). For Max1, F-Max1-GOMMA outperforms RELMA LAM not only with respect to recall (86% vs 78% averaged) but also for precision (75% vs 69%) and F-measure (80% vs 73%).
Multi-part strategy
The results are further improved by the proposed multi-part match strategy. A key to its success is the applied filtering with respect to conditional probability of concept parts, because it successfully avoids mapping to non-existing combinations of atomic LOINC concepts. Using the automatically computed order (MP-Max1-GOMMA-AO), we already achieved very good average precision (88%), recall (89%) and F-measure (88%). Based on expert knowledge, we manually optimized the order (MP-Max1-GOMMA-MO) leading to excellent match quality, with high precision (93% averaged), recall (91%) and F-measure (92%) results. Moreover, the generated recommendations leave only a small portion of the local terminologies unmapped (5% of SKH, 10% of Mackay, 13% of NTU) and thereby reduce the manual effort required to complete the mapping.
For comparison, we also tested other orders as well as multi-part matching without probability filtering, but observed significantly lower match quality as well as increased execution times (to 6–27 min) in all these cases. For example, the use of the LOINC-specific ordering of sub-ontologies (COM-PRO-TIM-SAM-SCA-MET) leads to a significantly lower F-measure value of 82%. For multi-part matching without probability filtering, the F-measure was even lower (71%). This is because independently matching concept parts often leads to concepts whose atomic concept combinations are non-existent in LOINC, so that only a few local concepts could be matched.
Unmappable local concepts
We classified the unmappable local concepts into several types. First, a local concept (eg, ‘Virus identified:Prid:Pt:?:NOM:Culture’) has a sample type (eg, Conjunctiva=‘Cnjt’), which is not included in the current LOINC. Next, the data representation of a test result (eg, for ‘Escherichia coli O157:H7:ACnc:Pt:Stool:?:Organism specific culture’) is different between local designers (eg, used nomial ‘Nom’) and the LOINC developer (eg, used ordinal ‘Ord’). Third, a few unmappable cases are that the combined local atomic concepts in PRO-SCA, PRO-SCA-MET or COM-PRO-SAM-MET parts are not contained in LOINC. We plan to further investigate the reasons for generating proposed LOINC concepts to help mapping adaptation for new LOINC versions.48
Limitations and future work
The proposed multi-part matching process uses a relatively strict Max1 selection for atomic concepts. In future work, we plan to provide users with the possibility to choose an atomic concept out of a list of suggestions. This could be useful in cases where experts have background knowledge on complex decisions for some sub-ontology. Similarly, we would like to add more user interaction during recommendations of proposed concepts.
We have proposed a simple and effective automatic ordering strategy for the multi-part strategy. However, we found that using expert knowledge to manually adapt this can lead to even better results. For future work, we thus plan to study further automatic techniques for determining optimized sub-ontology orders.
We analyzed datasets for three hospitals and were able to produce very good results. However, a limitation of this study is that only a small, specialized subset of LOINC was evaluated (813 local concepts associated with tests for 49 microbiology pathogens), making it difficult to generalize the conclusions to different classes of LOINC laboratory tests or the full LOINC terminology. We plan to evaluate other local laboratory datasets. In this study, we used translation for Traditional Chinese to English, but translation from other languages to English is also worth investigating. Moreover, we plan to evaluate our approaches to other pre-coordinated terminologies such as SNOMED CT and ICNP (International Classification for Nursing Practice).
Conclusion
A new multi-part matching strategy with conditional occurrence probability filtering for pre-coordinated LOINC terminology is proposed. The mapping quality of the proposed strategy is significantly better than RELMA LAM and the use of full name matching. The proposed recommendation of new combinations of atomic concepts has been shown to improve the mapping coverage for local terminologies and to automatically support post-coordination.
Supplementary Material
Footnotes
Contributors: L-HL, AG, and MH conceived and designed the study, performed the analysis, and wrote and edited the manuscript. L-HL created the strategy and evaluated the results. D-ML coordinated cooperative organizations, collected data, and input practical concerns to the study. ER coordinated the study and revised the manuscript critically for important intellectual content.
Funding: The work was supported by ‘NSC-DAAD Sandwich Program, 2012 Spring’ from the Taiwan National Science Council (NSC) and the German Academic Exchange Service (DAAD) under grant number NSC101-2911-I-010-001-2 and partly supported by Taiwan Centers for Disease Control (CDC) under grant number DOH101-DC-1014.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Obrst L. Ontological architectures. In: Poli R, Healy M, Kameas A, eds. Theory and applications of ontology: computer applications. Springer, 2010:27–66. [Google Scholar]
- 2.Henricks WH. “Meaningful use” of electronic health records and its relevance to laboratories and pathologists. J Pathol Inform 2011;2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canada Health Infoway. Interoperable EHR standard. https://http://www.infoway-inforoute.ca/index.php/programs-services/standards-collaborative/pan-canadian-standards/interoperable-ehr-standard (accessed 16 Apr 2013).
- 4.Regenstrief Institute. RELMA Regenstrief LOINC Mapping Assistant user manual. http://loinc.org/relma/ (accessed 10 Apr 2013).
- 5.Zunner C, Bürkle T, Prokosch HU, et al. Mapping local laboratory interface terms to LOINC at a German university hospital using RELMA V.5: a semi-automated approach. J Am Med Inform Assoc 2013;20:293–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abhyankar S, Demner-Fushman D, McDonald CJ. Standardizing clinical laboratory data for secondary use. J Biomed Inform 2012;45:642–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan AN, Griffith SP, Moore C, et al. Standardizing laboratory data by mapping to LOINC. J Am Med Inform Assoc 2006;13:353–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi J, Jenkins ML, Cimino JJ, et al. Toward semantic interoperability in home health care: formally representing OASIS items for integration into a concept-oriented terminology. J Am Med Inform Assoc 2005;12:410–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim H, El-Kareh R, Goel A, et al. An approach to improve LOINC mapping through augmentation of local test names. J Biomed Inform 2011;45:651–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun JY, Sun Y. A system for automated lexical mapping. J Am Med Inform Assoc 2006;13:334–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonald CJ, Huff SM, Suico JG, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem 2003;49:624–33 [DOI] [PubMed] [Google Scholar]
- 12.Huff SM, Rocha RA, McDonald CJ, et al. Development of the Logical Observation Identifier Names and Codes (LOINC) vocabulary. J Am Med Inform Assoc 1998;5:276–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vreeman DJ, McDonald CJ, Huff SM. LOINC(R)—A universal catalog of Individual clinical observations and uniform representation of enumerated collections. Int J Funct Inform Personal Med 2010;3:273–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenbloom ST, Miller RA, Johnson KB, et al. Interface terminologies: facilitating direct entry of clinical data into electronic health record systems. J Am Med Inform Assoc 2006;13:277–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin MC, Vreeman DJ, McDonald CJ, et al. Auditing consistency and usefulness of LOINC use among three large institutions—using version spaces for grouping LOINC codes. J Biomed Inform 2012;45:658–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan AN, Russell D, Moore C, et al. The map to LOINC project. AMIA Annu Symp Proc 2003;2003:890. [PMC free article] [PubMed] [Google Scholar]
- 17.Porter JP, Starmer J, King J, et al. Mapping laboratory test codes to LOINC for a regional health information exchange. AMIA Annu Symp Proc 2007;2007:1081. [PubMed] [Google Scholar]
- 18.Fidahussein M, Vreeman DJ. A corpus-based approach for automated LOINC mapping. J Am Med Inform Assoc 2014;21:64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiszman M, Shin D, Sneiderman CA, et al. A knowledge intensive approach to mapping clinical narrative to LOINC. AMIA Annu Symp Proc 2010;2010:227–31 [PMC free article] [PubMed] [Google Scholar]
- 20.Vreeman DJ, McDonald CJ. Automated mapping of local radiology terms to LOINC. AMIA Annu Symp Proc 2005;2005:769–73 [PMC free article] [PubMed] [Google Scholar]
- 21.Vreeman DJ, McDonald CJ. A comparison of Intelligent Mapper and document similarity scores for mapping local radiology terms to LOINC. AMIA Annu Symp Proc 2006;2006:809–13 [PMC free article] [PubMed] [Google Scholar]
- 22.Wade G, Rosenbloom ST. Experiences of mapping a legacy interface terminology to SNOMED CT. BMC Med Inform Decis Mak 2008;8(Suppl 1):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau LM, Johnson K, Monson K, et al. A method for the automated mapping of laboratory results to LOINC. Proc AMIA Symp Proc 2000;2000:472–6 [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald C, Huff S, Mercer K, et al. Logical Observation Identifiers Names and Codes (LOINC®) users’ guide 2011.
- 25.Zollo KA, Huff SM. Automated mapping of observation codes using extensional definitions. J Am Med Inform Assoc 2000;7:586–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gamache RE, Dixon BE, Grannis S, et al. Impact of selective mapping strategies on automated laboratory result notification to public health authorities. AMIA Annu Symp Proc 2012;2012:228–36 [PMC free article] [PubMed] [Google Scholar]
- 27.Canada Health Infoway. Pan-Canadian LOINC observation code database (pCLOCD) nomenclature standard. https://www.infoway-inforoute.ca/index.php/programs-services/standards-collaborative/pan-canadian-standards/pan-canadian-loinc-observation-code-database-pclocd-nomenclature-standard (accessed 27 Apr 2013).
- 28.Lee KN, Yoon JH, Min WK, et al. Standardization of terminology in laboratory medicine II. J Korean Med Sci 2008;23:711–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin MC, Vreeman DJ, Huff SM. Investigating the semantic interoperability of laboratory data exchanged using LOINC codes in three large institutions. AMIA Annu Symp Proc 2011;2011:805–14 [PMC free article] [PubMed] [Google Scholar]
- 30.Bodenreider O. Issues in mapping LOINC laboratory tests to SNOMED CT. AMIA Annu Symp Proc 2008;2008:51–5 [PMC free article] [PubMed] [Google Scholar]
- 31.Lin MC, Vreeman DJ, McDonald CJ, et al. A characterization of local LOINC mapping for laboratory tests in three large institutions. Methods Inf Med 2011;50:105–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vreeman DJ, Chiaravalloti MT, Hook J, et al. Enabling international adoption of LOINC through translation. J Biomed Inform 2012;45:667–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Regenstrief Institute. Logical Observation Identifiers Names and Codes (LOINC®). http://loinc.org/ (accessed 15 Nov 2013).
- 34.Euzenat J, Shvaiko P. Ontology matching. Berlin Heidelberg (DE): Springer-Verlag, 2007 [Google Scholar]
- 35.Bellahsene Z, Bonifati A, Rahm E. Schema matching and mapping. Berlin Heidelberg (DE): Springer-Verlag, 2011 [Google Scholar]
- 36.Tan H, Jakonienė V, Lambrix P, et al. Alignment of biomedical ontologies using life science literature. Lect Notes Comput Sc 2006;3886:1–17 [Google Scholar]
- 37.Rance B, Gibrat J-F, Froidevau C. An adaptive combination of matchers: application to the mapping of biological ontologies for genome annotation. DILS 2009;2009:113–26 [Google Scholar]
- 38.Zhang S, Bodenreider O. Alignment of multiple ontologies of anatomy: deriving indirect mappings from direct mappings to a reference. AMIA Annu Symp Proc 2005;2005:864–8 [PMC free article] [PubMed] [Google Scholar]
- 39.Kirsten T, Gross A, Hartung M, et al. GOMMA: a component-based infrastructure for managing and analyzing life science ontologies and their evolution. J Biomed Semantics 2011;2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rocha RA, Huff SM. Coupling vocabularies and data structures: lessons from LOINC. Proc AMIA Annu Fall Symp 1996;1996:90–4 [PMC free article] [PubMed] [Google Scholar]
- 41.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic acids research 2004;32(Database issue):D267–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Groß A, Hartung M, Kirsten T, et al. GOMMA Results for OAEI 2012. The Seventh International Workshop on Ontology Matching @ ISWC 2012; Boston, 2012 [Google Scholar]
- 43.Ontology Alignment Evaluation Initiative. Ontology Alignment Evaluation Initiative. http://oaei.ontologymatching.org/
- 44.MyMemory. MyMemory API technical specifications. http://mymemory.translated.net/doc/features.php (accessed 5 Sep 2012).
- 45.Ukkonen E. Approximate string-matching with q-grams and maximal matches. Theor Comput Sci 1992;92:191–211 [Google Scholar]
- 46.Adamson GW, Boreham J. The use of an association measure based on character structure to identify semantically related pairs of words and document titles. Inform Storage Ret 1974:253–60 [Google Scholar]
- 47.Do H-H, Rahm E. COMA: a system for flexible combination of schema matching approaches. Hong Kong, China: VLDB Endowment, 2002 [Google Scholar]
- 48.Groß A, Reis JCD, Hartung M, et al. Semi-automatic adaptation of mappings between life science ontologies. DILS 2013;2013:90–104 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.