ABSTRACT
The physiological state of natural competence for transformation allows certain bacteria to take up free DNA from the environment and to recombine such newly acquired DNA into their chromosomes. However, even though conserved components that are required to undergo natural transformation have been identified in several naturally competent bacteria, our knowledge of the underlying mechanisms of the DNA uptake process remains very limited. To better understand these mechanisms, we investigated the competence-mediated DNA transport in the naturally transformable pathogen Vibrio cholerae. Previously, we used a cell biology-based approach to experimentally address an existing hypothesis, which suggested the competence protein ComEA plays a role in the DNA uptake process across the outer membrane of Gram-negative bacteria. Here, we extended this knowledge by investigating the dynamics of DNA translocation across both membranes. More precisely, we indirectly visualized the transfer of the external DNA from outside the cell into the periplasm followed by the shuttling of the DNA into the cytoplasm. Based on these data, we conclude that for V. cholerae, the DNA translocation across the outer and inner membranes is spatially but not temporally coupled.
IMPORTANCE
As a mode of horizontal gene transfer, natural competence for transformation has contributed substantially to the plasticity of genomes and to bacterial evolution. Natural competence is often a tightly regulated process and is induced by diverse environmental cues. This is in contrast to the mechanistic aspects of the DNA translocation event, which are most likely conserved among naturally transformable bacteria. However, the DNA uptake process is still not well understood. We therefore investigated how external DNA reaches the cytosol of the naturally transformable bacterium V. cholerae. More specifically, we provide evidence that the DNA translocation across the membranes is spatially but not temporally coupled. We hypothesize that this model also applies to other competent Gram-negative bacteria and that our study contributes to the general understanding of this important biological process.
INTRODUCTION
Horizontal gene transfer (HGT) greatly contributes to the evolution of bacteria. As such, studying the regulatory networks and mechanistic aspects of HGT is of prime importance. One mode of HGT in bacteria is natural competence for transformation. Naturally competent bacteria are in a physiological state that allows them to take up free DNA from the environment and to incorporate this foreign DNA into their genome. Indeed, large pieces of DNA containing a series of genes can be transferred by transformation without the need for direct interaction with other microbes or the intercession of mobile genetic elements. This can quickly foster the spread of antibiotic resistance traits, adaptation of bacteria to new environmental niches, and emergence of new pathogens.
Although the regulation of natural competence differs widely between bacteria (reviewed in references 1–6), the core components of the DNA uptake machinery are often conserved, and the uptake process might be close to universal. However, major knowledge gaps exist with respect to the mechanistic aspects of the DNA uptake process, as also stated in a recent review by the groups of Patrice Polard and Jean-Pierre Claverys, who highlighted the fact that further investigations into this direction are needed (6). This is especially true for Gram-negative bacteria, where the transforming DNA needs to cross two membranes (and the periplasmic space) before entering the cytoplasm. The aim of our study was therefore to address the DNA uptake process in the naturally competent bacterium Vibrio cholerae and, more specifically, the transfer of transforming DNA (tDNA) from the periplasm into the cytoplasm.
Below, we describe a working model of how natural competence-driven DNA uptake might occur in Gram-negative bacteria. Variations of this model have existed for many years (see, for example, the model of the DNA uptake complex of Neisseria gonorrhoeae proposed by Fussenegger et al. in 1997 [7]). However, as is the case for many multiprotein complexes, all former and current DNA uptake models are derived from a combination of documented data combined with hypothetical assumptions, with the latter still requiring experimental validations. The gene/protein nomenclature used throughout the text is based on the V. cholerae genome annotation (8) and on proteins of V. cholerae that show similarities to proteins of other naturally competent bacteria (9, 10). We note that several excellent review articles are available with an emphasis on other naturally competent bacteria and these reviews offer a more detailed description of the putative components of the DNA uptake machineries (e.g., references 5, 6, and 11–19 and references therein).
With respect to the DNA uptake process, it is believed that a core part of the uptake machinery is composed of a (pseudo)pilus with similarities to type IV pili (Tfp) (20–22). Tfp are assembled by the polymerization of the major pilin protein subunit and can be several micrometers in length. The opposite process, namely, the depolymerization of the pilin subunits, has been suggested to occur through pilus retraction (23, 24) driven by the ATPase PilT (25–27). We recently demonstrated that V. cholerae indeed produces a specific Tfp structure after competence induction (9).
For Gram-negative bacteria, it is assumed that the Tfp crosses the outer membrane through a secretin pore formed by the PilQ protein (28–31). With respect to the DNA uptake machineries, such Tfp-associated secretins might also serve as the entry point of the incoming tDNA into the periplasm. We have recently demonstrated that PilQ-independent DNA uptake, which still resulted in naturally transformed bacteria, can occur in V. cholerae even though such events are extremely rare (9). Thus, assuming that in the majority of cases the DNA enters the periplasm through the secretin, it is still unclear which protein-protein complex pulls the DNA through this pore. To date, most DNA uptake models suggest that the force for the DNA uptake process might come from repeating cycles of pilus extension and pilus retraction. Indeed, the mechanical characteristics of the DNA uptake process in Bacillus subtilis (32) were reminiscent of pilus retraction in N. gonorrhoeae (23, 33). Yet, it was recently speculated (17, 19) or experimentally addressed that the DNA-binding protein ComEA (34, 35) might also be involved in DNA transport across the outer membrane, even though the exact mode of action of ComEA is still elusive. The majority of naturally competent bacteria contain at least one copy of comEA (the homologs are comE and comE1 in Neisseria spp. [36, 37] and Haemophilus influenzae [38], respectively). The four comE copies of Neisseria gonorrhoeae, which all encode the same ComE protein, were first identified in a seminal study by Chen and Gotschlich (37). The authors also further investigated this competence gene and demonstrated that a quadruple deletion strain that lacked all four copies of comE was severely impaired in DNA uptake and consequently also in natural transformation, despite a normal piliation and twitching motility phenotype. Moreover, they also suggested that ComE might be located in the periplasm (37). Indeed, Chen and Gotschlich used a signal peptide algorithm to predict the presence of a cleavable signal peptide, which was in accordance with the smaller size of the mature protein as observed by Western blot analysis of neisserial cell lysates (37). That purified ComE/ComEA binds DNA in vitro and in a sequence-unspecific manner was also demonstrated for other competent bacteria, including B. subtilis (35), N. gonorrhoeae (37), Campylobacter jejuni (39), Pasteurella multocida (40), and V. cholerae (41).
Based on these earlier data and supported by our recent study on ComEA in vivo (where we also experimentally confirmed its periplasmic localization within V. cholerae), we speculated that the protein might be compacting DNA (as demonstrated by heterologous expression of fluorescently tagged ComEA in Escherichia coli) and be pulling the tDNA into the periplasm of Gram-negative bacteria via a Brownian ratchet mechanism (41). Notably, while the present article was under review, Gangel et al. reported on the DNA translocation across the outer membrane of the Gram-negative bacterium N. gonorrhoeae and the involvement of ComE in this organism (42). This study nicely confirmed our earlier data in that it also established that tDNA is imported into the periplasm at random locations around the cell, that the incoming tDNA accumulated in the periplasm in a Tfp- and ComEA-dependent manner (visualized through the use of fluorescently labeled DNA), and that fluorescently tagged ComEA formed distinctive ComEA-tDNA clusters (41, 42). Notably, the authors even concluded that “ComE-dependent DNA-import into the periplasm can be considered a general mechanism for Gram-negative competent species” (42).
That the tDNA enters the cytoplasm of both Gram-positive and Gram-negative bacteria through the putative membrane channel protein ComEC (43) is widely accepted in the field. Next, and in accordance with what is known for Gram-positive bacteria, it is assumed that the incoming tDNA is single stranded and immediately decorated with single-stranded binding (Ssb) proteins and DprA to protect it from degradation and to promote loading of the RecA protein, which is required for homologous recombination with the chromosome (44, 45).
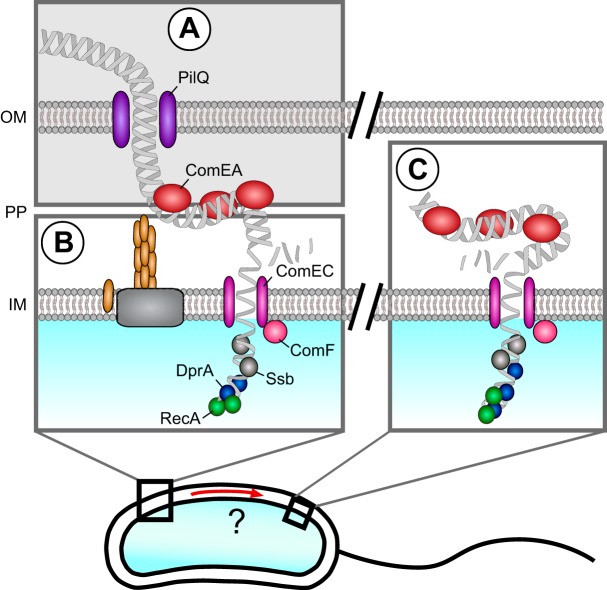
Here, we report on the translocation of tDNA across the inner membrane of naturally competent V. cholerae cells. We asked the following question (Fig. 1): does the tDNA enter the cytoplasm at the same position (Fig. 1B) where the translocation through the outer membrane occurred (Fig. 1A), or is the second translocation event spatially and temporally uncoupled from the first step (Fig. 1C)? To answer this question, we generated translational fusion constructs for ComEA and RecA, which were functional with respect to natural transformability. These fluorescently labeled protein markers allowed us to indirectly visualize the path of the tDNA, as both proteins bind to the tDNA in the course of natural transformation (Fig. 1). In summary, we provide evidence that the tDNA shuttling across the two membranes is spatially but not temporally coupled.
FIG 1 .
Putative positions of the DNA translocation events across the two membranes. The aim of this study was to assess the relative position of the DNA translocation across the inner membrane (IM) compared to the DNA passage across the outer membrane (OM). PP, periplasm. Two hypotheses were addressed in this study, namely, that the ComEA-assisted DNA uptake across the outer membrane (A [shaded in gray]) and the translocation of DNA into the cytosol coincide at one large DNA uptake complex (B) or that the outer and inner membrane transports are spatially uncoupled (C). Previous studies on naturally competent Gram-positive bacteria indicated that the inner membrane protein ComEC translocates the DNA across the cytoplasmic membrane (43), most likely in conjunction with the competence protein ComF, after which the single-stranded DNA is decorated with the proteins Ssb, DprA, and RecA.
RESULTS
Persistence of periplasmic ComEA-tDNA clusters differs significantly among competence-induced cells.
We have previously demonstrated that the periplasmic competence protein ComEA of V. cholerae is involved in DNA uptake across the outer membrane. More specifically, fluorescently tagged ComEA displayed high affinity for DNA binding in vivo and formed distinctive ComEA-tDNA clusters at a single site within the periplasm of competent cells (41). We therefore concluded that ComEA foci assemble upon translocation of the tDNA across the outer membrane, which most likely occurs at or in close proximity to the Tfp and the PilQ secretin (as no ComEA-tDNA clusters were detectable in the absence of PilQ or the major Tfp subunit PilA). Moreover, time-lapse microscopy movies suggested that the ComEA foci were resolved and that the even distribution pattern of ComEA was reconstituted over time (41). However, these downstream events were not further investigated.
To better understand what could initiate the resolution of the ComEA clusters, we now quantified the persistence of ComEA foci by measuring the time interval between the formation of the ComEA foci and the reconstitution of a uniform ComEA distribution pattern. Through time-lapse microscopy (40 min in total), we observed that the resolution of the ComEA foci was very heterogeneous, with some cells restoring the initial pattern within 2 min, whereas others took more than 20 min (Fig. 2A). Notably, we did not observe any difference between cells containing the putative inner membrane transporter protein (ComEC) and those that lacked it (Fig. 2B). Through the use of longer tDNA fragments, the persistence time of the ComEA foci was largely increased, with the majority of cells maintaining the ComEA-tDNA clusters for more than 20 min (e.g., 52% of cells) (Fig. 2C).
FIG 2 .
ComEA-mCherry focus persistence during DNA uptake. Competence-induced strains carrying chromosomal comEA-mCherry were exposed to 10.3-kb PCR fragments (A and B and D to F) or phage λ DNA (48.5 kb) (C) and imaged at 2-min intervals for 21 frames. n = 100 (B, C, D) or n = 200 (A, E, F) cells. Cells depicting early mCherry focus formation were analyzed for the duration of focus persistence. Frequencies (%) of the observed phenotypes are shown as bar plots. ComEA-mCherry focus persistence was quantified in a wild-type (WT) strain (A, C) or in strains lacking comEC (B), xds (D), dns (E), or xds and dns (F).
As the time distribution patterns for the resolution of the ComEA foci were indistinguishable between ComEC-positive and -negative cells (Fig. 2A and B), we reasoned that such resolution events might not or might only to a minor extent be caused by the transport of the tDNA across the inner membrane but could be due to another event that leads to the resolution of the complex. One option for such an effect would be the cleavage of one or both strands of the tDNA by nucleases. In this context, we have previously demonstrated that V. cholerae cells that lack the nucleases Dns and Xds are more transformable (46) and accumulated larger amounts of tDNA within the periplasm than nuclease-proficient strains (41). However, tDNA did not accumulate in these strains when ComEA was concomitantly absent, in which case the DNA uptake process seemed completely abolished. Thus, we suggested that ComEA’s major task was to pull tDNA into the periplasm rather than protecting the DNA against nucleases (41) or to modulate the nuclease activity, as has been suggested for Streptococcus pneumoniae (47). Here, we asked the question of whether these nucleases play a role in the resolution of the ComEA clusters. To test this question, we repeated the time-lapse microscopy experiments described above in V. cholerae strains that lacked either one or both nuclease genes (dns and xds). The absence of xds did not have an effect on ComEA focus persistence (Fig. 2D), which is in agreement with earlier transformation data (41, 46). In contrast, the ComEA clusters were maintained for at least 20 min in the vast majority of dns- and dns xds-negative cells (Fig. 2E and F). We therefore speculate that even though the expression of dns is downregulated at the time of competence induction in a quorum-sensing-dependent manner (as shown both on the transcriptional and cell-associated protein levels [46, 48–50]), the residual Dns protein is sufficient to degrade part of the periplasmic tDNA, thereby freeing ComEA from the complex.
Establishment of fluorescent protein markers to visualize the transport of the tDNA across the inner membrane.
Based on studies of other naturally competent bacteria, it is assumed that the transport of the tDNA across the inner membrane occurs through a putative channel formed by the ComEC protein (43). We have previously confirmed the importance of ComEC and ComF (a homolog of ComFC of B. subtilis [51]) in the DNA uptake process of V. cholerae (9, 41, 52). Thus, we reasoned that the point of DNA translocation across the inner membrane could be visualized through the use of ComEC or ComF translational fusions with the green fluorescent protein (GFP). After generating such constructs and using them to replace the indigenous native alleles of comEC and comF, respectively, we first assessed their functionality by scoring the natural transformability of the resulting strains. Indeed, both fusion proteins were fully proficient to drive natural transformation (see Table S1 in the supplemental material), whereas deletion strains lacking either comEC or comF were nontransformable (9). Similar results were obtained for a DprA-GFP fusion (see Table S1), which we also used to monitor entry of tDNA into the cytoplasm (Fig. 1). Unfortunately, the abundance of these three proteins in competence-induced cells (without or with external tDNA) was too low for detection by fluorescence microscopy. The low abundance of ComEC was in accordance with early expression data, which were based on transcriptional reporters and on quantification by quantitative reverse transcription-PCR (qRT-PCR) (48). Therefore, we decided to study the localization of another downstream-acting cytoplasmic protein, RecA, instead (Fig. 1). In fact, RecA localization has been previously studied in the noncompetent Gram-negative bacterium E. coli (53) and in the Gram-positive competent bacterium B. subtilis, where it colocalized with other competence proteins of the DNA uptake machinery at a single cell pole of the bacterium (54–57). Moreover, a direct interaction between DprA and RecA has been demonstrated using a yeast two-hybrid system and pulldown assays (45).
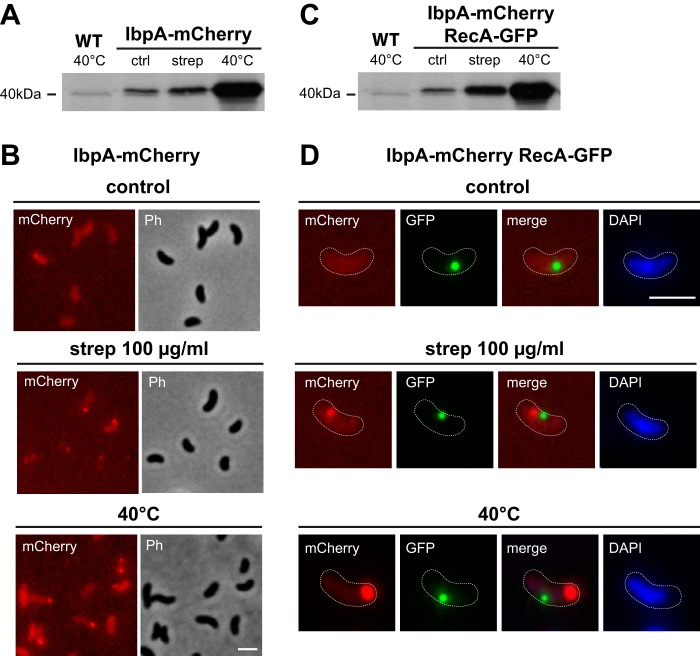
After replacing the chromosomal copy of recA with the recA-gfp allele in V. cholerae, we analyzed the growth behavior of the bacteria and their ability to be naturally transformed (Fig. 3). Notably, even though the growth of the RecA-GFP-expressing strain was impaired to a certain extent, the strain maintained extremely high levels of natural transformability (Fig. 3). Next, we studied the localization of RecA-GFP in non-competence-induced V. cholerae cells (Fig. 3C). Under those conditions, RecA-GFP was either evenly dispersed within the cytoplasm or clustered into distinctive and strongly fluorescent foci (Fig. 3C). We quantified the number of RecA-GFP foci per cell by using the MicrobeTracker software (58), and the analysis revealed that 58.6, 36.8, and 4.6% of cells contained none, one, or more than one fluorescence focus per cell, respectively (Fig. 3D). Renzette and colleagues previously showed that there were two types of RecA-GFP foci within E. coli cells: 4′,6-diamidino-2-phenylindole (DAPI)-sensitive clusters that contained DNA and DAPI-insensitive DNA-less foci (53). To better understand the type of RecA-GFP foci that were observable in V. cholerae, we tested whether their distribution within a population would change upon DAPI staining, which turned out not to be the case (Fig. 3D). Thus, the observed RecA-GFP clusters could act as DNA-less storage structures or, alternatively, be the result of nonfunctional protein aggregates. To verify whether the latter was the case, we constructed a translational fusion between the gene encoding inclusion body-associated protein A (IbpA) and mCherry. The small heat shock protein IbpA was originally identified in E. coli during the high-level production of heterologous proteins (59). Subsequently, IbpA has been shown to faithfully identify the localization of protein aggregates and inclusion bodies of E. coli and to assist chaperone-mediated protein disaggregation (60–62). Consequently, we replaced the ibpA gene of V. cholerae with an ibpA-mCherry translational fusion construct and studied its expression and localization under unstressed and stressed conditions (Fig. 4). Indeed, diverse stresses (exposure to either inhibitory concentrations of streptomycin [100 µg/ml] or a mild heat-shock at 40°C) induced the expression of IbpA-mCherry (Fig. 4A) and resulted in the formation of protein foci of IbpA-mCherry (Fig. 4B), as was the case for E. coli (60). Such an upregulation of IbpA-mCherry production upon external stress was equally observed in a V. cholerae strain that concomitantly produced RecA-GFP, indicating that the fusion protein per se does not induce ibpA-mCherry due to a putative aggregation effect (Fig. 4C). Importantly, whereas noncompetent V. cholerae strains often contained large and distinctive RecA-GFP foci (as described above) (Fig. 3), such foci did not attract the IbpA-mCherry fusion protein, indicating that the foci were not recognized as unfolded protein aggregates (Fig. 4D; see Table S2 in the supplemental material). Moreover, even after induction of IbpA-mCherry through antibiotic or heat stress, colocalization between IbpA-mCherry and RecA-GFP was only observed in <25% of the cells (Fig. 4D; see Table S2). We therefore concluded that the bright RecA-GFP foci depict DNA-less storage structures and not misfolded protein aggregates or inclusion bodies. Nonetheless, we cannot fully exclude that such bright and seemingly stable RecA-GFP foci, as also observed in E. coli (53), might be the result of the fluorescent tag, as recently addressed by Landgraf et al. (63). However, the dynamic and tDNA-dependent changes of the RecA-GFP localization as described below would not be expected in the latter case.
FIG 3 .

Functionality of chromosomally encoded RecA-GFP and localization of the protein. The native copy of recA on the chromosome of V. cholerae was replaced by a translational reporter fusion construct (recA-gfp). The functionality and localization of RecA-GFP were tested in live cells. (A) Colony morphology and size of a wild-type (WT) strain, ΔrecA strain, and recA-gfp strain. (B) Functionality of RecA-GFP in chitin-independent natural transformation assays. All strains contain the TntfoX transposon for competence induction. <d.l., below the detection limit. Shown are average transformation frequencies of at least three independent experiments ± SD. Statistical significance was tested using Welch’s t test on log-transformed values. **, P < 0.01. (C) Subcellular localization of RecA-GFP. Depicted is a representative fluorescence image of the green channel (GFP) and an overlay with the phase-contrast channel (merge). The inset was enhanced in contrast and brightness to visualize two cells that show homogenous RecA-GFP fluorescence. Scale bar, 2 µm. (D) Quantification of the number of GFP foci per cell in untreated (−DAPI) and DAPI-stained (+DAPI) cells.
FIG 4 .
RecA-GFP accumulations are not part of misfolded protein aggregates. The native copy of ibpA on the chromosome of V. cholerae was replaced by an ibpA-mCherry translational fusion. (A) The protein level of IbpA-mCherry was verified by Western blotting using polyclonal anti-mCherry antibodies. Cell lysates of a wild-type (WT) strain as well as lysates of bacteria carrying ibpA-mCherry were probed. Lysates of bacteria producing IbpA-mCherry were either prepared from an unstressed control culture (ctrl) or after exposure to streptomycin (strep) or to a mild heat shock (40°C). (B) The production and distribution of IbpA-mCherry were tested in unstressed bacterial cultures (control) or after exposure of the bacteria to streptomycin (strep [100 µg/ml]) or to a mild heat shock (40°C). Cells were imaged in the red (mCherry) or phase-contrast (Ph) channel. (C) Lysates of WT cells and bacteria expressing ibpA-mCherry and recA-gfp were probed by Western blotting as in panel A. (D) Absence of colocalization between IbpA-mCherry and RecA-GFP. Representative fluorescence images (mCherry and GFP) and overlays (merge) are shown. Chromosomal DNA was stained with DAPI. The cells are outlined with dashed lines. Scale bars, 2 µm.
DNA transport across the outer and inner membrane is spatially but not temporally coupled.
After validating the functionality of the RecA-GFP fusion and determining its subcellular distribution pattern in noncompetent bacteria, we aimed at using this construct to pinpoint the point of entry of the tDNA into the cytoplasm. The rationale behind this idea was that RecA would bind to ssDNA entering the cytoplasm, thereby changing its localization pattern to form protein clusters, which should be visible by fluorescence microscopy (Fig. 1). Indeed, earlier studies showed that RecA colocalized with the polar DNA uptake machinery of B. subtilis and that RecA filaments (threads) formed upon addition of tDNA, which seemingly emanated from the DNA uptake complex (54). Notably, the situation is different in V. cholerae as this bacterium does not contain polar competence machineries and the transport across the outer membrane does not strictly occur at the cell pole (9, 10, 41). Thus, to indirectly visualize the inner membrane transport process, we combined the recA-gfp and the comEA-mCherry alleles and imaged the resulting V. cholerae strain under competence-inducing conditions. In the absence of exogenous tDNA, competence-induced cells depicted a uniform distribution pattern of ComEA-mCherry, whereas the pattern of RecA-GFP distribution (see Fig. S1A and B in the supplemental material) was not significantly different from what was observed in competence-noninduced cells (Fig. 3C). However, when the competent cells were exposed to external tDNA, we frequently observed foci of ComEA-mCherry (see Fig. S1C to E) due to the uptake of tDNA into the periplasm. Notably, about half of the cells with ComEA-mCherry foci also contained clusters of RecA-GFP, and the two fluorescent signals colocalized often but not exclusively (see Fig. S1). We rarely observed foci from which RecA-GFP threads emanated, a finding that was in contrast to studies performed with B. subtilis (54, 64, 65). A hypothetical explanation for this difference could be that DNA transport across the inner membrane in V. cholerae proceeds in a 3′ to 5′ direction, in accordance with what has been demonstrated for H. influenzae (66), whereas the RecA protein polymerizes in the 5′ to 3′ direction (67). However, the absence of RecA filaments was not a concern for our study, as RecA-GFP primarily served as a marker protein to indicate the entry of the ssDNA into the cytoplasm.
Although we frequently detected an association between RecA-GFP and ComEA-mCherry foci in these fluorescence microscopy snapshots (see Fig. S1 in the supplemental material), the results were indicative but not conclusive and certainly not proof that incoming single-stranded tDNA caused RecA-GFP focus formation, especially as RecA-GFP foci were also abundant in noncompetent cells (as discussed above). Thus, to be able to distinguish between already existing versus newly formed RecA-GFP foci, we performed time-lapse experiments (at 2-min intervals). The cells, which depicted a uniform distribution pattern of both fusion proteins, followed by the formation of fluorescent foci of ComEA-mCherry and RecA-GFP, were most informative in this context (Fig. 5A), although other events, including ComEA-mCherry foci, which are indicative of DNA uptake across the outer membrane, were also considered (Fig. 5B to D). Indeed, under the conditions where RecA-GFP foci developed de novo throughout the time-lapse experiment, we exclusively observed colocalization of the foci with the ComEA-mCherry signal (Fig. 5E, row a). Furthermore, RecA proteins accumulated with a certain delay to ComEA (~8 min in Fig. 5A), even though the duration of this delay was not characteristic and varied between ≤2 min and 18 min (the shortest and longest possible delays according to the time-lapse settings). In contrast, if RecA foci were already present at the beginning of the experiment, they only colocalized with the ComEA signal in roughly one out of two cases (Fig. 5E, rows c and d). In the majority of cells (~50%), RecA did not form any clusters. This finding suggests that the tDNA might have never reached the cytoplasm in those cells within the tested time frame of 20 min. As a control, we repeated the quantification of the RecA-GFP and ComEA-mCherry foci in a comEC-deficient strain in which tDNA should not reach the cytoplasm. In this case, the number of cells that formed new RecA-GFP foci throughout the time-lapse experiment was extremely low and without any preference toward colocalization with ComEA-mCherry (Fig. 5F).
FIG 5 .
Time-lapse microscopy imaging of RecA-GFP ComEA-mCherry-expressing bacteria exposed to exogenous tDNA. Competence-induced V. cholerae strains expressing RecA-GFP and ComEA-mCherry were exposed to exogenous tDNA and imaged at 2-min intervals for a total of 20 min. Time points (min) are indicated above the images (A to D). Shown are examples of cells with distinctive ComEA-mCherry and RecA-GFP foci. Images taken in the fluorescence channels (mCherry, red; GFP, green) and overlays thereof (merge) are depicted. The cells are outlined with dashed lines. Observed phenotypes comprised cells that showed either de novo focus formation of mCherry and GFP (A), a preexisting mCherry focus (i.e., present at the beginning of the time-lapse experiment) and a newly formed GFP focus (B), a preexisting GFP focus and a newly formed mCherry focus (C), or preexisting foci of both constructs (D). (E) Quantification of the observed phenotypes, as exemplified in panels A to D. The arrow indicates the change of focus occurrence from a dispersed signal (−) to a focal distribution pattern (+). ± indicates cases of persisting or disappearing foci. Events with de novo RecA-GFP focus formation are shaded in gray. n.a., not applicable. (F to H) Quantification of observed phenotypes as in panel E, but for strains lacking comEC (F), dns (G), or comEC and dns (H). Scale bar, 1 µm.
Since we detected an increase in the duration of ComEA-mCherry focus persistence in the absence of Dns (Fig. 2), we wondered whether the nuclease could have an effect on the amount of tDNA that is imported into the cytosol and thus the de novo formation of RecA-GFP foci. Indeed, when we repeated the time-lapse experiment using a dns-negative strain, we observed a significant increase in de novo RecA-GFP focus formation (Fig. 5G) and an almost exclusive colocalization of these newly formed RecA clusters with the ComEA-mCherry signal (~94.4%). Importantly, a deletion of dns in a comEC-deficient background did not have such an effect (Fig. 5H).
DISCUSSION
As one of three modes of horizontal gene transfer, natural transformation is an important contributor to bacterial evolution. Nonetheless, the mechanistic basis of the DNA uptake process is still poorly understood. Current DNA uptake models suggest that most naturally competent bacteria contain a (pseudo)Tfp as part of their DNA uptake machinery, which is required for DNA transport across the outer membrane of Gram-negative bacteria or through the thick cell wall of Gram-positive bacteria (11, 20–22). It has been proposed that the DNA shuttling might therefore be driven by repeated cycles of pilus extension and retraction. Subsequently, single-stranded DNA reaches the bacterial cytoplasm after passage through the putative membrane channel formed by ComEC (43). The proteins that are involved in the DNA translocation process across the cytoplasmic membrane and the DNA-protein interactions taking place in the cytosol are most likely conserved among Gram-positive and Gram-negative bacteria (6, 11, 68); however, the cellular localization of the inner membrane transport has so far only been experimentally investigated in B. subtilis (54, 56). Notably, in this organism the majority of the competence proteins are localized to the cell pole, which is indicative of the presence of a large multiprotein complex composed of cytosolic and membrane-bound proteins (54–57). However, a strict polar localization of the Tfp and/or competence proteins was not observed in the case of S. pneumoniae (47, 69) or for the Gram-negative bacterium V. cholerae (9, 10, 41). Indeed, there is still a major knowledge gap with respect to the cellular localization of the DNA uptake complexes among bacterial species other than B. subtilis and especially in Gram-negative bacteria, where the tDNA has to cross two membranes. Notably, the localization might have an impact on the assembly and the functionality of the DNA uptake machines and therefore deserves further attention.
In the present study, we took a cell biology-based approach to indirectly visualize the DNA translocation across the outer and inner membranes of V. cholerae. Through time-lapse microscopy to visualize the fluorescently tagged ComEA and RecA proteins, both of which showed almost full functionality with respect to natural transformation, we demonstrated that the passage of the tDNA over the outer membrane and its translocation into the cytosol occurred in close proximity to each other (Fig. 1C and 5). Interestingly, the timing between both passages was not constant, and a significant delay was observable between the two crossing events, indicating that the two steps are spatially but not temporally coupled. Such a spatial restriction was not predictable as it was previously suggested that DNA uptake across the two membranes of Gram-negative bacteria might involve two independent steps (19). Indeed, DNA uptake in Gram-negative bacteria was initially studied in H. influenzae and Neisseria spp., where it was demonstrated that DNA uptake into a DNase-resistant state (e.g., into the periplasm or the “transformasome” [70]) still occurred in the absence of the inner membrane channel ComEC (homologs Rec2 in H. influenzae and ComA in Neisseria) (38, 71, 72) indicating that the outer and inner membrane translocation events are uncoupled. Such a two-step mechanism (e.g., distinguishing the transport events across the outer membrane and the cytoplasmic membrane) should not be confused with other two-step mechanisms previously proposed for naturally competent bacteria, which differentiated between initial DNA binding followed by the DNA uptake event (as mentioned, for example, by Chen and Gotschlich [37]). Indeed, it is well documented for Neisseria that DNA binding can be dissociated from DNA uptake. Notably, Neisseria species (as well as H. influenzae) preferentially take up genus-specific DNA fragments, which are recognized by the presence of so-called DNA uptake sequences (DUS) or uptake signal sequences (USS) (references 73 and 74, and recently reviewed by Mell and Redfield [68]). Such DUS/USS elements are highly overrepresented in these organisms. Interestingly, a minor pilin protein (ComP) of Neisseria meningitides has recently been shown to directly interact with DUS-containing DNA (75) thereby answering one of the many open questions in the field of natural transformation, namely, “which, if any, protein acts as a receptor for transforming DNA?” in Neisseria (13). However, comP mutants of N. gonorrhoeae, though impaired for natural transformation, were properly piliated and exhibited normal Tfp function (36). We previously demonstrated that V. cholerae does not differentiate between species-specific and species-nonspecific DNA at the level of the DNA uptake process and therefore also with respect to the initial binding event (52). Furthermore, no obvious ComP homologues exist in V. cholerae (9), suggesting that the initial DNA binding step might be different in other Gram-negative bacteria than what was reported for Neisseria spp.
That DNA uptake across the two membranes might be a two-step event was also suggested based in single-cell experiments performed on Helicobacter pylori (76), V. cholerae (41), and more recently also on N. gonorrhoeae (42). It should be noted though that H. pylori is considered the “black sheep” (6) of transformation as it uses a type IV secretion system rather than a Tfp for the DNA translocation across the outer membrane (77) and does not contain a ComEA homolog (19). H. pylori is therefore not widely representative of naturally competent Gram-negative bacteria. In these cell biology-based studies, the authors used fluorescently labeled DNA (and fluorescently tagged marker proteins) to visualize the outer membrane transport. However, none of these studies addressed the inner membrane transport, most likely due to experimental limitation (e.g., loss of fluorescence of intercalating dyes in the periplasm and inability of covalently labeled DNA to cross the cytoplasmic membrane [41, 42, 76]). In the present study, we overcame these obstacles by visualizing the participating proteins instead of the DNA itself (Fig. 1). Unfortunately, neither fluorescently tagged ComEC nor ComF resulted in strong enough signals that would allow live-cell imaging and time-lapse microscopy. This was consistent with our earlier studies that indicated low expression of comEC and that its overexpression resulted in cell death (9, 48). Notably, we refrained from overexpressing any of the reporter constructs as a recent study on the type II secretion system of V. cholerae indicated that this could result in severe localization artifacts (78). Instead, we targeted the downstream-acting proteins, namely, DprA and RecA (Fig. 1), with the latter resulting in sufficiently bright signals to perform live-cell imaging. Indeed, using a ComEA-mCherry RecA-GFP double-labeled strain of V. cholerae, we indirectly visualized the entry of the tDNA into the cytosol, which allowed us to conclude that the transport across the two membranes is spatially but not temporally coupled. Further studies are likely to indicate whether this also holds true for other naturally transformable Gram-negative bacteria such as Neisseria.
The DNA uptake mechanism might be different in Gram-positive bacteria. Indeed, Maier et al. used optical tweezers to investigate the DNA transport in B. subtilis. Such an experimental setting allowed the measurement of pulling forces exerted by the bacterium. Notably, no pulling of the tethered DNA occurred in a B. subtilis comEC mutant strain (32), even though DNA binding was readily observed in such mutants (32, 34). Based on these data, two hypothetical scenarios can be envisioned. (i) The transport of DNA in Gram-positive bacteria might be a one-step event where the pulling force is neither solely exerted by the pseudopilus, by ComEA, or a combination of both the pilus and ComEA but in addition requires the binding of cytoplasmic DNA-binding proteins (such as ComFA [79], Ssb, DprA, or RecA). (ii) The absence of ComEC might interfere with the full functionality of ComEA; thus, ComEA might still bind to the DNA, but the required ComEA clustering, which we previously suggested as being a prerequisite for pulling the tDNA toward the bacterium (41), might be inhibited.
Based on the present study, we hypothesize yet another role for ComEA and propose that the periplasmic ComEA-tDNA complexes might serve as storage reservoir for transforming DNA. This idea is based on the fact that the ComEA-tDNA complexes were not quickly resolved in most V. cholerae cells within the population but could be maintained over time, especially in the absence of nucleases (Fig. 2). It should be noted though that our current experimental setup does not allow us to exclude that some of the periplasmic tDNA might never reach the cytoplasm. If that were the case, then the periplasmic DNA might serve as a food source once the bacteria leave the biofilm state on the chitin surface (i.e., where competence is induced in V. cholerae [80]). Indeed, a switch from high cell density to low cell density would lead to an upregulation of the nuclease gene dns concomitantly with a discontinuation of comEA and comEC expression as these genes are regulated by quorum sensing (46, 48, 49, 52, 80, 81). That the nuclease Dns might foster rapid growth by providing nucleotides was suggested earlier by Blokesch and Schoolnik (46) and later experimentally confirmed by Seper et al. (82).
In line with the putative reservoir function of the periplasmic ComEA-tDNA clusters are recent findings concerning a cytoplasmic reservoir of ssDNA in naturally competent S. pneumoniae cells (83). Such ssDNA reservoirs were dependent on the second or alternative and competence-inducible single-stranded binding protein (SsbB) that is present in B. subtilis and S. pneumoniae but absent from many other bacteria, including V. cholerae. The SsbB protein of S. pneumoniae was produced at high levels upon competence induction to handle large quantities of incoming tDNA and to protect the ssDNA against degradation (83). Attaiech et al. speculated that such reservoirs would permit successive rounds of transformation, thereby contributing to the genetic plasticity of S. pneumoniae (83). We suggest that even though the two proteins SsbB and ComEA are not related to each other, they might fulfill a similar purpose in Gram-positive and Gram-negative bacteria, respectively, as they could form a complex with the tDNA, thereby creating a DNA reservoir that could transform the cells at a later time point (e.g., temporally uncoupled from the initial DNA uptake event). Interestingly, such a periplasmic reservoir function was also proposed by Gangel and colleagues in a recent study that was published while the manuscript for the present article was under review (42). Moreover, in this elegant study the authors provided evidence that the periplasm of N. gonorrhoeae is saturated with tDNA within minutes and that ComE/ComEA quantitatively controls the carrying capacity of the periplasm (42). However, further experiments will enable a better understanding of the fate of the periplasmic ComEA-tDNA clusters over time.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used throughout this study were derivatives of the V. cholerae El Tor strain A1552 (84) and are described in Table S3 in the supplemental material. Escherichia coli strain S17-1λpir (85) served as the donor for bacterial mating with V. cholerae. Bacterial cells were propagated in Luria-Bertani (LB) medium under shaking conditions and at 30 or 37°C unless otherwise stated. Solid LB agar plates contained 1.5% (wt/vol) agar. For chitin-independent competence induction in TntfoX-carrying strains, the medium was supplemented with 0.02% l-(+)-arabinose as described previously (48). Thiosulfate-citrate-bile salts-sucrose (TCBS) agar plates were prepared as instructed by the manufacturer (Fluka) and used to counterselect E. coli after bacterial conjugation. For sucrose-based sacB counterselection, NaCl-free LB-medium containing 6% (wt/vol) sucrose was used. The medium was supplemented with antibiotics at final concentrations of 50, 75, and 100 µg/ml for gentamicin, kanamycin, and ampicillin, respectively, if required. The ampicillin concentration was lowered to 50 µg/ml for competence-induced V. cholerae strains.
V. cholerae strain construction.
Genes were deleted from the parental strain A1552 (84) using either a gene disruption method based on the counterselectable suicide plasmid pGP704-Sac28 (86) or by natural transformation and FLP recombination, as recently described (TransFLP method [87–89]). Strains carrying genes coding for translational fusion proteins were generated by replacing the indigenous gene with a PCR-generated fusion construct using the TransFLP method (87–89). Standard molecular biology-based methods were used for DNA manipulations. For cloning purposes, the high-fidelity DNA polymerases Pwo (Roche) or KOD (Merck, Novagen) were used. Taq DNA polymerase (GoTaq; Promega) was used for colony PCRs. For the amplification of V. cholerae genes, the genomic DNA (gDNA) of strain A1552 served as the template. Modified DNA sequences were verified using Sanger sequencing (Microsynth, CH).
Natural transformation assays.
Transformation assays were performed as described (48) using gDNA of the A1552-lacZ-kan strain (87) as donor tDNA. Each independent experiment was repeated at least three times, and the averages of all experiments ± standard deviations (SD) are indicated. Statistical analysis of transformation frequencies was conducted in R (90). Differences were considered significant when P values of Welch’s t tests on log-transformed data were below 0.05 or 0.01.
Microscopy setup.
Microscopy images were obtained using a Zeiss Axio Imager M2 epifluorescence microscope. Details about the instrumentation, the configurations, and sample preparation and mounting, as well as sealing of the samples, are provided elsewhere (9, 41, 48). Bacterial colonies were imaged using a Leica DM IL LED Fluo microscope equipped with a HI PLAN 4×/0.10 objective and an ICC 50 HD camera module.
Protein (co)localization experiments using fluorescence microscopy.
To determine the duration of ComEA-mCherry focus persistence, the respective strains were grown aerobically in LB medium supplemented with 0.02% l-arabinose for ~7 h at 30°C. Cells were washed once in phosphate-buffered saline (PBS) buffer before being mixed with a 10.3-kb PCR fragments or phage λ DNA (48.5 kb; Roche). Time-lapse microscopy was then performed by acquiring images at 2-min intervals for 40 min (corresponding to 21 frames). Only cells that generated new ComEA-mCherry foci within 4 to 20 min were further analyzed. The times (i.e., number of frames) for which these cells maintained the ComEA clusters were counted manually.
RecA-GFP foci in non-competence-induced cells were observed after growing the bacteria in plain LB medium for 7 h and at 30°C. At that time, the cells were either directly visualized or washed and incubated for 15 min in PBS containing 5 µg/ml DAPI prior to imaging. The distribution of the RecA-GFP foci was evaluated using the MicrobeTracker software (58).
For the detection of IbpA-containing protein aggregates, strains carrying chromosomal ibpA-mCherry or ibpA-mCherry and recA-gfp were grown in plain LB medium for 6 h at 30°C. At this time point, cultures were either incubated in the absence (control) or the presence of 100 µg/ml streptomycin for 1 h at 30°C or incubated for 1 h at 40°C as indicated in the figures. The staining of chromosomal DNA was performed through the addition of DAPI (final concentration, 5 µg/ml) to bacterial cultures for at least 5 min.
To test for any colocalization between ComEA-mCherry and RecA-GFP, strains were grown under competence-inducing conditions as described previously (9, 41). Quantification in random fields of view was performed manually. Counts from three independent experiments were tested for consistency and pooled for statistical analysis. For time-lapse imaging, 50 µl of bacterial suspension was mixed with 1 µg of phage λ DNA (48.5 kb) and immediately mounted on agarose pads. Image acquisition was started within 5 to 10 min after initial exposure of bacteria to DNA (e.g., time needed for mounting the bacteria, sealing the coverslips, and defining random fields of view in the microscope). Time-lapse series were recorded at 2-min intervals for 20 min (11 frames). As for the analysis of snapshots, results from three independent experiments were checked for consistency and pooled for statistical analysis.
Analysis of microscopy images using MicrobeTracker.
The MicrobeTracker Suite (58) was used to determine the number of RecA-GFP foci per cell. The parameters of Algorithm 4 were altered slightly (splitThreshold set to 0.2 and split1 set to 1) to outline V. cholerae cells. Correct outlining was verified manually, and cells depicting severe morphological abnormalities were excluded. RecA-GFP foci were detected automatically in non-competence-induced cells using the spotFinderZ tool. Data from three biological replicates were checked for consistency and pooled for statistical analysis.
Image processing and annotation.
Image processing and manual quantification were performed using ImageJ. Linear adjustments of the contrast and brightness of microscopy images were applied to whole images and only used to optimally display gray values within the observed range. Adobe Illustrator was used for image annotation. Cells were outlined by Bézier curves according to the size and shape of the bacteria observed in the phase-contrast images.
Cell lysates, SDS-PAGE, and Western blot analysis.
For the preparation of cell lysates, bacteria expressing IbpA-mCherry were grown as described above. Cells were lysed in Laemmli buffer (adjusted for the optical density at 600 nm [OD600] of the bacterial culture) and incubated at 98°C for 15 min. Proteins were separated through SDS-PAGE using 11% acrylamide gels (91, 92) and transferred at 4°C onto polyvinylidene difluoride (PVDF) Western blotting membranes (Roche). Membrane-bound proteins were exposed to a primary antibody directed against mCherry (Biovision 5993-100; 1:5,000 diluted) and a secondary goat anti-rabbit antibody conjugated to peroxidase (Sigma A9169; 1:20,000 diluted). Immunoblots were developed using the Western Lightning-ECL enhanced chemiluminescence substrate (PerkinElmer), and signals were detected on chemiluminescence films (Amersham Hyperfilm ECL; GE Healthcare).
SUPPLEMENTAL MATERIAL
Distribution of ComEA-mCherry and RecA-GFP proteins in competence-induced bacteria. Shown are fluorescence microscopy images of competent, ComEA-mCherry- and RecA-GFP-expressing bacteria imaged in the red (mCherry), green (GFP), and phase-contrast (Ph) channels. Fluorescence channels were merged to analyze the distribution of foci (merge). (A and B) Representative images in the absence of exogenous tDNA. (C to E) Observed patterns of ComEA-mCherry and RecA-GFP foci for cells showing distinctive accumulations of both translational fusion proteins. Foci were found to colocalize (C), to cluster together without colocalization (D), or to appear nonclustered (E). The cells are outlined with dashed lines. Scale bar, 1 µm. Download
Transformation frequencies of strains expressing translational fusion constructs.
Quantification of IbpA-mCherry and RecA-GFP colocalization events.
V. cholerae strains used in this study.
ACKNOWLEDGMENTS
We acknowledge the technical assistance of Sandrine Borgeaud and Tiziana Scrignari and all members of the Blokesch lab for useful discussions. We thank Charles Van der Henst for advice on IbpA.
This work was supported by the Swiss National Science Foundation (31003 A_127029 and 31003 A_143356) and by the European Research Council (309064-VIR4ENV).
Footnotes
Citation Seitz P, Blokesch M. 2014. DNA transport across the outer and inner membranes of naturally transformable Vibrio cholerae is spatially but not temporally coupled. mBio 5(4):e01409-14. doi:10.1128/mBio.01409-14.
REFERENCES
- 1. Lorenz MG, Wackernagel W. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Macfadyen LP. 2000. Regulation of competence development in Haemophilus influenzae. J. Theor. Biol. 207:349–359. 10.1006/jtbi.2000.2179 [DOI] [PubMed] [Google Scholar]
- 3. Claverys JP, Prudhomme M, Martin B. 2006. Induction of competence regulons as a general response to stress in Gram-positive bacteria. Annu. Rev. Microbiol. 60:451–475. 10.1146/annurev.micro.60.080805.142139 [DOI] [PubMed] [Google Scholar]
- 4. Kline KA, Sechman EV, Skaar EP, Seifert HS. 2003. Recombination, repair and replication in the pathogenic Neisseriae: the 3 R’s of molecular genetics of two human-specific bacterial pathogens. Mol. Microbiol. 50:3–13. 10.1046/j.1365-2958.2003.03679.x [DOI] [PubMed] [Google Scholar]
- 5. Seitz P, Blokesch M. 2013. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol. Rev. 37:336–363. 10.1111/j.1574-6976.2012.00353.x [DOI] [PubMed] [Google Scholar]
- 6. Johnston C, Martin B, Fichant G, Polard P, Claverys JP. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12:181–196. 10.1038/nrmicro3199 [DOI] [PubMed] [Google Scholar]
- 7. Fussenegger M, Rudel T, Barten R, Ryll R, Meyer TF. 1997. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae—a review. Gene 192:125–134. 10.1016/S0378-1119(97)00038-3 [DOI] [PubMed] [Google Scholar]
- 8. Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483. 10.1038/35020000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seitz P, Blokesch M. 2013. DNA-uptake machinery of naturally competent Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 110:17987–17992. 10.1073/pnas.1315647110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Metzger LC, Blokesch M. 2014. Composition of the DNA-uptake complex of Vibrio cholerae. Mob. Genet. Elements 4:e28142. 10.4161/mge.28142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen I, Dubnau D. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241–249. 10.1038/nrmicro844 [DOI] [PubMed] [Google Scholar]
- 12. Chen I, Christie PJ, Dubnau D. 2005. The ins and outs of DNA transfer in bacteria. Science 310:1456–1460. 10.1126/science.1114021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hamilton HL, Dillard JP. 2006. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol. Microbiol. 59:376–385. 10.1111/j.1365-2958.2005.04964.x [DOI] [PubMed] [Google Scholar]
- 14. Johnsborg O, Eldholm V, Håvarstein LS. 2007. Natural genetic transformation: prevalence, mechanisms and function. Res. Microbiol. 158:767–778. 10.1016/j.resmic.2007.09.004 [DOI] [PubMed] [Google Scholar]
- 15. Claverys JP, Martin B, Polard P. 2009. The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol. Rev. 33:643–656. 10.1111/j.1574-6976.2009.00164.x [DOI] [PubMed] [Google Scholar]
- 16. Averhoff B. 2009. Shuffling genes around in hot environments: the unique DNA transporter of Thermus thermophilus. FEMS Microbiol. Rev. 33:611–626. 10.1111/j.1574-6976.2008.00160.x [DOI] [PubMed] [Google Scholar]
- 17. Burton B, Dubnau D. 2010. Membrane-associated DNA transport machines. Cold Spring Harb. Perspect. Biol. 2:a000406. 10.1101/cshperspect.a000406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Allemand JF, Maier B. 2009. Bacterial translocation motors investigated by single molecule techniques. FEMS Microbiol. Rev. 33:593–610. 10.1111/j.1574-6976.2009.00166.x [DOI] [PubMed] [Google Scholar]
- 19. Krüger NJ, Stingl K. 2011. Two steps away from novelty–principles of bacterial DNA uptake. Mol. Microbiol. 80:860–867. 10.1111/j.1365-2958.2011.07647.x [DOI] [PubMed] [Google Scholar]
- 20. Stone BJ, Kwaik YA. 1999. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 181:1395–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Graupner S, Frey V, Hashemi R, Lorenz MG, Brandes G, Wackernagel W. 2000. Type IV pilus genes pilA and pilC of Pseudomonas stutzeri are required for natural genetic transformation, and pilA can be replaced by corresponding genes from nontransformable species. J. Bacteriol. 182:2184–2190. 10.1128/JB.182.8.2184-2190.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen I, Dubnau D. 2003. DNA transport during transformation. Front. Biosci. 8:s544–s556. 10.2741/1047 [DOI] [PubMed] [Google Scholar]
- 23. Merz AJ, So M, Sheetz MP. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98–102. 10.1038/35024105 [DOI] [PubMed] [Google Scholar]
- 24. Skerker JM, Berg HC. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U. S. A. 98:6901–6904. 10.1073/pnas.121171698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wolfgang M, Park HS, Hayes SF, van Putten JP, Koomey M. 1998. Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U. S. A. 95:14973–14978. 10.1073/pnas.95.25.14973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wolfgang M, Lauer P, Park HS, Brossay L, Hébert J, Koomey M. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29:321–330. 10.1046/j.1365-2958.1998.00935.x [DOI] [PubMed] [Google Scholar]
- 27. Wolfgang M, van Putten JP, Hayes SF, Dorward D, Koomey M. 2000. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 19:6408–6418. 10.1093/emboj/19.23.6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tomb JF, el-Hajj H, Smith HO. 1991. Nucleotide sequence of a cluster of genes involved in the transformation of Haemophilus influenzae Rd. Gene 104:1–10. 10.1016/0378-1119(91)90457-M [DOI] [PubMed] [Google Scholar]
- 29. Drake SL, Koomey M. 1995. The product of the pilQ gene is essential for the biogenesis of type IV pili in Neisseria gonorrhoeae. Mol. Microbiol. 18:975–986. 10.1111/j.1365-2958.1995.18050975.x [DOI] [PubMed] [Google Scholar]
- 30. Collins RF, Davidsen L, Derrick JP, Ford RC, Tønjum T. 2001. Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J. Bacteriol. 183:3825–3832. 10.1128/JB.183.13.3825-3832.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Collins RF, Frye SA, Kitmitto A, Ford RC, Tønjum T, Derrick JP. 2004. Structure of the Neisseria meningitidis outer membrane PilQ secretin complex at 12 A resolution. J. Biol. Chem. 279:39750–39756. 10.1074/jbc.M405971200 [DOI] [PubMed] [Google Scholar]
- 32. Maier B, Chen I, Dubnau D, Sheetz MP. 2004. DNA transport into Bacillus subtilis requires proton motive force to generate large molecular forces. Nat. Struct. Mol. Biol. 11:643–649. 10.1038/nsmb783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maier B, Potter L, So M, Long CD, Seifert HS, Sheetz MP. 2002. Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. U. S. A. 99:16012–16017. 10.1073/pnas.242523299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Inamine GS, Dubnau D. 1995. ComEA, a Bacillus subtilis integral membrane protein required for genetic transformation, is needed for both DNA binding and transport. J. Bacteriol. 177:3045–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Provvedi R, Dubnau D. 1999. ComEA is a DNA receptor for transformation of competent Bacillus subtilis. Mol. Microbiol. 31:271–280. 10.1046/j.1365-2958.1999.01170.x [DOI] [PubMed] [Google Scholar]
- 36. Wolfgang M, van Putten JP, Hayes SF, Koomey M. 1999. The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol. Microbiol. 31:1345–1357. 10.1046/j.1365-2958.1999.01269.x [DOI] [PubMed] [Google Scholar]
- 37. Chen I, Gotschlich EC. 2001. ComE, a competence protein from Neisseria gonorrhoeae with DNA-binding activity. J. Bacteriol. 183:3160–3168. 10.1128/JB.183.10.3160-3168.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sinha S, Mell JC, Redfield RJ. 2012. Seventeen Sxy-dependent cyclic AMP receptor protein site-regulated genes are needed for natural transformation in Haemophilus influenzae. J. Bacteriol. 194:5245–5254. 10.1128/JB.00671-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jeon B, Zhang Q. 2007. Cj0011c, a periplasmic single- and double-stranded DNA-binding protein, contributes to natural transformation in Campylobacter jejuni. J. Bacteriol. 189:7399–7407. 10.1128/JB.01012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mullen LM, Bossé JT, Nair SP, Ward JM, Rycroft AN, Robertson G, Langford PR, Henderson B. 2008. Pasteurellaceae ComE1 proteins combine the properties of fibronectin adhesins and DNA binding competence proteins. PLoS One 3:e3991. 10.1371/journal.pone.0003991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seitz P, Pezeshgi Modarres H, Borgeaud S, Bulushev RD, Steinbock LJ, Radenovic A, Dal Peraro M, Blokesch M. 2014. ComEA is essential for the transfer of external DNA into the periplasm in naturally transformable Vibrio cholerae cells. PLoS Genet. 10:e1004066. 10.1371/journal.pgen.1004066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gangel H, Hepp C, Muller S, Oldewurtel ER, Aas FE, Koomey M, Maier B. 2014. Concerted spatio-temporal dynamics of imported DNA and ComE DNA uptake protein during gonococcal transformation. PLoS Pathog. 10:e1004043. 10.1371/journal.ppat.1004043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Draskovic I, Dubnau D. 2005. Biogenesis of a putative channel protein, ComEC, required for DNA uptake: membrane topology, oligomerization and formation of disulphide bonds. Mol. Microbiol. 55:881–896. 10.1111/j.1365-2958.2004.04430.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bergé M, Mortier-Barrière I, Martin B, Claverys JP. 2003. Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming DNA single strands. Mol. Microbiol. 50:527–536. 10.1046/j.1365-2958.2003.03702.x [DOI] [PubMed] [Google Scholar]
- 45. Mortier-Barrière I, Velten M, Dupaigne P, Mirouze N, Piétrement O, McGovern S, Fichant G, Martin B, Noirot P, Le Cam E, Polard P, Claverys JP. 2007. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell 130:824–836. 10.1016/j.cell.2007.07.038 [DOI] [PubMed] [Google Scholar]
- 46. Blokesch M, Schoolnik GK. 2008. The extracellular nuclease Dns and its role in natural transformation of Vibrio cholerae. J. Bacteriol. 190:7232–7240. 10.1128/JB.00959-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Berge MJ, Kamgoue A, Martin B, Polard P, Campo N, Claverys JP. 2013. Midcell recruitment of the DNA uptake and virulence nuclease, EndA, for pneumococcal transformation. PLoS Pathog. 9:e1003596. 10.1371/journal.ppat.1003596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lo Scrudato M, Blokesch M. 2012. The regulatory network of natural competence and transformation of Vibrio cholerae. PLoS Genet. 8:e1002778. 10.1371/journal.pgen.1002778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lo Scrudato M, Blokesch M. 2013. A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res. 41:3644–3658. 10.1093/nar/gkt041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Blokesch M. 2012. A quorum sensing-mediated switch contributes to natural transformation of Vibrio cholerae. Mob. Genet. Elements 2:224–227. 10.4161/mge.22284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Londoño-Vallejo JA, Dubnau D. 1993. comF, a Bacillus subtilis late competence locus, encodes a protein similar to ATP-dependent RNA/DNA helicases. Mol. Microbiol. 9:119–131. 10.1111/j.1365-2958.1993.tb01674.x [DOI] [PubMed] [Google Scholar]
- 52. Suckow G, Seitz P, Blokesch M. 2011. Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J. Bacteriol. 193:4914–4924. 10.1128/JB.05396-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Renzette N, Gumlaw N, Nordman JT, Krieger M, Yeh SP, Long E, Centore R, Boonsombat R, Sandler SJ. 2005. Localization of RecA in Escherichia coli K-12 using RecA-GFP. Mol. Microbiol. 57:1074–1085. 10.1111/j.1365-2958.2005.04755.x [DOI] [PubMed] [Google Scholar]
- 54. Kidane D, Graumann PL. 2005. Intracellular protein and DNA dynamics in competent Bacillus subtilis cells. Cell 122:73–84. 10.1016/j.cell.2005.04.036 [DOI] [PubMed] [Google Scholar]
- 55. Hahn J, Maier B, Haijema BJ, Sheetz M, Dubnau D. 2005. Transformation proteins and DNA uptake localize to the cell poles in Bacillus subtilis. Cell 122:59–71. 10.1016/j.cell.2005.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kramer N, Hahn J, Dubnau D. 2007. Multiple interactions among the competence proteins of Bacillus subtilis. Mol. Microbiol. 65:454–464. 10.1111/j.1365-2958.2007.05799.x [DOI] [PubMed] [Google Scholar]
- 57. Kaufenstein M, van der Laan M, Graumann PL. 2011. The three-layered DNA uptake machinery at the cell pole in competent Bacillus subtilis cells is a stable complex. J. Bacteriol. 193:1633–1642. 10.1128/JB.01128-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. 2011. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 80:612–627. 10.1111/j.1365-2958.2011.07579.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Allen SP, Polazzi JO, Gierse JK, Easton AM. 1992. Two novel heat shock genes encoding proteins produced in response to heterologous protein expression in Escherichia coli. J. Bacteriol. 174:6938–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. 2008. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl. Acad. Sci. U. S. A. 105:3076–3081. 10.1073/pnas.0708931105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ratajczak E, Zietkiewicz S, Liberek K. 2009. Distinct activities of Escherichia coli small heat shock proteins IbpA and IbpB promote efficient protein disaggregation. J. Mol. Biol. 386:178–189. 10.1016/j.jmb.2008.12.009 [DOI] [PubMed] [Google Scholar]
- 62. Van der Henst C, Charlier C, Deghelt M, Wouters J, Matroule JY, Letesson JJ, De Bolle X. 2010. Overproduced Brucella abortus PdhS-mCherry forms soluble aggregates in Escherichia coli, partially associating with mobile foci of IbpA-YFP. BMC Microbiol. 10:248. 10.1186/1471-2180-10-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J. 2012. Segregation of molecules at cell division reveals native protein localization. Nat. Methods 9:480–482. 10.1038/nmeth.1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kidane D, Carrasco B, Manfredi C, Rothmaier K, Ayora S, Tadesse S, Alonso JC, Graumann PL. 2009. Evidence for different pathways during horizontal gene transfer in competent Bacillus subtilis cells. PLoS Genet. 5:e1000630. 10.1371/journal.pgen.1000630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kidane D, Graumann PL. 2005. Dynamic formation of RecA filaments at DNA double strand break repair centers in live cells. J. Cell Biol. 170:357–366. 10.1083/jcb.200412090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Barany F, Kahn ME, Smith HO. 1983. Directional transport and integration of donor DNA in Haemophilus influenzae transformation. Proc. Natl. Acad. Sci. U. S. A. 80:7274–7278. 10.1073/pnas.80.23.7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Register JC, III, Griffith J. 1985. The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J. Biol. Chem. 260:12308–12312 [PubMed] [Google Scholar]
- 68. Mell JC, Redfield RJ. 2014. Natural competence and the evolution of DNA uptake specificity. J. Bacteriol. 196:1471–1483. 10.1128/JB.01293-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Laurenceau R, Péhau-Arnaudet G, Baconnais S, Gault J, Malosse C, Dujeancourt A, Campo N, Chamot-Rooke J, Le Cam E, Claverys JP, Fronzes R. 2013. A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae. PLoS Pathog. 9:e1003473. 10.1371/journal.ppat.1003473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kahn ME, Barany F, Smith HO. 1983. Transformasomes: specialized membranous structures that protect DNA during Haemophilus transformation. Proc. Natl. Acad. Sci. U. S. A. 80:6927–6931. 10.1073/pnas.80.22.6927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Barouki R, Smith HO. 1985. Reexamination of phenotypic defects in rec-1 and rec-2 mutants of Haemophilus influenzae Rd. J. Bacteriol. 163:629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Facius D, Meyer TF. 1993. A novel determinant (coma) essential for natural transformation competence in Neisseria gonorrhoeae and the effect of a comA defect on pilin variation. Mol. Microbiol. 10:699–712. 10.1111/j.1365-2958.1993.tb00942.x [DOI] [PubMed] [Google Scholar]
- 73. Danner DB, Deich RA, Sisco KL, Smith HO. 1980. An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene 11:311–318. 10.1016/0378-1119(80)90071-2 [DOI] [PubMed] [Google Scholar]
- 74. Goodman SD, Scocca JJ. 1988. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U. S. A. 85:6982–6986. 10.1073/pnas.85.18.6982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, Brady J, Baldwin GS, Lea SM, Matthews SJ, Pelicic V. 2013. Specific DNA recognition mediated by a type IV pilin. Proc. Natl. Acad. Sci. U. S. A. 110:3065–3070. 10.1073/pnas.1218832110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Stingl K, Müller S, Scheidgen-Kleyboldt G, Clausen M, Maier B. 2010. Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 107:1184–1189. 10.1073/pnas.0909955107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hofreuter D, Odenbreit S, Haas R. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41:379–391. 10.1046/j.1365-2958.2001.02502.x [DOI] [PubMed] [Google Scholar]
- 78. Lybarger SR, Johnson TL, Gray MD, Sikora AE, Sandkvist M. 2009. Docking and assembly of the type II secretion complex of Vibrio cholerae. J. Bacteriol. 191:3149–3161. 10.1128/JB.01701-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Londoño-Vallejo JA, Dubnau D. 1994. Mutation of the putative nucleotide binding site of the Bacillus subtilis membrane protein CoMFA abolishes the uptake of DNA during transformation. J. Bacteriol. 176:4642–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827. 10.1126/science.1120096 [DOI] [PubMed] [Google Scholar]
- 81. Antonova ES, Hammer BK. 2011. Quorum-sensing autoinducer molecules produced by members of a multispecies biofilm promote horizontal gene transfer to Vibrio cholerae. FEMS Microbiol. Lett. 322:68–76. 10.1111/j.1574-6968.2011.02328.x [DOI] [PubMed] [Google Scholar]
- 82. Seper A, Fengler VH, Roier S, Wolinski H, Kohlwein SD, Bishop AL, Camilli A, Reidl J, Schild S. 2011. Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol. Microbiol. 82:1015–1037. 10.1111/j.1365-2958.2011.07867.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Attaiech L, Olivier A, Mortier-Barrière I, Soulet AL, Granadel C, Martin B, Polard P, Claverys JP. 2011. Role of the single-stranded DNA-binding protein SsbB in pneumococcal transformation: maintenance of a reservoir for genetic plasticity. PLoS Genet. 7:e1002156. 10.1371/journal.pgen.1002156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yildiz FH, Schoolnik GK. 1998. Role of rpoS in stress survival and virulence of Vibrio cholerae. J. Bacteriol. 180:773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1:784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- 86. Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. 2004. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U. S. A. 101:2524–2529. 10.1073/pnas.0308707101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Marvig RL, Blokesch M. 2010. Natural transformation of Vibrio cholerae as a tool—optimizing the procedure. BMC Microbiol. 10:155. 10.1186/1471-2180-10-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. De Souza Silva O, Blokesch M. 2010. Genetic manipulation of Vibrio cholerae by combining natural transformation with FLP recombination. Plasmid 64:186–195. 10.1016/j.plasmid.2010.08.001 [DOI] [PubMed] [Google Scholar]
- 89. Blokesch M. 2012. TransFLP—a method to genetically modify V. cholerae based on natural transformation and FLP-recombination. J. Vis. Exp. 68:e3761. 10.3791/3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. R Development Core Team 2009. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 91. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 92. Sambrook J, Fritsch EF, Maniatis T. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of ComEA-mCherry and RecA-GFP proteins in competence-induced bacteria. Shown are fluorescence microscopy images of competent, ComEA-mCherry- and RecA-GFP-expressing bacteria imaged in the red (mCherry), green (GFP), and phase-contrast (Ph) channels. Fluorescence channels were merged to analyze the distribution of foci (merge). (A and B) Representative images in the absence of exogenous tDNA. (C to E) Observed patterns of ComEA-mCherry and RecA-GFP foci for cells showing distinctive accumulations of both translational fusion proteins. Foci were found to colocalize (C), to cluster together without colocalization (D), or to appear nonclustered (E). The cells are outlined with dashed lines. Scale bar, 1 µm. Download
Transformation frequencies of strains expressing translational fusion constructs.
Quantification of IbpA-mCherry and RecA-GFP colocalization events.
V. cholerae strains used in this study.