Abstract
Monitoring of renal graft status through peripheral blood (PB) rather than invasive biopsy is important as it will lessen the risk of infection and other stresses, while reducing the costs of rejection diagnosis. Blood gene biomarker panels were discovered by microarrays at a single center and subsequently validated and cross-validated by QPCR in gthe NIH SNSO1 randomized study from 12 US pediatric transplant programs. A total of 367 unique human PB samples, each paired with a graft biopsy for centralized, blinded phenotype classification, were analyzed (115 acute rejection (AR), 180 stable and 72 other causes of graft injury). Of the differentially expressed genes by microarray, Q-PCR analysis of a five gene-set (DUSP1, PBEF1, PSEN1, MAPK9 and NKTR) classified AR with high accuracy. A logistic regression model was built on independent training-set (n=47) and validated on independent test-set (n=198)samples, discriminating AR from STA with 91% sensitivity and 94% specificity and AR from all other non-AR phenotypes with 91% sensitivity and 90% specificity. The 5-gene set can diagnose AR potentially avoiding the need for invasive renal biopsy. These data support the conduct of a prospective study to validate the clinical predictive utility of this diagnostic tool.
Keywords: acute allograft rejection, transplantation genomics, transplantation, transplant rejection, translational research, renal transplantation, renal allograft rejection, biomarker, bioinformatics
The accurate and timely diagnosis of acute renal allograft rejection (AR) is necessary to optimize immunosuppressive drug management and preserve renal function in kidney transplant recipients. Unfortunately, the methods of diagnosis remain imperfect. Since many conditions other than AR lead to renal allograft dysfunction, the diagnosis of AR cannot be made on functional grounds alone and requires confirmation using a kidney biopsy. Although, the diagnostic biopsy criteria for AR have been codified over time (1), the diagnosis using biopsy process remains limited by sampling error, assessment variability, procedural morbidity and cost. Additionally, renal allograft dysfunction is a relatively insensitive means of detecting early AR; approximately 10% of patients with clinically normal renal function are found to have evidence of AR on surveillance biopsy (2). Ideally, a less-invasive means for diagnosing AR, could be used for surveillance of transplant recipients, thereby reducing the need for biopsy and providing a more efficient means of immune management of graft injury.
Transcriptional profiling studies on renal allograft biopsy specimens have demonstrated substantial, coordinated expression changes in many genes that uniquely identify patients with established AR, as well as other conditions in the differential diagnosis for allograft dysfunction(3)(4,5)(6). In general, these changes are related to the inflammatory infiltrate resident cells within the kidney, and associated transcriptional changes in renal tissue. However, when these studies have been applied to peripheral blood,(7)(8) the diagnostic changes related to AR have been less evident, presumably due to a reduced signal to noise ratio inherent in a site remote from the allograft (9).
In order to increase the sensitivity and specificity of detection for relatively rare biomarkers within molecularly heterogeneous samples such as peripheral blood, we employed a carefully designed methodological approach to integrate the transcriptional profiles of peripheral blood samples from patients with and without biopsy-proven AR from three different microarray platforms. Changes in peripheral blood transcriptional profiles were correlated with biopsy-proven AR, and used to distinguish AR from other common conditions arising in kidney transplant patients. The examination of changes across a highly regulated set of genes was used to assess their utility for the non-invasive diagnosis of AR and a diagnostic alternative to the invasive renal biopsy.
METHODS
Patient and Sample Information
367 peripheral blood (PB) samples from 236 unique pediatric and young adult kidney transplant recipients were enrolled (as shown in Figure 1). Within this cohort, 137 patients were enrolled from Stanford University for discovery and validation, and 99 patients from the NIH/NIAID prospective study from 12 US transplant centers, “Suppressing the Immune System With or Without Steroids in Children Who Have Received Kidney Transplants”(SNS01; NCT00141037; ClinicalTrials.gov) were enrolled for independent external validation (complete clinical data from the SNS study is discussed elsewhere in Sarwal et al (10). The study was governed by IRB approval and informed consent.
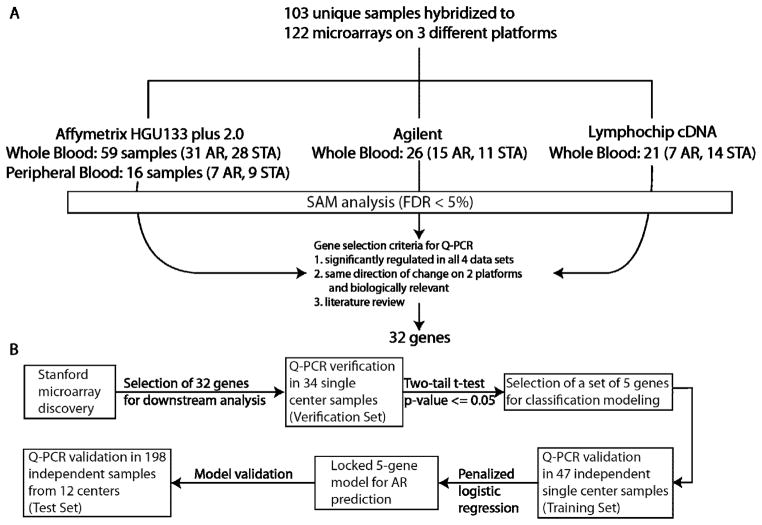
Figure 1. Summary of Study Design.
The gene-based biomarker discovery pipeline for an AR blood test follows a path of a) discovery by microarrays across 3 different platforms across a defined set (n=103) of AR and STA blood samples; followed by b) verification (n=34) and validation (n=47) on independent AR and STA blood samples; and c) a finally, prediction of AR (n=198) in other varying phenotypes of graft injury likely to be encountered in an outpatient clinical setting. Array data generated from the 3 platforms were compared by mapping the transcripts to Entrez Gene identifiers. Common genes regulated significantly in AR on each platform were identified using a common significance threshold (SAM; FDR<5%). 122 microarrays were run on 103 unique samples. 19 samples were used for correlation of within sample data, across the 3 different platforms. A 32-gene set was selected for initial verification on 34 samples (Verification Set containing 17 AR and 17 STA) chosen from the samples used on the microarrays and a significant set of 5 genes (p<0.05) were further validated in 47 independent samples from Stanford University for development of a 5-gene model by logistic regression analysis (Training Set containing 23 AR and 24 STA samples). This locked regression model generated using the 5 genes was applied to the second independent set of 198 samples (Test Set containing 32 AR, 94 STA and 72 nonAR/nonSTA clinical phenotypes) from SNSO1, for accurate AR classification. Raw microarray data are available in NCBI Gene Expression Omnibus (GEO) under Accession No. GSE14067.
Each PB sample in this study was paired with a contemporary renal allograft biopsy (within 48 hours) from the same patient. Surveillance biopsies were obtained from all patients at engraftment, 3, 6, 12 and 24 months post-transplantation and additionally at the times of suspected graft dysfunction (for SNS clinical study details see Sarwal et al (10); for SNS histology study details see Naesens et al (11)). Multiple PB-biopsy pairs from the same patient were utilized as long as each biopsy had a conclusive phenotypic diagnosis. Each biopsy was scored by the center pathologist for each enrolling clinical site; but given the possibility of discordance in biopsy reads across centers, all biopsies were blindly rescored by a single central pathologist using to the Banff (12) classification (complete SNS histology data in Naesens et al, 2012 (11)). The PB-biopsy pairs were categorized as “acute rejection” (AR; n=115), or as “stable” (STA, n=180), if there was absence of AR and any other substantial pathology. A third category of PB-biopsy pairs were characterized as “non-AR/non-STA” (n=72) if they exhibited no evidence of Banff graded AR, but either met the Banff criteria for “borderline” classification (n=12), had a diagnosis of chronic allograft nephropathy (CAN; samples had IFTA grade ≥ 1; n=37), or chronic calcineurin inhibitor toxicity (CNIT; n=16), or bacterial/viral infection or other undefined chronic graft injury (n=7).
Sample collection, RNA Extraction, Microarray Hybridization and Analysis
Blood was collected in 2.5 ml PAXgene™ Blood RNA Tubes (PreAnalytiX, Qiagen) or in Ficoll tubes for peripheral blood (PBL) isolation (the latter samples were only used for microarray discovery on Affymetrix). Total RNA was extracted using a previously published protocol9. Our goal was to maximize the power of discovering a robust gene-set for AR, and to minimize platform specific artifacts (e.g., issues of cross-hybridization (13), specificity of hybridization (14), globin gene effect9 of whole blood on the Affymetrix platform, differential stability of Cy dyes (15), platform specific bias). Furthermore, because each array platform uses different sets of genes that are represented by different probe set IDs, we used AILUN (http://ailun.stanford.edu)(16) to re-annotate the probe set IDs with the current Entrez Gene IDs. All Gene expression values were transformed to log2 for further analysis. We applied significance analysis of microarrays (SAM)(17) to identify differentially expressed genes for AR on all 3 platforms, with a threshold false discovery rate (FDR) < 5%.
Quantitative Polymerase Chain Reaction (Q-PCR)
Standard protocols were used for Q-PCR reactions on the ABI 7900 Sequence Detection System (Applied Biosystems, Foster City, CA) under standard cycle conditions (10 min at 95°C, 40 cycles of 15s 95°C, 30 s at 60°C), using gene expression assays (Applied Biosystems, Foster City, CA). The relative amount of RNA expression was calculated using a comparative CT method. Expression values were normalized to 18S using ribosomal RNA endogenous reference and universal RNA (Agilent Inc., Santa Clara; Cat #740000).
Biological Pathway and Cell Specific Expression Analysis
We used Ingenuity Pathway Analysis (IPA) to identify significant signaling pathways. We chose −log10P > 1.3 as a threshold for identifying significant pathways in IPA. We used BioGPS(18, 19) to identify the blood cell types in which the differentially expressed genes were highly expressed. A gene was highly expressed in a blood cell type if its expression in a given blood cell type was greater than 10 times its median expression over all tissues. We used hypergeometric test to determine whether the proportion of the highly expressed genes in each cell type was statistically significant or not. The p-values from hypergeometric test were corrected for multiple hypotheses using Benjamini-Hochberg correction.
Building a five-gene classification model for diagnosis of acute rejection
A schematic outline of the study is presented in Figure 1 and shows the number of samples used for discovery by microarrays (122 PB), verification by QPCR (34 PB), building an AR logistic regression model by penalized maximum likelihood method, in an independent sample set by QPCR (47 PB) and testing the performance of the model in the SNS clinical study (198 PB). Summary statistics for patient demographic and clinical variables are provided in Table 1. The 5-gene model was validated in a second independent cohort of 198 samples from SNS01 (Test Set). The Test set consisted of blood samples collected at the time of biopsy confirmed AR (n=32; (20)) with clinical graft dysfunction (greater than 10% increase from baseline serum creatinine values), and blood samples collected at the time of protocol biopsies with stable graft function (STA; n=94). There was an additional phenotype of samples within the SNSO1 sample set that was not used in the earlier process of single–center discovery and validation. These were PB collected at the time of biopsies where the diagnosis was not one of either Banff graded AR or one of normal renal histology; these samples were codified nonAR/nonSTA, and consisted of a collection of samples with different pathologies; n=72). In this latter category, many samples had clinical graft dysfunction and the different pathological categories were based on the centralized biopsy read-outs (12 borderline AR, 37 CAN, 16 CNIT and 7 other pathology).
Table 1.
Demographic Information of Peripheral Blood Samples for Microarray Experiments (n=122) and PCR Validation (n=106)*
Patient Demographics for all peripheral blood samples included in the microarray and Q-PCR studies. P values for age and post-transplant time were calculated using the T test with unequal variance. Probabilities of steroid usage, gender and donor source were calculated using Chi-Square analysis.
| Clinical Characteristics | Microarray Discovery | PCR Validation | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| AR (n=60) | STA (n=62) | p value | Training Set | Test Set | |||||
|
| |||||||||
| AR (n=23) | STA (n=24) | p value | AR (n=32) | Non-AR (n=166) | p value | ||||
| Recipients | |||||||||
| Gender, % females | 31.91% | 44.64% | 0.19 | 20.00% | 47.37% | 0.09 | 52.00% | 36.49% | 0.41 |
| Mean age, year | 12.49±5.04 | 10.94±6.01 | 0.17 | 10.31±5.24 | 14.24±5.87 | 0.05 | 11.91±5.57 | 11.85±5.69 | 0.78 |
| Immunosuppression, %SF | 51.06% | 57.14% | 0.54 | 66.67% | 73.68% | 0.67 | 44% | 44.6% | 1.00 |
| HLA match | 2.45±1.36 | 2.41±1.41 | 0.90 | 3.13±1.41 | 2.59±1.43 | 0.27 | 1.22±1.19 | 1.39±1.23 | 0.09 |
|
| |||||||||
| Donors | |||||||||
| Donor Source %LRD | 63.64% | 75.51% | 0.21 | 80.00% | 84.21% | 0.76 | 24.00% | 41.89% | 0.36 |
| Gender, % females | 53.49% | 48.00% | 0.60 | 46.67% | 63.16% | 0.35 | 40.00% | 41.89% | 1.00 |
| Mean age, year | 33.26±13.10 | 32.72±11.03 | 0.83 | 32.41±13.87 | 39.17±10.31 | 0.13 | 27.79±9.69 | 28.50±10.08 | 0.59 |
Values are means ± SD (Standard Deviation) AR, Acute rejection; STA, stable; SF, steroid free; txp, transplant; LRD, living related donor. The sample numbers for each dataset are shown. Discovery, and Training Set samples came from 137 unique Stanford patients; Test Set samples came from 99 unique SNSO1 patients. The Verification set samples came from a subset of samples used for Microarray Discovery.
Evaluation for Confounders
To examine if any demographic, clinical or immunosuppression confounders at baseline or at the time of sampling could have driven the segregation of the 5-gene set prediction score for AR, 18 different clinical confounders on the single-center samples were correlated with Q-PCR expression of each of the 5 genes in the single center data on 81 samples (34 Verification + 47 Training Set) using Pearson correlation. Additionally, we also performed univariate logistic regression for each clinical confounder with the risk of AR as well as a multivariate logistic regression model for a combination of all 18 clinical confounders and 5 genes’ expression values. The confounders were post-transplant time, recipient age, recipient gender, donor gender, donor source, donor age, steroid-free vs. steroid-based immunosuppression, total white blood cell count, hematocrit, CMV status, EBV status, BK virus infection, bacterial Infection, presence of donor-specific antibodies (DSA), panel reactive antibodies, use of induction therapy (either Daclizumab or T-cell depleting antibodies), use of calcineurin inhibitors (tacrolimus or cyclosporine), and use of anti-metabolites (mycophenolate mofetil or azathioprine).
RESULTS
Cross- Platform Microarray Discovery for AR Specific Genes in Peripheral Blood
From 122 PB, we identified 2382 differentially expressed genes (false discovery rate; FDR < 5%). All of the samples have been deposited at GSE14067 to NCBI Gene Expression Omnibus (GEO) database. These genes play a role in leukocyte extravasation, and chemokine, T-cell and B-cell receptor signaling (−log10P>1.3; IPA®; http://www.ingenuity.com). They are enriched (10× median intensity across all tissues; http://www.BioGPS.org) (19) in different blood cells, namely CD8+ T cells (126, p=3.80e-16), CD4+ T cells (118, p=5e-13), CD56+ NK cells (149, p=1.3e-9), CD33+ Myeloid cells (150, p=1.7e-8), Dendritic Cells (130, p=8.8e-8), CD14+ Monocytes (111, p=1.1e-4), CD34+ cells (119, p=4.8e-6) and CD19+ B cells (91, p=1.6e-3).
Verification of AR specific genes by Q-PCR
We chose 32 genes for QPCR verification (Figure 1A) that were differentially expressed in all microarray data sets, and were biologically relevant with enrichment of cell–specific immune responses in AR. These genes were DUSP1, IL1RAP, MCM7, NKTR, MAPK9, PSEN1, PTPRC, SLPI, STAT1, STAT3, CFLAR, IL32, PBEF1, PHLDA1, IFNGR1, IL8RA, ITGAX, PLCG1, PTPN11, TNFAIP6, ZAP70, GOLGA8A, RYBP, TLR8, RNF130, F2RL1, GRZYB, PFN1, FCGR1A, NFATC3 and IL6R. Given the recent research on the dual role of FOXP3 in rejection (21), (22) and tolerance (23, 24), it was also selected for verification. 15 genes were significantly differentially expressed between AR and STA (p-value < 0.05). Out of these 15 genes, five genes (F2RL1, STAT1, FOXP3, PTPRC and IL6R; p<0.05) have previously been shown to be involved in AR. Out of the remaining 10 genes, 8 genes were over-expressed in AR (CFLAR, p=0.0016; DUSP1, p=0.0013; IFNGR1, p=0.0062; ITGAX, p=0.0011; PBEF1, p=0.00008; PSEN1, p=0.00007; RNF130, p=0.0459; and RYBP, p=0.0012), and 2 genes were under-expressed in AR (MAPK9, p=0.0006; NKTR, p=0.0016).
Identification of the minimal discriminative gene set for AR
We applied logistic regression with best subset selection to the Verification Set in order to find the minimum number of genes necessary for the proper classification of biopsy-confirmed AR(25). Chi-square score for logistic regression models built using the 10 genes showed that in the data-set used, using five genes would have the same performance as a model using six or more genes. Additional selection criteria were used such as biological relevance and model performance (high statistical significance 10 p-value < 0.005 and low standard error of mean (SEM)), resulting in DUSP1, MAPK9, NKTR, PBEF1, and PSEN1.
Independent Validation of the 5 Genes in the Single-Center Training Set and Building the 5 Gene Diagnostic Model for AR
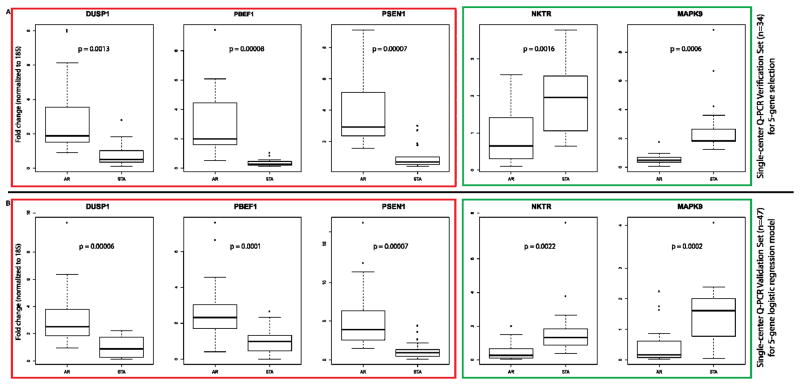
Expression of each of the five genes in an independent Training set of 47 Stanford samples (23 AR, 24 STA) was also significantly different (p-value < 0.05) (Figure 2A). This data was used to develop a logistic regression model with a penalized maximum likelihood method, which was a more robust estimation procedure than the usual maximum likelihood methods.(26, 27) In the 5 gene-set model, each of the regression coefficients describes the size of the contribution of that gene as a risk factor for diagnosing AR, where the larger the coefficient, the greater the influence of that gene in AR (Supplemental Table 1).
Figure 2. Single–center Verification and Validation of Gene Expression for the 5-Gene Set.
Box plots of the QPCR gene expression values are shown for the selected 5 genes: DUSP1, PBEF1 And PSEN1 are upregulated in AR (red outline); NKTR and MAPK9 are downregulated in AR (green outline) in the single center Verification Set (n=34; Figure 2A) and in the single center independent Training Set 1 (n=47; Figure 2B), for building the logistic regression model on the 5 gene-set. We applied logistic regression with best subset selection to the Verification Set in order to find the minimum number of genes necessary for the proper classification of biopsy-confirmed AR. Chi-square score for logistic regression models built using these 10 genes showed that increase in the score was minimal when more than five genes were used in the model. Chi-square score for logistic regression models built using all 10 genes showed that the increase in Chi-square score from a model with 1 gene to 3 genes is 7.70; from a model with 3 genes to 5 genes is 1.87; and from a model with 5 genes to a model with 6 is only an increase of 0.48. Hence, the logistic regression model using a set of 5 genes was selected based on the best performing 5-genes set (Chi-square score = 29.63) as DUSP1, PBEF1, PSEN1, MAPK9, and NKTR. The p values for comparison of gene expression data for each gene are shown in each dataset and each value is significant (p<0.05).
Evaluation for Confounders
To examine if any demographic, clinical or immunosuppression confounders at baseline or at the time of sampling could have driven the segregation of the 5-gene set prediction score for AR, 18 different clinical confounders on the single-center samples were correlated with Q-PCR expression of each of the 5 genes in the Training set of 47 samples (23 AR, 24 STA) using Pearson correlation. Univariate logistic regression was also done for each clinical confounder with the risk of AR as well as a multivariate logistic regression model for a combination of all 18 clinical confounders and 5 genes’ expression values. By t-test, all 5 genes had significant change in expression only with the presence of donor specific antibody (DSA; p<0.05). By univariate logistic regression model, all 5 genes were significantly associated with AR (p<0.0001; AUC from 0.829–0.938) and DSA positivity (p<0.0001; AUC=0.828) while there was no association with the histology grade or C4d positivity (p=0.80 for Banff score; p=0.79 for C4d positivity). These data thus underscore that the coordinated expression of the 5-gene set in peripheral blood can diagnose AR with high confidence, irrespective of the differences in patient characteristics, immunosuppression and rejection timing.
Independent validation of the 5-gene model in the multi-center SNSO1 sample set
The 5-gene model was validated in a second independent cohort of 198 samples (Test Set) collected in 12 different centers as part of the SNSO1 study (Figure 2B). The test set consisted of PB-biopsy pairs with AR, STA, and an additional phenotype of samples within the SNSO1 sample set that was not used in the earlier process of single–center discovery and validation. These PB samples were collected at the time of biopsies where the diagnosis was not one of either Banff graded AR or one of normal renal histology; these samples were codified nonAR/nonSTA, and consisted of a collection of samples with different pathologies; n=72; 12 borderline AR, 37 CAN, 16 CNIT and 7 other pathology
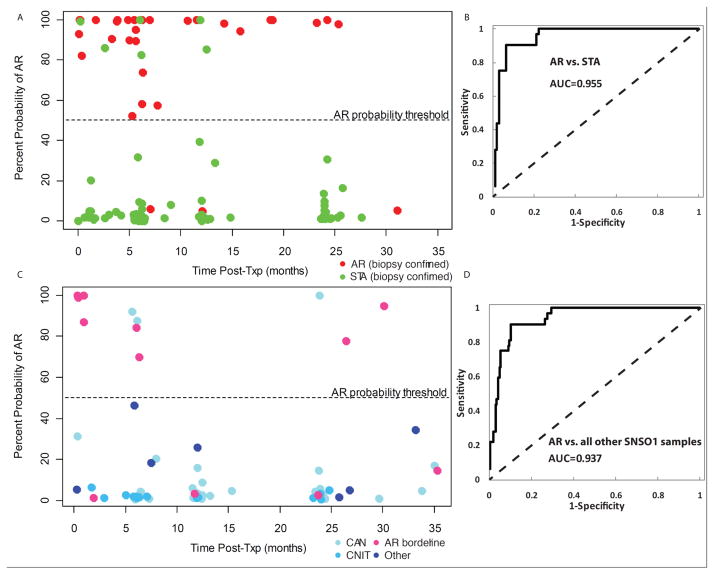
The accuracy of the 5-gene model was assessed by evaluating the sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) on the AR and STA samples, as well as the AR and non-AR in the Test Set (Figure 3A). The 5-gene model has 91% sensitivity, 94% specificity, 83% PPV, 97% NPV, and 92% accuracy, to separate AR from STA samples (AUC 0.955; Figure 3B); and 91% sensitivity, 90% specificity, 63% PPV, 98% NPV, and 90% accuracy to separate AR samples from all other phenotypes (STA and non-AR/non-STA; Figure 3C; AUC 0.937, Figure 3D). It is important to note that 8/12 of samples from patients classified as borderline rejection on biopsy were classified as AR by the 5-gene model (these have been classified as true negatives in the non-AR/non-STA cohort, but it can be argued that these samples could also be true positives). The high prediction of an AR phenotype in the borderline AR samples suggests that pre-clinical injury in AR may also be identified by Q-PCR analysis of a peripheral blood sample and suggest earlier treatment for the patient.
Figure 3. Multi-Center Validation of the QPCR Prediction Probability for AR by the 5-Gene Set.
A dot plot is shown for individual percent probability prediction score for AR on 198 independent samples over the course of the 3 year follow-up (time post-transplant in months on the Axis) in the SNSO1 multicenter study. Each blood sample is paired with a biopsy for blinded, centralized, histological diagnosis of the phenotype. Based on the Receiver Operating Characteristic (ROC) curve for the logistic regression model across DUSP1, PBEF1, PSEN1, MAPK9, and NKTR, a cutoff of θ = 0.52 was selected to have the best sensitivity and specificity to discriminate between AR and STA. In other words, the prediction probability has been derived from the logistic regression model across the 5 genes (Y Axis) and percent probability prediction score of >52% predicts the sample to have an AR phenotype. In Figure 3A the 32 AR samples are shown by red dots, with 3 misclassifications (91% accuracy within class); the 94 STA samples are shown by green dots with 6 misclassifications (92% accuracy). The ROC curve for AR vs STA class is shown in Figure 3B. In Figure 3C the 72 nonAR/nonSTA samples are shown, divided into 4 categories: 12 AR borderline (pink dots), 37 CAN (light blue dots), 16 CNIT (cyan dots) and 7 other diagnoses such as reflux nephropathy (n=2), BK nephropathy (n=1), FSGS recurrence (n=1) (dark blue dots) and ischemia (n=3). Within the AR borderline class 4 samples have <50% prediction scores for AR and misclassify, giving the within class accuracy of 67% (8/12 samples) for borderline AR. The ROC curve for AR vs nonAR/nonSTA class is shown in Figure 3D.
DISCUSSION
In the current study, we used a cross-platform, high-throughput, transcript profiling approach to identify a highly specific, biologically meaningful, concise gene set in peripheral blood whose expression correlates well with the AR/noAR status of contemporaneous biopsies collected from the same patients. A logistic regression model built on a set of 5 genes in peripheral blood and extensively validated by Q-PCR, accurately diagnosed rejection, with 91% sensitivity and 90% specificity, substantially improving on any current available method for specifically diagnosing AR. Importantly, though the 5-gene test was developed for a binary comparison of AR and STA samples, it was validated in an independent cohort that comprised of samples obtained at 12 different transplant centers, in patients with varying demographics and across multiple clinical phenotypes, such as CAN, CNIT, infection, and acute tubular necrosis, subclinical AR, clinical AR and STA. As the model was built using samples from a single center, and was validated in an independent multicenter cohort, general applicability of this test in real-world appears feasible where the patient population will contain heterogeneous graft conditions along the continuum from stable to AR.
The SNSO1 trial arbitrarily assigned borderline AR in the nonAR/nonSTA category, but in retrospect, this might not have been biologically accurate as most of the “misclassifications” were actually borderline AR, and their inclusion in the AR group would further enhance the PPV of the test. This suggests a longer biologic process than previously thought in immune changes leading to rejection. It would be important to evaluate serial samples from patients developing clinical AR episodes to examine if the 5-gene model can detect subclinical acute rejection, that is, acute rejection prior to its becoming clinically evident. Some of the Banff graded ARs were borderline (n=12) but were scored as AR biopsies in the SNSO1 validation sample to help compensate for the small number of AR events (n=32) that met the conventional AR criteria. Thus the 5-gene model was used to predict the set of AR and subclinical borderline AR biopsies in the SNS01 subjects.
Early minimally-invasive diagnosis of AR would be a significant advance over current practice standards that depend on biopsy for diagnosis and initiation of treatment. At present, by the time a clinical trigger is available to warrant doing a biopsy for rejection diagnosis, the rejection has evolved with its full humoral or cellular mechanisms. Having a clinical indication for the rejection episode, based on the high score on the 5-gene test, that is earlier than a rise in the serum creatinine, would be a significant advance for the management of patients, as it would result in the earlier diagnosis of rejection and provide an early trigger for performing an indicated biopsy, if warranted. Work is underway in our group to refine the performance of the larger gene-set for discriminating cellular from humoral rejection, clinically important for discriminating treatment for AR.
The excellent positive and negative predictive values of the 5-gene model suggest that a peripheral blood test based on these genes could be useful for screening patients for absence of AR. Given the excellent discrimination of this test, there is strong justification for a larger, more definitive follow-up study with a larger number of AR patients for study, to evaluate if a higher AR gene score translates into risk of more aggressive AR or humoral versus cellular AR. The strong negative predictive value of the model for diagnosing absence of AR opens the door for personalized therapy, where patients can be potentially screened serially by the 5-gene test, and in the absence of AR risk, have reduced follow-up, be candidates to avoid unnecessary protocol biopsies and, in the presence of graft dysfunction, be evaluated for alternative etiologies, such as infection, obstruction or toxicity.
The 5-gene blood test may also provide a new means to monitor for resolution of AR after treatment intensification. Additional samples will have to be evaluated from patients undergoing treatment of AR to examine if immunosuppression intensification causes a decrement of the 5-gene test prediction score, commensurate with histological resolution of the AR episode, perhaps guiding assessment of a patient’s response to therapy.
The PB genes most strongly associated with graft rejection, do not correlate with multiple demographic, clinical, treatment modality and bacterial/viral infection parameters. Although there is significant correlation with DSA positivity, our model predicts AR, irrespective of cellular or humoral AR. We are further analyzing our data to develop a blood gene-based model that can further distinguish humoral from cellular rejection. Even though this is a minimal set of 5 genes for AR classification, expanding out to other populations may require the inclusion of the 10 gene-set. The 5 genes are central to leukocyte trafficking and T/B cell activation, and are mostly expressed in by activated monocytes in the peripheral circulation, reflecting injury mechanisms relating to oxidative cellular stress responses (DUSP1), apoptosis (MAPK9), IL2 dependant activation of cytolytic genes (NKTR), increased cell adhesion via the e-cadherin/catenin complex (PSEN1), and vascular smooth muscle injury (PBEF1). It is likely that these genes play a pivotal role in the mechanism of cytolysis and graft microvasculature injury from activated monocytes in graft rejection (28, 29) The association of the gene profile of the selected genes in blood with DSA and peripheral trafficking of monocytes supports the growing recognition of DSA as a culprit in graft injury (30) (31) and monocytes as primary culprits in graft dysfunction (32, 33).
Serial performance of the 5-gene test proposed in the current study suggests a means to stratify patients as high or low risk for rejections, even in the presence of other histological injuries in the graft. It may be anticipated that the more frequent assessment of risk afforded by the minimally-invasive nature of this assay will facilitate more prompt therapeutic management which may alter the course of rejection, providing a critical, and as yet unavailable, new dimension of immunosuppression customization for a transplant patient. However, a couple of caveats should be noted. The sample numbers in the discovery set are limited, but are offset by the power of validating the discovery in the SNSO1 multicenter study. As this study was performed in children and young adults, the nature of the rejection may be more aggressive due to either the size mismatch of adult-sized organ and infant recipient, or the higher rate of treatment non-adherence adolescent recipient, both of which could result in stronger immune response signal. Additionally, none of the pediatric patients in this study received induction with anti-CD52 depletion therapy or with co-stimulatory blockade. Therefore, the performance of the 5-genes model should be further studied for its potential to diagnose rejection in patients of all ages, in larger sample cohorts and in different immunosuppressive regimens. The empirical results of the diagnostic potential of the selected 5-gene panel in this study suggest potential clinical utility and support the future development of a prospective clinical trial in children and extension of this work in adult renal transplant recipients to confirm clinical application.
Supplementary Material
Acknowledgments
We deeply appreciate the participation of patients at Stanford University and the NIH funded multicenter randomized study of steroid-avoidance versus steroid-based immunosuppression (SNSO1). We also are indebted to the support with patient recruitment and sample collections from transplant patients in the SNSO1 multicenter study centers. We thank Nancy Bridges and Daniel Rotrosen from NIH/NIAID for their continuous support and advice on the SNSO1 study. We thank Nikki Williams for the support throughout the manuscript preparation. The authors are grateful to Dr. Neeraja Kambham from Stanford University for her centralized, blinded reads of graft pathology and to Dr. Allan Kirk from Emory University for his support and suggestions to make the manuscript more meaningful. We are thankful to David Ikle, Michael Riggs, and Katie Poole in the validation phase of this project. Support from for this project was funded by NIH grants UO1AI055795 (OS awarded within the Cooperative Clinical Trials in Pediatric Transplantation Consortium), RO1AI061739 (MS), and ARRA funding 3UO1 AI077821-0351 (AK) awarded within the Cooperative Clinical Trials in Pediatric Transplantation Consortium.
ABBREVIATIONS
- AR
Acute Rejection
- AZA
Azathioprine
- BKV
BK Virus
- CMV
Cytomegalovirus
- CSA
cyclosporine A
- DSA
Donor Specific Antibodies
- EBV
Epstein-Barr virus
- FDR
false discovery rate
- FK
FK506
- HCT
Hematocrit
- LRD
living related donors
- MMF
Mycophenolate Mofetil
- PBL
Peripheral Blood Leukocytes
- PB
Peripheral Blood
- PAM
Prediction Analysis of Microarrays
- PRA
Panel Reactive Antibody
- Q-PCR
quantitative real time polymerase chain reaction
- ROC
Receiver Operating Characteristic
- SAM
Significance Analysis of Microarrays
- STA
stable
- WBC
White blood cell count
Footnotes
DISCLOSURE
Dr. Sarwal receives consulting and lecture fees from Bristol Meyers Squibb, Genentech and Astellas and has equity/ownership stock in Organ-I; Dr. Butte receives consulting fees from Johnson & Johnson, Genstruct, Lilly and Tercica, lecture fees from Siemens and Lilly and equity ownership/stock from Genstruct and NuMedii; Dr. Davis has equity ownership/stock in Affymetrix and Organ-I. VRD has received consulting fees from Bristol-Myers-Squibb and honoraria from Genzyme and Alexion.
Contributor Information
Li Li, Email: dlleely@gmail.com.
Purveshkumar Khatri, Email: pkhatri@stanford.edu.
Tara K. Sigdel, Email: sigdelt@cpmcri.org.
Tim Tran, Email: trantim@cpmcri.org.
Lihua Ying, Email: lihua.Ying@stanford.edu.
Matthew Vitalone, Email: vitalone@cpmcri.org.
Amery Chen, Email: amechen730@gmail.com.
Szu-chuan Hsieh, Email: Hsiehs@cpmcri.org.
Hong Dai, Email: Daih@cpmcri.org.
Meixia Zhang, Email: meixia_Zhang@yahoo.com.
Maarten Naesens, Email: maarten.naesens@uzleuven.be.
Valeriya Zarkhin, Email: lera_makarenkova@yahoo.com.
Poonam Sansanwal, Email: sansan@cpmcri.org.
Rong Chen, Email: Rchen1@stanford.edu.
Michael Mindrinos, Email: mindrinos@stanford.edu.
Wenzhong Xiao, Email: wzxiao@gmail.com.
Mark Benfield, Email: benfield@pednephal.com.
Robert Ettenger, Email: Rettenger@mednet.ucla.edu.
Vikas Dharnidharka, Email: vikasmd@ufl.edu.
Robert Mathias, Email: rmathias@nemours.org.
Anthony Portale, Email: aportale@peds.ucsf.edu.
Ruth McDonald, Email: eduruth.mcdonald@seattlechildrens.org.
William Harmon, Email: william.harmon@childrens.harvard.edu.
David Kershaw, Email: dkershaw@med.umich.edu.
V. Matti Vehaskari, Email: vvehas@lsuhsc.edu.
Elaine Kamil, Email: Elaine.Kamil@cshs.org.
H. Jorge Baluarte, Email: bBaluarte@email.chop.edu.
Brad Warady, Email: bwarady@cmh.edu.
Ron Davis, Email: dnamarkr@stanford.edu.
Atul J. Butte, Email: abutte@stanford.edu.
Oscar Salvatierra, Email: oscars@stanford.edu.
Minnie Sarwal, Email: sarwalm@cpmcri.org.
References
- 1.Racusen LC. The Banff schema and differential diagnosis of allograft dysfunction. Transplant Proc. 2004;36(3):753–4. doi: 10.1016/j.transproceed.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 2.Thierry A, Thervet E, Vuiblet V, Goujon JM, Machet MC, Noel LH, et al. Long-term Impact of Subclinical Inflammation Diagnosed by Protocol Biopsy One Year After Renal Transplantation. Am J Transplant. 2011 doi: 10.1111/j.1600-6143.2011.03695.x. Epub 2011/09/03. [DOI] [PubMed] [Google Scholar]
- 3.Sarwal M, Chua MS, Kambham N, Hsieh SC, Satterwhite T, Masek M, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349(2):125–38. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 4.Park W, Griffin M, Grande JP, Cosio F, Stegall MD. Molecular evidence of injury and inflammation in normal and fibrotic renal allografts one year posttransplant. Transplantation. 2007;83(11):1466–76. doi: 10.1097/01.tp.0000265501.33362.d3. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann SC, Hale DA, Kleiner DE, Mannon RB, Kampen RL, Jacobson LM, et al. Functionally significant renal allograft rejection is defined by transcriptional criteria. Am J Transplant. 2005;5(3):573–81. doi: 10.1111/j.1600-6143.2005.00719.x. Epub 2005/02/15. [DOI] [PubMed] [Google Scholar]
- 6.Mannon RB, Kirk AD. Beyond histology: novel tools to diagnose allograft dysfunction. Clin J Am Soc Nephrol. 2006;1(3):358–66. doi: 10.2215/CJN.01681105. Epub 2007/08/21. [DOI] [PubMed] [Google Scholar]
- 7.Deng MC, Eisen HJ, Mehra MR, Billingham M, Marboe CC, Berry G, et al. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. 2006;6(1):150–60. doi: 10.1111/j.1600-6143.2005.01175.x. [DOI] [PubMed] [Google Scholar]
- 8.Flechner SM, Kurian SM, Head SR, Sharp SM, Whisenant TC, Zhang J, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4(9):1475–89. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Ying L, Naesens M, Xiao W, Sigdel T, Hsieh S, et al. Interference of globin genes with biomarker discovery for allograft rejection in peripheral blood samples. Physiol Genomics. 2008;32(2):190–7. doi: 10.1152/physiolgenomics.00216.2007. [DOI] [PubMed] [Google Scholar]
- 10.Sarwal M, Ettenger R, Dharnidharka V, Benfield M, Mathias R, Portale A, et al. Complete steroid avoidance is effective and safe in children with renal transplants: a prospective multicenter randomized controlled trial with 3 year follow up. Am J Transplant. 2012 doi: 10.1111/j.1600-6143.2012.04145.x. Accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naesens M, Salvatierra O, Jr, Benfield M, Ettenger R, Dharnidharka V, Harmon W, et al. Steroid avoidance in pediatric kidney recipients does not influence subclinical inflamation or chronic renal allograft injury. Am J Transplant. 2012 doi: 10.1111/j.1600-6143.2012.04144.x. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mengel M, Sis B, Haas M, Colvin RB, Halloran PF, Racusen LC, et al. Banff 2011 Meeting Report: New Concepts in Antibody-Mediated Rejection. Am J Transplant. 2012;12(3):563–70. doi: 10.1111/j.1600-6143.2011.03926.x. Epub 2012/02/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naef F, Lim DA, Patil N, Magnasco M. DNA hybridization to mismatched templates: a chip study. Phys Rev E Stat Nonlin Soft Matter Phys. 2002;65(4 Pt 1):040902. doi: 10.1103/PhysRevE.65.040902. Epub 2002/05/15. [DOI] [PubMed] [Google Scholar]
- 14.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 15.Randolph JB, Waggoner AS. Stability, specificity and fluorescence brightness of multiply-labeled fluorescent DNA probes. Nucleic Acids Res. 1997;25(14):2923–9. doi: 10.1093/nar/25.14.2923. Epub 1997/07/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen R, Li L, Butte AJ. AILUN: reannotating gene expression data automatically. Nat Methods. 2007;4(11):879. doi: 10.1038/nmeth1107-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome biology. 2009;10(11):R130. doi: 10.1186/gb-2009-10-11-r130. Epub 2009/11/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101(16):6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Racusen LC, Solez K, Colvin RB, Bonsib SM, Castro MC, Cavallo T, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55(2):713–23. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- 21.Brown K, Moxham V, Karegli J, Phillips R, Sacks SH, Wong W. Ultra-localization of Foxp3+ T cells within renal allografts shows infiltration of tubules mimicking rejection. Am J Pathol. 2007;171(6):1915–22. doi: 10.2353/ajpath.2007.070396. Epub 2007/11/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan Z, Spencer JA, Lu Y, Pitsillides CM, Singh G, Kim P, et al. In vivo tracking of ‘color-coded’ effector, natural and induced regulatory T cells in the allograft response. Nat Med. 2010;16(6):718–22. doi: 10.1038/nm.2155. Epub 2010/05/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bestard O, Cruzado JM, Mestre M, Caldes A, Bas J, Carrera M, et al. Achieving donor-specific hyporesponsiveness is associated with FOXP3+ regulatory T cell recruitment in human renal allograft infiltrates. J Immunol. 2007;179(7):4901–9. doi: 10.4049/jimmunol.179.7.4901. Epub 2007/09/20. [DOI] [PubMed] [Google Scholar]
- 24.Graca L, Cobbold SP, Waldmann H. Identification of regulatory T cells in tolerated allografts. J Exp Med. 2002;195(12):1641–6. doi: 10.1084/jem.20012097. Epub 2002/06/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derksen S, Keselman HJ. Backward, forward and stepwise automated subset selection algorithms: Frequency of obtaining authentic and noise variables. British journal of mathematical & statistical psychology. 1992;45(2):265–82. [Google Scholar]
- 26.Heinze G, Schemper M. A solution to the problem of separation in logistic regression. Stat Med. 2002;21(16):2409–19. doi: 10.1002/sim.1047. Epub 2002/09/05. [DOI] [PubMed] [Google Scholar]
- 27.Heinze G. A comparative investigation of methods for logistic regression with separated or nearly separated data. Stat Med. 2006;25(24):4216–26. doi: 10.1002/sim.2687. Epub 2006/09/07. [DOI] [PubMed] [Google Scholar]
- 28.Steiniger B, Stehling O, Scriba A, Grau V. Monocytes in the rat: phenotype and function during acute allograft rejection. Immunol Rev. 2001;184:38–44. doi: 10.1034/j.1600-065x.2001.1840104.x. Epub 2002/06/28. [DOI] [PubMed] [Google Scholar]
- 29.Stehling O, Grau V, Steiniger B. Monocyte cytotoxicity during acute kidney graft rejection in rats. Int Immunol. 2004;16(1):101–10. doi: 10.1093/intimm/dxh007. Epub 2003/12/23. [DOI] [PubMed] [Google Scholar]
- 30.Zhu L, Lee PC, Everly MJ, Terasaki PI. Detailed examination of HLA antibody development on renal allograft failure and function. Clin Transpl. 2008:171–87. Epub 2008/01/01. [PubMed] [Google Scholar]
- 31.Terasaki P, Mizutani K. Antibody mediated rejection: update 2006. Clin J Am Soc Nephrol. 2006;1(3):400–3. doi: 10.2215/CJN.02311205. [DOI] [PubMed] [Google Scholar]
- 32.Zecher D, van Rooijen N, Rothstein DM, Shlomchik WD, Lakkis FG. An innate response to allogeneic nonself mediated by monocytes. J Immunol. 2009;183(12):7810–6. doi: 10.4049/jimmunol.0902194. Epub 2009/11/20. [DOI] [PubMed] [Google Scholar]
- 33.Fahim T, Bohmig GA, Exner M, Huttary N, Kerschner H, Kandutsch S, et al. The cellular lesion of humoral rejection: predominant recruitment of monocytes to peritubular and glomerular capillaries. Am J Transplant. 2007;7(2):385–93. doi: 10.1111/j.1600-6143.2006.01634.x. Epub 2007/02/07. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.