Background: Pex26p and AAA peroxins are required for the peroxisome biogenesis.
Results: Pex26p directly binds to Pex14p and acts as a scaffold protein to specify the molecular target of AAA peroxins.
Conclusion: AAA peroxins modulate the interaction between Pex26p and Pex14p on peroxisome membrane.
Significance: Functional modulation by Pex26p and AAA peroxins of the interaction between peroxins makes important contribution to peroxisome biogenesis.
Keywords: ATPase, Peroxisome, Protein Complex, Protein Targeting, Scaffold Protein, AAA Peroxins, Pex14p, Pex26p, Protein Import
Abstract
Pex1p and Pex6p are required for the relocation of the import receptor Pex5p from the peroxisomal membrane to the cytosol. We herein show that mammalian Pex26p directly binds to Pex14p, the initial docking receptor of Pex5p, and interacts with Pex5p via Pex14p. The binding affinity of Pex26p to Pex14p is altered by Pex5p. Further evidence suggests that the N-terminal region in Pex26p acts as a scaffold protein to recruit Pex14p·Pex5p complex together with Pex1p·Pex6p complexes on peroxisomes. Pex26p binding to Pex14p was suppressed by overexpression of Pex1p and Pex6p in an ATP-dependent manner, whereas Pex14p was not competed out by Pex1p and Pex6p from Pex26p mutant defective in peroxisomal matrix protein import. These results suggested that peroxisome biogenesis requires Pex1p- and Pex6p-regulated dissociation of Pex14p from Pex26p. Pex1p homo-oligomer directly binds to Pex5p as assessed by a surface plasmon resonance-based assay. Moreover, cytosolic Pex1p is likely to maintain the functional oligomer of Pex5p. Taken together, in the peroxisomal protein import, AAA peroxins modulate the interaction between Pex26p and Pex14p on peroxisome membrane as well as Pex5p oligomer in the cytosol.
Introduction
Peroxisomal matrix proteins are synthesized on free polyribosomes in the cytosol and then imported post-translationally into peroxisomes (1). They have two types of peroxisome targeting signals (PTSs)3 (2, 3): the C-terminal uncleavable tripeptides PTS1 and the N-terminal cleavable nonapeptides PTS2. More than 30 proteins, termed as PEX gene products or peroxins, are required for peroxisome assembly. Several peroxins have been delineated in their biological features. Pex5p and Pex7p are mobile receptors that shuttle between the cytosol and peroxisomes for import of PTS1 and PTS2 proteins, respectively (4, 5). Pex14p locates on peroxisome membranes and is suggested to be an initial docking site of cargo-mobile receptor complexes (6–8). Another peroxisomal integral membrane protein, Pex13p, binds to Pex14p and likewise participates in import of peroxisomal matrix proteins (9). Pex2p, Pex10p, and Pex12p, peroxisomal integral membrane proteins with RING finger at the C-terminal regions, play an important role in translocation of proteins across the peroxisome membrane (10). How these proteins cooperate in the import of matrix proteins across the peroxisomal membrane is still unresolved.
PEX26 encodes a 34-kDa type-II peroxisomal membrane protein with one C-terminal transmembrane segment, exposing its N-terminal part to the cytosol, and was identified as the last causal gene responsible for peroxisome biogenesis disorders (PBDs) of 14 complementation groups (CGs) thus far reported (11, 12). Pex26p interacts with Pex1p·Pex6p complexes and recruits them to peroxisomes (13). Yeast Pex15p, a functional homolog of mammalian Pex26p, forms a complex with Pex1p and Pex6p (14). Pex1p and Pex6p are members of the large AAA protein family and have two AAA cassettes, called D1 and D2. The AAA cassette is a 200–250-amino acid sequence conserved in AAA protein family and consists of Walker A, Walker B, and SRH (second region of homology) (15–17). The lysine residue in the Walker A is concerned with ATP binding, and the acidic residue in the Walker B is involved in ATP hydrolysis (18, 19). AAA peroxins, Pex1p and Pex6p, interact with each other in an ATP-dependent manner (20). We assigned the regions of these AAA peroxins involved in the interaction of Pex1p and Pex6p and elucidated pivotal roles of AAA cassettes in Pex1p·Pex6p interaction and peroxisome biogenesis (13). The N-terminal region of Pex26p is indispensable for the recruiting of Pex1p·Pex6p complex to peroxisomes and the transport of matrix proteins (13, 14). It is noteworthy that yeast and human AAA peroxins are required for the export of Pex5p from the peroxisomal membrane and shuttling back to the cytosol (21–23). However, it remains obscure how the AAA peroxins export Pex5p to the cytosol.
Rosenkranz et al. (24) showed that Pex15p is an incorporated component of the importomer on peroxisomal membrane. However, direct or indirect connections between Pex15p and the importomer complexes are not yet clear. As a further step to understanding the function of ternary complex of Pex26p, Pex6p, and Pex1p in peroxisomal protein import, we investigated the binding partner of Pex26p. Here we show by biochemical characterization that Pex26p forms a complex with Pex14p and Pex5p and acts as a scaffold protein for AAA peroxins. Moreover, we propose that peroxisome biogenesis requires the modulation of peroxin-peroxin interactions by AAA peroxins and Pex26p.
EXPERIMENTAL PROCEDURES
Cell Culture and DNA Transfection
Chinese hamster ovary (CHO) cells were cultured at 37 °C or 30 °C in Ham's F-12 medium supplemented with 10% fetal calf serum under 5% CO2 and 95% air (25). HEK293 cells were cultured at 37 °C in Dulbecco's modified Eagle's medium-high glucose supplemented with 10% fetal calf serum. DNA transfection to CHO cells was performed by lipofection method with Lipofectamine (Invitrogen).
Morphological Analysis
Peroxisomes were visualized by indirect immunofluorescence light microscopy using rabbit antibodies against PTS1 peptide (26). Antigen-antibody complexes were detected with fluorescein isothiocyanate-labeled goat antibodies against rabbit immunoglobulin G (MP Biomedicals-Cappel, Irvine, CA) under a Carl Zeiss Axioskop FL microscope.
Generation of Mutant Constructs
The mutations identified in the CG8 PBD patient PEX26, termed L45P, G255insT, intG231T (27), were introduced into pCMVSPORT/FLAG-HsPEX26 as described in Matsumoto et al. (28). FLAG-PEX26 variants truncated in the N- and C-terminal region were constructed as follows. To generate C-terminal deletion mutants, FLAG-PEX26-(1–133) and -(1–111) BglII-NotI fragments were prepared by PCR using pCMVSPORT/FLAG-HsPEX26 as a template with each set of primers, 1F (5′-AAGCTTGAGATCTCAAGAGCGATTCTTCGACC-3′) with 133R (5′-AGTAGCGGCCGCTCACTCTTGCATTTTGCT-3′) and 1F with 111R (5′-AGTAGCGGCCGCTCACTGGTAATACTGAAG-3′). The respective fragments were separately cloned into the BamHI-NotI sites of pCMVSPORT/FLAG-PEX26, resulting in pCMVSPORT/FLAG-PEX26-(1–133) and pCMVSPORT/FLAG-PEX26-(1–111), respectively. Likewise, the N-terminal deletion mutants FLAG-PEX26-(49–305), -(78–305), and -(97–305) were constructed by replacing the BamHI-NotI region of full-length FLAG-PEX26 in pCMVSPORT with BglII-NotI fragments of the PCR products amplified by a reverse primer, NotR (5′-GCCCGCGGCCGCTCAGTCACGGATGCGGAG-3′) and forward primers, 12F (5′-CTCTGAGATCTCTCTCAGGGGGCTCGGGGG-3′), 49F (5′-CTCCTGGAGATCTCTCTGGACTTCCGGGCG-3′), 78F (5′-CCCGAGATCTCTTCATTGGAGGTGAAGTGC-3′), and 97F (5′-CTGGAGATCTCTGATCGGTGGCAAGAAGTC-3′). Mutagenesis in the Walker motifs of Pex1p influenza virus hemagglutinin (HA) and Pex6p-HA was performed by overlap extension by PCR as described in Matsumoto et al. (11). All constructs were confirmed by nucleotide sequencing.
ATP Depletion Treatment
HEK293 cells were first incubated in ATP depletion medium (DMEM lacking glucose, supplemented 10 mm sodium azide, 10 mm sodium fluoride, and 50 mm 2-deoxy-d-glucose) for 15 min at 37 °C and then shifted to 16 °C for 2 h (29).
Co-immunoprecipitation Assay
HEK293 cells were lysed with lysis buffer containing 50 mm Tris-HCl, pH 7.4, 10% glycerol, 150 mm NaCl, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 1 mm EDTA, 1 mm dithiothreitol, and a protease inhibitor mixture. Pex26p was immunoprecipitated with antibody against Pex26pC conjugated to protein-A-Sepharose (GE Healthcare) (11). Peroxisome-deficient pex26 CHO mutant ZP167 and wild type CHO-K1 cells were transfected with normal or mutant FLAG-HsPEX26, pCMVSPORT/HsPEX1-HA, pCMVSPORT/HsPEX6-HA, and pCMVSPORT/HsPEX14. After a 2-day culture at 30 °C, cells were directly lysed with lysis buffer. FLAG-Pex26p variants were immunoprecipitated using agarose beads conjugated with anti-FLAG antibody (M2, Sigma). The immunocomplexes were analyzed by SDS-PAGE and immunoblotting (27).
In Vitro Binding Assay
Expression of GST-Pex26p, His-Pex14p, and His-Pex5p fusion proteins were done as described (30–32). Binding assays of GST-Pex26p to recombinant His-Pex14p and His-Pex5p were performed by GST pulldown using glutathione-linked Sepharose 6B beads (GE Healthcare) essentially as described in Matsumoto et al. (11). Proteins bound to the beads were analyzed by SDS-PAGE and immunoblot analysis.
Measurement of Binding Kinetics by Surface Plasmon Resonance (SPR)
The Biacore X100 system (Biacore, GE Healthcare) was used in this study. CM5 sensor chip was used to immobilize the His-Pex5p recombinant protein by an amine-coupling reaction. HBS-EP buffer (10 mm HEPES, 0.15 m NaCl, 3 mm EDTA, 0.005% Surfactant P20, pH 7.4) was used as the binding buffer. General procedures were according to the manufacturer's instructions. The immobilized amount of the ligand was ∼500 resonance units to ensure efficient binding. The control channel was treated in the same way as the assay channel but without His-Pex5p immobilized. The analyte was a FLAG-Pex1p protein. For isolation of FLAG-Pex1p, Sf9 cells were infected with baculoviruses harboring FLAG-PEX1. At 72 h post-infection, cells were lysed by sonication in buffer consisting of 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 25 μg/ml each of antipain and leupeptin, and 50 units of aprotinin (Sigma). FLAG-Pex1p was purified from the soluble fraction with anti-FLAG immunoglobulin G M2-agarose (Sigma). FLAG-Pex1p was isolated by elution with FLAG peptide (Sigma) (30). The titration was measured with purified FLAG-Pex1p from 5 to 40 μg/ml in HBS-EP. The binding was carried out at 25 °C with a flow rate at 30 μl/min, and data were collected for 1 min of association and 2 min of dissociation. To analyze the data, the assay channel was subtracted by the control channel to eliminate nonspecific interaction. Multiple sensorgrams from different concentrations of analyte were overlaid and aligned, and the kinetic constant was calculated by BIAevaluation 3.1 software with nonlinear fitting; the 1:1 (Langmuir) binding model was used, where KD = Kd/Ka.
Blue Native-PAGE Analysis
The compositions of blue native gels and gel buffers were as described (13). The resolving gel contained an acrylamide gradient from 4 to 16%. Sample (15 μl) containing 50 μg of protein was mixed with 15 μl of 10 × blue native-PAGE sample buffer containing 0.3 m aminocaproic acid, 2 mm EDTA, 0.2 m Bis-Tris, pH 7.0, 5% Coomassie Brilliant Blue G-250 (Nacalai Tesque, Kyoto, Japan) and incubated for 20 min on ice. The protein complexes were separated at 4 °C for 4 h at 100 V. For Western blot analysis, the protein complexes were transferred to a polyvinylidene difluoride membrane (Bio-Rad).
Other Methods
Western blot analysis on polyvinylidene difluoride membrane was done with primary rabbit antibodies against Pex5pN (33), Pex14pC (34), Pex26pC (11), and HA (13) and a second antibody, donkey anti-rabbit immunoglobulin G antibody conjugated with horseradish peroxidase, with ECL (enhanced chemiluminescence) Western blotting detection reagent (GE Healthcare). For the exogenously introduced oligomer rearrangement study, the purified His-Pex5p-FLAG protein was introduced into living CHO cells using the BioPORTER protein-loading reagent (Genlantis) and incubated for 4 h for the delivery of His-Pex5p-FLAG across the cell membrane.
RESULTS
Pex14p as the Interacting Partner of Pex26p
From our previous analyses of patient-derived Pex26p, we demonstrated that peroxisomal localization of Pex26p and Pex6p·Pex1p complexes is crucial for peroxisome biogenesis (11, 13). Pex26p is likely to be involved in a key regulatory step in Pex5p re-localization with Pex1p·Pex6p complexes. It is also possible that Pex26p may interact with other peroxins and act as a scaffold protein to specify the molecular target of chaperon-like function by Pex1p and Pex6p. Therefore, we searched for a putative binding partner of Pex26p using co-immunoprecipitation studies. FLAG-Pex26p was co-expressed in CHO-K1 cells or its pex mutant cells together with other peroxins, and cell extracts from these transformants were subjected to immunoprecipitation assay using anti-FLAG antibody. Pex14p was apparently co-immunoprecipitated with FLAG-Pex26p, whereas the other peroxins, except for Pex6p and Pex19p, showed barely detectable levels of the interaction with FLAG-Pex26p (Fig. 1A and data not shown). This result raised the question of whether endogenous Pex5p is recovered in the precipitates of FLAG-Pex26p, as Pex14p is an initial docking factor of Pex5p, the cytosolic receptor of PTS1-carrying protein (32). Therefore, the fraction bound to FLAG-Pex26p was analyzed by immunoblotting using anti-Pex5p antibody. Endogenous Pex5p was also co-immunoprecipitated with FLAG-Pex26p and Pex14p, implying that FLAG-Pex26p interacts with Pex5p through Pex14p (Fig. 1A).
FIGURE 1.
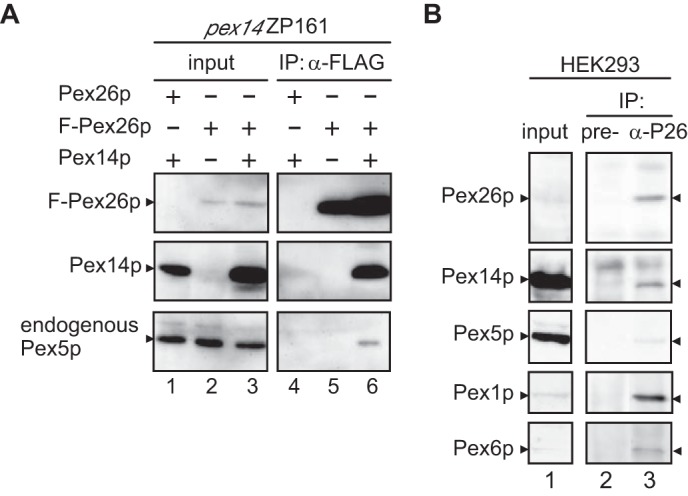
Interaction of Pex26p with Pex14p and Pex5p. A, immunoprecipitation assays of FLAG-Pex26p with Pex14p and Pex5p. FLAG-PEX26 or PEX26 was transfected to pex14 ZP161 cells together with or without PEX14 (lanes 1–6), and lysates were subjected to immunoprecipitation with antibody against FLAG. Input, 5% input used for immunoprecipitation (IP). Immunoprecipitated proteins were analyzed by SDS-PAGE and immunoblotting using antibodies to human Pex26pC (upper panel) and rat Pex14pC (middle panel). Endogenous Pex5p was detected with anti-HsPex5pN antibody (lower panel). B, Pex14p and Pex5p are binding partners of Pex26p. Lysates from HEK293 cells (2 × 106) were subjected to immunoprecipitation with pre-immune and antisera against Pex26pC (lanes 2 and 3). One-eighth aliquot of the input used for IP was loaded in lane 1. Immunoprecipitates were analyzed by immunoblotting with antibodies against Pex26pC, Pex14pC, Pex5pN, Pex1pC, and Pex6pN.
To confirm this finding, we performed immunoprecipitation studies using HEK293 cell lysates. Pex14p and Pex5p were co-immunoprecipitated together with Pex26p by using antibodies specific for a Pex26pC, but not with pre-immune antibody, indicating that Pex26p forms an endogenous complex with these peroxins (Fig. 1B). Pex1p·Pex6p complexes were also recovered with Pex26p as previously reported (11). The interaction of Pex26p with Pex14p and Pex5p was confirmed by glutathione S-transferase (GST) pulldown assays using recombinant proteins GST-Pex26p, His-Pex14p, and His-Pex5p. His-Pex14p was recovered with GST-Pex26p, but not GST, showing that Pex26p directly binds to Pex14p (Fig. 2A, lanes 2 and 3). His-Pex5p was not detectable in the fraction recovered with GST or GST-Pex26p (Fig. 2A, lanes 1 and 4), indicative of specific binding of Pex5p to Pex14p. Moreover, the addition of His-Pex5p to a pre-bound complex of GST-Pex26p·Pex14p showed significant interaction of the ternary complex, Pex26p·Pex14p·Pex5p, consistent with the results in co-immunoprecipitation studies of these peroxins (Fig. 2A, lane 7). In contrast, GST-Pex26p barely bound to the complexes of pre-mixed Pex14p·Pex5p complexes, and the ternary complex of Pex26p·Pex14p·Pex5p was not assembled with the addition of these three peroxins simultaneously (Fig. 2A, lanes 5 and 6). In addition, Pex5p titration studies revealed that formation of a ternary complex, Pex26p·Pex14p·Pex5p, was prevented in a Pex5p-dependent manner (Fig. 2B), implying that Pex5p binding to a part of Pex14p oligomer is sufficient to reduce the binding affinity to Pex26p. Taken together, it is possible that the oligomeric state of Pex14p for those two types of complexes is distinct, and the binding affinity of Pex14p to Pex26p is likely altered by the association with Pex5p. Therefore, PTS-protein importomer containing Pex14p·Pex5p may be protected from the interaction of Pex26p and Pex6p·Pex1p complexes. These results raise the interesting issue of how the binding affinity of Pex14p·Pex5p complexes to Pex26p is increased after the translocation of cargo proteins. The regulation of the Pex26p interaction with Pex14p·Pex5p complexes in Pex5p dislocation from peroxisomes remains to be defined.
FIGURE 2.
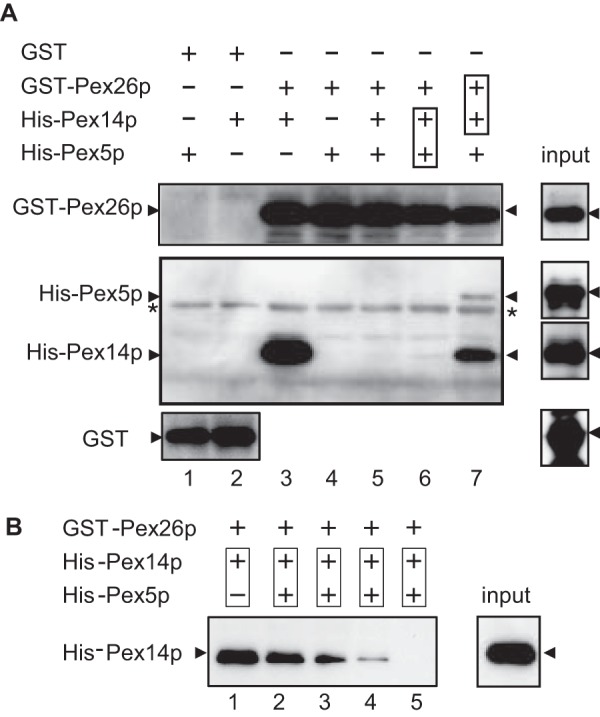
Ternary complex formation of Pex26p, Pex14p, and Pex5p. A, direct binding of Pex26p to Pex14p was assayed with GST (1.0 μg each, lanes 1 and 2), GST-Pex26p (0.5 μg each, lanes 3–7), His6-Pex14p (2.0 μg each, lanes 2, 3, and 5–7), and His6-Pex5pL (3.0 μg each, lanes 1 and 4–7). Purified His6-Pex14p and His6-Pex5p were separately incubated with glutathione-Sepharose beads conjugated to GST (lanes 1 and 2) or GST-Pex26p (lanes 3–7). Boxes in lanes 6 and 7 designate preincubation of the indicated proteins. GST-Pex26p in lane 6 was added to the preincubated mixture of His6-Pex14p and His6-Pex5p. In lane 7, pre-mixed GST-Pex26p and His6-Pex14p were further incubated with His6-Pex5p. Proteins bound to glutathione-Sepharose beads were assessed by immunoblotting with antibodies to Pex26pC (upper panel), Pex5pN plus Pex14pC (middle panel), and GST (lower panel). Input, 10% input. *, a nonspecific band. B, GST-Pex26p (0.5 μg each, lanes 1–5) was incubated with pre-mixed His6-Pex14p (2.0 μg each, lanes 1–5) and His6-Pex5p. Binding of GST-Pex26p to His6-Pex14p and His6-Pex5p was titrated with Pex5p (lane 2, 0.1 μg; lane 3, 0.2 μg; lane 4, 0.4 μg; lane 5, 0.8 μg). Input, 10% input.
Domain Mapping of Pex26p
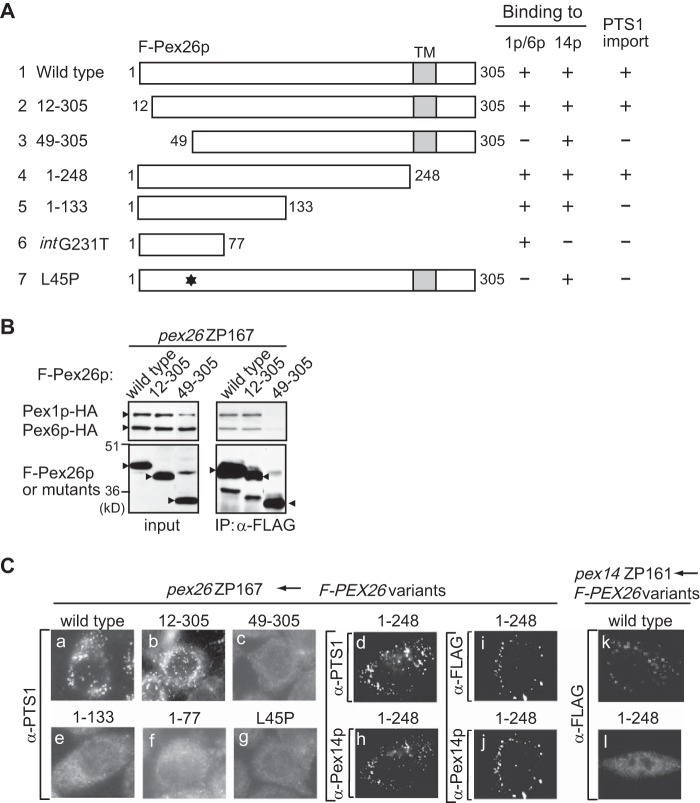
To define the region of Pex26p involved in the Pex26p·Pex14p interaction, we constructed various truncated FLAG-tagged Pex26p variants (Fig. 3A, 2∼6) and CG-8 PBD patient-derived Pex26p mutants (Fig. 3A, 7∼9) and verified them for interaction by co-immunoprecipitation assays. We earlier reported that a Pex26p-defective mutation, L45P, showed the dysfunction in the interaction with Pex1p·Pex6p complexes (27). Therefore, we used this mutant as a negative control. Pex14p was recovered in fractions bound to FLAG-Pex26p-(1–133), FLAG-Pex26p-(1–111), and FLAG-Pex26p-(1–85) at levels similar to that noted by using full-length FLAG-Pex26p, demonstrating that the Pex14p binding site resides at the N-terminal half of Pex26p (Fig. 3B, IP panel, lanes 1, 5, 6, and 8). FLAG-Pex26p-(49–305), FLAG-Pex26p-(78–305), and FLAG-Pex26pL45P showed the same level of binding to Pex14p as full-length FLAG-Pex26p, suggesting that the region including residues 78–85 is involved in binding to Pex14p (Fig. 3B, IP panel, lanes 1–3). Two more deletion variants, FLAG-Pex26p-(97–305) and FLAG-Pex26p-(1–77), did not interact with Pex14p (Fig. 3B, IP panel, lanes 4 and 9). The data described here were summarized in Fig. 3A, suggesting that the Pex26p binding to Pex14p requires the sequence at 78–85 in the N-terminal region of Pex26p.
FIGURE 3.
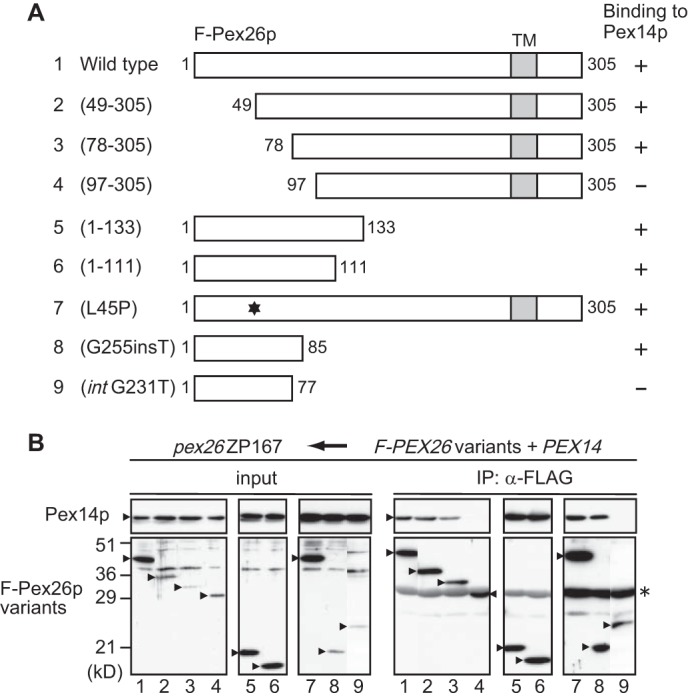
Domain mapping of Pex26p involved in interaction with Pex14p. A, schematic representation of human Pex26p variants. Numbers, amino acid residues of Pex26p; shaded boxes, transmembrane segments; *, L45P mutation. The results of F-Pex26p binding to Pex14p were summarized on the right. FLAG-Pex26pG255insT and FLAG-Pex26pintG231T encompass 113- and 159-amino acid-long sequences that contain only 85 and 77 authentic residues, respectively, from the N terminus of Pex26p (25). B, co-immunoprecipitation assay using FLAG-Pex26p mutants and Pex14p. pex26 ZP167 cells were transfected with plasmids encoding FLAG-Pex26p or its variants together with that for Pex14p. FLAG-Pex26p variants were immunoprecipitated from cell lysates using anti-FLAG antibody. Immunoprecipitation assays were performed as in Fig. 1A. Input, 5% input. *, IgG light chain.
In our earlier report we demonstrated that the mutations identified in PEX26 from CG8 PBD patients affected the Pex26p interaction with Pex1p·Pex6p complexes. Pex26p mutants FLAG-Pex26pG255insT and FLAG-Pex26pintG231T encompassed 113- and 159-amino acid-long sequences that contained only 85 and 77 authentic residues from the N terminus of Pex26p, respectively (Figs. 3A and 4A) (27, 28). These mutants bound to Pex6p as well as full-length Pex26p, suggesting that the N-terminal region of Pex26p is crucial for its binding to Pex1p·Pex6p complexes (27). We also reported that Pex26p-(del33–40) truncated in amino acid residues at 33–40 abolishes the recruiting of Pex1p·Pex6p complex to peroxisomes (13). In this study we, therefore, determined the region involved in Pex26p interaction with Pex1p·Pex6p by immunoprecipitation assays. Full-length FLAG-Pex26p and truncated mutants FLAG-Pex26p-(12–305) and FLAG-Pex26p-(49–305) were expressed together with Pex1p-HA and Pex6p-HA in ZP167 cells. Pex1p-HA and Pex6p-HA were detected in the immunoprecipitates of normal FLAG-Pex26p, consistent with earlier observations that Pex1p-HA and Pex6p-HA were co-immunoprecipitated with FLAG-Pex26p (Fig. 4B) (11, 35). Pex1p-HA and Pex6p-HA were similarly recovered with FLAG-Pex26p-(12–305), whereas the deletion of the N-terminal region, such as the 48-amino acid truncation in Pex26p-(49–305), showed a reduced and barely detectable level of the interaction with Pex6p and Pex1p (Fig. 4B and see Fig. 6). These results demonstrated that the N-terminal region of Pex26p, such as Pex26p-(12–48), is essential for sufficient association of Pex26p with Pex1p·Pex6p complex. Taken together, the binding regions for both the Pex1p·Pex6p complex and Pex14p are located in the N-terminal region of Pex26p but do not overlap. Next, peroxisome-restoring activity of truncated and patient-derived mutants was verified by expressing in the CHO PEX26-deficient cell line ZP167, which is pex26 null mutant. In ZP167 cells transfected with wild type FLAG-PEX26, FLAG-Pex26-(12–305), or FLAG-Pex26-(1–248), numerous PTS1-positive peroxisomes were detected in a punctate staining pattern, indicating the restoration of PTS1-protein import (Fig. 4C, a, b, and d). In contrast, in ZP167 cells expressing FLAG-Pex26p-(49–305), FLAG-Pex26p-(1–133), FLAG-Pex26pintG231T, or FLAG-Pex26pL45P, PTS1-positive particles were not discernible (Fig. 4C, c and e–g). Collectively, Pex1p·Pex6p recruitment to peroxisomes was required for the complementing activity. It is of interest to note that FLAG-Pex26p-(1–248) with deletion of the transmembrane domain was localized to peroxisomes in pex26 ZP167 and was apparently competent in PTS1 protein import (Fig. 4C, d and h–j), whereas this deletion mutant was barely detected in a punctate staining pattern in pex14 ZP161 (Fig. 4C, k and l), implying that Pex14p interacts with Pex26p and aids in the peroxisomal localization of Pex26p. We previously reported that almost all mutations in PEX26 from patients with PBD CG8 were located at N-terminal half portion of Pex26p. These results strongly suggested the importance of the N-terminal region for its function.
FIGURE 4.
The region of Pex26p required for binding to Pex1p and Pex6p. A, schematic representation of Pex26p variants was described as in Fig. 3A. The results of F-Pex26p binding to Pex14p or Pex1p/Pex6p and the complementing activity of PTS1 import were summarized at the right (25) (this study). B, co-immunoprecipitation assay using FLAG-Pex26p mutants with Pex1p/Pex6p. FLAG-Pex26p, Pex26p-(12–305), and Pex26p-(49–305) were separately expressed in pex26 ZP167 cells together with Pex1p-HA and Pex6p-HA (25). Immunoprecipitates using anti-FLAG antibody were analyzed by SDS-PAGE and immunoblotting with antibodies to HA (upper panel) and FLAG (lower panel). C, functional domain mapping of Pex26p. Wild type and variants of Pex26p were transiently expressed in pex26 ZP167 and pex14 ZP161. Cells were stained using antibody to PTS1 peptide, and complementing activity was verified by restoration of PTS1 protein import in ZP167 (a–g). Cells were stained with antibody to Pex14p (h and j) and FLAG (i, k, and l). Bar, 20 μm.
FIGURE 6.
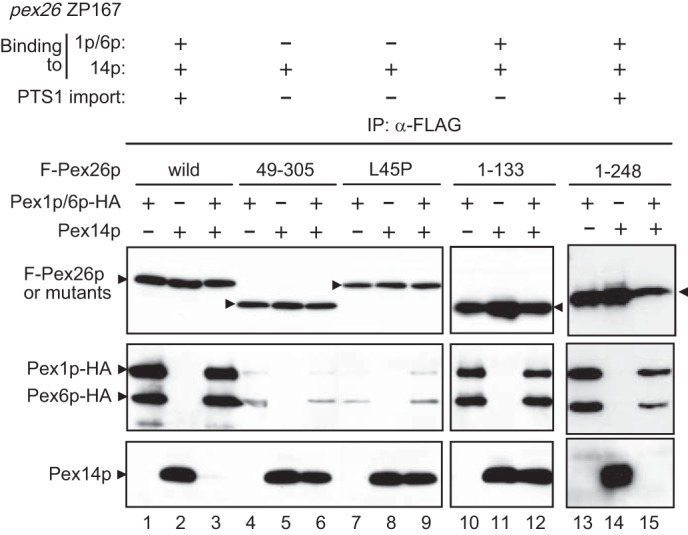
Pex1p·Pex6p complex dissociates Pex26p bound to Pex14p. Wild type or mutants of FLAG-Pex26p were expressed in pex26 ZP167 together (+) with Pex1p-HA and Pex6p-HA (lanes 1, 4, 7, 10, and 13), Pex14p (lanes 2, 5, 8, 11, and 14), or Pex1p-HA and Pex6p-HA plus Pex14p (lanes 3, 6, 9, 12, and 15). Lysates from these cells were subjected to immunoprecipitation of FLAG-Pex26p with anti-FLAG antibody, and co-immunoprecipitated proteins were assessed by immunoblotting with antibodies to Pex26pC (upper panel), HA (middle panel), and Pex14pC (lower panel). The results obtained here with respect to the F-Pex26p binding to Pex14p or Pex1p/Pex6p and the complementing activity toward PTS1 import are summarized at the top (this study), which were consistent with Tamura et al. (25).
ATP Depletion Enhances Pex26p·Pex14p Binding
Previous studies demonstrated that Pex5p was accumulated on peroxisomes in human skin fibroblast cells under the condition of ATP depletion and low temperature (29). Therefore, we examined whether such a treatment influences the Pex26p interaction with Pex14p. After ATP depletion and low temperature treatment to HEK293 cells, the postnuclear supernatant fraction was separated into cytosolic and organelle fractions. Endogenous Pex5p was detected by immunoblot in the cytosol and less in the organelles under the normal conditions. In contrast, the treatment of ATP depletion showed a clearly increased level of Pex5p in the organelle, suggesting accumulation of Pex5p on peroxisomes in good agreement with previous study (Fig. 5A). Co-immunoprecipitation assays by use of anti-Pex26p antibody showed that the recovery of Pex14p and Pex5p was enhanced by the treatment of ATP depletion (Fig. 5B). Taken together, Pex26p binding to Pex14p is likely to be dissociated in an ATP-dependent manner, implying that AAA peroxins Pex1p and Pex6p are involved in the modulation of peroxin-peroxin interaction.
FIGURE 5.
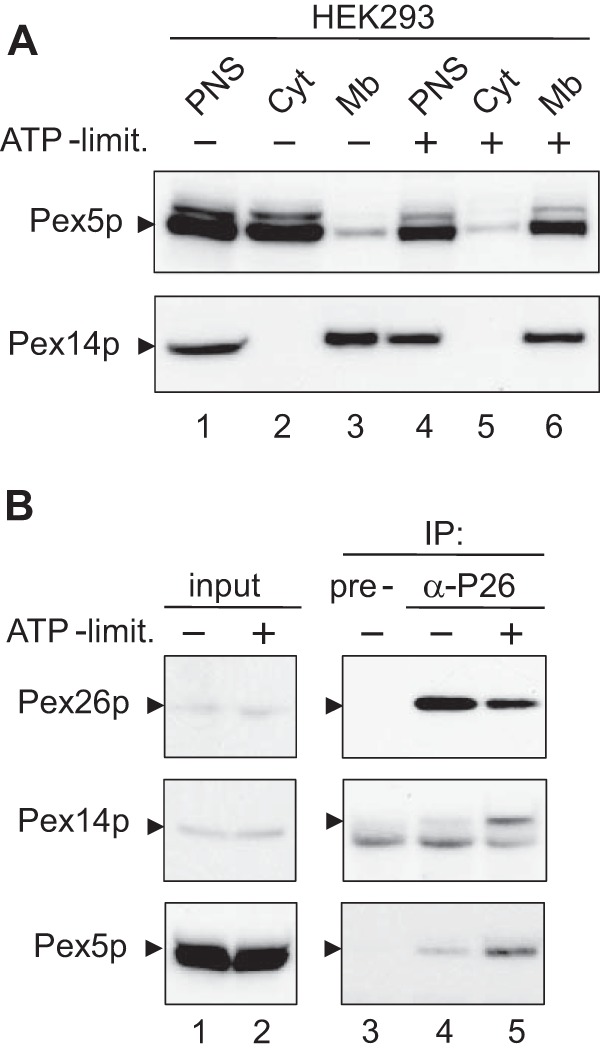
ATP depletion and low temperature enhance the binding affinity of Pex26p to Pex14p·Pex5p complex. A, subcellular fractionation. HEK293 cells were cultured under normal conditions (−) or in ATP-limited conditions (+) and separated into an organelle pellet and cytosolic supernatant fractions from postnuclear supernatant (PNS) fractions. Both fractions were analyzed by SDS-PAGE and immunoblotting using antibodies to Pex5pN (upper panel) and Pex14pC (lower panel). PNS (lanes 1 and 4); Cyt, cytosolic fraction (lanes 2 and 5); Mb, organelle fraction (lanes 3 and 6). B, in vivo binding assay. Lysates from HEK293 cells that had been cultured in normal medium (lanes 3 and 4) or in ATP-limited medium (lane 5) were subjected to immunoprecipitation with pre-immune (lane 3) and antisera against Pex26pC (lanes 4 and 5). Pex26p, Pex14p, and Pex5p were probed with antibodies to Pex26pC (upper panel), Pex14pC (middle panel), and Pex5pN (lower panel). Note that the amount of co-immunoprecipitated Pex14p with anti-Pex26p antibody was increased under the ATP-limited condition. Input, 5% input (lanes 1 and 2).
Pex1p and Pex6p Modulate Pex26p the Interaction with Pex14p·Pex5p Complexes
We showed that the N-terminal region of Pex26p provided the binding site for the Pex1p·Pex6p complex and Pex14p. In addition, it is also possible that ATP hydrolysis is involved in the regulation of Pex26p binding to Pex14p·Pex5p complexes. Next, we investigated whether AAA peroxins Pex1p and Pex6p influence the Pex26p binding to Pex14p. FLAG-Pex26p and Pex14p were expressed in pex26 ZP167 cells with or without Pex1p-HA and Pex6p-HA. FLAG-Pex26p was co-immunoprecipitated with Pex1p-HA and Pex6p-HA complexes sufficiently, consistent with our earlier results (Figs. 4B and 6, lane 1) (27). Pex14p was recovered in the precipitate of FLAG-Pex26p from ZP167 cells transfected with FLAG-PEX26 and PEX14 (Fig. 6, lane 2). Interestingly, the addition of Pex1p-HA and Pex6p-HA dramatically reduced FLAG-Pex26p binding to Pex14p, whereas Pex1p-HA and Pex6p-HA apparently bound to FLAG-Pex26p (Fig. 6, lane 3). These results strongly suggested that Pex1p-HA and Pex6p-HA dissociated Pex14p from Pex26p. Next, FLAG-Pex26p variants were verified for binding to Pex14p on co-expression with Pex1p-HA and Pex6p-HA. FLAG-Pex26p-(49–305) and FLAG-Pex26pL45P showed a clearly reduced level, <20% of wild type Pex26p, of the binding to Pex1p-HA and Pex6p-HA. Although these mutants, FLAG-Pex26p-(49–305) and FLAG-Pex26pL45P, apparently bound to Pex14p, co-expression of Pex1p-HA and Pex6p-HA failed to dissociate Pex14p from these Pex26p mutants (Fig. 6, lane 4–9). On the other hand, the C-terminal deletion variants, FLAG-Pex26p-(1–133) and FLAG-Pex26p-(1–248), bound to both Pex1p·Pex6p complexes and Pex14p, whereas FLAG-Pex26p-(1–133) failed to dissociate Pex14p from Pex26p. These results suggested that the region 134–248 of Pex26p is likely involved in the Pex14p release from Pex26p (Fig. 6, lanes 10–15) and that the AAA peroxins, Pex1p and Pex6p, likely modulate Pex26p interaction with Pex14p·Pex5p. Moreover, the binding domains for both Pex1p·Pex6p and Pex14p do not overlap in the N-terminal region of Pex26p (Fig. 6, lane 12).
We reported earlier that AAA cassettes, called D1 and D2 domains, of Pex1p and Pex6p are essential for the Pex1p·Pex6p interaction and peroxisome-restoring activity of Pex1p and Pex6p (13). Mutation of the lysine residue in the P-loop of the Walker A eliminates the ATP binding, whereas that of the acidic residues in the Walker B blocks the ATP hydrolysis (13). We verified these Walker motif mutants, such as Pex1pA1(K605E), Pex1pB1(D662N), Pex1pA2(K887E), Pex1pB2(D940N), Pex6pA1(K476E), Pex6pA2(K750E), and Pex6pB2(D803N), for the function of Pex14p dissociation from Pex26p as in Fig. 6. With respect to B1 in Pex6p, the conserved aspartic acid residue in the Walker B motif is not present. Therefore, we did not assess the Pex14p-releasing activity of the Pex6p B1 mutant. Mutant Pex6pA1(K476E), Pex1pB1(D662N). and Pex1pB2(D940N) showed a normal or a little lower level of the Pex14p dissociation from Pex26p as compared with wild type (Fig. 7A, lanes 5, 7, and 8). In contrast, the Pex1p mutants, A1(K605E) and A2(K887E), and the Pex6p mutants, A2(K750E) and B2(D803N), increased Pex14p binding to FLAG-Pex26p (Fig. 7A, lanes 4, 6, 9, and 10). In these mutant forms, Pex1pA2(K887E), Pex6pA2(K750E), and Pex6pB2(D803N) significantly reduced the dissociating activity toward Pex14p·Pex26p complex (Fig. 7A, lanes 6, 9, and 10, and B). In regard to the binding of Walker-motif mutants of Pex1p and Pex6p, these mutants appear to interact with Pex26p in the co-immunoprecipitation assays. We earlier reported that AAA cassettes D1 and D2 were essential for peroxisome-restoring activity of Pex1p and Pex6p (13). Collectively, these results are consistent with our earlier report on peroxisome-restoring activity of the Walker motif mutants of Pex1p and Pex6p. On the other hand, the D1 domain of Pex6p is required for PTS1-protein import, whereas this domain is not likely to be involved in the dissociation of Pex14p from Pex26p. Therefore, it is more likely that the D1 and D2 of Pex1p and D2 of Pex6p are essential for the modulation of Pex26p interaction with Pex14p by these AAA peroxins.
FIGURE 7.
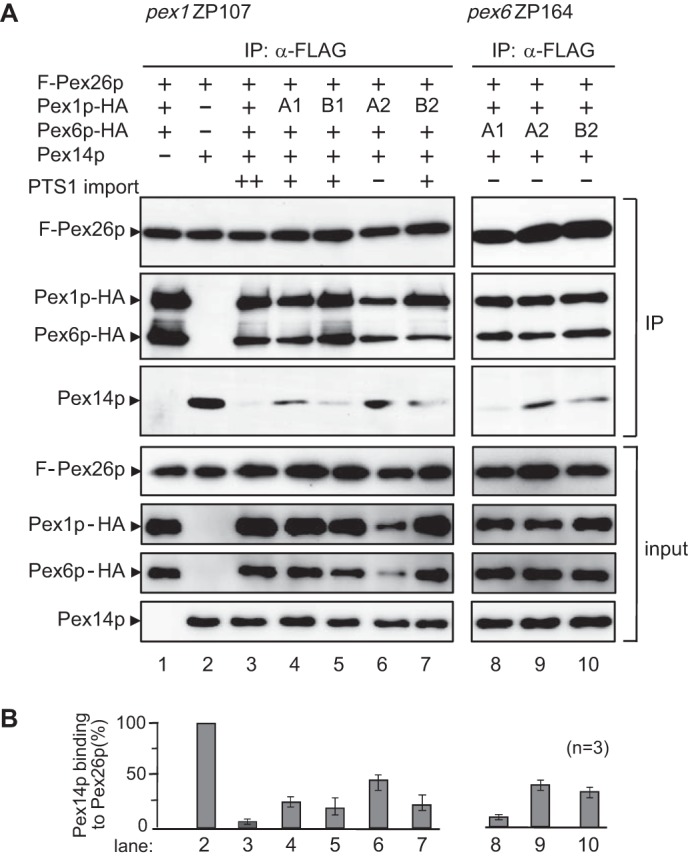
AAA cassettes of Pex1p and Pex6p are involved in the dissociation of the Pex26p·Pex14p complex. A, FLAG-Pex26p was expressed in pex1 ZP107 together with Pex1p-HA and Pex6p-HA (lane 1) as well as Pex14p (lane 2) as indicated at the top. AAA cassette mutants of Pex6p were expressed in pex6 ZP164 together with FLAG-Pex26p, Pex1p-HA, and Pex14p (lanes 8–10). Proteins recovered with anti-FLAG antibody-conjugated beads were analyzed by SDS-PAGE and immunoblotting with antibodies to Pex26pC (IP, upper panel), HA (IP, middle panel), and Pex14pC (IP, lower panel). Input, 5% input. B, Pex14p bound to FLAG-Pex26p was quantified by densitometric scanning, normalized as a ratio of Pex14p to Pex26p density, and represented by taking as 1 that in the absence of Pex1p·Pex6p (lane 2). Filled column, Pex14p. Values are the means ± S.D. from three experiments.
Pex1p Homo-oligomer Interacts with Pex5p
Pex1p was suggested to possess two distinct oligomeric forms, a homo-oligomer in the cytosol and a hetero-oligomer on peroxisome membranes, possibly playing distinct functions in peroxisome biogenesis (13). Next, we attempt to characterize the role of cytosolic Pex1p in the modulation of Pex5p oligomer formation. As a first step, we investigated direct interaction between Pex1p and Pex5p by the technique based on SPR biosensor assays. Purified recombinant F-Pex1p, His-Pex5p, and His-Pex14p were found to be at a purity sufficient for SPR assays (Fig. 8Aa). To define the oligomer structure, we analyzed the recombinant F-Pex1p by blue native-PAGE. F-Pex1p was largely detected as a complex of 450 kDa, indicating that Pex1p is in homo-trimer (Fig. 8Ab). This result is consistent with our earlier report on the cytosolic Pex1p from HEK293 cells by blue native-PAGE. His-Pex5p was immobilized to sensor chip as the ligand and then tested against the F-Pex1p as the analyte. This binding assay showed a typical association and dissociation response by comparison to the control channel, which only reflected fluctuations in analyte concentration. The subtracted sensorgrams at a medium concentration of each analyte were overlaid and shown in Fig. 8B. Sensorgrams showed specific F-Pex1p binding to His-Pex5p, suggesting that Pex1p directly interacts with Pex5p in the cytosol. The calculated equilibrium affinity (KD) for F-Pex1p binding to His-Pex5p was 1.0 μm, showing a lower binding affinity. In contrast, preloading of His-Pex14p abolished specific F-Pex1p binding to Pex5p. This may be due to conformational change of His-Pex5p in response to binding to His-Pex14p.
FIGURE 8.
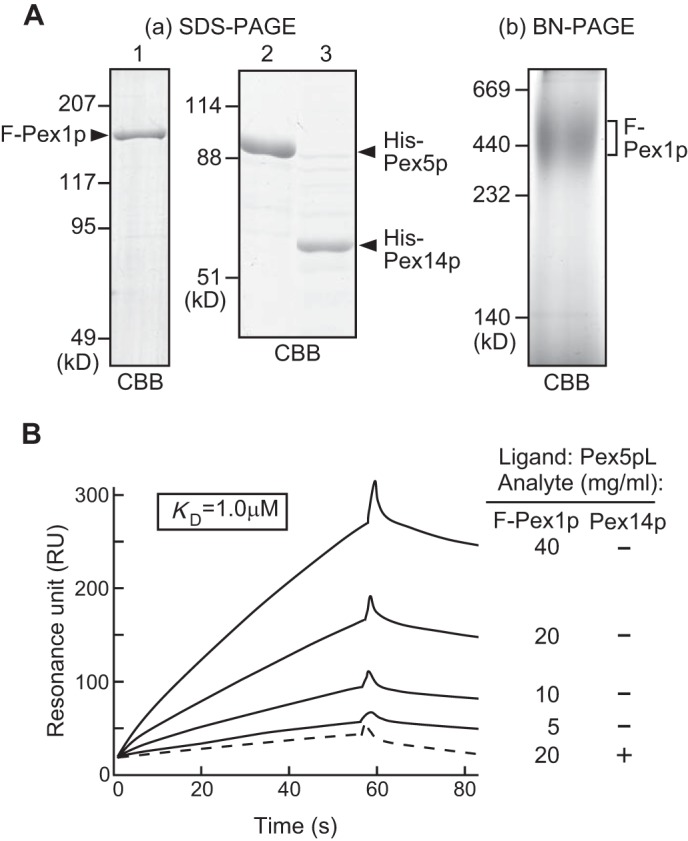
Direct binding of Pex1p to Pex5p. Aa, SDS-PAGE analysis of purified FLAG-Pex1p (lane 1), His-Pex5p (lane 2), and His-Pex14p (lane 3). b, homo-oligomer of recombinant FLAG-Pex1p. FLAG-Pex1p was verified for oligomerization by BN-PAGE. Staining was done with Coomassie Brilliant Blue (CBB). B, SPR-based assay. Overlay of the sensorgrams from different concentrations of FLAG-Pex1p. His-Pex5p was immobilized on a sensor chip as a ligand, and FLAG-Pex1p and His-Pex14p were tested as analytes. Specific binding was identified by subtraction of the control channel from the assay channel. Broken line, 20 μg/ml His-Pex14p was preloaded to sensor chip.
Although SPR assays evaluated the Pex1p binding to Pex5p, the role of cytosolic Pex1p is still obscure in peroxisome biogenesis. One possible common feature of AAA proteins is a force-induced structural rearrangement of client proteins in an ATP-dependent manner. To define the role of cytosolic Pex1p, we analyzed the Pex5p oligomer structure as a target protein of cytosolic Pex1p. The cytosolic fractions from CHO-K1, pex14 ZP161, and pex1 ZP107 cells were subjected to blue native-PAGE and immunoblot using anti-Pex5p antibody. Pex5p from CHO-K1 and pex14 ZP161 cells were detected in two distinct protein complexes with 180 and 160 kDa as the likely homo- or hetero-dimers (Fig. 9A). However, it was interesting to note that the 180-kDa complex was barely detectable in pex1 ZP107 cells. Together, cytosolic Pex1p was shown to interact with Pex5p oligomers, and it is more likely that Pex1p is involved in structural rearrangement of Pex5p homo- or hetero-oligomers. Furthermore, we assessed the oligomer rearrangement activity of Pex1p by delivering FLAG-tagged His-Pex5p recombinant proteins into CHO cells. His-Pex5p-FLAG recombinant protein was purified by affinity and size exclusion chromatography (data not shown). Our rationale was that such a delivered oligomer His-Pex5p-FLAG would be rearranged by chaperon-like activity of cytosolic Pex1p and co-immunoprecipitated with endogenous Pex5p or other proteins by using anti-FLAG antibody-conjugated agarose beads. His-Pex5p-FLAG was delivered into CHO-K1, pex14 ZP161, pex1 ZP107, pex6 ZP164, pex26 ZP167, and pex5 ZP101 cells. His-Pex5p-FLAG was immunoprecipitated from each cell lysate and immunoblotted with antibodies to Pex5p. Endogenous Pex5p bound to His-Pex5p-FLAG was discernible in the immunoprecipitates from ZP161, ZP164, and ZP167 cells and at a lower level in CHO-K1 cells (Fig. 9B). On the other hand, co-immunoprecipitated Pex5p was barely detected in pex1 ZP107 cells (Fig. 9B, lane 3), suggesting that Pex1p is involved in rearrangement of Pex5p oligomer structure.
FIGURE 9.
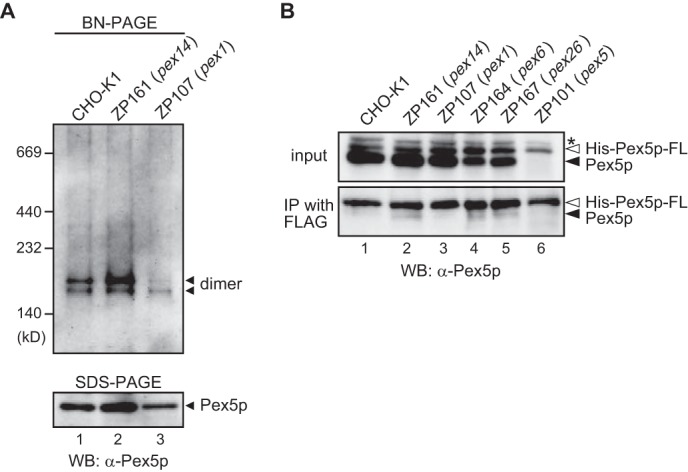
In vivo oligomerization of Pex5p is impaired in pex1 ZP107 cells. A, cytosolic fractions each from CHO-K1 (lane 1), pex14 ZP161 (lane 2), and pex1 ZP107 (lane 3) were separated by blue native PAGE. Protein complexes were detected by immunoblotting with anti-Pex5p antibody. Molecular mass markers are on the left. BN, blue native; WB, Western blot. B, purified recombinant His-Pex5p-FLAG was introduced into CHO-K1 (lane 1), pex14 ZP161 (lane 2), pex1 ZP107 (lane 3), pex6 ZP164 (lane 4), pex26 ZP167 (lane 5), and pex5 ZP101 (lane 6) cells. Cells were then cultured for 24 h at 37 °C. His-Pex5p-FLAG was immunoprecipitated from cell lysates using anti-FLAG antibody. Immunoprecipitates were analyzed by SDS-PAGE and immunoblotting with anti-Pex5p antibody. Input, 10% input. Solid and open arrowheads indicate endogenous Pex5p and His-Pex5p-FLAG, respectively. *, a nonspecific band.
DISCUSSION
We reported earlier that the dysfunction of PEX26 is responsible for PBD of CG8 (11) and Pex26p recruits Pex1p·Pex6p complexes to peroxisomes via Pex6p (13). It was proposed that Pex1p, Pex6p, and Pex26p are involved in the export of Pex5p from peroxisomes (21–23). However, the underlying molecular mechanisms of Pex5p shuttling between the cytoplasm and peroxisomes remain to be delineated. The data presented here provide an indication that mammalian Pex26p directly interacts with Pex14p, which is the initial docking receptor for Pex5p and is in good agreement with the finding in binding assays of the large complexes of peroxisomal importomer and Pex15p, the functional homolog of Pex26p in Saccharomyces cerevisiae (24). We assigned the binding region responsible for the interaction between Pex26p and Pex14p as well as Pex1p·Pex6p. The N-terminal region of Pex26p is involved in the interaction with both Pex1p·Pex6p complexes and Pex14p by immunoprecipitation assay using truncated variants of Pex26p. The regions encompassing the residues at 13–48 and 78–85 of Pex26p are required for interaction with Pex1p·Pex6p and Pex14p, respectively. We previously reported that Pex26p-(del1–40) failed to recruit Pex1p and Pex6p complexes on the peroxisome membrane (30). These results suggested that Pex26p acts as a scaffold protein on the peroxisomal membrane to specify the target of ATPase activity of Pex1p·Pex6p complexes, facilitating the interaction of AAA peroxins with Pex14p·Pex5p in a functional importomer. Moreover, PEX26 mutations identified in CG8 patients are mostly located at the region encoding the N-terminal part of Pex26p, implying that this region plays a crucial role in peroxisomal protein import, especially in interacting with Pex1p·Pex6p and Pex14p. It is noteworthy that yeast and human AAA peroxins were suggested to be involved in the export of Pex5p from peroxisomes. It has been reported that Pex5p and the AAA peroxins are in a complex at the peroxisome membrane (23), whereas no direct interaction of Pex5p·Pex14p complexes with either Pex1p or Pex6p was reported by pulldown or immunoprecipitation assays and SPR-based binding assays. The mechanism of substrate recognition by Pex1p·Pex6p was not understood yet. In this report we demonstrate two lines of evidence that Pex1p and Pex6p interact with Pex5p. First, Pex26p acts as a bridge between Pex14p·Pex5p and Pex1p·Pex6p complexes, allowing Pex1p·Pex6p to interact with Pex14p·Pex5p mediated by Pex26p on the peroxisomal membrane; second, a SPR-based assay demonstrated that Pex1p homo-oligomer binds directly to Pex5p, although at a low affinity. Additionally, cytosolic Pex1p is likely to modulate or maintain functional oligomer structure of Pex5p at the final step of Pex5p recycling. These results are consistent with our earlier report that Pex1p possesses two distinct oligomeric forms, a homo-oligomer in the cytosol and a hetero-oligomer with Pex6p on peroxisome membranes (13), possibly playing distinct roles in peroxisome biogenesis.
We also showed that two AAA cassette structures (D1 and D2) of Pex1p and D2 of Pex6p are required for dissociation of Pex14p from Pex26p. To assess the role of ATP for Pex14p·Pex26p dissociation, two different mutations were chosen to inactivate the D1 and D2 cassettes of Pex1p and Pex6p in analogy to the reported mutations of N-ethylmaleimide-sensitive fusion protein (35), one preventing ATP binding (A1 and A2) and the other preventing ATP hydrolysis (B1 and B2). The data presented here demonstrate the importance of Walker motifs in Pex1p and Pex6p, and such a Pex14p dissociation requires both ATP binding and hydrolysis on D1 and D2 of Pex1p and D2 of Pex6p. Moreover, we showed that FLAG-Pex26pL45P and FLAG-Pex26p-(49–305) failed to associate with Pex1p·Pex6p and that such Pex26p mutants were apparently abrogated in the Pex14p dissociation from Pex26p. On the other hand, FLAG-Pex26p-(1–133) was incompetent of Pex14p dissociation even under the co-expression of Pex1p and Pex6p. Collectively these findings demonstrate the importance of the AAA cassette of Pex1p·Pex6p together with N-terminal part of Pex26p for Pex14p dissociation.
Moreover, we also showed that the treatment of HEK293 cells with ATP depletion and low temperature accumulated Pex5p on peroxisomes and enhanced endogenous binding of Pex26p to Pex14p·Pex5p complexes. It has been reported that Pex5p associated with Pex14p was more detectable in ATP-limiting conditions (29). We (23) and Platta et al. (22) demonstrated that dysfunction of Pex1p or Pex6p or treatment with ATP depletion caused failure in peroxisomal export of Pex5p. Hence it is conceivable that Pex5p accumulated in peroxisomes is in a large complex comprising Pex5p·Pex14p, Pex26p, and Pex1p·Pex6p and that Pex14p dissociation from Pex26p correlates with Pex5p dislocation from peroxisomes. These are the first observations revealing how Pex1p·Pex6p interacts with Pex5p·Pex14p complexes. It is more likely that PBD patient-derived Pex26p mutants showing insufficient binding to Pex1p·Pex6p complexes failed to dissociate Pex26p·Pex14p complex, hence inferring that Pex5p is abrogated in the dislocation from peroxisome membrane. Although Pex26p, like Pex1p and Pex6p, is involved in dislocation of Pex5p from peroxisomes to the cytosol, the molecular details of Pex5p dissociation from Pex14p need to be defined.
We reported earlier that Pex14p forms homo-oligomers and interacts with its binding partners in a manner dependent on homo-oligomer sizes (36) where the Pex14p monomer or dimer interacted with Pex5p and Pex19p and binding to Pex13p gave rise to high molecular mass hetero-oligomeric complexes. Conversely, Pex5p, but not the cargo-loaded Pex5p, mediated disassembly of Pex14p oligomer, resulting in its no longer binding to Pex13p, in good agreement with the finding in binding assays that Pex5p affects a potential complex of Pex14p with Pex13p in a cargo-dependent manner (33). In this study we verified the involvement of Pex26p in the export step of Pex5p. Intriguingly, we revealed that GST-Pex26p·Pex14p complexes are competent to bind to Pex5p, whereas GST-Pex26p no longer binds to pre-bound complexes of Pex14p·Pex5p. Because molecular structures of Pex14p homo-oligomers vary depending upon the binding partner (36), Pex26p is likely to interact with the specific form of Pex14p. These findings strongly suggest that the cargo-unloaded Pex5p mediates disassembly of Pex14p homo-oligomers, resulting in a decrease of the binding affinity to Pex26p. In other words, pre-bound Pex14p·Pex5p complexes may require the conformational changes to interact with Pex26p. The question has to be addressed of how Pex26p distinguish Pex5p·Pex14p complexes destined for Pex5p dislocation from cargo-loaded Pex5p·Pex14p in cargo protein translocation. Most interestingly, the PTS1 receptor Pex5p of S. cerevisiae and rat are monoubiquitinated (37–39). The functional role of mono-ubiquitination of Pex5p constitutes the export signal, whereas polyubiquitination seems to provide a signal for the quality control pathway of Pex5p (40). The attachment of mono-ubiquitin may induce conformational changes within Pex5p·Pex14p complexes to expose the binding sites to Pex26p. Moreover, Pex14p is a target for post-translational modifications. Both Hansenula polymorpha and Pichia pastoris Pex14p could be detected in a phosphorylated and an unphosphorylated state (41, 42). Accordingly, it is possible that phosphorylation is a post-translational modification that leads to a conformational change of Pex14p·Pex5p complexes and an increase of Pex14p binding affinity to Pex26p. In addition, these modifications by mono-ubiquitination and phosphorylation may regulate the peroxisomal matrix protein import. The direct mechanistic roles of these modifications on the peroxin-peroxin interactions and Pex5p export reaction remain to be investigated.
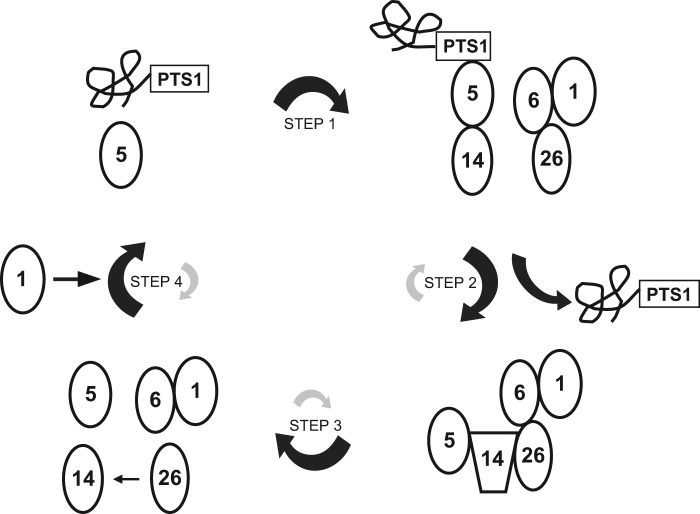
Here we propose a model for the role of Pex26p in peroxisomal matrix protein import (Fig. 10). The Pex5p-PTS1 protein complex targets to peroxisomes through its binding to the docking complex containing Pex14p and Pex13p at the first step. During step 2, PTS1 proteins are unloaded from Pex5p inside or at the surface of peroxisomes, and Pex14p might change its structure to be competent to form a recycling complex with Pex5p, Pex26p, Pex6p, and Pex1p. It is also possible that cargo release from Pex5p results in conformational change of Pex14p. Moreover, Pex26p is in a recycling complex with other peroxins in the importomer, such as Pex13p, Pex10p and Pex12p, as reported by Rosenkranz et al. (24), Pex15p associates with these peroxins in yeast. At step 3, Pex1p·Pex6p disassembles the recycling complex by ATP hydrolysis-driven and Pex26p-mediated conformational change of Pex14p. In the absence of PTS1 proteins, Pex13p·Pex14p docking complexes are unable to associate with Pex5p (33). Conformational change of Pex14p might lead to Pex5p release from Pex14p. Because Walker motif B2 in Pex6p is reported to be involved in its dissociation from Pex26p (13), ATP hydrolysis by Pex6pB2 is likely linked to the dissociation from Pex26p·Pex14p complexes. In the last step, released Pex5p oligomer interacts with cytosolic Pex1p and modulates its oligomeric form. Thereby, Pex5p binds to PTS1-proteins, and the disassembled Pex14p is available to interact with other Pex5p·cargo protein complexes in the cytosol and acts as the initial docking site of Pex5p again. It is also possible that the post-translational modifications including ubiquitination and phosphorylation regulate these reactions toward adequate directions of respective reaction equilibrium in steps 2, 3, and 4. Further investigation addressing the disassembly of recycling complex and the dynamism of disassembled peroxins such as Pex5p and Pex14p would elucidate a more detailed molecular mechanism of peroxisomal matrix protein import.
FIGURE 10.
A model for the role of Pex26p in Pex5p shuttling. Pex5p-PTS1 complexes formed in the cytosol dock onto Pex14p (step 1). PTS1 proteins are released at the inner surface and/or inside of peroxisomes, and then conformational change of Pex5p enables Pex14p·Pex5p complexes to interact with Pex26p (step 2). Pex14p is dissociated from Pex26p by chaperon-like activities coupled with ATP hydrolysis of Pex1p·Pex6p (step 3). The disassembled Pex14p releases PTS1-unloaded Pex5p, and then Pex5p shuttles back to the cytosol (step 4), where cytosolic Pex1p modulates the oligomeric forms of Pex5p.
Acknowledgments
We thank T. Umemoto for technical assistance, K. Shimizu for preparing the figures, and the other members of our laboratory for discussions.
This work was supported in part by Grants-in-aid for Scientific Research 21570116 and 24570134 (to S. T.) and 20370039, 24247038, 25116717, 25112518, and 26116007 (to Y. F.); by the Global Center of Excellence (GCOE) program and Grants for Excellent Graduate Schools from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; and by a CREST (Core Research for Evolutional Science and Technology) grant (to Y. F.) from the Science and Technology Agency of Japan, and grants (to Y. F.) from Takeda Science Foundation and Japan Foundation for Applied Enzymology.
- PTS1 and PTS2
- peroxisome targeting signal types 1 and 2
- CG
- complementation group
- PBD
- peroxisome biogenesis disorder
- SPR
- surface plasmon resonance
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- IP
- immunoprecipitation.
REFERENCES
- 1. Lazarow P. B., Fujiki Y. (1985) Biogenesis of peroxisomes. Ann. Rev. Cell Biol. 1, 489–530 [DOI] [PubMed] [Google Scholar]
- 2. Fujiki Y. (1997) Molecular defects in genetic diseases of peroxisomes. Biochim. Biophys. Acta 1361, 235–250 [DOI] [PubMed] [Google Scholar]
- 3. Subramani S. (1997) PEX genes on the rise. Nat. Genet. 15, 331–333 [DOI] [PubMed] [Google Scholar]
- 4. Subramani S., Koller A., Snyder W. B. (2000) Import of peroxisomal matrix and membrane proteins. Annu. Rev. Biochem. 69, 399–418 [DOI] [PubMed] [Google Scholar]
- 5. Fujiki Y. (2000) Peroxisome biogenesis and peroxisome biogenesis disorders. FEBS Lett. 476, 42–46 [DOI] [PubMed] [Google Scholar]
- 6. Albertini M., Rehling P., Erdmann R., Girzalsky W., Kiel J. A., Veenhuis M., Kunau W.-H. (1997) Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell 89, 83–92 [DOI] [PubMed] [Google Scholar]
- 7. Will G. K., Soukupova M., Hong X., Erdmann K. S., Kiel J. A., Dodt G., Kunau W.-H., Erdmann R. (1999) Identification and characterization of the human orthologue of yeast Pex14p. Mol. Cell. Biol. 19, 2265–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang C.-C., Warren D. S., Sacksteder K. A., Gould S. J. (1999) PEX12 interacts with PEX5 and PEX10 and acts downstream of receptor docking in peroxisomal matrix protein import. J. Cell Biol. 147, 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Erdmann R., Blobel G. (1996) Identification of Pex13p, a peroxisomal membrane receptor for the PTS1 recognition factor. J. Cell Biol. 135, 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Okumoto K., Abe I., Fujiki Y. (2000) Molecular anatomy of the peroxin Pex12p: RING finger domain is essential for Pex12p function and interacts with the peroxisome-targeting signal type 1-receptor Pex5p and a RING peroxin, Pex10p. J. Biol. Chem. 275, 25700–25710 [DOI] [PubMed] [Google Scholar]
- 11. Matsumoto N., Tamura S., Fujiki Y. (2003) The pathogenic peroxin Pex26p recruits the Pex1p·Pex6p AAA ATPase complexes to peroxisomes. Nat. Cell Biol. 5, 454–460 [DOI] [PubMed] [Google Scholar]
- 12. Thoms S., Gärtner J. (2012) First PEX11β patient extends spectrum of peroxisomal biogenesis disorder phenotypes. J. Med. Genet. 49, 314–316 [DOI] [PubMed] [Google Scholar]
- 13. Tamura S., Yasutake S., Matsumoto N., Fujiki Y. (2006) Dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p. J. Biol. Chem. 281, 27693–27704 [DOI] [PubMed] [Google Scholar]
- 14. Birschmann I., Stroobants A. K., van den Berg M., Schäfer A., Rosenkranz K., Kunau W.-H., Tabak H. F. (2003) Pex15p of Saccharomyces cerevisiae provides a molecular basis for recruitment of the AAA peroxin Pex6p to peroxisomal membranes. Mol. Biol. Cell 14, 2226–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ogura T., Wilkinson A. J. (2001) AAA+ superfamily ATPases: common structure-diverse function. Genes Cells 6, 575–597 [DOI] [PubMed] [Google Scholar]
- 16. Vale R. D. (2000) AAA proteins: lords of the ring. J. Cell Biol. 150, F13–F19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lupas A. N., Martin J. (2002) AAA proteins. Curr. Opin. Struct. Biol. 12, 746–753 [DOI] [PubMed] [Google Scholar]
- 18. Beyer A. (1997) Sequence analysis of the AAA protein family. Protein Sci. 6, 2043–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanson P. I., Whiteheart S. W. (2005) AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6, 519–529 [DOI] [PubMed] [Google Scholar]
- 20. Faber K. N., Heyman J. A., Subramani S. (1998) Two AAA family peroxins, PpPex1p and PpPex6p, interact with each other in an ATP-dependent manner and are associated with different subcellular membranous structures distinct from peroxisomes. Mol. Cell. Biol. 18, 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Collins C. S., Kalish J. E., Morrell J. C., McCaffery J. M., Gould S. J. (2000) The peroxisome biogenesis factors Pex4p, Pex22p, Pex1p, and Pex6p act in the terminal steps of peroxisomal matrix protein import. Mol. Cell. Biol. 20, 7516–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Platta H. W., Grunau S., Rosenkranz K., Girzalsky W., Erdmann R. (2005) Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat. Cell Biol. 7, 817–822 [DOI] [PubMed] [Google Scholar]
- 23. Miyata N., Fujiki Y. (2005) Shuttling mechanism of peroxisome targeting signal type 1 receptor Pex5: ATP-independent import and ATP-dependent export. Mol. Cell. Biol. 25, 10822–10832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rosenkranz K., Birschmann I., Grunau S., Girzalsky W., Kunau W.-H., Erdmann R. (2006) Functional association of the AAA complex and the peroxisomal importomer. FEBS J. 273, 3804–3815 [DOI] [PubMed] [Google Scholar]
- 25. Tamura S., Matsumoto N., Imamura A., Shimozawa N., Suzuki Y., Kondo N., Fujiki Y. (2001) Phenotype-genotype relationships in peroxisome biogenesis disorders of PEX1-defective complementation group 1 are defined by Pex1p·Pex6p interaction. Biochem. J. 357, 417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Otera H., Okumoto K., Tateishi K., Ikoma Y., Matsuda E., Nishimura M., Tsukamoto T., Osumi T., Ohashi K., Higuchi O., Fujiki Y. (1998) Peroxisome targeting signal type 1 (PTS1) receptor is involved in import of both PTS1 and PTS2: studies with PEX5-defective CHO cell mutants. Mol. Cell. Biol. 18, 388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Furuki S., Tamura S., Matsumoto N., Miyata N., Moser A., Moser H. W., Fujiki Y. (2006) Mutations in the peroxin Pex26p responsible for peroxisome biogenesis disorders of complementation group 8 impair its stability, peroxisomal localization, and interaction with the Pex1p·Pex6p complex. J. Biol. Chem. 281, 1317–1323 [DOI] [PubMed] [Google Scholar]
- 28. Matsumoto N., Tamura S., Furuki S., Miyata N., Moser A., Shimozawa N., Moser H. W., Suzuki Y., Kondo N., Fujiki Y. (2003) Mutations in novel peroxin gene PEX26 that cause peroxisome-biogenesis disorders of complementation group 8 provide a genotype-phenotype correlation. Am. J. Hum. Genet. 73, 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dodt G., Gould S. J. (1996) Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J. Cell Biol. 135, 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nashiro C., Kashiwagi A., Matsuzaki T., Tamura S., Fujiki Y. (2011) Recruiting mechanism of the AAA peroxins, Pex1p and Pex6p, to Pex26p on peroxisome membrane. Traffic 12, 774–788 [DOI] [PubMed] [Google Scholar]
- 31. Otera H., Nishimura M., Setoguchi K., Mori T., Fujiki Y. (2001) Biogenesis of nonspecific lipid transfer protein and sterol carrier protein x: studies using peroxisome assembly-defective pex cell mutants. J. Biol. Chem. 276, 2858–2864 [DOI] [PubMed] [Google Scholar]
- 32. Otera H., Harano T., Honsho M., Ghaedi K., Mukai S., Tanaka A., Kawai A., Shimizu N., Fujiki Y. (2000) The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates Pex7p-PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J. Biol. Chem. 275, 21703–21714 [DOI] [PubMed] [Google Scholar]
- 33. Otera H., Setoguchi K., Hamasaki M., Kumashiro T., Shimizu N., Fujiki Y. (2002) Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: conserved Pex5p WXXXF/Y motifs are critical for matrix protein import. Mol. Cell. Biol. 22, 1639–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shimizu N., Itoh R., Hirono Y., Otera H., Ghaedi K., Tateishi K., Tamura S., Okumoto K., Harano T., Mukai S., Fujiki Y. (1999) The peroxin Pex14p: cDNA cloning by functional complementation on a Chinese hamster ovary cell mutant, characterization, and functional analysis. J. Biol. Chem. 274, 12593–12604 [DOI] [PubMed] [Google Scholar]
- 35. Weller S., Cajigas I., Morrell J., Obie C., Steel G., Gould S. J., Valle D. (2005) Alternative splicing suggests extended function of PEX26 in peroxisome biogenesis. Am. J. Hum. Genet. 76, 987–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Itoh R., Fujiki Y. (2006) Functional domains and dynamic assembly of the peroxin Pex14p, the entry site of matrix proteins. J. Biol. Chem. 281, 10196–10205 [DOI] [PubMed] [Google Scholar]
- 37. Williams C., van den Berg M., Sprenger R. R., Distel B. (2007) A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 282, 22534–22543 [DOI] [PubMed] [Google Scholar]
- 38. Platta H. W., El Magraoui F., Schlee D., Grunau S., Girzalsky W., Erdmann R. (2007) Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 177, 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grou C. P., Carvalho A. F., Pinto M. P., Wiese S., Piechura H., Meyer H. E., Warscheid B., Sá-Miranda C., Azevedo J. E. (2008) Members of the E2D (UbcH5) family mediate the ubiquitination of the conserved cysteine of Pex5p, the peroxisomal import receptor. J. Biol. Chem. 283, 14190–14197 [DOI] [PubMed] [Google Scholar]
- 40. Kiel J. A., Emmrich K., Meyer H. E., Kunau W.-H. (2005) Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J. Biol. Chem. 280, 1921–1930 [DOI] [PubMed] [Google Scholar]
- 41. Komori M., Kiel J. A., Veenhuis M. (1999) The peroxisomal membrane protein Pex14p of Hansenula polymorpha is phosphorylated in vivo. FEBS Lett. 457, 397–399 [DOI] [PubMed] [Google Scholar]
- 42. Johnson M. A., Snyder W. B., Cereghino J. L., Veenhuis M., Subramani S., Cregg J. M. (2001) Pichia pastoris Pex14p, a phosphorylated peroxisomal membrane protein, is part of a PTS-receptor docking complex and interacts with many peroxins. Yeast 18, 621–641 [DOI] [PubMed] [Google Scholar]