Morphological, functional, and molecular analyses including single-cell gene expression profiling revealed that keratinocytes from transgene-free human induced pluripotent stem cell (hiPSC) lines were more similar to normal human keratinocytes than those from transgene-residual hiPSC lines, which may be partly explained by reactivation of residual transgenes upon induction of keratinocyte differentiation. These results suggest that transgene-free hiPSC lines should be chosen for therapeutic purposes.
Keywords: Human, Induced pluripotent stem cell, Keratinocyte, Differentiation, Transgene
Abstract
Human induced pluripotent stem cell (hiPSC) lines have a great potential for therapeutics because customized cells and organs can be induced from such cells. Assessment of the residual reprogramming factors after the generation of hiPSC lines is required, but an ideal system has been lacking. Here, we generated hiPSC lines from normal human dermal fibroblasts with piggyBac transposon bearing reprogramming transgenes followed by removal of the transposon by the transposase. Under this condition, we compared the phenotypes of transgene-residual and -free hiPSCs of the same genetic background. The transgene-residual hiPSCs, in which the transcription levels of the reprogramming transgenes were eventually suppressed, were quite similar to the transgene-free hiPSCs in a pluripotent state. However, after differentiation into keratinocytes, clear differences were observed. Morphological, functional, and molecular analyses including single-cell gene expression profiling revealed that keratinocytes from transgene-free hiPSC lines were more similar to normal human keratinocytes than those from transgene-residual hiPSC lines, which may be partly explained by reactivation of residual transgenes upon induction of keratinocyte differentiation. These results suggest that transgene-free hiPSC lines should be chosen for therapeutic purposes.
Introduction
Reprogramming of differentiated somatic cells into a pluripotent state has been previously carried out by cell fusion or nuclear transfer [1]. The molecular basis of reprogramming has been revealed by exogenous expression of combinations of transcription factors. Recently, four factors, namely OCT4, SOX2, KLF4, and cMYC, which are highly expressed in embryonic stem cells (ESCs), have been shown to reprogram both mouse and human somatic cells into ESC-like pluripotent cells, named induced pluripotent stem cells (iPSCs) [2, 3].
ESCs have an unlimited proliferative capacity and extensive differentiation capability and are thought to be a powerful cell source for regenerative medicine [4]. However, there are both ethical and biological concerns for the clinical use of ESCs that are related to their derivation from embryos and potential for immunological rejection, respectively. In addition, it is difficult to generate patient- or disease-specific ESCs that are required for their effective application. These disadvantages can be at least partially avoided by the alternative use of iPSCs.
Induction of reprogramming by defined factors has been mostly carried out by coinfection with retroviral vectors [2, 3]. The major problems of this retrovirus-based method are its oncogenicity and mutagenesis. Reactivation of the proviral Myc oncogene is one of the reasons for the oncogenicity of iPSCs [5]. However, although three-factor (Oct4, Sox2, and Klf4) iPSC-derived mice do not develop tumors [6], ectopic expression of any one of these genes may have deleterious consequences. For example, ectopic expression of Oct4 in the skin or intestine causes tumor development [7]. Overexpression of Klf4 induces dysplasia of the skin [8]. Furthermore, retroviral integration itself causes insertional mutagenesis and may alter the expression pattern of nearby genes [9]. Therefore, transgene integration-free iPSCs are necessary for their clinical use.
To achieve such iPSCs, we used the piggyBac transposon system to deliver the reprogramming factors. The piggyBac transposon is a moth-derived DNA transposon [10] that is highly active in mammalian cells and that has been used for gene delivery and mutagenesis [11]. The advantage over viral integration is that transposons can be easily removed from the host genome. Among the various DNA transposons, the piggyBac transposon does not leave “footprint” mutations upon excision [12]. The TTAA integration sites used by piggyBac transposons are repaired to the original sequence upon excision [12], resulting in removal of transposons from the host genome without changing any nucleotide sequences. Using this piggyBac transposon system, transgene integration-free and mutation-free mouse iPSCs have already been generated and reported by some groups, including our own [13–15].
In this study, by exploiting this unique property of the piggyBac transposon system, we generated transgene-free (Tg−) human induced pluripotent stem cell (hiPSCs) from transgene-retaining (Tg+) parental hiPSCs, thereby preserving the common isogenic genetic background. We generated epidermal keratinocytes from both Tg− and Tg+ hiPSCs in vitro and directly compared their tissue reconstitution potentials, allowing precise evaluation of the net effect of residual transgenes in hiPSCs and their derivatives.
Materials and Methods
Plasmid Construction
Five human reprogramming factors (POU5F1, SOX2, KLF4, cMYC, and LIN28) were amplified by polymerase chain reaction (PCR) with fusion of the first and second half of the T2A sequence in the C and N termini, respectively, except for the N terminus of POU5F1 and the C terminus of LIN28. PCR products were cloned into pCR4 using a Zero Blunt Topo PCR cloning kit (Life Technologies, Rockville, MD, http://www.lifetech.com), resulting in pCR-O, pCR-S, pCR-K, pCR-M, and pCR-L, respectively. The sequences of the inserts were verified by capillary sequencing. To combine all five factors into a single coding sequence, the BglII-SalI fragment containing the SOX2-coding sequence in pCR-S was first cloned into the BamHI-SalI site of pCR-O, resulting in pCR-OS. KLF4, MYC, and LIN28 fragments were sequentially inserted in the same manner, resulting in the final construct, pCR-OSKML (h5F). The EcoRI-SalI fragment of h5F was cloned into the EcoRI-SalI site of pBluescript, resulting in pBS-h5F. The BamHI-SalI fragment of pBS-h5F was cloned into the BglII-XhoI site of the piggyBac transposon vector, pPB-CAG.EBNXN, resulting in PB-CAG-h5F. Finally, the negative selection marker PGK-puΔtk cassette was excised from pFlexible [16] by XhoI digestion and then inserted into the SalI site of pPB-CAG-h5F, resulting in pPB-CAG-h5F-puroTK. Primers for construction of the vectors are listed in supplemental online Table 1.
Cell Culture
Mouse embryonic fibroblasts (MEFs) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (Life Technologies), 2 mM l-glutamine, 1× nonessential amino acids (Life Technologies), and 0.1 mM 2-mercaptoethanol. Normal human dermal fibroblasts (NHDFs) (Lonza, Walkersville, MD, http://www.lonza.com) from neonatal male skin were cultured in the same medium. Normal human epidermal keratinocytes (Lonza), also from neonatal male skin, and keratinocytes derived from iPSCs (iKCs) were cultured in serum-free keratinocyte-specific medium (CnT57; CELLnTEC, Bern, Switzerland, http://cellntec.com). Human iPSC lines and ESC lines (KhES-1 and KhES-3; Kyoto University, Kyoto, Japan, http://www.kyoto-u.ac.jp/en [17]) were cultured on mitomycin C-treated MEFs in serum-free human ESC (hESC) medium consisting of DMEM/F-12 (Life Technologies) with 20% knockout serum replacement (Life Technologies), 2 mM l-glutamine, 1× nonessential amino acids (Life Technologies), 0.1 mM 2-mercaptoethanol, and 5 ng/ml basic fibroblast growth factor (bFGF) (Katayama Chemical Industries Co., Ltd., Osaka, Japan, http://www.katayamakagaku.co.jp).
Generation of hiPSCs
Generation of hiPSCs was conducted according to the protocol described in Figure 1. NHDFs were plated in six-well plates and grown to 60%–70% confluence. On day 0, 2.5 μg of pCMV-mPBase and plasmids containing the piggyBac transposon carrying the reprogramming factors (pPB-CAG-h5F-puroTK) were simultaneously transfected into cells using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol. On day 5, transfected NHDFs were trypsinized and replated onto feeder cells (1 × 105 MEFs per 60-mm dish) in serum-free hESC medium. The medium was refreshed every other day. On days 10–14, ESC-like colonies appeared in the dishes, and at approximately day 21, colonies were counted, picked, and expanded further.
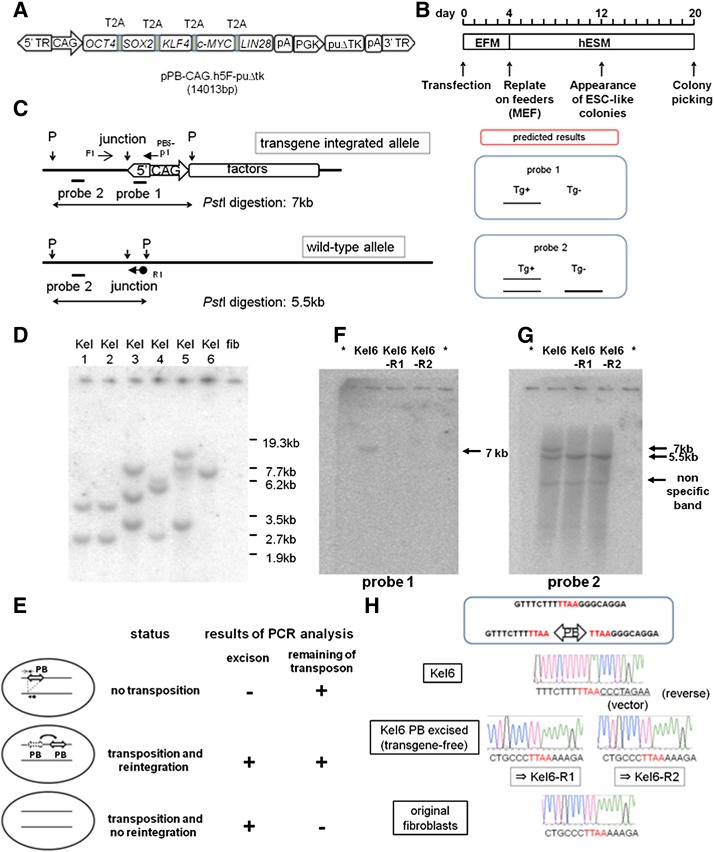
Figure 1.
Generation of human induced pluripotent stem cells (hiPSCs) using the piggyBac transposon system and establishment of transgene-free hiPSCs. (A): Schematic representation of the piggyBac transposon vector carrying reprogramming factors (pPB-CAG.h5F-puΔtk, 14,013 bp). Genes encoding five factors were linked by sequences encoding 2A peptides. Expression of these factors was driven by constitutively active CAG promoters. (B): Timeline of reprogramming. (C): Representative restriction enzyme map and positions of primers used to evaluate transposon removal in the hiPSC genome at the site of transposon vector integration. Probes 1 and 2 were designed as indicated by the thick line. A schematic representation of the predicted results of Southern blot analysis using probe 1/2 is shown (right). The thick line indicates a higher density band. F1, R1, and PB5-p1 are primers used to evaluate transposon removal. Primer sequences are listed in supplemental online Table 1. (D): Southern blot analysis of primary hiPSCs. Genomic DNA was digested with PstI and hybridized with probe 1. Lanes 1–6, primary hiPSCs generated by transposon-mediated reprogramming. Lane 7, original fibroblast genomic DNA as a negative control. (E): Schematic representation of the strategy of transposon removal. Each arrow indicates a designated primer used for a two-step PCR screening (Fig. 1C; supplemental online Fig. 2). Representative PCR results for each status are shown in supplemental online Fig. 3. (F, G): The results of Southern blot analysis indicating transposon-free hiPSCs were confirmed by PCR screening. (F) and (G) show the results using probes 1 and 2, respectively. Lanes marked by asterisks contained sample loading buffer only (without DNA). (H): Sequencing analysis of the integration site on chromosome 7 in Kel6 and transposon-excised KeI6 (KeI6-R1 and KeI6-R2). The reference sequences (top) indicate the transposon integration sites where TTAA sequences were duplicated at both ends of the transposon. The electrophoretograms demonstrate that the sequences in KeI6-R1 and KeI6-R2 are identical to the original sequences in the fibroblasts and reference sequences. Abbreviations: EFM, embryonic fibroblast medium; ESC, embryonic stem cell; fib, fibroblast; hESM, human embryonic stem cell medium; junction, junction site of the genome and transposon vector; MEF, mouse embryonic fibroblast; p, PstI; PB, piggyBac transposon vector; PCR, polymerase chain reaction; puΔtk, puΔtk expression cassette; Tg+, transgene-residual; Tg−, transgene-free; 3′TR, 3′ terminal repeat of the piggyBac transposon; 5′TR, 5′ terminal repeat of the piggyBac transposon.
Preparation of Splinkerettes
Splinkerettes were prepared by annealing Spl-top and Spl-blunt (to generate blunt ends). The sequences of these oligonucleotides are listed in supplemental online Table 1. After heat denaturation at 95°C for 10 minutes, annealing was performed by cooling down the mixture of 11 pmol of each strand in 10 mM Tris-HCl (pH 7.4) and 5 mM MgCl2 in a total volume of 100 μl.
Determination of Transposon Integration Sites
Transposon integration sites were determined by splinkerette PCR. Genomic DNA from primary iPSCs was digested by appropriate restriction enzymes (HaeIII and RsaI) for 2 hour in 20-μl reactions. After heat inactivation, 2 μl of the digestion mixture was ligated with the 0.5 μl of splinkerette adaptors in 20-μl reactions. One microliter of the ligation mixture was then subjected to nested PCR. A primer pair, Spl-P1 and PB5-P1, was used for the first PCR. In the second PCR, the Spl-P2 and PB5-P2 primer pair was used (primer sequences are listed in supplemental online Table 1). Finally, PCR products were directly sequenced to determine the genomic sequences flanking the piggyBac terminal repeats. Sequences were analyzed by Blat searching the UCSC Genome Browser.
Bisulfite Sequencing Analysis
Bisulfite conversion of DNA was performed using a MethylEasy Xceed Rapid DNA Bisulphite Modification Kit (Genetic Signatures, Randwick, Australia, http://geneticsignatures.com) according to the manufacturer’s protocol, and then sequencing was performed. Primers for amplification of bisulfited DNA are listed in supplemental online Table 1.
Southern Blot Analysis
To detect the copy number of integrated transposons, genomic DNA from each hiPSC line was digested and hybridized with partial sequences of the piggyBac transposon 5′ terminal repeat as the probe. Primer sequences for generating the probe are listed in supplemental online Table 1.
Transposon Removal From Primary hiPSCs
Fifteen micrograms of the piggyBac transposase expression vector with the blasticidin (bsd) selection cassette was electroporated into 1 × 106 dissociated hiPSCs. A total of 2 × 105 cells were seeded onto 6-cm dishes containing feeder cells (MEFs) in hESC medium containing 10 μM ROCK inhibitor. From the following day, 2 µg/ml bsd was added to the culture medium, and selection was continued for 3 days. After an additional 7 days of culture without bsd, the resulting colonies were dissociated to single cells and expanded in hESC medium containing 10 μM ROCK inhibitor. Transposon removal was screened by two-step PCR analysis with the primers listed in supplemental online Table 1. The results of the PCR screening were confirmed by Southern blot analysis using the 5′ terminal repeat of the piggyBac transposon as one of the probes and genomic sequences just outside of the transposon integration site as the other probe. Primer sequences for generating the probe are listed in supplemental online Table 1.
Keratinocyte Induction From hiPSCs
We used the differentiation protocol described in Results to obtain keratinocytes from hiPSCs and hESCs. One week of random differentiation was induced in differentiation medium (hESC medium without bFGF). Then the culture medium was changed to serum-free keratinocyte-specific medium, and the cells were cultured for an additional 4–5 weeks.
Immunostaining
Cells were fixed in 3% formaldehyde in phosphate-buffered saline (PBS) for 15 minutes at room temperature and then permeabilized in 100% methanol for 10 minutes at −20°C when needed. Then the cells were blocked in serum-free protein block (Dako, Glostrup, Denmark, http://www.dako.com) for 1 hour at room temperature. The cells were incubated with antibodies against NANOG (Cell Signaling Technology, Beverly, MA, http://www.cellsignal.com; 1:200, rabbit polyclonal), OCT4 (Cell Signaling Technology; 1:200, rabbit polyclonal), SOX2 (Cell Signaling Technology, 1:200, rabbit polyclonal), SSEA4 (Cell Signaling Technology; 1:200, mouse IgG3 monoclonal), TRA-1-60 (Cell Signaling Technology; 1:200, mouse IgM monoclonal), TRA-1-81 (Cell Signaling Technology; 1:200, mouse IgM monoclonal), human keratin 5/14 (Covance, Princeton, NJ, http://www.covance.com; 1:500, rabbit IgG polyclonal), and each negative control antibody (Dako) overnight at 4°C. After washing with PBS (three times for 5 minutes), cells were labeled with Alexa Fluor 488- or Alexa Fluor 555-conjugated secondary antibodies (Life Technologies) for 1 hour at room temperature. After washing with PBS (three times for 5 minutes), specimens were analyzed under a fluorescence microscope (BZ-9000 BIOREVO; Keyence, Osaka, Japan, http://www.keyence.com).
Deparaffinized and rehydrated tissue sections were immersed in preheated (∼100°C) target retrieval solution (Dako) for 10 minutes. After cooling at room temperature, the sections were washed three times with PBS, and nonspecific binding sites were blocked by incubation with serum-free protein block for 1 hour at room temperature. Then the sections were incubated with antibodies against human keratin 14 or involucrin (GeneTex, San Antonio, TX, http://www.genetex.com; 1:500, rabbit IgG polyclonal) at 4°C overnight. After three washes with PBS, tissue sections were reacted with an Alexa Fluor 555-conjugated secondary antibody for 1 hour at room temperature. After further washing, the sections were mounted with Fluoprep (bioMérieux, Marcy l’Etoile, France, http://www.biomerieux.com).
Real-Time PCR
Total RNA was extracted using an SV Total RNA Isolation System (Promega, Madison, WI, http://www.promega.com). One microgram of total RNA was reverse transcribed using random primers by Moloney murine leukemia virus reverse transcriptase (Life Technologies) and then subjected to PCR using the primers listed in supplemental online Table 1. Quantitative RT-PCR was performed using Power SYBR Green PCR Master Mix (Life Technologies) and an ABI7900HT sequence detector (Life Technologies) according to the manufacturer’s protocols. Quantification of gene expression was based on the ΔCt method and normalized to β-actin expression. Melting curve and electrophoresis analyses were undertaken to verify the specificities of the PCR products and to exclude nonspecific amplification.
Single-Cell Gene Expression Analysis (Fluidigm Dynamic Array)
Single-cell gene expression profiling was performed using a Biomark dynamic array (Fluidigm, South San Francisco, CA, http://www.fluidigm.com) according to the manufacturer’s protocol. Briefly, the cells were single-cell sorted using a BD FACSAria (BD Biosciences, San Diego, CA, http://www.bdbiosciences.com) into 2× CellsDirect buffer (Life Technologies). Cells were subjected to target-specific reverse transcription and 18 cycles of PCR preamplification with a mix of primers specific to the target genes (STA). STA products were then processed for real-time PCR analysis on Biomark 48:48 dynamic array integrated fluidic circuits (Fluidigm). The results obtained from the analysis were conducted with the R statistics package and output as a heat map. The primers for the analysis are listed in supplemental online Table 2.
Results
Successful Establishment of hiPSCs Using the piggyBac Transposon System
To deliver the reprogramming factors to recipient cells, we constructed piggyBac transposon-based reprogramming vectors as described in Materials and Methods (Fig. 1A). cDNAs constituting the open reading frames of OCT4, SOX2, KLF4, cMYC, and LIN28 were combined into a single open reading frame separated by sequences encoding 2A peptides and placed under the constitutively active CAG promoter. The ∼20-amino acid 2A peptides from the Thosea asigna virus (T2A) work as self-cleaving signals and enable the expression of several gene products from a single transcript [18], thereby facilitating multigene delivery to target cells.
NHDFs were transfected with the above-mentioned piggyBac transposon vector carrying the five factors to induce reprogramming. The protocol we used is illustrated in Figure 1B. We performed cationic lipofection to introduce 2.5 μg of piggyBac transposon and 2.5 μg of piggyBac transposase expression vector into ∼70% confluent NHDFs cultured in six-well plates and maintained the cells in embryonic fibroblast medium for 4 days. At day 5, the transfected NHDFs were replated onto feeder cells (mitomycin C-treated MEFs) and maintained in human ESC (hESC) medium. At approximately day 14, small ESC-like colonies appeared. At day 20, colonies were picked up and passaged. In summary, we transfected 1 × 105 NHDFs with a transfection efficiency of approximately 10%, which was confirmed with the preliminary results using green fluorescent protein as reporter, and obtained approximately 100 colonies. Therefore, the overall reprogramming efficiency was approximately 1%, which is virtually equivalent to that of the retroviral method.
Derivation of Transgene Integration-Free and Mutation-Free hiPSCs by piggyBac Transposon Vector Removal
We investigated the copy number of the integrated transposons by Southern blot analysis with a probe designed in the 5′ repeat of the piggyBac transposon (probe 1 in Fig. 1C). As shown in Figure 1D, most hiPSCs colonies had multiple insertions. Among the colonies, we obtained a colony with single-copy integration (named KeI6). In the following experiments, our established hiPSC clone, KeI6, or its derivatives were mainly used.
To obtain transgene-free hiPSCs, we first used splinkerette PCR to identify the integration sites and found that the transposon was integrated at chromosome 7 (supplemental online Fig. 1). Next, we attempted to eliminate transposons from the hiPSC genome. As described in Materials and Methods, we used a two-step PCR screening system to select transposon-free iPSCs after transfection of the piggyBac transposase expression plasmid (Fig. 1E). The first PCR was conducted with a primer pair that detected excision of the transposon from the original site (Fig. 1C). The second PCR was conducted with a primer pair that detected the transposon itself (supplemental online Fig. 2). Representative results of this PCR screening are shown in supplemental online Figure 3.
After transient transposase expression and bsd selection, because transposase expression vector possesses bsd selection cassette, we picked colonies and screened them by PCR as described above. We obtained 10 clones of 55 colonies from the transposase-transfected Kel6 line, in which excision of the transposon had occurred (positive in the first step PCR analysis). After the second PCR analysis, we obtained two transposon-free candidate clones of the 10 clones. We then expanded two transposon-free clones (named KeI6-R1 and KeI6-R2) and confirmed them by Southern blot analysis using the 5′ repeat of the piggyBac transposon as the probe (probe 1 in Fig. 1C). We confirmed the loss of transposons in both clones that we identified as transposon-free by PCR (Fig. 1F). We also performed further Southern blot analysis using a probe designed just outside of the transposon-genome junction (probe 2 in Fig. 1C), which confirmed the results described above (Fig. 1G). Thus, we concluded that the transposons had been successfully removed in KeI6-R1 and KeI6-R2.
The piggyBac transposon does not leave a footprint at the excised site. To confirm that the transgene-free hiPSCs had no footprint mutations, we amplified the genomic regions encompassing the integration excision sites by PCR and then sequenced the PCR products directly. If there was a footprint mutation (e.g., a nucleotide change, insertion, or deletion), the sequence after TTAA, the target site of piggyBac integration, would be a mixture of sequences from the wild-type and excised alleles. The signals from all integration sites were identical to those from the original genomic sequence (Fig. 1H), indicating the absence of footprint mutations. Thus, we “cured” transgene-harboring hiPSCs by removing the transposon.
Characteristics of the Established hiPSCs
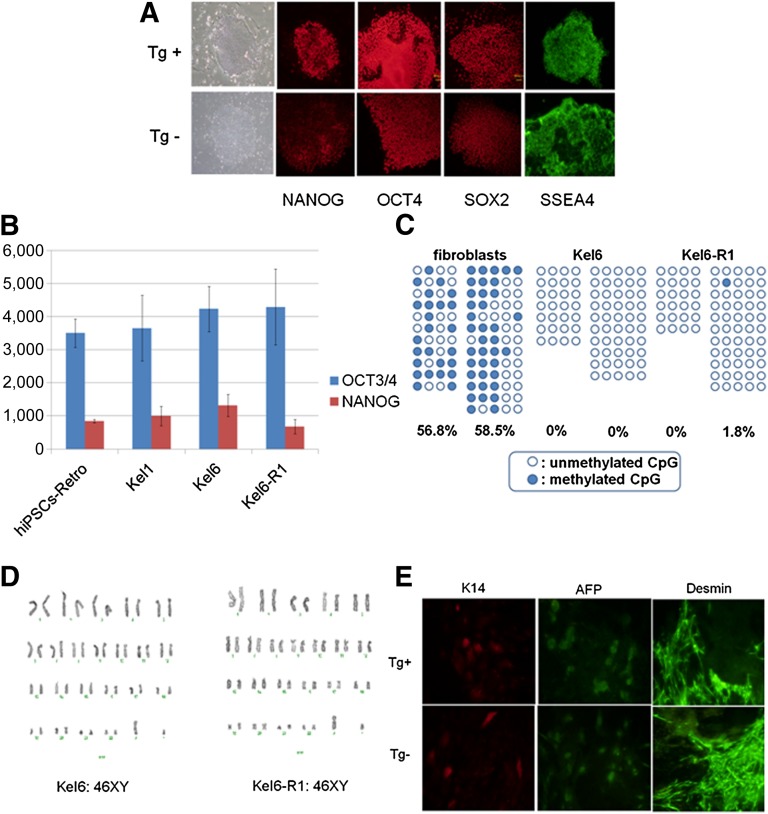
Next, we investigated the characteristics of the established hiPSCs. Immunofluorescence staining showed that both transgene-residual and -free hiPSC lines uniformly expressed stem cell markers such as OCT4, SOX2, NANOG, and SSEA4 (Fig. 2A). We also conducted RT-PCR analysis of OCT4 and NANOG gene expression and found that these genes were expressed in our hiPSCs at approximately the same levels as those in retrovirus-based hiPSCs, whereas gene expression associated with the transposon appeared to be silenced (Fig. 2B; supplemental online Fig. 2). To evaluate the epigenetic reprogramming status, we investigated the DNA methylation status at OCT4 promoter regions by bisulfite sequencing analysis and found that these promoter regions were almost completely unmethylated in the hiPSCs, which was in contrast to the status of the original fibroblasts (58% of the CpGs were methylated; Fig. 2C). This result indicated that the epigenetic reprogrammed status was maintained in our established hiPSCs even after the transgenes were eliminated. As shown in Figure 2D, karyotypic analysis revealed that both KeI6 and KeI6-R1 had a normal karyotype, 46XY. To confirm the pluripotency of the hiPSCs, we performed in vitro differentiation analysis. Both transgene-residual and -free hiPSC lines successfully differentiated into the three germ layers, namely ectoderm (K14-positive cells), mesoderm (desmin-positive cells), and endoderm (α-fetoprotein-positive cells) (Fig. 2E). These results suggested that the transgene-residual and -free hiPSC lines had phenotypes similar to those of pluripotent cells in terms of gene expression, epigenetic modifications, and the potential for differentiation into the three germ layers.
Figure 2.
Characteristics of the established hiPSCs. (A): Immunofluorescence analysis of NANOG, OCT4, SOX2, and SSEA4 expression in transgene-residual (KeI6) and transgene-free (KeI6-R1) hiPSCs. (B): Quantitative RT-PCR analysis of ESC-specific genes (OCT3/4 and NANOG) in transgene-residual (KeI1 and KeI6) and transgene-free (KeI6-R1) hiPSCs. Each bar indicates the means and SEM. (C): Bisulfite sequencing of the OCT4 promoter region. Open and closed circles indicate unmethylated and methylated CpG dinucleotides, respectively. (D): Normal karyotype of the transgene-residual (KeI6) and transgene-free (KeI6-R1) hiPSCs. (E): We confirmed the differentiation capabilities of transgene-residual (KeI6) and transgene-free (KeI6-R1) hiPSCs into all three germ layers by direct differentiation in vitro: K14, ectoderm; AFP, endoderm; and desmin, mesoderm. Abbreviations: AFP, α-fetoprotein; hiPSCs-Retro, retrovirus-mediated reprogrammed human induced pluripotent stem cells (MRC-5); Tg+, transgene-residual; Tg−, transgene-free.
Keratinocyte Induction From hiPSCs
We next attempted to differentiate the established hiPSCs into epidermal keratinocytes. There are few reports that describe protocols for differentiation of hiPSCs or hESCs into keratinocytes, in which retinoic acid (RA) and/or bone morphogenetic protein 4 (BMP4) are preferably used [19, 20]. In the present study, as described in Materials and Methods, we used a modified protocol in which neither RA nor BMP4 were used; this protocol is briefly illustrated in Figure 3A. At 4–5 weeks after induction of the differentiation, we obtained cells that had a keratinocyte-like morphology. These cells proliferated and were passaged at least five times in serum-free keratinocyte medium without feeder cells. Moreover, these cells were positive for K5/K14 as indicated by immunohistochemistry (Fig. 3D), suggesting successful differentiation of keratinocytes from hiPSCs. We called these cells induced keratinocytes (iKCs).
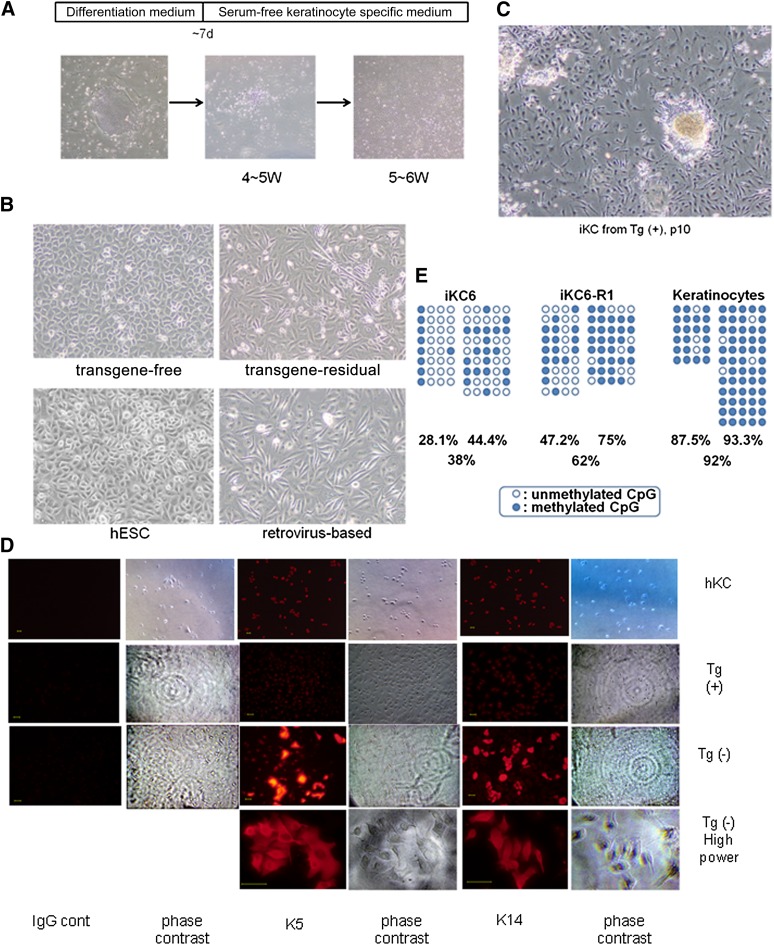
Figure 3.
Keratinocytes induced from established hiPSCs. (A): Timeline of keratinocyte differentiation. Lower panels show representative photographs of cells at each stage (×40). (B): Morphology of cells derived from hiPSCs or hESCs indicated in panels, respectively (×100). (C): Morphology of cells derived from transgene-residual hiPSCs at a later passage (p10, ×40). (D): Immunofluorescence analysis of K5 and K14 expression in primary keratinocytes (hKCs) and iKCs derived from transgene-residual (KeI6) and transgene-free (KeI6-R1) hiPSCs. Scale bars = 50 μm. (E): Bisulfite sequencing of the OCT4 promoter region in iKCs and primary human keratinocytes. Open and closed circles indicate unmethylated and methylated CpG dinucleotides, respectively. Abbreviations: cont, control; d, days; hESC, human embryonic stem cell; hKC, human keratinocyte; IgG cont, IgG control; iKC, induced keratinocyte; Tg(+), transgene-residual; Tg(−), transgene-free; W, weeks.
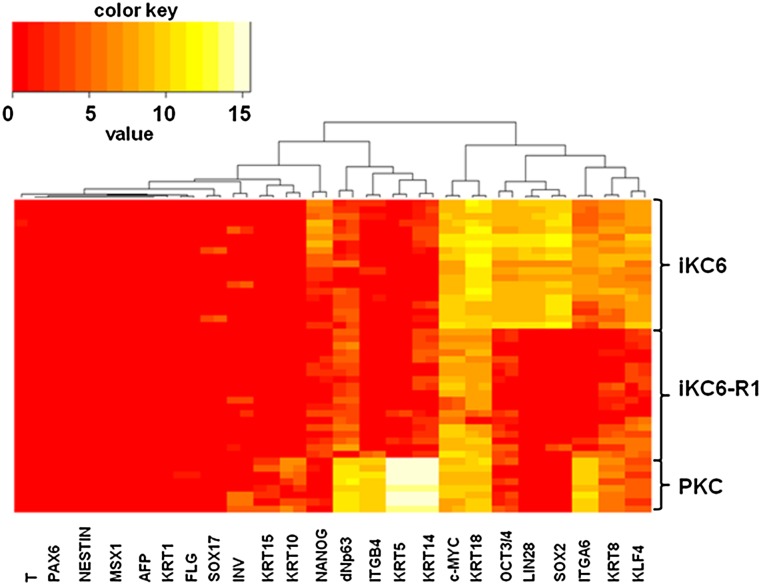
Comparing our established transgene-residual iKCs with transgene-free iKCs, there were obvious morphological differences in which transgene-residual iKCs showed a somewhat spindle-shaped morphology (Fig. 3B). Subsequent culturing resulted in transgene-residual iKCs showing a more spindle-shaped morphology. Moreover, some morphologically undifferentiated colonies were developing in the culture (Fig. 3C). Therefore, we checked the epigenetic reprogramming status by the expression of stem cell markers, including OCT4 and NANOG, and various differentiation markers in these iKCs by bisulfite sequencing analysis and real-time PCR, respectively. As expected, the DNA methylation status at OCT4 promoter regions showed hypomethylation in the original hiPSCs (KeI6 and KeI6-R1) and hypermethylation in iKCs (iKC6 and iKC6-R1) (Fig. 3E). However, by comparing transgene-residual iKCs (iKC6) with transgene-free iKCs (iKC6-R1), the levels of DNA methylation were clearly higher in transgene-free iKCs (38% vs. 62%, respectively; Fig. 3E). To clarify the characteristics of iKCs, we performed single-cell gene expression profiling using Fluidigm dynamic arrays [21]. In line with the results of bisulfite sequencing analysis, stem cell markers, including OCT4, SOX2, KLF4, MYC, LIN28, and NANOG, were obviously expressed in transgene-residual iKCs (though at lower levels than those in hiPSCs), indicating that residual transgenes may have been reactivated in iKCs (Fig. 4; supplemental online Figs. 2, 4). Examination of the expression of keratinocyte-specific genes, including K5, K14, and dNp63, revealed higher expression in iKCs derived from transgene-free iPSCs than in those derived from transgene-residual iPSCs (both transposon- and retrovirus-based hiPSCs) (Fig. 4; supplemental online Fig. 5). On the other hand, K8 and K18, which are expressed during the early stage of epidermal cell lineages, were strongly positive in transgene-residual iKCs (Fig. 4). Based on these results, residual transgenes in hiPSCs could influence epidermal differentiation by reactivation, resulting in partial differentiation into K5/K14-positive keratinocytes.
Figure 4.
Single-cell gene expression analysis using the Fluidigm dynamic array. Single-cell gene expression profiling using Fluidigm dynamic arrays was performed. Heat maps depict the expression of genes differentially expressed between iKC6 (transgene residual) and iKC6-R1 (transgene-free) hiPSCs and primary human keratinocytes (control). Rows represent individual cells, and columns represent the evaluated genes. Genes highly expressed are shown in white, and genes with low expression are shown in red. Abbreviations: AFP, α-fetoprotein; dNp63, Δ N p63; iKC, induced keratinocyte; INV, involucrin; ITGA6, α6 integrin; ITGB4, β4 integrin; KRAT, keratin; PKC, primary human keratinocytes from neonatal male skin.
For further examination, retrovirus-based hiPSCs (transgene residual) and hESCs (no transgenes) were differentiated into keratinocytes using the same protocols described in Figure 3A. At 4–5 weeks of differentiation, cells that could be maintained in keratinocyte-specific medium and that were positive for K5/K14 were obtained. Interestingly, iKCs derived from retrovirus-based hiPSCs morphologically resembled those derived from transgene-residual transposon-based hiPSCs, which showed a spindle-shaped morphology (Fig. 3B). The expression levels of stem cell marker genes, including OCT4 and NANOG, in iKCs derived from retrovirus-based hiPSCs were as low as those in normal human somatic tissues (supplemental online Fig. 4). However, gene expression from the residual retrovirus vector was found to be reactivated in iKCs derived from retrovirus-based hiPSCs (supplemental online Fig. 6). Additionally, two other independent hiPSC lines, KeI1 and KeI5 (Fig. 1D), both of which retain reprogramming transgenes, were subjected to the keratinocyte differentiation protocol. The KeI1- and KeI5-derived iKCs (iKC1 and iKC5, respectively) showed spindle-shaped morphologies similar to those of transgene-residual iKCs (iKC6) (Fig. 3B; data not shown).
Functional Properties of iKCs
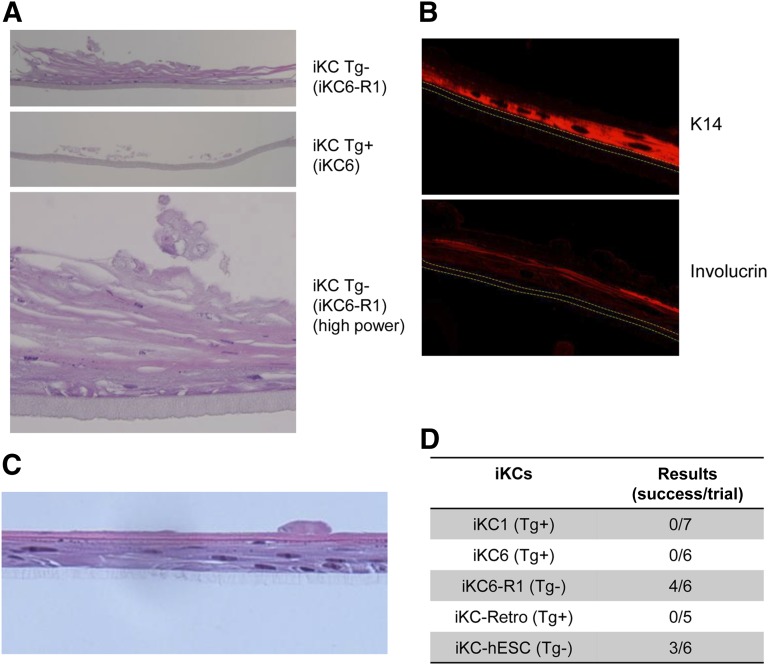
As described above, the morphological and molecular phenotypes of iKCs derived from transgene-free hiPSCs or hESCs appeared to be more similar to those of normal human keratinocytes. Finally, we compared the tissue reconstitution potential of these iKCs using a three-dimensional (3D) culturing system for keratinocytes (CELLnTEC). According to the manufacturer’s protocols, we performed the 3D culture experiments with both transgene-residual (iKC6) and -free (iKC6-R1) cell lines for approximately 21 days. As representatively shown in Figure 5A, iKC6-R1 cells successfully formed pluristratified epidermal structures in four of six experiments (67%), whereas no such structures were formed by iKC6 cells in any of the six experiments. In 3D culture, keratinocyte differentiation was evaluated by immunofluorescence staining of K14 and involucrin proteins (Fig. 5B). Lower layers of the pluristratified structure were positive for K14, whereas the upper layers were positive for involucrin, indicating in vitro reproduction of normal keratinocyte differentiation in vivo. A pluristratified structure was also formed by transgene-free hESCs in three of six experiments (50%) (Fig. 5C). In contrast, no such structures were formed by any of the transgene-residual hiPSC lines including iKC1 (seven experiments) and iKC-Retro (five experiments) (Fig. 5D). These results indicated that residual transgenes and their reactivation upon differentiation of keratinocytes from hiPSCs could critically influence not only the cellular phenotypes of such keratinocytes but also the functional properties of these cells for potential therapeutic use in regenerative medicine.
Figure 5.
Functional analysis of iKCs from hiPSCs. (A): Representative cross-sectional views of three-dimensional (3D) culture of iKCs using a 3D epidermal culture system (CELLnTEC). iKC6 and iKC6-R1 keratinocytes were generated from KeI6 (Tg+) and KeI6-R1 (Tg−) hiPSCs, respectively. Hematoxylin and eosin staining. Magnification is ×40 (top and middle) and ×200 (bottom). (B): Immunofluorescence analysis of K14 and involucrin expression in the reconstituted epidermis shown in (A) (×100). (C): Representative cross-sectional view of 3D culture of iKCs from hESCs. Hematoxylin and eosin staining is shown at ×100 magnification. (D): Results of 3D culture-based functionality tests of iKCs derived from various hiPSC lines or a hESC line. Abbreviations: hESC, human embryonic stem cell; iKC, induced keratinocyte; iKC-hESC, iKCs from hESCs; Tg+, transgene-residual; Tg−, transgene-free.
Discussion
In this study, we present a direct comparison of the effects of excising reprogramming transgenes from hiPSCs in terms of cellular and molecular phenotypes and their potential for differentiation into keratinocytes in vitro. Comparison between hiPSCs generated by different induction methods, e.g., retrovirus-based (transgene-residual) hiPSCs versus Sendai virus-based (transgene-free) hiPSCs, did not reveal any effects of residual transgenes in hiPSCs. However, our piggyBac transposon-based method allowed generation of an isogenic pair of hiPSC lines with or without retention of the reprogramming transgenes, leading to a precise evaluation of the effects of residual transgenes in hiPSCs and their derivatives.
The present study showed that, in the presence of the residual transgenes that had been silenced in a pluripotent state before differentiation induction, the iPSCs underwent transcriptional reactivation of the exogenous genes and showed less efficient differentiation into keratinocytes. In fact, we compared the phenotypes and function of transgene-residual and -free hiPSCs and their derivatives (iKCs). In the undifferentiated state, there appeared to be no characteristic differences between transgene-residual and -free hiPSCs. Next, we differentiated these established hiPSCs into epidermal keratinocytes using a modified method published elsewhere [19, 20]. In morphological, gene expression, and functional analyses, we found that transgene-residual hiPSCs did not fully differentiate into keratinocytes. Single-cell analysis using the Fluidigm dynamic array revealed that the cells derived from transgene-residual hiPSCs remained in an early developmental stage of keratinocyte differentiation, i.e., the K8/K18-positive stage, which may be related to residual transgene reactivation and subsequent activation of endogenous pluripotency genes such as NANOG (Fig. 4). On the other hand, cells that resembled normal human keratinocytes were effectively induced from transgene-free hiPSCs (Figs. 3–5). Single-cell analysis showed that our keratinocyte differentiation method successfully induced epidermal lineage cells, because cell types belonging to other lineages were not detected as shown in Figure 4. However, the expression of keratinocyte-specific genes such as K5, K14, and dNP63 was still weak even in cells differentiated from transgene-free hiPSCs compared with that in primary human keratinocytes, indicating that better methods need to be established for keratinocyte differentiation. Another explanation is that a minor population (∼10%) of iKC cells that expressed an extremely high level of K14 as shown in Figure 3D was included in the bulk analysis but excluded from the single-cell analysis by random selection (Fig. 4).
In terms of the relationship between residual transgenes and iPSC function, we found conflicting results in the present study. Previous reports have suggested that impaired silencing of transgenes in hiPSCs results in poor differentiation [22, 23]. Once the transgenes were silenced in iPSCs, these cells appeared to show a normal differentiation ability. Major et al. [24] reported that there are no differences between neuronal cell differentiation from transgene-residual or -free hiPSCs (induced by a lentiviral vector with Cre-loxp-mediated transgene excision system), and in that study, transgenes were silenced in hiPSCs. On the other hand, Toivonen et al. [25] reported that the reactivation of transgene in retrovirally generated hiPSCs affected the differentiation ability of these cells. The major problems of these previous contradicted reports were that hiPSCs generated from different methods (i.e., retrovirus-based and Sendai virus-based hiPSCs) had been compared. However, with our present situation, we could compare the phenotypes of transgene-residual and -free hiPSCs of the same genetic background, and this led to a precise evaluation of the effects of residual transgenes in hiPSCs and their derivatives. Thus, in our study, although the transgenes appeared to be silenced in hiPSCs, reactivation of the transgenes was obvious upon keratinocyte differentiation, leading to poor keratinocyte differentiation.
From another point of view, Itoh et al. [20] also reported that they selected cells in which the transgenes were not reactivated when evaluating their keratinocytes induced from retrovirus-based hiPSCs. Based on these results, including our own, the differentiation abilities of transgene-residual hiPSCs may be impaired, and this phenomenon may be more easily detected depending on the induced cell type. Specifically, residual transgenes tended to be reactivated more easily upon keratinocyte differentiation.
To further investigate whether reactivation of residual transgenes in hiPSCs was related to the method of derivation, namely transposon system specific, we differentiated retrovirus-based hiPSCs that were already confirmed to be pluripotent [26] (transgene residual) and hESCs (no transgenes) into epidermal keratinocytes using the same protocols and performed characteristic analyses of these cells. Morphologically, as expected, the cells differentiated from retrovirus-based hiPSCs showed a spindle shape resembling that of the differentiated transposon-based transgene-residual hiPSCs. On the other hand, cells differentiated from hESCs showed a cobblestone appearance that resembled the morphology of transposon-based transgene-free hiPSCs. Moreover, we clearly observed transgene reactivation in cells differentiated from retrovirus-based hiPSCs. These results strongly suggest that residual transgenes in hiPSCs can affect the differentiation ability, at least when differentiating into keratinocytes, through the reactivation of residual transgenes.
Moreover, the piggyBac transposon-based system for cellular reprogramming allowed efficient removal of reprogramming transgenes without residual exogenous sequences or any footprint mutations in the hiPSC genome. Even for establishment of human disease models in vitro, proper quality and safety precautions may be required because of the use of hiPSCs with viral transgene integration. Reactivation of any integrated transgene is one of the reasons for the oncogenicity of iPSCs [5–8]. Furthermore, transgene integration itself causes insertional mutagenesis [9]. Therefore, several methods have been developed to generate iPSCs, other than retrovirus-based methods, including those using plasmids [27], recombinant proteins [28], episomal viral vectors [29], and mRNA [30, 31] for derivation of transgene integration-free iPSCs. Among these methods, although the Sendai viral vector is now widely used to generate transgene integration-free iPSCs, it can be quite difficult to show that there is no residual virus in the cells. Therefore, among the various methods, we selected the piggyBac transposon system to generate hiPSCs.
Conclusion
Our results have significant implications for the clinical use (or even laboratory use) of hiPSCs. Specifically, we confirmed that transgene-residual hiPSCs are not suitable for clinical use and that transgene integration-free hiPSCs are necessary. The timing of integrated transgene reactivation cannot be predicted, and thus, transgene-free hiPSCs are more appropriate not only for clinical use and also laboratory use; otherwise the results may be affected. In addition, our piggyBac transposon system for the creation of hiPSCs may be a powerful approach, especially for clinical use.
Supplementary Material
Acknowledgments
We thank K. Nishida for the histological analysis and M. Tokunaga for single-cell sorting. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This work was also supported in part by a grant from the Takeda Science Foundation. Human embryonic stem cells were used following the guidelines for use of human embryonic stem cells of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author Contributions
K.I. and J.T.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; C.K. and K.H.: collection and/or assembly of data, data analysis and interpretation, manuscript writing; K. Yusa: construction of reprogramming vectors, manuscript writing; Y.Y., K. Yamauchi, and H.S.: collection and/or assembly of data; H.Y. and I.K.: conception and design; M.T., N.K., H.O., Y.M., H.A., and A.U.: provision of retroviral-based hiPSCs.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Jones JM, Thomson JA. Human embryonic stem cell technology. Semin Reprod Med. 2000;18:219–223. doi: 10.1055/s-2000-12560. [DOI] [PubMed] [Google Scholar]
- 5.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa M, Koyanagi M, Tanabe K, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 7.Hochedlinger K, Yamada Y, Beard C, et al. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 8.Foster KW, Liu Z, Nail CD, et al. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene. 2005;24:1491–1500. doi: 10.1038/sj.onc.1208307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nair V. Retrovirus-induced oncogenesis and safety of retroviral vectors. Curr Opin Mol Ther. 2008;10:431–438. [PubMed] [Google Scholar]
- 10.Cary LC, Goebel M, Corsaro BG, et al. Transposon mutagenesis of baculoviruses: Analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989;172:156–169. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]
- 11.Ding S, Wu X, Li G, et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 12.Fraser MJ, Ciszczon T, Elick T, et al. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol Biol. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 13.Yusa K, Rad R, Takeda J, et al. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods. 2009;6:363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaji K, Norrby K, Paca A, et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woltjen K, Michael IP, Mohseni P, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Weyden L, Adams DJ, Harris LW, et al. Null and conditional semaphorin 3B alleles using a flexible puroDeltatk loxP/FRT vector. Genesis. 2005;41:171–178. doi: 10.1002/gene.20111. [DOI] [PubMed] [Google Scholar]
- 17.Suemori H, Yasuchika K, Hasegawa K, et al. Efficient establishment of human embryonic stem cell lines and long-term maintenance with stable karyotype by enzymatic bulk passage. Biochem Biophys Res Commun. 2006;345:926–932. doi: 10.1016/j.bbrc.2006.04.135. [DOI] [PubMed] [Google Scholar]
- 18.Szymczak AL, Workman CJ, Wang Y, et al. Correction of multi-gene deficiency in vivo using a single “self-cleaving” 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 19.Guenou H, Nissan X, Larcher F, et al. Human embryonic stem-cell derivatives for full reconstruction of the pluristratified epidermis: A preclinical study. Lancet. 2009;374:1745–1753. doi: 10.1016/S0140-6736(09)61496-3. [DOI] [PubMed] [Google Scholar]
- 20.Itoh M, Kiuru M, Cairo MS, et al. Generation of keratinocytes from normal and recessive dystrophic epidermolysis bullosa-induced pluripotent stem cells. Proc Natl Acad Sci USA. 2011;108:8797–8802. doi: 10.1073/pnas.1100332108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Citri A, Pang ZP, Südhof TC, et al. Comprehensive qPCR profiling of gene expression in single neuronal cells. Nat Protoc. 2011;7:118–127. doi: 10.1038/nprot.2011.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sommer CA, Sommer AG, Longmire TA, et al. Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem Cells. 2010;28:64–74. doi: 10.1002/stem.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramos-Mejía V, Montes R, Bueno C, et al. Residual expression of the reprogramming factors prevents differentiation of iPSC generated from human fibroblasts and cord blood CD34+ progenitors. PLoS ONE. 2012;7:e35824. doi: 10.1371/journal.pone.0035824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Major T, Menon J, Auyeung G, et al. Transgene excision has no impact on in vivo integration of human iPS derived neural precursors. PLoS ONE. 2011;6:e24687. doi: 10.1371/journal.pone.0024687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toivonen S, Ojala M, Hyysalo A, et al. Comparative analysis of targeted differentiation of human induced pluripotent stem cells (hiPSCs) and human embryonic stem cells reveals variability associated with incomplete transgene silencing in retrovirally derived hiPSC lines. Stem Cells Translational Medicine. 2013;2:83–93. doi: 10.5966/sctm.2012-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makino H, Toyoda M, Matsumoto K, et al. Mesenchymal to embryonic incomplete transition of human cells by chimeric OCT4/3 (POU5F1) with physiological co-activator EWS. Exp Cell Res. 2009;315:2727–2740. doi: 10.1016/j.yexcr.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 27.Okita K, Nakagawa M, Hyenjong H, et al. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 28.Zhou H, Wu S, Joo JY, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fusaki N, Ban H, Nishiyama A, et al. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyoshi N, Ishii H, Nagano H, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Subramanyam D, Lamouille S, Judson RL, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.