Cold-induced, unstructured chloroplast proteins increase plant freezing tolerance by stabilizing membranes, but not enzymes, through folding and binding.
Abstract
Freezing can severely damage plants, limiting geographical distribution of natural populations and leading to major agronomical losses. Plants native to cold climates acquire increased freezing tolerance during exposure to low nonfreezing temperatures in a process termed cold acclimation. This involves many adaptative responses, including global changes in metabolite content and gene expression, and the accumulation of cold-regulated (COR) proteins, whose functions are largely unknown. Here we report that the chloroplast proteins COR15A and COR15B are necessary for full cold acclimation in Arabidopsis (Arabidopsis thaliana). They protect cell membranes, as indicated by electrolyte leakage and chlorophyll fluorescence measurements. Recombinant COR15 proteins stabilize lactate dehydrogenase during freezing in vitro. However, a transgenic approach shows that they have no influence on the stability of selected plastidic enzymes in vivo, although cold acclimation results in increased enzyme stability. This indicates that enzymes are stabilized by other mechanisms. Recombinant COR15 proteins are disordered in water, but fold into amphipathic α-helices at high osmolyte concentrations in the presence of membranes, a condition mimicking molecular crowding induced by dehydration during freezing. X-ray scattering experiments indicate protein-membrane interactions specifically under such crowding conditions. The COR15-membrane interactions lead to liposome stabilization during freezing. Collectively, our data demonstrate the requirement for COR15 accumulation for full cold acclimation of Arabidopsis. The function of these intrinsically disordered proteins is the stabilization of chloroplast membranes during freezing through a folding and binding mechanism, but not the stabilization of chloroplastic enzymes. This indicates a high functional specificity of these disordered plant proteins.
Low temperature is a major environmental factor affecting plant growth, development, and geographical distribution, as well as crop yield. Freezing of tissues can lead to plant death, mediated for example by excessive reactive oxygen species production, inactivation of enzymes, and damage to cellular membranes (Steponkus, 1984; Guy, 1990). Plants native to cold climates increase their freezing tolerance during exposure to low but nonfreezing temperatures in a process termed cold acclimation (Levitt, 1980; Steponkus, 1984; Guy, 1990; Thomashow, 1999). This involves complex remodeling of the plant transcriptome, proteome, metabolome, and lipidome (for review, see Guy et al., 2008; Hincha et al., 2012). A major signal transduction pathway in cold acclimation involves the C-repeat binding factors (Thomashow, 2010). These cold-induced transcription factors activate downstream target genes encoding enzymes involved in compatible solute biosynthesis and cold-regulated (COR) proteins, many of which belong to the group of late-embryogenesis abundant (LEA) proteins (Thomashow, 1999; Hundertmark and Hincha, 2008). LEA proteins have been found not only in plants but also in freezing and desiccation-tolerant invertebrates (Browne et al., 2002; Hand et al., 2011).
Most LEA proteins are intrinsically disordered proteins (IDPs) under fully hydrated conditions that gain a predominantly α-helical structure during dehydration or in the presence of membranes (for review, see Tunnacliffe et al., 2010; Hincha and Thalhammer, 2012). From a large number of in vitro studies, the protection of enzymes and the stabilization of membranes under stress conditions have emerged as the predominant potential functions of these proteins in various organisms, although other functions have also been suggested.
One of the best characterized cold-induced LEA proteins is the Arabidopsis (Arabidopsis thaliana) protein COR15A. Constitutive expression of COR15A increases the freezing tolerance of Arabidopsis chloroplasts in leaves and of isolated protoplasts by 1°C to 2°C (Artus et al., 1996). The recombinant protein prevents phospholipid vesicle fusion, but not leakage of soluble content (Uemura et al., 1996), and protects the activity of the enzyme lactate dehydrogenase (LDH) during freezing in liquid nitrogen (Lin and Thomashow, 1992a; Nakayama et al., 2007, 2008). In addition, COR15A prevents aggregation of the isolated chloroplast enzyme Rubisco during drying (Nakayama et al., 2007). The closest homolog of COR15A is COR15B, with 82% nucleic acid sequence identity (Wilhelm and Thomashow, 1993). The genes are localized in the nuclear genome, where they are arranged as a tandem repeat. The encoded proteins are targeted to the chloroplast stroma via signal peptides (Lin and Thomashow, 1992b; Nakayama et al., 2007; Candat et al., 2013) resulting in mature proteins of about 9 kD. Both mature proteins are highly hydrophilic and predominantly unstructured in solution, whereas they fold into amphipathic α-helices during drying. In the dry state, they interact specifically with the chloroplast galactolipid monogalactosyldiacylglycerol (MGDG; Thalhammer et al., 2010).
Although the function of COR15A has been previously studied, there is no information available about the role of COR15B and many questions remain for both proteins. Although it was shown that constitutive expression of COR15A leads to increased freezing tolerance in nonacclimated Arabidopsis plants (Artus et al., 1996), the contribution of the COR15 proteins to the increased freezing tolerance during cold acclimation has not been investigated. Likewise, there is evidence that COR15 proteins function as both membrane (Artus et al., 1996; Uemura et al., 1996; Steponkus et al., 1998) and enzyme stabilizers (Lin and Thomashow, 1992; Nakayama et al., 2007, 2008). However, it is unclear whether any chloroplast enzymes are inactivated during freezing in vivo, or whether cold acclimation has any effect on enzyme stability in vivo. It is also unknown whether any such effects may be related to the accumulation of COR15 proteins in the chloroplast stroma. Finally, both COR15 proteins are IDPs that fold and interact with membranes during drying (Thalhammer et al., 2010). However, these proteins do not function in the dry state; rather, they function in cells that are only partially dehydrated because of extracellular ice formation at comparatively mild freezing temperatures. Therefore, their folded state after desiccation is not of direct physiological relevance to explain structure-function relationships in COR15 proteins. Moreover, evidence for direct protein-membrane interaction in a physiologically relevant dehydration state is lacking. In this study, we attempted to provide answers to these open questions and thereby contribute to a functional understanding of intrinsically disordered stress proteins.
RESULTS
COR15A and COR15B Are Necessary for Arabidopsis to Attain Full Freezing Tolerance during Cold Acclimation
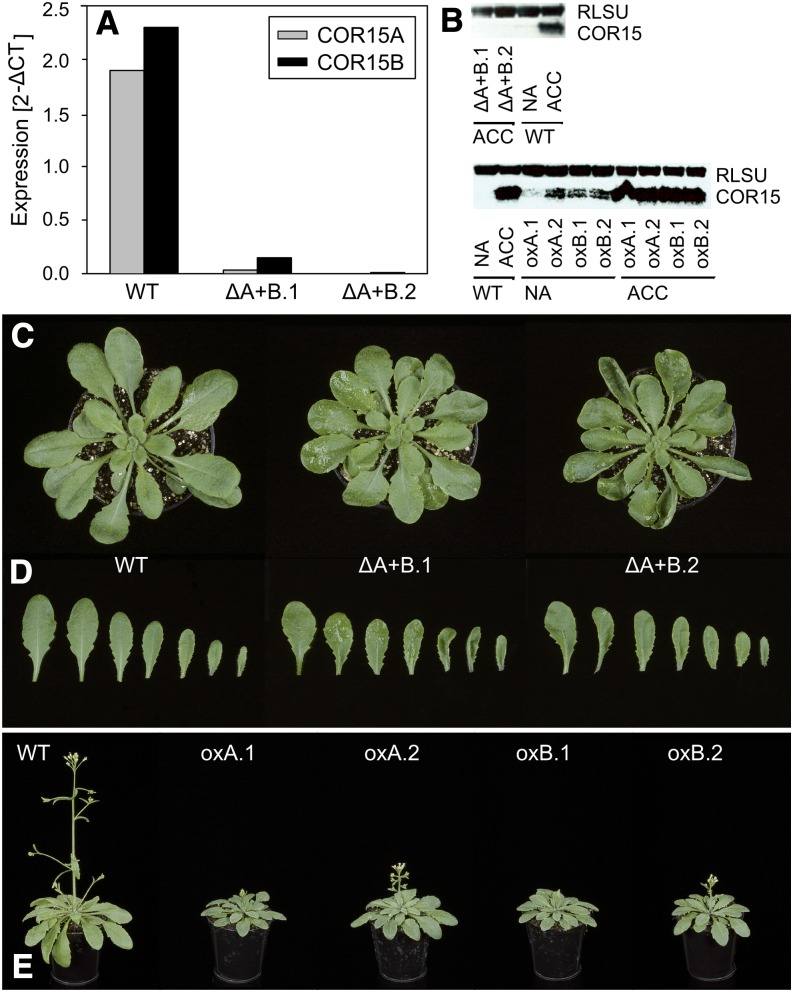
We used RNA interference (RNAi) to simultaneously silence both the COR15A and COR15B genes and identified two independent transformants that showed severe transcriptional silencing after 12 h of cold acclimation at 4°C (Fig. 1A). In addition, we used plants constitutively overexpressing either COR15A or COR15B (Yang et al., 2011). All transgenic lines were validated by quantitative reverse transcription (qRT)-PCR and by western-blot analysis using an anti-COR15A antibody (Lin and Thomashow, 1992b) that reacts with both proteins (Thalhammer et al., 2010). Although COR15 proteins were clearly detectable in cold-acclimated leaves of wild-type plants, no accumulation of the proteins was observed in the RNAi lines (Fig. 1B; Supplemental Fig. S1 shows the quantification). Conversely, in the COR15-overexpressing lines, COR15 protein bands were already visible under nonacclimating conditions, although protein abundance was less than in leaves of cold-acclimated wild-type plants. Under cold-acclimated conditions, COR15-overexpressing and wild-type plants showed similar COR15 protein levels (Fig. 1B; Supplemental Fig. S1 shows the quantification).
Figure 1.
Molecular and phenotypic characterization of COR15 gain-of-function and loss-of-function lines. A, COR15 transcript levels of cold-acclimated plants in the T3 generation were determined by qRT-PCR using COR15A-specific (gray) and COR15B-specific (black) primers and are shown as relative expression values normalized to the average of two housekeeping genes (2-ΔCT). B, Western blots of total soluble protein extracts of cold-acclimated COR15 RNAi (top) and nonacclimated and cold-acclimated COR15-overexpressing plants (bottom) were probed with anti-COR15 antibody. Unspecific binding of anti-COR15 antibody to the large subunit of Rubisco facilitated its use as a loading control. Total soluble protein extracts of nonacclimated and cold acclimated wild-type plants are shown for comparison. Distinct morphological effects of COR15 RNAi silencing and overexpression were recorded under standard growth conditions. C and D, Effects of simultaneous RNAi silencing of both COR15 genes on growth (C) and leaf phenotypes (D) compared with the wild type 35 d after sowing. E, Morphological effects of COR15A and COR15B overexpression compared with the wild type 45 d after sowing. ACC, Cold-acclimated; NA, nonacclimated; RLSU, large subunit of Rubisco; WT, wild type.
The alterations in COR15 gene expression had only a mild effect on plant growth and morphology. Silencing of both COR15 genes yielded plants with slightly reduced rosette size (Fig. 1C) and spoon-shaped leaves (Fig. 1D), whereas overexpression resulted in growth inhibition (Fig. 1E), but no alteration in leaf shape.
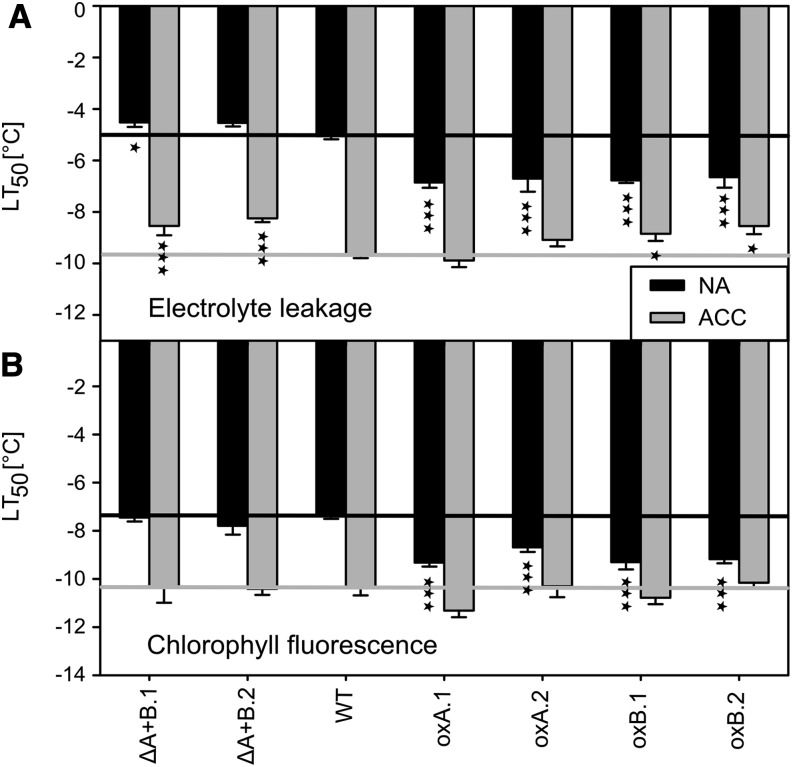
The effect of altered COR15 protein levels on Arabidopsis freezing tolerance was tested using two independent approaches: an electrolyte leakage assay to assess freezing damage to the plasma membrane and the tonoplast (Rohde et al., 2004), and a chlorophyll fluorescence imaging assay to obtain information on the inactivation of photosynthesis and the intactness of chloroplast membranes (Ehlert and Hincha, 2008). As shown in Figure 2, freezing tolerance is expressed as the temperature at which 50% damage occurred or LT50. In wild-type plants, the LT50 of cold-acclimated leaves was about 4°C lower than that of nonacclimated leaves, in agreement with previously published values for the accession Columbia-0 of Arabidopsis (Col-0; Rohde et al., 2004; Zuther et al., 2012). Under nonacclimated conditions, no significant effect of COR15 silencing on leaf freezing tolerance was observed. However, electrolyte leakage experiments revealed a significant impairment of the freezing tolerance of cold-acclimated leaves of the silencing lines compared with the wild type. The effect of cold acclimation was reduced by about 30% to 40% as a result of the lack of COR15 proteins in the chloroplasts. Interestingly, this was not reflected in the chlorophyll fluorescence measurements, indicating that the freezing tolerance of thylakoid membranes in the acclimated state was supported by other factors than COR15 proteins. In contrast, the freezing tolerance of COR15A-and COR15B-overexpressing lines under nonacclimated conditions was significantly higher compared with the wild type in both assays. This accounted for about 30% to 50% of the total effect of cold acclimation. Cold-acclimated COR15 overexpression lines were not further improved in their freezing tolerance compared with the wild type, in line with the similar amounts of COR15 proteins that were observed (Fig. 1B).
Figure 2.
Freezing tolerance of COR15 RNAi silencing and overexpression lines before (NA) and after (ACC) 14 d of cold acclimation at 4°C. Freezing tolerance was determined from electrolyte leakage (A) and chlorophyll fluorescence (B) measurements and is indicated as the LT50. Error bars represent ±sem from at least six biological replicates, with each replicate including three leaves from different plants. Asterisks indicate the ANOVA P values compared with the wild type (*0.05 < P ≤ 0.01, **0.01 < P ≤ 0.001, and ***P < 0.001). ACC, Cold-acclimated; NA, nonacclimated; WT, wild type.
Cold Acclimation Can Enhance the Stability of Enzymes during Freezing in Vivo, But COR15 Proteins Play No Role in This Process
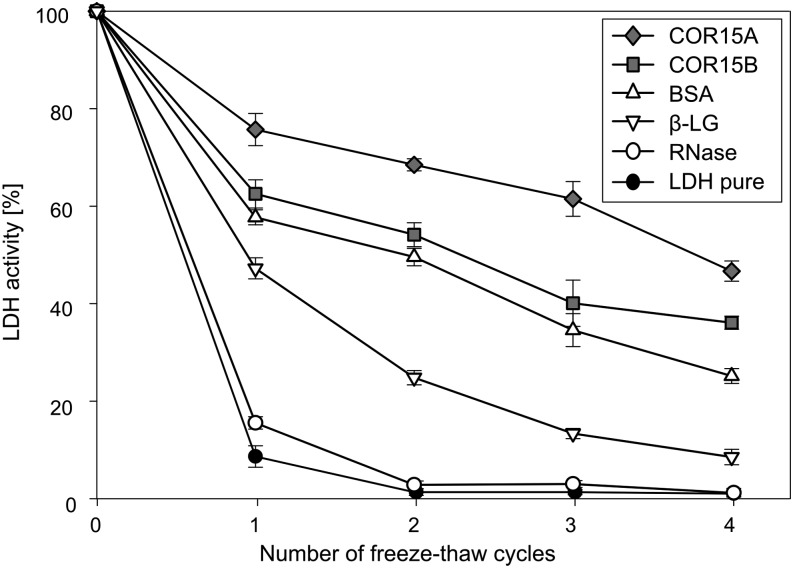
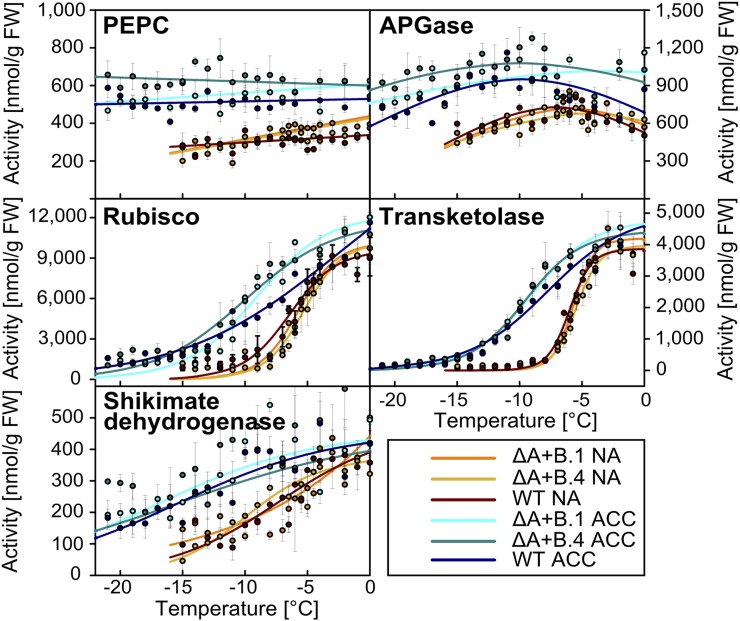
Several LEA proteins from both plants and invertebrates are able to stabilize sensitive enzymes such as LDH during freezing or drying in vitro (Battaglia et al., 2008; Tunnacliffe et al., 2010). Because such an activity was previously reported for COR15A as well (Lin and Thomashow, 1992a; Nakayama et al., 2007, 2008), we tested it for the recombinant COR15A and COR15B proteins previously described (Thalhammer et al., 2010). The assay involved freezing of enzyme samples in liquid nitrogen and thawing at room temperature. This freeze-thaw cycle was repeated for up to four times and after thawing, enzyme activity was determined relative to unfrozen control samples. Figure 3 shows that both COR15 proteins efficiently protected this sensitive enzyme from inactivation. The three tested reference proteins were less effective, although bovine serum albumin (BSA) in particular also provided some protection. However, an assay using a mammalian enzyme frozen in liquid nitrogen may not adequately reflect the in vivo situation. In addition, it was unclear whether any Arabidopsis chloroplast enzymes were actually inactivated at physiologically relevant freezing temperatures in planta. We therefore determined the total activity of four soluble chloroplast localized enzymes (ADP-Glc pyrophosphorylase [AGPase], Rubisco, transketolase, and shikimate dehydrogenase) and one cytosolic enzyme (PEP carboxylase [PEPC]) in protein extracts from nonacclimated and cold-acclimated Arabidopsis leaves frozen and thawed under the same conditions as used to determine LT50 values. Strong variation in activity was found as a function of freezing temperature among the enzymes after a freeze-thaw cycle (Fig. 4). PEPC and AGPase were highly stable even when leaves were frozen to temperatures below the LT50, whereas shikimate dehydrogenase showed a moderate, almost linear loss of activity with decreasing temperatures. Transketolase and Rubisco, on the other hand, showed a strong temperature-dependent loss of activity during freezing. Interestingly, the loss of activity of these last three enzymes was clearly influenced by cold acclimation, which shifted their inactivation to lower temperatures.
Figure 3.
Effect of COR15 proteins on the activity of LDH during multiple freeze-thaw cycles. Residual LDH activity after up to four freeze-thaw cycles between liquid nitrogen (−196°C) and room temperature normalized to unfrozen controls. LDH was frozen in the presence of COR15A or COR15B, or a set of control proteins (BSA, β-LG, and RNase) in a 1:10 m ratio. Error bars represent ±sem from 10 replicate measurements.
Figure 4.
Stability of candidate enzymes during an in vivo freeze-thaw cycle. Effect of simultaneous RNAi silencing of COR15A and COR15B on the activity of selected enzymes after a freeze-thaw cycle to different temperatures. Leaves were sampled from nonacclimated and cold-acclimated transgenic and wild-type plants. Lines were fitted to the data by linear (PEPC) or nonlinear regression analysis (all other enzymes) and were drawn to guide the eye. ACC, Cold-acclimated; FW, fresh weight; NA, nonacclimated; WT, wild type.
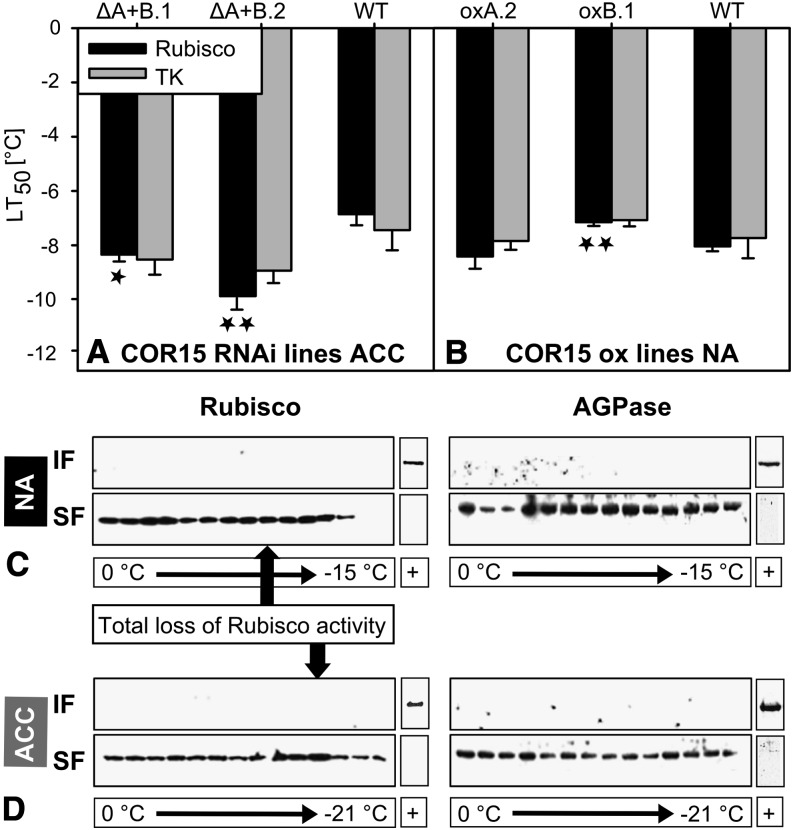
We used the RNAi lines lacking COR15 proteins in the acclimated state and the overexpression lines that contain the proteins already in the nonacclimated state described above to test the hypothesis that the increased in vivo stability of Rubisco and transketolase after cold acclimation was the result of COR15 accumulation. To make quantitative comparisons, we fitted a logistic model to the curves (Supplemental Table S1 shows r2 values of fitted curves) to calculate LT50 values for the acclimated RNAi lines (Fig. 5A), the nonacclimated overexpression lines (Fig. 5B), and the wild type under both conditions. The data provided no evidence for a function of COR15 in enzyme stabilization in intact leaves (i.e. LT50 was not increased in RNAi lines or decreased in overexpression lines compared with the wild type). In fact, the LT50 of Rubisco was significantly reduced in both silencing lines compared with the wild type after cold acclimation, whereas COR15 overexpression did not have a significant effect in nonacclimated leaves.
Figure 5.
Effect of COR15 proteins on enzyme stability during an in vivo freeze-thaw cycle. A and B, LT50 values of Rubisco (black) and transketolase (gray) activity indicate 50% residual activity after freezing and thawing of leaves from cold-acclimated COR15 RNAi silencing and wild-type plants (A) and from nonacclimated COR15 overexpression and wild-type plants (B). LT50 values were derived from curve fitting using a logistic regression model. Error bars represent ±sem from four biological replicates, each including three leaves from different plants. Asterisks indicate the ANOVA P values compared with the wild type (WT). (*0.05 < P ≤ 0.01, **0.01 < P ≤ 0.001). C and D, Western blots of protein extracts from nonacclimated (C) and cold-acclimated (D) wild-type leaves frozen to different temperatures. After thawing, the insoluble and soluble protein fractions of the leaves at each temperature step were probed using anti-Rubisco (left) and anti-AGPase (right) antibodies. Protein extracts were prepared from three leaves of different plants for each temperature step and condition. Arrows indicate the temperature at which total loss of Rubisco activity had occurred. Protein extracts from heat-treated leaves were used as positive controls (+). ACC, Cold-acclimated; IF, insoluble protein fraction; NA, nonacclimated; SF, soluble protein fraction.
It is widely assumed that a central function of LEA proteins is to protect target enzymes from aggregation during drying or freezing (Tunnacliffe et al., 2010). However, evidence for enzyme aggregation during freezing of leaves is lacking. We therefore used immunoblotting to assess the distribution of the sensitive enzyme Rubisco and the tolerant enzyme AGPase between the soluble and insoluble fraction of total leaf extracts as a function of freezing temperature. Under both nonacclimated (Fig. 5C) and cold-acclimated conditions (Fig. 5D), no protein was found in the insoluble fractions over the whole temperature range. Strikingly, Rubisco remained in the soluble fraction even at temperatures at which the enzyme was completely inactivated. On the other hand, heat treatment of both nonacclimated and cold-acclimated leaves resulted in the expected appearance of both enzymes in the insoluble fraction, confirming the validity of our experimental approach.
Solute-Induced Crowding and the Presence of Membranes Induce Folding in COR15A and COR15B
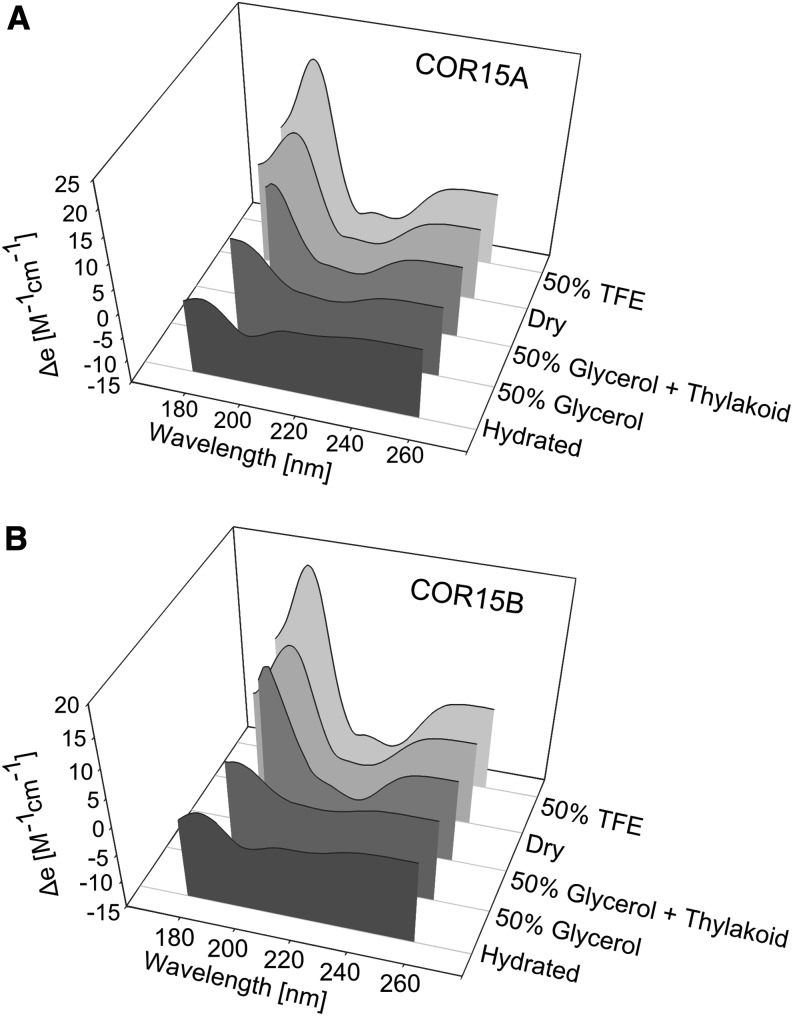
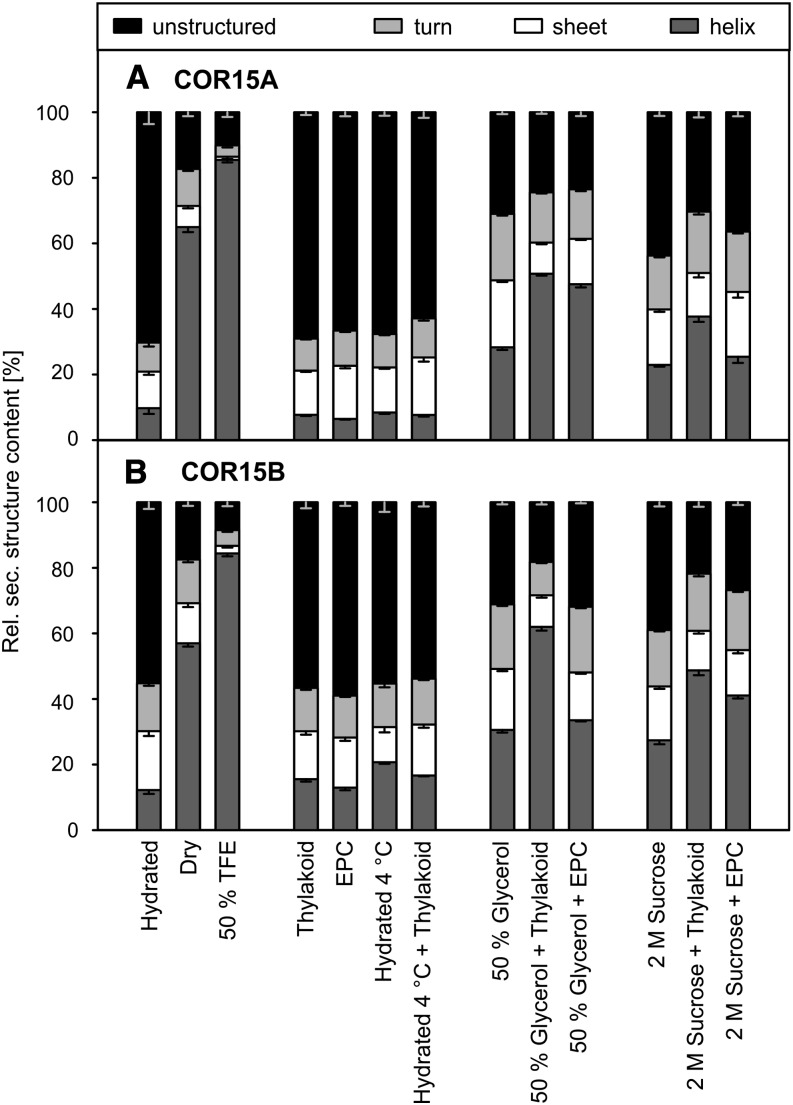
Recombinant COR15A and COR15B fold into α-helices upon desiccation (Thalhammer et al., 2010). However, these proteins have no function in desiccation tolerance, but effectively stabilize chloroplast membranes at mild subzero temperatures (Fig. 2). We therefore investigated the folding behavior of both proteins by circular dichroism (CD) spectroscopy (Figs. 6 and 7). Both the original CD spectra (Fig. 6) and the contributions of different types of secondary structure elements to the total structure content of the proteins, calculated from such spectra (Fig. 7), show significant folding after drying and in the presence of 50% (v/v) trifluoroethanol (TFE), an additive known to induce α-helicity in proteins (Greff et al., 1976). The addition of membranes or exposure of the proteins to low temperature (4°C) during the measurements was ineffective, either alone or in combination (Fig. 7). This was true for pure phosphatidylcholine membranes and for membranes with a lipid composition similar to the possible target membranes of the COR15 proteins in chloroplasts (Hincha et al., 1998).
Figure 6.
Far-UV CD spectra of COR15A (A) and COR15B (B) under different experimental conditions. All spectra were averaged over at least three replicate measurements. Proteins (final concentration of about 0.75 mg/mL) were either dissolved in pure water, in 50% (v/v) glycerol, or in 50% (v/v) TFE. Liposomes containing 40% MGDG, 30% DGDG, 15% SQDG, and 15% EPG (Thylakoid) in a 1:5 protein:lipid weight ratio were added to samples containing 50% glycerol.
Figure 7.
Quantitative analysis of secondary structure of COR15 proteins by CD spectroscopy. The secondary structure composition of COR15A (A) and COR15B (B) was calculated from CD spectra (compare Fig. 6). Error bars represent ±sem from at least three replicate measurements, using three calculation algorithms and two different reference protein sets. Large unilamellar liposomes were prepared from 100% EPC or from 40% MGDG, 30% DGDG, 15% SQDG, and 15% EPG (Thylakoid).
When leaves are frozen, ice crystallizes in the extracellular spaces, leading to partial dehydration of the cells (Levitt, 1980; Steponkus, 1984). Thus, the high solute concentrations reached in the cells of frozen leaves may be sufficient to induce folding in the COR15 proteins. To test this hypothesis, we obtained CD spectra of the proteins with high concentrations of glycerol or Suc in the absence or presence of liposomes (Fig. 6). Suspending the proteins in 50% (v/v) glycerol or 2 m Suc induced 25% to 30% α-helicity in both proteins (Fig. 7). The addition of liposomes resulted in a further increase of secondary structure, although egg phosphatidylcholine (EPC) liposomes were not effective in all cases.
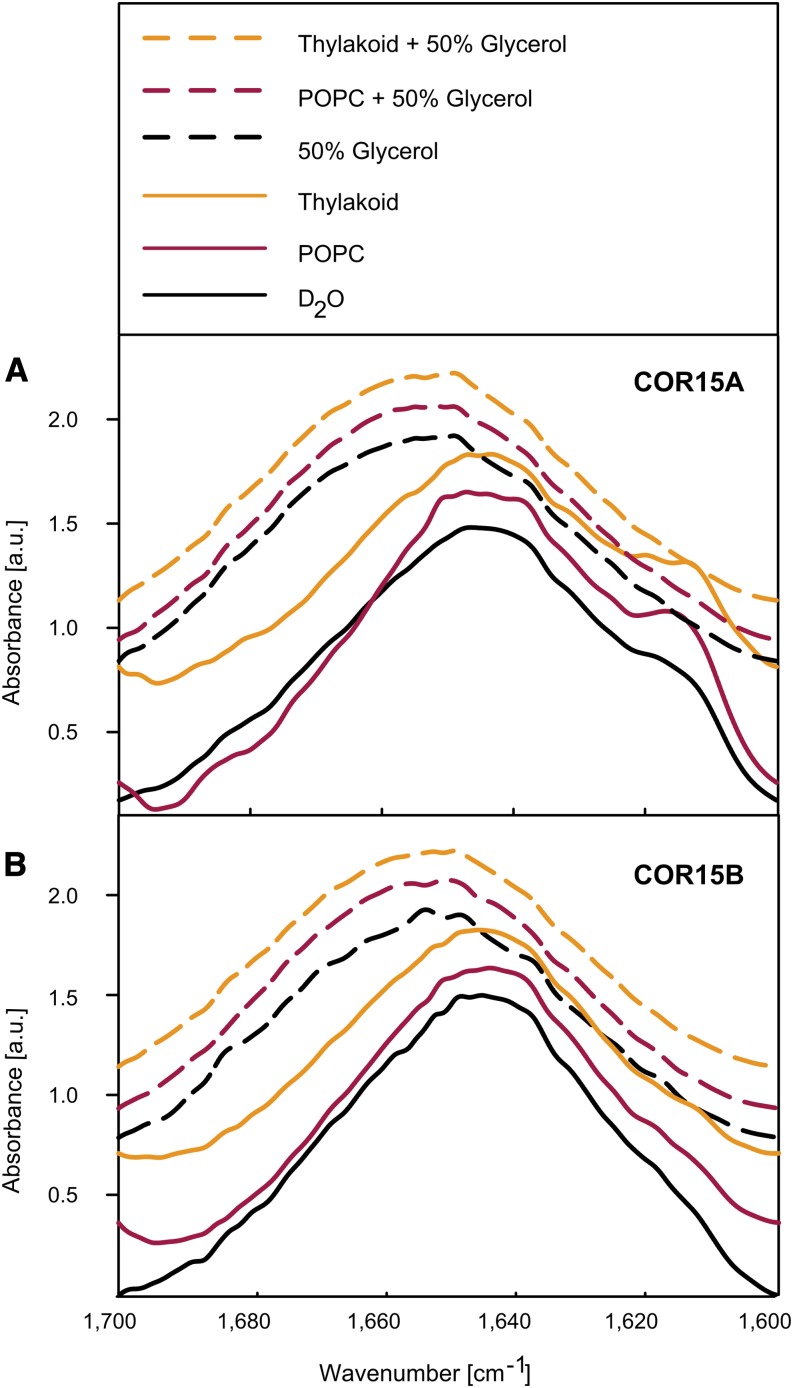
Because far-UV CD measurements may be influenced by high solute concentrations or the presence of membranes, we used Fourier transform infrared (FTIR) spectroscopy to validate these results (Fig. 8). The broad Amide I peak in FTIR spectra (1700–1600 cm−1) is composed of overlapping component peaks indicating the presence of secondary structures such as α-helices (1660–1650 cm−1) and unordered regions (1650–1640 cm−1; Susi and Byler, 1986; Barth, 2007). Under fully hydrated conditions, both proteins showed peaks around 1640 cm−1 in the presence and absence of liposomes in agreement with the predominantly unstructured character of COR15A and COR15B. In 50% (v/v) glycerol in the absence or presence of membranes, the Amide I peak was shifted to around 1650 cm−1, confirming the gain of α-helical structure under these conditions.
Figure 8.
Analysis of COR15 protein secondary structure by FTIR spectroscopy. Normalized Amide I peaks from FTIR spectra of COR15A (A) and COR15B (B) were obtained in the presence and absence of liposomes and 50% glycerol. The different peaks in each panel are offset from each other for better visibility. Large unilamellar liposomes were prepared from 100% EPC or from 40% MGDG, 30% DGDG, 15% SQDG, and 15% EPG (Thylakoid). Proteins were dissolved in D2O, or contained 50% glycerol, 50% TFE, or 2 m Suc in the same solvent. Where indicated, liposomes were added at a 1:5 protein:lipid mass ratio. a.u., Arbitrary units.
The COR15 Proteins Directly Interact with Lipid Membranes
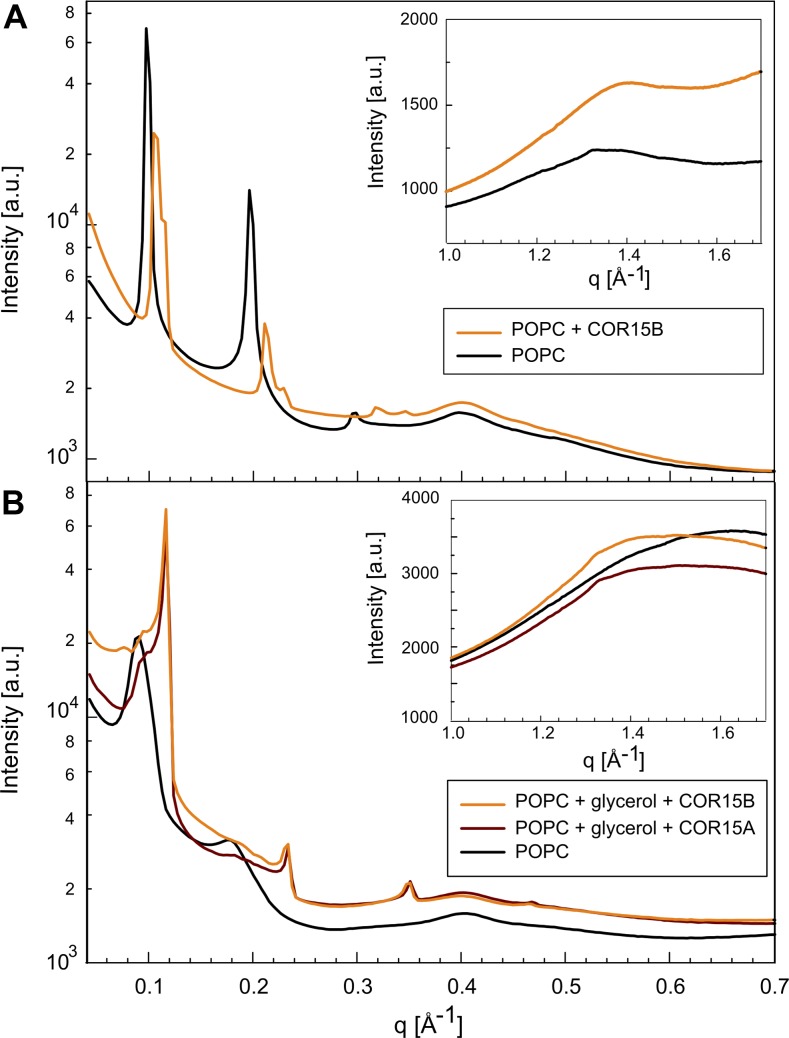
We used small-angle x-ray scattering (SAXS) and wide-angle x-ray scattering (WAXS) to further characterize COR15 membrane systems in water or 50% (v/v) glycerol. Supplemental Figure S2 shows a schematic drawing that visualizes the terminology of the corresponding results. We used 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC) liposomes instead of liposomes made from thylakoid lipids because these samples yielded scattering patterns that could not be interpreted in a straightforward manner as a result of the complex lipid composition. Further experimental characterization of the pure thylakoid lipid system will be necessary before it can be used to investigate interactions with proteins.
Distances between membranes were determined from the peak positions in the SAXS region (Fig. 9, main graphs), whereas distances between the lipid chains in the bilayers were derived from the peaks in the WAXS region (Fig. 9, insets). The lipids were in the fluid phase in all samples, as indicated by the broad WAXS peak at scattering vector values around q = 1.4 Å−1. POPC in water showed a repeat spacing of 64.0 ± 0.2 Å, which was reduced in the presence of COR15B protein to 59.0 ± 0.2 Å. However, in the wide-angle regime, the broad fluid peak moved to a lower q value in the presence of the protein, corresponding to an increase in the average chain distance from approximately 4.5 to 4.7 Å. This indicates that in the presence of COR15, the lipid bilayer thickness was slightly smaller and that the interchain distances were slightly larger than in the absence of the protein, implying that the COR15 protein slightly fluidized the membranes.
Figure 9.
X-ray scattering results indicate protein-membrane interaction. SAXS (main) and WAXS (insets) profiles of POPC liposomes suspended in water (A) or 50% glycerol (B) in the absence (black lines) or presence of COR15A (red lines) or COR15B (orange lines). a.u., Arbitrary units.
The repeat spacing increased to 70.0 ± 0.8 Å in the presence of glycerol, which was considerably larger than in water, whereas the interchain spacing was reduced to about 3.9 Å. The presence of COR15 proteins in 50% (v/v) glycerol drastically reduced the primary repeat spacing to 54.4 ± 0.5 Å. Moreover, both COR15A and COR15B increased the interchain distance to about 4.3 Å, indicating penetration of the proteins into the membranes. In addition, the graphs show evidence of lipid phase separation in the presence of glycerol, with small peaks occurring approximately at the same position as in the system without glycerol. More experiments will be needed to examine this process.
The COR15 Proteins Stabilize Liposomes during Freezing
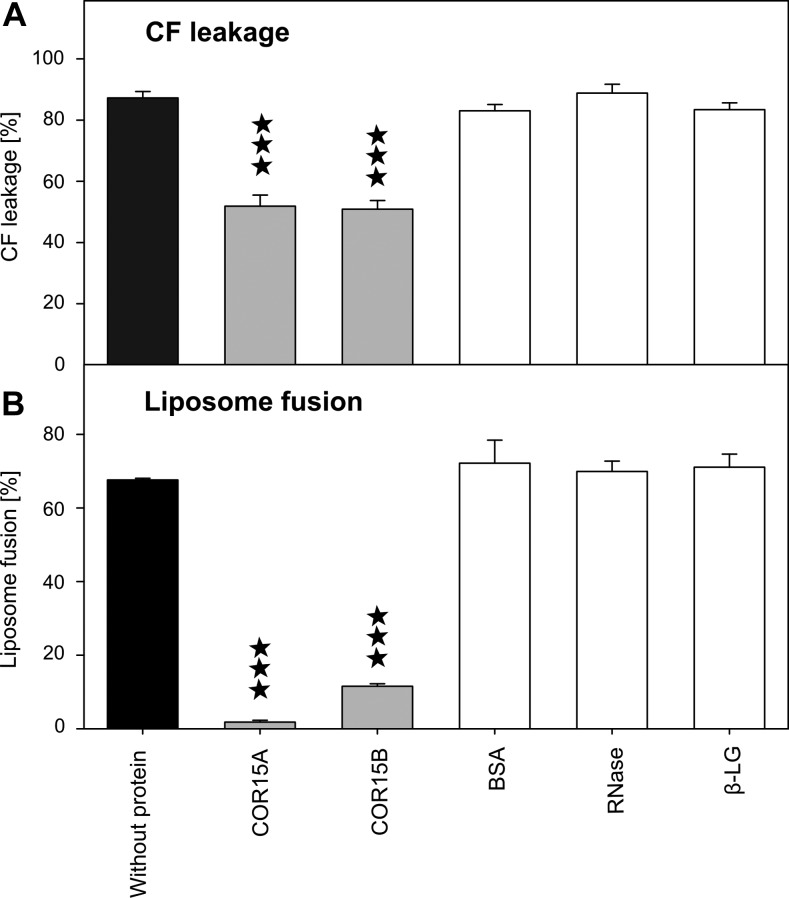
To investigate the potential of the COR15 proteins to protect membrane integrity during freezing, we assessed their effects on solute leakage and membrane fusion in liposomes mimicking the lipid composition of chloroplast membranes. Liposomes were frozen in the presence of COR15 proteins and a set of reference proteins. Only COR15A and COR15B significantly stabilized the liposomes, either in terms of freeze-induced solute leakage (Fig. 10A) or membrane vesicle fusion (Fig. 10B).
Figure 10.
Effect of freezing on liposome stability. Intactness of large unilamellar liposomes mimicking the lipid composition of chloroplast membranes (40% MGDG, 30% DGDG, 15% SQDG, and 15% EPG) in the absence or presence of COR15A or COR15B, or control proteins at a 1:25 protein:lipid weight ratio after freezing and thawing. Preservation of liposomes was assayed either as leakage of the soluble marker CF (A), or as membrane fusion (B). Error bars represent ±sem from at least three replicate measurements. Asterisks indicate ANOVA P values compared with samples without protein (***P < 0.001).
DISCUSSION
Although a large number of gene expression studies have documented the association of LEA proteins and other IDPs with exposure to environmental stress factors, there is only very limited genetic evidence for a significant contribution of such proteins to the stress tolerance of eucaryotic cells. In yeast (Saccharomyces cerevisiae), the knockout and overexpression of two different hydrophilins strongly influences cell survival after desiccation (Dang and Hincha, 2011), whereas the heat shock protein12 protects the cells from heat damage (Welker et al., 2010). In Caenorhabditis elegans, RNAi knockdown of an LEA gene reduces the desiccation tolerance of the animals (Gal et al., 2004). In plants, simultaneous knockdown of three LEA genes reduces long-term survival of seeds (Hundertmark et al., 2011). Heterologous expression of various LEA genes leads to increased tolerance to stresses such as drought and freezing (Tunnacliffe et al., 2010) and increased freezing tolerance was also found in nonacclimated Arabidopsis plants constitutively expressing the COR15A gene (Artus et al., 1996). This could be verified for COR15A and extended to COR15B in this study. Furthermore, knockdown of both genes indicated the necessity of COR15 proteins for the plants to attain full freezing tolerance after cold acclimation.
Several lines of evidence suggest that COR15 proteins function by stabilizing membranes, including previous results from NMR (Steponkus et al., 1998) and FTIR (Thalhammer et al., 2010) spectroscopy and their effects on electrolyte leakage from leaves reported here and in a previous study (Artus et al., 1996), carboxyfluorescein (CF) leakage from liposomes, and the SAXS results presented here. On the other hand, a role of COR15A in the stabilization of enzymes during freezing was suggested based on the protection of LDH and the prevention of Rubisco aggregation in vitro (Lin and Thomashow, 1992a; Nakayama et al., 2007, 2008). Although membrane and enzyme protection are not necessarily mutually exclusive properties of LEA proteins, they were found to be associated with two distinct, but closely related, LEA proteins from a desiccation-tolerant rotifer in in vitro experiments with purified proteins (Pouchkina-Stantcheva et al., 2007).
However, it was unclear whether any enzymes are inactivated during freezing in vivo and whether LEA proteins are able to protect them under these conditions. Determination of the temperature-dependent inactivation of several enzymes after an in vivo freeze-thaw cycle indicated that the selected Arabidopsis enzymes differed strongly in their stability and that the stability of freezing-sensitive enzymes was improved by plant cold acclimation. However, we found no improvement in the activity levels of the freezing-sensitive enzymes Rubisco and transketolase in the COR15 overexpression lines and no reduction in the COR15 RNAi lines compared with the wild type under nonacclimated or cold-acclimated conditions, respectively, excluding the possibility that the COR15 proteins are responsible for the observed effect of cold acclimation on enzyme stability. In addition, western-blot analysis provided no evidence for the presence of insoluble enzyme aggregates, even at temperatures below the complete inactivation of Rubisco. Collectively, these data strongly argue against a role of COR15 proteins in the stabilization of chloroplastic enzymes during freezing in vivo and also provide a note of caution for the interpretation of in vitro enzyme protection assays.
Many LEA proteins fold into α-helices upon desiccation (Tunnacliffe et al., 2010; Hincha and Thalhammer, 2012), but not all LEA proteins function in desiccation tolerance. The physiological role of the COR15 proteins is to stabilize chloroplast membranes at mild freezing temperatures between approximately −5°C and −15°C, at which extracellular ice crystallization leads to partial cell dehydration and increased molecular crowding. Macromolecular crowding, applied either through high concentrations of globular proteins or of polymers such as dextran, has generally only limited effects on IDP structure (Mouillon et al., 2008; Szasz et al., 2011; Goldenberg and Argyle, 2014). However, osmolyte-induced crowding was also ineffective to induce folding in some plant LEA proteins (Mouillon et al., 2008), although it can induce refolding of denatured globular proteins (Davis-Searles et al., 2001). COR15A and COR15B, on the other hand, showed significant α-helicity in the presence of 2 m Suc or 50% glycerol, corresponding to a freezing point depression of approximately −10°C and −28°C, respectively, indicating that partial folding may take place in a physiologically relevant temperature range.
This conclusion is emphasized by the fact that folding increased further in glycerol or Suc solutions with the addition of lipid membranes. This membrane-induced folding was specific to the crowded conditions, because no folding was induced in the proteins by membranes in the absence of osmolytes. Apparently, the presence of at least some hydrophobic protein surface resulting from the crowding-induced formation of an amphipathic helix is necessary to allow a protein-membrane interaction that will then drive additional folding. Although this folding was not strictly specific to a particular membrane type, pure POPC membranes were less effective under some conditions than membranes containing chloroplast lipids, indicating the possibility of a preference of the COR15 proteins for interaction with chloroplast-type membranes.
Direct evidence for COR15-membrane interactions was provided by SAXS and WAXS measurements that showed an increase in lipid spacing in the membranes in the presence of COR15 proteins only in a solution containing 50% glycerol. These results suggest an orientation of the amphipathic helices parallel with the membrane surface and partially embedded between the lipid headgroups, increasing interlipid spacing. Such interactions are favored by the presence of nonbilayer lipids such as MGDG (Webb and Green, 1991) in the membranes (Strandberg et al., 2012). At the same time, the negative curvature stress induced in the membranes by the presence of a nonbilayer lipid can be reduced through the interaction with the amphipathic α-helices that preferentially take place where lipid-packing defects allow access of the protein into the membrane surface (Vamparys et al., 2013). By this mechanism, membrane destabilization during freezing, which leads to solute leakage from liposomes and membrane fusion, was prevented by the COR15 proteins.
The presented data allow us to propose a mechanism for the function of an intrinsically disordered stress protein in cellular freezing tolerance. In the cold, the COR15 proteins are accumulated in the chloroplast stroma where they are unstructured in the absence of freezing. When temperatures drop below the freezing point and ice crystallizes in the extracellular spaces of the leaves, the resulting crowding effects lead to a partial folding of the proteins and their association with the inner chloroplast envelope membrane, which is rich in the nonbilayer lipid MGDG (Uemura and Steponkus, 1997). Binding of the proteins stabilizes the inner envelope membrane against fusion and hexagonal II phase formation also involving the outer envelope and the plasma membrane, ultimately leading to reduced electrolyte leakage from the cells, as previously proposed (Steponkus et al., 1998). Thylakoid membranes and soluble chloroplast enzymes are presumably stabilized by other cold-induced protectants such as the sugars Suc and raffinose, which are also accumulated in this compartment during cold acclimation (Schneider and Keller, 2009). This is in agreement with the finding that a knockdown of COR15A and COR15B had no effect on either chloroplastic enzyme or thylakoid membrane stabilities in cold-acclimated plants.
MATERIALS AND METHODS
Cloning, Plant Transformation, and Expression of Recombinant Proteins
Full-length complementary DNA clones of COR15A (At2g42540) and COR15B (At2g42530) were obtained from the RIKEN Arabidopsis Full-Length clone database (Seki et al., 2002; Sakurai et al., 2005). The complementary DNA sequences encoding either the precursor proteins or the mature proteins lacking the N-terminal signal peptides were amplified by PCR and inserted into the Gateway pENTR/SD/D-TOPO vector (Invitrogen). Primer sequences for the precursor proteins were as follows: CACCATGGCGATGTCTTTCTCAGG (COR15Afw), CTACTTTGTGGCATCCTTAGCC (COR15Arv), CACCATGGCGATGTCTTTATCAGGA (COR15Bfw), and TCAGGACTTTGTGGCATTCT (COR15Brv). The identity of the inserts was checked by restriction analysis and sequencing. Sequences encoding the precursor proteins were transformed into the RNAi vector pJawohl8 (designed at the MPI for Plant Breeding Research) via LR reactions. The identity of the inserts was checked by restriction analysis and colony PCR and the plasmids were transferred to the Agrobacterium tumefaciens strain GV:3101pmp90:RK for transformation of Arabidopsis (Arabidopsis thaliana) Col-0 plants using the floral-dip method (Clough and Bent, 1998). The mature proteins were expressed in Escherichia coli and the His-tagged proteins were purified as previously described (Thalhammer et al., 2010). Seeds of COR15A- and COR15B-overexpressing Arabidopsis Col-0 plants (Yang et al., 2011) were kindly provided by Chung-Mo Park (University of Seoul).
Plant Cultivation and Selection of COR15 Knockdown Plants
The Arabidopsis accession Col-0 was used for all experiments. Plants were grown in soil in a greenhouse at 16-h day length with light supplementation to reach at least 200 µE m−2 s−1 and temperatures of 20°C during the day and 18°C during the night until 42 d after sowing (nonacclimated plants). For cold acclimation, plants were transferred to a 4°C growth cabinet (16-h day/8-h night cycle, 4°C) with 90 μE m−2 s−1 for an additional 14 d (Rohde et al., 2004). Phosphotricine resistance was used as a selectable marker to identify homozygous transgenic plant lines. Expression of the COR15A and COR15B genes was assessed by qRT-PCR as previously described (Zuther et al., 2012) using the primer sequences from Hundertmark and Hincha (2008).
SDS-PAGE and Western-Blot Analysis
Total soluble leaf protein was extracted from homogenized plant material in 50 mm HEPES (pH 7.2) containing 25 mg/mL polyvinylpolypyrrolidone and Complete Protease Inhibitor Cocktail (Roche) on ice and cell debris was removed by centrifugation. Proteins were separated by SDS-PAGE (Schägger and von Jagow, 1987) and protein was quantified in the sample buffer using Amidoblack (Schaffner and Weissmann, 1973). Proteins were electrophoretically transferred to nitrocellulose membranes (porablot NCP; Macherey-Nagel) and 5% nonfat dry milk was used to block nonspecific binding sites. Membranes were probed with an antibody raised against recombinant COR15A, kindly provided by Prof. Michael F. Thomashow (Michigan State University). COR15 bands were visualized with horseradish peroxidase coupled to anti-Protein A antibody (Thermo Fisher Scientific) as described (Lin and Thomashow, 1992b), using the ECL Plus kit (GE Healthcare) according to manufacturer’s instructions. Bands were quantified using ImageJ software. Pixel density of COR15 bands was normalized to the pixel density of the corresponding Rubisco large subunit band.
To assess the aggregation of proteins after freezing and thawing, total soluble protein from three homogenized leaves was extracted in 500 µL of buffer (0.1 m KPO4, pH 7.8, 1 mm EDTA, 1% (v/v) Triton X-100, 10% (v/v) glycerol, and 1 mm dithiothreitol) and cell debris was removed by centrifugation (21,000g for 10 min at 4°C). The pellet fractions were filled up with SDS sample buffer to the original extraction volume. For control purposes, total soluble protein was extracted from leaves incubated at 100°C for 10 min in sealed plastic bags and the soluble and insoluble fractions were separated as described above. Both supernatant and pellet fractions were then diluted with SDS sample buffer in a 1:8 ratio for Rubisco and in a 1:2 ratio for AGPase. Both enzymes were detected by western-blot analysis using antibodies from Agrisera (http://www.agrisera.com) and a goat-antirabbit IgG coupled to horseradish peroxidase (Thermo Fisher Scientific).
Determination of Leaf Freezing Tolerance
Briefly, series of detached leaves were frozen in pools of three to temperatures ranging from −1°C to −21°C at a rate of 4°C/h. Unfrozen control samples were kept on ice during the duration of the experiment. Leaves were slowly thawed on ice. For electrolyte leakage measurements, leaves were immersed in distilled water and placed on a shaker for 16 h at 4°C. Electrolyte leakage was determined as the ratio of conductivity measured in the water before and after boiling and normalized to the unfrozen controls as previously described in detail (Rohde et al., 2004; Zuther et al., 2012). Chlorophyll fluorescence imaging analysis was performed on dark adapted leaves directly after thawing (i.e. omitting the incubation in distilled water), with freezing and thawing conditions identical to those described above, integrating measurements over the whole leaf area (Ehlert and Hincha, 2008). The LT50 was calculated as the logEC50 value derived from sigmoidal curves using GraphPad Prism3 software (Rohde et al., 2004).
Determination of Enzyme Activity after Leaf Freezing
Total soluble proteins were extracted from homogenized leaf material (Nunes-Nesi et al., 2007) and enzyme activities were measured in 96-well plates using the cycling assay system previously described (Gibon et al., 2004). Enzyme reactions were pipetted using a Janus robot (PerkinElmer). Final reaction products were quantified spectrophotometrically in a microplate reader. Rubisco activity was determined according to Sulpice et al. (2007) and AGPase, transketolase, shikimate dehydrogenase, and PEPC were determined according to Gibon et al. (2004). Enzyme activities were normalized to leaf fresh weight. The LT50 value for enzyme inactivation was derived from nonlinear regression analysis using a logistic regression model with the following equation: f = a/{1 + exp[−(x − x0)/b]}.
LDH Activity Assay
Assay proteins (COR15A and COR15B) and reference proteins (BSA, ribonuclease A [RNase], and β-lactoglobulin [β-LG]; Sigma) were randomly distributed on a 96-well plate in five replicates (1.5 µL of 5 µm protein in Tris, pH 7.0). Five plates were prepared and 1.5 µL of 0.5 µm (tetramer) LDH from rabbit muscle (Roche) in Tris, pH 7.0, was added per well. LDH activity in one plate was measured immediately after adding freshly prepared assay buffer (100 mm sodium phosphate, pH 8.0, 100 µm NADH, and 2 mm pyruvate). Absorbance was recorded on a Synergy HT microplate reader (Bio-Tek) at 340 nm every 15 s for a period of 5 min and reaction rates were determined with KC4 software (Bio-Tek). The remaining plates were frozen in liquid nitrogen for 1.5 min and thawed at room temperature (approximately 20°C). This was repeated up to four times. Remaining LDH activity was calculated as the percentage of LDH activity = (mean reaction rate of frozen samples/mean reaction rate of unfrozen samples) × 100.
Preparation of Liposomes
EPC, egg phosphatidylglycerol, and POPC were obtained from Avanti Polar Lipids. MGDG, digalactosyldiacylglycerol, and sulfoquinovosyldiacylglycerol were purchased from Lipid Products. Lipids dissolved in chloroform were mixed at the appropriate mass ratios in a glass tube, dried under a stream of N2, and subsequently kept under vacuum overnight to remove residual solvent. Lipids were rehydrated with either water for CD or D2O for FTIR experiments to reach concentrations of 6 mg/mL in experiments involving Suc or 2 mg/mL in all other experiments. For solute leakage experiments, lipids were rehydrated in 100 mm CF, 10 mm TES, and 0.1 mm EDTA, pH 7.4 (Hincha et al., 1998; Oliver et al., 1998). Liposomes for fusion assays (Struck et al., 1981) contained an additional 1 mol % each of the fluorescent probe pair N-(7-nitro-2,1,3-benzoxadiazol-4-yl) dipalmitoyl-phosphatidylethanolamine and N-(lissamine Rhodamine B sulfonyl) dipalmitoyl-phosphatidylethanolamine and were prepared in a buffer containing 10 mm TES, 0.1 mm EDTA, and 50 mm NaCl, pH 7.4 (Hincha et al., 1998; Oliver et al., 1998). All liposomes were formed by extrusion through two 100-nm polycarbonate membranes, using a hand-held extruder (Avestin; MacDonald et al., 1991).
CF Leakage and Liposome Fusion Experiments
Liposomes (10 mg/mL) were mixed with COR15 or reference proteins (BSA, RNase, and β-LG) at a ratio of 25:1 (w/w) and were frozen in an ethylene glycol bath at −20°C for 2 h in three replicates.
Liposomes containing CF were separated from free CF and transferred into TES, EDTA, NaCl buffer as above by gel filtration chromatography (NAP-5 columns; GE Healthcare). The leakage experiments are based on incapsulating CF in liposomes at a high, self-quenching concentration. Liposome destabilization leads to leakage of the dye into the surrounding medium, where it is diluted sufficiently to allow fluorescence emission. CF leakage was determined with a Fluoroscan Ascent fluorescence plate reader (excitation wavelength of 444 nm and emission wavelength of 555 nm; Labsystems) before and after liposome disruption by the addition of Triton X-100 (Hincha et al., 2002).
For membrane fusion measurements, two liposome samples were prepared, one containing 1 mol % of NBD-PE and Rh-PE each, the other only with unlabeled lipids. After extrusion, both samples were mixed at a 1:9 labeled:unlabeled ratio. Liposome fusion after a freeze-thaw cycle was determined with a Kontron SFM 25 fluorometer (Bio-Tek Instruments) at an excitation wavelength of 450 nm and an emission wavelength of 530 nm before and after liposome disruption with Triton X-100 (Hincha et al., 1998; Oliver et al., 1998).
CD Spectroscopy
CD spectra were obtained with a Jasco-715 spectropolarimeter (Jasco Instruments) as previously described (Thalhammer et al., 2010). Spectra were analyzed with CDPro software (Sreerama et al., 2000) using three different algorithms: CONTINLL, CDSSTR, and SELCON3. Sets of reference spectra containing denatured proteins were chosen for the analysis. Because the results were similar in each case for the three parallel samples and all algorithms, averages are shown. Proteins (final concentration of about 0.75 mg/mL) were either dissolved in water, 50% (v/v) glycerol, 50% (v/v) TFE, or 2 m Suc and measured in the absence or presence of liposomes in a 1:5 protein:lipid weight ratio. Spectra were either taken at room temperature (approximately 22°C) or at 4°C by cooling the sample holder with a circulating water bath. Reference spectra of the pure solvents with or without liposomes were recorded for background subtraction.
FTIR Spectroscopy
For FTIR spectroscopy, proteins were dissolved in D2O or in 50% glycerol or 50% TFE in D2O at a concentration of 2 mg/mL and measured in the absence or presence of liposomes at a 1:5 protein:lipid weight ratio. Samples (40 µL) were mounted in a PerkinElmer GX2000 FTIR spectrometer between two CaF2 windows separated by a 0.01-mm Teflon spacer. The sample compartment was continuously purged with N2 and 216 spectra were coadded from each sample and analyzed as recently described (Popova et al., 2011).
SAXS and WAXS Experiments
Experiments were conducted at the Australian Synchrotron SAXS/WAXS beamline with a 12-keV beam of wavelength 1.0322 Å. Diffraction patterns were recorded on a 2D Dectris Pilatus 1M detector giving a scattering vector range from 0.002 to 1.8 Å−1, covering the length scales of interest for the primary repeat distance and the wide-angle reflection. The detailed beamline properties are described in Kirby et al. (2013). Samples were prepared as described above, placed into 1.5-mm-diameter quartz capillaries, and sealed with epoxy resin. Samples were put in a multisample changer and placed in the sample position. After each exposure (duration 1.0 s), the samples were translated vertically to illuminate a different part of the sample. At least five independent measurements were made on each sample. The scattering from the pinhole geometry is radially symmetric, producing a series of concentric rings as a result of the Bragg scattering from structures within the sample. These rings are radially averaged to produce intensity versus scattering vector data, as shown in Figure 9. The analysis is similar to that reported previously for interactions between membranes and sugars (Lenné et al., 2009, 2010; Kent et al., 2010; Garvey et al., 2013). The peaks are fit with a Gaussian function to determine the repeat spacings that are further illustrated in Supplemental Figure S2. For samples with pure water in the absence or presence of protein, the values and errors are the averages and sds from at least five independent measurements. For the samples with 50% glycerol, good fits were not obtained, so the values derived from the peaks of the scattering curves are quoted (with larger errors).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Quantification of COR15 protein levels in COR15 RNAi silencing and overexpression lines.
Supplemental Figure S2. Schematic of a lipid bilayer membrane.
Supplemental Table S1. Determination of goodness of fit for enzyme activity curves.
Acknowledgments
We thank Michael F. Thomashow and Sarah J. Gilmour (Michigan State University) for the COR15 antiserum, Chung-Mo Park (University of Seoul) for COR15 overexpression lines, Beatrice Encke and Nicole Krohn (Max Planck Institute Potsdam) for excellent support with enzyme activity measurements, and Mark Aurel Schöttler (Max Planck Institute Potsdam) for the CD spectrometer.
Glossary
- LEA
late-embryogenesis abundant
- IDP
intrinsically disordered protein
- LDH
lactate dehydrogenase
- MGDG
monogalactosyldiacylglycerol
- RNAi
RNA interference
- qRT
quantitative reverse transcription
- LT50
median lethal temperature (causing 50% death)
- Col-0
ecotype Columbia-0 of Arabidopsis
- BSA
bovine serum albumin
- AGPase
ADP-Glc pyrophosphorylase
- PEPC
PEP carboxylase
- CD
circular dichroism
- TFE
trifluoroethanol
- FTIR
Fourier transform infrared
- SAXS
small-angle x-ray scattering
- WAXS
wide-angle x-ray scattering
- CF
carboxyfluorescein
- RNase
ribonuclease A
- β-LG
β-lactoglobulin
Footnotes
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin C, Thomashow MF. (1996) Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci USA 93: 13404–13409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth A. (2007) Infrared spectroscopy of proteins. Biochim Biophys Acta 1767: 1073–1101 [DOI] [PubMed] [Google Scholar]
- Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA. (2008) The enigmatic LEA proteins and other hydrophilins. Plant Physiol 148: 6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne J, Tunnacliffe A, Burnell A. (2002) Anhydrobiosis: plant desiccation gene found in a nematode. Nature 416: 38. [DOI] [PubMed] [Google Scholar]
- Candat A, Poupart P, Andrieu JP, Chevrollier A, Reynier P, Rogniaux H, Avelange-Macherel MH, Macherel D. (2013) Experimental determination of organelle targeting-peptide cleavage sites using transient expression of green fluorescent protein translational fusions. Anal Biochem 434: 44–51 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dang NX, Hincha DK. (2011) Identification of two hydrophilins that contribute to the desiccation and freezing tolerance of yeast (Saccharomyces cerevisiae) cells. Cryobiology 62: 188–193 [DOI] [PubMed] [Google Scholar]
- Davis-Searles PR, Saunders AJ, Erie DA, Winzor DJ, Pielak GJ. (2001) Interpreting the effects of small uncharged solutes on protein-folding equilibria. Annu Rev Biophys Biomol Struct 30: 271–306 [DOI] [PubMed] [Google Scholar]
- Ehlert B, Hincha DK. (2008) Chlorophyll fluorescence imaging accurately quantifies freezing damage and cold acclimation responses in Arabidopsis leaves. Plant Methods 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal TZ, Glazer I, Koltai H. (2004) An LEA group 3 family member is involved in survival of C. elegans during exposure to stress. FEBS Lett 577: 21–26 [DOI] [PubMed] [Google Scholar]
- Garvey CJ, Lenné T, Koster KL, Kent B, Bryant G. (2013) Phospholipid membrane protection by sugar molecules during dehydration - insights into molecular mechanisms using scattering techniques. Int J Mol Sci 14: 8148–8163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M. (2004) A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg DP, Argyle B. (2014) Minimal effects of macromolecular crowding on an intrinsically disordered protein: a small-angle neutron scattering study. Biophys J 106: 905–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greff D, Toma F, Fermandjian S, Löw M, Kisfaludy L. (1976) Conformational studies of corticotropin1-32 and constitutive peptides by circular dichroism. Biochim Biophys Acta 439: 219–231 [DOI] [PubMed] [Google Scholar]
- Guy C, Kaplan F, Kopka J, Selbig J, Hincha DK. (2008) Metabolomics of temperature stress. Physiol Plant 132: 220–235 [DOI] [PubMed] [Google Scholar]
- Guy CL. (1990) Cold acclimation and freezing stress tolerance: role of protein metabolism. Annu Rev Plant Physiol Plant Mol Biol 41: 187–223 [Google Scholar]
- Hand SC, Menze MA, Toner M, Boswell L, Moore D. (2011) LEA proteins during water stress: not just for plants anymore. Annu Rev Physiol 73: 115–134 [DOI] [PubMed] [Google Scholar]
- Hincha DK, Espinoza C, Zuther E. (2012) Transcriptomic and metabolomic approaches to the analysis of plant freezing tolerance and cold acclimation. In Tuteja N, Gill SS, Tiburcio AF, Tuteja R, eds, Improving Crop Resistance to Abiotic Stress, Vol 1, Berlin, Wiley-Blackwell, pp 255–287 [Google Scholar]
- Hincha DK, Oliver AE, Crowe JH. (1998) The effects of chloroplast lipids on the stability of liposomes during freezing and drying. Biochim Biophys Acta 1368: 150–160 [DOI] [PubMed] [Google Scholar]
- Hincha DK, Thalhammer A. (2012) LEA proteins: IDPs with versatile functions in cellular dehydration tolerance. Biochem Soc Trans 40: 1000–1003 [DOI] [PubMed] [Google Scholar]
- Hincha DK, Zuther E, Hellwege EM, Heyer AG. (2002) Specific effects of fructo- and gluco-oligosaccharides in the preservation of liposomes during drying. Glycobiology 12: 103–110 [DOI] [PubMed] [Google Scholar]
- Hundertmark M, Buitink J, Leprince O, Hincha DK. (2011) The reduction of seed-specific dehydrins reduces seed longevity in Arabidopsis thaliana. Seed Sci Res 21: 165–173 [Google Scholar]
- Hundertmark M, Hincha DK. (2008) LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent B, Garvey CJ, Lenné T, Porcar L, Haramus VM, Bryant G. (2010) Measurement of glucose exclusion from fully hydrated DOPE inverse hexagonal phase. Soft Matter 6: 1197–1202 [Google Scholar]
- Kirby NM, Mudie ST, Hawley AM, Cookson DJ, Mertens HDT, Cowieson N, Samardzic-Boban V. (2013) A low-background-intensity focusing small angle X-ray scattering undulator beamline. J Appl Cryst 46: 1670–1680 [Google Scholar]
- Lenné T, Garvey CJ, Koster KL, Bryant G. (2009) Effects of sugars on lipid bilayers during dehydration—SAXS/WAXS measurements and quantitative model. J Phys Chem B 113: 2486–2491 [DOI] [PubMed] [Google Scholar]
- Lenné T, Garvey CJ, Koster KL, Bryant G. (2010) Kinetics of the lamellar gel-fluid transition in phosphatidylcholine membranes in the presence of sugars. Chem Phys Lipids 163: 236–242 [DOI] [PubMed] [Google Scholar]
- Levitt J. (1980) Responses of Plants to Environmental Stresses. Volume I: Chilling, Freezing, and High Temperature Stresses, Ed 2, Orlando, Florida, Academic Press [Google Scholar]
- Lin C, Thomashow MF. (1992a) A cold-regulated Arabidopsis gene encodes a polypeptide having potent cryoprotective activity. Biochem Biophys Res Commun 183: 1103–1108 [DOI] [PubMed] [Google Scholar]
- Lin C, Thomashow MF. (1992b) DNA sequence analysis of a complementary DNA for cold-regulated Arabidopsis gene cor15 and characterization of the COR15 polypeptide. Plant Physiol 99: 519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald RC, MacDonald RI, Menco BPM, Takeshita K, Subbarao NK, Hu LR. (1991) Small-volume extrusion apparatus for preparation of large, unilamellar vesicles. Biochim Biophys Acta 1061: 297–303 [DOI] [PubMed] [Google Scholar]
- Mouillon JM, Eriksson SK, Harryson P. (2008) Mimicking the plant cell interior under water stress by macromolecular crowding: Disordered dehydrin proteins are highly resistant to structural collapse. Plant Physiol 148: 1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Okawa K, Kakizaki T, Honma T, Itoh H, Inaba T. (2007) Arabidopsis Cor15am is a chloroplast stromal protein that has cryoprotective activity and forms oligomers. Plant Physiol 144: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Okawa K, Kakizaki T, Inaba T. (2008) Evaluation of the protective activities of a late embryogenesis abundant (LEA) related protein, Cor15am, during various stresses in vitro. Biosci Biotechnol Biochem 72: 1642–1645 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR. (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Oliver AE, Hincha DK, Crowe LM, Crowe JH. (1998) Interactions of arbutin with dry and hydrated bilayers. Biochim Biophys Acta 1370: 87–97 [DOI] [PubMed] [Google Scholar]
- Popova AV, Hundertmark M, Seckler R, Hincha DK. (2011) Structural transitions in the intrinsically disordered plant dehydration stress protein LEA7 upon drying are modulated by the presence of membranes. Biochim Biophys Acta 1808: 1879–1887 [DOI] [PubMed] [Google Scholar]
- Pouchkina-Stantcheva NN, McGee BM, Boschetti C, Tolleter D, Chakrabortee S, Popova AV, Meersman F, Macherel D, Hincha DK, Tunnacliffe A. (2007) Functional divergence of former alleles in an ancient asexual invertebrate. Science 318: 268–271 [DOI] [PubMed] [Google Scholar]
- Rohde P, Hincha DK, Heyer AG. (2004) Heterosis in the freezing tolerance of crosses between two Arabidopsis thaliana accessions (Columbia-0 and C24) that show differences in non-acclimated and acclimated freezing tolerance. Plant J 38: 790–799 [DOI] [PubMed] [Google Scholar]
- Sakurai T, Satou M, Akiyama K, Iida K, Seki M, Kuromori T, Ito T, Konagaya A, Toyoda T, Shinozaki K. (2005) RARGE: a large-scale database of RIKEN Arabidopsis resources ranging from transcriptome to phenome. Nucleic Acids Res 33: D647–D650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W, Weissmann C. (1973) A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem 56: 502–514 [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Schneider T, Keller F. (2009) Raffinose in chloroplasts is synthesized in the cytosol and transported across the chloroplast envelope. Plant Cell Physiol 50: 2174–2182 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al. (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Sreerama N, Venyaminov SY, Woody RW. (2000) Estimation of protein secondary structure from circular dichroism spectra: inclusion of denatured proteins with native proteins in the analysis. Anal Biochem 287: 243–251 [DOI] [PubMed] [Google Scholar]
- Steponkus PL. (1984) Role of the plasma membrane in freezing injury and cold acclimation. Annu Rev Plant Physiol 35: 543–584 [Google Scholar]
- Steponkus PL, Uemura M, Joseph RA, Gilmour SJ, Thomashow MF. (1998) Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 14570–14575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandberg E, Tiltak D, Ehni S, Wadhwani P, Ulrich AS. (2012) Lipid shape is a key factor for membrane interactions of amphipathic helical peptides. Biochim Biophys Acta 1818: 1764–1776 [DOI] [PubMed] [Google Scholar]
- Struck DK, Hoekstra D, Pagano RE. (1981) Use of resonance energy transfer to monitor membrane fusion. Biochemistry 20: 4093–4099 [DOI] [PubMed] [Google Scholar]
- Sulpice R, Tschoep H, von Korff M, Büssis D, Usadel B, Höhne M, Witucka-Wall H, Altmann T, Stitt M, Gibon Y. (2007) Description and applications of a rapid and sensitive non-radioactive microplate-based assay for maximum and initial activity of D-ribulose-1,5-bisphosphate carboxylase/oxygenase. Plant Cell Environ 30: 1163–1175 [DOI] [PubMed] [Google Scholar]
- Susi H, Byler DM. (1986) Resolution-enhanced Fourier transform infrared spectroscopy of enzymes. Methods Enzymol 130: 290–311 [DOI] [PubMed] [Google Scholar]
- Szasz CS, Alexa A, Toth K, Rakacs M, Langowski J, Tompa P. (2011) Protein disorder prevails under crowded conditions. Biochemistry 50: 5834–5844 [DOI] [PubMed] [Google Scholar]
- Thalhammer A, Hundertmark M, Popova AV, Seckler R, Hincha DK. (2010) Interaction of two intrinsically disordered plant stress proteins (COR15A and COR15B) with lipid membranes in the dry state. Biochim Biophys Acta 1798: 1812–1820 [DOI] [PubMed] [Google Scholar]
- Thomashow MF. (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Thomashow MF. (2010) Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol 154: 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe A, Hincha DK, Leprince O, Macherel D. (2010) LEA proteins: versatility of form and function. In Lubzens E Cerda J Clark M, eds, Sleeping Beauties - Dormancy and Resistance in Harsh Environments, Berlin, Springer, pp 91–108 [Google Scholar]
- Uemura M, Gilmour SJ, Thomashow MF, Steponkus PL. (1996) Effects of COR6.6 and COR15am polypeptides encoded by COR (cold-regulated) genes of Arabidopsis thaliana on the freeze-induced fusion and leakage of liposomes. Plant Physiol 111: 313–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura M, Steponkus PL. (1997) Effect of cold acclimation on the lipid composition of the inner and outer membrane of the chloroplast envelope isolated from rye leaves. Plant Physiol 114: 1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamparys L, Gautier R, Vanni S, Bennett WF, Tieleman DP, Antonny B, Etchebest C, Fuchs PFJ. (2013) Conical lipids in flat bilayers induce packing defects similar to that induced by positive curvature. Biophys J 104: 585–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb MS, Green BR. (1991) Biochemical and biophysical properties of thylakoid acyl lipids. Biochim Biophys Acta 1060: 133–158 [Google Scholar]
- Welker S, Rudolph B, Frenzel E, Hagn F, Liebisch G, Schmitz G, Scheuring J, Kerth A, Blume A, Weinkauf S, et al. (2010) Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function. Mol Cell 39: 507–520 [DOI] [PubMed] [Google Scholar]
- Wilhelm KS, Thomashow MF. (1993) Arabidopsis thaliana cor15b, an apparent homologue of cor15a, is strongly responsive to cold and ABA, but not drought. Plant Mol Biol 23: 1073–1077 [DOI] [PubMed] [Google Scholar]
- Yang SD, Seo PJ, Yoon HK, Park CM. (2011) The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuther E, Schulz E, Childs LH, Hincha DK. (2012) Clinal variation in the non-acclimated and cold-acclimated freezing tolerance of Arabidopsis thaliana accessions. Plant Cell Environ 35: 1860–1878 [DOI] [PubMed] [Google Scholar]