Abstract
The use of supervised classification to extract markers from primary flow cytometry data is an emerging field that has made significant progress, spurred by the growing complexity of multidimensional flow cytometry. Whether the markers are extracted without supervision or by conventional gate and region methods, the number of candidate variables identified is typically larger than the number of specimens (p < n) and many variables are highly intercorrelated. Thus, comparison across groups or treatments to determine which markers are significant is challenging. Here, we utilized a data set in which 86 variables were created by conventional manual analysis of individual listmode data files, and compared the application of five multivariate classification methods to discern subtle differences between the stem/progenitor content of 35 non-small cell lung cancer and adjacent normal lung specimens. The methods compared include elastic-net, lasso, random forest, diagonal linear discriminant analysis, and best single variable (best-1). We described a broadly applicable methodology consisting of: (1) variable transformation and standardization; (2) visualization and assessment of correlation between variables; (3) selection of significant variables and modeling; and (4) characterization of the quality and stability of the model. The analysis yielded both validating results (tumors are aneuploid and have higher light scatter properties than normal lung), as well as leads that require followup: Cytokeratin+ CD133+ progenitors are present in normal lung but reduced in lung cancer; diploid (or pseudo-diploid) CD117+CD44+ cells are more prevalent in tumor. We anticipate that the methods described here will be broadly applicable to a variety of multidimensional cytometry problems.
Keywords: multivariate analysis, p < n problem, elastic net, lasso, random forest, diagonal linear discriminant analysis, nonsmall cell lung cancer, normal lung, stem cells
Analysis of flow cytometry data is usually performed in two steps. In primary analysis, variables such as the proportion of events having given properties (e.g., percent CD3+ of CD45+) are extracted from individual Flow Cytometry Standard (FCS) files. Secondary analysis involves statistical comparison of the tabulated variables between groups or treatments. Although the use of statistical classification methods to automate the extraction of markers from raw or minimally processed FCS is an emerging field (1–4), the application of the similar methodologies to secondary data analysis has not been exploited. Regardless of the method of primary analysis, the number of candidate variables detected in multidimensional cytometry data may exceed the number of specimens to be compared. Thus, simple bivariate comparison, one variable at a time, lacks statistical power to detect between group differences and is blind to correlations that may exist between variables. Clearly, a multivariate approach is needed for the secondary analysis of the large number of variables that are routinely extracted from multidimensional FCS files. When the number of specimens is limited, the conventional approach of nonoverlapping training and validation data sets is not feasible. In such cases, a naïve multivariate analysis may overfit the data, and then optimistically estimate the magnitude of the association between the variables and the group or treatment, resulting in findings that cannot be replicated. Further, small changes in the data or classification method may change those variables are identified as significant when intercorrelation is significant. There are a number of suitable contemporary computational methods for variable selection and modeling, but they have fundamentally different theoretical motivations, require tuning and can choose different variables as "significant." Analysts who have success with one method on one data set tend to apply that method to subsequent data sets. Thus, the results of a particular analysis may become reified without a sense of their sensitivity to different variable selection methods or random variation.
In this report, we propose an analysis workflow to address these issues. Given a previously published set of variables assessed on a limited number of specimens (5), our goal is to identify an appropriate analytical method to select variables related to a dichotomous condition (nonsmall cell lung cancer versus normal lung), to build a model of the condition as a mathematical function of the variables that can be used to classify subsequent specimens and to characterize the robustness of the analysis. By focusing on a flow cytometry data set previously analyzed using simple bivariate comparisons between tumor and adjacent lung specimens, the present analysis will address hazards particular to flow cytometry data: (1) highly correlated variables result in competing models that are difficult to compare; (2) statistical tests summarized by p-values do not necessarily produce sets of variables that reproducibly discriminate between conditions; (3) estimates of sensitivity and specificity must be adjusted when the number of specimens is insufficient to form independent training and testing sets. The proposed workflow, summarized here, and presented in detail as online Supporting Information, provides a practical approach to the secondary analysis of multidimensional flow cytometry data.
METHODS
Terminology
Because the statistical and cytometry literature often attribute different meanings to the same terms (e.g., parameter, sample), we have used the following terminology. A flow cytometry assay assesses a limited number of features (e.g., forward light scatter, 525/40 fluorescence intensity) on hundreds of thousands or millions of events (d, e.g., cells) that are typically acquired from a limited number (n) of specimens (e.g., lung tumor specimens). Feature expression can either be treated as continuous (e.g., fluorescence intensity of an event), or dichotomous (present or absent on a given event). These features are then combined through a gating process to define a number of markers (e.g., CD90+/CD44+). Variables (p) are derived from markers as either: (1) ratios of the numbers of events having or not having these markers as determined by a gating process (e.g., proportion of CD90+/CD44+ cells among cytokeratin+ cells); (2) absolute number (events per volume) of events having a given marker; or (3) the numerical value of a feature in a population of events defined by a marker (e.g., the mean fluorescence intensity of CD90-PE among CD90+/CD44+/cytokeratin+ events).
Acquisition of Data by Flow Cytometry
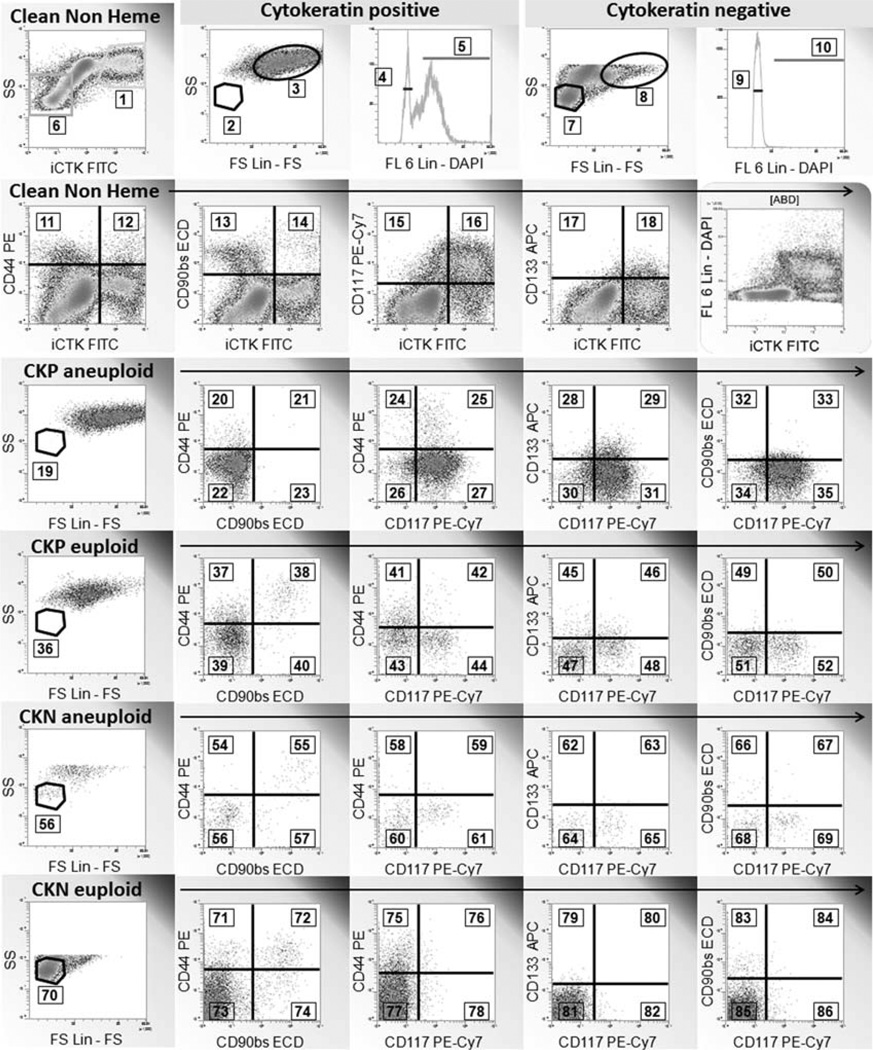
Eight-color, 11-feature data were collected for 16 paired nonsmall cell lung tumors/normal lung specimens, plus three additional tumor specimens. All specimens were collected under IRB approval (UPCI 99-053). Specimens, specimen processing, data acquisition, and variable extraction using conventional (gate and region) analysis have been reported in detail, including a MIFlowCyt checklist (5). Following artifact removal (6,7), the analysis was focused on the comparison of stem/progenitor marker expression (CD44, CD90, CD117, CD133) on tumor cells and cells from adjacent normal lung. After limiting the analysis to CD45−/CD14−/CD33−/glycophorin A-cells, we subseted based on cytokeratin expression (epithelial versus nonepithelial or pre-epithelial) and ploidy (2N vs.>2N), yielding four classes of cells on which to assess variables (proportion of cells positive for stem/progenitor markers, proportion with low versus high light scatter, Fig. 1). The reason for using DNA content as a grouping variable is that, in tumor specimens, we could be certain that the majority of aneuploid cells were of bona fide tumor origin (as opposed to normal stromal or epithelial cells). The analytical regions in Figure 1, numbered 1–86, are linked to variable names and descriptions in Table 1. The data for all listmode files were analyzed using VenturiOne software (Applied Cytometry Systems, Dinnington, UK) in one session to standardize the placement of analytical regions. Region data (event counts) were exported to a comma-separated variable (.csv) file for statistical analysis. The data are available to interested investigators by request to the corresponding author.
Figure 1.
Regions and gates used in mutivariate analysis. This example shows a lung adenocarcinoma. Logical gates applied to each histogram are shown above the histogram frame. The “Clean Non Heme” gate was created on CD45-/CD14-/CD33-/glycophorin A-singlet events with DNA content ≥ 2N (not shown). The region numbers indicate the markers used for multivariate analysis and are keyed to Table 1. CKP + cytokeratin+, CKN = cytokeratin negative Euploid = gated on region 4 or 9. Aneuploid = gated on region 5 or 10. The euploid and low light scatter regions were matched to tissue infiltrating lymphocytes (not shown).
Table 1.
Variable names and descriptions keyed to the analytical regions in Figure 1
| REGION | VARIABLE NAME | DESCRIPTION (MARKER) | DENOMINATOR (MARKER) |
|---|---|---|---|
| 1 | CKP | Cytokeratin+ | Singlet ≥ 2N DNA events |
| 2 | CKP_SM | Cytokeratin+ lymphoid scatter | Cytokeratin+ |
| 3 | CKP_LG | Cytokeratin +, > lymphoid scatter | Cytokeratin+ |
| 4 | CKP_2N | Cytokeratin+ euploid | Cytokeratin+ |
| 5 | CKP_GT2N | Cytokeratin+ aneuploid | Cytokeratin+ |
| 6 | CKN | Cytokeratin negative | Singlet ≥ 2N DNA events |
| 7 | CKN_SM | Cytokeratin negative lymphoid scatter | Cytokeratin negative |
| 8 | CKN_LG | Cytokeratin negative > lymphoid scatter | Cytokeratin negative |
| 9 | CKN_2N | Cytokeratin negative euploid | Cytokeratin negative |
| 10 | CKN_GT2N | Cytokeratin negative aneuploid | Cytokeratin negative |
| 11 | CKN_44P | Cytokeratin negative CD44+ | Cytokeratin negative |
| 12 | CKP_44P | Cytokeratin+ CD44+ | Cytokeratin+ |
| 13 | CKN_90P | Cytokeratin negative CD90+ | Cytokeratin negative |
| 14 | CKP_90P | Cytokeratin+ CD90+ | Cytokeratin+ |
| 15 | CKN_117P | Cytokeratin negative CD117+ | Cytokeratin negative |
| 16 | CKP_117P | Cytokeratin+ CD117+ | Cytokeratin+ |
| 17 | CKN_133P | Cytokeratin negative CD133+ | Cytokeratin negative |
| 18 | CKP_133P | Cytokeratin+ CD133+ | Cytokeratin+ |
| 19 | CPG2_SMALL | Cytokeratin+ aneuploid lymphoid scatter | Cytokeratin+ aneuploid |
| 20 | CPG290N44P | Cytokeratin+ aneuploid CD90 negative CD44+ | Cytokeratin+ aneuploid |
| 21 | CPG290P44P | Cytokeratin+ aneuploid CD90+ CD44+ | Cytokeratin+ aneuploid |
| 22 | CPG290N44N | Cytokeratin+ aneuploid CD90 negative CD44 negative | Cytokeratin+ aneuploid |
| 23 | CPG290P44N | Cytokeratin+ aneuploid CD90+ CD44 negative | Cytokeratin+ aneuploid |
| 24 | CPG2117N44P | Cytokeratin+ aneuploid CD117 negative CD44+ | Cytokeratin+ aneuploid |
| 25 | CPG2117P44P | Cytokeratin+ aneuploid CD117+ CD44+ | Cytokeratin+ aneuploid |
| 26 | CPG2117N44N | Cytokeratin+ aneuploid CD117 negative CD44 negative | Cytokeratin+ aneuploid |
| 27 | CPG2117P44N | Cytokeratin+ aneuploid CD117+ CD44 negative | Cytokeratin+ aneuploid |
| 28 | CPG2117N133P | Cytokeratin+ aneuploid CD117 negative CD133+ | Cytokeratin+ aneuploid |
| 29 | CPG2117P133P | Cytokeratin+ aneuploid CD117+ CD133+ | Cytokeratin+ aneuploid |
| 30 | CPG2117N133N | Cytokeratin+ aneuploid CD117 negative CD133 negative | Cytokeratin+ aneuploid |
| 31 | CPG2117P133N | Cytokeratin+ aneuploid CD117+ CD133 negative | Cytokeratin+ aneuploid |
| 32 | CPG2117N90P | Cytokeratin+ aneuploid CD117 negative CD90+ | Cytokeratin+ aneuploid |
| 33 | CPG2117P90P | Cytokeratin+ aneuploid CD117+ CD90+ | Cytokeratin+ aneuploid |
| 34 | CPG2117N90N | Cytokeratin+ aneuploid CD117 negative CD90 negative | Cytokeratin+ aneuploid |
| 35 | CPG2117P90N | Cytokeratin+ aneuploid CD117+ CD90 negative | Cytokeratin+ aneuploid |
| 36 | CP2_SMALL | Cytokeratin+ euploid lymphoid scatter | Cytokeratin+ euploid |
| 37 | CP90N44P | Cytokeratin+ euploid CD90 negative CD44+ | Cytokeratin+ euploid |
| 38 | CP90P44P | Cytokeratin+ euploid CD90+ CD44+ | Cytokeratin+ euploid |
| 39 | CP90N44N | Cytokeratin+ euploid CD90 negative CD44 negative | Cytokeratin+ euploid |
| 40 | CP90P44N | Cytokeratin+ euploid CD90+ CD44 negative | Cytokeratin+ euploid |
| 41 | CP2117N44P | Cytokeratin+ euploid CD117 negative CD44+ | Cytokeratin+ euploid |
| 42 | CP2117P44P | Cytokeratin+ euploid CD117+ CD44+ | Cytokeratin+ euploid |
| 43 | CP2117N44N | Cytokeratin+ euploid CD117 negative CD44 negative | Cytokeratin+ euploid |
| 44 | CP2117P44N | Cytokeratin+ euploid CD117+ CD44 negative | Cytokeratin+ euploid |
| 45 | CP2117N133P | Cytokeratin+ euploid CD117 negative CD133+ | Cytokeratin+ euploid |
| 46 | CP2117P133P | Cytokeratin+ euploid CD117+ CD133+ | Cytokeratin+ euploid |
| 47 | CP2117N133N | Cytokeratin+ euploid CD117 negative CD133 negative | Cytokeratin+ euploid |
| 48 | CP2117P133N | Cytokeratin+ euploid CD117+ CD133 negative | Cytokeratin+ euploid |
| 49 | CP2117N90P | Cytokeratin+ euploid CD117 negative CD90+ | Cytokeratin+ euploid |
| 50 | CP2117P90P | Cytokeratin+ euploid CD117+ CD90+ | Cytokeratin+ euploid |
| 51 | CP2117N90N | Cytokeratin+ euploid CD117 negative CD90 negative | Cytokeratin+ euploid |
| 52 | CP2117P90N | Cytokeratin+ euploid CD117+ CD90 negative | Cytokeratin+ euploid |
| 53 | CNG2_SMALL | Cytokeratin negative aneuploid lymphoid scatter | Cytokeratin negative aneuploid |
| 54 | CNG290N44P | Cytokeratin negative aneuploid CD90 negative CD44+ | Cytokeratin negative aneuploid |
| 55 | CNG290P44P | Cytokeratin negative aneuploid CD90+ CD44+ | Cytokeratin negative aneuploid |
| 56 | CNG290N44N | Cytokeratin negative aneuploid CD90 negative CD44 negative | Cytokeratin negative aneuploid |
| 57 | CNG290P44N | Cytokeratin negative aneuploid CD90+ CD44 negative | Cytokeratin negative aneuploid |
| 58 | CNG2117N44P | Cytokeratin negative aneuploid CD117 negative CD44+ | Cytokeratin negative aneuploid |
| 59 | CNG2117P44P | Cytokeratin negative aneuploid CD117+ CD44+ | Cytokeratin negative aneuploid |
| 60 | CNG2117N44N | Cytokeratin negative aneuploid CD117 negative CD44 negative | Cytokeratin negative aneuploid |
| 61 | CNG2117P44N | Cytokeratin negative aneuploid CD117+ CD44 negative | Cytokeratin negative aneuploid |
| 62 | CNG2117N133P | Cytokeratin negative aneuploid CD117 negative CD133+ | Cytokeratin negative aneuploid |
| 63 | CNG2117P133P | Cytokeratin negative aneuploid CD117+ CD133+ | Cytokeratin negative aneuploid |
| 64 | CNG2117N133N | Cytokeratin negative aneuploid CD117 negative CD133 negative | Cytokeratin negative aneuploid |
| 65 | CNG2117P133N | Cytokeratin negative aneuploid CD117+ CD133 negative | Cytokeratin negative aneuploid |
| 66 | CNG2117N90P | Cytokeratin negative aneuploid CD117 negative CD90+ | Cytokeratin negative aneuploid |
| 67 | CNG2117P90P | Cytokeratin negative aneuploid CD117+ CD90+ | Cytokeratin negative aneuploid |
| 68 | CNG2117N90N | Cytokeratin negative aneuploid CD117 negative CD90 negative | Cytokeratin negative aneuploid |
| 69 | CNG2117P90N | Cytokeratin negative aneuploid CD117+ CD90 negative | Cytokeratin negative aneuploid |
| 70 | CN2_SMALL | Cytokeratin negative euploid lymphoid scatter | Cytokeratin negative euploid |
| 71 | CN90N44P | Cytokeratin negative euploid CD90 negative CD44+ | Cytokeratin negative euploid |
| 72 | CN90P44P | Cytokeratin negative euploid CD90+ CD44+ | Cytokeratin negative euploid |
| 73 | CN90N44N | Cytokeratin negative euploid CD90 negative CD44 negative | Cytokeratin negative euploid |
| 74 | CN90P44N | Cytokeratin negative euploid CD90+ CD44 negative | Cytokeratin negative euploid |
| 75 | CN2117N44P | Cytokeratin negative euploid CD117 negative CD44+ | Cytokeratin negative euploid |
| 76 | CN2117P44P | Cytokeratin negative euploid CD117+ CD44+ | Cytokeratin negative euploid |
| 77 | CN2117N44N | Cytokeratin negative euploid CD117 negative CD44 negative | Cytokeratin negative euploid |
| 78 | CN2117P44N | Cytokeratin negative euploid CD117+ CD44 negative | Cytokeratin negative euploid |
| 79 | CN2117N133P | Cytokeratin negative euploid CD117 negative CD133+ | Cytokeratin negative euploid |
| 80 | CN2117P133P | Cytokeratin negative euploid CD117+ CD133+ | Cytokeratin negative euploid |
| 81 | CN2117N133N | Cytokeratin negative euploid CD117 negative CD133 negative | Cytokeratin negative euploid |
| 82 | CN2117P133N | Cytokeratin negative euploid CD117+ CD133 negative | Cytokeratin negative euploid |
| 83 | CN2117N90P | Cytokeratin negative euploid CD117 negative CD90+ | Cytokeratin negative euploid |
| 84 | CN2117P90P | Cytokeratin negative euploid CD117+ CD90+ | Cytokeratin negative euploid |
| 85 | CN2117N90N | Cytokeratin negative euploid CD117 negative CD90 negative | Cytokeratin negative euploid |
| 86 | CN2117P90N | Cytokeratin negative euploid CD117+ CD90 negative | Cytokeratin negative euploid |
Data Analysis Workflow
We analyzed the data according to the following steps: (1) transformation and standardization; (2) visualization and assessment of correlation between variables; (3) selection of significant markers and modeling; (4) characterization of the quality and stability of the model. All of the analytic steps applied to the data in the .csv file were performed using the open-source statistical package R (8). A diagram of the workflow (Fig. S1) and the associated R code is included in the online Supporting Information.
Transformation and Standardization
Because the variables analyzed in this data set were proportions (e.g., proportion of cytokeratin+ viable cells), it was necessary to address their statistical properties. First, when a population of cells is divided into a set and its complement, the two proportions are perfectly correlated (r = −1). Similarly, large fractions are often highly correlated. For example, cytokeratin+ and cytokeratin negative, expressed as a proportion of nucleated cells, are highly inversely correlated. Second, mid-range proportions (e.g., aneuploid tumor cells as a proportion of cytokeratin positive cells) tend to be normally distributed, but proportions that are close to 0 or 1 have asymmetric distributions over the specimens. Third, proportion estimates on small denominators (<50 cells) are unreliable. Finally, zero proportions do not necessarily indicate an impossible event, but, possibly, a rare, yet informative event, especially when denominators are small. We addressed these issues by unzeroing, stabilization, and standardization.
Unzeroing
Some numerical transformations (e.g., logarithmic) cannot operate on zeros or ones, and some statistical classifiers perform worse on highly skewed data. We replaced zeros by a random value between zero and one-tenth the smallest nonzero value and ones by a random value between 1 and 1 − (1 − m)/10, where m is the largest value less than 1. Additional details are presented in the Supporting Information.
Stabilization
Some statistical classification methods assume that the variables have Gaussian distributions (e.g., Fisher’s linear discriminant analysis) or do not accommodate highly skewed data well. We used the logit transform in our analysis because it is symmetric and its range is all positive and all negative numbers. The logit and inverse logit transformations are shown in Supporting Information Figure S2, where merits of various transformations are discussed.
Standardization
For each variable, we subtracted the mean and divided by the standard deviation, so that, over specimens from all conditions, mean equals 0 and the observed standard deviation equals 1. This was required because some computational algorithms are unstable when operating simultaneously on values with multilog differences in scale.
Visualization of Data and Assessment of Correlation Between Variables
The transformed, standardized data were scanned for artifacts, influence points, and obvious classifiers before analysis. Outliers, in the usual sense of very large (positive or negative) values, are not present in transformed, standardized proportions. However, influence points (single values that induce correlations), such as clusters of 0 and 1 counts, were evident in the data and required unzeroing as described above.
Initial Assessment of Variables for Classification
We performed comparisons of the individual variables versus the condition by t-tests and/or rank sum tests to determine whether modeling was feasible (Table 2). Because a number of significant differences were observed, we proceeded with the analysis. Although a small p-value does not necessarily indicate a good classifier, if none of the p-values were significant, we would have terminated the analysis.
Table 2.
P-values of unpaired t- and rank-sum tests on tumor versus normal lung specimens. The 32 variables with the smallest P-values (t-test) are shown
| P-VALUES | ||
|---|---|---|
| VARIABLE | T-TEST | RANK SUM TEST |
| CKP_GT2N | 0.00044 | 0.00099 |
| CPG2117N133P | 0.00055 | 0.00175 |
| CKP_2N | 0.00056 | 0.0014 |
| CNG2117P44N | 0.00576 | 0.02728 |
| CKP_SM | 0.00637 | 0.00934 |
| CP2117P44P | 0.01037 | 0.00417 |
| CP2117N90N | 0.01227 | 0.0113 |
| CP2117P90N | 0.01378 | 0.0254 |
| CP90P44N | 0.01406 | 0.04156 |
| CNG2117P90N | 0.02459 | 0.13389 |
| CNG2117P133N | 0.0254 | 0.32797 |
| CN2117P90P | 0.02604 | 0.07043 |
| CP2117P90P | 0.02838 | 0.02851 |
| CN2117P90N | 0.03012 | 0.08804 |
| CN2117P44N | 0.03157 | 0.02228 |
| CP2117P44N | 0.04039 | 0.10825 |
| CP90P44P | 0.04301 | 0.08205 |
| CP2117P133N | 0.04342 | 0.04512 |
| CKP_117P | 0.04358 | 0.10827 |
| CP2117N90P | 0.04361 | 0.07645 |
| CKN_LG | 0.04424 | 0.02766 |
| CKP_LG | 0.0519 | 0.01636 |
| CP2_SMALL | 0.05602 | 0.0417 |
| CKN_GT2N | 0.05917 | 0.08813 |
| CN2117P133N | 0.06373 | 0.14972 |
| CPG2117P133N | 0.06616 | 0.16938 |
| CNG2117P90P | 0.08081 | 0.35498 |
| CP2117P133P | 0.0812 | 0.08487 |
| CKN_117P | 0.08168 | 0.26727 |
| CN2117N133P | 0.09289 | 0.07113 |
| CPG2117N90N | 0.09721 | 0.16938 |
| CKN_2N | 0.14364 | 0.3625 |
Selection of Significant Variables and Modeling of Condition
We tested a number of methods in a resampling framework to determine if: (1) some methods classify the specimens better than others; (2) there is agreement between the methods on which variables are important; (3) there are any specimens that have particular influence over the variables selected as important and/or coefficient estimates; and (4) the estimated coefficients are sensitive to small perturbations in the data. We used two resampling methods to address these issues: cross-validation and bootstrapping.
Cross-Validation
The data were partitioned into 10 disjoint subsets. At each of 10 iterations, a different subset was set aside and modeling parameters were estimated from the other 9 subsets, pooled together. We estimated the sensitivity and specificity, and set tuning parameters using the set-aside test set.
Bootstrapping
In the nonparametric bootstrap (9), a sample of specimens, the same size as the original sample specimen data set, was constructed by random draws from the original sample with replacement. That is, once an observation was selected for the bootstrap sample, it was replaced in the pool of eligible observations where it could be sampled again. All of the methods were trained on the bootstrap sample, and the specimens that were excluded from the sample (equal, on average, to (1 − 1/n)n ≈ 0.37 of the sample size, n) were treated as the test set. These excluded samples are referred to as out-of-bag (OOB). The bootstrapping process was repeated 500 times, each bootstrap iteration testing the full set of methods on different training and test sets, both of which were drawn from the full sample. Once the bootstrap iterations were completed, the average and the variability of the cross-validated sensitivity, specificity, accuracy, selected variables, influential specimens, and estimated coefficients of the different methods were estimated by the empirical distribution of bootstrap estimates. Further discussion and software code appears in the online Supporting Information.
Variable Importance
Within each bootstrap iteration, variables were chosen as either significant contributors to the discriminator, or nonsignificant. The proportion of iterations in which a given variable was considered significant is a measure of the robustness of its power as a discriminator. The correlation of selection between variables (where each variable is coded 1 if it is selected, and 0 otherwise) was interpreted as a descriptor of structure; variables whose selection indices are negatively correlated are possibly associated with a pathway.
Influential Observations
We also calculated the proportion of bootstrap iterations in which each test specimen was correctly classified as tumor or normal lung. Specimens that are consistently misclassified by one method, but not another, may indicate that a nonlinear partition may be required, or that the specimen is dissimilar from other specimens with the same condition. A specimen that is misclassified in close to ½ of the bootstrap iterations probably lies close to the partition boundary.
Candidate Classifiers
We focused on three methods that reflect different philosophies of model building: diagonal linear discrimination (DLDA), random forests, and elastic net. DLDA is frequently used in very high-dimensional analyses; it is structurally very simple, and reduces computationally complexity by ignoring interactions between the variables. Random forest is a nonparametric machine learning method that itself uses a resampling method to form a partition of the variable space that may be highly nonlinear. Elastic net is a “regularized” version of logistic regression that is designed for stability in the presence of highly correlated variables and has a built-in variable selection scheme. We also used the lasso (which is a special case of elastic net), and a naïve classifier, best-single marker. All of these methods were embedded in the bootstrap loop; all methods were applied to each bootstrap sample, and then the results were compared. Details and code for all these methods are in the online Supporting Information.
RESULTS
Visualization and Assessment of Correlation Within Subjects and Between Variables
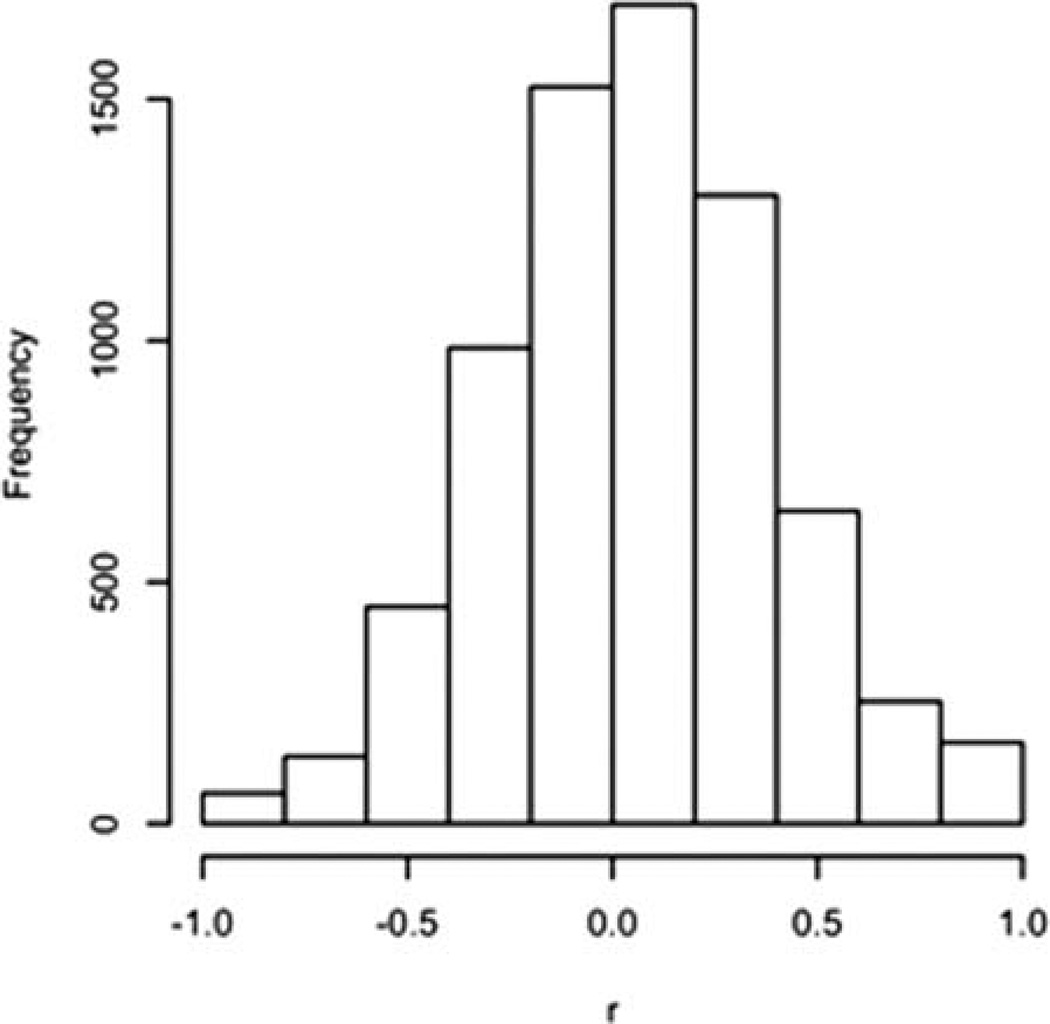
Before logit transformation, centering, and scaling, we jittered 0 values to between 1/10 of the smallest non-zero value and 0. Values of 1 were treated analogously. The intrasubject correlation (between tumor and normal lung) on the 86 variables was measured on the 16 pairs of specimens (three tumor specimens did not have paired normal tissue). The median of the 86 correlation coefficients was 0.22, and ranged from −0.63 to 0.87. The low mean of the correlations indicated that an analysis based on differences within subject was unlikely to be useful. We decided not to analyze the data as paired (tumor and normal specimens), but to treat the specimens as independent. Scatterplots of variables (tumor versus normal) with large coefficients indicated that many of the largest correlations (either positive or negative) were artifactual. The distribution of the 86 × (86 − 1)/2 = 3655 pairwise correlation coefficients (tumor vs. normal lung) is illustrated in Figure 2. A heat map and dendrogram of the normal lung and tumor specimens in the lung data set did not immediately indicate a dominant cluster of discriminating variables (Fig. S3). The closest two single variables are CKN_44P (cytokeratin negative/CD44+) and CN2117N44P (cytokeratin negative, euploid, CD117 negative/CD44+), which are highly correlated (r = 0.96) because the latter is a subset of the former.
Figure 2.
Distribution of between-variable correlations in the example data set.
Initial Assessment of Variables for Classification
Table 2 displays the p-values of Student’s t and Wilcoxon rank sum tests comparing the distributions between normal lung and tumor specimens for all of the variables, from most significant (Student’s t) to least significant. It is seen that there are some significant differences, so that further classification analysis is warranted, but that the distributions of most variables in normal lung and tumor specimens are similar, so the final discriminators are not expected to include many variables.
Sensitivity and Specificity
The sensitivity, specificity, and accuracy of the discriminators, estimated over the 500 bootstrap samples, are presented in Table 3. The bootstrapped estimates are calculated by classifying the OOB samples (averaged over all bootstrap iterations), whereas the resubstitution estimates are obtained by classifying the training samples (also averaged over all bootstrap iterations). DLDA, random forests, elastic net, and the lasso produce essentially equivalent accuracy; random forest is different from the other three in that its estimated specificity is larger than its sensitivity. The differences between sensitivity and specificity in these methods are more likely due to peculiarities of the example set than the methods themselves. Best-1 accuracy is lower than the other four methods, which is not surprising, given that it is restricted to a single variable. The bootstrapped estimates of sensitivity, specificity, and accuracy are much less optimistic than the resubstitution estimates. Resubstitution estimates of the operating characteristics of Best-1 were not as optimistic, but the bootstrapped estimates were not as good as the other methods.
Table 3.
Sensitivity, specificity, and accuracy of the five methods
| E-NET | RANDOM FOREST | DLDA | LASSO | BEST-1 | ||
|---|---|---|---|---|---|---|
| Bootstrapped Estimates | Sensitivity | 0.696 | 0.623 | 0.691 | 0.694 | 0.659 |
| Specificity | 0.659 | 0.692 | 0.617 | 0.658 | 0.548 | |
| Accuracy | 0.679 | 0.654 | 0.658 | 0.678 | 0.609 | |
| Resubstitution Estimates | Sensitivity | 0.895 | 1 | 0.895 | 0.895 | 0.684 |
| Specificity | 0.938 | 1 | 0.875 | 0.938 | 0.750 | |
| Accuracy | 0.914 | 1 | 0.886 | 0.914 | 0.714 |
Sensitivity is defined as true positives (TP)/(TP + false negatives (FN)), Specificity = true NEGATIVES (TN)/(TN + False positives (FP)), Accuracy = (TP+TN)/(TP+TN+FP+FN).
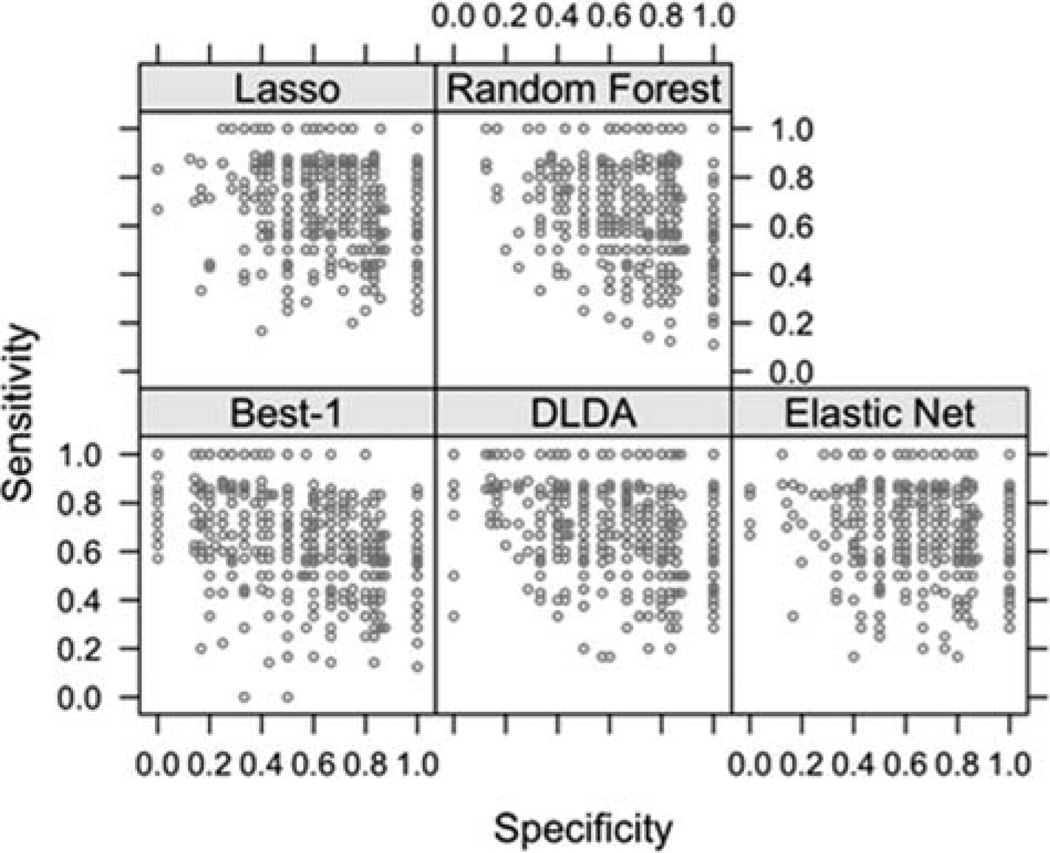
Figure 3 is a scatterplot of the estimates of sensitivity and specificity over all 500 bootstrap samples for the five methods. This plot demonstrates the variability in sensitivity and specificity that could occur when sampling from the population from which the study sample was drawn. If there was no evaluation of the variability of estimation, then the observed sensitivity and specificity could be any one of the graphed values, and it would not be possible to determine how typical or atypical those estimates might be. It is seen that the observed sensitivities and specificities (Table 3) are roughly at the modes in Figure 3.
Figure 3.
Specificity and sensitivity of 500 bootstrap samples.
Variables Selected
Table 4 presents the importance index of each variable on a scale of 0–500 (the number of bootstrap iterations). The values for elastic net, random forest, DLDA, and lasso are much larger than those for Best-1 discrimination because the total number of variables chosen over 500 bootstrap iterations of Best-1 is fixed at a total of 500. The table is ordered by the average importance index over the five methods, where the top variables are, on average, most important. There is a fairly large drop in importance after the fifth most important variable, CKN_SM (cytokeratin negative, lymphoid scatter). Except for CKN_SM and CPG2117N133P (cytokeratin+ aneuploid CD117 negative CD133+), the five most important variables do not cluster together (Fig. S3), suggesting that they represent different processes. Figure 4 shows the first two principal components of the top five variables. The variables commonly chosen are mostly coherent on the average, but different methods can be fairly discordant on the same bootstrap sample. Table 5 displays the concordance of the five methods in choosing CKP_GT2N (cytokeratin+ aneuploid), the variable with the highest mean importance index, across the bootstrap samples. Even though elastic net and lasso are similar in concept, they are discordant in 86/500 = 17.2% of the bootstrap samples. Best-1, which is not smoothed or regularized in any way, is usually discordant with the other methods.
Table 4.
Importance of variables on a scale of 0–500
| E-NET | RANDOM FOREST |
DLDA | LASSO | BEST-1 | |
|---|---|---|---|---|---|
| CKP_GT2N | 408 | 321 | 391 | 342 | 84 |
| CPG2117N133P | 386 | 293 | 378 | 350 | 130 |
| CP2117P44P | 357 | 246 | 229 | 318 | 33 |
| CKP_2N | 265 | 289 | 366 | 180 | 67 |
| CKP_SM | 335 | 254 | 270 | 246 | 42 |
| CNG2117P44N | 116 | 158 | 328 | 76 | 35 |
| CP2117P90P | 264 | 60 | 125 | 231 | 20 |
| CP2_SMALL | 193 | 201 | 139 | 114 | 1 |
| CP90P44N | 192 | 94 | 188 | 156 | 15 |
| CN2117P44N | 197 | 162 | 121 | 142 | 1 |
| CKN_GT2N | 213 | 127 | 93 | 164 | 1 |
| CP2117N90N | 120 | 167 | 203 | 93 | 9 |
| CN2117P90P | 129 | 131 | 149 | 106 | 9 |
| CP2117P90N | 105 | 118 | 195 | 84 | 9 |
| CKN_133P | 238 | 33 | 30 | 207 | 0 |
| CN2117P90N | 103 | 121 | 121 | 71 | 2 |
| CNG2117P90N | 89 | 57 | 174 | 76 | 9 |
| CKN_LG | 83 | 135 | 105 | 59 | 3 |
| CKP_LG | 49 | 144 | 145 | 35 | 5 |
| CP2117N90P | 86 | 117 | 75 | 53 | 1 |
| CP90P44P | 83 | 75 | 84 | 66 | 1 |
| CPG2117P90P | 146 | 13 | 26 | 119 | 0 |
| CPG2_SMALL | 57 | 167 | 46 | 30 | 0 |
| CNG2117N133P | 107 | 83 | 10 | 78 | 1 |
| CNG2117P90P | 24 | 107 | 90 | 21 | 7 |
| CN2117N133P | 46 | 89 | 67 | 33 | 1 |
| CNG2_SMALL | 83 | 63 | 25 | 62 | 0 |
| CNG2117P133N | 32 | 23 | 151 | 20 | 1 |
| CPG2117P133P | 99 | 24 | 13 | 80 | 0 |
| CKN_SM | 88 | 32 | 31 | 63 | 0 |
| CN2117P133N | 44 | 84 | 45 | 36 | 0 |
| CNG2117P133P | 106 | 6 | 14 | 80 | 0 |
| CP2117N44N | 76 | 32 | 40 | 55 | 2 |
| CN90P44N | 95 | 22 | 12 | 69 | 0 |
| CN2117P44P | 70 | 36 | 30 | 43 | 1 |
| CP90N44N | 88 | 8 | 17 | 55 | 0 |
| CN2_SMALL | 45 | 50 | 36 | 33 | 1 |
| CP2117P44N | 18 | 50 | 86 | 7 | 0 |
| CPG290P44N | 63 | 11 | 37 | 47 | 0 |
| CN2117P133P | 54 | 21 | 34 | 45 | 3 |
| CKN_117P | 52 | 21 | 33 | 47 | 0 |
| CN2117N133N | 54 | 50 | 6 | 38 | 0 |
| CKP_117P | 44 | 17 | 56 | 25 | 1 |
| CP2117P133P | 25 | 38 | 64 | 10 | 2 |
| CKN | 49 | 27 | 26 | 36 | 0 |
| CKN_2N | 47 | 27 | 40 | 22 | 0 |
The variables are sorted in order of their average importance across all five methods. Variables chosen as significant (modal number) by the different methods are shaded. Red shading indicates that the value of the variable is greater in tumor than in normal, green shading indicates the converse. The most important 32 variables are shown.
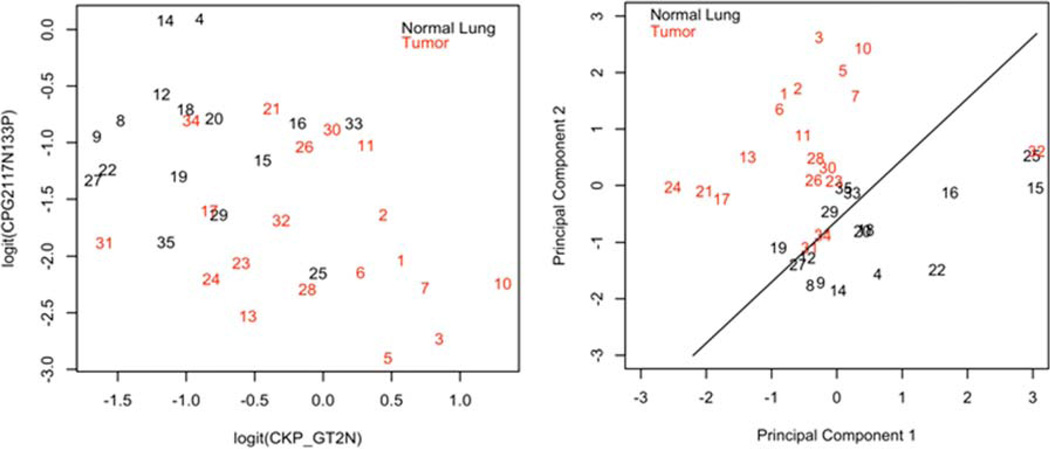
Figure 4.
Two most important variables versus principal component analysis discriminating between tumor and normal lung. The plot on the left shows ability of the two most predictive variables (cytokeratin+ aneuploid and cytokeratin+ aneuploid CD117 negative CD133+) to distinguish between tumor and normal lung. The plot on the right shows the two principal components of five most important variables (Table 4). Numbers refer to the specimen ID column in Table 6. The case number indicates the frequency with which the case was correctly classified (lower number more frequently classified correctly). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Table 5.
Concordance between methods in choosing CKP_GT2N (cytokeratin+ aneuploid cells) as a significant classifying variable over 500 bootstrap iterations
| RF | DLDA | LASSO | BEST-1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | Y | N | Y | N | Y | N | Y | ||
| EN | N | 46 | 35 | 35 | 46 | 75 | 6 | 80 | 1 |
| Y | 107 | 312 | 63 | 356 | 80 | 339 | 309 | 110 | |
| RF | N | 68 | 85 | 65 | 88 | 142 | 11 | ||
| Y | 30 | 317 | 90 | 257 | 247 | 100 | |||
| DLDA | N | 51 | 47 | 98 | 0 | ||||
| Y | 104 | 298 | 291 | 111 | |||||
| Lasso | N | 148 | 7 | ||||||
| Y | 241 | 104 | |||||||
Two by two tables are shown for each pair of methods. “Y” indicates the given model included CKP_GT2N, “N” indicates that it was excluded.
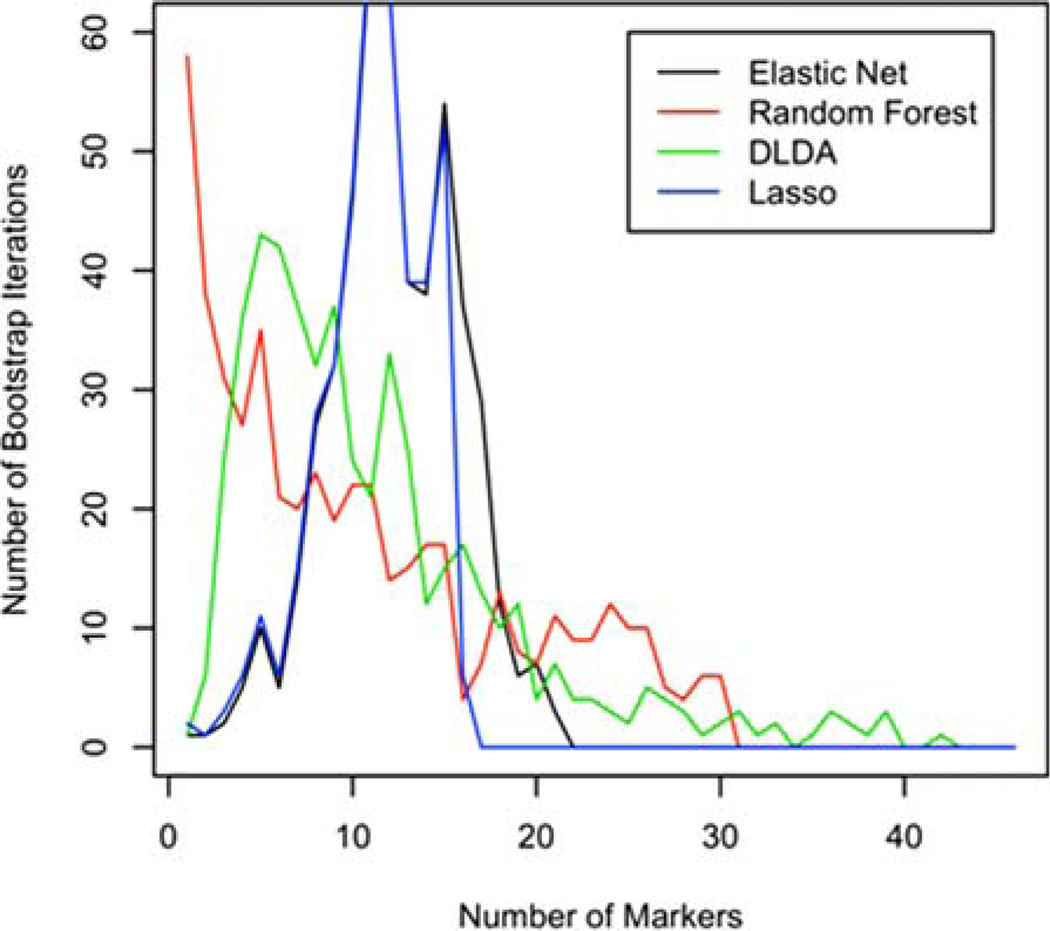
Figure 5 displays the distributions of the numbers of variables selected by the different methods across the bootstrap samples (Best-1 is not included because it always chooses exactly one variable). As expected, the numbers of variables in the elastic net models with the most variables were larger than similar numbers associated with the lasso (Fig. 5). Reducing the mixing parameter α shifts the elastic net curve to the right, capturing more variables, but not increasing the accuracy of classification (data not shown). Although DLDA’s number of variables is smaller than the first two, it has a longer tail, describing a few runs where over 20 variables were selected. Random forest tends to build more parsimonious models, but also sometimes will indicate that 20 or even 30 variables are useful in classifying specimens.
Figure 5.
Numbers of variables chosen by individual method (Best-1 always chooses one variable). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Classification of Individual Specimens
Table 6 shows the frequency with which each specimen was correctly classified (tumor or normal lung) by each method over 500 bootstrap iterations. The column labeled c.v. is a measure of concordance of the methods, with low coefficients of variation indicating better concordance. All of the methods have difficulty with specimens 27 through 35.
Table 6.
Proportion of correct classifications by method for each specimen
| ID | E-NET | RANDOM FOREST | DLDA | LASSO | BEST-1 | C.V. | |
|---|---|---|---|---|---|---|---|
| LungCATSC076/TUMOR | 1 | 0.9941 | 0.9822 | 0.9941 | 1.0000 | 0.9704 | 0.0120 |
| LungCATSC094/TUMOR | 2 | 0.9830 | 0.9830 | 0.9943 | 0.9886 | 0.9830 | 0.0052 |
| CircLungCATSC028/TUMOR | 3 | 1.0000 | 0.9593 | 0.9884 | 1.0000 | 0.9651 | 0.0196 |
| LungCATSC098/NL | 4 | 0.9830 | 0.9886 | 0.9886 | 0.9830 | 0.9148 | 0.0328 |
| LungCATSC072/TUMOR | 5 | 0.9887 | 0.9605 | 0.9774 | 0.9774 | 0.9379 | 0.0205 |
| LungCATSC101/TUMOR | 6 | 0.9672 | 0.9672 | 0.9836 | 0.9672 | 0.8962 | 0.0359 |
| LungCATSC074/TUMOR | 7 | 0.9763 | 0.8817 | 0.9941 | 0.9704 | 0.7988 | 0.0893 |
| CircLungCATSC029/NL | 8 | 1.0000 | 0.9846 | 0.9026 | 1.0000 | 0.7026 | 0.1384 |
| CircLungCATSC028/NL | 9 | 0.9748 | 1.0000 | 0.8994 | 0.9686 | 0.7233 | 0.1232 |
| CircLungCATSC025/TUMOR | 10 | 0.9697 | 0.7727 | 0.9596 | 0.9747 | 0.8081 | 0.1095 |
| CircLungCATSC023/TUMOR | 11 | 0.9711 | 0.9653 | 0.9538 | 0.9595 | 0.6012 | 0.1816 |
| LungCATSC092/NL | 12 | 0.8497 | 0.9016 | 0.9378 | 0.8549 | 0.7772 | 0.0701 |
| LungCATSC087/TUMOR | 13 | 0.9055 | 0.8358 | 0.9204 | 0.9005 | 0.6915 | 0.1113 |
| LungCATSC081/NL | 14 | 0.9322 | 0.7627 | 0.6836 | 0.9322 | 0.6328 | 0.1762 |
| LungCATSC087/NL | 15 | 0.7988 | 0.9290 | 0.8462 | 0.7929 | 0.4970 | 0.2115 |
| CircLungCATSC023/NL | 16 | 0.7354 | 0.7725 | 0.7778 | 0.7460 | 0.4497 | 0.1996 |
| LungCATSC121/TUMOR | 17 | 0.7472 | 0.5506 | 0.7921 | 0.7247 | 0.6517 | 0.1364 |
| LungCATSC089/NL | 18 | 0.5529 | 0.8235 | 0.6706 | 0.5412 | 0.6765 | 0.1754 |
| LungCATSC101/NL | 19 | 0.6705 | 0.7443 | 0.5568 | 0.6818 | 0.5966 | 0.1137 |
| LungCATSC072/NL | 20 | 0.6374 | 0.8304 | 0.4678 | 0.6374 | 0.5965 | 0.2051 |
| LungCATSC098/TUMOR | 21 | 0.6213 | 0.8107 | 0.6272 | 0.5799 | 0.4793 | 0.1926 |
| CircLungCATSC027/NL | 22 | 0.5774 | 0.6845 | 0.4881 | 0.5952 | 0.6488 | 0.1255 |
| LungCATSC089/TUMOR | 23 | 0.6850 | 0.5750 | 0.5350 | 0.6650 | 0.5050 | 0.1335 |
| CircLungCATSC029/TUMOR | 24 | 0.6973 | 0.4162 | 0.4919 | 0.6919 | 0.4703 | 0.2380 |
| CircLungCATSC024/NL | 25 | 0.6923 | 0.5621 | 0.6864 | 0.6450 | 0.1479 | 0.4187 |
| LungCATSC081/TUMOR | 26 | 0.6382 | 0.2613 | 0.4422 | 0.6884 | 0.4925 | 0.3359 |
| LungCATSC121/NL | 27 | 0.5000 | 0.4202 | 0.4681 | 0.5000 | 0.4787 | 0.0693 |
| LungCATSC115/TUMOR | 28 | 0.4780 | 0.3297 | 0.4560 | 0.4835 | 0.6099 | 0.2113 |
| LungCATSC115/NL | 29 | 0.4036 | 0.3193 | 0.3373 | 0.3855 | 0.4036 | 0.1059 |
| CircLungCATSC024/TUMOR | 30 | 0.2047 | 0.2632 | 0.4971 | 0.2281 | 0.4854 | 0.4277 |
| PE89/TUMOR | 31 | 0.2934 | 0.1976 | 0.2695 | 0.2814 | 0.3653 | 0.2128 |
| CircLungCATSC027/TUMOR | 32 | 0.0851 | 0.1064 | 0.1436 | 0.0798 | 0.6170 | 1.1189 |
| CircLungCATSC025/NL | 33 | 0.1910 | 0.2079 | 0.1180 | 0.1966 | 0.2416 | 0.2371 |
| LungCATSC092/TUMOR | 34 | 0.0663 | 0.1327 | 0.1990 | 0.0663 | 0.2500 | 0.5692 |
| LungCATSC094/NL | 35 | 0.0684 | 0.1632 | 0.0421 | 0.0737 | 0.2789 | 0.7767 |
A value close to 1 indicates the specimen was almost always classified correctly. The c.v. column indicates the coefficient of variation of the five methods; higher values indicate more variability in the classification of the given observation between methods. Specimen IDs correspond to Figure 4.
DISCUSSION
The secondary analysis of multidimensional flow cytometry data can be daunting, in part because there are arbitrary many ways that data can be parsed when conventional gate/region-based analysis is used. During exploratory data analysis, such as the example presented here, there is usually a fundamental objective that drives the analysis. In this case, it was to discern any differences in the expression of stem/ progenitor markers between non-small cell lung tumors and adjacent normal lung. As tumors are heterogeneous and also contain much non-neoplastic tissue (stromal, vascular, and immune cells) our objectives were to: (1) First, examine cytokeratin (a definitive epithelial marker) versus the stem/ progenitor markers, one at a time; and then (2) break the data into four classes (cytokeratin + euploid, cytokeratin + aneuploid, cytokeratin negative euploid and cytokeratin negative aneuploid) to study the light scatter properties and the coexpression of the stem/progenitor markers in a pairwise fashion. As the latter categories are subsets of the former, it is expected that many of the variables derived from them will be correlated. Another problem inherent to multivariate cytometry data sets is that the number of variables (derived from proportions of analytical regions) is great, often larger than the number of specimens. In this example, we had 86 variables (p) and only 35 specimens (n). Conventional bivariate analysis would require adjustment for the number of comparisons, greatly diluting the statistical power to detect differences.
In this report, we offer an objective method to deal with multidimensional data sets where there is the potential for highly correlated variables and where p >> n. We used five different methods and found two, elastic net and the lasso, particularly useful for this data set, because they consistently chose a manageable number of variables (Fig. 5) that resulted in excellent discrimination between tumor and normal (Fig. 4). Random forest and DLDA were also useful and might prove superior for other data sets. Only best-1 was inadequate to the purpose of this analysis, because it inherently chooses the single best discriminating variable (Fig. 3). An area which was not addressed in this study is the effect of the variance of individual measurements. As raw event counts vary between variables and between samples, not all measured results are equally certain. Future analytical models can take this into account.
Classifying multivariate data against known categories (tumor and normal) can accomplish two important objectives: picking out differences of potentially biological importance for further exploration, and model building for prospective classification of unknowns. Requirements for accuracy are more relaxed for the first application, and considerably more stringent if the model is going to be used prospectively to classify unknown individual cases. In the present data set the model performed excellently for the first application, but consistently misclassified specific individual cases (Table 6). Given that we evaluated 86 variables covering the expression and coexpression of four stem/progenitor associated markers, the similarity in expression patterns between tumor and normal was striking (Fig. S3), suggesting that even among the most deranged tumor cells, these proteins serve important functions that are conserved. Among the most important markers that did distinguish tumor and normal were those in routine use by pathologists, such as cytokeratin + aneuploid cells in tumors, and cytokeratin + small (low scatter), and euploid cells in normal lung. In addition to these known discriminators, selection of which validates the methodology proposed here, there are also several interesting leads (Table 4) such as the expression of the stem/progenitor marker CD133 (CPG2117N133P) and the coexpression of CD117 and CD44 (CP2117P44P). Indeed, CD133, coexpressed with the proliferation marker Ki67 has recently been proposed as a marker of poor prognosis in non-small cell lung cancer (19), but our comparison with normal tissue reveals a greater prevalence of cytokeratin+ cycling CD117 negative/CD133+ cells in normal lung (chosen by all 5 methods, Table 4). Similarly coexpression of CD117, a lung stem cell-associated growth factor receptor (20), in euploid (or pseudo-diploid) cells and CD44 a principal marker associated with tumorigenicity in breast cancer (7,21) also distinguished tumor from normal. Although small data sets such as ours cannot by their nature offer conclusive proof that such markers are of mechanistic, diagnostic or prognostic significance, they provide a sound rational for prospective studies and a model for confirmatory data analysis.
Supplementary Material
Acknowledgments
The authors like to acknowledge our clinical collaborators Drs. James D. Luketich and Rodney J. Landreneau, as well as Dr. Ludovic Zimmerlin, Melanie Pfeifer, James Arbore, and E. Michael Meyer for their assistance in compiling the data set used in this analysis. Biostatistical analysis was supported by P30CA047904 and P50CA090440. The authors have no conflicts of interest to declare.
Grant sponsor: Department of Defense; Grant number: BC032981, BC044784; Grant sponsor: NIH/NCI, P30CA047904, P30CA047904, P50CA090440; Grant sponsor: Commonwealth of Pennsylvania, the Hillman Foundation, the Glimmer of Hope Foundation.
Footnotes
Additional Supporting Information may be found in the online version of this article.
LITERATURE CITED
- 1.Linderman MD, Bjornson Z, Simonds EF, Qiu P, Bruggner R, Sheode K, Meng TH, Plevritis SK, Nolan GP. CytoSPADE: High-Performance Analysis and Visualization of High-Dimensional Cytometry Data. Bioinformatics. 2012;28:2400–2401. doi: 10.1093/bioinformatics/bts425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qiu P, Simonds EF, Bendall SC, Gibbs KD, Jr, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashashati A, Brinkman RR. A survey of flow cytometry data analysis methods. Adv Bioinfo. 2009:584603. doi: 10.1155/2009/584603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hokanson JA, Rosenblatt JI, Leary JF. Some theoretical and practical considerations for multivariate statistical cell classification useful in autologous stem cell transplantation and tumor cell purging. Cytometry. 1999;36:60–70. doi: 10.1002/(sici)1097-0320(19990501)36:1<60::aid-cyto8>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 5.Donnenberg VS, Landreneau RJ, Pfeifer ME, Donnenberg AD. Flow cytometric determination of stem/progenitor content in epithelial tissues: An example from nonsmall lung cancer and normal lung. Cytometry A. 83A doi: 10.1002/cyto.a.22156. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmerlin L, Donnenberg VS, Pfeifer ME, Meyer EM, Peault B, Rubin JP, Donnen-berg AD. Stromal vascular progenitors in adult human adipose tissue. Cytometry A. 2010;77A:22–30. doi: 10.1002/cyto.a.20813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnenberg VS, Donnenberg AD, Zimmerlin L, Landreneau RJ, Bhargava R, Wetzel RA, Basse P, Brufsky AM. Localization of CD44 and CD90 positive cells to the invasive front of breast tumors. Cytometry B Clin Cytom. 2010;78B:287–301. doi: 10.1002/cyto.b.20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 9.Efron B. Bootstrap Methods: Another Look at the Jackknife. Ann Stat. 1979;7:1–26. [Google Scholar]
- 19.Woo T, Okudela K, Mitsui H, Yazawa T, Ogawa N, Tajiri M, Yamamoto T, Rino Y, Kitamura H, Masuda M. Prognostic value of CD133 expression in stage I lung adenocarcinomas. Int J Clin Exp Pathol. 2010;4:32–42. [PMC free article] [PubMed] [Google Scholar]
- 20.Kajstura J, Rota M, Hall SR, Hosoda T, D’Amario D, Sanada F, Zheng H, Ogorek B, Rondon-Clavo C, Ferreira-Martins J, et al. Evidence for human lung stem cells. N Engl J Med. 2011;364:1795–1806. doi: 10.1056/NEJMoa1101324. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.