Abstract
Members of the Parvoviridae utilize glycan receptors for cellular attachment and subsequent interactions determine transduction efficiency or pathogenic outcome. This review focuses on the identity of the glycan receptors utilized, their capsid binding footprints, and a discussion of the overlap of these sites with tropism, transduction, and pathogenicity determinants. Despite high sequence diversity between the different genera, most parvoviruses bind to negatively charged glycans, such as sialic acid and heparan sulfate, abundant on cell surface membranes. The capsid structure of these viruses exhibit high structural homology enabling common regions to be utilized for glycan binding and at the same time the sequence diversity at the common footprints allows for binding of different glycans or differential binding of the same glycan.
Introduction
Viruses are durable nanomachines evolved to utilize an assortment of strategies to manipulate a host cell's replication machinery for successful infection. The key initial step in this process is the attachment to cell surface receptors. This is followed by internalization into the cytoplasm and delivery of the viral genome to the appropriate replication compartment; the cytoplasm for most RNA packaging viruses and the nucleus for those that package DNA. Initial binding is often mediated by ‘attachment factors’ that concentrate the virus on the cell surface and prime it to interact with secondary receptors or co-receptors for internalization.
Glycans and glycoconjugates, displayed on cell surface, serve in communication as well as primary receptors for many viruses. The variability of glycan structures expressed in different species and in different tissues within the same species creates diversity in viral tissue tropism [1]. Mostly, the glycoepitopes consist of negatively charged terminal sialic acid (SIA) or sulfated oligosaccharide motifs of glycosaminoglycans (e.g. heparan sulfate (HS)) and thus mediate electrostatic interactions with the viral capsid. The virus capsid receptor binding motif can be projections or depressions conformed on the assembled capsid surface of non-enveloped viruses, or glycoproteins decorating the lipid membrane of enveloped viruses.
The Parvoviridae, a family of ssDNA viruses, have evolved to ‘hijack’ the interaction functionality of glycans for gaining cellular entry during infection. Receptor-mediated attachment and entry are essential first steps in their infection [2-4]. Following an introduction to the family, this review will discuss current knowledge on (I) glycan receptors utilized for cellular entry and (II) mapped glycan receptor binding footprints with mention of overlaps with transduction efficiency (for non-pathogenic members being exploited as gene delivery vectors), and pathogenesis (for autonomous members) determinants.
The Parvoviridae
The Parvoviridae, small (∼260 Å diameter) non-enveloped T=1 icosahedral viruses, package linear ssDNA genomes of 4 to 6 kb [5]. The family is divided into two subfamilies based on host range: the Parvovirinae infect vertebrates and the Densovirinae infect insects and arthropods [6]. Due to limited information on the Densovirinae with respect to receptor utilization, this review will focus on the Parvovirinae. The Parvovirinae is further subdivided into five genera: Amdovirus, Bocavirus, Dependovirus, Erythrovirus, and Parvovirus, with type members Aleutian mink Disease Virus (AMDV), Bovine Parvovirus (BPV), Adeno-associated virus serotype 2 (AAV2), Human Parvovirus B19 (B19), Minute Virus of Mice (MVM), respectively, based on genomic architecture and protein sequence-based phylogenetic analyses [6]. Their capsid open reading frame (cap) encodes two or three (depending on virus) overlapping structural viral proteins (VP) which assemble the T=1 capsid [7]. The dependovirues rely on co-infection with a complex helper virus (such as Adenovirus, Herpes Simplex Virus or Papillomavirus) for productive infection [8-13] and are non-pathogenic. The other genera replicate independently of helper virus function (‘autonomously replicating’) and contain nonpathogenic and pathogenic members [14-17].
The capsid structures for several parvoviruses, including the type member for each Parvovirinae genera (Fig. 1), have been determined by X-ray crystallography and/or cryo-electron microscopy and image reconstruction (cryo-reconstruction) (reviewed in [18,19] and unpublished data). Despite low sequence similarity (e.g. 14% to 36% between genera), the ordered VP region (VP2 or VP3 depending on virus) is highly conserved with a superposable core eight-stranded β-barrel and αA helix (Fig. 1A). The tops of the loops between these conserved regions are varied in sequence and structure (even within each genus) and defined as variable regions (VRs) I-IX or VR1-8 for dependo and autonomous parvoviruses, respectively [20,21]. The capsid surface is characterized by depressions at the 2-fold axes (dimple) and surrounding a cylindrical channel at the 5-fold axes (canyon), and protrusions at or surrounding the 3-fold axes (Fig. 1B-F). A wall is located between the depressions at the 2-fold axes and surrounding the 5-fold channel, the “2/5-fold wall” (Fig. 1B-F) [18,19]. The VRs cluster at the 5-fold axes, the 3-fold protrusions, and depression at the 2-fold axes to create local variations of the characteristic capsid surface features exhibited by members of each genus. Mutagenesis, biochemical, and structural studies demonstrate that residues in these VRs play important roles in viral life infection, including viral-receptor binding (reviewed in [14,19,22]).
Fig.1.
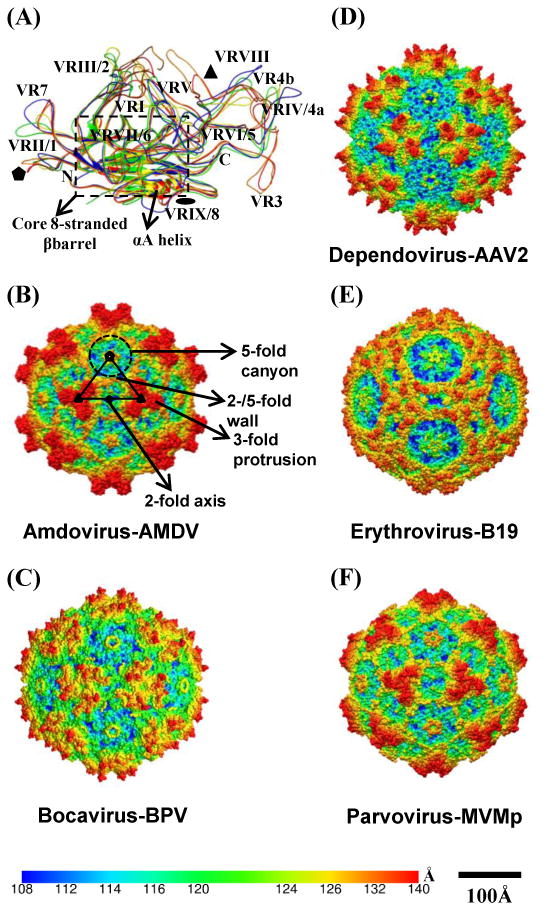
Parvovirinae capsid structure. (A) Structure superposition of the structurally ordered VP region for type members of the Parvovirinae subfamily: amdovirus - ADV (orange); bocavirus – BPV (yellow); dependovirus - AAV2 (blue); erythrovirus - B19 (green); and parvovirus – MVM (red). The N-terminus (N), C-terminus and variable regions (VRI-IX, VR1-8) are labeled, the conserved core eight-stranded β-barrel and αA are delineated by a dashed box. (B-F) Depth cued (from capsid center to surface: blue-green-yellow-red) capsid surface representation of the viruses shown in (A). A viral asymmetric unit (AU, black triangle), bounded by a 5-fold axis (filled pentagon) and two 3-fold axes (filled triangles) separated by 2-fold axis (filled oval), is shown on the AMDV capsid image in (B). The topological features of the parvovirus capsid, such as 3-fold protrusion, 2-fold depression, 5-fold canyon and 2/5-fold wall are labeled on the AMDV image. A horizontal color bar for radial distance (Å) from the center of the capsid and a horizontal scale bar (100Å) for diameter measurements are provided. Panel (A) was generated using the PyMol program [112] and (B-F) were generated using the UCSF-Chimera program [113].
Glycan receptor utilization by the dependoviruses and capsid recognition sites
Currently, over one hundred AAV genomic isolates have been reported [23-33], with thirteen (AAV1-13) serotypes described for the human and non-human primate sequences. Due to their ability to package and delivery foreign genes to different tissue types and the lack of associated disease, AAVs are being developed and used as gene delivery vectors, and serve as the first approved gene therapy treatment [34-40]. AAVs share ∼60 to 99% identity and display differential cell and tissue tropisms as well as transduction efficiency in vitro and in vivo [23,41]. For several of these viruses, cell infectivity and transduction is reported to be dictated by the ability of their capsids to recognize cell surface glycans (Table 1) as primary attachment receptors. AAV1 binds both α2-3 and α2-6 N-linked SIA [42-44]; AAV2, AAV3, and AAV13 bind heparan sulfate proteoglycan (HSPG) [27,45-48]; AAV4 and AAV5 bind α2-3 O-linked and α2-3 N-linked sialic acids, respectively, ([42,49-52] and our unpublished data); AAV6, a tissue culture adapted recombinant between AAV1 and AAV2, binds both HS and α2-3 and α2-6 N-linked SIA [44,52-54], with SIA being the determinant of transduction; AAV9 binds terminal N-linked galactose (GAL) [55,56]. The nature of the glycosylation (if any) required for cellular recognition by AAV7, AAV8, and AAV10-12 are not known. Thus, the human and non-human primate AAVs fall into 3 groups with respect to their glycan receptor usage: HSPG for AAV2, AAV3, AAV6, and AAV13; SIA for AAV1, AAV4, AAV5, and AAV6; GAL for AAV9. Bovine AAV (bAAV) utilizes terminal gangliosides, more commonly observed in glycolipids, and chitotriose, a trimer of β1,4-linked N-acetyl-D-glucosamine, for cellular infection [57,58].
Table 1. Parvoviruses, their hosts and glycan receptors.
| Virus | Host | Glycan Receptors | Phenotype | Residues | References |
|---|---|---|---|---|---|
| AAV1 | Humans | α2-3 and α2-6 N-linked sialic acid (SIA) | SIA binding | unpublished | [42-44], unpublished data |
| AAV2 | Humans | Heparan sulfate proteoglycan (HSPG) | HSPG binding/Transduction | R484, R487, K532, R585, R588 | [45,46,48] |
| AAV3b | Humans | HSPG | HSPG binding | R594 (A593) | [47,61,62] |
| AAV4 | NHP* | α2-3 O-linked SIA | SIA binding/Transduction | K492 (S498), K503 (T503), M523 (M523), G581 (N582), Q583 (Q584), N585 (G586) | [49,51,65] |
| AAV5 | Humans | α2-3 and α2-6 N-linked SIA | SIA binding/Transduction | A581 (A591) | [42,49-52,66,67] |
| AAV6 | Humans | α2-3 and α2-6 N-linked SIA, HSPG | HSPG binding/Transduction | K459 (S458), K493 (S492), K531 (E530), R576 (Q575) | [44,52-54,63,64] |
| AAV9 | Humans | Galactose (GAL) | GAL binding/Transduction | D271 (D269), N272 (N270), Y446 (Y444), N470 (D469), W503 (W502) | [55,56] |
| AAV13 | NHP | HSPG | HSPG binding/Transduction | K528 (E530) | [27] |
| Bovine AAV | Bovine | Gangliosides, Chitotriose | Receptor Binding | N/D | [57,58] |
| BPV | Bovine | α2,3 O-linked SIA (Glycophorin A) | SIA Binding | N/D | [69-71] |
| B19 | Humans | Erythrocyte P antigen (Globoside) | Globoside binding | Q399, Q400, Y401, T402, D403, Q404, E406 | [72-74,94] |
| MVM | Rodents | α2-3 and α2-8 N-linked SIA | SIA binding/Host tropism/pathogenicity | K241, M243, I362, K368, Y396, W398, D399, T401, F403, D553, Y558, T578 | [4,81,82,103-109] |
| H-1PV | Rodents | α2-3 N-linked SIA | SIA binding | I368, H374 | [89], |
| CPV and FPV | Canine, | α2-3 and α2-6 N-linked Neu5Gc | SIA binding | R377, E396, R397 | [95-97] |
| Host range/pathogenicity/Transferrin receptor binding | N93, G299, A300, N323 | [77,98-100] | |||
| PPV | Swine | α2-3 and α2-6 N and O- linked SIA | SIA Binding | N/D | [93] |
NHP=Non-Human Primate;
N/D: not defined; Residues in parentheses are AAV2 VP1 numbering; residues in bold are overlapping glycan binding and transduction determinants; residues in italics are overlapping glycan binding and pathogenicity determinants; residues underlined are overlapping glycan binding and tropism determinants.
Cell binding, transduction assays, mutagenesis, and structural approaches have been utilized to identify the capsid amino acid residues involved in receptor recognition for most of the AAVs for which glycan receptors are known (Table 1; Fig. 2A-C), with the interaction between AAV2 and HSPG the best characterized. Mutagenesis identified the AAV2 HS binding footprint as five basic residues: R484, R487, K532, R585, and R588 (Fig. 2A-C), with R585 and R588 (VP1 numbering) the most critical [45,46]. Cryo-reconstruction of the AAV2-HS complex confirmed this binding site, as adjacent residues, at the inner wall of the protrusions surrounding the 3-fold axis on AAV2 capsid (Fig. 2A-C) [59,60]. These residues are located on VR-V, VR-VI, and VR-VIII. Structural and biochemical approaches identified the HS binding site on AAV3B, a minor variant of AAV3, as R594 (in VR-VIII) located also on the inner wall of the 3-fold protrusion but not overlapping with the AAV2 footprint (Fig. 2A-C) [61,62]. For AAV6, two separate mutagenesis studies identified two non-adjacent residues as being important for HS recognition; K459 (VR-IV) located close to the top of the 3-fold protrusion and K531 (VR-VI) located at the base of 3-fold protrusion facing the 2-fold axis (Fig. 2A-C) [53,63,64]. Random capsid VP mutagenesis and in vivo assays suggest that the SIA binding properties of AAV4 are controlled by residues K492, K503, M523, G581, Q583, and N585. These residues are located at the top of the 3-fold protrusion: K492 and K503 within VR-V, and G581, Q583, and N585 within VR-VIII. [65]. Mutagenesis studies for AAV5 suggest the involvement of A581 (VR-VIII) in SIA binding and transduction [66,67]. This residue is also located on the inner wall of the 3-fold protrusion (Fig. 2A-C). By mutagenesis and computational molecular docking, the GAL binding residues on AAV9 were identified as D271 (VR-I), N272 (VR-I), Y446, N470, and W503 (VR-V) that form a pocket at the base of the 3-fold protrusion facing the 5-fold axis (Fig. 2A-C) [56]. Residues Y466, N470, and W503, are the 3/26 reported to directly or indirectly affect AAV9 GAL binding by AAV Barcode-Seq [68]. An analogous pocket on AAV1 has been identified as its SIA binding site by structural mapping (unpublished data). Residue K528 in AAV13, which is structurally equivalent to K531 in AAV6, dictates its HS interaction [27]. Thus regardless of the glycan recognized, the 3-fold protrusion serves as the binding region on several AAV capsids (Fig. 2A-C). These sites, mostly located in VRs, overlap residues reported to affect the transduction properties of the AAVs (Table 1) (reviewed in [22]).
Fig.2.

Capsid surface images for AAV2 and B19. (A) Capsid surface image of AAV2 on which the residues involved in glycan receptor binding for AAV2, AAV3B, AAV4, AAV5, AAV6, AAV9, and AAV13 are highlighted along with residues determining transduction phenotypes. The view is approximately down the icosahedral 2-fold axis. (B) A zoom of the receptor binding footprints in the section delineated by a dashed box in (A). (C) Stereographic Roadmap projection [114] of the glycan footprint shown in (A) viewed down the threefold axis. Residues are labeled by type (three letter code) and number (VP1 numbering). The amino acid residues that are exposed on the capsid exterior are visible in this image. The boundary for each residue is shown in black, and a color key is provided. The AU is depicted as in Fig. 1B. (D, E, and F) Capsid surface image of B19, zoom of the dashed box in (D) and Roadmap projection, respectively, of the globoside footprint at the 3-fold axis. The image labels are as in (A-C). Panels (A, B, D, and E) were generated using the UCSF-Chimera program [113] and (C and F) were generated using the RIVEM program [114].
Glycans receptor utilization by the autonomous parvoviruses and capsid recognition sites
Pathogenic members of the autonomous parvoviruses are associated with serious diseases in the young of the species infected and immunocompromised adults, while nonpathogenic members establish asymptomatic but persistent infections (reviewed in [17]). The glycans involved in cellular recognition are known for most of the type members of the autonomous Parvovirinae, except amdovirus, and for genus parvovirus this information is available for several members. BPV (bocavirus) binds to glycophorin A via O-linked α2,3-linked sialic acid [69-71]. B19 (erythrovirus), a human pathogen, binds to the neutral glycosphingolipid erythrocyte P antigen (globoside/Gb4) on erythroid progenitor cells [72,73], and another structurally and biosynthetically unrelated glycolipid called paragloboside (neolactotetraglycosylceramide or nLc4) [74]. However, Kaufmann and coworkers [75] reported that recombinant B19 capsids do not bind to globoside Gb4 in vitro.
For several members of the parvovirus genus, terminal SIA serves as a glycan receptor (Table 1), although for Canine Parvovirus (CPV) and Feline Panleukopenia Virus (FPV), SIA recognition is not essential for infection, but rather utilized during hemagglutination (HA) of erythrocytes [76]. CPV and FPV recognize both α2-3 and α2-6 linked N-glycolyl neuraminic acid, a non-human SIA. Host cellular infection by CPV and FPV requires recognition of their respective host transferrin receptor type-1 with differential N-glycosylation dictating viral host specificity ([77-80]. For other members, for example H-1 Parvovirus (H-1PV), LuIII, MVM, and Porcine Parvovirus (PPV), SIA binding is essential for infection and the recognition is to N-acetyl neuraminic acid. MVM utilizes terminal SIA on an unknown glycoprotein for cellular recognition [4] and binds specifically to α2-3 N-linked sialylated glycans including the s(Lex)3 motif [81,82]. This Lewis X motif is over expressed in cancer cells and likely dictates MVM's oncotropism [83]. MVM exists in several highly homologous strains, including the prototype strain (MVMp) and the immunosuppressive strain (MVMi) which share 97% sequence identity. These viruses are reciprocally restricted for growth in each other's permissive cell type, although both viruses are able to infect the SV40 transformed human fibroblast cell line NB324K [84]. MVMp is non-pathogenic and results in an asymptomatic infection [85], while MVMi causes a lethal infection and is neurotropic [86,87]. Additional recognition of α2-8 linked multi-SIA glycans, over-expressed in neuronal cells, by MVMi likely mediates its neurotropism [81,88]. Similar to MVM, H-1PV, and LuIII also recognize α2-3 linked SIA glycans, especially the sLex motif ([89] and unpublished data). Significantly, these rodent parvoviruses (MVM, H-1PV, and LuIII) exhibit tumor specific cytotoxic activity in vitro and oncosuppressive activity in vivo and are under development as viral vectors for anticancer gene therapy (reviewed in [90-92]). PPV binds to SIAs on cell surface glycoproteins yet to be identified [93].
Similar to the dependovirues, a multitude of approaches have been utilized to characterize the glycan recognition site for a number of the autonomous parvoviruses. Cryo-reconstruction localized the B19 globoside footprint to the depression between the protrusions surrounding the 3-fold axes of the capsid [94], similar to the HS and SIA sites for the AAVs. The globoside footprint includes residues Q399, Q400, Y401, T402, D403, Q404, and E406 (VP2 numbering, Table 1, Fig. 2D-F). For CPV, mutations of residues R377K, E396K, R397G/E on the wall of the depression surrounding the 2-fold axis of the capsid reduced or abolished HA, hence, SIA recognition [95-97]. Residues N93, G299, A300, and N323 control CPV/FPV host range and pathogenicity determination as well as transferrin receptor attachment [77,98-100]. These residues are located at the top and shoulder of the 3-fold protrusions [101,102] (Fig. 3A-C). Mutagenesis, cell binding assays, and structural studies indicate that the H-1PV SIA footprint contains I368 and H374 located at the 2-fold depression ([89] and unpublished data). For MVM, mutagenesis, and structural studies have also identified the depression at the 2-fold axis as the SIA binding site [103]. The binding pocket is shallow and surrounded by charged and hydrophobic residues that include K241, M243, I362, K368, Y396, W398, D399, T401, F403, D553, Y558, and I578. Significantly, the residues determining in vitro tropism (T317 and G321), conferring fibrotropism on MVMi (D399,S 460, D553, Y558), in vivo pathogenicity (V325, I362, K368), and those associated with the development of leukopenia (G321, A551, V575) are localized in the vicinity of this SIA binding pocket [104-109] (Fig. 3D-F). In addition, tissue tropism and pathogenicity determinants for ADV, CPV, and PPV overlap with this region [14]. There is no information on residues controlling BPV, LuIII, or PPV capsid – SIA interactions. The available information identifies the 2-fold (CPV and MVM) and 3-fold (B19) depressions as important for glycan receptor engagement for the autonomous viruses.
Fig. 3.

Capsid surface images for CPV and MVM. (A-C) Capsid surface image of CPV, zoom of the dashed box in (A), and Roadmap projection, respectively, of the residues involved in SIA binding and host tropism/pathogenicity/transferrin receptor binding. (D-F) Capsid surface image of MVM, zoom of the dashed box in (D), and Roadmap projection, respectively, of the residues involved in SIA binding, tissue tropism, and pathogenicity. The image labels are as in Fig. 2. Panels (A, B, D, and E) were generated using the UCSF-Chimera program [113] and (C and F) were generated using the RIVEM program [114].
Commonality in utilization and binding region are features of Parvovirinae glycan interactions, although recognition determinants differ
A role for glycan recognition in dictating successful cellular infection and as a determinant of tissue tropism is well established for the Parvovirinae. Evidence points to a role in dictating transduction efficiency and host pathogenicity for dependo and autonomous parvoviruses, respectively, due to overlap of the capsid residues involved. However, this possibility requires further investigation given the ubiquity of SIA and HS which are the most commonly recognized glycans. Thus parvovirus tissue (especially for the AAVs) and host specificity likely requires post cell entry interactions, including the recognition of specific internalization receptors/co-receptors which are often glycosylated [110,111]. Regardless of the outcome of infection, pathogenic or non-pathogenic, commonality in utilization and binding region is a feature of Parvovirinae glycan interactions, despite the sequence diversity that exists between genus members (14-36%) and within each genera (e.g. 50-66% for parvovirus genus).
The known interaction sites involve residues at the 3-fold depression (e.g. AAVs and B19); pocket at base of 3-fold protrusion (AAV1 and AAV9), or 2-fold depression (e.g. CPV and MVM) (Figs. 2 and 3). Significantly, both the 2- and 3-fold depressions are utilized to bind SIA, e.g. in MVM and AAV4/AAV5, respectively, despite low sequence and structure similarity at these capsid regions. In addition, analogous capsid regions/pockets are used to bind different glycans, e.g. SIA, globoside, and HS at the 3-fold depression, and SIA and GAL at the base of the 3-fold protrusions. Furthermore, while 3/5 AAV2 HS binding footprint residues are conserved in most of the AAV serotypes, the critical R585 and R588 are not. For AAV3B and AAV6/AAV13, their HS binding residues, R594 and K531/K528, respectively, are not conserved in other HS binding AAVs. For SIA binding, 4/6 SIA binding residues in AAV4 (K492, K503, G581, and N585) are not conserved in AAV1 and AAV5, the serotypes that also bind SIA and while AAV5's A581 position is structurally conserved in other AAVs, including AAV2, the adjacent residues and structure are not. With respect to the common pocket for SIA and GAL binding by AAV1 and AAV9, respectively, N470, the most critical residue for the AAV9 phenotype, is G470 in AAV1. In B19, the globoside binding pocket residues are not conserved in other genus type members. For MVM, the SIA binding footprint residues are conserved in H-1PV consistent with a common pocket, but not for SIA binding dependovirus members, likely leading to their use of the 3-fold region for this interaction. Importantly, these glycan recognition sites overlap with transduction and pathogenicity determinants. These observations suggest a parvovirus capsid evolution to establish host recognition niches rather than conservation of glycan binding pockets. Similar binding site localization points to structural motifs evolved to serve an essential functional role and begin to establish a receptor binding pattern that could inform efforts to use these capsids for targeted therapies or develop viral infection inhibitors.
Highlights.
Glycan interactions are an essential first step in successful host infection by the Parvovirinae subfamily of the Parvoviridae.
The binding sites for glycans overlap with determinants of transduction efficiency and pathogenicity for non-pathogenic and pathogenic members, respectively.
The Parvovirinae exhibit commonality in the glycans recognized across genera and they utilize common capsid regions for binding to disparate glycans.
Identifies capsid regions that can inform engineering efforts for tissue targeted gene delivery therapies or development of viral infection inhibitors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Olofsson S, Bergstrom T. Glycoconjugate glycans as viral receptors. Ann Med. 2005;37:154–172. doi: 10.1080/07853890510007340. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett JS, Wilcher R, Samulski RJ. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J Virol. 2000;74:2777–2785. doi: 10.1128/jvi.74.6.2777-2785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basak S, Turner H. Infectious entry pathway for canine parvovirus. Virology. 1992;186:368–376. doi: 10.1016/0042-6822(92)90002-7. [DOI] [PubMed] [Google Scholar]
- 4.Cotmore SF, Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 5.Berns K, Parrish CR. Parvoviridae. In: Knipe DM, Howley PM, editors. Fields Virology. Fifth. Lippincott Williams and Wilkins; 2007. pp. 2437–2478. [Google Scholar]
- 6.Bergoin M, Kleinschmidt J, Almendral JM, Hedman K, Li Y, Agbandje-McKenna M, Tattersall P, Tijssen P, Pintel DJ, Flegel TW. Ninth Report of the International Committee on Taxonomy of Viruses. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy. Elsevier; 2011. [Google Scholar]
- 7.Cotmore SF, Tattersall R. Structure and Organization of the Viral Genome. In: Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hodder Arnold; 2006. p. 73. [Google Scholar]
- 8.Weitzman MD. The Parvovirus Life Cycle: An Introduction to Molecular Interactions Important for Infection. In: Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hodder Arnold; 2006. pp. 143–156. [Google Scholar]
- 9.Bowles DE, Rabinowitz JE, S RJ. The Genus Dependovirus. In: Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hodder Arnold; 2006. pp. 15–23. [Google Scholar]
- 10.Alazard-Dany N, Nicolas A, Ploquin A, Strasser R, Greco A, Epstein AL, Fraefel C, Salvetti A. Definition of herpes simplex virus type 1 helper activities for adeno-associated virus early replication events. PLoS Pathog. 2009;5:e1000340. doi: 10.1371/journal.ppat.1000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geoffroy MC, Salvetti A. Helper functions required for wild type and recombinant adeno-associated virus growth. Curr Gene Ther. 2005;5:265–271. doi: 10.2174/1566523054064977. [DOI] [PubMed] [Google Scholar]
- 12.McCaffrey AP, Fawcett P, Nakai H, McCaffrey RL, Ehrhardt A, Pham TT, Pandey K, Xu H, Feuss S, Storm TA, et al. The host response to adenovirus, helper-dependent adenovirus, and adeno-associated virus in mouse liver. Mol Ther. 2008;16:931–941. doi: 10.1038/mt.2008.37. [DOI] [PubMed] [Google Scholar]
- 13.Walz C, Deprez A, Dupressoir T, Durst M, Rabreau M, Schlehofer JR. Interaction of human papillomavirus type 16 and adeno-associated virus type 2 co-infecting human cervical epithelium. J Gen Virol. 1997;78(Pt 6):1441–1452. doi: 10.1099/0022-1317-78-6-1441. [DOI] [PubMed] [Google Scholar]
- 14.Agbandje-McKenna M, Chapman MS. Correlating structure with function in viral capsid. In: Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hodder Arnold; 2006. pp. 125–139. [Google Scholar]
- 15.Brown KE. The Genus Erythrovirus. In: Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hodder Arnold; 2006. pp. 25–45. [Google Scholar]
- 16.Parrish CR. Autonomous Parvovirus Variation and Evolution. In: Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hodder Arnold; 2006. pp. 47–53. [Google Scholar]
- 17.Tattersall P. The Evolution of Parvovirus Taxonomy. In: Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hodder Arnold; 2006. pp. 5–14. [Google Scholar]
- 18.Chapman MS, Agbandje-McKenna M. Atomic structure of viral particles. In: Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hodder Arnold; 2006. pp. 107–123. [Google Scholar]
- 19.Halder S, Ng R, Agbandje-McKenna M. Parvoviruses: structure and infection. Future Virology. 2012;7:253–278. [Google Scholar]
- 20.Govindasamy L, Padron E, McKenna R, Muzyczka N, Kaludov N, Chiorini JA, Agbandje-McKenna M. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J Virol. 2006;80:11556–11570. doi: 10.1128/JVI.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kontou M, Govindasamy L, Nam HJ, Bryant N, Llamas-Saiz AL, Foces-Foces C, Hernando E, Rubio MP, McKenna R, Almendral JM, et al. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J Virol. 2005;79:10931–10943. doi: 10.1128/JVI.79.17.10931-10943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agbandje-McKenna M, Kleinschmidt J. AAV capsid structure and cell interactions. Methods Mol Biol. 2011;807:47–92. doi: 10.1007/978-1-61779-370-7_3. [DOI] [PubMed] [Google Scholar]
- 23.Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, Wilson JM. Clades of Adeno-associated viruses are widely disseminated in human tissues. J Virol. 2004;78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori S, Wang L, Takeuchi T, Kanda T. Two novel adeno-associated viruses from cynomolgus monkey: pseudotyping characterization of capsid protein. Virology. 2004;330:375–383. doi: 10.1016/j.virol.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt M, Katano H, Bossis I, Chiorini JA. Cloning and characterization of a bovine adeno-associated virus. J Virol. 2004;78:6509–6516. doi: 10.1128/JVI.78.12.6509-6516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt M, Voutetakis A, Afione S, Zheng C, Mandikian D, Chiorini JA. Adeno-associated virus type 12 (AAV12): a novel AAV serotype with sialic acid- and heparan sulfate proteoglycan-independent transduction activity. J Virol. 2008;82:1399–1406. doi: 10.1128/JVI.02012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt M, Govindasamy L, Afione S, Kaludov N, Agbandje-McKenna M, Chiorini JA. Molecular characterization of the heparin-dependent transduction domain on the capsid of a novel adeno-associated virus isolate, AAV(VR-942) J Virol. 2008;82:8911–8916. doi: 10.1128/JVI.00672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arbetman AE, Lochrie M, Zhou S, Wellman J, Scallan C, Doroudchi MM, Randlev B, Patarroyo-White S, Liu T, Smith P, et al. Novel caprine adeno-associated virus (AAV) capsid (AAV-Go1) is closely related to the primate AAV-5 and has unique tropism and neutralization properties. J Virol. 2005;79:15238–15245. doi: 10.1128/JVI.79.24.15238-15245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bello A, Tran K, Chand A, Doria M, Allocca M, Hildinger M, Beniac D, Kranendonk C, Auricchio A, Kobinger GP. Isolation and evaluation of novel adeno-associated virus sequences from porcine tissues. Gene Ther. 2009;16:1320–1328. doi: 10.1038/gt.2009.82. [DOI] [PubMed] [Google Scholar]
- 30.Bossis I, Chiorini JA. Cloning of an avian adeno-associated virus (AAAV) and generation of recombinant AAAV particles. J Virol. 2003;77:6799–6810. doi: 10.1128/JVI.77.12.6799-6810.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hess M, Paul G, Kling S, Monreal G. Molecular characterization of two strains of the avian adeno-associated virus (AAAV) Arch Virol. 1995;140:591–598. doi: 10.1007/BF01718434. [DOI] [PubMed] [Google Scholar]
- 32.Brown KE, Green SW, Young NS. Goose parvovirus--an autonomous member of the dependovirus genus? Virology. 1995;210:283–291. doi: 10.1006/viro.1995.1345. [DOI] [PubMed] [Google Scholar]
- 33.Farkas SL, Zadori Z, Benko M, Essbauer S, Harrach B, Tijssen P. A parvovirus isolated from royal python (Python regius) is a member of the genus Dependovirus. J Gen Virol. 2004;85:555–561. doi: 10.1099/vir.0.19616-0. [DOI] [PubMed] [Google Scholar]
- 34.Alexander JJ, Hauswirth WW. Prospects for retinal cone-targeted gene therapy. Drug News Perspect. 2008;21:267–271. doi: 10.1358/dnp.2008.21.5.1219012. [DOI] [PubMed] [Google Scholar]
- 35.Brantly ML, Chulay JD, Wang L, Mueller C, Humphries M, Spencer LT, Rouhani F, Conlon TJ, Calcedo R, Betts MR, et al. Sustained transgene expression despite T lymphocyte responses in a clinical trial of rAAV1-AAT gene therapy. Proc Natl Acad Sci USA. 2009;106:16363–16368. doi: 10.1073/pnas.0904514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cideciyan AV, Hauswirth WW, Aleman TS, Kaushal S, Schwartz SB, Boye SL, Windsor EA, Conlon TJ, Sumaroka A, Pang JJ, et al. Human RPE65 gene therapy for Leber congenital amaurosis: persistence of early visual improvements and safety at 1 year. Hum Gene Ther. 2009;20:999–1004. doi: 10.1089/hum.2009.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.High K. AAV-mediated gene transfer for hemophilia. Genet Med. 2002;4:56S–61S. doi: 10.1097/00125817-200211001-00012. [DOI] [PubMed] [Google Scholar]
- 38.Mandel RJ. CERE-110, an adeno-associated virus-based gene delivery vector expressing human nerve growth factor for the treatment of Alzheimer's disease. Curr Opin Mol Ther. 2010;12:240–247. [PubMed] [Google Scholar]
- 39.Nathwani AC, Tuddenham EGD, Rangarajan S, Rosales C, McIntosh J, Linch DC, Chowdary P, Riddell A, Pie J, Harrington C, O'Beirne J, Smith K, Pasi J, Glader B, Rustagi P, Ng CYC, Kay MA, Zhou J, Spence Y, Morton CL, Allay J, Coleman J, Sleep S, Cunningham JM, Srivastava D, Basner-Tschakarjan E, Mingozzi F, High KA, Gray JT, Reiss UM, Nienhuis AW, Davidoff AM. Adenovirus-Associated Virus Vector–Mediated Gene Transfer in Hemophilia B. the New England Journal of Medicine. 2011 doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kastelein JJ, Ross CJ, Hayden MR. From mutation identification to therapy: discovery and origins of the first approved gene therapy in the Western world. Hum Gene Ther. 2013;24:472–478. doi: 10.1089/hum.2013.063. [DOI] [PubMed] [Google Scholar]
- 41.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 42.Chen S, Kapturczak M, Loiler SA, Zolotukhin S, Glushakova OY, Madsen KM, Samulski RJ, Hauswirth WW, Campbell-Thompson M, Berns KI, et al. Efficient transduction of vascular endothelial cells with recombinant adeno-associated virus serotype 1 and 5 vectors. Hum Gene Ther. 2005;16:235–247. doi: 10.1089/hum.2005.16.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rabinowitz JE, Rolling F, Li C, Conrath H, Xiao W, Xiao X, Samulski RJ. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J Virol. 2002;76:791–801. doi: 10.1128/JVI.76.2.791-801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Z, Miller E, Agbandje-McKenna M, Samulski RJ. Alpha2,3 and alpha2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6. J Virol. 2006;80:9093–9103. doi: 10.1128/JVI.00895-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kern A, Schmidt K, Leder C, Muller OJ, Wobus CE, Bettinger K, Von der Lieth CW, King JA, Kleinschmidt JA. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J Virol. 2003;77:11072–11081. doi: 10.1128/JVI.77.20.11072-11081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Opie SR, Warrington KH, Jr, Agbandje-McKenna M, Zolotukhin S, Muzyczka N. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J Virol. 2003;77:6995–7006. doi: 10.1128/JVI.77.12.6995-7006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Handa A, Muramatsu S, Qiu J, Mizukami H, Brown KE. Adeno-associated virus (AAV)-3-based vectors transduce haematopoietic cells not susceptible to transduction with AAV-2-based vectors. J Gen Virol. 2000;81:2077–2084. doi: 10.1099/0022-1317-81-8-2077. [DOI] [PubMed] [Google Scholar]
- 48.Summerford C, Samulski RJ. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaludov N, Brown KE, Walters RW, Zabner J, Chiorini JA. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J Virol. 2001;75:6884–6893. doi: 10.1128/JVI.75.15.6884-6893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walters RW, Yi SM, Keshavjee S, Brown KE, Welsh MJ, Chiorini JA, Zabner J. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J Biol Chem. 2001;276:20610–20616. doi: 10.1074/jbc.M101559200. [DOI] [PubMed] [Google Scholar]
- 51.Walters RW, Pilewski JM, Chiorini JA, Zabner J. Secreted and transmembrane mucins inhibit gene transfer with AAV4 more efficiently than AAV5. J Biol Chem. 2002;277:23709–23713. doi: 10.1074/jbc.M200292200. [DOI] [PubMed] [Google Scholar]
- 52.Seiler MP, Miller AD, Zabner J, Halbert CL. Adeno-associated virus types 5 and 6 use distinct receptors for cell entry. Hum Gene Ther. 2006;17:10–19. doi: 10.1089/hum.2006.17.10. [DOI] [PubMed] [Google Scholar]
- 53.Wu Z, Asokan A, Grieger JC, Govindasamy L, Agbandje-McKenna M, Samulski RJ. Single amino acid changes can influence titer, heparin binding, and tissue tropism in different adeno-associated virus serotypes. J Virol. 2006;80:11393–11397. doi: 10.1128/JVI.01288-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Halbert CL, Allen JM, Miller AD. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J Virol. 2001;75:6615–6624. doi: 10.1128/JVI.75.14.6615-6624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen S, Bryant KD, Brown SM, Randell SH, Asokan A. Terminal N-linked galactose is the primary receptor for adeno-associated virus 9. J Biol Chem. 2011;286:13532–13540. doi: 10.1074/jbc.M110.210922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bell CL, Gurda BL, Van Vliet K, Agbandje-McKenna M, Wilson JM. Identification of the galactose binding domain of the adeno-associated virus serotype 9 capsid. J Virol. 2012;86:7326–7333. doi: 10.1128/JVI.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Di Pasquale G, Kaludov N, Agbandje-McKenna M, Chiorini JA. BAAV transcytosis requires an interaction with beta-1-4 linked- glucosamine and gp96. PLoS One. 2010;5:e9336. doi: 10.1371/journal.pone.0009336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt M, Chiorini JA. Gangliosides are essential for bovine adeno-associated virus entry. J Virol. 2006;80:5516–5522. doi: 10.1128/JVI.02393-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levy HC, Bowman VD, Govindasamy L, McKenna R, Nash K, Warrington K, Chen W, Muzyczka N, Yan X, Baker TS, et al. Heparin binding induces conformational changes in Adeno-associated virus serotype 2. J Struct Biol. 2009;165:146–156. doi: 10.1016/j.jsb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O'Donnell J, Taylor KA, Chapman MS. Adeno-associated virus-2 and its primary cellular receptor--Cryo-EM structure of a heparin complex. Virology. 2009;385:434–443. doi: 10.1016/j.virol.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lerch TF, Xie Q, Chapman MS. The structure of adeno-associated virus serotype 3B (AAV-3B): insights into receptor binding and immune evasion. Virology. 2010;403:26–36. doi: 10.1016/j.virol.2010.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lerch TF, Chapman MS. Identification of the heparin binding site on adeno-associated virus serotype 3B (AAV-3B) Virology. 2012;423:6–13. doi: 10.1016/j.virol.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ng R, Govindasamy L, Gurda BL, McKenna R, Kozyreva OG, Samulski RJ, Parent KN, Baker TS, Agbandje-McKenna M. Structural characterization of the dual glycan binding adeno-associated virus serotype 6. J Virol. 2010;84:12945–12957. doi: 10.1128/JVI.01235-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xie Q, Lerch TF, Meyer NL, Chapman MS. Structure-function analysis of receptor-binding in adeno-associated virus serotype 6 (AAV-6) Virology. 2011;420:10–19. doi: 10.1016/j.virol.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen S, Troupes AN, Pulicherla N, Asokan A. Multiple roles for sialylated glycans in determining the cardiopulmonary tropism of adeno-associated virus 4. J Virol. 2013;87:13206–13213. doi: 10.1128/JVI.02109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Excoffon KJ, Koerber JT, Dickey DD, Murtha M, Keshavjee S, Kaspar BK, Zabner J, Schaffer DV. Directed evolution of adeno-associated virus to an infectious respiratory virus. Proc Natl Acad Sci USA. 2009;106:3865–3870. doi: 10.1073/pnas.0813365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dickey DD, Excoffon KJ, Koerber JT, Bergen J, Steines B, Klesney-Tait J, Schaffer DV, Zabner J. Enhanced sialic acid-dependent endocytosis explains the increased efficiency of infection of airway epithelia by a novel adeno-associated virus. J Virol. 2011;85:9023–9030. doi: 10.1128/JVI.05154-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adachi K, Enoki T, Kawano Y, Veraz M, Nakai H. Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nat Commun. 2014;5:3075. doi: 10.1038/ncomms4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blackburn SD, Cline SE, Hemming JP, Johnson FB. Attachment of bovine parvovirus to O-linked alpha 2,3 neuraminic acid on glycophorin A. Arch Virol. 2005;150:1477–1484. doi: 10.1007/s00705-005-0496-y. [DOI] [PubMed] [Google Scholar]
- 70.Thacker TC, Johnson FB. Binding of bovine parvovirus to erythrocyte membrane sialylglycoproteins. J Gen Virol. 1998;79(Pt 9):2163–2169. doi: 10.1099/0022-1317-79-9-2163. [DOI] [PubMed] [Google Scholar]
- 71.Johnson FB, Fenn LB, Owens TJ, Faucheux LJ, Blackburn SD. Attachment of bovine parvovirus to sialic acids on bovine cell membranes. J Gen Virol. 2004;85:2199–2207. doi: 10.1099/vir.0.79899-0. [DOI] [PubMed] [Google Scholar]
- 72.Brown KE, Cohen BJ. Haemagglutination by parvovirus B19. J Gen Virol. 1992;73(Pt 8):2147–2149. doi: 10.1099/0022-1317-73-8-2147. [DOI] [PubMed] [Google Scholar]
- 73.Brown KE, Anderson SM, Young NS. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science. 1993;262:114–117. doi: 10.1126/science.8211117. [DOI] [PubMed] [Google Scholar]
- 74.Cooling LL, Koerner TA, Naides SJ. Multiple glycosphingolipids determine the tissue tropism of parvovirus B19. J Infect Dis. 1995;172:1198–1205. doi: 10.1093/infdis/172.5.1198. [DOI] [PubMed] [Google Scholar]
- 75.Kaufmann B, Baxa U, Chipman PR, Rossmann MG, Modrow S, Seckler R. Parvovirus B19 does not bind to membrane-associated globoside in vitro. Virology. 2005;332:189–198. doi: 10.1016/j.virol.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 76.Lofling J, Michael Lyi S, Parrish CR, Varki A. Canine and feline parvoviruses preferentially recognize the non-human cell surface sialic acid N-glycolylneuraminic acid. Virology. 2013;440:89–96. doi: 10.1016/j.virol.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hafenstein S, Palermo LM, Kostyuchenko VA, Xiao C, Morais MC, Nelson CD, Bowman VD, Battisti AJ, Chipman PR, Parrish CR, et al. Asymmetric binding of transferrin receptor to parvovirus capsids. Proc Natl Acad Sci USA. 2007;104:6585–6589. doi: 10.1073/pnas.0701574104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goodman LB, Lyi SM, Johnson NC, Cifuente JO, Hafenstein SL, Parrish CR. Binding site on the transferrin receptor for the parvovirus capsid and effects of altered affinity on cell uptake and infection. J Virol. 84:4969–4978. doi: 10.1128/JVI.02623-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Palermo LM, Hafenstein SL, Parrish CR. Purified feline and canine transferrin receptors reveal complex interactions with the capsids of canine and feline parvoviruses that correspond to their host ranges. J Virol. 2006;80:8482–8492. doi: 10.1128/JVI.00683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Palermo LM, Hueffer K, Parrish CR. Residues in the apical domain of the feline and canine transferrin receptors control host-specific binding and cell infection of canine and feline parvoviruses. J Virol. 2003;77:8915–8923. doi: 10.1128/JVI.77.16.8915-8923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nam HJ, Gurda-Whitaker B, Gan WY, Ilaria S, McKenna R, Mehta P, Alvarez RA, Agbandje-McKenna M. Identification of the sialic acid structures recognized by minute virus of mice and the role of binding affinity in virulence adaptation. J Biol Chem. 2006;281:25670–25677. doi: 10.1074/jbc.M604421200. [DOI] [PubMed] [Google Scholar]
- 82.Halder S, Cotmore S, Heimburg-Molinaro J, Smith DF, Cummings RD, Chen X, Trollope AJ, North SJ, Haslam SM, Dell A, et al. Profiling of glycan receptors for minute virus of mice in permissive cell lines towards understanding the mechanism of cell recognition. PLoS One. 2014;9:e86909. doi: 10.1371/journal.pone.0086909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kannagi R. Molecular mechanism for cancer-associated induction of sialyl Lewis X and sialyl Lewis A expression-The Warburg effect revisited. Glycoconj J. 2004;20:353–364. doi: 10.1023/B:GLYC.0000033631.35357.41. [DOI] [PubMed] [Google Scholar]
- 84.Tattersall P, Bratton J. Reciprocal productive and restrictive virus-cell interactions of immunosuppressive and prototype strains of minute virus of mice. J Virol. 1983;46:944–955. doi: 10.1128/jvi.46.3.944-955.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kimsey PB, Engers HD, Hirt B, Jongeneel CV. Pathogenicity of fibroblast- and lymphocyte-specific variants of minute virus of mice. J Virol. 1986;59:8–13. doi: 10.1128/jvi.59.1.8-13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramirez JC, Fairen A, Almendral JM. Parvovirus minute virus of mice strain i multiplication and pathogenesis in the newborn mouse brain are restricted to proliferative areas and to migratory cerebellar young neurons. J Virol. 1996;70:8109–8116. doi: 10.1128/jvi.70.11.8109-8116.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Segovia JC, Gallego JM, Bueren JA, Almendral JM. Severe leukopenia and dysregulated erythropoiesis in SCID mice persistently infected with the parvovirus minute virus of mice. J Virol. 1999;73:1774–1784. doi: 10.1128/jvi.73.3.1774-1784.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sato C, Fukuoka H, Ohta K, Matsuda T, Koshino R, Kobayashi K, Troy FA, 2nd, Kitajima K. Frequent occurrence of pre-existing alpha 2-->8-linked disialic and oligosialic acids with chain lengths up to 7 Sia residues in mammalian brain glycoproteins. Prevalence revealed by highly sensitive chemical methods and anti-di-, oligo-, and poly-Sia antibodies specific for defined chain lengths. J Biol Chem. 2000;275:15422–15431. doi: 10.1074/jbc.275.20.15422. [DOI] [PubMed] [Google Scholar]
- 89.Allaume X, El-Andaloussi N, Leuchs B, Bonifati S, Kulkarni A, Marttila T, Kaufmann JK, Nettelbeck DM, Kleinschmidt J, Rommelaere J, et al. Retargeting of Rat Parvovirus H-1PV to Cancer Cells through Genetic Engineering of the Viral Capsid. J Virol. 2012;86:3452–3465. doi: 10.1128/JVI.06208-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rommelaere J, Cornelis JJ. Antineoplastic activity of parvoviruses. J Virol Methods. 1991;33:233–251. doi: 10.1016/0166-0934(91)90024-t. [DOI] [PubMed] [Google Scholar]
- 91.Rommelaere J, Geletneky K, Angelova AL, Daeffler L, Dinsart C, Kiprianova I, Schlehofer JR, Raykov Z. Oncolytic parvoviruses as cancer therapeutics. Cytokine Growth Factor Rev. 2010;21:185–195. doi: 10.1016/j.cytogfr.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 92.Blechacz B, Russell SJ. Parvovirus vectors: use and optimisation in cancer gene therapy. Expert Rev Mol Med. 2004;6:1–24. doi: 10.1017/S1462399404008026. [DOI] [PubMed] [Google Scholar]
- 93.Boisvert M, Fernandes S, Tijssen P. Multiple pathways involved in porcine parvovirus cellular entry and trafficking toward the nucleus. J Virol. 2010;84:7782–7792. doi: 10.1128/JVI.00479-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chipman PR, Agbandje-McKenna M, Kajigaya S, Brown KE, Young NS, Baker TS, Rossmann MG. Cryo-electron microscopy studies of empty capsids of human parvovirus B19 complexed with its cellular receptor. Proc Natl Acad Sci USA. 1996;93:7502–7506. doi: 10.1073/pnas.93.15.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barbis DP, Chang SF, Parrish CR. Mutations adjacent to the dimple of the canine parvovirus capsid structure affect sialic acid binding. Virology. 1992;191:301–308. doi: 10.1016/0042-6822(92)90192-r. [DOI] [PubMed] [Google Scholar]
- 96.Chang SF, Sgro JY, Parrish CR. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J Virol. 1992;66:6858–6867. doi: 10.1128/jvi.66.12.6858-6867.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tresnan DB, Southard L, Weichert W, Sgro JY, Parrish CR. Analysis of the cell and erythrocyte binding activities of the dimple and canyon regions of the canine parvovirus capsid. Virology. 1995;211:123–132. doi: 10.1006/viro.1995.1385. [DOI] [PubMed] [Google Scholar]
- 98.Govindasamy L, Hueffer K, Parrish CR, Agbandje-McKenna M. Structures of host range-controlling regions of the capsids of canine and feline parvoviruses and mutants. J Virol. 2003;77:12211–12221. doi: 10.1128/JVI.77.22.12211-12221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hueffer K, Govindasamy L, Agbandje-McKenna M, Parrish CR. Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J Virol. 2003;77:10099–10105. doi: 10.1128/JVI.77.18.10099-10105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hueffer K, Parker JS, Weichert WS, Geisel RE, Sgro JY, Parrish CR. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J Virol. 2003;77:1718–1726. doi: 10.1128/JVI.77.3.1718-1726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Agbandje M, McKenna R, Rossmann MG, Strassheim ML, Parrish CR. Structure determination of feline panleukopenia virus empty particles. Proteins. 1993;16:155–171. doi: 10.1002/prot.340160204. [DOI] [PubMed] [Google Scholar]
- 102.Xie Q, Chapman MS. Canine parvovirus capsid structure, analyzed at 2.9 A resolution. J Mol Biol. 1996;264:497–520. doi: 10.1006/jmbi.1996.0657. [DOI] [PubMed] [Google Scholar]
- 103.Lopez-Bueno A, Rubio MP, Bryant N, McKenna R, Agbandje-McKenna M, Almendral JM. Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J Virol. 2006;80:1563–1573. doi: 10.1128/JVI.80.3.1563-1573.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lopez-Bueno A, Segovia JC, Bueren JA, O'Sullivan MG, Wang F, Tattersall P, Almendral JM. Evolution to pathogenicity of the parvovirus minute virus of mice in immunodeficient mice involves genetic heterogeneity at the capsid domain that determines tropism. J Virol. 2008;82:1195–1203. doi: 10.1128/JVI.01692-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rubio MP, Lopez-Bueno A, Almendral JM. Virulent variants emerging in mice infected with the apathogenic prototype strain of the parvovirus minute virus of mice exhibit a capsid with low avidity for a primary receptor. J Virol. 2005;79:11280–11290. doi: 10.1128/JVI.79.17.11280-11290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Agbandje-McKenna M, Llamas-Saiz AL, Wang F, Tattersall P, Rossmann MG. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure. 1998;6:1369–1381. doi: 10.1016/s0969-2126(98)00137-3. [DOI] [PubMed] [Google Scholar]
- 107.Ball-Goodrich LJ, Tattersall P. Two amino acid substitutions within the capsid are coordinately required for acquisition of fibrotropism by the lymphotropic strain of minute virus of mice. J Virol. 1992;66:3415–3423. doi: 10.1128/jvi.66.6.3415-3423.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gardiner EM, Tattersall P. Mapping of the fibrotropic and lymphotropic host range determinants of the parvovirus minute virus of mice. J Virol. 1988;62:2605–2613. doi: 10.1128/jvi.62.8.2605-2613.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maxwell IH, Spitzer AL, Maxwell F, Pintel DJ. The capsid determinant of fibrotropism for the MVMp strain of minute virus of mice functions via VP2 and not VP1. J Virol. 1995;69:5829–5832. doi: 10.1128/jvi.69.9.5829-5832.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weigel-Kelley KA, Yoder MC, Srivastava A. Alpha5beta1 integrin as a cellular coreceptor for human parvovirus B19: requirement of functional activation of beta1 integrin for viral entry. Blood. 2003;102:3927–3933. doi: 10.1182/blood-2003-05-1522. [DOI] [PubMed] [Google Scholar]
- 111.Parker JS, Murphy WJ, Wang D, O'Brien SJ, Parrish CR. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J Virol. 2001;75:3896–3902. doi: 10.1128/JVI.75.8.3896-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.DeLano WL. The PyMOL Molecular Graphic System. DeLano Scientific; 2002. [Google Scholar]
- 113.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 114.Xiao C, Rossmann MG. Interpretation of electron density with stereographic roadmap projections. J Struct Biol. 2007;158:182–187. doi: 10.1016/j.jsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]


