Abstract
Numerous members of Ascomycota and Basidiomycota produce only poorly differentiated arthroconidial asexual morphs in culture. These arthroconidial fungi are grouped in genera where the asexual-sexual connections and their taxonomic circumscription are poorly known. In the present study we explored the phylogenetic relationships of two of these ascomycetous genera, Arthrographis and Arthropsis. Analysis of D1/D2 sequences of all species of both genera revealed that both are polyphyletic, with species being accommodated in different orders and classes. Because genetic variability was detected among reference strains and fresh isolates resembling the genus Arthrographis, we carried out a detailed phenotypic and phylogenetic analysis based on sequence data of the ITS region, actin and chitin synthase genes. Based on these results, four new species are recognised, namely Arthrographis chlamydospora, A. curvata, A. globosa and A. longispora. Arthrographis chlamydospora is distinguished by its cerebriform colonies, branched conidiophores, cuboid arthroconidia and terminal or intercalary globose to subglobose chlamydospores. Arthrographis curvata produced both sexual and asexual morphs, and is characterised by navicular ascospores and dimorphic conidia, namely cylindrical arthroconidia and curved, cashewnut-shaped conidia formed laterally on vegetative hyphae. Arthrographis globosa produced membranous colonies, but is mainly characterised by doliiform to globose arthroconidia. Arthrographis longispora also produces membranous colonies, but has poorly differentiated conidiophores and long arthroconidia. Morphological variants are described for A. kalrae and our results also revealed that Eremomyces langeronii and A. kalrae, traditionally considered the sexual and asexual morphs of the same species, are not conspecific.
Keywords: arthroconidial fungi, Arthrographis, Arthropsis, Eremomyces, phylogeny, taxonomy
INTRODUCTION
The arthroconidial genus Arthrographis was proposed by Cochet (1939) with A. langeronii as the type species, but it was invalid because it lacked a Latin diagnosis, which was at that time still required as prerequisite by the International Code of Botanical Nomenclature. Sigler & Carmichael (1976) subsequently validated the genus name based on Oidiodendron kalrae. In addition to the type species, A. kalrae, the genus currently includes three other taxa, A. alba, A. lignicola and A. pinicola (Sigler & Carmichael 1976, 1983, Sigler et al. 1990, Gené et al. 1996). Other species previously included in the genus were A. cuboidea and A. sulphurea. While the former species was transferred to the genus Scytalidum (Kang et al. 2010), A. sulphurea was considered a possible synonym of Pachysolen tannophilus (Saccharomycetes) (von Arx 1985).
Apart from A. kalrae, which was traditionally associated with the sexual morph Eremomyces langeronii, other ascomycetes have been described with unnamed Arthrographis morphs, i.e., Leucothecium coprophilum, L. emdenii and Faurelina indica (von Arx & Samson 1973, von Arx 1978, von Arx et al. 1981, Malloch & Sigler 1988, Valldosera et al. 1991).
Arthrographis species have been isolated from air, compost, marine sediments, soil, wood and, occasionally, from opportunistic infections in humans (de Hoog et al. 2011). Morphologically, they are recognised by a slow growth rate and by the presence of 1-celled, hyaline, smooth-walled, cylindrical arthroconidia released schizolytically from dendritic conidiophores (Sigler & Carmichael 1976). A particular feature of A. kalrae is the presence of a trichosporiella-like synasexual morph characterised by solitary, globose to subglobose conidia, which grow laterally and sessile on undifferentiated vegetative hyphae (Sigler & Carmichael 1983). Recently, a phylogenetic study based on sequences analysis of SSU, ITS and RPB2, revealed the polyphyly of Arthrographis (Kang et al. 2010).
Another arthroconidial genus morphologically similar to Arthrographis is Arthropsis. The genus comprises four species, i.e., Arthropsis cirrhata, A. hispanica, A. microsperma and A. truncata (Sigler et al. 1982, Sigler & Carmichael 1983, van Oorschot & de Hoog 1984, Ulfig et al. 1995). These fungi are usually reported from plant material, but A. hispanica, which was only known from marine sediments, has recently been isolated from clinical specimens (Giraldo et al. 2013). Arthropsis shows pigmented or non-pigmented arthroconidia, joined by adjacent connectives, released rhexolytically from undifferentiated conidiophores and occasionally has a Humicola synasexual morph (Sigler et al. 1982, van Oorschot & de Hoog 1984). Van Oorschot & de Hoog (1984) questioned the distinction between Arthrographis and Arthropsis, and suggested transferring Arthrographis species, excluding the type species A. kalrae, to the genus Arthropsis. Other authors, however, rejected this proposal (Malloch & Sigler 1988, Sigler et al. 1990).
In the present study we compared the D1/D2 sequences of the available types of Arthrographis and Arthropsis spp. with those of taxa retrieved from GenBank to clarify their taxonomy, and to determine their phylogenetic relationships. By combining morphological observations with multilocus DNA sequence analysis, several novel cryptic species of Arthrographis were delineated, which are newly described in this study.
MATERIALS AND METHODS
Isolates
The fungal isolates and DNA sequences included in the study are shown in Table 1. Twenty-six clinical Arthrographis isolates were provided by the Fungus Testing Laboratory at the University of Texas Health Science Center (UTHSC), the majority previously identified as A. kalrae by Giraldo et al. (2013). Because these isolates varied in morphology and their DNA sequence data, all isolates were re-examined in the present study. The type strains from the new species described here were deposited in the CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands.
Table 1.
Strains included in this study.
| Species | Strains1 | Origin | Previous identification | GenBank accession no.2 |
|||
|---|---|---|---|---|---|---|---|
| 28S rDNA | ITS | ACT1 | CHS1 | ||||
| Acarospora smaragdula | – | Unknown, Sweden | Acarospora smaragdula | AY853354 | – | – | – |
| Ajellomyces dermatitidis | ATCC 18187 T | Human, unknown | Ajellomyces dermatitidis | AY176704 | – | – | – |
| Amauroascus albicans | NRRL 5141 T | Soil, Honduras | Amauroascus albicans | – | – | – | – |
| Apinisia graminicola | CBS 156.77 | Skin lesion in dog, USA | Apinisia graminicola | AB040696 | – | – | – |
| CBS 721.68 T | Grass, United Kingdom | AY176709 | – | – | – | ||
| Aquaphila albicans | BCC 3520 | On wooden test block (Acacia oblonga), Thailand | Aquaphila albicans | DQ341102 | – | – | – |
| BCC 3543 | On wooden test block (Dipterocarpus alatus), Thailand | DQ341101 | – | – | – | ||
| Arachnomyces minimus | CBS 324.70 T | Decayed wood, Canada | Arachnomyces minimus | FJ358274 | – | – | – |
| Arachnotheca glomerata | CBS 348.71 T | Unknown, Central African Republic | Arachnotheca glomerata | AB075352 | – | – | – |
| Arthroderma cajetanum | UAMH 2937 | Single ascospore isolate from a gymnothecium on soil, unknown | Arthroderma cajetanum | AB075326 | – | – | – |
| Arthroderma ciferrii | CBS 272.66 T | Soil, USA | Arthroderma ciferrii | AB040681 | – | – | – |
| Arthrographis alba | CBS 370.92 T | Marine sediments, Spain | Arthrographis alba | HG004546 | AB213434 | – | – |
| Arthrographis arxii | CBS 203.78 T | Dung of herbivore, India | Eremomyces langeronii | AB213426 | GQ272638 | HG316563 | HG316582 |
| Arthrographis chlamydospora | UTHSC 06-1053 T | Urine, USA | Arthrographis sp. III | HG004543 | HG004554 | HG316560 | HG316580 |
| Arthrographis curvata | FMR 4032 | Marine sediments, Spain | Arthrographis sp. I | HG004539 | HG004557 | HG316557 | HG316577 |
| UTHSC 11-1163 T | Nails, USA | Arthrographis sp. I | HG004542 | HG004556 | HG316558 | HG316578 | |
| Arthrographis globosa | UTHSC 11-757 T | Bronchial wash, USA | Arthrographis sp. IV | HG004541 | HG004553 | HG316561 | HG316581 |
| Arthrographis kalrae | CBS 693.77 T | Sputum, India | Arthrographis kalrae | AB116544 | AB116536 | HG316544 | HG316564 |
| UTHSC 01-2742 | Artificial pulmonary valve, USA | – | HG004570 | – | – | ||
| UTHSC 04-2580 | Blood, USA | – | HG004569 | HG316545 | HG316565 | ||
| UTHSC 04-3423 | Toe nail, USA | – | HG004568 | HG316546 | HG316566 | ||
| UTHSC 05-17 | Blood, USA | – | HG004567 | HG316547 | HG316567 | ||
| UTHSC 06-982 | Pleural fluid, USA | – | HG004571 | – | – | ||
| UTHSC 06-3158 | Toe nail, USA | – | HG004572 | – | – | ||
| UTHSC 07-2450 | Eye, USA | – | HG004566 | HG316548 | HG316568 | ||
| UTHSC 08-527 | Lung tissue, USA | – | HG004573 | – | – | ||
| UTHSC 08-786 | Lung biopsy, USA | – | HG004565 | HG316549 | HG316569 | ||
| UTHSC 08-1699 | Bronchial wash, USA | – | HG004574 | – | – | ||
| UTHSC 08-1804 | Nails, USA | – | HG004564 | HG316550 | HG316570 | ||
| UTHSC 08-2107 | Leg, USA | – | – | – | – | ||
| UTHSC 08-3547 | Sputum , USA | – | HG004575 | – | – | ||
| UTHSC 09-141 | Lung biopsy, USA | – | HG004563 | HG316551 | HG316571 | ||
| UTHSC 09-2903 | Bronchial wash, USA | – | HG004576 | – | – | ||
| UTHSC 10-1652 | Cornea, USA | – | HG004562 | HG316552 | HG316572 | ||
| UTHSC 10-1719 | Cornea, USA | – | HG004577 | – | – | ||
| UTHSC 10-2021 | Catheter tip, USA | – | HG004561 | HG316553 | HG316573 | ||
| UTHSC 10-2583 | Urine, USA | – | HG004560 | HG316554 | HG316574 | ||
| UTHSC 10-2729 | Nasal sinus, USA | – | HG004559 | HG316555 | HG316575 | ||
| Arthrographis kalrae | UTHSC 11-1256 | Bronchial wash, USA | – | HG004558 | HG316556 | HG316576 | |
| UTHSC 11-302 | Eye, USA | – | – | – | – | ||
| Arthrographis lignicola | CBS 689.83 T | Gymnosperm wood chips and bark, Canada | Arthrographis lignicola | HG004547 | – | – | – |
| Arthrographis longispora | UTHSC 05-3220 T | Foot, USA | Arthrographis sp. II | HG004540 | HG004555 | HG316559 | HG316579 |
| Arthrographis pinicola | CBS 653.89 T | Gallery of Ips latidens in Pinus contorta, Canada | Arthrographis pinicola | HG004548 | – | – | – |
| Arthropsis cirrhata | CBS 628.83 T | Wall, The Netherlands | Arthropsis cirrhata | HG004549 | – | – | – |
| Arthropsis hispanica | CBS 351.92 T | Bottom of water deposit, Spain | Arthropsis hispanica | HE965759 | – | – | – |
| UTHSC 09-3174 | Bronchial wash, USA | HE965757 | – | – | – | ||
| Arthropsis microsperma | UAMH 4290 | Grass, England | Arthropsis microsperma | HG004551 | – | – | – |
| Arthropsis truncata | CBS 584.82 T | Leaf litter, Perú | Arthropsis truncata | HG004550 | – | – | – |
| Chalara longipes | NBRC 100564 | Decaying fir needles, Japan | Chalara longipes | – | – | – | – |
| Chlamydotubeufia huaikangplaensis | – | – | Chlamydotubeufia huaikangplaensis | JN865198 | – | – | – |
| Coniochaeta velutina | UAMH 10912 | Ex gametophytes of Hylcomium splendens, Canada | Coniochaeta velutina | EU999180 | – | – | – |
| Ctenomyces serratus | CBS 187.61 T | Soil, Australia | Ctenomyces serratus | AB040683 | – | – | – |
| Eremascus albus | CBS 975.69 | Unknown, USA | Eremascus albus | FJ358283 | – | – | – |
| Eremomyces bilateralis | CBS 781.70 T | Dung of pack rat, USA | Eremomyces bilateralis | HG004545 | – | HG316562 | – |
| Eurotium herbariorum | CBS 516.65 | Unpainted board, USA | Eurotium herbariorum | JF922029 | – | – | – |
| Faurelina indica | CBS 126.78 | Dung of cow, India | Faurelina indica | GU180654 | – | – | – |
| CBS 301.78 | Dung of goat, India | GU180653 | – | – | – | ||
| Geomyces pannorum | UAMH 10473 | Ex biofilm on soil, United Kingdom | Geomyces pannorum | GU951697 | – | – | – |
| Gymnascella aurantiaca | CBS 655.71 T | Clay soil, USA | Gymnascella aurantiaca | AB040684 | – | – | – |
| Gymnascella hyalinospora | CBS 548.72 T | Dung of Guinea pig, India | Gymnascella hyalinospora | AB040687 | – | – | |
| Gymnoascoideus petalosporus | UAMH 3593 | Tinea pedis, human, USA | Gymnoascoideus petalosporus | AB359428 | – | – | |
| Gymnoascus reesii | CBS 410.72 T | Soil, USA | Gymnoascus reesii | JF922021 | – | – | – |
| Helicomyces macrofilamentosus | HKUCC 10235 | – | Helicomyces macrofilamentosus | AY849942 | – | – | – |
| Hyalodendriella betulae | CBS 261.82 | Alnus glutinosa, The Netherlands | Hyalodendriella betulae | EU040232 | – | – | – |
| Hysterographium fraxini | CBS 109.43 | Unknown, Switzerland | Hysterographium fraxini | FJ161171 | – | – | – |
| CBS 242.34 | Unknown, Canada | FJ161189 | – | – | – | ||
| Lambertella brunneola | NBRC 6894 | Aucuba japonica, Japan | Lambertella brunneola | – | – | – | – |
| Leucothecium emdenii | CBS 576.73 T | Agricultural soil, The Netherlands | Leucothecium emdenii | FJ358286 | – | – | – |
| Malbranchea aurantiaca | CBS 127.77 T | Culture contaminant, USA | Malbranchea aurantiaca | AB040704 | – | – | – |
| Malbranchea cinnamomea | CBS 960.72 | Unknown, France | Malbranchea cinnamomea | JF922020 | – | – | – |
| Mallochia reticulata | CBS 392.61 T | Rhizosphere of Musa sapientum, Honduras | Mallochia reticulata | AB075320 | – | – | – |
| Monascus lunisporas | CBS 113675 | Soil, Brazil | Monascus lunisporas | JF922026 | – | – | – |
| Monascus ruber | FRR 2447 T | Soil, India | Monascus ruber | JF922025 | – | – | – |
| Onygena corvina | JCM 9546 | Decaying bone, Japan | Onygena corvina | AB075355 | – | – | – |
| Onygena equina | CBS 947.70 | Cow hoof, Germany | Onygena equina | AB075356 | – | – | – |
| Ostreichnion curtisii | CBS 198.34 | On Quercus sp., USA | Ostreichnion curtisii | FJ161176 | – | – | – |
| Polytolypa hystricis | UAMH 7299 | Ex porcupine dung, Canada | Polytolypa hystricis | AY176718 | – | – | – |
| Porosphaerella borinquensis | ICMP 15117 | Wood, New Zealand | Porosphaerella borinquensis | EF063573 | – | – | – |
| Pseudoarachniotus trochleosporus | CBS 591.71 | Soil, USA | Pseudoarachniotus trochleosporus | AB075344 | – | – | – |
| Pycnora xanthococca | – | Unknown, Sweden | Pycnora xanthococca | AY853388 | – | – | – |
| Rhexothecium globosum | CBS 955.73 T | Desert soil, Egypt | Rhexothecium globosum | HG004544 | – | – | – |
| Rutstroemia cuniculi | NBRC 9671 | Dung of rabbit, England | Rutstroemia cuniculi | – | – | – | – |
| Rutstroemia paludosa | NBRC 9672 | On Symplocarpus foetidus, USA | Rutstroemia paludosa | – | – | – | – |
| Sarcoleotia globosa | – | – | Sarcoleotia globosa | AY789409 | – | – | – |
| Sarea resinae | – | – | Sarea resinae | AY640965 | – | – | – |
| Scleromitrula shiraiana | NBRC 30255 | On Morus bombycis, unknown | Scleromitrula shiraiana | – | – | – | – |
| Scytalidium cuboideum | UAMH 7144 | Ex lingula specimen, USA | Scytalidium cuboideum | AB213427 | – | – | – |
| UAMH 8435 | Bronchial washing, USA | AB213428 | – | – | – | ||
| Shanorella spirotricha | CBS 304.56 | Dung of rabbit, USA | Shanorella spirotricha | FJ358288 | – | – | – |
| Stromatinia gladioli | NBRC 7169 | – | Stromatinia gladioli | – | – | – | – |
| Trichophyton ajelloi var. ajelloi | – | – | Trichophyton ajelloi var. ajelloi | AB075329 | – | – | – |
1 ATCC: American Type Culture Collection, Manassas, VA, USA; BCC: Biotec Culture Collection, National Center for Genetic Engineering and Biotechnology (BIOTEC), Bangkok, Thailand; CBS: CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands; FFR: culture collection of CSIRO, Australia; FMR: Faculty of Medicine Reus, Spain; HKUCC, Hong Kong University Culture Collection, Department of Ecology and Biodiversity, Hong Kong, China; ICMP: International Collection of Microorganisms, Landcare Research, Auckland, New Zealand; JCM: Japanese collection of microorganism; NBRC: NITE Biological Resource Center, Japan; UAMH: University of Alberta Microfungus Collection and Herbarium; Edmonton, Canada; UTHSC: Fungus Testing Laboratory, University of Texas Health Science Center, San Antonio, TX, USA; T = Ex-type strain.
2 Accession numbers of sequences newly determined in this study are indicated in bold. ITS: internal transcribed spacer regions of the nrDNA and intervening 5.8S nrDNA; ACT1: partial actin gene; CHS1: chitin synthase gene.
Phenotypic studies
Isolates were studied following the criteria of Sigler & Carmichael (1976, 1983) and Ulfig et al. (1995). Morphological features were examined on potato dextrose agar (PDA; Pronadisa, Madrid, Spain), 2 % malt extract agar (MEA; BD Difco™, Franklin Lakes, NJ, USA), potato carrot agar (PCA; potatoes, 20 g; carrot, 20 g; agar, 20 g; distilled water to final volume of 1 000 mL) and oatmeal agar (OA; filtered oat flakes after 1 h of simmering, 30 g; agar, 20 g; distilled water to final volume of 1 000 mL). Cultures were incubated at 25 °C in the dark for 4 wk. Colony diameters were measured after 14 d of incubation and rated according to the colour charts of Kornerup & Wanscher (1978). Microscopic features were examined and measured in either 85 % lactic acid or lactophenol cotton blue under a light microscope Olympus CH-2 (Olympus Corporation, Tokyo, Japan). Photomicrographs were obtained with a Zeiss Axio-Imager M1 light microscope (Zeiss, Oberkochen, Germany), using phase contrast and Nomarski differential interference.
The ability of the fungi to grow at 15, 20, 25, 30, 35, 37, 40, 42 and 45 °C was determined on PDA. To determine the resistance to cycloheximide, isolates were transferred to Petri dishes containing PCA supplemented with chloramphenicol (200 mg/L) and cycloheximide at a final concentration of 2 g/L, and incubated at 25 °C for 2 wk. All tests were performed in duplicate. To evaluate the ability of isolates to convert to the yeast phase, a portion from a fresh culture on PDA was transferred to tubes with Brain Heart Infusion broth (BHI; Becton Dickinson & Company, Franklin Lakes, NJ, USA) and incubated at 37 °C for 2 wk. Subsequently, several transfers to BHI broth were performed.
DNA extraction, amplification and sequencing
Isolates were grown on yeast extract sucrose agar (YES; yeast extract, 20 g; sucrose, 150 g; agar, 20 g; distilled water to final volume of 1 000 mL) for 10 d at 25 °C and DNA extracted using PrepMan Ultra sample preparation reagent (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s protocol. The DNA was quantified using NanoDrop 3000 (ThermoScientific, Asheville, NC, USA). The internal transcribed spacer (ITS) regions and D1/D2 domains of the 28S rDNA were amplified with the primer pairs ITS5/ITS4, NL1/NL4b and LR0R/LR5 (Vilgalys & Hester 1990, White et al. 1990, O’Donnell 1993). A portion of the actin gene (ACT1) was amplified using the primer set Act1/Act4 (Voigt & Wöstemeyer 2000) and a chitin synthase gene (CHS1) using the primers CHS-79F/CHS-354R (Carbone & Kohn 1999). PCR products were purified and sequenced at Macrogen Europe (Amsterdam, The Netherlands). The program SeqMan v. 7.0.0 (DNASTAR, Madison, WI, USA) was used to obtain consensus sequences of each isolate. In addition, numerous D1/D2 sequences, corresponding to different classes, orders and families of ascomycetes retrieved from GenBank or NITE/NRBC databases were included in the phylogenetic study (Table 1). Most of these sequences were published by different authors (Sugiyama et al. 1999, Sugiyama & Mikawa 2001, Untereiner et al. 2002, Reeb et al. 2004, Xi et al. 2004, Murata et al. 2005, Wang et al. 2005, Wedin et al. 2005, Kodsueb et al. 2006, Réblová & Seifert 2007, Tsui et al. 2007, Gueidan et al. 2008, Boehm et al. 2009, Sugiyama et al. 2002, Boonmee et al. 2011, Pettersson et al. 2011, Réblová et al. 2011, Giraldo et al. 2013). The selection of these sequences was based on the results of a BLAST search using the D1/D2 and ITS sequences from each of the ex-type strains of the different species of Arthrographis and Arthropsis.
Phylogenetic analysis
Sequences were aligned using Clustal X v. 1.8 (Thompson et al. 1997) with default parameters, followed by manual adjustments with a text editor. The phylogenetic relationship between Arthrographis and Arthropsis species with other genera was determined through the analysis of D1/D2 sequences. Since genetic and also morphological variability was detected among isolates of Arthrographis, a multi-locus sequence analysis was carried out to confirm the results obtained from D1/D2 data. This analysis included a fragment of the ACT1 gene, the CHS1 gene and the ITS region. Phylogenetic analyses were performed with MEGA v. 5.05 (Tamura et al. 2011), using the Maximum Composite Likelihood (ML). The selection of the best nucleotide substitution model (Tamura-Nei with Gamma distribution) was made using the model selection analysis under MEGA v. 5.05. Gaps or missing data were treated as partial deletion with a site coverage cut-off of 95 % and Nearest-Neighbour-Interchange (NNI) used as Heuristic method. The internal branch support was assessed by a search of 1 000 bootstrapped sets of data. DNA sequence data were deposited in GenBank (Table 1), the alignment and trees in TreeBASE (http://www.treebase.org) and taxonomic novelties in MycoBank (http://www.MycoBank.org; Crous et al. 2004).
RESULTS
Phylogenetic analyses
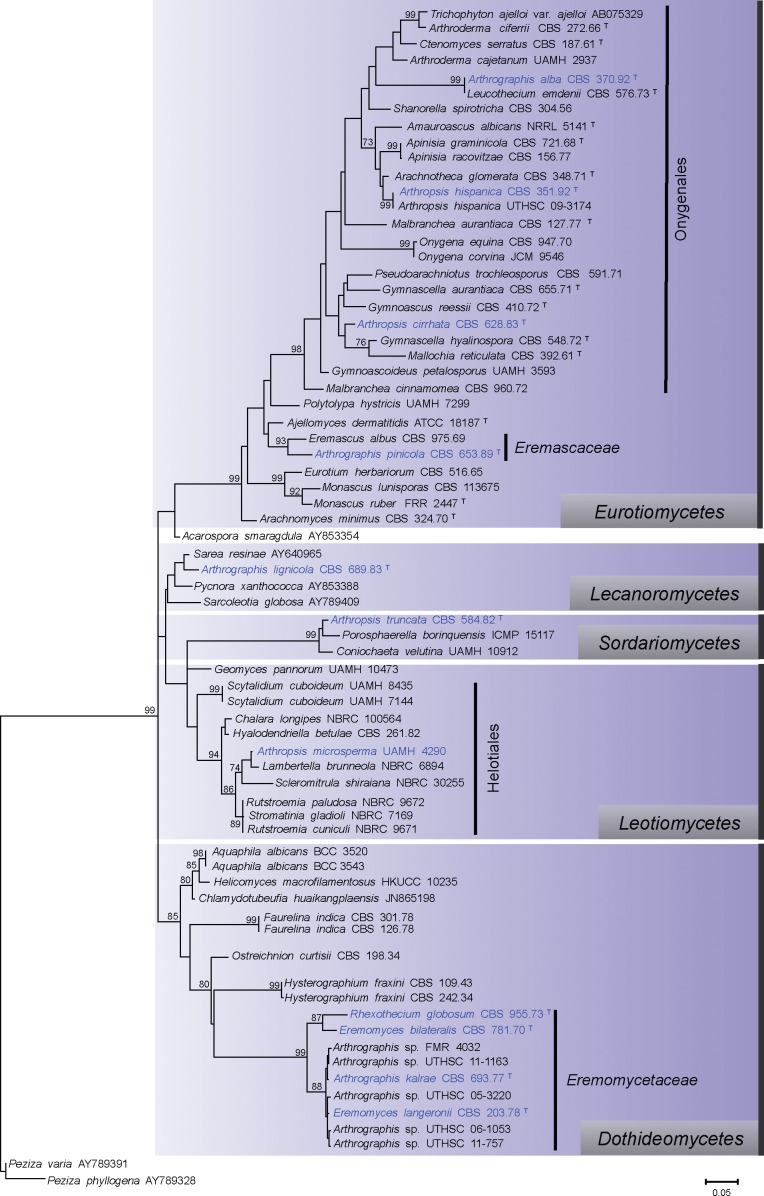
The D1/D2 phylogenetic tree that included the ex-type strains of the different species of Arthrographis and Arthropsis and representative members of different fungal classes and orders revealed that both genera are polyphyletic (Fig. 1). The type species of Arthrographis, A. kalrae, was included in a well-supported clade within the Dothideomycetes (85 % bootstrap, bs), forming together a highly supported subclade (99 % bs) with Rhexothecium globosum, Eremomyces bilateralis, E. langeronii and four unidentified species of Arthrographis. While the ex-type strain of Arthrographis lignicola was related to different genera of the Lecanoromycetes, such as Sarea and Pycnora, the ex-type strains of Arthrographis pinicola and A. alba were associated with the Eurotiomycetes (99 % bs). Arthrographis pinicola and Eremascus albus (Eremascaceae) formed a well-supported clade (93 % bs), while A. alba and Leucothecium emdenii formed a well-supported clade (99 % bs) within the Onygenales.
Fig. 1.
Maximum-likelihood (ML) tree constructed with sequences of the D1/D2 domains of the 28S rRNA gene. Bootstrap support values above 70 % are indicated at the nodes. The phylogenetic tree was rooted to Peziza varia and Peziza phyllogena. T = Ex-type strain.
The type species of Arthropsis, A. truncata, clustered with Porosphaerella borinquensis and Coniochaeta velutina (99 % bs), both members of Sordariomycetes. Arthropsis cirrhata and A. hispanica were accommodated within the Onygenales. The only available reference strain of A. microsperma grouped with different members of the Helotiales (94 % bs).
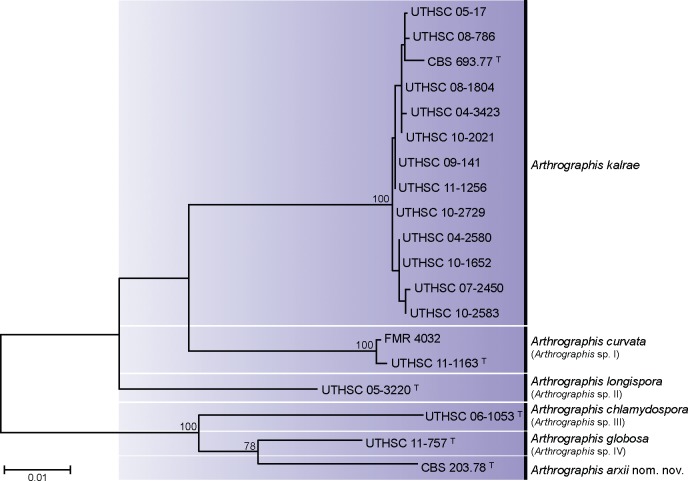
The multilocus sequence analysis was carried out with the ex-type strains of Arthrographis kalrae (CBS 693.77) and Eremomyces langeronii (CBS 203.78), 12 isolates identified as A. kalrae and five isolates identified as an Arthrographis sp. Due to the low intra-specific variability detected in the ITS sequences among the 22 isolates identified as A. kalrae (98.5–100 % similarity), we selected 12 isolates that represented the most characteristic morphological variants observed.
With the primers used, we were able to amplify and sequence 300–350 bp, 450–500 bp and 750–820 bp of the CHS1 gene, the ITS region and ACT1 gene, respectively. The topology of the combined ML tree was similar to trees based on individual genes (data not shown). The combined tree included 1 544 bp and showed four main lineages (Fig. 2). The largest lineage was represented by a clade with 12 clinical strains of A. kalrae, including the ex-type strain. Sequences within the clade were practically identical, showing similarities of 98.5–100 % for each over the three loci. The second lineage (Arthrographis sp. I, 100 % bs) included one strain from marine sediments (FMR 4032) and another from nails (UTHSC 11-1163), with an intra-specific similarity of 99–100 %. The third lineage (Arthrographis sp. II) comprised only one strain (UTHSC 05-3220) from clinical origin (foot). Finally, the fourth lineage comprised a clade with three strains separated from each other by a considerable genetic distance. They were the clinical strains UTHSC 06-1053 (Arthrographis sp. III) and UTHSC 11-757 (Arthrographis sp. IV) and CBS 203.78, the ex-type strain of E. langeronii from herbivore dung. The latter two strains formed a well-supported subclade and showed genetic similarities that ranged from 93.8 % for ITS region to 96–96.7 % for CHS1 and ACT1 genes. Surprisingly, the ex-type strains of A. kalrae and E. langeronii were located in two different clades, showing genetic similarities of 94.1 % for ITS region and 92.8 % and 88.1 % for ACT1 and CHS1 genes, respectively.
Fig. 2.
Maximum-likelihood (ML) tree obtained from the combined DNA sequence data from three loci (ITS, ACT1, CHS1). Bootstrap support values above 70 % are indicated at the nodes. T = Ex-type strain.
Phenotypic studies
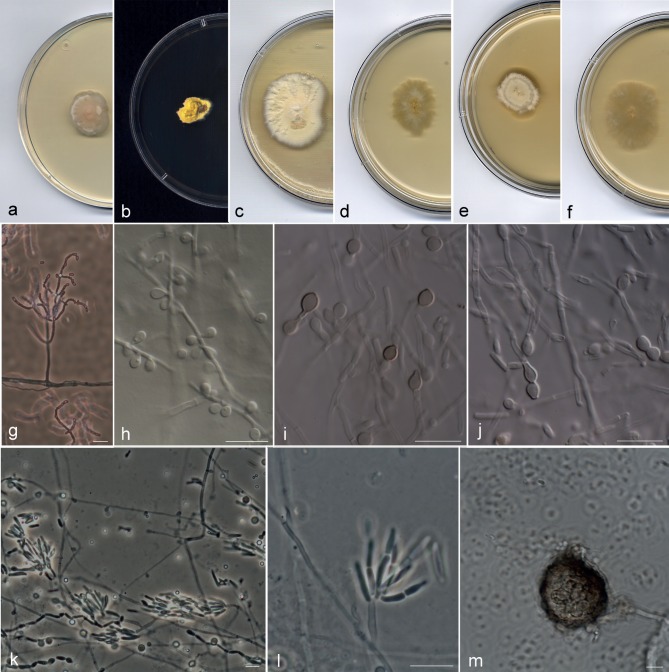
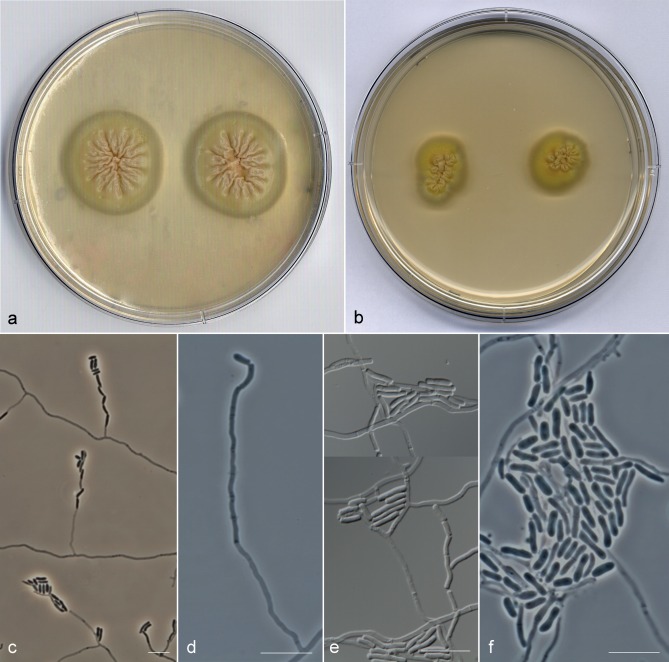
Most of the strains included in the A. kalrae clade (Fig. 2) showed the typical phenotypic characters described for the species; i.e., colonies at 25 °C with slow to moderate growth (up to 10–21 mm diam after 10 d on PDA), flat to slightly folded, initially beige and moist with a yeast-like appearance, becoming tan or yellowish and powdery to granular (Fig. 3a–f); conidiophores hyaline and usually branched (Fig. 3g); conidiogenous hyphae hyaline, simple or branched; arthroconidia 1-celled, hyaline, smooth-walled, cylindrical with truncate ends, 2.5–9 × 1–2 μm. All strains formed a trichosporiella-like synasexual morph with sessile, globose to subglobose, hyaline, thin and smooth-walled conidia, 2–4 × 2–3 μm (Fig. 3h). Several strains showed some atypical characters not previously described for this species. The strains UTHSC 02-1022, UTHSC 06-982, UTHSC 07-2450, UTHSC 08-1804, UTHSC 08-2107, UTHSC 10-1652, UTHSC 10-2583 and UTHSC 11-1256 produced intercalary or terminal chlamydospores with smooth or slightly rugose walls. While in most of these isolates the chlamydospores were hyaline to subhyaline, those of strain UTHSC 11-1256 turned brown on PDA and OA (Fig. 3i, j) giving a dark pigmentation to the colony. The UTHSC 05-17 strain showed a predominance of small conidiophores (up to 70 μm long) composed of a terminal whorl of numerous short chains of clavate or cylindrical arthroconidia with rounded ends (Fig. 3k, l); in old cultures (12 wk) this isolate developed immature ascomata submerged in the agar of all media tested. These ascomata were spherical, non-ostiolate, 37–70 μm diam, with a dark brown, pseudoparenchymatous peridium of textura angularis, surrounded by brown hyphae (Fig. 3m).
Fig. 3.
Arthrographis kalrae. a–c. Colonies on PDA after 21 d at 25 °C; d–f. colonies on MEA 2 % at 25 °C after 21 d; g. branched conidiophores; h. lateral sessile conidia; i, j. pigmented chlamydospores and hyaline arthroconidia; k, l. whorls of short arthroconidial chains; m. sterile ascoma (a, d. CBS 693.77; b, e. UTHSC 09-141; c, f–h, k–m. UTHSC 05-17; i, j. UTHSC 11-1256). — Scale bars = 10 μm.
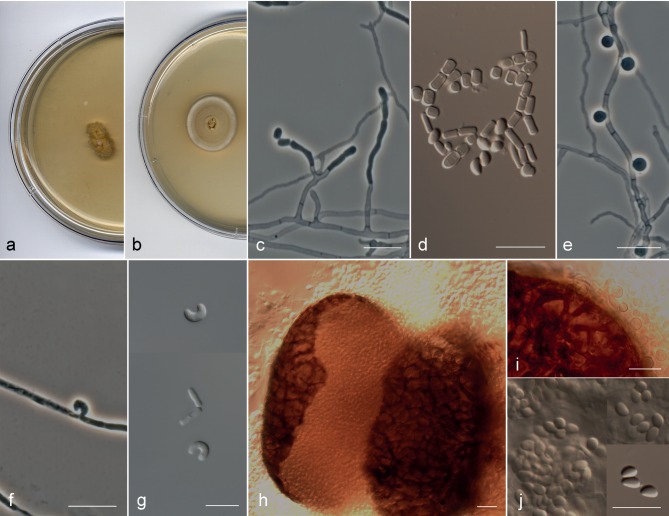
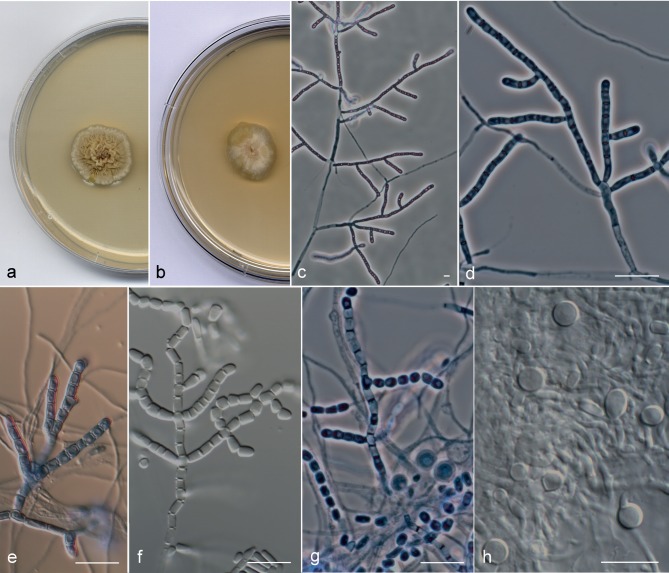
Arthrographis sp. I (FMR 4032 and UTHSC 11-1163) (Fig. 5a–j) showed similar morphological characteristics to those of the A. kalrae clade, but differed in the following features: the colonies on MEA 2 % were orange-yellow (4B8) and showed a very slow growth (6–7 mm diam in 14 d) (Fig. 5a); in addition to the trichosporiella-like synasexual morph (Fig. 5e), both strains produced on PDA at 25 °C and BHI at 37 °C curved and cashewnut-shaped sessile conidia formed laterally on undifferentiated hyphae (Fig. 5f, g); and the strain UTHSC 11-1163 produced superficial spherical ascomata with evanescent asci and navicular ascospores (Fig. 5h–j).
Fig. 5.
Arthrographis curvata. a, b. Colonies on MEA 2 % and PDA, respectively, after 21 d at 25 °C. — c–g. Asexual morph: c. simple or poorly branched conidiophores; d. arthroconidia and ascospores; e. lateral sessile conidia. f, g. curved conidia produced in BHI at 37 °C. — h–j. Sexual morph: h. ascoma; i. dark brown hyphae on the peridium; j. ascospores (a–c, e, g. FMR 4032; d, f, h–j. UTHSC 11-1163). — Scale bars = 10 μm.
The lineage representing Arthrographis sp. II (UTHSC 05-3220), produced membranous colonies in all the media tested (Fig. 7a, b), conidiophores poorly differentiated (Fig. 7c, d) and arthroconidia longer (5–10(–13) μm) than those of the members of A. kalrae clade (Fig. 7e, f). In this strain, as in Arthrographis sp. III and Arthrographis sp. IV, the production of a trichosporiella-like synasexual morph was not observed.
Fig. 7.
Arthrographis longispora UTHSC 05-3220. a, b. Colonies on PDA and MEA 2 %, respectively, after 21 d at 25 °C; c, d. poorly differentiated conidiophores; e, f. cylindrical arthroconidia with truncate or rounded ends. — Scale bars = 10 μm.
Arthrographis sp. III displayed umbonate, cerebriform and velvety colonies on PDA (Fig. 4a), branched conidiophores (Fig. 4c, d), cylindrical, cubic and doliiform arthroconidia (Fig. 4e–g) and terminal or intercalary globose chlamydospores (Fig. 4h).
Fig. 4.
Arthrographis chlamydospora UTHSC 06-1053. a, b. Colonies on PDA and MEA 2 %, respectively, after 21 d at 25 °C; c, d. branched conidiophores; e, f. conidiogenous hyphae fragmenting schizolytically; g. cylindrical and doliiform arthroconidia; h. chlamydospores. — Scale bars = 10 μm.
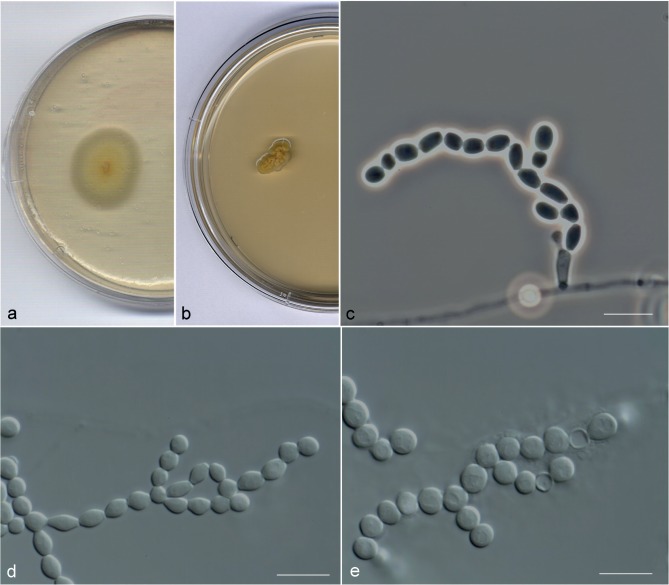
Finally, the most representative morphological characters observed in Arthrographis sp. IV were the production of membranous colonies (Fig. 6a, b), poorly differentiated conidiophores (Fig. 6c) and doliiform, ellipsoidal, slightly fusiform or globose arthroconidia (Fig. 6d, e).
Fig. 6.
Arthrographis globosa UTHSC 11-757. a, b. Colonies on PDA and MEA 2 %, respectively, after 21 d at 25 °C; c. poorly differentiated conidiophores; d. conidiogenous hyphae fragmenting schizolytically producing ellipsoidal, doliiform, slightly fusiform and globose arthroconidia; e. globose arthroconidia. — Scale bars = 10 μm.
All strains grouped in the clade of A. kalrae were able to grow at all the temperatures tested, attaining up to 30 mm diam at 40 °C and 5–15 mm at 45 °C on PDA after 14 d. Arthrographis sp. I and Arthrographis sp. III grew well at 37 °C (13–16 mm diam after 14 d), but at 40 °C the growth of both species was restricted (6–7 mm diam after 14 d). Conversely, Arthrographis sp. II and sp. IV were not able to grow at 37 °C. All isolates tolerated high doses of cycloheximide (2 g/L). Only isolates of A. kalrae were able to convert to a yeast phase, producing oval to ellipsoidal (2.5 × 4 μm) yeast-like budding cells at 37 °C after several transfers in BHI broth.
Taxonomy
On the basis of the morphological features observed, which correlated with the phylogenetic analysis, we concluded that Arthrographis spp. I–IV are different from the taxa currently accepted in this genus and are therefore described here as new. These species are named A. chlamydospora, A. curvata, A. globosa and A. longispora. In addition, the new name Arthrographis arxii is proposed for the ascomycete Eremomyces langeronii.
Arthrographis arxii Guarro, Giraldo, Gené & Cano, nom. nov. — MycoBank MB804634
Basionym. Pithoascus langeronii Arx, Persoonia 10: 24. 1978.
≡Pithoascina langeronii (Arx) Valmaseda, T.A. Martínez & Barrasa, Canad. J. Bot. 65: 1805. 1987.
≡Eremomyces langeronii (Arx) Malloch & Sigler, Canad. J. Bot. 66: 1931. 1988.
Etymology. The specific epithet is given in honour of the mycologist Josef Adolf von Arx (1922–1988), who actively published on this group of fungi.
Notes — Since our results demonstrated that A. kalrae and E. langeronii are not conspecific, and the name A. langeronii was occupied, a new name is proposed for E. langeronii.
Arthrographis chlamydospora Giraldo, Deanna A. Sutton,
Gené & Madrid, sp. nov. — MycoBank MB804632; Fig. 4
Etymology. Referring to the presence of chlamydospores.
Colonies on PDA at 25 °C attaining 15–16 mm diam after 14 d, pale to greyish orange (5A–B3) with whitish margin, umbonate, cerebriform, velvety. On OA and PCA at 25 °C attaining 23–25 mm and 15–16 mm diam, respectively, after 14 d, orange-white (5A2), flat, powdery or granulose. On MEA 2 % at 25 °C attaining 14–15 mm diam in 14 d, orange-yellow (4B8), flat, radially striated, granulose. At 37 °C on PDA the colonies attaining 12–13 mm diam after 14 d, brownish orange (6C3–4), cerebriform, velvety. Vegetative hyphae septate, hyaline, smooth- and thin-walled, 1.5–2 μm wide. Conidiophores mostly repeatedly branched, erect, up to 350 μm long, hyaline, smooth-walled. Conidiogenous hyphae simple or laterally branched, 1.5–2.5 μm wide, thick-walled, forming septa basipetally to form arthroconidia released via schizolythic secession. Arthroconidia unicellular, cylindrical, cuboid or doliiform, straight, 3–6(–7) × 1.5–2.5 μm, hyaline to subhyaline, thick- and smooth-walled. Chlamydospores terminal or intercalary, solitary, unicellular, globose or subglobose, 5–6 × 5–6 μm, hyaline, rough- and thick-walled, strongly chromophilic. Sexual morph and trichosporiella-like synasexual morph not observed.
Cardinal temperature for growth — Optimum 25–30 °C, maximum 42 °C, minimum 15 °C. The fungus was unable to grow at 45 °C.
Specimen examined. USA, Florida, from human urine, D.A. Sutton (holotype CBS H-21346, cultures ex-type CBS 135396, FMR 12129, UTHSC 06-1053).
Arthrographis curvata Giraldo, Gené, Deanna A. Sutton & Cano, sp. nov. — MycoBank MB804630; Fig. 5
Etymology. Referring to the presence of curved conidia.
Colonies on PDA at 25 °C attaining 17–19 mm diam in 14 d, pale to greyish orange (5A–B3) with whitish margin, umbonate at centre and flat toward the periphery, powdery. On OA and PCA at 25 °C attaining 19–20 mm and 24–26 mm diam, respectively, after 14 d, whitish, flat, dusty. On MEA 2 % at 25 °C attaining 6–7 mm diam in 14 d, orange-yellow (4B8), elevated, cerebriform, membranous. At 37 °C on PDA the colonies attaining 15–16 mm diam after 14 d, orange-grey (5B2), flat, powdery. Vegetative hyphae septate, hyaline, smooth- and thin-walled, 1.5–2 μm wide. Ascomata cleistothecial, superficial, spherical, brown, 52–132 μm diam, peridium pseudoparenchymatous with textura angularis, surrounded by dark brown, thick-walled hyphae. Asci evanescent, globose, thin-walled. Ascospores unicellular, navicular in lateral view, ellipsoidal in front view, thin- and smooth-walled, without germ pores, 2.8–3.8 × 1.4–2 μm, hyaline to pale brown in mass. Conidiophores poorly differentiated, erect, simple or slightly branched, up to 35 μm long, hyaline, smooth-walled. Conidiogenous hyphae simple or branched, 1–2 μm wide, thin-walled, forming septa basipetally to form arthroconidia released by schizolythic secession. Arthroconidia unicellular, cylindrical or short-cylindrical, straight or slightly curved, 3–4.5(–7) × 1–2 μm, hyaline to subhyaline, thin- and smooth-walled. Synasexual morph trichosporiella-like with conidia growing directly on undifferentiated hyphae, lateral, sessile, globose, 2–3 μm diam, hyaline and smooth-walled. On PDA and BHI at 25 °C and 37 °C, respectively, conidia were occasionally observed to be unicellular, curved, cashewnut-shaped, hyaline and smooth-walled, 3.5–6 × 1.5–2 μm, growing solitary and sessile on vegetative hyphae.
Cardinal temperature for growth — Optimum 25–30 °C, maximum 42 °C, minimum 15 °C. The fungus was unable to grow at 45 °C.
Specimens examined. SPAIN, Amposta, Ebro river, from river bank, K. Ulfig (CBS 135934, FMR 4032). – USA, Colorado, from human nails, D.A. Sutton (holotype CBS H-21344, cultures ex-type CBS 135933, FMR 12125, UTHSC 11-1163).
Notes — The GenBank sequences AB128973.1 (ITS region) and AB128975 (28S rDNA), corresponding to the isolate E. langeronii UAMH 7600 from a fingernail, were 99.6 and 100 % (ITS region and 28S rDNA, respectively) similar to those of the type species of A. curvata.
Arthrographis globosa Giraldo, Deanna A. Sutton, Cano & Guarro, sp. nov. — MycoBank MB804633; Fig. 6
Etymology. Referring to the presence of globose conidia.
Colonies on PDA at 25 °C attaining 22–24 mm diam after 14 d, buttercup yellow (4A7), flat, membranous. On OA and PCA at 25 °C attaining 14–15 mm diam after 14 d, whitish, flat, at first glabrous becoming slightly powdery. On MEA 2 % at 25 °C attaining 4–5 mm diam in 14 d, orange-yellow (4A8), cerebriform, membranous. Vegetative hyphae septate, hyaline, with golden pigment accumulation inside, smooth- and thin-walled, 1.5 μm wide. Conidiophores absent or poorly differentiated, hyaline, smooth-walled. Conidiogenous hyphae simple or branched, 1–1.5 μm wide, thin-walled, forming septa basipetally to form arthroconidia released via schizolythic secession. Arthroconidia unicellular, doliiform, ellipsoidal, slightly fusiform or globose, 3–5(–6.5) × 2–4 μm, hyaline, thick- and smooth-walled. Sexual morph, trichosporiella-like synasexual morph and chlamydospores not observed.
Cardinal temperature for growth — Optimum 25–30 °C, maximum 35 °C, minimum 15 °C. The fungus was unable to grow at 37 °C.
Specimen examined. USA, Texas, from human bronchial wash, D.A. Sutton (holotype CBS H-21347, cultures ex-type CBS 135397, FMR 12124, UTHSC 11-757).
Arthrographis longispora Giraldo, Deanna A. Sutton, Cano & Guarro, sp. nov. — MycoBank MB804631; Fig. 7
Etymology. Referring to the length of the arthroconidia.
Colonies on PDA at 25 °C attaining 18–21 mm diam after 14 d, yellowish orange (4A7), radially folded or rugose at centre and flat toward the periphery, membranous. On OA and PCA at 25 °C attaining 18–21 mm and 8–9 mm diam, respectively, after 14 d, whitish, flat, at first glabrous becoming slightly powdery. On MEA 2 % at 25 °C attaining 11–12 mm diam in 14 d, orange-yellow (4B8), cerebriform at centre and flat toward the periphery, membranous. Vegetative hyphae septate, hyaline, with golden pigment accumulation inside, smooth- and thin-walled, 1.5–2 μm wide. Conidiophores poorly differentiated, erect, up to 60 μm long, hyaline, smooth-walled. Conidiogenous hyphae, simple, occasionally slightly branched, 1–1.5 μm wide, thin-walled, septating basipetally to form arthroconidia released by schizolythic secession. Arthroconidia unicellular, cylindrical with truncated or rounded ends, straight or slightly curved, 5–10(–13) × 1–1.5 μm, hyaline, thin- and smooth-walled. Sexual morph, trichosporiella-like synasexual morph and chlamydospores not observed.
Cardinal temperature for growth — Optimum 25–30 °C, maximum 35 °C, minimum 15 °C. The fungus was unable to grow at 37 °C.
Specimen examined. USA, Utah, from human foot, D.A. Sutton (holotype CBS H-21345, cultures ex-type CBS 135935, FMR 12101, UTHSC 05-3220).
DISCUSSION
The genus Arthrographis was traditionally considered a member of the Eremomycetaceae, Dothideomycetes (Malloch & Sigler 1988). However, our D1/D2 analysis demonstrated that only the type species, A. kalrae, and the new taxa proposed here (i.e., A. arxii, A. chlamydospora, A. curvata, A. globosa and A. longispora) are members of the family, and that the name Arthrographis should be restricted to these species. The other species previously attributed to the genus are phylogenetically distant from the type. Arthrographis lignicola belongs to the Lecanoromycetes, forming a weakly supported clade with Sarea resinae, Pycnora xanthococca and Sarcoletia globosa. Although a BLAST search using D1/D2 and ITS sequences of A. lignicola showed close relationships with other members of that class, we could not include more sequences of Lecanoromycetes in our phylogenetic analysis due to the difficulties in performing a reliable alignment. Arthrographis pinicola and A. alba are accommodated in the Eurotiomycetes, more particularly, the former in Eremascaceae, closely related to Eremascus albus, and the latter in the Gymnoascaceae, closely related to Leucothecium emdenii. Arthrographis alba was described based on several isolates from different origins and, although some morphological similarity with the anamorph of L. emdenii was already mentioned, none of those isolates developed the sexual morph (Gené et al. 1996). The present study reveals that sequences from D1/D2 and ITS of both species are practically identical (data not shown), which indicates that A. alba must be considered the asexual morph of L. emdenii. The genus Leucothecium was described by von Arx & Samson (1973) to accommodate ascomycetes with yellowish globose ascomata, hyaline peridium, bivalve-lenticular ascospores and asexual morphs with hyaline arthroconidia. Currently, the genus comprises two species, L. emdenii, the type species, and L. coprophilum, both traditionally included in Gymnoascaceae on the basis of the morphology of the asexual morph and ascospores (von Arx & Samson 1973, Valldosera et al. 1991).
The ascomycete Faurelina indica also produces an arthrographis-like asexual morph similar to the Arthrographis anamorph of E. langeronii (von Arx 1978, von Arx et al. 1981). The genus Faurelina, with a coprophilous habitat, was originally included in the family Chadefaudiellaceae, Microascales (Locquin-Linard 1975, Cannon & Kirk 2007). This genus was characterised by pustulate or hemispherical ascomata, a peridium composed of vertical rows of dark cells, asci arranged in vertical chains and striate, and pale brown ascospores (Guarro et al. 2012). Our D1/D2 analysis revealed a well-supported relationship of F. indica with the Dothideomycetes, although distantly related to the genus Arthrographis and other members of Eremomycetaceae. Réblová et al. (2011), based on LSU sequences analysis, demonstrated the relationship of F. indica with the Didymellaceae, including it in the Pleosporales. The exclusion of F. indica from Microascales correlates with the morphological features of the asexual morph, since in that order the asexual morphs are characterised by percurrently proliferating conidiogenous cells (annellides) usually belonging to the genera Scopulariopsis, Graphium, Scedosporium, Cephalotrichum and Wardomycopsis (Valmaseda et al. 1986, Zhang et al. 2006, Réblová et al. 2011).
Eremomyces langeronii was traditionally considered to be the sexual morph of A. kalrae (von Arx 1978, Malloch & Sigler 1988). However, this connection was questioned by Sigler & Carmichael (1983), and later by Gené et al. (1996), arguing that both species produced different RFLP patterns. The present study confirms that E. langeronii and A. kalrae are not conspecific. Eremomyces langeronii was initially described as Pithoascus langeronii (von Arx 1978), later being transferred to the genus Pithoascina by Valmaseda et al. (1986). Malloch & Sigler (1988) accommodated this species in the genus Eremomyces (Eremomycetaceae) (Malloch & Cain 1971, Malloch & Sigler 1988) together with E. bilateralis and Rhexothecium globosum. Members of the Eremomycetaceae are characterised by cleistothecial ascomata, clavate to ovoid, evanescent asci, unicellular, hyaline to pale yellow-brown ascospores, arthrographis-like or trichosporiella-like asexual morphs and a coprophilous habitat (Malloch & Sigler 1988). Similar morphological features such as non-ostiolate dark ascomata and hyaline, unicellular ascospores can be also found in species of Pseudeurotiaceae (incertae sedis, Lumbsch & Huhndorf 2010), but the members of this family display pale-brown or olive-brown ascospores at maturity and asexual morphs with poorly differentiated conidiophores sympodially producing subspherical to ovoidal conidia. Our study demonstrated that the family Eremomycetaceae encompasses the genera Arthrographis s.str., Rhexothecium and Eremomyces (91.4–95.3 % inter-generic similarity in D1/D2 sequences). The latter now is restricted only to E. bilateralis, which is the type species of the genus. Eremomyces bilateralis is distinguished from Arthrographis s.str. and Rhexothecium by DNA sequence data (92.8 %, 89 % and 76.4 % similarity in D1/D2, ACT1 and ITS sequences, respectively) and by its cephalothecoid peridium, dark coloured colonies and the absence of an asexual morph (Malloch & Cain 1971).
The multilocus sequence analysis revealed the existence of four new species in Arthrographis, A. curvata being the only one that showed both sexual and asexual morphs in culture. Its ascomata and ascospores are similar to those of A. arxii; however, in A. arxii the ascomata are immersed, and the ascomata and ascospores are larger (75–160 μm diam and 2.7–5 × 1.8–2.6 μm, respectively). The asexual morph of A. curvata differs from A. arxii and A. kalrae mainly by less differentiated and poorly branched conidiophores, the presence of curved, sessile conidia and a restricted growth at 40 °C. Another fungus that also produces curved, cashewnut shaped conidia is the dermatophyte Trichophyton phaseoliforme, but this species is a member of Eurotiomycetes, produces pycnidium-like conidiomata and cigar-shaped macroconidia in clusters (de Hoog et al. 2011). Arthrographis arxii differs from A. kalrae in producing shorter and wider (3.5–5 × 2–2.5 μm) 1-septate arthroconidia.
Arthrographis chlamydospora is characterised by the production of repeatedly branched conidiophores, cuboid or doliiform arthroconidia and numerous chlamydospores. The species morphologically closest to A. chlamydospora is A. kalrae, but the latter differs by exhibiting yellowish to tan colonies, thin-walled arthroconidia, good growth at 40 °C and the presence of the trichosporiella-like synasexual morph. The other two new species, A. globosa and A. longispora, share several phenotypic features, i.e. absence of trichosporella-like synasexual morph, membranous colonies, inability to grow at 37 °C and resistance to high doses of cycloheximide. Arthrographis globosa can easily be distinguished by its globose to doliiform arthroconidia, and A. longispora by its poorly differentiated conidiophores producing large cylindrical arthroconidia. Other species of Arthrographis s.lat. unable to grow at 37 °C are A. alba, A. lignicola and A. pinicola. Arthrographis alba produces white colonies, pseudodichotomously branched conidiophores and, in our study, this species was susceptible to high doses of cycloheximide (2 g/L); A. lignicola can be distinguished by its lemon-yellow to olive-green colonies with a diffusible brown pigment, narrow branched conidiophores and yellow arthroconidia; and A. pinicola produces floccose conidiomata composed by repeatedly branched conidiophores and is susceptible to low doses of cycloheximide (Sigler & Carmichael 1983, Sigler et al. 1990, Gené et al. 1996).
In this study we observed some morphological variability in A. kalrae, with the presence of some characteristics not previously reported for this species. Such variations, however, did not correlate with genetic differences in any of the three loci sequenced. Several isolates showed chlamydospores that were terminal or intercalary, solitary or catenulate, hyaline or pigmented. In the protologue of Oidiodendron kalrai, based in the strain CBS 693.77, Tewari & Macpherson (1971) reported the occasional presence of oval to round, thick-walled chlamydospores; however, Sigler & Carmichael (1976) did not mention these structures and only reported the sessile conidia of the trichosporiella-like synasexual morph. The UTHSC 05-17 isolate produced infertile ascomata morphologically similar to the ascomata produced by A. arxii and A. curvata, but in that isolate these structures were smaller (37–70 μm diam). That isolate also produced abundant conidiophores with whorls of short chains of clavate or cylindrical arthroconidia. The clavate conidia was reported by von Arx (1978) in the description of the asexual morph of E. langeronii, but probably this description was based on a single strain of this species and not on the ex-type strain of A. kalrae.
The genus Arthropsis was established by Sigler et al. (1982) with A. truncata as the type species, to accommodate species with dark arthroconidia, joined by adjacent connectives and developed from undifferentiated conidiogenous hyphae. Until now the species of this genus have not been associated to any sexual morph. Our D1/D2 sequence analysis demonstrates that Arthropsis is polyphyletic and unrelated to Arthrographis s.str. Arthropsis hispanica and A. cirrhata fall into the Onygenales, as do other species previously included in Arthrographis. Other arthroconidial anamorphs of the Onygenales are included in the genus Malbranchea. However, Malbranchea is morphologically distinguished by its branched and arcuate fertile hyphae, straight in some species, that produce alternate arthroconidia (Sigler & Carmichael 1976). Our analysis placed the only available living strain of A. microsperma (UAMH 4290) in the Helotiales (Leotiomycetes). Arthropsis microsperma was originally described by Berkeley & Broome (1873) as Oidium microspermum and later transferred to Arthropsis by Sigler & Carmichael (1983) based on its arthroconidial ontogeny. Therefore, the name of this species should be reconsidered because Oidium anamorphs are currently associated with members of the Leotiomycetes (Braun & Cook 2012). Finally, Arthropsis truncata is related to members of the Sordariomycetes. Although such type of asexual morphs have not been described in that class, humicola-like asexual morphs similar to the Humicola synasexual morph of A. truncata are present in some species of Chaetomium (Gené & Guarro 1996, Seifert et al. 2011). Further studies with a greater number of taxa of Sordariomycetes are needed to ascertain a defined position for A. truncata within this class.
Acknowledgments
We are grateful to Lynne Sigler from the University of Alberta Microfungus Collection for providing us the reference strain of Arthropsis microsperma (UAMH 4290). This study was supported by the Spanish Ministerio de Economía y Competitividad, grant CGL 2011-27185.
REFERENCES
- Arx JA von. 1978. Notes on Microascaceae with the description of two new species. Persoonia 10: 23–31 [Google Scholar]
- Arx JA von. 1985. Sporotrichum sulphureum, possibly an older name for Pachysolen tannophilus. Mycotaxon 23: 251–252 [Google Scholar]
- Arx JA von, Mukerji KG, Singh N. 1981. Faurelina indica spec. nov. Sydowia 34: 39–41 [Google Scholar]
- Arx JA von, Samson RA. 1973. Two new genera of the Eurotiales. Persoonia 7: 377–380 [Google Scholar]
- Berkeley MJ, Broome CE. 1873. Notices of British fungi. Annals and Magazine of Natural History 11: 339–349 [Google Scholar]
- Boehm EWA, Schoch CL, Spatafora JW. 2009. On the evolution of the Hysteriaceae and Mytilinidiaceae (Pleosporomycetidae, Dothideomycetes, Ascomycota) using four nuclear genes. Mycological Research 113: 461–479 [DOI] [PubMed] [Google Scholar]
- Boonmee S, Zhang Y, Chomnunti P, Chukeatirote E, Tsui CKM, et al. 2011. Revision of lignicolous Tubeufiaceae based on morphological reexamination and phylogenetic analysis. Fungal Diversity 51: 63–102 [Google Scholar]
- Braun U, Cook RTA. 2012. Taxonomic manual of the Erysiphales (Powdery Mildews). CBS Biodiversity Series 11. CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands [Google Scholar]
- Cannon PF, Kirk PM. 2007. Fungal families of the world. CAB International, Wallingford, United Kingdom [Google Scholar]
- Carbone L, Kohn LM. 1999. A method for designing primer sets for speciation in filamentous ascomycetes. Mycologia 91: 553–556 [Google Scholar]
- Cochet G. 1939. Sur un nouveau champignon arthrospore (Arthrographis langeronii) agent d’une onychomycose humaine n.g., n.sp. Annales de Parasitologie Humaine et Comparee 17: 98–101 [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. 2004. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22 [Google Scholar]
- Gené J, Guarro J. 1996. A new Chaetomium from Thailand. Mycological Research 100: 1005–1009 [Google Scholar]
- Gené J, Guillamón JM, Ulfig K, Guarro J. 1996. Studies on keratinophilic fungi. X. Arthrographis alba sp. nov. Canadian Journal of Microbiology 42: 1185–1189 [Google Scholar]
- Giraldo A, Sutton D, Gené J, Fothergill A, Cano J, Guarro J. 2013. Rare arthroconidial fungi in clinical samples: Scytalidium cuboideum and Arthropsis hispanica. Mycopathologia 157: 115–121 [DOI] [PubMed] [Google Scholar]
- Guarro J, Gené J, Stchigel AM, Figueras MJ. 2012. Atlas of soil ascomycetes. CBS Biodiversity Series 10. CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands [Google Scholar]
- Gueidan C, Villasenor CR, Hoog GS de, Gorbushina AA, Untereiner WA, Lutzoni F. 2008. A rock-inhabiting ancestor for mutualistic and pathogen-rich fungal lineages. Studies in Mycology 61: 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoog GS de, Guarro J, Gené J, Figueras MJ. 2011. Atlas of clinical fungi. CD-ROM version 3.1. CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands [Google Scholar]
- Kang HJ, Sigler L, Lee J, Gibas CF, Yun SH, Lee YW. 2010. Xylogone ganodermophthora sp. nov., an ascomycetous pathogen causing yellow rot on cultivated mushroom Ganoderma lucidum in Korea. Mycologia 102: 1167–1184 [DOI] [PubMed] [Google Scholar]
- Kodsueb R, Jeewon R, Vijaykrishna D, McKenzie EHC, Lumyong P, et al. 2006. Systematic revision of Tubeufiaceae based on morphological and molecular data. Fungal Diversity 21: 105–130 [Google Scholar]
- Kornerup A, Wanscher JH. 1978. Methuen handbook of colour. 3rd ed Eyre Methuen, London [Google Scholar]
- Locquin-Linard M. 1975. Faurelina, nouveau genre d’Ascomycetes. Revue Mycologique 39: 125–129 [Google Scholar]
- Lumbsch HT, Huhndorf SM. 2010. Myconet Volume 14. Part One. Outline of Ascomycota – 2009. Fieldiana, Life and Earth Sciences 1: 1–42 [Google Scholar]
- Malloch D, Cain RF. 1971. Four new genera of cleistothecial Ascomycetes with hyaline ascospores. Canadian Journal of Botany 49: 847–854 [Google Scholar]
- Malloch D, Sigler L. 1988. The Eremomycetaceae (Ascomycotina). Canadian Journal of Botany 66: 1929–1932 [Google Scholar]
- Murata Y, Sano A, Nishimura K, Kamei K. 2005. The first isolation of Arthrographis kalrae from the oral cavity of a canine in Japan. Proceedings of the Annual Meeting of the Mycological Society of Japan. Abstracts of Submitted Papers 49: 168 [Google Scholar]
- O’Donnell K. 1993. Fusarium and its near relatives. In: Reynolds DR Taylor JW (eds), The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics: 225–233. CAB International, Wallingford [Google Scholar]
- Oorschot CAN van, Hoog GS de. 1984. Some hyphomycetes with thallic conidia. Mycotaxon 20: 129–132 [Google Scholar]
- Pettersson OV, Leong SL, Lantz H, Rice T, Dijksterhuis J, et al. 2011. Phylogeny and intraspecific variation of the extreme xerophile, Xeromyces bisporus. Fungal Biology 115: 1100–1111 [DOI] [PubMed] [Google Scholar]
- Réblová M, Gams W, Seifert KA. 2011. Monilochaetes and allied genera of the Glomerellales, and a reconsideration of families in the Microascales. Studies in Mycology 68: 163–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová M, Seifert KA. 2007. A new fungal genus, Teracosphaeria, with a phialophora-like anamorph (Sordariomycetes, Ascomycota). Mycological Research 111: 287–298 [DOI] [PubMed] [Google Scholar]
- Reeb V, Lutzoni F, Roux C. 2004. Contribution of RPB2 to multilocus phylogenetic studies of the euascomycetes (Pezizomycotina, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Molecular Phylogenetics and Evolution 32: 1036–1060 [DOI] [PubMed] [Google Scholar]
- Seifert K, Morgan-Jones G, Gams W, Kendrick B. 2011. The genera of Hyphomycetes. CBS Biodiversity Series 9. CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands [Google Scholar]
- Sigler L, Carmichael JW. 1976. Taxonomy of Malbranchea and some other Hyphomycetes with arthroconidia. Mycotaxon 4: 349–488 [Google Scholar]
- Sigler L, Carmichael JW. 1983. Redisposition of some fungi referred to Oidium microspermum and a review of Arthrographis. Mycotaxon 18: 495–507 [Google Scholar]
- Sigler L, Dunn MT, Carmichael JW. 1982. Arthrocristula and Arthropsis, two new Hyphomycetes with dematiaceous arthroconidia. Mycotaxon 15: 409–419 [Google Scholar]
- Sigler L, Yamaoka Y, Hiratsuka Y. 1990. Taxonomy and chemistry of a new fungus from bark beetle infested Pinus contorta var. latifolia. Part 1. Arthrographis pinicola sp. nov. Canadian Journal of Microbiology 36: 77–82 [Google Scholar]
- Sugiyama M, Mikawa T. 2001. Phylogenetic analysis of the non-pathogenic genus Spiromastix (Onygenaceae) and related onygenalean taxa based on large subunit ribosomal DNA sequences. Mycoscience 42: 413–421 [Google Scholar]
- Sugiyama M, Ohara A, Mikawa T. 1999. Molecular phylogeny of onygenalean fungi based on small subunit ribosomal DNA (SSU rDNA) sequences. Mycoscience 40: 251–258 [Google Scholar]
- Sugiyama M, Summerbell RC, Mikawa T. 2002. Molecular phylogeny of onygenalean fungi based on small subunit (SSU) and large subunit (LSU) ribosomal DNA sequences. Studies in Mycology 47: 5–23 [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari RP, Macpherson CR. 1971. A new dimorphic fungus, Oidiodendron kalrai: morphological and biochemical characteristics. Mycologia 63: 602–611 [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The Clustal X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui CK, Sivichai S, Rossman AY, Berbee ML. 2007. Tubeufia asiana, the teleomorph of Aquaphila albicans in the Tubeufiaceae, Pleosporales, based on cultural and molecular data. Mycologia 99: 884–894 [DOI] [PubMed] [Google Scholar]
- Ulfig K, Gené J, Guarro J. 1995. Studies on keratinophilic fungi VI. A new Arthropsis (Fungi imperfecti) from marine sediments. Mycotaxon 54: 281–286 [Google Scholar]
- Untereiner WA, Scott JA, Naveau FA, Currah RS, Bachewich J. 2002. Phylogeny of Ajellomyces, Polytolypa and Spiromastix (Onygenaceae) inferred from rDNA sequence and non-molecular data. Studies in Mycology 47: 25–35 [Google Scholar]
- Valmaseda M, Martinez T, Barrasa JM. 1986. Annellidic conidiogenesis in Pithoascus schumacheri and redefinition of Pithoascus and related fungi. Canadian Journal of Botany 65: 1802–1805 [Google Scholar]
- Valldosera M, Guarro J, Figueras MJ. 1991. Two coprophilous fungi from Spain. Mycological Research 95: 243–256 [Google Scholar]
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt K, Wöstemeyer J. 2000. Reliable amplification of actin genes facilitates deep-level phylogeny. Microbiological Research 155: 179–195 [DOI] [PubMed] [Google Scholar]
- Wang Z, Binder M, Hibbett DS. 2005. Life history and systematics of the aquatic discomycete Mitrula (Helotiales, Ascomycota) based on cultural, morphological, and molecular studies. American Journal of Botany 92: 1565–1574 [DOI] [PubMed] [Google Scholar]
- Wedin M, Wiklund E, Crewe A, Döring H, Ekman S, et al. 2005. Phylogenetic relationships of Lecanoromycetes (Ascomycota) as revealed by analyses of mtSSU and nLSU rDNA sequence data. Mycological Research 109: 159–172 [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA Gelfand DH Sninsky JJ White TJ (eds), PCR protocols: a guide to the methods and applications: 315–322. Academic Press, Inc., New York [Google Scholar]
- Xi L, Fukushima K, Changming L, Takizawa K, Liao R, Nishimura K. 2004. First case of Arthrographis kalrae Ethmoid Sinusitis and Ophthalmitis in the People’s Republic of China. Journal of Clinical Microbiology 42: 4828–4831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Castlebury L, Miller A, Huhndorf SM, Schoch CL, et al. 2006. An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia 98: 1076–1087 [DOI] [PubMed] [Google Scholar]