Abstract
Circadian clocks serve as internal pacemakers that influence many basic homeostatic processes; consequently, the expression and function of their components are tightly regulated by intricate networks of feedback loops that fine-tune circadian processes. Our knowledge of these components and pathways is far from exhaustive. In recent decades, the nuclear envelope has emerged as a global gene regulatory machine, although its role in circadian regulation has not been explored. We report that transcription of the core clock component BMAL1 is positively modulated by the inner nuclear membrane protein MAN1, which directly binds the BMAL1 promoter and enhances its transcription. Our results establish a novel connection between the nuclear periphery and circadian rhythmicity, therefore bridging two global regulatory systems that modulate all aspects of bodily functions.
DOI: http://dx.doi.org/10.7554/eLife.02981.001
Research organism: D. melanogaster, mouse
eLife digest
If rodents, or indeed humans, are kept in constant darkness for a number of days, they continue to show patterns of sleep and wakefulness that repeat roughly every 24 hr. This internal ‘circadian rhythm’ controls many aspects of animal physiology, including body temperature, blood pressure, and hormone levels. It does so by regulating the expression of key genes: this means that the activity of the proteins encoded by these genes also varies in accordance with the circadian rhythm.
A second mechanism used by the body to coordinate gene expression on a large scale entails making adjustments to the membrane that surrounds the cell nucleus. This ‘nuclear envelope’ consists mainly of lipids, but it also contains proteins that bind DNA. These proteins regulate gene expression by controlling how easy it is for other proteins that activate or repress genes to gain access to specific DNA sequences.
Lin et al. now reveal that these mechanisms work together. The first evidence for this was the discovery that the levels of three specific nuclear envelope proteins influence, and are influenced by, circadian rhythms. In particular, two of these proteins control the activity of the third, which is known as MAN1. This protein in turn triggers the expression of a gene called BMAL1, which is one of the small number of ‘clock genes’ that are responsible for generating the internal circadian rhythm.
As well as adding to our knowledge of circadian biology and the nuclear envelope, this study reveals a mechanism by which cells can orchestrate the expression of large numbers of genes. Such mechanisms allow a wide range of physiological and behavioral processes to be co-ordinated.
Introduction
Most organisms, ranging from cyanobacteria to humans, are governed by their circadian rhythms: endogenous and self-sustained oscillations with a period of roughly 24 hr, which manifest in diverse metabolic, physiological, and behavioral processes (Ueda et al., 2005). This internal pacemaker is charged with two important roles: to perpetuate its own rhythms and to regulate the expression of genes that are under circadian control. In mammals, this internal pacemaker consists of a complex network of transcriptional regulations, at the core of which is transcription activators BMAL1 (also known as ARNTL1 or MOP3) and CLOCK, which form heterodimers and regulate gene expression. Up to 15% of the organism's genome is regulated in a circadian manner (Panda et al., 2002; Emery and Reppert, 2004; Zhang and Kay, 2010). Well-studied examples include the transcription repressors (PERIODs and CRYPTOCRHOMEs) that bind to CLOCK/BMAL1 and suppress their own transcription, thereby forming a feedback loop. Since the identification and cloning of the first mammalian clock gene, CLOCK, two decades ago (Vitaterna et al., 1994), the field of chronobiology has uncovered many additional players that regulate circadian rhythms on a transcriptional and/or post-transcriptional level, and many more such candidates are currently being evaluated.
Recently, mutations in nuclear envelope (NE) proteins have been shown to cause a surprisingly broad range of inherited diseases, thus shedding light on roles played by the NE in global regulations at cellular and organismal levels (Padiath et al., 2006; Dauer and Worman, 2009). These diseases (often referred to as nuclear envelopathies or laminopathies) can impact muscle, nerve, fat metabolism, bone formation, and others. NE consists of outer and inner nuclear membranes (connected by nuclear pore complexes) and nuclear lamina. The inner nuclear membrane proteins (such as MAN1, LBR, LAP2, etc) include approximately 60 putative transmembrane proteins specifically retained in the inner nuclear membrane and most of them are poorly characterized (Schirmer et al., 2005). In metazoan, nuclear lamina is a protein mesh-like structure composed of type V intermediate filament proteins lamins (including A, C, B1, B2, and B3 types) and sits primarily underneath the inner nuclear membrane (Zwerger and Medalia, 2013). The idea of nuclear envelope components as transcription regulators in mammals is relatively new, conceived from the observation that gene-rich chromosomes are generally located in more internal nuclear regions, whereas gene-poor chromosomes are relegated to the periphery (Spector, 2003). Many NE components such as inner nuclear membrane proteins, nuclear lamina, and the nuclear pore complex, harbor DNA-binding domains that are involved in anchoring chromatin to the periphery (Ulbert et al., 2006; Mekhail and Moazed, 2010). Functional relevance of these positional distinctions became apparent as studies with yeast and flies revealed that the NE can sequester factors that affect gene transcription in both repressive and, surprisingly, activating manners (Akhtar and Gasser, 2007). Although recent findings highlight the important functions of the nuclear periphery, its relationship with the circadian clock has not been probed. Given the increasing awareness of the global roles that these two systems play in myriad pathways, we set out to investigate the possibility that these seemingly separate pathways are connected and can work synergistically in regulating diverse functions.
Results
Nuclear envelope participates in circadian regulation
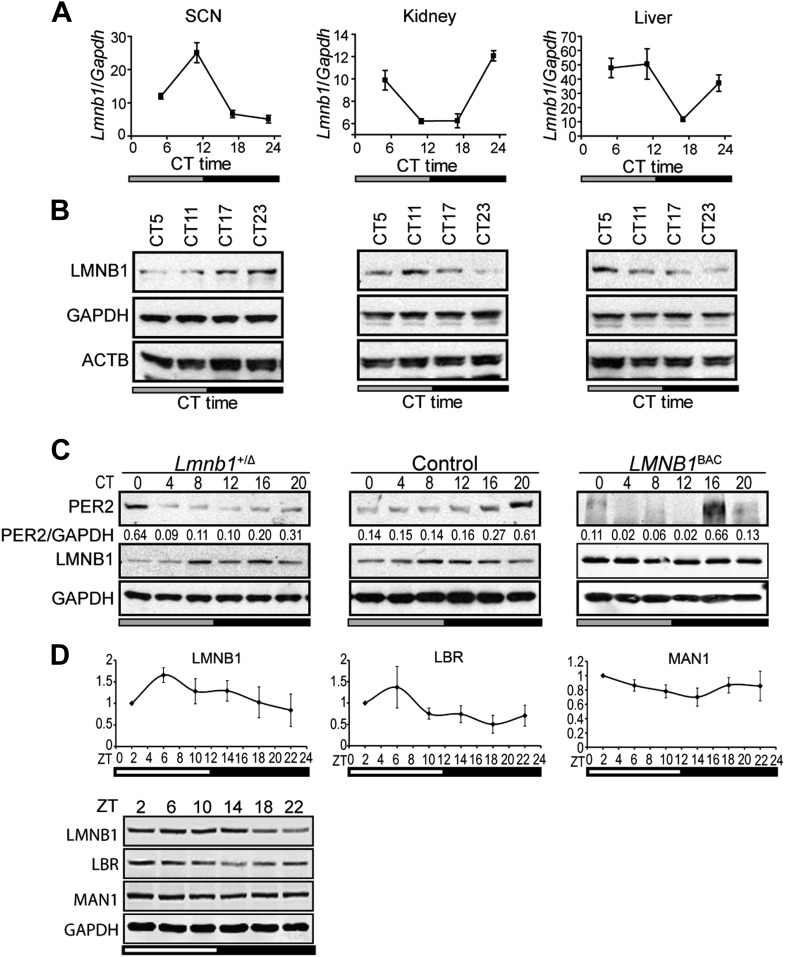
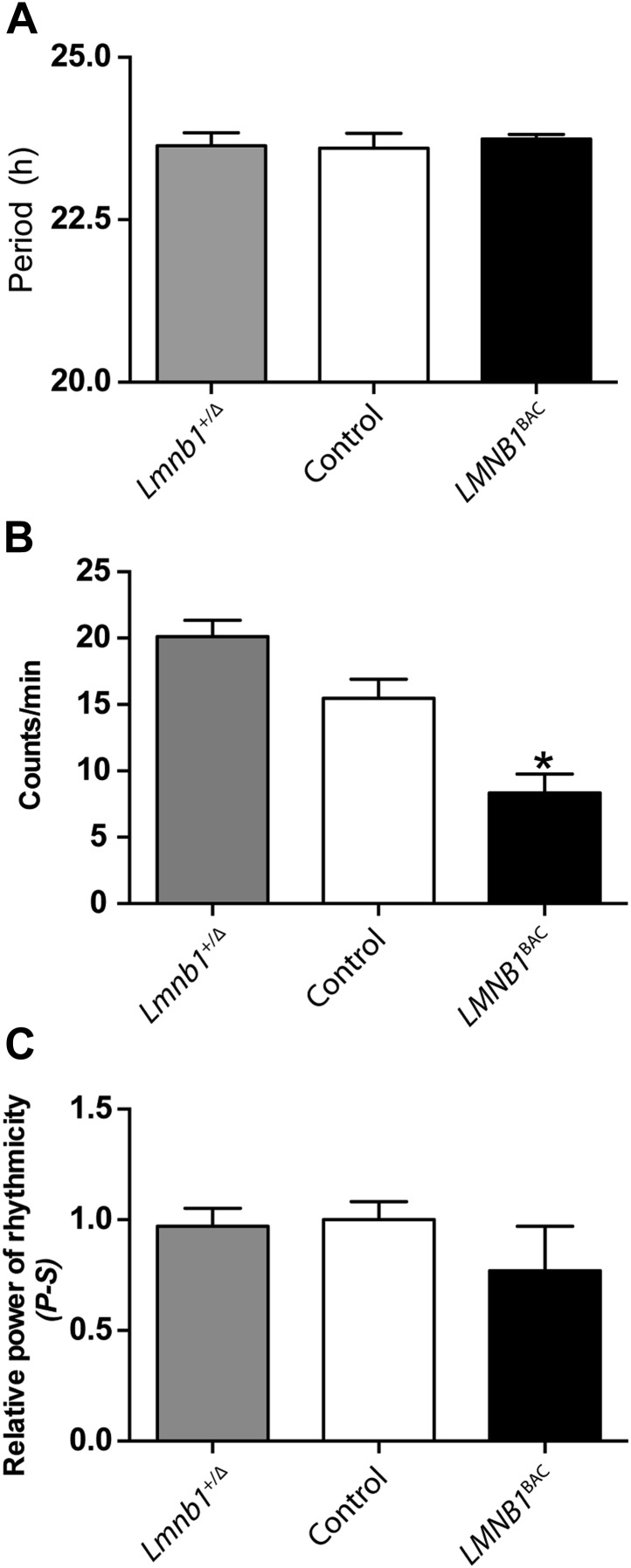
In order to investigate whether NE proteins are involved in circadian regulation, we began by focusing on lamin B1 since it has been shown to play a role in transcriptional regulation (Hutchison, 2002; Shevelyov et al., 2009). In vivo oscillation of lamin B1 (Lmnb1) expression patterns (both RNA and protein levels) were confirmed using mouse tissues from suprachiasmatic nuclei (SCN), kidney, and liver (Figure 1A,B). To test whether the level of lamin B1 affects the molecular clock, we examined the protein expression patterns for the core clock gene PERIOD2 (PER2) using Lmnb1 heterozygous knock out (homozygosity is lethal) and LMNB1 wild-type BAC transgenic mice (Vergnes et al., 2004; Heng et al., 2013). Oscillating PER2 expression patterns were phase delayed in Lmnb1 heterozygous knock out mice and phase advanced in LMNB1 BAC transgenic mice (overexpression) when compared to wild-type control mice (Figure 1C), suggesting that the level of lamin B1 can modulate circadian clock. However, neither Lmnb1 heterozygous knock out mice nor LMNB1 BAC transgenic mice demonstrated significant output behavioral change (Figure 1—figure supplement 1). To expand the investigation, we chose to include two additional NE proteins that are known to associate with lamin B1, LBR, and MAN1. We found that LBR and MAN1 expression also oscillate, albeit mildly for MAN1 (Figure 1D).
Figure 1. Lamin b1 regulates the circadian clock.
Expression levels of lamin b1 from SCN, kidney, and liver extracts in C57BL/6J mice (A and B). (A) mRNA levels of Lmnb1 and Gapdh were assayed at indicated circadian times (CT, n = 4). (B) Representative immunoblots show the levels of LMNB1, GAPDH, and β-ACTIN. (C) Representative immunoblots show PER2 (with intensity values indicated at the bottom) and LMNB1 abundance in Lmnb1+/Δ, wild-type and LMNB1BAC liver extracts. (D) Expression patterns of LMNB1, LBR, and MAN1 in C57BL/6 mouse livers at indicated Zeitgeber times (ZT) (n = 3). Quantifications (top panels) of Western blots (bottom panel) were obtained by using GAPDH as a loading control. Data represent means ± SD.
Figure 1—figure supplement 1. Mice with altered LMNB1 levels do not exhibit altered behavioral rhythms.
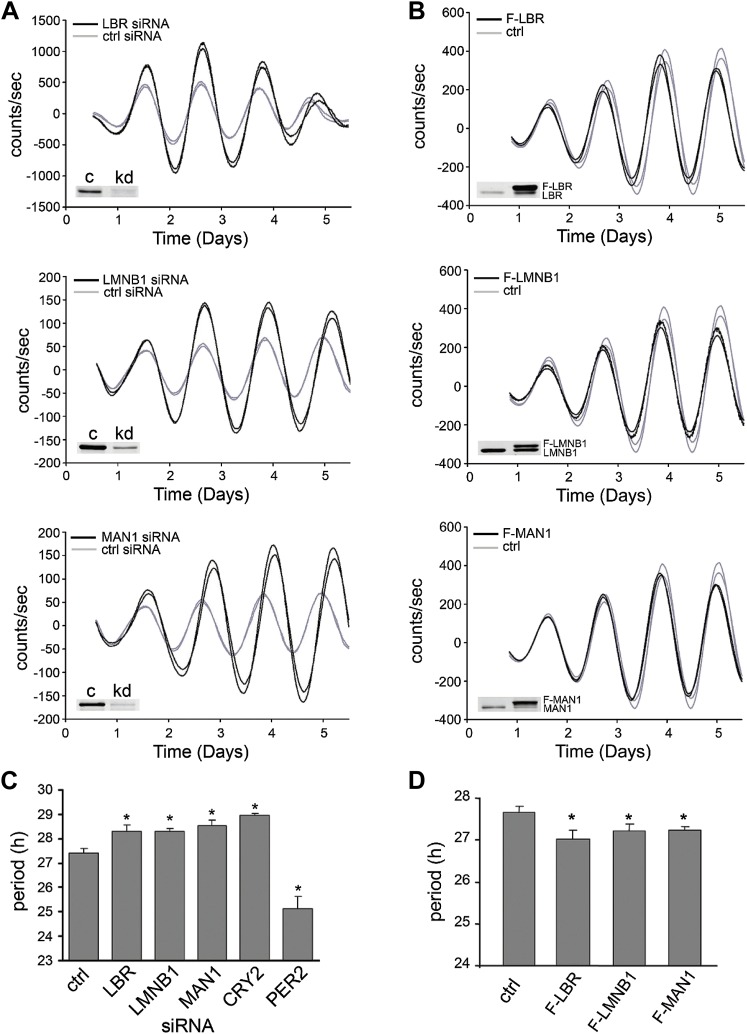
To determine if these NE genes passively receive cues from the core clock apparatus or if their protein products also actively play a role in maintaining circadian rhythms, we altered their protein levels in human osteosarcoma U2OS cells that express a luciferase reporter gene under the control of mouse Bmal1 promoter (Bmal1-Luc) and examined circadian period in cell culture (Vollmers et al., 2008). siRNA-induced reduction of LMNB1, LBR, and MAN1 in this cell-based system resulted in a longer circadian period (τ) (Figure 2A), whereas the over-expression of all three led to a shorter τ (Figure 2B). Cells transfected with LBR, LMNB1, and MAN1 siRNA lengthened τ by 54–69 min (n ≥ 4, *p < 0.05), when compared with control siRNA (τ = 27.39 ± 0.22 hr, n = 8) (Figure 2C). On the other hand, overexpression of FLAG-tagged LBR, LMNB1, or MAN1 shortened τ by 25.8–37.8 min (n ≥ 4, *p < 0.05) compared to empty vector controls (τ = 27.7 ± 0.15, Figure 2D). These changes in τ together with the altered phase of Lmnb1 heterozygous knock out mice and over-expressing mice suggest that these NE proteins participate in modulating circadian clock and therefore could impose significant impacts on downstream biological pathways.
Figure 2. LBR, LMNB1, and MAN1 are necessary for normal circadian rhythms.
Two representative traces of real-time bioluminescence analyses are shown for each, and Western blot verification of down-regulation or over-expression is demonstrated in the inset images. (A) Period was lengthened when LBR, LMNB1, or MAN1 was knocked down. (B) Over-expression of FLAG-tagged LBR (F-LBR), LMNB1 (F-LMNB1), or MAN1 (F-MAN1) shortened period compared to cells transfected with empty vector (ctrl). (C and D) Summary of period in (A and B). CRYPTOCRHOME2 (CRY2) and PER2 siRNA knockdowns served as positive controls. Data represent means ± SD.
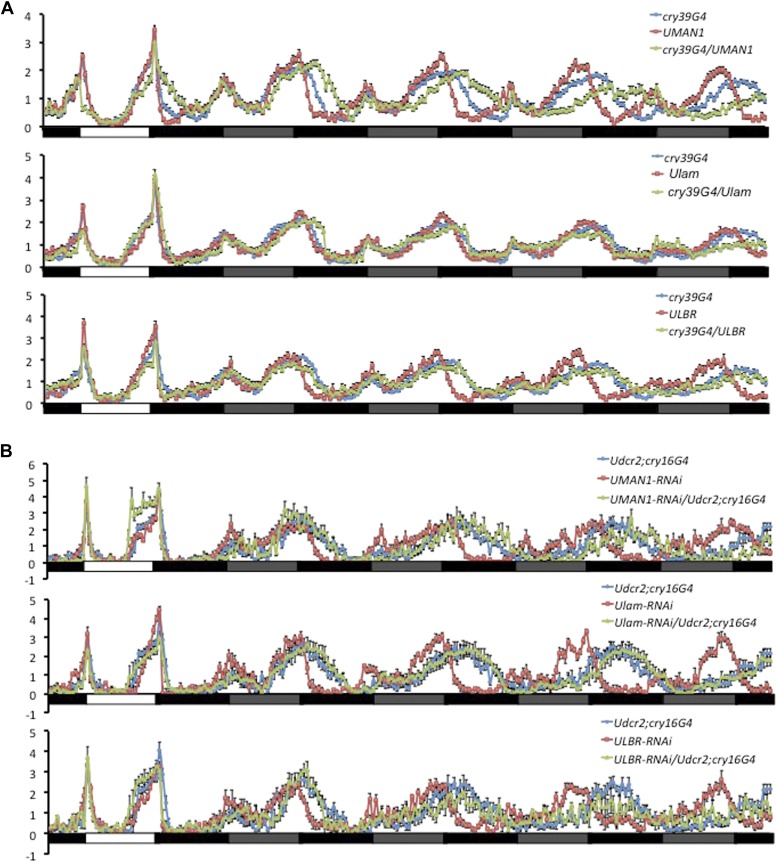
Figure 2—figure supplement 1. Over-expressing or knocking down nuclear envelope components alters circadian rhythms in flies.
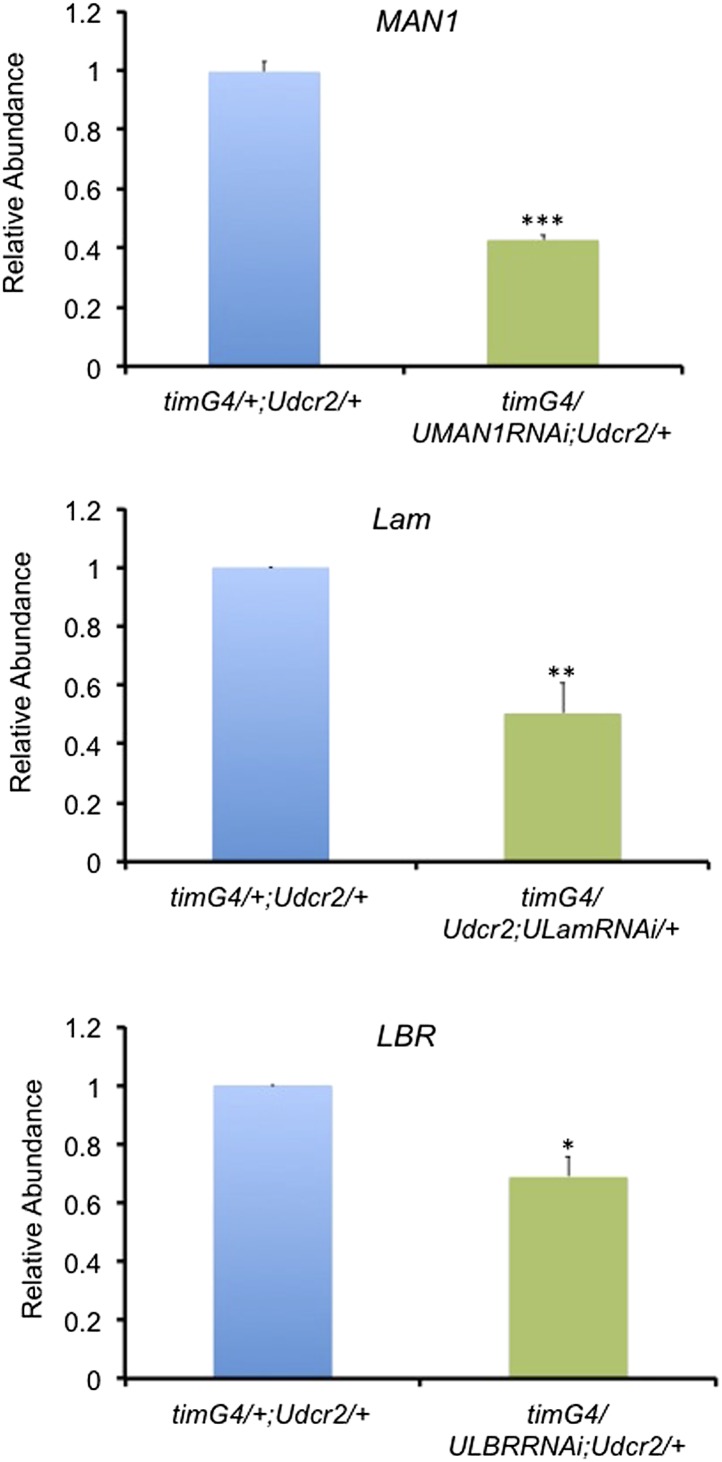
Figure 2—figure supplement 2. The mRNA levels of nuclear envelope genes are reduced in the corresponding knockdown flies.
The effects of NE proteins on Drosophila circadian clock were also examined. Consistent with the mammalian data, over-expressing dLamin (dLam) (Padiath et al., 2006) in circadian neurons with cryptochrome (cry)GAL4-39 and cryGAL4-16 (Emery et al., 2000) in vivo resulted in substantially shortened periods of behavioral rhythms in constant darkness compared to GAL4 controls (but not to UASLMNB1/+, Table 1; Figure 2—figure supplement 1A). Knocking down dLam in circadian neurons lengthened the period (Table 2; Figure 2—figure supplement 1B). On the other hand, over-expressing dMAN1 and dLBR lengthened the period (Table 1; Figure 2—figure supplement 1A), while knocking down dMAN1 also lengthened period (Table 2; Figure 2—figure supplement 1B). Besides altering the period, most of these manipulations reduced the amplitude of behavioral rhythms as indicated by the reduced power values. In addition, we have assessed the mRNA levels of dMAN1, dLam, and dLBR to confirm knockdown (Figure 2—figure supplement 2). Taken together, these results indicate that NE proteins also participate in the regulation of fly clock.
Table 1.
Over-expressing NE genes alters the behavioral period in flies
| Genotype | Period (hr) | Power | Rhythmic% | N |
|---|---|---|---|---|
| UASMAN1/+ | 23.8 ± 0.0 | 90 ± 5 | 96 | 76 |
| cryGAL4-39/UASMAN1 | 26.4 ± 0.3*,† | 31 ± 6*,† | 64 | 28 |
| UASMAN1/+; cryGAL4-16/+ | 26.8 ± 0.2*,† | 40 ± 7*,† | 66 | 32 |
| UASLam/+ | 23.9 ± 0.0 | 87 ± 6 | 93 | 55 |
| cryGAL4-39/UASLam | 23.8 ± 0.2† | 33 ± 7*,† | 73 | 22 |
| UASLMNB1/+; cryGAL4-16/+ | 24.4 ± 0.2† | 5 ± 2*,† | 26 | 31 |
| UASLBR/+ | 23.7 ± 0.0 | 59 ± 5 | 79 | 67 |
| cryGAL4-39/UASLBR | 25.6 ± 0.1*,† | 44 ± 6† | 79 | 33 |
| UASLBR/+; cryGAL4-16/+ | N/A | 1 ± 0*,† | 0 | 32 |
| cryGAL4-39/+ | 24.8 ± 0.1 | 86 ± 5 | 93 | 57 |
| cryGAL4-16/+ | 25.6 ± 0.1 | 86 ± 5 | 94 | 64 |
One-way ANOVA compared to UAS control lines, p < 0.001.
One-way ANOVA compared to GAL4 control lines, p < 0.001.
Table 2.
Knocking-down NE genes lengthens the behavioral period in flies
| Genotype | Period (hr) | Power | %Rhythmic | N |
|---|---|---|---|---|
| UASMAN1RNAi | 24.4 ± 0.1 | 110 ± 10 | 100 | 14 |
| UASMAN1RNAi;cryGAL4-39/+; UASdcr2/+ | 26.3 ± 0.3*,† | 29 ± 6*,† | 80 | 15 |
| UASMAN1RNAi; UASdcr2/+;cryGAL4-16/+ | 27.9 ± 0.4*,† | 39 ± 8*,† | 69 | 16 |
| UASLamRNAi/+ | 23.6 ± 0.1 | 129 ± 12 | 100 | 16 |
| cryGAL4-39/+; UASLamRNAi/UASdcr2 | 25.1 ± 0.4* | 26 ± 17*,† | 50 | 14 |
| UASLamRNAi/UASdcr2; cryGAL4-16/+ | 26.8 ± 0.1*,† | 126 ± 17 | 100 | 13 |
| UASLBRRNAi/+ | 23.5 ± 0.0 | 112 ± 14 | 100 | 15 |
| cryGAL4-39/UASLBRRNAi; UASdcr2/+ | 24.9 ± 0.1 | 61 ± 12* | 80 | 15 |
| UASLBRRNAi/UASdcr2; cryGAL4-16/+ | 24.6 ± 1.7 | 12 ± 4*,† | 44 | 16 |
| cryGAL4-39/+; UASdcr2/+ | 24.9 ± 0.1 | 69 ± 12 | 88 | 16 |
| UASdcr2/+;cryGAL4-16/+ | 26.1 ± 0.1 | 87 ± 10 | 94 | 16 |
One-way ANOVA compared to UASRNAi control lines, p < 0.05.
One-way ANOVA compared to control lines with GAL4 and UASdcr2, p < 0.05.
dicer2 (dcr2) is co-expressed to enhance the effects of RNAi.
Lamin B1 and LBR likely act through MAN1
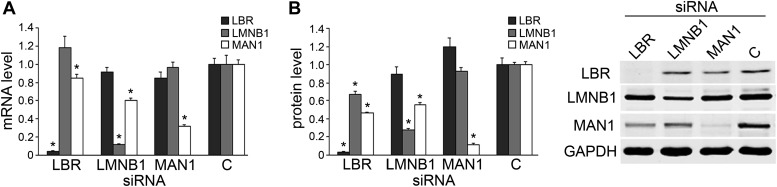
We next explored the relationship of LBR, LMNB1, and MAN1 by examining mRNA and protein levels while knocking them down one at a time. Both LBR and LMNB1 knockdown significantly decreased the transcript level of MAN1 (by 15% and 40%, respectively) (Figure 3A). The effects of LBR or LMNB1 knockdown on MAN1 expression are even more dramatic at the protein level, with 54% and 44% reductions, respectively (Figure 3B). Moreover, knockdown of LBR expression reduces the amount of LMNB1 protein by 32%, which is consistent with the observation that reduction of LBR expression in the fibroblasts of patients harboring a heterozygous LBR mutation results in the abolition of LMNB1 protein (Gaudy-Marqueste et al., 2010), whereas a decrease in LMNB1 does not significantly affect LBR expression. MAN1 knockdown also does not change the expression of LBR and LMNB1, either at the mRNA or protein level (Figure 3). These results suggest that MAN1 is modulated by LBR and LMNB1, and thus the effects of LBR and LMNB1 on the clock are at least partially through MAN1. Therefore, we further investigated the effects of MAN1 on the molecular clock.
Figure 3. Knocking down LBR/LMNB1 reduces MAN1 mRNA and protein levels but not vice versa.
Assessing mRNA (A) and protein (B) levels of LBR, LMNB1, and MAN1 while knocking them down one at a time in U2OS cells via RNAi. (A) mRNA levels of LBR, LMNB1, and MAN1 in each of the three knockdown conditions were quantified using qRT-PCR (n = 14, *p < 0.05). (B) MAN1 was significantly down-regulated when LBR or LMNB1 was knocked down (n = 14 *p < 0.001). The error bars represent SEM (left panel). Representative immunoblots show the protein levels of LBR, LMNB1 and MAN1 (right panel).
MAN1 regulates the clock by promoting BMAL1 expression
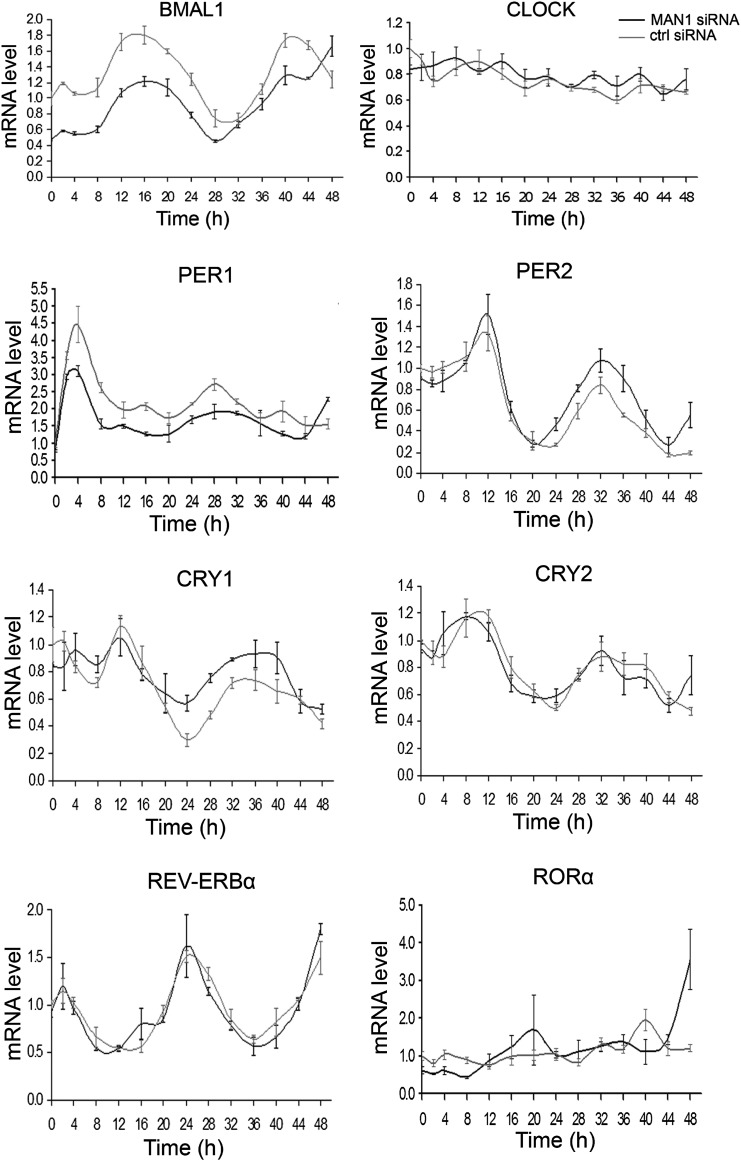
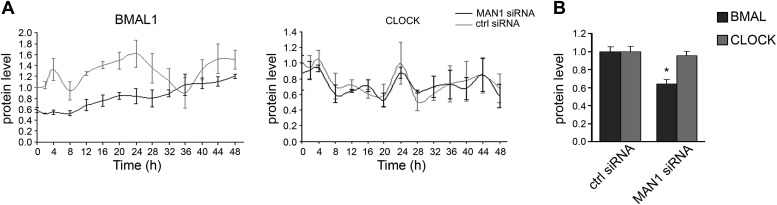
A lengthened period due to decreased MAN1 may arise from altered regulation of core clock proteins and/or altered transcription of core clock genes. Either of these effects would result in disruptions of the stoichiometry and temporal control of the dynamics of the core circadian feedback loops. Given what is known regarding the role of NE proteins in transcriptional regulation, we tested whether reductions in MAN1 expression would affect the transcription of clock genes. We examined the circadian oscillation of core clock genes at the mRNA level and found that only BMAL1 showed a clear difference wherein overall mRNA levels were down-regulated to half the levels of controls (Figure 4). Western blots also showed lower expression of BMAL1 when MAN1 was knocked down (Figure 4—figure supplement 1). The non-oscillatory CLOCK showed no significant change of either transcript or protein levels (Figure 4, Figure 4—figure supplement 1). The conserved mRNA levels of REV-ERBα and RORα in MAN1 knockdown cells suggest that the reduced BMAL1 expression is not caused by altered transcriptional activation of REV-ERBα, a BMAL1 repressor, or transcriptional repression of RORα, a BMAL1 activator.
Figure 4. Knocking down MAN1 reduces the levels of BMAL1 mRNA.
Each graph shows cells transfected with MAN1 siRNA vs ctrl siRNA. Time 0 indicates the moment that U2OS cells were treated with dexamethasone (100 nM). Data are presented as means ± SEM, n = 3.
Figure 4—figure supplement 1. Knocking down MAN1 reduces BMAL1 protein levels.
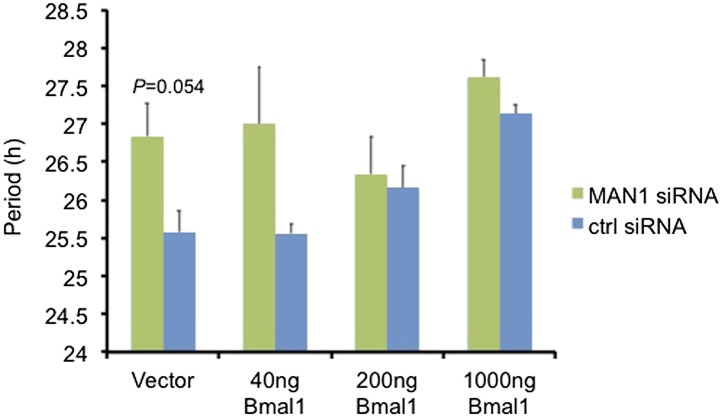
Figure 4—figure supplement 2. Over-expressing Bmal1 suppresses the period lengthening effect of MAN1 knockdown.
To confirm MAN1 regulates the clock by targeting BMAL1, we over-expressed Bmal1 in MAN1 knockdown U2OS Bmal1-Luc cells. Knocking down MAN1 lengthened the period in control cells as described above (Figure 2), whereas cells over-expressing sufficient Bmal1 did not demonstrate period lengthening compared to cells without MAN1 knockdown (Figure 4—figure supplement 2), suggesting that the lengthened period caused by MAN1 deficiency is due to reduction of BMAL1. Together, these results indicate that MAN1 functions to promote BMAL1 expression, and thus exerting effects on the clock.
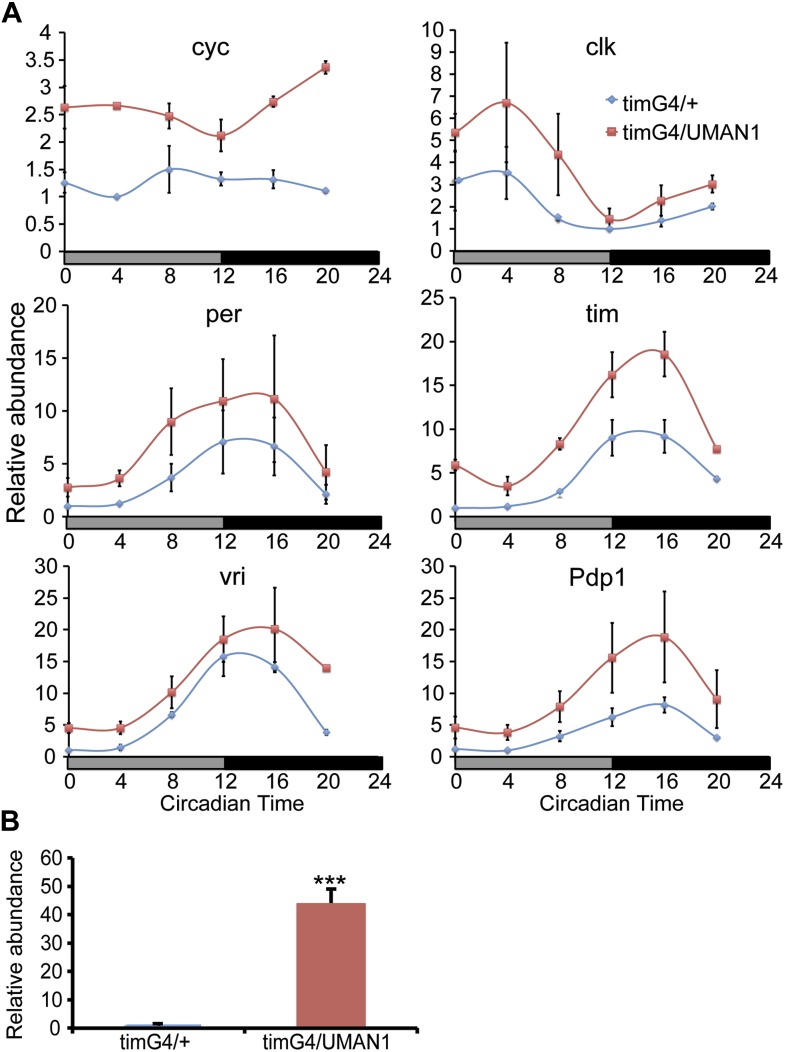
Consistent with the cell culture data, over-expressing MAN1 in all clock cells in flies using a timeless (tim) GAL4 driver (Emery et al., 1998) resulted in a significantly increased level of cycle (cyc) mRNA, the Drosophila BMAL1 homologue (Rutila et al., 1998; Figure 5A). The mRNA level of core clock gene tim was also significantly elevated. In addition, we have assessed the levels of MAN1 mRNA to confirm over-expression (Figure 5B). We also examined the effect of knocking down MAN1 in clock cells but did not observe altered cyc mRNA levels (Figure 5—figure supplement 1).
Figure 5. MAN1 increases cyc mRNA levels.
(A) Plots of relative mRNA abundance vs CT for clock genes from whole head extracts of timGAL4/+ and timGAL4/UASMAN1 flies during the first day of DD determined by qRT-PCR (n = 2). Gray box indicates subjective light period and black box indicates dark period. Significant effect of genotypes (Two-way ANOVA) were found for cyc (p = 0.0278) and tim (p = 0.0161). For each time series, the value of the lowest time point was set to 1. (B) Plot of relative mRNA abundance for MAN1 from whole head extracts of timGAL4/+ and timGAL4/UASMAN1 flies determined by qRT-PCR (n = 6, Student's t test, ***p < 0.001). The value of timGAL4/+ in one experiment was set to 1. Error bars represent SEM.
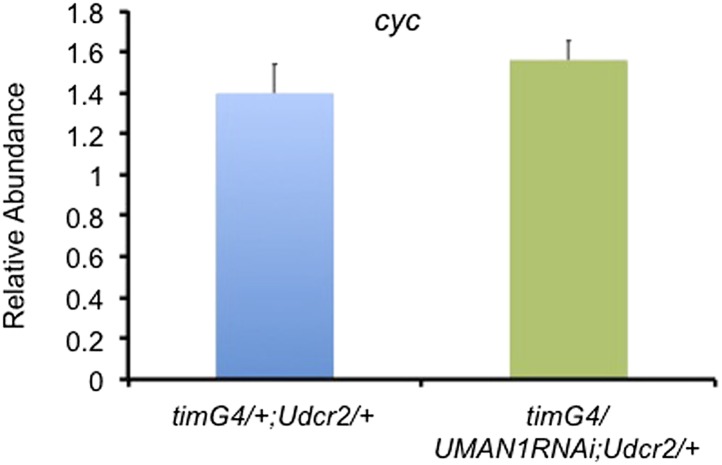
Figure 5—figure supplement 1. cyc transcript level is not altered in MAN1 knock-down flies.
MAN1 enhances BMAL1 transcription
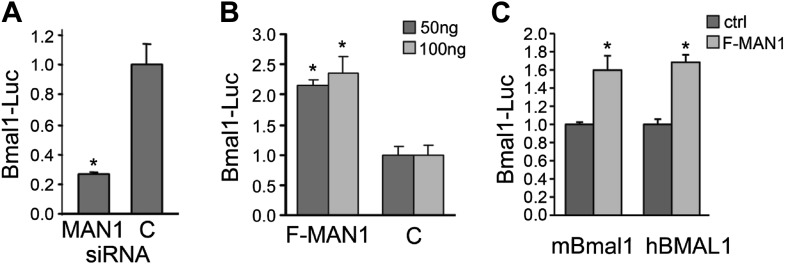
A luciferase reporter assay using HEK293 cells was used to further investigate the effect of MAN1 on BMAL1 transcription. MAN1 knockdown decreased Bmal1-Luc activity by 72% (Figure 6A), whereas overexpression of FLAG-MAN1 increased the luciferase activity by more than twofold vs cells transfected with empty vector (Figure 6B). Similar results were obtained with a longer, human BMAL1 promoter (Figure 6C). These data indicate that MAN1 may play a role in circadian regulation by activating the promoter of BMAL1.
Figure 6. MAN1 promotes BMAL1 transcriptional activity.
(A) Reduction of MAN1 transcripts (13 nM siRNA) reduced Bmal1 promoter activity (n = 3, *p < 0.001). (B) Over-expression of FLAG-tagged MAN1 (F-MAN1) enhanced Bmal1-Luc activity (n = 3, *p < 0.001). (C) Over-expression of FLAG-tagged MAN1 (F-MAN1) enhanced luciferase activities driven by mBmal1 promoter (530 bp) or hBMAL1 promoter (3.4 kb) (n = 3, *p < 0.05). Error bars represent SD.
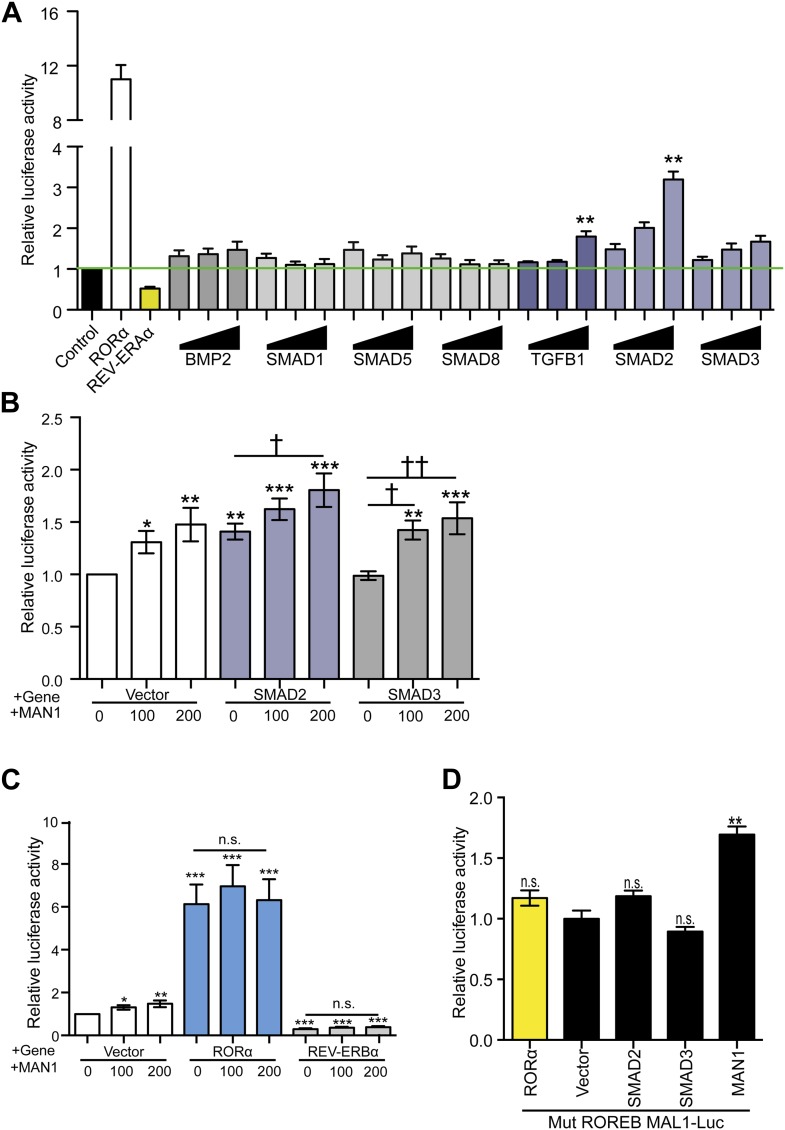
Previously, MAN1 has been shown to exert antagonistic regulatory functions on signal transduction through its binding to R-SMADs (Osada et al., 2003; Raju et al., 2003; Hellemans et al., 2004; Lin et al., 2005; Pan et al., 2005; Cohen et al., 2007) and two types of R-SMADs are found in mammals: TGFβ-responsive (SMAD2 and SMAD3) and BMP-responsive (SMAD1, SMAD5, and SMAD8). To determine whether R-SMADs have an effect on BMAL1 transcription, we first expressed R-SMADs individually in HEK293 cells transfected with BMAL1-Luc to determine which R-SMAD/s is/are involved in regulating BMAL1 transcription. Expressing SMAD1, SMAD5, SMAD8, and SMAD3 had no significant effect on BMAL1 transcription but SMAD2 showed significant enhancing effect, suggesting a possible regulatory function by SMAD2 in BMAL1 regulation (Figure 7A). The enhancing action of SMAD2 was then examined together with MAN1 to determine whether there is interplay between MAN1 and SMAD2 on BMAL1 promoter activity. Intriguingly, MAN1 further augmented the enhancing effect of SMAD2 on BMAL1 in an additive manner, indicating that the positive regulatory function of MAN1 and SMAD2 on BMAL1 might be independent of each other (Figure 7B).
Figure 7. MAN1 and SMAD2 enhance BMAL1 transcription.
(A–D) Luciferase reporter activities in transfected HEK293 cells. Cells transfected with indicated constructs in the presence of the 3.4 kb hBMAL1 promoter for 48 hr and relative luciferase activities were measured in extracts and normalized to Renilla luciferase activities. Relative luciferase activities were shown on the y-axis. Values are means ± SEM, n = 3, **p < 0.01 compared to control. †p < 0.05, ††p < 0.01 compared to MAN1 0, one-way ANOVA with Newman–Keuls test.
Since BMAL1 transcription is regulated by RORα and REV-ERBα, we wondered whether the effect of MAN1 on BMAL1 is influenced by RORα/REV-ERBα. MAN1 increases BMAL1 transcription in the HEK293 luciferase reporter assay but this effect was overshadowed by the presence of either RORα or REV-ERBα, and the impact of RORα and REV-ERBα on BMAL1 was not influenced by the presence of MAN1 (Figure 7C). In addition, the effect of MAN1 was also not significantly altered by mutating the RORE sequence (+3 ∼ +13 and +39 ∼ +48) in the BMAL1 promoter (Figure 7D), which serves as the DNA binding target of RORα and REV-ERBα. Together, these data suggest that the effect of MAN1 on BMAL1 transcription is not through the RORE and does not require or impact RORα and REV-ERBα.
MAN1 binds to the BMAL1 promoter
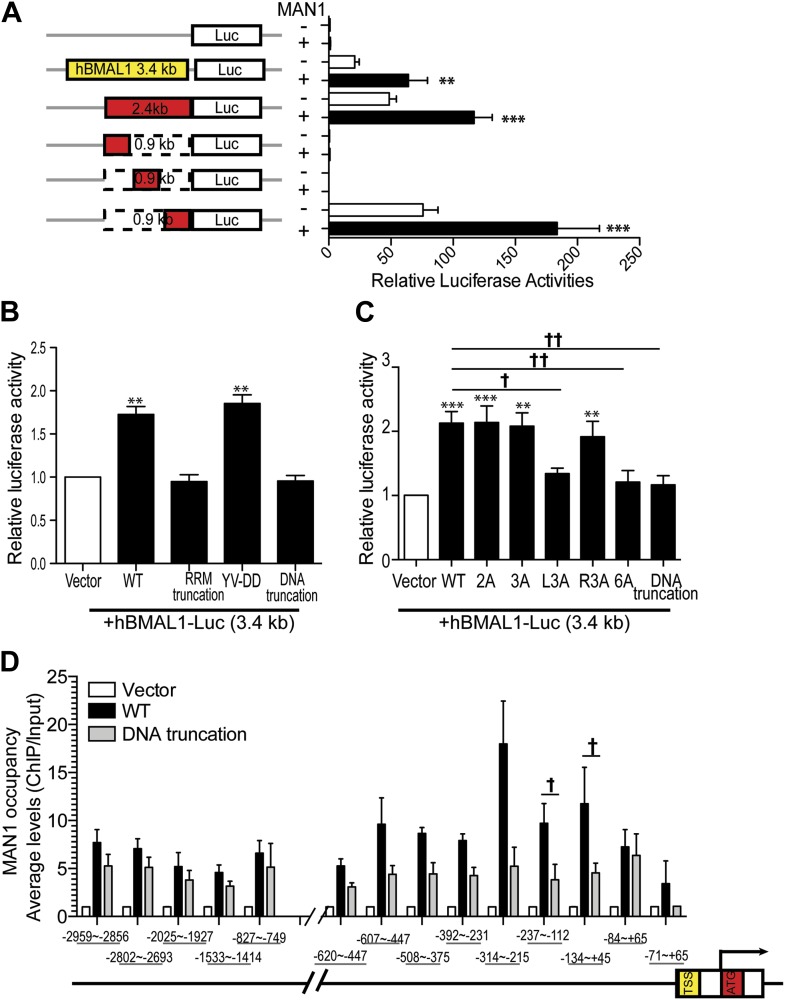
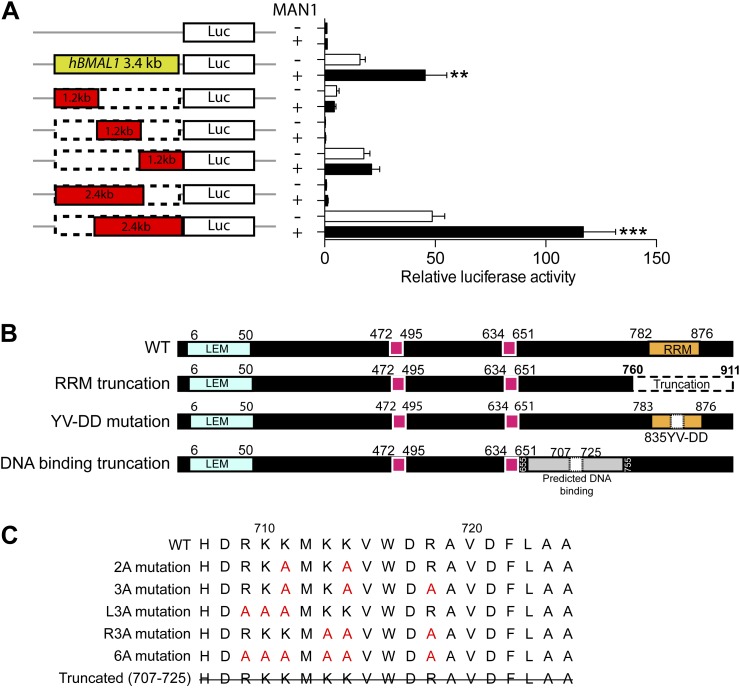
Since MAN1 does not execute its function through RORE, we investigated the promoter region of BMAL1 to determine what is necessary for the enhancing effect of MAN1. A series of deletion constructs of the BMAL1 promoter were generated for luciferase assays and a 900 bp region (−795 ∼ +106) was identified to be the region harboring the necessary DNA sequence for the regulatory effect of MAN1 on BMAL1 (Figure 8A, Figure 8—figure supplement 1A).
Figure 8. MAN1 binds to the BMAL1 promoter to enhance its transcription.
(A) Luciferase activities of deleted hBMAL1-promoter constructs in the absence or presence of MAN1 expression vectors. n = 3, Student's t test, **p < 0.01, ***p < 0.001. (B and C) Luciferase activities of the 3.4 Kb hBMAL1-Luc in the presence of MAN1 constructs as indicated. n = 3, **p < 0.01, ***p < 0.001 compared to control; †p < 0.05; ††p < 0.001 compared to WT MAN1. (D) ChIP analysis of MAN1 (WT or DNA binding truncation) for 14 segments of hBMAL1 promoter region. Data represent pull-down relative to input. n = 6, †p < 0.05, compared to WT MAN1. One-way ANOVA with Newman–Keuls test. All data are presented as ratio of means ± SEM.
Figure 8—figure supplement 1. Domain-specific interactions between MAN1 and the hBMAL1 promoter.
MAN1 has an N-terminal LAP2-Emerin-MAN1 (LEM) domain, two transmembrane segments in the middle, a unique DNA binding domain, and a C-terminal RNA recognition motif (RRM) that is required for its binding with R-SMADs (Caputo et al., 2006). We next examined the domain of MAN1 necessary for its effect on BMAL1. Two truncation constructs of MAN1 were generated, one without the DNA binding domain (amino acids 707–725) and the other without the RRM domain (amino acids 760–911) (Pan et al., 2005; Figure 8—figure supplement 1B). In addition, we utilized a substitution mutant of MAN1 (YV-DD) (Pan et al., 2005), containing two amino acid alterations in the RRM that nullify the ability of MAN1 to antagonize R-SMADs. These constructs were then used to test their transcriptional enhancing effect on BMAL1 promoters (either full length 3.4 kb or 2.4 kb [−2300 ∼ +105]). Intriguingly, the RRM truncation mutation lost the enhancing effect on the BMAL1 promoter (Figure 8B). The DNA binding domain truncation mutation also lost the activating effect on BMAL1, and this effect can be produced by simply mutating three positively charged amino acids (RKK) at amino acids 709–711 (Figure 8C, Figure 8—figure supplement 1C). These results imply that the effect of MAN1 on BMAL1 transcription requires potential DNA binding ability of MAN1 as well as interaction with a protein partner (possibly SMAD2) through RRM.
Since the DNA binding domain is required for the effect of MAN1 on the BMAL1 promoter, we next tested whether MAN1 directly binds to the BMAL1 promoter. Chromatin immunoprecipitation (ChIP) analysis revealed that the region −237 bp to +45 bp from transcriptional start site was pulled down by MAN1 indicating a direct interaction (Figure 8D). All together, these data suggest that MAN1 binds to the BMAL1 promoter region (−237 bp to +45 bp) to enhance its transcription.
Discussion
The nuclear envelope plays essential roles in diverse cellular functions including global regulation of gene expression. Interestingly, another global regulatory mechanism is the molecular clock that modulates our body and cellular daily rhythm. We sought to see whether there is cross talk between these two regulatory mechanisms. Our studies revealed that some components of nuclear envelope do show daily oscillations, indicating that nuclear envelope is subject to clock control. On the other hand, we found that one of the major transcription activators of the molecular clock, BMAL1, is regulated by one of the nuclear envelope proteins, MAN1. Thus, there is reciprocity between these two global regulatory mechanisms.
The nuclear envelope physically separates genomic DNA from the cytoplasm and functions as a signaling control center. An increasing number of human diseases are recognized to be caused by mutations in genes encoding nuclear envelope proteins and hence, termed ‘envelopathies’ (Dauer and Worman, 2009). Several inner nuclear membrane proteins are known to regulate critical signaling pathways and act as intranuclear regulators of signaling pathways that receive and transduce signals from extracellular cues. The nuclear lamina provides structural support for the nucleus and the nuclear envelope; however, lamins and their associated proteins are also involved in most of the nuclear processes. Lamin B1 is essential for brain development and is required for the integrity of the nuclear lamina (Padiath et al., 2006; Coffinier et al., 2011). Interestingly, no abnormality has been reported for the heterozygous Lmnb1 knock out mice (Stewart et al., 2007; Coffinier et al., 2011), and it is estimated that these heterozygous mice still express 70% of lamin b1 (SG Young, personal communication). Consistent with these reports, we did not find significant phenotype including circadian behavior change in these mice despite the clear phase shift on PER2 oscillation in the liver. It is possible that the oscillation of one or more additional core clock components are also altered by reduced lamin b1 level and these additional alterations can compensate the effect of PER2 phase shift on output behavior. Alternatively, it is also possible that the SCN clock is resilient to perturbations caused by reduced lamin b1 level, hence we can only observe alterations in peripheral clocks. Further investigation is necessary to reveal the mechanism leading to the findings we report here.
MAN1 belongs to the inner nuclear membrane LEM protein family (Worman, 2006; Bengtsson, 2007). LEM domains mediate the interaction with a chromatin-binding protein, barrier-to-autointegration factor (BAF), which has the ability to bind dsDNA, chromatin, histones, lamin binding proteins, and various transcription factors (Liu et al., 2003; Worman, 2006; Bengtsson, 2007). Therefore, LEM proteins have roles in gene regulation, chromatin organization, regulation of transcription factor activity at the nuclear periphery, and regulation of specific signal pathways. Both amino and carboxyl termini of MAN1 are nucleoplasmic domains. The amino-terminal nucleoplasmic region of MAN1 (including LEM) binds to the nuclear lamins and emerin in addition to BAF. It is also necessary for efficient localization of MAN1 to the inner nuclear membrane (Mansharamani and Wilson, 2005). The carboxyl-terminal nucleoplasmic region (residues 655–911) exhibits two globular domains (Pan et al., 2005; Caputo et al., 2006). The first globular domain contains a winged helix (including the sequence RKKMKKVWDR) which is mainly used for DNA binding and recognition. The second domain (amino acids 782–911) is an RRM-like protein interaction domain where it can interact with R-SMADs. The entire carboxy-terminal region of MAN1 was shown to participate in DNA binding, and this interaction is synergistic to the binding of MAN1 to different transcriptional regulators, including R-SMADs (Osada et al., 2003; Raju et al., 2003; Lin et al., 2005; Pan et al., 2005). Consistently, we found that both RRM and DNA binding domains are required for the activation of the BMAL1 promoter by MAN1. Also congruent with previous findings, we found that mutating three of the highly positively charged and conserved amino acids within the winged helix region is sufficient to dampen the activation efficiency of MAN1. In contrast to previous reports, we found that MAN1 further augments (but not antagonizes) the positive effect of SMAD2 on BMAL1. Consistent with this result, we also found that the substitution mutation YV-DD of MAN1 does not influence its effect on BMAL1 (though RRM domain is required). Collectively, these results suggest that the effects of MAN1 and SMAD2 on BMAL1 might not be completely independent to each other and that MAN1 might interact with SMAD2 in more than one way (presumably through other protein partners) to modulate transcription of target genes.
Many of our body functions manifest a daily rhythm which is maintained by the rhythmic regulation of approximately 15% of genes by the core molecular clock (Vollmers et al., 2009; Menet et al., 2012). However, cells must be flexible enough to allow for responses to exogenous and endogenous stimuli. This regulation is likely to be mediated not solely by the molecular clock, but also by many additional global and local mechanisms including at the level of chromatin and genome organization (Aguilar-Arnal et al., 2013; Hubner et al., 2013). Genetic loci associated with the nuclear lamina through large regions of chromatin (lamin associated domains–LADs) are associated with changes in transcriptional status (Peric-Hupkes et al., 2010). Circadian genes or genes involved in rhythmic processes display robust rhythmic expression patterns at the level of nascent mRNA and mRNA (Menet et al., 2012; Rodriguez et al., 2012), suggesting prominent contribution of transcriptional regulation to clock gene expression. Interestingly, genes harboring this expression pattern are dramatically enriched for specific function in transcriptional regulation and chromatin organization (Menet et al., 2012). These unbiased genome-wide transcriptome reports raised the possibility that components of nuclear envelope may modulate oscillation of clock genes at transcriptional levels. A recent study has also demonstrated that the molecular clock drives circadian changes in spatial and temporal chromosomal organization (Aguilar-Arnal et al., 2013). Indeed, our study links the nuclear periphery with circadian regulation via the regulatory effects of MAN1 on BMAL1 through transcription.
This effect is evident not only in mammalian systems, but also in flies, as over-expressing MAN1 resulted in significantly increased levels of cyc mRNA. MAN1 over-expression also increased tim mRNA levels, which may be at least partially due to increased cyc. Knocking down MAN1 did not alter cyc levels, possibly because the residual MAN1 is still sufficient to maintain normal cyc levels. However, this manipulation lengthened behavioral period, suggesting that MAN1 may target other clock components in addition to cyc. Over-expressing MAN1 and LBR in flies led to a lengthened behavioral period, while in U2OS cells, these manipulations resulted in moderate shortening of the period. On the other hand, over-expressing and knocking down Lam in flies shortened and lengthened the period, respectively, which is consistent with the mammalian data. These discrepancies reflect the differences between mammalian and insect clock, which has been implied in previous work as well (Lowrey et al., 2000; Preuss et al., 2004; Xu et al., 2005). Nevertheless, our results indicate that there is a conserved role for NE components in setting the clock in organisms ranging from invertebrates to humans.
Although only 22–30% of cycling mRNA is driven transcriptionally (Koike et al., 2012; Menet et al., 2012), demonstration of involvement of the nuclear envelope in regulation of the molecular circadian clock suggests one pathway through which the nuclear envelope may globally and temporally regulate large numbers of genes. Interestingly, expression of the genes for many nuclear envelope proteins also oscillates. This finding sheds new light on the interconnectedness of these biological processes and provides further insight into the mechanism whereby cellular, metabolic, physiological, and behavioral processes that oscillate are modulated in a highly coordinated manner.
Materials and methods
Plasmids
To generate FLAG-tagged proteins, human LBR, LMNB1, and MAN1 genes were subcloned into pCMV-Tag2A (Stratagene; La Jolla, CA). HA-tagged SMAD1, FLAG-MAN1 (YV-DD), and FLAG-MAN1 (1–759) were gifts from Dr Kunxin Luo (Pan et al., 2005). mBmal1-Luc was generated in Dr Satchidananda Panda's lab and kindly provided by Dr John Hogenesch (Vollmers et al., 2008), while the human BMAL1-luc construct was a generous gift from Dr Toru Takumi (Akashi and Takumi, 2005). Syrian hamster Bmal1 in pcDNA3.1 vector was provided by Dr David Weaver (Kume et al., 1999). pRL-TK was purchased from Promega (Madison, WI). Mutations of expression constructs were introduced by PCR, and all constructs used in this study were verified by sequencing.
LumiCycle analysis
A stable U2OS-B6 cell line that expresses a destabilized firefly luciferase gene under the control of the mBmal1 promoter was obtained from Dr Satchidananda Panda (Vollmers et al., 2008). siRNAs targeted to LBR, LMNB1, MAN1, or SMAD1 (10 nM, Invitrogen; Carlsbad, CA; see Supplementary file 1A) were individually transfected into 35-mm culture dishes using Lipofectamine RNAiMAX (Invitrogen). For overexpression of FLAG-tagged constructs, plasmid (2 µg) was distributed into each well along with FuGENE HD (4 μl, Roche; Switzerland). For co-transfection of MAN1 siRNA and Bmal1, Lipofectamine 2000 (Invitrogen) was used. 24 hr after transfection, cells were synchronized with 100 nM dexamethasone in serum-free DMEM containing 10 mM HEPES (pH 7.5) at 37°C for 2 hr. Following synchronization, the media were replaced with phenol red-free DMEM supplemented with 10 mM HEPES and 40 µM Luciferin-EF (Promega). Cells sealed with coverslips were incubated in a 32-channel LumiCycle (Actimetrics; Evanston, IL) to monitor real-time bioluminescence for 5 days. Data were analyzed using Lumicycle Analysis (Actimetrics).
Fly behavior experiments and analysis
To over-express NE, cryGAL4-39 and cryGAL4-16 (Emery et al., 2000) were crossed to MAN1GS2297 (Kyoto Stock Center; Japan), UASLam (Padiath et al., 2006) and LBRGS2162 (Kyoto Stock Center). To knock down NE, cryGAL4-39;UASdcr2 and UASdcr2;cryGAL4-16 were crossed to UASMAN1RNAi (3167R-1, NIG; Japan), UASLamRNAi (45,635, VDRC; Vienna) and UASLBRRNAi (KK110508, VDRC). For controls, the UAS and GAL4 lines were crossed to w1118 or yw strains. Male progenies were assayed for behavior.
Locomotor activity levels of flies were monitored using Trikinetics Activity Monitors (Waltham, MA) for 7 days of 12 hr light-12 hr dark (LD) conditions followed by 7 days of constant darkness (DD). For DD rhythmicity, chi-squared periodogram analyses were performed using Clocklab (Actimetrics). Rhythmic flies were defined as those in which the chi-squared power was ≥10 above the significance line. Period calculations also considered all flies with rhythmic power ≥10.
Quantitative real-time PCR (qRT-PCR)
RNeasy Mini Kit (Qiagen) was used to isolate total RNAs from synchronized U2OS-B6 cells that were collected at interval of 4 hr over the course of 48 hr. Purified RNA (2 µg) was applied in 20-μl reactions for RT primed with Oligo(dT)20 using Super-Script III First-Strand Synthesis System (Invitrogen). All qPCR reactions were carried out on a Rotor-Gene RG-3000 (Corbett Research; Netherland)/or 7900HT Fast Real-Time PCR System (Life technologies; Carlsbad, CA) using FastStart SYBR Green Master (Rox) (Roche). The templates were denatured at 95°C for 10 min, followed by forty cycles with 15 s at 95°C, 10 s at 58°C (Rotor-Gene) or 60 s at 60°C (HT7900 system and data acquisition at the end of this step), or 40 s at 72°C, and an additional 2 s for data acquisition (Rotor-Gene). The standard curve and delta–delta CT methods were used for quantification (Applied Biosystems; Carlsbad, CA). Primers used for expression analysis are listed in Supplementary file 1B. Primers used for ChIP assay are labeled as the nucleotide distance from the transcriptional start site (TSS) and +1 indicates the starting of TSS.
Fly heads were isolated at the indicated time points and total RNA was isolated with TRIzol reagent (Invitrogen). After the removal of contaminating genomic DNA by RQ1 DNase (Promega) digestion, total RNA was directly amplified with the QuantiTect SYBR green RT-PCR kit (QIAGEN). The following primers were used: for cyc, cyc_110.f 5′-GAGGTCTTCGTCGGAAAGG-3′ and cyc_347.r 5′-AAAGCACATGGGAATCATGG-3′; for tim, tim.f 5′-CTGGGGAGTGACCATGG-3′ and tim.r 5′-GCTGGAATCGCCACTG-3′; for dMAN1, dMAN1_148.f 5′-ATTTTGGCCTGTGACACTGC-3′ and dMAN1_303.r 5′-GAAGCCGCTCTGGATTAGC-3′; for dLam, dLam_446.f 5′-CGAGGAGCTCAAGAACAAGC-3′ and dLam_675_r. 5′-GCGACAGTGTCTCCTGTTCC-3′; for dLBR, dLBR_645.f 5′-CATTGACCACCAACACATCC-3′ and dLBR_825.r 5′-GTTATGCGTTTGCGAATGG-3′; for dActin, dActin.f1 5′-CTAACCTCGCCCTCTCCTCT-3′ and dActin.r1 5′-GCAGCCAAGTGTGAGTGTGT-3′. All other primers used for fly tissues are previously published (Lim et al., 2007; Kilman et al., 2009).
Mouse behavior experiment and analyses
Lmnb1+/Δ mice with a targeted disruption of the Lmnb1 gene (Vergnes et al., 2004) or LMNB1BAC mouse model overexpressing lamin B1 (Heng et al., 2013) was generated as previous described. The animals used here were derived from these mice and have been backcrossed to a C57BL/6J background for at least 10 generations. Wild-type littermates were used in pairs for subsequent experiments. Mice housed in light-tight, sound-attenuated cabinets were entrained to LD cycle for 14 days and then released into DD. Wheel-running activity of mice were monitored using Clocklab (Actimetrics). For DD rhythmicity, chi-squared periodogram analyses were performed using Clocklab. Experiments were approved by the Institutional Animal Care and Use Committee at University of California, San Francisco.
Western blot
Brains or livers were collected from mice that were entrained in LD cycle for 14 days and were then released into DD. Total cellular proteins were extracted from mouse brain or liver using RIPA buffer (150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 20 mM Tris, pH 7.5, and 5 mM EDTA). Protein lysates from cells were prepared in SDS-PAGE loading buffer. Equal amounts of protein were resolved on 8% SDS-PAGE gels and then transferred to nitrocellulose membrane. After incubation with primary antibody at 4°C overnight, membranes were incubated with secondary antibodies at room temperature for 1 hr. The primary antibodies were anti-LBR rabbit polyclonal antibody (1:500; Abcam), anti-LMNB1 rabbit polyclonal antibody (1:1000; Abcam; England), anti-MAN1 rabbit polyclonal antibody (1:3000; from Dr Kunxin Luo) (Pan et al., 2005), anti-mPER2 rabbit polyclonal antibody (1:500; Alpha Diagnostic International; San Antonio, TX), anti-GAPDH mouse monoclonal antibody (1:5000; Chemicon; Billerica, MA), anti-BMAL1 goat polyclonal antibody (1:500; Santa Cruz; Dallas, TX), anti-CLOCK rabbit polyclonal antibody (1:1000; Santa Cruz), and anti-FLAG M2 antibody (1:5000; Sigma; St Louis, MO). The MAN1 antibody was generated by immunizing rabbits with C-terminal peptide (SHLRLRTGLTNSQGSS) of human MAN1 (1:1000; Covance and Agbio, Inc; Princeton, NJ). Secondary antibodies were conjugated either with IRDye 680 or IRDye 800 (LI-COR Biosciences; Lincoln, NE) and visualized with an Odyssey Infrared Imaging System (LI-COR Biosciences).
Luciferase assays
HEK293 cells were cultured in 24-well plates in DMEM containing 10% fetal bovine serum 24 hr prior to transient transfection with FuGENE HD (Roche) for overexpression (50–200 ng cDNA constructs), or Lipofectamine 2000 (Invitrogen) for siRNA knockdown (8–13 pmol, Invitrogen). All transfection mixtures included a Renilla luciferase plasmid (pRL-TK; 0.7 ng), as well as a reporter construct consisting of firefly luciferase driven by mouse Bmal1 or human BMAL1 promoter (50 ng). We assayed the Bmal1/BMAL1 promoter luciferase activity using the Dual-Luciferase Reporter Assay System (Promega), modifying the protocol to use 30 μl of luciferase substrate and Stop-n-Glo/substrate mix for each reaction. The luciferase activity was quantified with a TECAN GENios Pro Microplate Reader (TECAN; Switzerland) 48 hr after the initial transfection. Luciferase reporter vector used is pGL3-basic.
Chromatin immunoprecipitation (ChIP) assay
We performed ChIP assays using Millipore's EZ-ChIP assay kit (cat. # 17–371; Millipore; Billerica, MA) and protein-G sepharose. In brief, HEK293 cells were transfected with hBMAL1-luciferase (3.4 Kb) plus vector, FLAG-tagged WT or truncated DNA-binding constructs of MAN1 as indicated. Cell lysates were sonicated on ice using Branson digital sonifier #250 and 1% of cell lysate was taken as input sample. After incubation with FLAG M2 antibody (Sigma), antibody-loaded protein G agarose beads were washed with cold wash buffer six times followed by low-salt buffer, high salt wash buffer, LiCl wash buffer, and then once with TE (10 mM Tris–HCl at pH 8.0, 1 mM EDTA at pH 8.0). After washing, the beads were re-suspended in 100 μl of ChIP elution buffer supplemented with proteinase K and incubated for 2 hr at 65°C followed by 10 min at 95°C. The beads were spun down and the supernatant was saved. DNA was recovered from the spin column and resuspended in 50 μl of TE, and a 1 μl portion was used for qRT-PCR. The PCR products were analyzed by qRT-PCR and quantitated using 7900HT Fast Real-Time PCR System (Life technologies).
Statistics
Statistical analyses were performed using the unpaired Student's t test, one-way ANOVA with Newman–Keuls test, or two-way ANOVA (Prism5, GraphPad; La Jolla, CA). Data are presented as Mean ± SEM or SD. Significant differences (p < 0.05) are marked with asterisks in figures.
Acknowledgements
We thank Drs S Panda and J Hogenesch for mBmal1-luc construct; Dr T Takumi for hBMAL1-luc construct; Dr D Weaver for pcDNA3.1-Bmal1 construct; and Dr K Luo for MAN1 constructs, HA-SMAD1 construct, and MAN1 antibody.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Information
This paper was supported by the following grants:
National Institutes of Health FundRef identification ID: http://dx.doi.org/10.13039/100000002 HL059596 to Louis Ptáček, Ying-Hui Fu.
National Institutes of Health FundRef identification ID: http://dx.doi.org/10.13039/100000002 GM079180 to Louis Ptáček, Ying-Hui Fu.
National Institutes of Health FundRef identification ID: http://dx.doi.org/10.13039/100000002 NS072360 to Louis Ptáček, Ying-Hui Fu.
Howard Hughes Medical Institute FundRef identification ID: http://dx.doi.org/10.13039/100000011 to Louis Ptáček.
Additional information
Competing interests
LP: Reviewing editor, eLife.
The other authors declare that no competing interests exist.
Author contributions
S-TL, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents.
LZ, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents.
XL, Conception and design, Acquisition of data, Analysis and interpretation of data, Contributed unpublished essential data or reagents.
LCZ, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
VEG, Acquisition of data, Analysis and interpretation of data.
C-WT, Acquisition of data, Analysis and interpretation of data.
LP, Conception and design, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents.
Y-HF, Conception and design, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents.
Ethics
Animal experimentation: All experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by Institutional Animal Care and Use Committee of University of California San Francisco. The protocol was approved by the Committee on the Ethics of Animal Experiments of the University of California San Francisco (AN089663-01).
Additional files
(A) Sequences of siRNAs used in the study. (B) qRT-PCR primer sequences for human clock genes.
References
- Aguilar-Arnal L, Hakim O, Patel VR, Baldi P, Hager GL, Sassone-Corsi P. 2013. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nature Structural & Molecular Biology 20:1206–1213. doi: 10.1038/nsmb.2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi M, Takumi T. 2005. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nature Structural & Molecular Biology 12:441–448. doi: 10.1038/nsmb925 [DOI] [PubMed] [Google Scholar]
- Akhtar A, Gasser SM. 2007. The nuclear envelope and transcriptional control. Nature Reviews Genetics 8:507–517. doi: 10.1038/nrg2122 [DOI] [PubMed] [Google Scholar]
- Bengtsson L. 2007. What MAN1 does to the Smads. TGFbeta/BMP signaling and the nuclear envelope. The FEBS Journal 274:1374–1382. doi: 10.1111/j.1742-4658.2007.05696.x [DOI] [PubMed] [Google Scholar]
- Caputo S, Couprie J, Duband-Goulet I, Konde E, Lin F, Braud S, Gondry M, Gilquin B, Worman HJ, Zinn-Justin S. 2006. The carboxyl-terminal nucleoplasmic region of MAN1 exhibits a DNA binding winged helix domain. The Journal of Biological Chemistry 281:18208–18215. doi: 10.1074/jbc.M601980200 [DOI] [PubMed] [Google Scholar]
- Coffinier C, Jung HJ, Nobumori C, Chang S, Tu Y, Barnes RH, II, Yoshinaga Y, de Jong PJ, Vergnes L, Reue K, Fong LG, Young SG. 2011. Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Molecular Biology of the Cell 22:4683–4693. doi: 10.1091/mbc.E11-06-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TV, Kosti O, Stewart CL. 2007. The nuclear envelope protein MAN1 regulates TGFbeta signaling and vasculogenesis in the embryonic yolk sac. Development 134:1385–1395. doi: 10.1242/dev.02816 [DOI] [PubMed] [Google Scholar]
- Dauer WT, Worman HJ. 2009. The nuclear envelope as a signaling node in development and disease. Developmental Cell 17:626–638. doi: 10.1016/j.devcel.2009.10.016 [DOI] [PubMed] [Google Scholar]
- Emery P, Reppert SM. 2004. A rhythmic Ror. Neuron 43:443–446. doi: 10.1016/j.neuron.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Emery P, So WV, Kaneko M, Hall JC, Rosbash M. 1998. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95:669–679. doi: 10.1016/S0092-8674(00)81637-2 [DOI] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Helfrich-Forster C, Emery-Le M, Hall JC, Rosbash M. 2000. Drosophila CRY is a deep brain circadian photoreceptor. Neuron 26:493–504. doi: 10.1016/S0896-6273(00)81181-2 [DOI] [PubMed] [Google Scholar]
- Gaudy-Marqueste C, Roll P, Esteves-Vieira V, Weiller PJ, Grob JJ, Cau P, Levy N, De Sandre-Giovannoli A. 2010. LBR mutation and nuclear envelope defects in a patient affected with Reynolds syndrome. Journal of Medical Genetics 47:361–370. doi: 10.1136/jmg.2009.071696 [DOI] [PubMed] [Google Scholar]
- Hellemans J, Preobrazhenska O, Willaert A, Debeer P, Verdonk PC, Costa T, Janssens K, Menten B, Van Roy N, Vermeulen SJ, Savarirayan R, Van Hul W, Vanhoenacker F, Huylebroeck D, De Paepe A, Naeyaert JM, Vandesompele J, Speleman F, Verschueren K, Coucke PJ, Mortier GR. 2004. Loss-of-function mutations in LEMD3 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. Nature Genetics 36:1213–1218. doi: 10.1038/ng1453 [DOI] [PubMed] [Google Scholar]
- Heng MY, Lin ST, Verret L, Huang Y, Kamiya S, Padiath QS, Tong Y, Palop JJ, Huang EJ, Ptacek LJ, Fu YH. 2013. Lamin B1 mediates cell-autonomous neuropathology in a leukodystrophy mouse model. The Journal of Clinical Investigation 123:2719–2729. doi: 10.1172/JCI66737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner MR, Eckersley-Maslin MA, Spector DL. 2013. Chromatin organization and transcriptional regulation. Current Opinion in Genetics & Development 23:89–95. doi: 10.1016/j.gde.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CJ. 2002. Lamins: building blocks or regulators of gene expression? Nature Reviews Molecular Cell Biology 3:848–858. doi: 10.1038/nrm950 [DOI] [PubMed] [Google Scholar]
- Kilman VL, Zhang L, Meissner RA, Burg E, Allada R. 2009. Perturbing dynamin reveals potent effects on the Drosophila circadian clock. PLOS ONE 4:e5235. doi: 10.1371/journal.pone.0005235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. 2012. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338:349–354. doi: 10.1126/science.1226339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193–205. doi: 10.1016/S0092-8674(00)81014-4 [DOI] [PubMed] [Google Scholar]
- Lim C, Chung BY, Pitman JL, McGill JJ, Pradhan S, Lee J, Keegan KP, Choe J, Allada R. 2007. Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Current Biology 17:1082–1089. doi: 10.1016/j.cub.2007.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Morrison JM, Wu W, Worman HJ. 2005. MAN1, an integral protein of the inner nuclear membrane, binds Smad2 and Smad3 and antagonizes transforming growth factor-beta signaling. Human Molecular Genetics 14:437–445. doi: 10.1093/hmg/ddi040 [DOI] [PubMed] [Google Scholar]
- Liu J, Lee KK, Segura-Totten M, Neufeld E, Wilson KL, Gruenbaum Y. 2003. MAN1 and emerin have overlapping function(s) essential for chromosome segregation and cell division in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of USA 100:4598–4603. doi: 10.1073/pnas.0730821100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. 2000. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288:483–492. doi: 10.1126/science.288.5465.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansharamani M, Wilson KL. 2005. Direct binding of nuclear membrane protein MAN1 to emerin in vitro and two modes of binding to barrier-to-autointegration factor. The Journal of Biological Chemistry 280:13863–13870. doi: 10.1074/jbc.M413020200 [DOI] [PubMed] [Google Scholar]
- Mekhail K, Moazed D. 2010. The nuclear envelope in genome organization, expression and stability. Nature Reviews Molecular Cell Biology 11:317–328. doi: 10.1038/nrm2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. 2012. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife 1:e00011. doi: 10.7554/eLife.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada S, Ohmori SY, Taira M. 2003. XMAN1, an inner nuclear membrane protein, antagonizes BMP signaling by interacting with Smad1 in Xenopus embryos. Development 130:1783–1794. doi: 10.1242/dev.00401 [DOI] [PubMed] [Google Scholar]
- Padiath QS, Saigoh K, Schiffmann R, Asahara H, Yamada T, Koeppen A, Hogan K, Ptacek LJ, Fu YH. 2006. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nature Genetics 38:1114–1123. doi: 10.1038/ng1872 [DOI] [PubMed] [Google Scholar]
- Pan D, Estevez-Salmeron LD, Stroschein SL, Zhu X, He J, Zhou S, Luo K. 2005. The integral inner nuclear membrane protein MAN1 physically interacts with the R-Smad proteins to repress signaling by the transforming growth factor-{beta} superfamily of cytokines. The Journal of Biological Chemistry 280:15992–16001. doi: 10.1074/jbc.M411234200 [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. 2002. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109:307–320. doi: 10.1016/S0092-8674(02)00722-5 [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, Reinders M, Wessels L, van Steensel B. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Molecular Cell 38:603–613. doi: 10.1016/j.molcel.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss F, Fan JY, Kalive M, Bao S, Schuenemann E, Bjes ES, Price JL. 2004. Drosophila doubletime mutations which either shorten or lengthen the period of circadian rhythms decrease the protein Kinase activity of casein kinase I. Molecular and Cellular Biology 24:886–898. doi: 10.1128/MCB.24.2.886-898.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju GP, Dimova N, Klein PS, Huang HC. 2003. SANE, a novel LEM domain protein, regulates bone morphogenetic protein signaling through interaction with Smad1. The Journal of Biological Chemistry 278:428–437. doi: 10.1074/jbc.M210505200 [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Menet JS, Rosbash M. 2012. Nascent-seq indicates widespread cotranscriptional RNA editing in Drosophila. Molecular Cell 47:27–37. doi: 10.1016/j.molcel.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. 1998. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93:805–814. doi: 10.1016/S0092-8674(00)81441-5 [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Florens L, Guan T, Yates JR, III, Gerace L. 2005. Identification of novel integral membrane proteins of the nuclear envelope with potential disease links using subtractive proteomics. Novartis Foundation Symposium 264:63–76. doi: 10.1002/0470093765.ch6 [DOI] [PubMed] [Google Scholar]
- Shevelyov YY, Lavrov SA, Mikhaylova LM, Nurminsky ID, Kulathinal RJ, Egorova KS, Rozovsky YM, Nurminsky DI. 2009. The B-type lamin is required for somatic repression of testis-specific gene clusters. Proceedings of the National Academy of Sciences of USA 106:3282–3287. doi: 10.1073/pnas.0811933106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. 2003. The dynamics of chromosome organization and gene regulation. Annual Review of Biochemistry 72:573–608. doi: 10.1146/annurev.biochem.72.121801.161724 [DOI] [PubMed] [Google Scholar]
- Stewart CL, Kozlov S, Fong LG, Young SG. 2007. Mouse models of the laminopathies. Experimental Cell Research 313:2144–2156. doi: 10.1016/j.yexcr.2007.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. 2005. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nature Genetics 37:187–192. doi: 10.1038/ng1504 [DOI] [PubMed] [Google Scholar]
- Ulbert S, Platani M, Boue S, Mattaj IW. 2006. Direct membrane protein-DNA interactions required early in nuclear envelope assembly. The Journal of Cell Biology 173:469–476. doi: 10.1083/jcb.200512078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnes L, Peterfy M, Bergo MO, Young SG, Reue K. 2004. Lamin B1 is required for mouse development and nuclear integrity. Proceedings of the National Academy of Sciences of USA 101:10428–10433. doi: 10.1073/pnas.0401424101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. 1994. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264:719–725. doi: 10.1126/science.8171325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. 2009. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proceedings of the National Academy of Sciences of USA 106:21453–21458. doi: 10.1073/pnas.0909591106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C, Panda S, DiTacchio L. 2008. A high-throughput assay for siRNA-based circadian screens in human U2OS cells. PLOS ONE 3:e3457. doi: 10.1371/journal.pone.0003457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ. 2006. Inner nuclear membrane and regulation of Smad-mediated signaling. Biochimica et Biophysica Acta 1761:626–631. doi: 10.1016/j.bbalip.2006.02.010 [DOI] [PubMed] [Google Scholar]
- Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptacek LJ, Fu YH. 2005. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature 434:640–644. doi: 10.1038/nature03453 [DOI] [PubMed] [Google Scholar]
- Zhang EE, Kay SA. 2010. Clocks not winding down: unravelling circadian networks. Nature Reviews Molecular Cell Biology 11:764–776. doi: 10.1038/nrm2995 [DOI] [PubMed] [Google Scholar]
- Zwerger M, Medalia O. 2013. From lamins to lamina: a structural perspective. Histochemistry and Cell Biology 140:3–12. doi: 10.1007/s00418-013-1104-y [DOI] [PubMed] [Google Scholar]