Abstract
Natural populations of widely distributed organisms often exhibit genetic clinal variation over their geographical ranges. The European anchovy, Engraulis encrasicolus, illustrates this by displaying a two-clade mitochondrial structure clinally arranged along the eastern Atlantic. One clade has low frequencies at higher latitudes, whereas the other has an anti-tropical distribution, with frequencies decreasing towards the tropics. The distribution pattern of these clades has been explained as a consequence of secondary contact after an ancient geographical isolation. However, it is not unlikely that selection acts on mitochondria whose genes are involved in relevant oxidative phosphorylation processes. In this study, we performed selection tests on a fragment of 1044 bp of the mitochondrial cytochrome b gene using 455 individuals from 18 locations. We also tested correlations of six environmental features: temperature, salinity, apparent oxygen utilization and nutrient concentrations of phosphate, nitrate and silicate, on a compilation of mitochondrial clade frequencies from 66 sampling sites comprising 2776 specimens from previously published studies. Positive selection in a single codon was detected predominantly (99%) in the anti-tropical clade and temperature was the most relevant environmental predictor, contributing with 59% of the variance in the geographical distribution of clade frequencies. These findings strongly suggest that temperature is shaping the contemporary distribution of mitochondrial DNA clade frequencies in the European anchovy.
Keywords: positive selection, Engraulis encrasicolus, adaptation, sea temperature, high metabolism, mitochondrial cytochrome b gene
1. Introduction
I have called the principle, by which each slight variation, if useful, is preserved by the term of Natural Selection. [1, pp. 60–61]
Mitochondrial DNA (mtDNA) has been widely used in evolutionary biology research over the past 20 years under the implicit assumption of neutrality [2]. However, there is strong evidence that this molecule may be under positive selection, often related to thermal adaptation and aerobic capacity ([3] and references therein). The assumption that mtDNA polymorphisms are neutral has been tested in the historical demographic context, but rarely have these tests been taken further. Genetic variation affected by selection and not chiefly by demography can compromise mtDNA markers' usefulness to correctly estimate demographic changes, population structure or to date biogeographic events. In this event, molecular markers under selection may be useful to understand the processes that shape species distribution patterns and local adaptation.
Mitochondrial genes are involved in oxidative phosphorylation processes (OXPHOS complex) by which means the electron transport chain (ETC) creates a trans-membrane proton gradient that generates adenosine triphosphate (ATP) [4]. The ETC is formed by protein complexes of subunits that are encoded in either nuclear or mtDNA. Non-synonymous single nucleotide polymorphisms in any of the genes encoding ETC subunits can potentially affect the quality of electron flow or influence other relevant binding sites, such as that of coenzyme Q or CoQ [5]. It is therefore plausible that non-synonymous changes in the mtDNA will impact the fitness of organisms given the pivotal role of mitochondrial bioenergetics on adaptation to environmental variability [6].
An increasing number of studies have detected positive selective sweeps in the mitochondria, including adaptation to extreme O2 requirements of flying capacity in bats [7], low energy diet in large body mammals [8], high-altitude resistance in alpacas and monkeys [8–10] and climate-mediated adaption in humans [11–13]. Although there are few studies of mtDNA selection in marine fishes, selection in mitochondria has been invoked to explain patterns of genetic variation in the slippery-dick labrid (Halichoeres bivittatus) [14], the association between the distribution of mitochondrial lineages and sea surface temperature in the walleye pollock (Theragra chalcogramma) [15] and the cause of mito-nuclear coevolution that increases aerobic capacity and swimming performances in billfishes (Xiphiidae and Istiophoridae families [16]). Selection is also suspected to have promoted amino acid changes in proton pumping that influenced fitness in Pacific salmon species [17] and the Atlantic herring (Clupea harengus) [18].
The European anchovy provides an ideal system to investigate adaptive selection. It is distributed throughout tropical, subtropical and cold-temperate coastal areas (ca 60° N–40° S), facing contrasting environmental features, which implies an impressive tolerance to a broad range of temperatures (2–30°C) and salinities (5–41‰). This species also shows both morphological and genetic variability across its distributional area, displaying a dual-clade mitochondrial structure, arranged into a clinal frequency in the eastern Atlantic [19]. Clade A is present throughout the whole geographical distribution, but with lower frequencies at higher latitudes, whereas clade B has an anti-tropical distribution, with frequencies decreasing towards the tropics. This structure may reflect post-glacial secondary contact after an ancient isolation [20]. Nevertheless, one cannot exclude the relevance of other processes such as sex-biased dispersal, nuclear allelic convergence, incomplete mtDNA lineage sorting, adaptive introgression, demographic disparities, gamete incompatibility or, as considered in this work, adaptive selection [21,22] in promoting the observed genetic divergence.
Recently, a mito-genomic survey of a widely distributed marine mammal, the killer whale, showed high levels of amino acid conservation and only two positively selected codons, both in the cytochrome b (Cytb) gene, correlated with temperature adaptation [23]. Here, we focus on this gene to explore a putative instance of positive selection shaping the distribution of the two European anchovy Engraulis encrasicolus genetic lineages. We suggest that the Cytb of the European anchovy may be potentially affected by positive selective regimes that influence metabolism and constrain the distribution of mtDNA clades. We expect to detect non-synonymous substitutions that would provide selective advantage to one of the clades, possibly altering the function of the protein, promoting a better adaptation to local environment. We correlate the present-day distribution of mtDNA lineages of anchovies with various environmental factors. Our study is part of an emerging effort to better understand the role of natural selection in shaping the geographical distribution of genetic variation of organisms and their adaptation to changing environments.
2. Material and methods
(a). Sample collection, DNA extraction and amplification and sequence alignment
We collected 455 eastern Atlantic specimens of the European anchovy E. encrasicolus, from 18 locations, from Norway to South Africa, the Baltic and the Mediterranean Seas, covering most of the geographical distribution of the species (figure 1; electronic supplementary material, table S1). Fish were purchased at small coastal fish markets, as artisanal fisherman do not venture far, to ensure the correct origin of fish, or were collected on scientific cruises (see Acknowledgements). A small portion of white muscle or fin tissue were preserved in 96% ethanol and stored at −20°C. DNA extraction, polymerase chain reaction, purification and sequencing were performed for a 1044 bp fragment of the mitochondrial Cytb as described by Silva et al. [19]. Sequences were deposited in GenBank (see Data accessibility). The sequences were aligned using Clustal W [24] and visually inspected in Geneious v. 5.4 [25].
Table 1.
Sample summary information. (Map code numbers are represented in figure 1a. Sampling points = number of independent sampling points in each region; for complete information, see the electronic supplementary material, table S1. N, number of individuals; A, clade A absolute frequencies; B, clade B absolute frequencies.)
| sea basin | country | location | map code | N | A | B | sampling points |
|---|---|---|---|---|---|---|---|
| Baltic | Poland | Gdansk | 1 | 9 | 2 | 7 | 1 |
| Baltic | Germany | Kiel | 2 | 28 | 3 | 25 | 1 |
| Atlantic | Denmark | Denmark | 3 | 15 | 1 | 14 | 1 |
| Atlantic | Norway | Oslo | 4 | 24 | 2 | 22 | 1 |
| Atlantic | Scotland | Scotland | 5 | 35 | 6 | 29 | 2 |
| Atlantic | Germany | Germany | 6 | 11 | 1 | 10 | 1 |
| Atlantic | France | English Channel | 7 | 52 | 8 | 44 | 2 |
| Atlantic | France/Spain | Bay of Biscay | 8 | 336 | 170 | 166 | 11 |
| Atlantic | Spain | Galicia | 9 | 29 | 26 | 3 | 1 |
| Atlantic | Portugal | Portugal North | 10 | 93 | 81 | 12 | 2 |
| Atlantic | Portugal | Portugal South/Gulf of Cadiz | 11 | 236 | 212 | 24 | 5 |
| Atlantic | Spain | Canary islands | 12 | 82 | 76 | 6 | 2 |
| Atlantic | Senegal | Dakar | 13 | 34 | 32 | 2 | 1 |
| Atlantic | Guinea-Bissau | Guinea-Bissau | 14 | 20 | 20 | 0 | 1 |
| Atlantic | Ghana | Accra | 15 | 25 | 25 | 0 | 1 |
| Atlantic | Angola | Angola | 16 | 24 | 20 | 4 | 1 |
| Atlantic | Namibia | Namibia | 17 | 24 | 5 | 19 | 1 |
| Atlantic | South Africa | South Africa | 18 | 59 | 24 | 35 | 3 |
| Mediterranean | Spain | Alboran Sea | 19 | 115 | 106 | 9 | 2 |
| Mediterranean | Spain | Balearic Sea | 20 | 75 | 27 | 48 | 3 |
| Mediterranean | France | Gulf of Lion | 21 | 72 | 26 | 46 | 2 |
| Mediterranean | Italy | Livorno | 22 | 55 | 27 | 28 | 1 |
| Mediterranean | Tunisia | Tunis | 23 | 28 | 16 | 12 | 1 |
| Mediterranean | Italy | Adriatic Sea North | 24 | 114 | 12 | 102 | 2 |
| Mediterranean | Italy/Croatia | Adriatic Sea South | 25 | 188 | 31 | 157 | 4 |
| Mediterranean | Greece | Aegean Sea/ Ionian Sea | 26 | 967 | 598 | 369 | 12 |
| Mediterranean | Israel | Telaviv | 27 | 26 | 26 | 0 | 1 |
| grand total | 2776 | 1583 | 1193 | 66 |
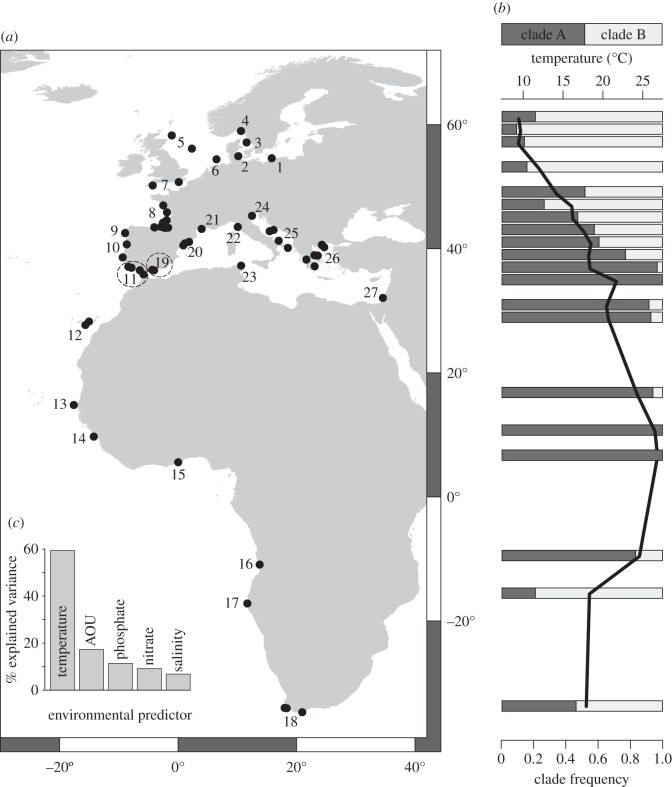
Figure 1.
(a) Locations used for environmental correlates of mitochondrial clades frequencies; numbers correspond to map code in table 1 and the electronic supplementary material, table S1. (b) Climatological sea temperature from World Ocean Atlas 2009 between 0 and 10 m (black line) and clade frequency (clade A, dark grey; clade B, light grey). (c) Importance of environmental variables after hierarchical partitioning analysis; AOU, apparent oxygen utilization.
(b). Tests of recombination and selection
We tested the alignment for evidence of mitochondrial Cytb recombinants using Genetic Algorithms for Recombination Detection (GARD) analysis [26] implemented in the online interface www.datamonkey.org [27]. To assess whether selection was acting on Cytb, the Z-test [28] was performed in Mega v. 5 [29]. We further implemented likelihood and Bayesian-based methods to identify site-specific Cytb positive selection where the rate of non-synonymous substitution (dN) is greater than the rate of synonymous substitution (dS). We applied a suit of different algorithms for selection inference to our data: single likelihood ancestor counting (SLAC), fixed effects likelihood (FEL) [30], internal fixed effects likelihood (IFEL) [31], fast unconstrained Bayesian approximation (FUBAR) [32] and mixed effects model of evolution (MEME) [33] to our data. SLAC is based on the reconstruction of the ancestral sequences and the counts of dS and dN at each codon position of the phylogeny. The FEL method estimates the ratio of non-synonymous to synonymous substitutions on a site-by-site basis for the entire tree, whereas IFEL does the same estimation only on the interior branches. FUBAR detects positive selection much faster than the other methods by using a likelihood instead of a Bayesian approximation. MEME is capable of identifying instances of both episodic and pervasive positive selection at the level of an individual site. Sites with p-values less than 0.05 for SLAC, FEL, IFEL and MEME, posterior probability of more than 0.9 for FUBAR, were considered as being under selection. We applied all these methods to prevent against our results being an artefact of a particular methodology or a set of assumptions.
(c). Biochemical sources of intrinsic variation
The Cytb dataset was aligned to a reference sequence available on GenBank (ACC number: NC_009581). Additionally, we used TreeSAAP [34] to categorize 539 biochemical/structural physico-chemical changes owing to amino acid replacements into eight magnitude categories and determine whether the observed magnitude of amino acid changes deviates significantly from neutral expectations. We run analyses according to Woolley et al. [34] and considered only amino acid replacements with significant magnitude categories six to eight (p < 0.001). The crystallographic structure of the cytochrome bc1 complex interacting with cytochrome c was taken from the Protein Databank, PDB 3CX5 [35]. The homology model for the E. encrasicolus structure, based on a sequence variant with a Methionine residue at position 368 (the yeast sequence has a Valine residue at the homologous position), was obtained from the ModBase database of homology models [36].
(d). Environmental correlates of mitochondrial clade frequencies
We compiled E. encrasicolus mitochondrial clade frequencies comprising 66 sampling sites from previous studies [19,20,37–41] (electronic supplementary material, table S1) and used general linear models (GLM) of the binomial family (logit) to evaluate the correlation between clade frequencies and a variety of environmental variables. Data on temperature, salinity, apparent oxygen utilization and nutrient concentrations (phosphate, nitrate and silicate) for depths less than 10 m were obtained from the World Ocean Atlas 2009, one-degree objectively analysed climatology datasets [42] in NetCDF format and imported as geo-referenced layers into R 2.15.3 [43] using the ncdf [44] and raster [45] packages (electronic supplementary material, table S1). The package hier.part [46] was used to quantify the independent correlation of each predictor variable with the clade frequency, a method called hierarchical partitioning [47,48]. Hierarchical partitioning is a regression technique in which all possible linear models are jointly considered in an attempt to identify the most likely causal factors, providing a measure of the effect of each variable that is largely independent from that of other variables [47,48].
3. Results
(a). Recombination and selection
From the 455 individuals analysed for the Cytb fragment, 246 polymorphic sites yielded 316 haplotypes, seven amino acid variable sites and 11 amino acid type sequences (figure 2). No evidence for recombination was found with GARD. Positive selection over all sites was detected on Cytb (clade A: −5.81; p = 1; clade B: −8.75; p = 1). In total, four amino acid changes were identified as being under selection, three under purifying selection and one under positive selection (codon 368) (table 2). However, the significant positive selected site located in the ninth trans-membrane helix of the Cytb, was detected only in clade B with FUBAR, IFEL and FEL methods. Codon 368 contains a Valine or an Alanine in 379 individuals, 25% belonging to clade B, and a Methionine in 66 individuals, 99% of which belonging to clade B (figure 2).
Figure 2.
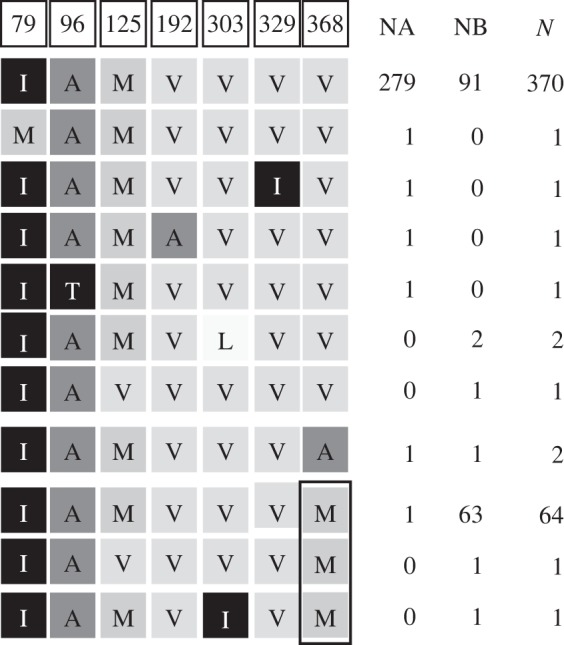
Amino acid substitutions in the mitochondrial fragment of cytochrome b of E. encrasicolus. NA, number of clade A individuals; NB, number of clade B individuals; N, total number of individuals. Shading is meant to evidence differences and similarities between amino acid positions. Box indicates amino acid change under selection.
Table 2.
Positively and negatively selected sites in the cytochrome b gene estimated by FUBAR (*bpp ≥ 0.9; **bpp ≥ 0.95; ***bpp ≥ 0.99; ****bpp ≥ 0.999) and SLAC, IFEL, FEL and MEME models (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
| alignment position |
278 | 302 | 812 | 1007 | |
|---|---|---|---|---|---|
| codon |
125 | 133 | 303 | 368 | |
| selection type |
purifying | purifying | purifying | positive | |
| all | FUBAR | * | *** | * | |
| SLAC | ** | * | |||
| IFEL | * | * | |||
| FEL | * | ** | ** | ||
| MEME | |||||
| clade A | FUBAR | *** | *** | ** | |
| SLAC | *** | * | |||
| IFEL | |||||
| FEL | *** | ** | * | ||
| MEME | |||||
| clade B | FUBAR | **** | |||
| SLAC | ** | ||||
| IFEL | *** | ||||
| FEL | **** | ||||
| MEME | *** | ||||
(b). Adaptation at the molecular level
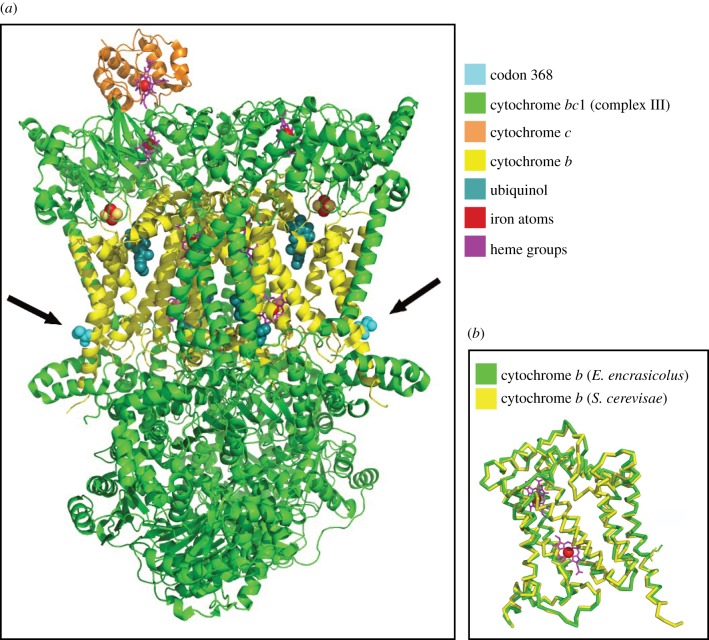
The Cytb sequences of yeast Saccharomyces cerevisae and E. encrasicolus were aligned, showing 50% identity. Alignment of the model with the Cytb monomer in the 3CX5 produced an root mean square deviation of 0.7 Å and allowed identification of the yeast structural homologue of Met 368 in the Engraulis sequence (Valine 369 in yeast) (figure 3). TreeSAAP identified 15 significant physico-chemical properties potentially influenced by positive selection in codon 368 (electronic supplementary material, table S2).
Figure 3.
(a) Crystal structure of yeast (S. cerevisae) cytochrome bc1 (complex III) complexed with cytochrome c. The black arrows point to the two Valine 369 residues (368 in the E. encrasicolus sequence); (b) Backbone alignment of the yeast (S. cerevisae) and E. encrasicolus cytochrome b structures.
(c). Environmental correlates of mitochondrial clades frequency
The clade frequencies shifted smoothly along latitudinal gradients in the Atlantic Ocean, the Baltic and the Mediterranean (figure 1). Clade A was found in all the sampling locations, whereas clade B was absent from locations off Senegal, Guinea, Ghana and Israel. Clade A was present in higher frequencies mostly at lower latitudes (off Portugal, Morocco, Canary Islands, Senegal, Guinea, Ghana, Angola, northern and central Aegean and Israel), whereas clade B was present in higher frequencies at higher latitudes in the Atlantic Ocean (from the Norwegian coast to the Bay of Biscay and from the Namibia coast to South African waters), the Baltic Sea and in most of northern Mediterranean locations (off Gulf of Lion, Adriatic, Ionian and southern Aegean). Locations in the Bay of Biscay, Ligurian Sea and off Tunisia presented ratios between 0.4 and 0.6. From the six tested environmental variables, silicate was not considered in the final GLM because its inclusion did not significantly improve the model (comparison of the six variable versus the five variable model: χ21 = 1.39, p = 0.24). The coefficients for the remaining variables are shown in table 3. Sea surface temperature was the best clade frequency predictor, with a relative importance of 58.7%, followed by apparent oxygen utilization, phosphate, nitrate and salinity, with relative importance of 14.6%, 11.2%, 8.9% and 6.5%, respectively (figure 1c).
Table 3.
Results of the GLMs relating clade frequency with predictor variables from World Ocean Atlas 2009. (Silicate is not listed because it did not contribute significantly to the model (see Results). n.s., not significant.)
| parameters | estimate | s.e. | z-value | Pr(>|z|) | sign. |
|---|---|---|---|---|---|
| intercept | −0.93091 | 0.95578 | −0.974 | 0.725 | n.s. |
| temperature | −0.34154 | 0.02762 | −12.367 | <2 × 10−16 | <0.001 |
| nitrate | 0.29028 | 0.05215 | −5.566 | 2.61 × 10−8 | <0.001 |
| salinity | 0.15407 | 0.02378 | 6.479 | 9.23 × 10−11 | <0.001 |
| phosphate | 4.02129 | 0.60220 | 6.678 | 2.43 × 10−11 | <0.001 |
| apparent oxygen utilization (AOU) | 4.01298 | 0.66952 | 5.994 | 2.05 × 10−9 | <0.001 |
4. Discussion
Our results contribute to a better understanding of the role of natural selection in shaping the distribution of marine organisms, in particular the influence of sea temperature on the distribution of mitochondrial lineages in the European anchovy. Here, we identified one putatively adaptive change in the mitochondrial Cytb gene, associated with clade B, more abundant in low-temperature environments, suggesting that selection is acting on the E. encrasicolus mito-genome. Although dN/dS methods are generally biased against detecting positive selection in conservative gene sequences even when single amino acid changes can turn out to be adaptive [23], all applied methods were capable of detecting selection on a single amino acid change, which strengthens the nonetheless circumstantial evidence of the inference.
(a). Genetic clines and environmental correlates of mitochondrial clade frequency
The European anchovy is widely distributed implying an adaptation to distinct environmental features, such as the steep thermal cline in the eastern Atlantic or salinity gradients between the Baltic Sea and the Atlantic Ocean. The two mtDNA clades found in the European anchovy are sympatric over most of the distribution range and exhibit a remarkable latitudinal cline in the eastern Atlantic [19]. Previous studies [21,22,40,41] assumed the observed two-clade pattern as a consequence of ancient isolations followed by secondary contact. However, genetic clines may represent a balance between selection, genetic drift and dispersal, across time and space [49,50]. These clines are present in different small pelagic fish species and have been related to both historical factors and hydrographic barriers to dispersal in sardines [51] or maintained by selective pressures in the Atlantic herring [18]. One possible explanation for the origin and persistence of the dual-clade structure in the European anchovy may be an adaptation to the physical properties of the environment, in particular sea temperature, as suggested by the GLM (figure 1 and table 3). Temperature along the distribution range of the European anchovy varies clinally (figure 1) and contributes 58.7% to the model, accounting for five to six times more variance in the geographical distribution of clade frequencies than any other environmental predictor. The second best predictor for the distribution of the mtDNA clade frequencies was apparent oxygen utilization (14.6%). Oxygen availability is of extreme importance in ectothermic small pelagic fishes, especially at higher latitudes where water temperature is low and consequently metabolism decreases. Although anchovies have high capacity for migration, environmental affinity precludes dispersal, contributing to population structure [19,40,41,52]. Temperature has been identified as one of the major selective forces acting on mtDNA [53]. Temperature-mediated selection was found in humans [11–13], where genetic differentiation between pairs of populations is correlated to difference in temperature [11]. In the marine realm, the influence of temperature in shaping mitochondrial diversity was described in the walleye pollock (T. chalcogramma) where sea surface temperature and mitochondrial lineages were significantly correlated, showing a latitudinal clinal distribution and higher genetic diversity than under a mutation-drift equilibrium model [15].
(b). Adaptation at the molecular level
Small pelagic fishes are ectothermic and metabolic rates increase with increasing water temperature [54]. Anchovies have high-metabolic requirements and more than 95% of the myofibrils are adjacent to mitochondria, suggesting a high dependence on aerobic metabolism [55]. When water temperature decreases, body temperature and metabolic rates decrease, probably affecting swimming performance [56], muscle-associated energetic needs [55,57], egg size and fecundity [58].
We hypothesized that the substitution of a Valine for a Methionine in codon 368 could play a role in the ETC, enhancing the electron transfer process from Cytb to Cytc. However, analysis of the crystal structure of the Cytb (cytochrome bc1, complex III) shows that the Valine replacement by Methionine at position 368 is not likely to affect efficiency of the electron transfer mechanism (figure 3), for the following reasons. First, a direct effect is unlikely, owing to the marginal positioning of residue 368, well clear of the Cytb electron pathways. Second, conformational effects are also unlikely, because a single replacement of Valine by Methionine should not cause major structural changes, particularly at the protein surface, as is the case here. The latter consideration is supported by the similarity between the crystal structure and the ModBase model at this position (figure 3). Other possible causes may be related to protein trafficking, membrane integration or stability of the putative cytochrome c reductase–oxidase super-complex as indicated by the distribution of amino acid residues in the 18 non-redundant families of thermophilic proteins (KUMS000101; electronic supplementary material, table S2). A change in heat capacity (HUTJ700101; electronic supplementary material, table S2) could be related to a variation in the entropic component, the free energy of folding of Cytb (hydrophobic effect), but its significance depends on the magnitude of the heat capacity change, which should be very small for a single residue replacement. Also this effect tends to be much less important for a protein working in a non-polar environment like a lipid membrane. Another plausible explanation for positive selection in codon 368 is related to a response to oxidative stress. Chemically reactive molecules containing oxygen, known as ROS (reactive oxygen species), are formed as a natural by-product of the normal metabolism of oxygen [59]. In ectothermic animals, ROS increases at low temperatures, causing oxidative damage to cells [60]. Methionine residues may act as catalytic antioxidants, protecting both the protein and other macromolecules [61]. Cells with decreased Methionine content in Eschirichia coli were more susceptible to oxidative stress and exhibited lower rates of survival [61]. The biochemical intricacy of the phosphorilative oxidation processes precludes us from predicting the exact functional implications of the substitution, and we are limited to the suggestion that it will have an impact on the overall ATP production by the respiratory chain, and consequently on the overall metabolic performance of clade B.
(c). Neutrality versus non-neutrality of mitochondrial DNA in marine fishes
The vast majority of population genetics and phylogeography studies in marine fishes do not reveal any evidence of potential climate selection in the mtDNA. This observation may be explained because deviations to neutrality are usually performed in an historical demography context, with tests such as Tajima D [62], or Fu's Fs test [63]. The studies report many instances of deviations, but those deviations are mostly interpreted as population expansions [64]. Interestingly, negative D values can also indicate natural selection, but this possibility is seldom explored. The result is that most phylogeography studies over the past 30 years relied on mitochondrial markers assuming that selection was not acting on the molecule. However, the panorama is changing, and the neutrality assumption was contested by a meta-analysis of over 1600 animal species [65]. In marine fishes, we accounted for several instances where mtDNA non-neutrality was invoked to explain the genetic patterns detected [14–18]. It is possible that many more instances of selection may be hidden in the demographic analysis and remained unreported. Neutrality assumptions may have compromised studies' interpretations and biased conclusions. This paper shows that the role-played by selection may reflect the effect of environmental drivers on geographical/genetic patterns and not the species historical demography. It is therefore relevant that future studies properly address the deviations of neutrality commonly found in demographic inferences, to evaluate the putative role of selection.
5. Conclusion
Over the past 30 years, most genetic studies performed in marine fish species assumed that mtDNA was neutral and did not account for the influence of selective pressures in the estimation of population structure. Our results contribute to the growing body of evidence that mtDNA of natural populations is affected by selective pressures that need to be accounted for in historical interpretations of biogeographic scenarios. Moreover, our findings strongly suggest that the contemporary distribution of mtDNA clade frequencies in the European anchovy is being shaped by temperature. Additionally, molecular adaptations to different metabolic requirements may be the key to understand how species will adapt to future climate change.
Supplementary Material
Acknowledgements
This work was only possible owing to the collaboration of colleagues, institutions and research vessels for collecting and shipping samples: E. Torstensen (IMR, Norway), S. Vaz, F. Coppin, Y. Verin, D. Leroy and B. Liorzou (IFREMER, France), I. Zarraonaindia (UPV/EHU, Spain), R. Ben-Hamadou and K. Erzini (CCMAR, Portugal), A. Brito, A. Guerra and A. Dias (Portugal), J. Tornero and J. Alarcón (IEO, Spain), M. Santamaria (Canary Islands), A. Hervé (Senegal), L. Fonseca (Guinea-Bissau), H. Dankwa (WRI, Ghana), Rob Leslie (South Africa) and S. Marcato (Italy). We also would like to thank to J. Neiva, J. Macedo, R. Ben-Hamadou, C. Patrão, António M. Santos and W. S. Grant for fruitful discussions.
Data accessibility
DNA sequences: GenBank accession nos. JQ716609–JQ716625; JQ716629–JQ716748; JX683020–JX683113; KF601435–KF601545. Data information of samples used for environmental correlates analyses and number of individuals sequenced used for selection tests uploaded as electronic supplementary material.
Funding statement
G.S.'s research was supported by a doctoral grant from FCT (SFRH/BD/36600/2007). F.P.L.'s research was supported by FCT through an Investigator FCT contract (IF/00043/2012).
References
- 1.Darwin C. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life, 1st edn London, UK: John Murray. [PMC free article] [PubMed] [Google Scholar]
- 2.Avise JC. 2000. Phylogeography: the history and formation of species. Cambridge, MA: Harvard University Press. [Google Scholar]
- 3.Galtier N, Nabholz B, Giémin S, Hurst GDD. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol. Ecol. 18, 4541–4550. ( 10.1111/j.1365-294X.2009.04380.x) [DOI] [PubMed] [Google Scholar]
- 4.Mitchell P. 2011. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biochim. Biophys. Acta Bioenerg. 1807, 1507–1538. ( 10.1016/j.bbabio.2011.09.018) [DOI] [PubMed] [Google Scholar]
- 5.Beckstead WA, Ebbert MTW, Rowe MJ, McClellan DA. 2009. Evolutionary pressure on mitochondrial cytochrome b is consistent with a role of CytbI7T affecting longevity during caloric restriction. PLoS ONE 4, e5836 ( 10.1371/journal.pone.0005836) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gershoni M, Templeton AR, Mishmar D. 2009. Mitochondrial bioenergetics as a major motive force of speciation. BioEssays 31, 642–650. ( 10.1002/bies.200800139) [DOI] [PubMed] [Google Scholar]
- 7.Shen Y-Y, Liang L, Zhu Z-H, Zhou W-P, Irwin DM, Zhang Y-P. 2010. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc. Natl Acad. Sci. USA 107, 8666–8671. ( 10.1073/pnas.0912613107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Fonseca R, Johnson W, O'Brien S, Ramos M, Antunes A. 2008. The adaptive evolution of the mammalian mitochondrial genome. BMC Genomics 9, 119 ( 10.1186/1471-2164-9-119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hochachka PW, Stanley C, Merkt J, Sumar-Kalinowski J. 1983. Metabolic meaning of elevated levels of oxidative enzymes in high altitude adapted animals: an interpretive hypothesis. Resp. Physiol. 52, 303–313. ( 10.1016/0034-5687(83)90087-7) [DOI] [PubMed] [Google Scholar]
- 10.Yu L, Wang X, Ting N, Zhang Y. 2011. Mitogenomic analysis of Chinese snub-nosed monkeys: evidence of positive selection in NADH dehydrogenase genes in high-altitude adaptation. Mitochondrion 11, 497–503. ( 10.1016/j.mito.2011.01.004) [DOI] [PubMed] [Google Scholar]
- 11.Balloux F, Handley L-JL, Jombart T, Liu H, Manica A. 2009. Climate shaped the worldwide distribution of human mitochondrial DNA sequence variation. Proc. R. Soc. B 276, 3447–3455. ( 10.1098/rspb.2009.0752) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishmar D, et al. 2003. Natural selection shaped regional mtDNA variation in humans. Proc. Natl Acad. Sci. USA 100, 171–176. ( 10.1073/pnas.0136972100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. 2004. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 303, 223–226. ( 10.1126/science.1088434) [DOI] [PubMed] [Google Scholar]
- 14.Haney RA, Silliman BR, Rand DM. 2010. Effects of selection and mutation on mitochondrial variation and inferences of historical population expansion in a Caribbean reef fish. Mol. Phylogenet. Evol. 57, 821–828. ( 10.1016/j.ympev.2010.07.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant WS, Spies IB, Canino MF. 2006. Biogeographic evidence for selection on mitochondrial DNA in north Pacific walleye pollock Theragra chalcogramma. J. Hered. 97, 571–580. ( 10.1093/jhered/esl033) [DOI] [PubMed] [Google Scholar]
- 16.Dalziel AC, Moyes CD, Fredriksson E, Lougheed SC. 2006. Molecular evolution of cytochrome c oxidase in high-performance fish (Teleostei: Scombroidei). J. Mol. Evol. 62, 319–331. ( 10.1007/s00239-005-0110-7) [DOI] [PubMed] [Google Scholar]
- 17.Garvin MR, Bielawski JP, Gharrett AJ. 2011. Positive Darwinian selection in the piston that powers proton pumps in complex I of the mitochondria of Pacific salmon. PLoS ONE 6, e24127 ( 10.1371/journal.pone.0024127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teacher A, Andre C, Merila J, Wheat C. 2012. Whole mitochondrial genome scan for population structure and selection in the Atlantic herring. BMC Evol. Biol. 12, 248 ( 10.1186/1471-2148-12-248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva G, Horne JB, Castilho R. 2014. Anchovies go north and west without losing diversity: post-glacial range expansions in a small pelagic fish. J. Biogeogr. 41, 1171–1182. ( 10.1111/jbi.12275). [DOI] [Google Scholar]
- 20.Magoulas A, Tsimenides N, Zouros E. 1996. Mitochondrial DNA phylogeny and the reconstruction of the population history of a species: the case of the European anchovy (Engraulis encrasicolus). Mol. Biol. Evol. 13, 178–190. ( 10.1093/oxfordjournals.molbev.a025554) [DOI] [PubMed] [Google Scholar]
- 21.Grant WS. 2005. A second look at mitochondrial DNA variability in European anchovy (Engraulis encrasicolus): assessing models of population structure and the Black Sea isolation hypothesis. Genetica 125, 293–309. ( 10.1007/s10709-005-0717-z) [DOI] [PubMed] [Google Scholar]
- 22.Kristoffersen JB, Magoulas A. 2008. Population structure of anchovy Engraulis encrasicolus L. in the Mediterranean Sea inferred from multiple methods. Fish. Res. 91, 187–195. ( 10.1016/j.fishres.2007.11.024) [DOI] [Google Scholar]
- 23.Foote AD, Morin PA, Durban JW, Pitman RL, Wade P, Willerslev E, Gilbert MTP, da Fonseca RR. 2010. Positive selection on the killer whale mitogenome. Biol. Lett. 7, 116–118. ( 10.1098/rsbl.2010.0638) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. ( 10.1093/nar/22.22.4673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drummond A, et al. 2011. Geneious v. 5.4. Auckland, New Zealand: Biomatters Ltd; (http://www.geneious.com). [Google Scholar]
- 26.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22, 3096–3098. ( 10.1093/bioinformatics/btl474) [DOI] [PubMed] [Google Scholar]
- 27.Delport W, Poon AFY, Frost SDW, Kosakovsky Pond SL. 2010. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26, 2455–2457. ( 10.1093/bioinformatics/btq429) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. ( 10.1093/Molbev/Msr121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosakovsky Pond SL, Frost SDW. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22, 1208–1222. ( 10.1093/molbev/msi105) [DOI] [PubMed] [Google Scholar]
- 31.Kosakovsky Pond SL, Frost SDW, Grossman Z, Gravenor MB, Richman DD, Brown AJL. 2006. Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput. Biol. 2, e62 ( 10.1371/journal.pcbi.0020062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. 2013. FUBAR: a fast, unconstrained Bayesian approximation for inferring selection. Mol. Biol. Evol. 30, 1196–1205. ( 10.1093/molbev/mst030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8, e1002764 ( 10.1371/journal.pgen.1002764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA. 2003. TreeSAAP: selection on amino acid properties using phylogenetic trees. Bioinformatics 19, 671–672. ( 10.1093/bioinformatics/btg043) [DOI] [PubMed] [Google Scholar]
- 35.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The protein data bank. Nucleic Acids Res. 28, 235–242. ( 10.1093/nar/28.1.235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pieper U, Eswar N, Stuart AC, Ilyin VA, Sali A. 2002. ModBase, a database of annotated comparative protein structure models. Nucleic Acids Res. 30, 255–259. ( 10.1093/nar/30.1.255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borrell YJ, Piñera JA, Sánchez Prado JA, Blanco G. 2012. Mitochondrial DNA and microsatellite genetic differentiation in the European anchovy Engraulis encrasicolus L. ICES J. Mar. Sci. 69, 1357–1371. ( 10.1093/icesjms/fss129) [DOI] [Google Scholar]
- 38.Bouchenak-Khelladi Y, Durand JD, Magoulas A, Borsa P. 2008. Geographic structure of European anchovy: a nuclear-DNA study. J. Sea Res. 59, 269–278. ( 10.1016/j.seares.2008.03.001) [DOI] [Google Scholar]
- 39.Grant WS, Leslie RW, Bowen BW. 2005. Molecular genetic assessment of bipolarity in the anchovy genus Engraulis. J. Fish Biol. 67, 1242–1265. ( 10.1111/j.1095-8649.2005.00820.x) [DOI] [Google Scholar]
- 40.Magoulas A, Castilho R, Caetano S, Marcato S, Patarnello T. 2006. Mitochondrial DNA reveals a mosaic pattern of phylogeographical structure in Atlantic and Mediterranean populations of anchovy (Engraulis encrasicolus). Mol. Phylogenet. Evol. 3, 734–746. ( 10.1016/j.ympev.2006.01.016) [DOI] [PubMed] [Google Scholar]
- 41.Zarraonaindia I, Iriondo M, Albaina A, Pardo MA, Manzano C, Grant WS, Irigoien X, Estonba A. 2012. Multiple SNP markers reveal fine-scale population and deep phylogeographic structure in European anchovy (Engraulis encrasicolus L.). PLoS ONE 7, e42201 ( 10.1371/journal.pone.0042201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyer TP, et al. 2009. World Ocean Database. Washington, DC: NOAA Atlas NESDIS 66, U.S. Government Printing Office. [Google Scholar]
- 43.R Development Core Team. 2013. R: a language and environment for statistical computing. Viennia, Austria: R Foundation for Statistical Computing.
- 44.Pierce D. 2011. ncdf: interface to Unidata netCDF data files. R package v. 1.6.6. See http://cran.r-project.org/web/packages/ncdf.
- 45.Hijmans RJ. 2013. Raster: geographic data analysis and modeling. R package v. 2.1–48. See http://cran.r-project.org/web/packages/raster/.
- 46.Walsh C, Nally RM. 2013. hier.part: hierarchical partitioning. R package v. 1.0–4. See http://cran.r-project.org/web/packages/hier.part/.
- 47.Chevan A, Sutherland M. 1991. Hierarchical partitioning. Am. Stat. 45, 90–96. ( 10.1080/00031305.1991.10475776) [DOI] [Google Scholar]
- 48.Nally RM. 1996. Hierarchical partitioning as an interpretative tool in multivariate inference. Aust. J. Ecol. 21, 224–228. ( 10.1111/j.1442-9993.1996.tb00602.x) [DOI] [Google Scholar]
- 49.Barton NH, Hewitt GM. 1985. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 16, 113–148. ( 10.1146/annurev.es.16.110185.000553) [DOI] [Google Scholar]
- 50.Barton NH, Gale KS. 1993. Genetic analysis of hybrid zones. In Hybrid zones and the evolutionary process (ed. Harrison RG.), pp. 13–45. New York, NY: Oxford University Press. [Google Scholar]
- 51.Chlaida M, Laurent V, Kifani S, Benazzou T, Jaziri H, Planes S. 2009. Evidence of a genetic cline for Sardina pilchardus along the northwest African coast. ICES J. Mar. Sci. 66, 264–271. ( 10.1093/icesjms/fsn206) [DOI] [Google Scholar]
- 52.Viñas J, Sanz N, Peñarrubia L, Araguas R-M, García-Marín J-L, Roldán M-I, Pla C. 2013. Genetic population structure of European anchovy in the Mediterranean Sea and the Northeast Atlantic Ocean using sequence analysis of the mitochondrial DNA control region. ICES J. Mar. Sci. 71, 391–397. ( 10.1093/icesjms/fst132) [DOI] [Google Scholar]
- 53.Ballard JWO, Whitlock MC. 2004. The incomplete natural history of mitochondria. Mol. Ecol. 13, 729–744. ( 10.1046/j.1365-294X.2003.02063.x) [DOI] [PubMed] [Google Scholar]
- 54.Elliott JM. 1976. The energetics of feeding, metabolism and growth of brown trout (Salmo trutta L.) in relation to body weight, water temperature and ration size. J. Anim. Ecol. 45, 923–948. ( 10.2307/3590) [DOI] [Google Scholar]
- 55.Johnston IA. 1982. Quantitative analyses of ultrastructure and vascularization of the slow muscle fibres of the anchovy. Tissue Cell 14, 319–328. ( 10.1016/0040-8166(82)90030-1) [DOI] [PubMed] [Google Scholar]
- 56.Blier PU, Guderley HE. 1993. Mitochondrial activity in rainbow trout red muscle: the effect of temperature on the ADP-dependence of ATP synthesis. J. Exp. Biol. 176, 145–158. [Google Scholar]
- 57.Johnston IA, Dunn J. 1987. Temperature acclimation and metabolism in ectotherms with particular reference to teleost fish. In Symposia of the society for the experimental biology (eds Bowler K, Fuller BJ.), pp. 67–93. Cambridge, UK: Cambridge University Press. [PubMed] [Google Scholar]
- 58.Ballard JWO, Rand DM. 2005. The population biology of mitochondrial DNA and its phylogenetic implications. Annu. Rev. Ecol. Evol. Syst. 36, 621–642. ( 10.1146/annurev.ecolsys.36.091704.175513) [DOI] [Google Scholar]
- 59.Dowling DK, Friberg U, Lindell J. 2008. Evolutionary implications of non-neutral mitochondrial genetic variation. Trends Ecol. Evol. 23, 546–554. ( 10.1016/j.tree.2008.05.011) [DOI] [PubMed] [Google Scholar]
- 60.Lalouette L, Williams CM, Hervant F, Sinclair BJ, Renault D. 2011. Metabolic rate and oxidative stress in insects exposed to low temperature thermal fluctuations. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 158, 229–234. ( 10.1016/j.cbpa.2010.11.007) [DOI] [PubMed] [Google Scholar]
- 61.Luo S, Levine RL. 2009. Methionine in proteins defends against oxidative stress. FASEB J. 23, 464–472. ( 10.1096/fj.08-118414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tajima F. 1989. Statistical testing for the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fu YX. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cox NL, Zaslavskaya NI, Marko PB. 2013. Phylogeography and trans-Pacific divergence of the rocky shore gastropod Nucella lima. J. Biogeogr. 41, 615–627. ( 10.1111/jbi.12217) [DOI] [Google Scholar]
- 65.Bazin E, Glémin S, Galtier N. 2006. Population size does not influence mitochondrial genetic diversity in animals. Science 312, 570–572. ( 10.1126/science.1122033) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences: GenBank accession nos. JQ716609–JQ716625; JQ716629–JQ716748; JX683020–JX683113; KF601435–KF601545. Data information of samples used for environmental correlates analyses and number of individuals sequenced used for selection tests uploaded as electronic supplementary material.