Abstract
Dysregulation of the insulin-like growth factor type I receptor (IGF1R) has been implicated in the progression and therapeutic resistance of malignancies. In acute myeloid leukemia (AML) cells, IGF1R is one of the most abundantly phosphorylated receptor tyrosine kinases, promoting cell growth through the PI3K/Akt signaling pathway. However, little is known regarding the molecular mechanisms underlying IGF1R gene dysregulation in cancer. We discovered a novel intragenic long noncoding RNA (lncRNA) within the IGF1R locus, named IRAIN, which is transcribed in an antisense direction from an intronic promoter. The IRAIN lncRNA was expressed exclusively from the paternal allele, with the maternal counterpart being silenced. Using both reverse transcription-associated trap and chromatin conformation capture assays, we demonstrate that this lncRNA interacts with chromatin DNA and is involved in the formation of an intrachromosomal enhancer/promoter loop. Knockdown of IRAIN lncRNA with shRNA abolishes this intrachromosomal interaction. In addition, IRAIN was downregulated both in leukemia cell lines and in blood obtained from high-risk AML patients. These data identify IRAIN as a new imprinted lncRNA that is involved in long-range DNA interactions.
INTRODUCTION
Dysregulation of the genes encoding members of the insulin-like growth factor axis, including the receptor IGF1R and the ligands IGF1 and IGF2, can contribute to the progression and metastasis of human cancers (1–5). In cells from patients with acute myeloid leukemia (AML), IGF-I and IGF-II promote cell growth and survival via IGF1R receptor-mediated activation of the PI3K/Akt signaling pathway (6–8). In clinical samples, AML blasts contain high concentrations of IGF-I/II, resulting in a growth and survival advantage and increasing the survival of leukemia cells through autocrine and paracrine loops (9). IGF1R is one of the most abundantly phosphorylated receptor tyrosine kinases in leukemia cells (10–12), and phosphorylation is increased in leukemia cells with Ara-C resistance (13,14) The IGF1R inhibitor BMS-536924 substantially inhibited growth and proliferation of both mouse and human leukemia cells in vitro (15).
Numerous clinical cancer trials have been performed that target IGF1R (16–20), including those with drugs that inhibit the IGFIR tyrosine kinase using monoclonal antibodies and small molecules (21). However, little is known regarding the mechanism by which IGFIR becomes dysregulated in tumors. Using a novel R3C (RNA-guided Chromatin Conformation Capture) method recently developed in our lab (Supplementary Figure S1) (22), we demonstrate the presence of a novel long noncoding RNA (lncRNA) originating from the IGF1R promoter. lncRNAs have been implicated in a number of regulatory functions in eukaryotic genomes (23–25), including the epigenetic regulation in cis and in trans of a cluster of genes within large chromosomal domains (26–30). In this communication, we characterize the allelic expression of IRAIN lncRNA and its role in the formation of interchromosomal interactions in normal and tumor cells.
MATERIALS AND METHODS
Cell lines
Leukemia cell lines used in this study, K562, KG-1, KG-1a, HL60 and TF1, were purchased from ATCC. Cells were grown in RP1640 Media, supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin.
AML and peripheral blood cell samples
The protocol was approved by the Human Medical Ethical Review Committee from Jilin University First Hospital and informed consent was obtained from each AML patient and normal subject. Bone marrow samples were obtained from 34 AML patients at diagnosis and 10 healthy volunteers in Jilin University First Hospital (Supplementary Table S1) in Changchun City, China. AML patients were classified into high-risk and low-risk groups by cytogenetics and molecular abnormalities according to the NCCN guidelines (version 2.2013). The low-risk group (n = 18) was defined as patients with t(8;21) or RUNX1-RUNX1T1, inv(16), t(16;16) or CEBF-MYH11, normal karyotype with NPM1 mutation and without FLT3-ITD mutation, and normal karyotype with isolated biallelic CEBPA mutation (normal karyotype). The high-risk group (n = 16) included patients with inv(3), t(3;3) or RPN1-EVI1, t(6;9) or DEK-NUP214, t(9;22) or BCR-ABL, t(v;11) (v;q23), MLL rearranged, −5 or del (5q), −7 or del (7q), complex karyotype, monosomal karyotype, normal karyotype with FLT3-ITD mutation (Supplementary Table S2).
Leukocyte fractions from AML samples and normal bone marrow specimens were isolated by Ficoll-Hypaque (Sigma, MO) centrifugation and then cryopreserved. After thawing, total RNA was extracted by RNeasy Kit (Qiagen, CA) for qPCR quantitation.
Reverse transcription-PCR analysis
Total RNA was extracted from tissues by TRI-REAGENT (Sigma, CA), according to the manufacturer's guide, and cDNA was synthesized with RNA reverse transcriptase as previously described (31,32). Briefly, 1 μg of total RNA was used, and polymerase chain reaction (PCR) was carried out under liquid wax in a 6 μl reaction containing 2 μl of 3× Klen-Taq I Mix, 2 μl cDNA and 1 μl of each 2.5 μM primer. After incubation at 95°C for 2 min, IRAIN cDNA was amplified by 32 cycles of 95°C for 30 s, 65°C for 30 s of annealing and 72°C for 35 s of extension, and finally with extension at 72°C for 5 min. Amplified PCR products of the expected size were quantified by densitometric measurements and normalized to ‘β-actin’ values.
Gene strand-specific RT-PCR
A strand-specific PCR (SSRT) assay was used to map the transcription of the IRAIN lncRNA. Total RNA was extracted from tissues by TRI-REAGENT (Sigma, CA), according to the manufacturer's guide, and cDNA was synthesized with reverse transcriptase using gene specific primers. Briefly, 400 ng total RNA was reverse transcribed with the IRAIN 5′- or 3′-primers instead of random hexamers. The RT reaction was performed with Maxima Reverse Transcriptase (Thermo Fisher Scientific, CA) at 60? for 50 min, followed by 85? for 5 min to inactivate the transcriptase. After 10-fold dilution, PCR was carried out under liquid wax in a 6 μl reaction containing 2 μl of 3 × Klen-Taq I Mix, 2 μl cDNA and 1 μl of each 2.5 μM downstream PCR primer set. After initial denaturing at 95°C for 2 min, IRAIN cDNA was amplified by 32 cycles at 95°C for 30 s, 65°C for 30 s of annealing and 72°C for 35 s of extension, followed by incubation at 72°C for 5 min. PCR products with the expected size were quantified by densitometric measurements and normalized to ‘β-actin’ values.
Characterization of the IRAIN lncRNA by 5′ racing
The full length of IRAIN lncRNA was characterized by Marathon cDNA Amplification Kit (Clonthech, CA) (33). Total RNA was reverse transcribed into cDNA. The 5′- and 3′-ends were raced using two primers that cover a unique Bam H1 site in the middle of the IRAIN lncRNA: JH1095 (forward): GGCTCGCTGAAGGTCACAGC and JH986 (reverse): AGGCTGGGGCTCTTGTTTACCA. The 5′- and 3′-RACE products were cloned into pJet vector (Thermo Fisher Scientific, CA) and sequenced. The full-length IRAIN lncRNA was constructed by joining these two RACE products in a pCMV-MS-pEF1-coGFP/Puro lentiviral vector using two steps of Xba1-BamH1 and BamH1-EcoR1 cloning. The 5′- and 3′-end of the IRAIN lncRNA was determined by sequencing.
Gene expression by real-time qPCR
After removing genomic DNA contamination with DNase I (Sigma), M-MLV Reverse Transcriptase (Invitrogen, CA) was used to synthesize cDNA (31,34). For qPCR, cDNA samples were amplified using CFX96TM real-time system (BIO-RAD) by SYBR PrimeScript™ RT-PCR Kit (TaKaRa) (35). The mRNA expression level of IRAIN and IGF1R was quantitated by normalizing with β-actin (housekeeping gene) as previously described (34,36). PCR primers used for qPCR included (1) IRAIN: 5′-CGACACATGGTCCAATCACTGTT-3′ (forward) and 5′-AGACTCCCCTAGGACTGCCATCT-3′ (reverse); and (2) IGF1R: 5′-GAAGTCTGGCTCCGGAGGAGGGTC-3′ (forward) and 5′-ATGTGGAGGTAGCCCTCGATCAC-3′ (reverse); and β-actin: 5′-AGATCAAGATCATTGCTCCTCCTGA-3′ (forward) and 5′-ATACTCCTGCTTGCTGATCCACATC-3′.
Northern analysis
Total RNA from breast cancer samples was separated by electrophoresis on a 1.5% (w/v) denaturing agarose gel, transferred to a Hybond-N nylon membrane (Amersham, UK) and cross-linked with UV light. The probe was prepared from the IRAIN clone DNA with 32P-dCTP labeling using Megaprime DNA Labelling Kit (Amersham, UK). The membrane was prehybridized in Rapid-hyb buffer (Amersham) for 30 min, followed by hybridization with the labeled probe at 65°C for 2 h. The membrane was exposed to X-ray film overnight and the image was scanned by a densitometer.
Reverse transcription-associated trap assay
A ‘reverse transcription-associated trap’ (RAT) assay was developed for the measurement of RNA–DNA interactions (Supplementary Figure S2). Briefly, 1.0 × 107 cells were cross-linked with 2% formaldehyde and lysed with cell lysis buffer (10 mM Tris [pH 8.0], 10 mM NaCl, 0.2% NP-40, 1× protease inhibitors). Nuclei were collected, suspended in 1× reverse transcription buffer in the presence of 0.3% sodium dodecyl sulfate (SDS) and incubated at 37°C for 1 h. Triton X-100 was then added to a final concentration of 1.8% to sequester the SDS. An aliquot of nuclei (3 × 106) was used for gene strand-specific reverse transcription in the presence of biotin-dCTP. For the parallel controls, we established three groups (5′-prime RT, 3′-prime RT and non-prime RT group). After 50 min of reverse transcription of IRAIN lncRNA with Maxima Reverse Transcriptase (Thermo Fisher Scientific, CA) at 60°C, the reaction was stopped by heating at 85°C for 5 min. The biotinylated-cDNA/chromatin DNA complex was diluted with 200 μl 1× NEB EcoRI buffer and digested at 37°C for 2.5 h with rotation by 600 μl EcoRI (New England BioLabs, CA). Biotin-streptavidin magic beads (Invitrogen, CA) were used to pull down the biotinylated-cDNA/DNA complex. After washing, the pull-down sample was treated with 10 mg/ml proteinase K at 65°C overnight to reverse the cross-links. Following incubation with 0.4 μg/ml RNase A for 30 min at 37°C, DNA was extracted and used for PCR amplification of the genomic DNA that interacts with the lncRNA.
Chromosome conformation capture
The chromosome conformation capture (3C) assay was performed as previously described (37). Briefly, 1.0 × 107 cells were cross-linked with 2% formaldehyde and lysed with cell lysis buffer (10 mM Tris [pH 8.0], 10 mM NaCl, 0.2% NP-40, protease inhibitors). Nuclei were collected, suspended in 1× restriction enzyme buffer in the presence of 0.3% SDS and incubated at 37°C for 1 h. Triton X-100 was then added to a final concentration of 1.8% to sequester the SDS. An aliquot of nuclei (2 × 106) was digested with 800 U of restriction enzyme Hind III at 37°C overnight. After stopping the reaction by adding 1.6% SDS and incubating the mixture at 65°C for 20 min, chromatin DNA was diluted with NEB ligation reaction buffer, and 2 μg DNA was ligated with 4000 U of T4 DNA ligase (New England BioLabs, CA) at 16°C for 4 h (final DNA concentration, 2.5 μg/ml). After treatment with 10 mg/ml proteinase K at 65°C overnight to reverse cross-links and with 0.4 μg/ml RNase A for 30 min at 37°C, DNA was extracted with phenol-chloroform, ethanol precipitated and used for PCR amplification of the ligated DNA products.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (38,39). Briefly, 5 million cells were fixed with 1% formaldehyde and sonicated for 180 s (10 s on and 10 s off) on ice with a Branson sonicator with a 2-mm microtip at 40% output control and 90% duty cycle settings. The sonicated chromatin (1 ml) was clarified by centrifugation, aliquoted and snap-frozen in liquid nitrogen. To perform ChIP, sonicated chromatin (150 μl) was diluted 10-fold and purified with specific antiserum (2–5 μl) and protein G-agarose (60 μl). ChIP antibodies were obtained from Abcam (Cambridge, MA), including anti-trimethyl-H3-K4 (#ab8580), trimethyl-H3-K9 (#ab8898) and trimethyl-H3-K27 (#ab24684). DNA that was released from the bound chromatin after cross-linking reversal and proteinase K treatment was precipitated and diluted in 100 μl of low-TE buffer (1 mM Tris, 0.1 mM EDTA). PCR reactions were performed under liquid wax in a reaction containing 1 μl ChIP (or input) DNA, 0.5 μM appropriate primer pairs, 50 μM deoxynucleotide triphosphate and 0.2 U Klen-Taq I (Ab Peptides, MO). Standard PCR conditions were 95°C for 2 min, followed by 32 cycles of 95°C for 15 s, 65°C for 30 s of annealing and 72°C for 30 s of extension. The PCR products were separated on a 5% polyacrylamide–urea gel and quantified by a Phosphorimager (Molecular Dynamics, CA).
Examination of genomic imprinting
Total RNA extraction and cDNA synthesis were performed as previously described (31). Allelic expression of IRAIN and IGF1R was examined by PCR in cDNA samples using primers specific for polymorphic restriction enzymes. Allelic expression of IRAIN was assessed by polymorphic restriction enzymes Alu II and Sac II, and of IGF1R by Rsa II and Pvu II. In some cases, DNA sequencing of genomic DNA and cDNA PCR products was used to determine allelic expression of IRAIN. PCR primers used to assess allelic expression are listed in Supplementary Table S3.
α-Amanitin treatment of KG-1 cells
KG-1 cells were cultured in RPMI1640 medium to 70–80% confluence. As previously described (40,41), cells were treated with α-amanitin (75 μg/ml) (Sigma, MO) for 5 h to block nascent mRNA synthesis. After depletion of pre-mRNA, cells were collected by centrifuge at 1000 rpm for 5 min and saved for RAT assay.
Knockdown of IRAIN lncRNA by shRNA
Two IRAIN shRNAs (shIRAIN-1 and shIRAIN-2) were synthesized by GeneChem (Shanghai, China) and were transfected into MB-MDA231 cells. Forty-eight hours after transfection, cells were selected by puromycin and were collected for both IRAIN lncRNA quantitation using qPCR and intrachromosomal interaction using 3C assay.
Lentiviral expression of IRAIN lncRNA
The full-length 5.4 kb IRAIN lncRNA was amplified with PCR primers containing the Xba1 and EcoRV restriction sites. The PCR products were gel-purified, cut by restriction enzymes and ligated into the pCMV-MS-EF1-copGFP/Puro vector constructed in the lab. The IRAIN lncRNA clone was confirmed by sequencing and then packaged in 293SF-PacLV packing cells (42) using the method described in our lab (22,35). After transduction, cell clones were selected by 1 μg/μl puromycin and used for cell migration assay (35).
Statistical analysis
All experiments were performed in triplicate, and the data were expressed as mean ± SD. The data were analyzed with Student's t-test or by one-way analysis of variance, and results were considered statistically significant at P ≤ 0.05.
RESULTS
Identification of a novel antisense noncoding RNA in the IGF1R locus
IGFIR is frequently overexpressed in both solid tumors and hematopoietic malignancies, participating in the regulation of cancer cell proliferation, survival, metabolism and metastasis. To address the mechanisms underlying the dysregulation of IGF1R in leukemia cells, we used a novel R3C method recently developed in our lab (Supplementary Figure S1) (22), and detected the presence of RNA molecules within the IGF1R promoter and intron 1, where the IGF1R mRNA is not transcribed. To characterize this novel RNA molecule, we designed a series of PCR primers covering the IGF1R promoter region and mapped its transcription in the IGF1R locus (Figure 1A). Using PCR, we detected the transcription of this noncoding RNA from the IGF1R promoter and intron 1 (Figure 1B, from C to H sites, lanes 3–8). No PCR products were detected further upstream (sites A, B; lanes 1–2) or the region near exon 2 (sites I, J; lanes 9–10).
Figure 1.
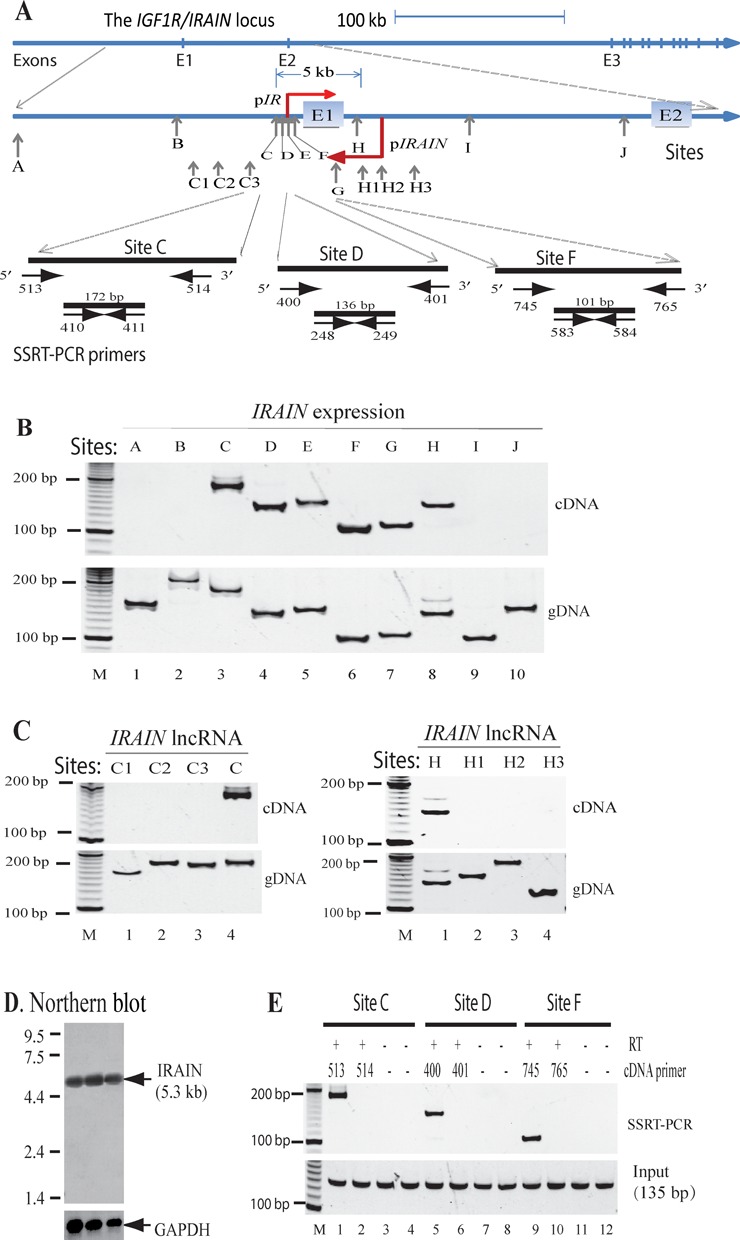
Characterization of IRAIN as an antisense lncNRA. (A) The diagram of the IRAIN/IGF1R locus. pIRAIN: IRAIN lncRNA promoter; pIR: IGF1R coding RNA promoter. Vertical arrows: the location of lncRNA PCR primers. (B and C) Mapping of the IRAIN lncRNA in K562 leukemia cells. gDNA: genomic DNA used as the control to test the efficiency of the PCR primers. M: 100 bp marker. (D) Northern blot of the IRAIN lncRNA in breast cancer tissues. Total RNA from three breast cancer tumors was separated on a 1.5% (w/v) denaturing agarose gel and was hybridized with the 32P-dCTP labeled IRAIN cDNA clone probe. GAPDH was used as the control. E: IRAIN lncRNA is an antisense lncRNA. Horizontal arrows: SSRT-PCR primers used to map the orientation of IRAIN lncRNA. The strand-specific cDNAs were synthesized using either the 5′- or the 3′-oligonucleotides at sites C, D and F. A pair of PCR primers located between two cDNA oligonucleotides was then used to determine the transcription orientation of the IRAIN lncRNA. M: 100 bp marker; input: total RNA collected before SSRT-PCR; RT: reverse transcriptase.
The transcription of this new RNA was further mapped using sequential primers covering the upstream C1–C3 sites and the downstream H1–H3 sites (Figure 1A, middle panel). No PCR products were detected using PCR primers at these sites (Figure 1C), indicating that this noncoding RNA is located between the C3 and H1 sites covering all of the IGF1R promoter and exon 1. Using a 5′-racing approach, we characterized IRAIN as a 5366 bp noncoding RNA (Supplementary Figure S2). The presence of full-length IRAIN lncRNA was further confirmed by northern blotting in three breast cancer samples (Figure 1D).
We then used strand-specific RT-PCR (SSRT) to examine whether this RNA is transcribed in a sense or antisense direction. Total RNA was extracted from leukemia K562 cells. Strand-specific reverse transcription (SSRT) cDNA was synthesized by thermo-stable reverse transcriptase (Thermo Fisher Scientific, PA) utilizing a 5′-specific oligonucleotide or a 3′-specific oligonucleotide, respectively. After SSRT, a pair of PCR primers designed to detect regions downstream of the 5′- or 3′-oligonucleotide was used to amplify the strand-specific cDNA. With this assay, only the lncRNA that is complementary to the 5′- or 3′-oligonucleotide was reverse transcribed in the subsequent PCR using the downstream primers (Figure 1E, top panel).
Using this SSRT assay, we found that this noncoding RNA was detected only when cDNA was synthesized using 5′-oligonucleotides (#513, #400 and #744) (Figure 1E, lanes 1, 5 and 9). No PCR products were amplified when the 3′ oligonucleotides were used (#514, #401 and #764; lanes 2, 6 and 10) or in the RT-minus controls (lanes 3–4, 7–8 and 11–12). These data indicate that this noncoding RNA is transcribed in the antisense direction as compared with the IGF1R coding RNA. The transcription was initiated from a promoter located in the IGF1R intron 1. We therefore refer to the noncoding RNA as IRAIN (IGF1R antisense intragenic noncoding RNA).
IRAIN lncRNA is imprinted in hematopoietic cells
The expression pattern of the IGF1R/IRAIN locus is quite similar to the Igf2r/Airn imprinting locus in the mouse, where Airn is imprinted and the lncRNA Airn regulates in cis the allelic expression of the Igf2r coding RNA (43–46). We therefore examined if IRAIN uses a similar epigenetic mechanism to regulate genes. We first examined if IRAIN was monoallelically expressed in tissues. After screening several single nucleotide polymorphisms (SNPs) within the IRAIN locus, we took advantage of polymorphic Alu I and Sac II sites to examine allelic expression of the IRAIN lncRNA (Figure 2A).
Figure 2.
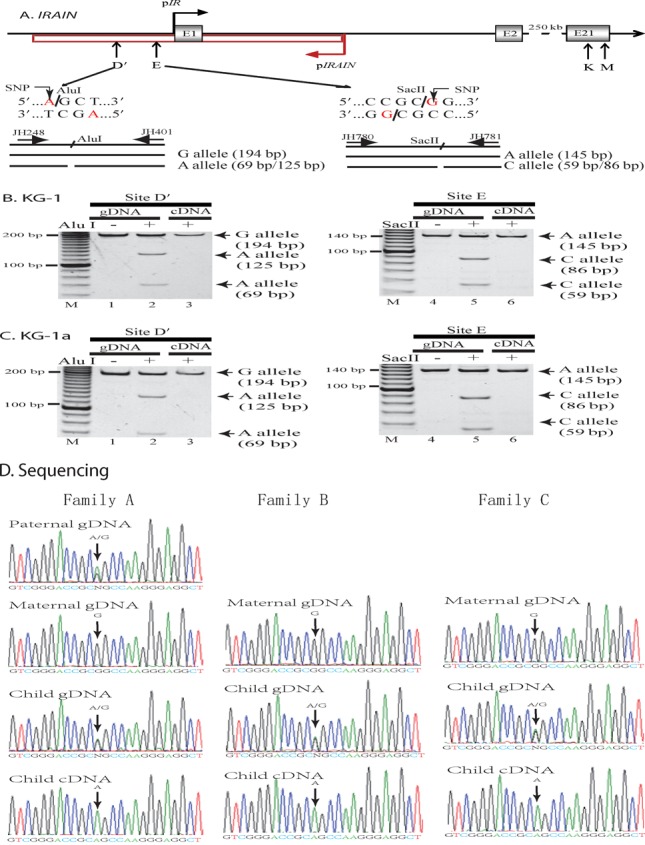
The IRAIN lncRNA is imprinted in hematopoietic cells. (A) Polymorphic restriction enzymes used to distinguish the two parental alleles. SNP: single nucleotide polymorphism. The IRAIN lncRNA was reverse transcribed into cDNA using SSRT oligonucleotides. The PCR products were digested by polymorphic Alu I and Sac II. (B and C) Allelic expression of IRAIN lncRNA at the E site in KG-1 and KG-1a leukemia cells. gDNA: heterozygous genomic DNA. Note the single ‘A’ allele or the single ‘G’ allele expression of IRAIN lncRNA. (D) Parental imprinting of IRAIN lncRNA by tracking allelic expression in three families. Genomic DNA and cDNA were amplified from peripheral blood cells and the PCR products were sequenced for the A/G alleles. Note the monoallelic expression of IRAIN lncRNA from the paternal allele.
Leukemia KG-1 cells are informative for both Alu I and Sac II. After restriction enzyme digestion, both the A and G (or C) alleles were detected in the genomic DNA (Figure 2B, lanes 2 and 5). In cDNA samples, however, only the ‘A’ allele was detected (lanes 3 and 6), indicating that IRAIN lncRNA is monoallelically transcribed in KG-1 leukemia cells. Similarly, KG-1a cells also expressed the IRAIN lncRNA in a monoallelic manner, as only the ‘A’ allele was detected in both the Alu I and Sac II sites (Figure 2C, lanes 3 and 6).
To determine which parental allele transcribes this noncoding RNA, we collected peripheral white blood cells from three families and tracked the expression pattern of the IRAIN lncRNA. In family A, the father carried both the A and G alleles and the mother had the G allele. The child was informative at the polymorphic site, carrying both the A and G alleles in genomic DNA. In the cDNA sample, however, we detected the expression of IRAIN lncRNA only from the A allele that was inherited from the father (Figure 2D, left panel), demonstrating that this lncRNA is paternally expressed and maternally suppressed. We also confirmed the paternal expression in two other families. In both family B and family C, the mothers were homozygous for the G allele, while the children were heterozygotes, carrying both the A and G alleles; only the paternal A allele was expressed (middle and right panels). Together, these data suggest that IRAIN is an imprinted gene, with the paternal allele expressed and the maternal allele suppressed.
Allelic regulation of IRAIN by epigenetic mechanisms
Imprinted genes are often regulated by differentially methylated regions (DMRs) in an imprinting control region (ICR) (26,47,48). As the IRAIN promoter is very rich in CpG dinucleotides (Figure 3A), we analyzed the status of DNA methylation in the IRAIN promoter. Genomic DNA was treated with sodium bisulfite to convert unmethylated cytosines into uracils while methylated cytosines are left unchanged. The treated DNA was amplified with methylation-specific primers, cloned in pJet vector and sequenced.
Figure 3.
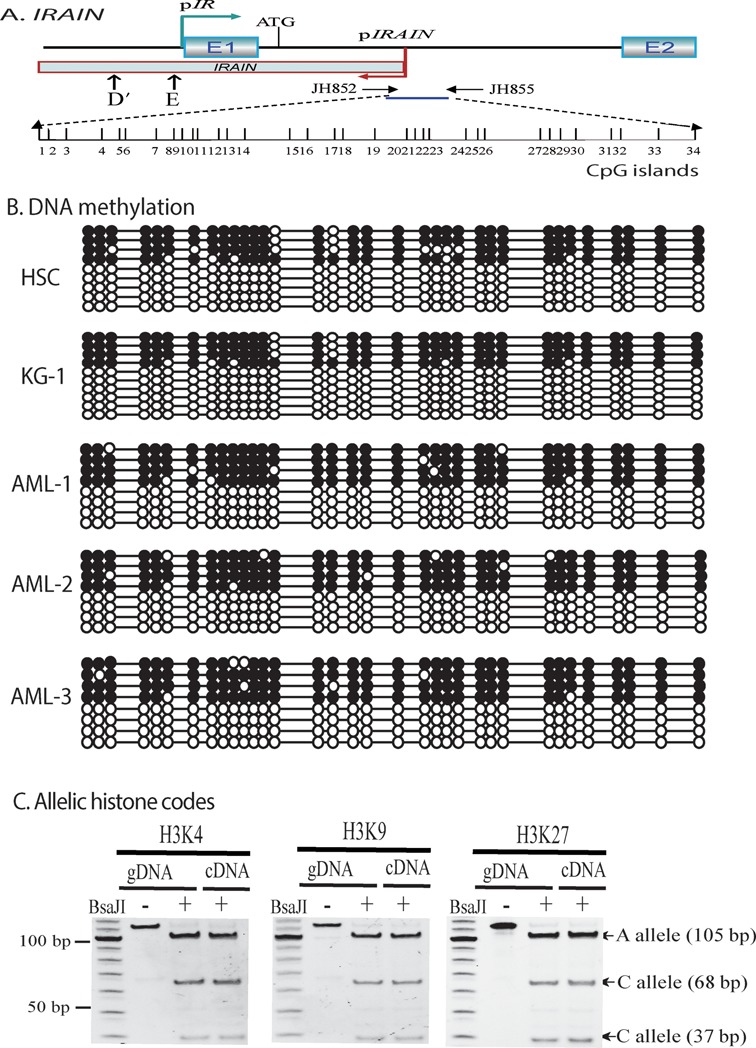
Epigenetic regulation of the IRF1R/IRAIN locus. (A) Schematic diagram of CpG islands in the IRAIN promoter. Vertical lines: location of CpG islands. (B) The status of CpG island DNA methylation in the IRAIN promoter. Genomic DNAs were extracted from leukemia cells and AML cells collected from three patients. After treatment with sodium bisulfite, the IRAIN promoter DNA was amplified and sequenced. Open circles: unmethylated CpGs; solid circles: methylated CpGs. (C) Allelic histone codes in the IRAIN promoter. Chromatin complex DNAs were pulled down by immunoprecipitation. After PCR, the two parental alleles were separated by polymorphic restriction enzyme BsaJ1 for histone methylation at H3K4, H3K9 and H3k27. Input: chromatin DNAs collected before antibody immunoprecipitation.
Using bisulfite sequencing, we found that the IRAIN promoter is hemi-methylated in normal hematopoietic stem cells (HSCs) (Figure 3B). A similar hemi-methylation pattern was also observed in the IRAIN promoter in KG-1 leukemia cells and in three AML samples. Although we could not find a SNP in this region to distinguish the two parental alleles in this DMR, the hemi-methylation pattern is typical of a DMR in an ICR, suggesting that the monoallelic expression of IRAIN may be associated with the status of DNA methylation in the gene promoter. However, K562 leukemia cells showed complete methylation in the promoter (Supplementary Figure S3). Thus, the epigenotype in the IRAIN promoter may be aberrantly altered in this tumor line.
We then utilized ChIP to examine the histone methylation in the IRAIN promoter. After immunoprecipitation using antibodies against methylated H3-K4, H3-K9 and H3-K27, PCR was used to amplify the chromatin DNAs. A BsaJ1 polymorphic restriction site, located ∼300 bp downstream of the differentially methylated promoter region, was used. We did not detect an allelic difference in the histone methylation products between the two alleles (Figure 3C), excluding histone methylation in the promoter as the determinant in the control of IRAIN allelic expression.
Interaction of IRAIN lncRNA with chromatin DNAs
As several lncRNA molecules regulate gene function by directly binding to their target chromatin DNAs (28–30,49,50), we determined if IRAIN lncRNA bound to IGF1R chromatin DNA, particularly the promoter region. We developed a ‘RAT’ assay that overcomes the high noise to signal background of existing methodologies to detect RNA/DNA interactions (Figure 4A). Cells were treated with formaldehyde to fix the structure of the lncRNA-chromatin conformations. The chromatin DNA-interacting lncRNA was then reverse transcribed into cDNA using strand-specific oligonucleotides and a thermo-stable reverse transcriptase in the presence of biotin-dCTP. After digestion with restriction enzyme or sonication to shear chromatin DNAs, the biotinylated IRAIN cDNA–DNA complex was then separated from other DNA–DNA products by streptavidin pull-down using paramagnetic Dynabeads (Dynal, Invitrogen) and analyzed by PCR using interacting DNA-specific primers.
Figure 4.
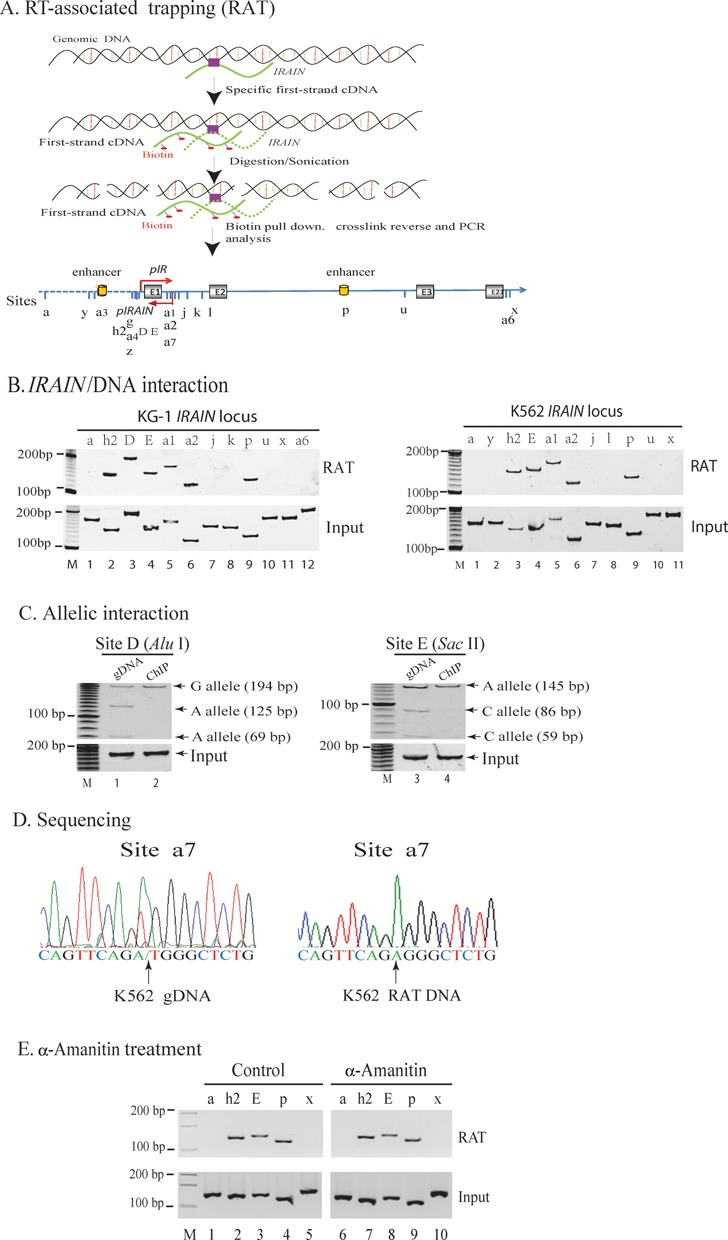
Chromatin DNA interaction of the IRAIN lncRNA. (A) Detection of IRAIN lncRNA–DNA interaction. A reverse transcription-associated trap (RAT) assay was used to detect the IRAIN lncRNA-specific interaction with IGF1R promoter DNAs. Alphabetic numbers: location of lncRNA binding sites. (B) IRAIN lncRNA–DNA interaction in KG-1 and K562 leukemia cells. Note the enriched binding of IRAIN lncRNA at IGF1R promoter (a4 and a5 sites) and intragenic enhancer (p site). Input: RNA–chromatin complex aliquots collected before RAT. (C) Allelic interaction of the IRAIN lncRNA with chromatin DNAs. Two polymorphic restriction enzymes Alu I and Sac II were used to distinguish the paternal and maternal alleles. Note the monoallelic binding of the IRAIN lncRNA. (D) Sequencing confirmation of allelic IRAIN lncRNA binding with the IGF1R promoter chromatin DNA. Left: K562 genomic DNA with both ‘A/T’ alleles; right: the IRAIN ChIP DNA with the single ‘A’ allele. (E) The lncRNA/DNA interaction after depletion of nascent RNA with α-amanitin. Cells were pretreated with the RNA polymerase inhibitor α-amanitin. After depletion of the nascent RNA, cells were fixed and used to detect DNA interaction with the RAT assay. The α-amanitin treatment does not interfere with lncRNA/DNA interaction.
Using this approach, we detected an interaction of IRAIN lncRNA with chromatin DNAs in the IGF1R promoter (Figure 4B, sites h2–a2, lanes 2–6) and in an intronic enhancer (site P, lane 9) in KG-1 cells. Taking advantage of polymorphic Alu I and Sac II sites, we observed that the IRAIN lncRNA interacted with chromatin DNA in a monoallelic manner, with only the A allele detected by both polymorphic restriction enzymes (Figure 4C, lanes 2 and 4).
A similar interacting pattern was also detected in the leukemia K562 cell line (Figure 4B, lanes 3–6, 9). By sequencing, it was also determined that these PCR products were derived from the A allele alone (Figure 4D, bottom panel) as compared with the genomic DNA that contains both the A and T alleles (top panel).
To further validate this chromatin DNA binding, we treated cells with the RNA polymerase inhibitor α-amanitin (40,41). As the half-life of pre-RNA is very short, usually on the order of minutes (51), we treated cells with α-amanitin (75 μg/ml) for 5 h. To examine the efficiency of α-amanitin treatment, we extracted chromatin RNA by sucrose gradient centrifugation (52) and examined target preRNAs and RNAs using RT-PCR. As previously reported (53,54), α-amanitin treatment not only permitted preRNAs to mature, but also significantly reduced the level of target RNAs (Supplementary Figure S4). After depletion of the pre-mRNA, the RAT assay revealed that it was the IRAIN lncRNA that interacted with genomic DNAs (Figure 4E). Further studies are still needed to exclude the presence of the trace amount of the IRAIN pre-RNA formed during transcription in the chromatin complex.
Long-range intrachromosomal loop orchestrated by IRAIN lncRNA
Since IRAIN lncRNA interacts with both the IGF1R promoter and the intronic enhancer DNA, which are about 150 kb apart, we hypothesized that the IRAIN lncRNA may participate in scaffolding these long-distance DNA regions to form an intrachromosomal loop. We therefore used 3C (37) to assess the potential chromatin interactions between these two DNA regions.
KG-1 cells were fixed with 2% formaldehyde, digested with restriction enzyme Hind III and then ligated with T4 DNA ligase to examine the remote interaction between these two DNA regions that are 150 kb apart (Figure 5A). Using primers from these remote regions, we found that the IGF1R promoter DNA interacted with the putative intronic enhancer DNA (Figure 5B, lane 4). In addition, a local DNA interaction was also detected between sites d′ and h′ (lane 2). Both interactions were confirmed by DNA sequencing (Figure 5C). ChIP assay also showed that both the IGF1R promoter (h2′) and the enhancer (p′) region sites were associated with the active enhancer marker, H3K4 methylation (Supplementary Figure S5). These data suggest that IRAIN lncRNA may be actively involved in the interaction of two remote regions of the IGF1R gene.
Figure 5.
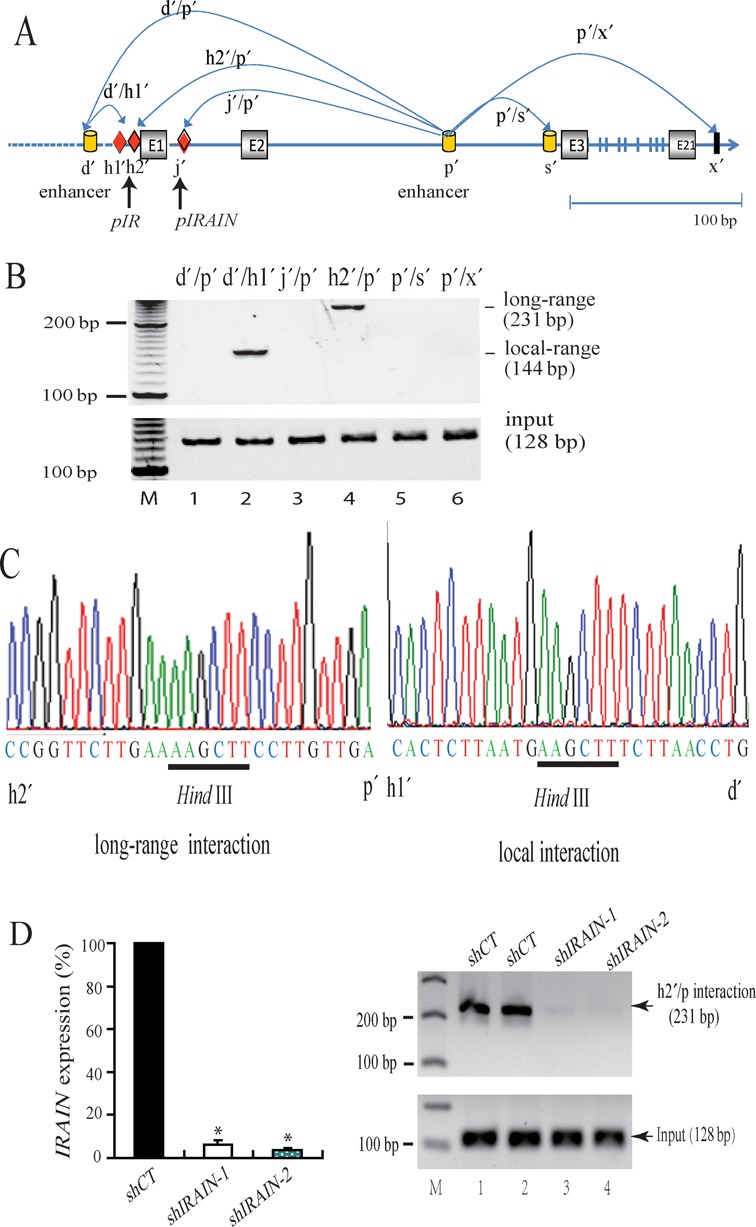
Intrachromosomal loop between IGF1R promoter and the intragenic enhancer. (A) Schematic diagram of the Hind III sites in the IGF1R/IRAIN gene locus used for the 3C assay. (B) Detection of chromatin interactions in KG-1 leukemia cells as detected by 3C assay. M: 100 bp markers, input: chromatin complex aliquots collected before 3C assay. (C) Sequencing confirmation of the 3C PCR products. Left: the Hind III site in the 3C DNA is flanked by DNA sequences from the IGF1R promoter and enhancer; right: local chromatin interaction. (D) IRAIN lncRNA knockdown abolishes the intrachromosomal DNA interaction. Cells were transfected with two shRNAs. After IRAIN knockdown, cells were fixed and used to detect DNA interaction with the 3C assay. Note the requirement for IRAIN in the maintenance of the IGF1R promoter/enhancer DNA interaction.
To further study the role of the lncRNA in this intrachromosomal interaction, we knocked down IRAIN lncRNA with two shRNAs. Both shRNAs significantly decreased the IRAIN lncRNA (Figure 5D, left panel). In the mock shRNA (shCT) treated cells, there was an interaction between the IGF1R promoter and the enhancer (Figure 5D, right panel, lanes 1 and 2). However, this long-range intrachromosomal interaction was abolished in the IRAIN knockdown cells (lanes 3 and 4). These data suggest the involvement of the lncRNA in the formation and/or maintenance of the long-range intrachromosomal loop.
Downregulation of IRAIN lncRNA in hematopoietic malignancies
IGF1R is frequently overexpressed in human tumors. We were curious if IRAIN was dysregulated in tumors as well. We first used real-time PCR to compare the abundance of IRAIN lncRNA and IGF1R coding RNA transcripts in hematopoietic cell lines. Using a normal hematopoietic stem cell line HSC2 as a standard, we found that IRAIN was downregulated in leukemia cell lines as compared with the IGF1R sense coding RNA (Figure 6A).
Figure 6.
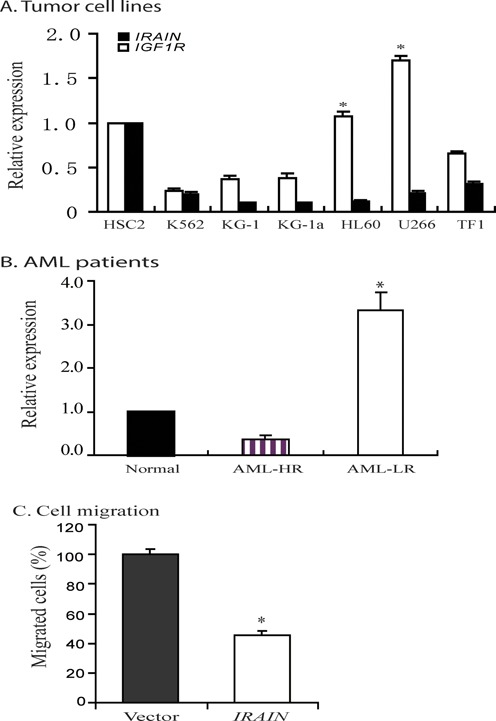
Downregulation of the IRAIN lncRNA in tumor cell lines and AML tissues. (A) Expression patterns of IRAIN and IGF1R in tumor cell lines. β-Actin was used in the internal PCR control for qPCR. Data are represented as mean ± SD. The relative expression was determined by normalizing the qPCR signals over that of normal HSC2 cells. *P < 0.05 as compared with the IGF1R coding RNA. (B) Expression of IRAIN lncRNA in AML patients. HR, LR: AML with high and low risk (AML-HR, n = 16; AML-LR, n = 18; and normal, n = 5). Data are represented as mean ± SD. *P < 0.05 as compared with the AML high-risk patients. (C) Inhibition of cell migration by 5.4 kb IRAIN lncRNA. Lentiviruses containing the full-length lncRNA and the vector control were transduced into MDA 231 cells. Stable cell clones were used to test the cell migration using transwell assay. Data are represented as mean ± SD. *P < 0.01 as compared with the vector control.
We then collected bone marrow cells from high- and low-risk AML patients. Real-time PCR was used to quantitate the transcript abundance of both coding and noncoding RNAs. We found that IRAIN abundance was low in the high-risk group and high in the lower risk group (Figure 6B).
As a first step to explore the role of IRAIN, we virally expressed the full-length 5.4 kb IRAIN lncRNA in MDA231 tumor cells. The stable cell clones were then tested for their migration by transwell assay (35). As compared with the vector control, expression of the full-length 5.4 kb IRAIN lncRNA significantly inhibited tumor cell migration (Figure 6C). We also found that exposure of cultured cells to the chemotherapeutic agent cytarabine (AraC) enhanced IRAIN expression (Supplementary Figure S6).
DISCUSSION
IGF1R is a validated tumor target for developing small molecule inhibitors and antibody therapies that block its tyrosine kinase activity. In examining the mechanism accounting for IGF1R dysregulation in tumors, we have identified a novel antisense noncoding RNA, IRAIN, that is expressed in antisense orientation and in a parent-of-origin-specific manner. The full-length IRAIN transcript is a 5.4 kb noncoding RNA; no large open reading frames can be identified using software prediction programs. Allelic expression of IRAIN is correlated with the status of CpG DNA methylation in the gene promoter (Figure 3B). We have not been able to find a SNP in the sequencing region to distinguish the two parental alleles. There is a BsaJ1 polymorphic site in the 3′-region of the promoter. However, the DNA sequence containing this BsaJ1 site is very GC-rich. After sodium bisulfite treatment, the fragment could not be amplified. Future studies using interspecific Mus spretus/Mus musculus F1 mice (45,55) may be used to examine the role of DNA methylation in IRAIN parental imprinting.
Gene ‘expression competition’ and ‘transcriptional interference’ have been proposed as mechanisms by which noncoding RNAs regulate allelic transcription. The best-studied examples are X chromosome inactivation and Igf2r/Airn allelic expression (47,56–59). Initiation of both imprinted and random X chromosome inactivation is dependent on a unique, noncoding RNA (Xist). The Xist lncRNA triggers the silencing of the X chromosome in cis through distinct domains (60) using a ‘chromosome coating’ mechanism (61). Similarly, short synthetic hairpin RNAs that are complementary to gene promoters are also able to modulate gene expression (62–64). In this study, we demonstrate that the IRAIN lncRNA is transcribed in an antisense orientation using a promoter located in the IGF1R intron 1. It is possible that IRAIN may use a similar ‘coating’ approach to regulate the transcription of the coding IGF1R RNA. By overlapping with the IGF1R promoter in antisense, the IRAIN lncRNA may directly compete with IGF1R in cis for the transcriptional machinery in the host. Downregulation of this lncRNA, as observed in high-risk hematopoietic malignancies (Figure 6), may relax the ‘transcription competition’ control and thus activate the IGF1R gene, leading to growth advantage and tumor progression. However, we found that both IRAIN shRNA knockdown and IRAIN overexpression approaches did not significantly affect the level of IGF1R RNA in treated cells (Supplementary Figure S7). It should be emphasized that both methods affect IRAIN lncRNA at the post-transcriptional level. It could be possible that like Airn, IRAIN lncRNA may use a ‘cis’ mechanism to regulate gene expression. If so, the overexpressed ‘trans’ lncRNA may not significantly affect the allelic expression of IGF1R. For this reason, a gene knock-out model may be needed to address a potential ‘cis’ role of IRAIN lncRNA.
Several lncRNAs regulate their target genes by directly binding their promoters and enhancers, including Kcnq1ot1, Xist and HOTAIR (28,29,65,66). HOTAIR can serve as a scaffolding factor, providing a binding surface to assemble histone modifying enzymes (67). In this study, we demonstrate that IRAIN interacts directly with regulatory chromatin DNAs of the IGF1R enhancer and promoter (Figure 5). While HOTAIR and IRAIN do not contain any similar sequences, they may share structural features that could allow them to regulate gene expression using a common epigenetic mechanism (68).
Long-range intrachromosomal interaction is a common epigenetic mechanism that juxtaposes a promoter with a remote regulatory element (34,39,69,70). For example, Kcnq1ot1 lncRNA is a bidirectional silencer that regulates genes in cis over ∼1 Mb in the Kcnq1 imprinting domain (29,30). KvDMR1, the region that carries the imprinting signal to control imprinting, is ∼200 kb away from Kcnq1. Our recent data demonstrate that using intrachromosomal looping, Kcnq1ot1 recruits the histone H3K27 methylase EZH2, a component of the polycomb repressive complex 2 (PRC2), to the Kcnq1 promoter target sequence, where the allele-specific histone H3-K27 methylation turns off the expression of the paternal Kcnq1. In Kcnq1ot1 deficient cells, the absence of intrachromosomal looping leads to biallelic expression of Kcnq1 (22). The data in this study demonstrate that IRAIN lncRNA is involved in the formation of an intrachromosomal interaction between the IGF1R promoter and a distant intragenic enhancer (Figure 6). Knockdown of this lncRNA abolishes this intrachromosomal interaction. Further studies are needed to delineate if this intrachromosomal interaction is also involved in the regulation of IGF1R expression in cis as in the case of Airn lncRNA in the IGF2R locus.
It is interesting to note that IRAIN is transcribed monoallelically. By tracking the allelic expression in three families, we demonstrate that IRAIN lncRNA is transcribed from the paternal chromosome, while the maternal allele is suppressed (Figure 2D). A single parental IRAIN allele is transcribed in KG-1 and KG-1a tumor cells (Figure 2B and C). This study thus adds IRAIN lncRNA to the list of monoallelically expressed transcripts (48).
Equally interesting is the finding that the expression of both the antisense IRAIN lncRNA and the sense IGF1R coding RNA is uncoupled in tumors. We found that IRAIN is monoallelically expressed (Figure 2). The IGF1R coding mRNA, however, is biallelically expressed (Supplementary Figure S8) in accordance with previously published reports (71,72). In several well-studied imprinting loci, the allelic expression of the sense and antisense RNAs is coupled via a cis transcription competition mechanism. For example, allelic expression of genes in the mouse Igf2r/Airn loci is coupled (56,73,74) and tightly associated with the status of DNA methylation in the Airn promoter (32) (Supplementary Figure S9, right panel). The maternal Airn promoter is hypermethylated and is silenced. Lack of the Airn lncRNA cis-competition leads to the active Igf2r transcription from the maternal allele. In contrast, the unmethylated paternal Airn promoter is expressed. The transcribed noncoding RNA silences in cis the Igf2r promoter (32,75). In the IGF1R/IRAIN locus, however, IRAIN is paternally expressed, but its sense IGF1R coding RNA is biallelically expressed (Supplementary Figure S9, left panel). Currently, it is unclear how these two RNAs are expressed in a divergent manner. Nonetheless, the fact that both IRAIN antisense lncRNA and IGF1R sense RNA are transcribed from the paternal chromosome without transcription competition or inhibition may provide a unique model to study imprinting mechanisms as seen in the Igf2r/Airn locus (56,74).
IRAIN is a newly identified lncRNA. We know very little about the specific function of IRAIN. Expression analyses reveal that IRAIN is downregulated in leukemia cell lines. The AML low-risk patients had greater IRAIN lncRNA expression than did the AML high-risk patients. Viral expression of the 5.4 kb lncNRA inhibits tumor cell migration, suggesting a tumor suppressor function. However, the low-risk patients also had greater IRAIN expression than did the normal subjects (Figure 6). The low-risk patients still exhibit the malignant leukemia characteristics and are prone to convert to high-risk AML. Thus, further studies are needed to address the specific role of the IRAIN lncRNA in tumors.
In summary, we have identified IRAIN as a novel lncRNA that is downregulated in leukemia cell lines and in patients with high-risk AML. The lncRNA is imprinted and directly interacts with the IGF1R promoter and enhancer chromatin DNA sequences. IRAIN also participates in the orchestration of an intrachromosomal loop between the IGF1R promoter and enhancer. Further studies are needed to delineate the specific role of this newly identified lncRNA in the upregulation of the IGF pathway in malignancies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We are grateful to Julia Heidmann and Zhentong Wei for assistance in collecting tissues.
Footnotes
These authors are senior authors of this report.
FUNDING
National Institutes of Health [1R43 CA103553-01]; California Institute of Regenerative Medicine (CIRM) [RT2-01942]; Jilin International Collaboration [20120720, 20130413010GH]; National Natural Science Foundation of China [81272294 to J.F.H, 81372835 to W.L.]; Jilin Great Science Grant [11ZDGG003 to W.L.]; Key Project of Chinese Ministry of Education [311015 to C.J.]; Jilin University First Hospital Junior Scientist Grant [JDYY42013021 to J.S.]; Medical Research Service of the Department of Veterans Affairs [to A.R.H.]. Source of open access funding: California Institute of Regenerative Medicine (CIRM) [RT2-01942].
Conflict of interest statement. None declared.
REFERENCES
- 1.Seccareccia E., Brodt P. The role of the insulin-like growth factor-I receptor in malignancy: an update. Growth Horm. IGF Res. 2012;22:193–199. doi: 10.1016/j.ghir.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat. Rev. Cancer. 2012;12:159–169. doi: 10.1038/nrc3215. [DOI] [PubMed] [Google Scholar]
- 3.Ozkan E.E. Plasma and tissue insulin-like growth factor-I receptor (IGF-IR) as a prognostic marker for prostate cancer and anti-IGF-IR agents as novel therapeutic strategy for refractory cases: a review. Mol. Cell. Endocrinol. 2011;344:1–24. doi: 10.1016/j.mce.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Capoluongo E. Insulin-like growth factor system and sporadic malignant melanoma. Am. J. Pathol. 2011;178:26–31. doi: 10.1016/j.ajpath.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallagher E.J., LeRoith D. The proliferating role of insulin and insulin-like growth factors in cancer. Trends Endocrinol. Metab. 2010;21:610–618. doi: 10.1016/j.tem.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapuis N., Tamburini J., Cornillet-Lefebvre P., Gillot L., Bardet V., Willems L., Park S., Green A.S., Ifrah N., Dreyfus F., et al. Autocrine IGF-1/IGF-1R signaling is responsible for constitutive PI3K/Akt activation in acute myeloid leukemia: therapeutic value of neutralizing anti-IGF-1R antibody. Haematologica. 2010;95:415–423. doi: 10.3324/haematol.2009.010785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grandage V.L., Gale R.E., Linch D.C., Khwaja A. PI3-kinase/Akt is constitutively active in primary acute myeloid leukaemia cells and regulates survival and chemoresistance via NF-kappaB, Mapkinase and p53 pathways. Leukemia. 2005;19:586–594. doi: 10.1038/sj.leu.2403653. [DOI] [PubMed] [Google Scholar]
- 8.Xu Q., Simpson S.E., Scialla T.J., Bagg A., Carroll M. Survival of acute myeloid leukemia cells requires PI3 kinase activation. Blood. 2003;102:972–980. doi: 10.1182/blood-2002-11-3429. [DOI] [PubMed] [Google Scholar]
- 9.Doepfner K.T., Spertini O., Arcaro A. Autocrine insulin-like growth factor-I signaling promotes growth and survival of human acute myeloid leukemia cells via the phosphoinositide 3-kinase/Akt pathway. Leukemia. 2007;21:1921–1930. doi: 10.1038/sj.leu.2404813. [DOI] [PubMed] [Google Scholar]
- 10.Ge N.L., Rudikoff S. Insulin-like growth factor I is a dual effector of multiple myeloma cell growth. Blood. 2000;96:2856–2861. [PubMed] [Google Scholar]
- 11.Sprynski A.C., Hose D., Caillot L., Reme T., Shaughnessy J.D., Jr, Barlogie B., Seckinger A., Moreaux J., Hundemer M., Jourdan M., et al. The role of IGF-1 as a major growth factor for myeloma cell lines and the prognostic relevance of the expression of its receptor. Blood. 2009;113:4614–4626. doi: 10.1182/blood-2008-07-170464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sprynski A.C., Hose D., Kassambara A., Vincent L., Jourdan M., Rossi J.F., Goldschmidt H., Klein B. Insulin is a potent myeloma cell growth factor through insulin/IGF-1 hybrid receptor activation. Leukemia. 2010;24:1940–1950. doi: 10.1038/leu.2010.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abe S., Funato T., Takahashi S., Yokoyama H., Yamamoto J., Tomiya Y., Yamada-Fujiwara M., Ishizawa K., Kameoka J., Kaku M., et al. Increased expression of insulin-like growth factor i is associated with Ara-C resistance in leukemia. Tohoku J. Exp. Med. 2006;209:217–228. doi: 10.1620/tjem.209.217. [DOI] [PubMed] [Google Scholar]
- 14.Wahner Hendrickson A.E., Haluska P., Schneider P.A., Loegering D.A., Peterson K.L., Attar R., Smith B.D., Erlichman C., Gottardis M., Karp J.E., et al. Expression of insulin receptor isoform A and insulin-like growth factor-1 receptor in human acute myelogenous leukemia: effect of the dual-receptor inhibitor BMS-536924 in vitro. Cancer Res. 2009;69:7635–7643. doi: 10.1158/0008-5472.CAN-09-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medyouf H., Gusscott S., Wang H., Tseng J.C., Wai C., Nemirovsky O., Trumpp A., Pflumio F., Carboni J., Gottardis M., et al. High-level IGF1R expression is required for leukemia-initiating cell activity in T-ALL and is supported by Notch signaling. J. Exp. Med. 2011;208:1809–1822. doi: 10.1084/jem.20110121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karamouzis M.V., Papavassiliou A.G. Targeting insulin-like growth factor in breast cancer therapeutics. Crit. Rev. Oncol. Hematol. 2012;84:8–17. doi: 10.1016/j.critrevonc.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Tognon C.E., Sorensen P.H. Targeting the insulin-like growth factor 1 receptor (IGF1R) signaling pathway for cancer therapy. Expert Opin. Ther. Targets. 2012;16:33–48. doi: 10.1517/14728222.2011.638626. [DOI] [PubMed] [Google Scholar]
- 18.Rowinsky E.K., Schwartz J.D., Zojwalla N., Youssoufian H., Fox F., Pultar P., Novosyadlyy R., Cosaert J., Ludwig D.L. Blockade of insulin-like growth factor type-1 receptor with cixutumumab (IMC-A12): a novel approach to treatment for multiple cancers. Curr. Drug Targets. 2011;12:2016–2033. doi: 10.2174/138945011798829401. [DOI] [PubMed] [Google Scholar]
- 19.Buck E., Mulvihill M. Small molecule inhibitors of the IGF-1R/IR axis for the treatment of cancer. Expert Opin. Investig. Drugs. 2011;20:605–621. doi: 10.1517/13543784.2011.558501. [DOI] [PubMed] [Google Scholar]
- 20.Heidegger I., Pircher A., Klocker H., Massoner P. Targeting the insulin-like growth factor network in cancer therapy. Cancer Biol. Ther. 2011;11:701–707. doi: 10.4161/cbt.11.8.14689. [DOI] [PubMed] [Google Scholar]
- 21.Moreau P., Cavallo F., Leleu X., Hulin C., Amiot M., Descamps G., Facon T., Boccadoro M., Mignard D., Harousseau J.L. Phase I study of the anti insulin-like growth factor 1 receptor (IGF-1R) monoclonal antibody, AVE1642, as single agent and in combination with bortezomib in patients with relapsed multiple myeloma. Leukemia. 2011;25:872–874. doi: 10.1038/leu.2011.4. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H., Zeitz M.J., Wang H., Niu B., Ge S., Li W., Cui J., Wang G., Qian G., Higgins M.J., et al. Long noncoding RNA-mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus. J. Cell Biol. 2014;204:61–75. doi: 10.1083/jcb.201304152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasanth K.V., Spector D.L. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 25.Magistri M., Faghihi M.A., St Laurent G., III, Wahlestedt C. Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends Genet. 2012;28:389–396. doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin J.Y., Fitzpatrick G.V., Higgins M.J. Two distinct mechanisms of silencing by the KvDMR1 imprinting control region. EMBO J. 2008;27:168–178. doi: 10.1038/sj.emboj.7601960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercer T.R., Mattick J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 28.Lee J.T. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 29.Mancini-DiNardo D., Steele S.J., Ingram R.S., Tilghman S.M. A differentially methylated region within the gene Kcnq1 functions as an imprinted promoter and silencer. Hum. Mol. Genet. 2003;12:283–294. doi: 10.1093/hmg/ddg024. [DOI] [PubMed] [Google Scholar]
- 30.Thakur N., Tiwari V.K., Thomassin H., Pandey R.R., Kanduri M., Gondor A., Grange T., Ohlsson R., Kanduri C. An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol. Cell. Biol. 2004;24:7855–7862. doi: 10.1128/MCB.24.18.7855-7862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu J.F., Vu T.H., Hoffman A.R. Promoter-specific modulation of insulin-like growth factor II genomic imprinting by inhibitors of DNA methylation. J. Biol. Chem. 1996;271:18253–18262. doi: 10.1074/jbc.271.30.18253. [DOI] [PubMed] [Google Scholar]
- 32.Hu J.F., Oruganti H., Vu T.H., Hoffman A.R. Tissue-specific imprinting of the mouse insulin-like growth factor II receptor gene correlates with differential allele-specific DNA methylation. Mol. Endocrinol. 1998;12:220–232. doi: 10.1210/mend.12.2.0062. [DOI] [PubMed] [Google Scholar]
- 33.Yao X.M., Hu J.F., Daniels M., Li T., Yang Y.W., Li Z.H., Vu T.H., Hoffman A.R. Epigenetic regulation of the taxol resistance-associated gene TRAG-3 in human tumors. Cancer Genet. Cytogenet. 2004;151:1–13. doi: 10.1016/j.cancergencyto.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 34.Li T., Hu J.F., Qiu X., Ling J., Chen H., Wang S., Hou A., Vu T.H., Hoffman A.R. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex-2 intrachromosomal loop. Mol. Cell. Biol. 2008;28:6473–6482. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S., Zhong B., Chen M., Yang G., Li Y., Wang H., Wang G., Li W., Cui J., Hoffman A.R., et al. Epigenetic reprogramming reverses the malignant epigenotype of the MMP/TIMP axis genes in tumor cells. Int. J. Cancer. 2014;134:1583–1594. doi: 10.1002/ijc.28487. [DOI] [PubMed] [Google Scholar]
- 36.Chen H.L., Li T., Qiu X.W., Wu J., Ling J.Q., Sun Z.H., Wang W., Chen W., Hou A., Vu T.H., et al. Correction of aberrant imprinting of IGF2 in human tumors by nuclear transfer-induced epigenetic reprogramming. EMBO J. 2006;25:5329–5338. doi: 10.1038/sj.emboj.7601399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H., Jiao W., Sun L., Fan J., Chen M., Wang H., Xu X., Shen A., Li T., Niu B., et al. Intrachromosomal looping is required for activation of endogenous pluripotency genes during reprogramming. Cell Stem Cell. 2013;13:30–35. doi: 10.1016/j.stem.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H., Niu B., Hu J.F., Ge S., Wang H., Li T., Ling J., Steelman B.N., Qian G., Hoffman A.R. Interruptionof intrachromosomal looping by CCCTC binding factor decoy proteins abrogatesgenomic imprinting of human insulin-like growth factorII. J. Cell Biol. 2011;193:475–487. doi: 10.1083/jcb.201101021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kedinger C., Gniazdowski M., Mandel J.L., Jr, Gissinger F., Chambon P. Alpha-amanitin: a specific inhibitor of one of two DNA-pendent RNA polymerase activities from calf thymus. Biochem. Biophys. Res. Commun. 1970;38:165–171. doi: 10.1016/0006-291x(70)91099-5. [DOI] [PubMed] [Google Scholar]
- 41.Lindell T.J., Weinberg F., Morris P.W., Roeder R.G., Rutter W.J. Specific inhibition of nuclear RNA polymerase II by alpha-amanitin. Science. 1970;170:447–449. doi: 10.1126/science.170.3956.447. [DOI] [PubMed] [Google Scholar]
- 42.Broussau S., Jabbour N., Lachapelle G., Durocher Y., Tom R., Transfiguracion J., Gilbert R., Massie B. Inducible packaging cells for large-scale production of lentiviral vectors in serum-free suspension culture. Mol. Ther. 2008;16:500–507. doi: 10.1038/sj.mt.6300383. [DOI] [PubMed] [Google Scholar]
- 43.Stoger R., Kubicka P., Liu C.G., Kafri T., Razin A., Cedar H., Barlow D.P. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- 44.Wutz A., Smrzka O.W., Schweifer N., Schellander K., Wagner E.F., Barlow D.P. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 45.Hu J.F., Balaguru K.A., Ivaturi R.D., Oruganti H., Li T., Nguyen B.T., Vu T.H., Hoffman A.R. Lack of reciprocal genomic imprinting of sense and antisense RNA of mouse insulin-like growth factor II receptor in the central nervous system. Biochem. Biophys. Res. Commun. 1999;257:604–608. doi: 10.1006/bbrc.1999.0380. [DOI] [PubMed] [Google Scholar]
- 46.Hu J.F., Pham J., Dey I., Li T., Vu T.H., Hoffman A.R. Allele-specific histone acetylation accompanies genomic imprinting of the insulin-like growth factor II receptor gene. Endocrinology. 2000;141:4428–4435. doi: 10.1210/endo.141.12.7857. [DOI] [PubMed] [Google Scholar]
- 47.Bartolomei M.S. Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev. 2009;23:2124–2133. doi: 10.1101/gad.1841409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skaar D.A., Li Y., Bernal A.J., Hoyo C., Murphy S.K., Jirtle R.L. The human imprintome: regulatory mechanisms, methods of ascertainment, and roles in disease susceptibility. ILAR J. 2012;53:341–358. doi: 10.1093/ilar.53.3-4.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mercer T.R., Mattick J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 50.Nagano T., Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 51.Wang J., Shen L., Najafi H., Kolberg J., Matschinsky F.M., Urdea M., German M. Regulation of insulin preRNA splicing by glucose. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4360–4365. doi: 10.1073/pnas.94.9.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mondal T., Rasmussen M., Pandey G.K., Isaksson A., Kanduri C. Characterization of the RNA content of chromatin. Genome Res. 2010;20:899–907. doi: 10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen V.T., Giannoni F., Dubois M.F., Seo S.J., Vigneron M., Kedinger C., Bensaude O. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 1996;24:2924–2929. doi: 10.1093/nar/24.15.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferrai C., Xie S.Q., Luraghi P., Munari D., Ramirez F., Branco M.R., Pombo A., Crippa M.P. Poised transcription factories prime silent uPA gene prior to activation. PLoS Biol. 2010;8:e1000270. doi: 10.1371/journal.pbio.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu J., Vu T., Hoffman A. Differential biallelic activation of three insulin-like growth factor II promoters in the mouse central nervous system. Mol. Endocrinol. 1995;9:628–636. doi: 10.1210/mend.9.5.7565809. [DOI] [PubMed] [Google Scholar]
- 56.Barlow D.P. Competition—a common motif for the imprinting mechanism. EMBO J. 1997;16:6899–6905. doi: 10.1093/emboj/16.23.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Latos P.A., Pauler F.M., Koerner M.V., Senergin H.B., Hudson Q.J., Stocsits R.R., Allhoff W., Stricker S.H., Klement R.M., Warczok K.E., et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 58.Pauler F.M., Koerner M.V., Barlow D.P. Silencing by imprinted noncoding RNAs: is transcription the answer? Trends Genet. 2007;23:284–292. doi: 10.1016/j.tig.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shearwin K.E., Callen B.P., Egan J.B. Transcriptional interference—a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wutz A., Rasmussen T.P., Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 61.Panning B., Dausman J., Jaenisch R. X chromosome inactivation is mediated by Xist RNA stabilization. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- 62.Chu Y., Kalantari R., Dodd D.W., Corey D.R. Transcriptional silencing by hairpin RNAs complementary to a gene promoter. Nucleic Acid Ther. 2012;22:147–151. doi: 10.1089/nat.2012.0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Janowski B.A., Huffman K.E., Schwartz J.C., Ram R., Hardy D., Shames D.S., Minna J.D., Corey D.R. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat. Chem. Biol. 2005;1:216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 64.Morris K.V. The emerging role of RNA in the regulation of gene transcription in human cells. Semin. Cell Dev. Biol. 2011;22:351–358. doi: 10.1016/j.semcdb.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Splinter E., de Wit E., Nora E.P., Klous P., van de Werken H.J., Zhu Y., Kaaij L.J., van Ijcken W., Gribnau J., Heard E., et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25:1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sleutels F., Zwart R., Barlow D.P. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 67.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsai C.L., Rowntree R.K., Cohen D.E., Lee J.T. Higher order chromatin structure at the X-inactivation center via looping DNA. Dev. Biol. 2008;319:416–425. doi: 10.1016/j.ydbio.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qiu X., Vu T.H., Lu Q., Ling J.Q., Li T., Hou A., Wang S.K., Chen H.L., Hu J.F., Hoffman A.R. A complex deoxyribonucleic Acid looping configuration associated with the silencing of the maternal igf2 allele. Mol. Endocrinol. 2008;22:1476–1488. doi: 10.1210/me.2007-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murrell A., Heeson S., Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 71.Howard T.K., Algar E.M., Glatz J.A., Reeve A.E., Smith P.J. The insulin-like growth factor 1 receptor gene is normally biallelically expressed in human juvenile tissue and tumours. Hum. Mol. Genet. 1993;2:2089–2092. doi: 10.1093/hmg/2.12.2089. [DOI] [PubMed] [Google Scholar]
- 72.Ogawa O., McNoe L.A., Eccles M.R., Morison I.M., Reeve A.E. Human insulin-like growth factor type I and type II receptors are not imprinted. Hum. Mol. Genet. 1993;2:2163–2165. doi: 10.1093/hmg/2.12.2163. [DOI] [PubMed] [Google Scholar]
- 73.Barlow D.P., Stoger R., Hermann B.G., Saito K., Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 74.Barlow D.P. Gametic imprinting in mammals. Science. 1995;270:1610–1613. doi: 10.1126/science.270.5242.1610. [DOI] [PubMed] [Google Scholar]
- 75.Wutz A., Barlow D.P. Imprinting of the mouse Igf2r gene depends on an intronic CpG island. Mol. Cell. Endocrinol. 1998;140:9–14. doi: 10.1016/s0303-7207(98)00022-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


