Abstract
Higher order chromatin structures across the genome are maintained in part by the architectural proteins CCCTC binding factor (CTCF) and the cohesin complex, which co-localize at many sites across the genome. Here, we examine the role of these proteins in mediating chromatin structure at the cystic fibrosis transmembrane conductance regulator (CFTR) gene. CFTR encompasses nearly 200 kb flanked by CTCF-binding enhancer-blocking insulator elements and is regulated by cell-type-specific intronic enhancers, which loop to the promoter in the active locus. SiRNA-mediated depletion of CTCF or the cohesin component, RAD21, showed that these two factors have distinct roles in regulating the higher order organization of CFTR. CTCF mediates the interactions between CTCF/cohesin binding sites, some of which have enhancer-blocking insulator activity. Cohesin shares this tethering role, but in addition stabilizes interactions between the promoter and cis-acting intronic elements including enhancers, which are also dependent on the forkhead box A1/A2 (FOXA1/A2) transcription factors (TFs). Disruption of the three-dimensional structure of the CFTR gene by depletion of CTCF or RAD21 increases gene expression, which is accompanied by alterations in histone modifications and TF occupancy across the locus, and causes internalization of the gene from the nuclear periphery.
INTRODUCTION
The generation of higher order chromatin structures, which establish domains of transcription and bring cell-specific enhancers into close association with their target promoters, is critical for gene expression. These mechanisms are particularly important for clusters of linked loci that are coordinately regulated such as the homeobox or globin genes and also for large genes with multiple intragenic cis-regulatory elements, for example the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Previous results showed that intronic enhancers within CFTR coordinate epithelial-specific looping of the active locus (1,2). Specifically, an intestinal-selective enhancer element located in the middle of the locus was shown to interact directly with the gene promoter by quantitative chromosome conformation capture (q3C), despite its genomic location 100 kb away. Direct interactions were also evident between the CFTR promoter and enhancer-blocking insulator elements 5′ (2) and 3′ to the locus (1,3), around 200 kb from the promoter. We also suggested a role for cohesin in stabilizing the looped structure of the CFTR locus, since the cohesin component RAD21 was enriched at the insulator elements 5′ and 3′ to the gene (1). These data were consistent with previous observations that CCCTC binding factor (CTCF) bound to enhancer-blocking insulator elements flanking the CFTR gene (3,4), since CTCF and cohesin are known to interact at insulator elements (5–7). Most recently, we identified a transcriptional network involving the pioneer factors forkhead box A1/A2 (FOXA1/A2), hepatocyte nuclear factor 1 (HNF1) and caudal-type homeobox 2 (CDX2) that mediate the function of several intestinal-selective enhancers, which are involved in the looping of active CFTR (8).
The role of CTCF in the function of insulators and chromatin barriers is well-established (9–11). Genome-wide, CTCF binds to 45,000–65,000 sites, of which 46% are intergenic, 34% are intragenic (within introns or exons) and 20% are promoter proximal (12,13). While 40–60% of CTCF-occupied sites are ubiquitous, the others are important for cell and tissue-specific regulation (14). Moreover, recent data generated by 3C-carbon copy (5C, (15)) suggest that enhancer-blocking or chromatin barrier activity may not be the main functions of CTCF, since 79% of long-range interactions circumvent one or more CTCF binding sites (16). The identification of many different roles for CTCF in regulating genome organization led to its classification as an architectural protein (17).
Here, we use siRNA-mediated depletion of CTCF, cohesin components and FOXA1/A2, followed by q3C to show that though each of these proteins contributes to maintenance of the three-dimensional (3D) looped structure of this genomic region, the underlying mechanisms are different. Moreover, rather than repressing its transcription, we observe that disruption of this looped structure increases CFTR expression through changes in chromatin structure, accessibility, transcription factor (TF) occupancy and nuclear localization of the locus.
MATERIALS AND METHODS
Cell culture
The following human cell lines were grown by standard methods: Caco2 colon carcinoma, Calu-3 lung carcinoma, 16HBE14o- immortalized bronchial epithelial cells.
Transient siRNA knockdown experiments
Stealth™ CTCF or RAD21 (5) and non-targeting medium GC negative control siRNA were transfected with Lipofectamine® 2000 or RNAiMAX™ (Life Technologies (LT)) and 20 nM hFOXA1 (Santa Cruz Biotechnology (SCB) sc-37930), hFOXA2 (SCB sc-35569) or 40 nm control (SCB sc-37007) siRNA as described (8). Cells were harvested after 72 h.
Western blot analysis
Standard protocols were used and protein levels assayed with antibodies against CFTR (570, a generous gift from T Jensen and JR Riordan, (18)), CTCF (Millipore 07–729), RAD21 (Millipore 05–908) and β-tubulin (Sigma–Aldrich T4026). Protein quantification was performed using ImageJ software (NIH) (http://rsb.info.nih.gov/ij/).
RT-qPCR (Reverse transcription-quantitative PCR)
Total RNA was extracted using TRIzol (LT). CFTR mRNA was assayed as described previously using a TaqMan primer/probe set spanning CFTR exons 5 and 6, and normalized to endogenous 18S rRNA (19).
Chromatin immunoprecipitation (ChIP)
ChIP was performed using standard methods as previously described (1,8). Antibodies were against FOXA1 (Abcam ab5089), CDX2 (Bethyl Laboratories A300–691A), FOXA2 (SCB sc-6554x), histone H3 (ab1791), H3K9Me3 (ab8898), H3K9Ac (Millipore 07–352), H3K27ac (ab4729), CTCF (Millipore 07–729), RAD21 (ab992), SMC1 (Bethyl Laboratories A300–055A), normal goat IgG (SCB sc-2028), or normal rabbit IgG (Millipore 12–370). qPCR was performed using SYBR® Green reagents with primers listed in Supplementary Table S1.
Quantitative chromosome conformation capture (q3C)
q3C was performed as described previously (3,20) using primers listed in Supplementary Table S1.
Fluorescence in situ hybridization (FISH)
The FISH protocol was described previously (21) and used a bacterial artificial chromosome (BAC) spanning the CFTR locus (22). Probes were biotin-labeled by nick translation and signals were detected using an anti-biotin antibody conjugated to fluorescein (Jackson Laboratories). DAPI was used as the nuclear stain.
Nuclear Positioning Analysis
Nuclear positioning analysis used a set of scripts written in MATLAB (Neems et al. unpublished). Briefly, binary masks of nuclear volume were created using a semi-automated method based on Otsu's method of image segmentation. Next, FISH signal masks were created using manual segmentation methods and their intensity-weighted centroids used for the analysis. FISH signals were considered to be diffraction-limited spots and thus only single points (centroids) were considered in calculations. For the analysis in three dimensions, the shortest distance from the FISH signal to the nuclear periphery was calculated by determining all the pairwise distances and finding the minimum distance within the nuclear mask. This value was reported as the minimum distance to the nuclear periphery in micrometers.
RESULTS
Architectural protein occupancy across the CFTR locus
To determine the contribution of CTCF and cohesin to the 3D structure of the CFTR locus we first identified sites occupied by these factors. Inspection of ENCODE data (23) from multiple cell types revealed binding sites for CTCF and RAD21 (SCC1) and these were confirmed by ChIP in Caco2 intestinal carcinoma cells, which express abundant CFTR. CTCF/RAD21 binding was seen at −80.1 kb (I) 5′ to the translational start site (TSS) of the CFTR locus, within an intron of ASZ1 (corresponding to the -79.5 kb DNase I hypersensitive site (DHS) reported earlier (24)) and also at sites in the CTTNBP2 gene, +48.9 kb (IV) and +83.7 kb (V) from translational stop site of CFTR (Figure 1A and Table 1). The ENCODE data also confirmed CTCF occupancy at enhancer-blocking insulator elements −20.9 kb (II) 5′ to the TSS that we described previously (3,4). CTCF does not bind to an element at +15.6 kb that we showed to be a CTCF independent enhancer-blocking insulator (4). Modest enrichment of CTCF and RAD21 at the CFTR promoter is likely a result of indirect interactions with distal CTCF binding sites, since no CTCF binding motifs are predicted in silico or observed in genome-wide CTCF ChIP-seq data (23) within the promoter region. Moreover, we saw no evidence for CTCF occupancy at the site shown in intron 2 in ENCODE data. RAD21 occupancy was also seen in Caco2 cells at the majority of the CTCF sites (I, II, IV and V). CTCF and RAD21 occupancy at each of these sites was next confirmed by ChIP in airway cell lines (Calu-3 and 16HBE14o-), which are known to exhibit different mechanisms of CFTR regulation than are seen in intestinal cells (Supplementary Figure S1, (1,25,26)). The CTCF/RAD21 binding sites at −80.1 kb (I), +48.9 kb (IV) and +83.7 kb (V) are ubiquitously occupied, while those at −20.9 kb (II) and +6.8 kb (III) show variable occupancy in the different cell lines. The most marked difference between the airway cells and Caco2 is the prominence of CTCF/RAD21 occupancy at site I and relatively low levels at site IV in the airway.
Figure 1.
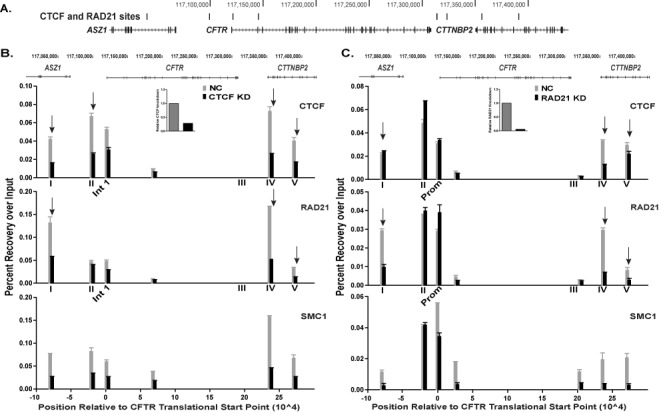
CTCF and cohesin binding are interdependent at the CFTR locus. (A) UCSC Genome Browser graphic of the CFTR locus showing CTCF/cohesin binding sites (ENCODE data) marked by vertical black bars. (B) and (C) ChIP for CTCF, RAD21 and SMC1 in Caco2 cells transfected with negative control (NC) siRNA (gray) or siRNA targeting CTCF (B) or RAD21 (C) (black) for knockdown (KD). Data from one representative experiment are shown as percent recovery over input, n ≥ 2. qPCR was performed in duplicate at each site and error bars are ± SEM. Arrows denote sites of particular interest discussed in the text. Inset panels show relative siRNA-mediated depletion of CTCF or RAD21.
Table 1. Genomic locations of CTCF and cohesin sites on ENCODE.
| CTCF/Cohesin Sites | Location chr 7 (hg19) |
|---|---|
| −80.1 kb (I) | 117039873 - 117040255 |
| −20.9 kb (II) | 117099046 - 117099445 |
| Intron 1 | 117121084 - 117121463 |
| Intron 2 | 117145360 - 117145679 |
| +6.8 kb (III) | 117313933 - 117314336 |
| +48.9kb (IV) | 117355840 - 117356209 |
| +83.7 kb (V) | 117390670 - 117391129 |
CTCF and cohesin may have different roles at individual elements across the CFTR locus
CTCF and RAD21 co-occupancy was observed at all the sites assayed by ChIP across the locus. However, to determine whether the two proteins were functionally interdependent at each site we used an siRNA approach in Caco2 cells. CTCF or RAD21 were depleted and occupancy of both factors and SMC1, another cohesin component, was measured at each site by ChIP (Figure 1B and C). Knockdown of CTCF caused about a 60% loss of CTCF occupancy at sites I, II, IV and V (marked by arrows) and a 40% reduction at a site in intron 1 (Figure 1B). Depletion of CTCF greatly reduced occupancy of the cohesin components at sites I and IV and caused moderate decreases at sites II and V. RAD21 depletion followed by ChIP showed loss of the cohesin components, RAD21 and SMC1, at sites I, IV, and V flanking the gene (Figure 1C). However, the decrease in RAD21 only reduced CTCF occupancy by 60% at sites IV and 30% at site V and had no impact on site I. Additionally, whereas CTCF loss depleted CTCF and the cohesin components at site II, RAD21 knockdown had no effect at that site (Figure 1B and C). Together these data suggest different roles of recruitment for CTCF and the cohesin complex at sites of co-occupancy 5′ and 3′ to the CFTR gene and that these factors are not always functionally interdependent.
CTCF and cohesin mediate distinct conformational interactions
CTCF and cohesin have important roles in the establishment and maintenance of long-range chromatin interactions, which are required for normal gene expression. To determine if CTCF and RAD21 are critical for chromatin looping across the CFTR locus, q3C was performed after siRNA-mediated depletion of CTCF or RAD21 in Caco2 cells. No marked changes in interactions between the CFTR promoter (bait marked as a dotted line) and the cis-regulatory elements within and flanking the gene were seen after CTCF knockdown (Figure 2A). Minor reductions were evident in promoter interactions with the −20.9 kb (II), +6.8 kb/+15.6 kb (III) enhancer-blocking insulators closest to the locus (marked by arrows). However, with a 3C bait at the −20.9 kb insulator (dotted line, Figure 2B), depletion of CTCF was seen to decrease interactions with a site 3′+20 kb (marked by arrow) and significantly with the +48.9 kb (IV) site (Figure 2B, marked ∧). In contrast, RAD21 depletion caused a partial loss of interactions between the CFTR promoter and all of the cis-acting elements across the locus (Figure 2C). These included both insulator elements flanking the locus (II and III) and intronic enhancers, such as the one at DHS11. A similar decrease in interactions across CFTR was evident using the 3C bait at −20.9 kb, including between this insulator element and the promoter, and also the enhancer at DHS11 (Figure 2D, significant sites marked ∧). However, the reduction in RAD21 did not alter the interactions between the −20.9 kb bait and elements 3′ to CFTR, particularly the +48.9 kb site (IV) which is impacted by CTCF knockdown (Figure 2D). In combination, these q3C data after CTCF or RAD21 depletion suggest that CTCF has a dominant role in the chromatin looping that draws together insulator elements 5′ and 3′ to the CFTR locus and brings them close to the promoter. In contrast, though the cohesin complex, evaluated by RAD21, also contributes to the maintenance of CTCF-mediated interactions across the locus, it appears to have an additional role in stabilizing the association between intronic enhancers and the gene promoter.
Figure 2.
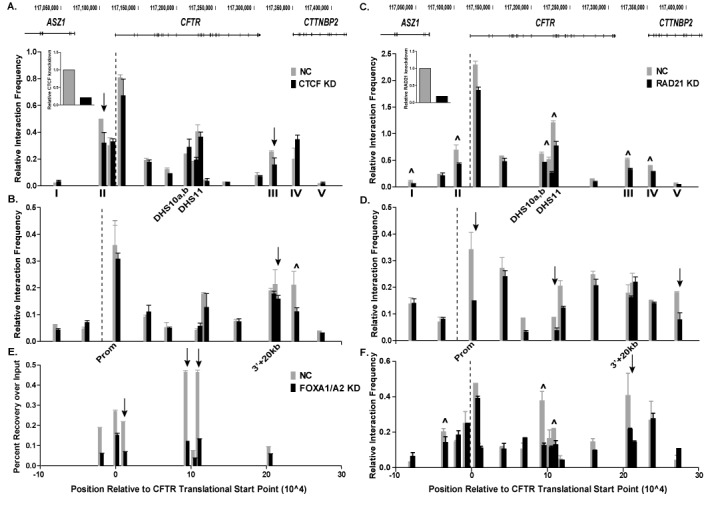
Depletion of CTCF or RAD21 alters long-range interactions across the CFTR locus. (A)–(D) q3C interactions in cells treated with NC siRNA (gray) or siRNA targeting CTCF (A) and (B) (black), or RAD21 (C) and (D) (black). The 3C bait (dotted line) is in a HindIII fragment at the CFTR promoter (A) and (C) or at the −20.9 kb insulator element (B) and (D). (E) ChIP for FOXA2 on cells transfected with NC (gray) or FOXA1/2 siRNA (black). Data are shown as percent recovery over input. (F) q3C interactions in NC (gray) or FOXA1/2 siRNA treated cells (black). The 3C bait (dotted line) is in a HindIII fragment at the CFTR promoter. Multiple sites were tested for interactions within distal HindIII fragments across the CFTR locus. x-axis = position relative to translational start site of CFTR, y-axis = interaction frequency relative to that between two fragments in the ubiquitously expressed ERCC3 gene. Data shown are from one representative q3C experiment, n = 3. Error bars represent the ± SEM of duplicate qPCRs for each fragment. ∧ indicate sites that are statistically different after knockdown as determined by an unpaired, two-tailed Student's t-test, n = 3, P-values listed in Supplementary Table S2. Other sites of interest marked with arrows. Inset panels show relative siRNA-mediated depletion of CTCF or RAD21.
The FOXA1/A2 pioneer transcription factors influence the conformation of CFTR
The distinct role of cohesin in mediating the interaction of cis-acting enhancers with the CFTR promoter led us to investigate the contribution of tissue-specific TFs, which bind cis-elements in introns 10 and 11, to chromatin looping. We showed previously that FOXA1/A2 interact in vivo with multiple CFTR intronic regulatory elements including DHS10a,b and DHS11 in intestinal cell lines (8). Depletion of FOXA1 and FOXA2 using specific siRNAs reduced FOXA1/A2 protein levels by 80–90% and led to a ∼40% reduction in CFTR mRNA expression in Caco2 cells (27). SiRNA-mediated depletion of FOXA1/A2 was shown by ChIP to reduce FOXA2 enrichment at several sites across the CFTR locus in Caco2 cells (Figure 2E). The −20.9 kb insulator element and DHS in introns 1, 10 (DHS10a,b (19,28)), and 11 (marked by arrows) showed ∼30–70% reduction in FOXA2 occupancy. Next, we examined the effect of FOXA1/A2 depletion on long-range interactions across the CFTR locus in the same cells. A significant decrease in interaction frequency was observed between the CFTR promoter bait and an upstream enhancer at -35 kb and the middle of the locus (introns 10–11) (Figure 2F, significant sites marked with ∧). Moreover, a decrease in interactions between the promoter and the 3′ insulators (III) was also detected. These data show that FOXA1/A2 are required for the establishment and/or maintenance of long-range interactions across the CFTR locus, particularly for the recruitment of intronic enhancers to the promoter. This is consistent with data shown in Figure 2A–D, which suggest a CTCF-independent role for cohesin in stabilizing interactions of cis-acting intronic enhancers in CFTR with the gene promoter.
Depletion of both CTCF and cohesin diminishes interactions across the CFTR locus
To further delineate the function of CTCF and cohesin at the CFTR locus, both CTCF and RAD21 were targeted for simultaneous knockdown. Depletion of CTCF and RAD21 led to a loss of interactions across the locus between all the cis-regulatory elements and the two baits at the CFTR promoter and −20.9 kb as measured by q3C (Figure 3A and B). A significant decrease in associations was observed between the promoter and CTCF/cohesin binding sites I, II, IV and V (Figure 3A, marked by ∧). Moreover, a significant loss of interactions was also detected between the promoter and cis-regulatory elements within introns 10, 11 and 19, some of which are intestinal-selective enhancers (8). The most dramatic change in interactions with the −20.9 kb bait was at the +48.9 kb (IV) site, where a 50% loss in looping was detected (Figure 3B, marked by ∧).
Figure 3.
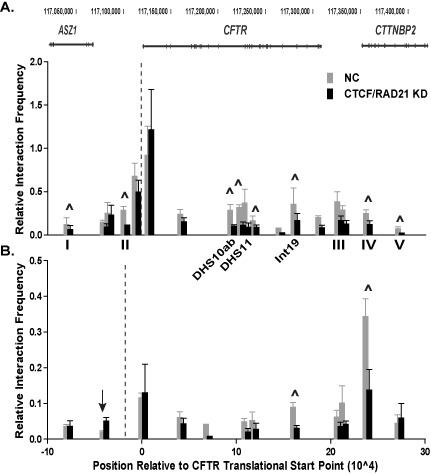
Altered long-range interactions after CTCF and RAD21 depletion. (A) and (B) q3C interactions in cells treated with NC siRNA (gray) or siRNAs targeting CTCF and RAD21 (black), n = 3. The 3C bait (dotted line) is either at the CFTR promoter (A) or the −20.9 kb site (B). ∧ indicate sites that are statistically different after knockdown as determined by an unpaired, two-tailed Student's t-test, P-values listed in Supplementary Table S2. Other sites of interest are marked with arrows. Experimental details are in Figure 2 legend.
Disruption of the looped architecture of the active CFTR locus enhances expression
To determine the effect of disruption of looping across the CFTR locus on gene expression, CFTR mRNA levels were measured after siRNA-mediated depletion of CTCF (Figure 4A and B) or RAD21 (Figure 4D and E) in Caco2 cells. A greater than 80% reduction of both architectural proteins significantly increased CFTR mRNA levels 1.4 - (CTCF, Figure 4C) and 1.6- (RAD21, Figure 4F) fold in comparison to a non-targeting control siRNA, when measured by reverse transcription and qPCR (RT-qPCR). Depletion of both CTCF and RAD21 together (Figure 4G) led to 2.8-fold increase in CFTR expression (Figure 4H), showing an additive effect, which is consistent with data showing distinct roles of CTCF and cohesin at the CFTR locus (Figure 2). To ensure the increase in CFTR expression was gene-specific and not simply an artifact of generalized disruption of chromatin architecture, we analyzed the expression of several other genes that are regulated by CTCF and cohesin. Depletion of RAD21 in HeLa cells repressed the solute carrier family 35, member C2 (SLC35C2) gene (5) and we observed the same effect in Caco2 cells after RAD21 knockdown (Supplementary Figure S2). In previous work, we also identified CTCF as a regulator of the gel-forming mucin genes on 11p15.5 (29) and after siRNA-mediated depletion of CTCF we observed a decrease in MUC2 and an increase in MUC6 mRNA (Supplementary Figure S2). These data confirm that the increased expression of CFTR that is seen after CTCF and RAD21 knockdown is likely a gene-specific effect. Next, we sought to determine if the observed increase in CFTR mRNA levels also occurred at the protein level. Western blots for CFTR after depletion of CTCF, RAD21, or both factors led to a marked increase in CFTR protein expression (Figure 4I–K). This was confirmed by quantification of the scanned western blots, showing a significant, ∼ 3-fold increase in CFTR protein levels after combined CTCF and RAD21 depletion, a 2-fold increase after RAD21 reduction alone, and a 1.5-fold increase after CTCF depletion alone (Figure 4L).
Figure 4.
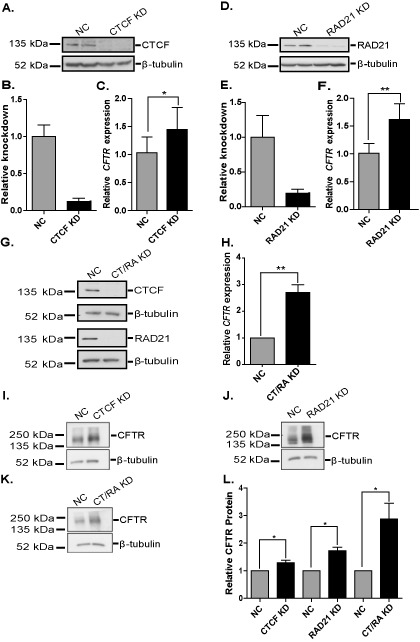
siRNA-mediated depletion of architectural proteins augments CFTR expression. (A), (D) and (G) Western blots show siRNA-mediated depletion of CTCF (A), RAD21 (D), or both (G) in Caco2 cells with β-tubulin as the loading control. (B) and (E) ImageJ quantification of western blots shown in (A) and (D), respectively. (C), (F) and (H) RT-qPCR for CFTR expression in Caco2 cells after knockdown of CTCF (C), RAD21 (F) or both (H) (black) compared to NC siRNA treated cells (gray). (I)–(K) Western blots show CFTR levels after loss of CTCF (I), RAD21 (J) or both factors (K) in Caco2 cells with β-tubulin as the loading control. (L) ImageJ quantification of the CFTR western blots, n ≥ 3. *P < 0.05; **P < 0.01 as determined by an unpaired, two-tailed Student's t-test. CT/RA KD is short for CTCF and RAD21 knockdown.
We also extended our investigation of the role of architectural proteins in regulating CFTR expression to airway cell lines, which use a different set of cis-regulatory elements from the intestinal elements to drive promoter activity (1,8,26). Specifically, though the same enhancer-blocking insulator elements flanking the gene are important, the cis-elements in the middle of the locus (introns 10 and 11) are not evident in airway cells and a pair of elements associated with DHS at -35 kb and -44 kb to the gene promoter seem critical. The DHS at -35 kb contains a strong airway-selective enhancer (25,26). q3C in two airway cell lines that express high levels of CFTR (Calu-3 and 16HBE14o-) confirmed distinct cell-type-specific interactions (Supplementary Figure S3A). No interactions are seen between a bait at the promoter and DHS 11, though strong, cell-type dependent interactions are seen with the −20.9 kb DHS (II) (16HBE14o-), DHS -35 kb (Calu-3 and 16HBE14o-) and -44 kb (Calu-3 only). CTCF depletion in 16HBE14o- cells decreased interactions between the CFTR promoter (dotted line) and the −20.9 kb (II) site and caused a minor reduction in interactions with the +6.8 kb/+15.6 kb (III) and +48.9 kb (IV) CTCF/cohesin binding sites (Supplementary Figure S3B). A significant increase in interactions with the -35 kb enhancer (marked ∧) and a slight increase with the -44 kb element was also seen. Consistent with this alteration in the profile of interactions of 5′ elements with the promoter (−20.9 kb (II), -35 kb and -44 kb), siRNA-mediated depletion of CTCF in 16HBE14o- cells caused a significant increase in CFTR expression (Supplementary Figure S3C and D), similar to the change in Caco2 cells. In contrast, RAD21 depletion had little effect on CFTR mRNA levels in 16HBE14o- cells (Supplementary Figure S3E and F). This result likely reflects the different CFTR regulatory mechanisms in the intestinal and airway cells.
Fine-tuning of CFTR expression in the looped conformation
Since looping of the CFTR locus is associated with active gene expression, perturbation of the 3D structure by depletion of the architectural proteins was predicted to repress, not increase, CFTR expression. To investigate the mechanism underlying the increase in CFTR expression after depletion of CTCF and RAD21, we evaluated changes in the chromatin landscape of the locus. ChIP for three histone modifications, H3K9me3, H3K9ac and H3K27ac, was performed after depletion of CTCF in Caco2 cells (Figure 5A–C). H3K9me3 is found at repressed or inactive regions of chromatin including at promoters, silenced enhancers, and in the gene body (30). H3K9ac is generally associated with active or transcribed regions of the genome, specifically at promoters, transcription start sites and regulatory elements (31,32) and H3K27ac is a mark of active enhancers (30). Enrichment of each histone mark was calculated as recovery over percent input and normalized to total histone H3 at each site. Loss of CTCF decreased enrichment of the H3K9me3 mark at −20.9 kb (site II), the promoter (-0.5 kb), and within the gene body at a site in intron 14a that is not associated with any known cis-regulatory element (Figure 5A, marked by arrows). Correspondingly, H3K9ac increased at the −20.9 kb site, though the modification remained unchanged at all other regions tested (Figure 5B). The greatest changes in histone marks after CTCF depletion were in H3K27ac enrichment, which increased significantly at the known, active enhancers in introns 1 and 11 and a cis-element in intron 10 (Figure 5C, marked by arrows and ∧).
Figure 5.
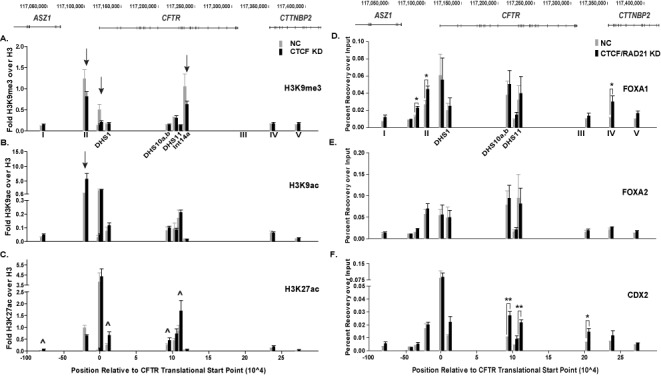
Depletion of architectural proteins alters histone modifications and transcription factor occupancy. (A)–(C) ChIP for H3K9me3 (A), H3K9ac (B) and H3K27ac (C) in Caco2 cells transfected with NC siRNA (gray) or a siRNA targeting CTCF (black). Data are shown as percent recovery over input and each histone modification is normalized to recovery over input of unmodified histone H3 at each site, n = 3. ∧ indicate sites that are statistically different after knockdown as determined by an unpaired, two-tailed Student's t-test, P-values are listed in Supplementary Table S2. Other sites of interest are marked with arrows. (D)–(F) ChIP for FOXA1 (D), FOXA2 (E) and CDX2 (F) in Caco2 cells transfected with NC siRNA (gray) or siRNAs targeting CTCF and RAD21 (black). Data are shown as percent recovery over input, n = 3. *P < 0.05; **P < 0.01 as determined by an unpaired, two-tailed Student's t-test.
We next investigated if the changes in histone modifications across the locus altered TF occupancy. We focused our analyses on TFs previously shown to bind to cis-regulatory elements in the CFTR locus and regulate its expression (8,27). ChIP for three TFs, FOXA1, FOXA2 and CDX2 was performed after depletion of both CTCF and RAD21, since this had the greatest impact on CFTR expression (Figure 4G and H). FOXA1 occupancy, after CTCF/RAD21 reduction, significantly increased at the 5′ airway-selective enhancer element at -35 kb and two CTCF/cohesin binding sites (II, IV) (Figure 5D, marked by *). Interestingly, this was specific to FOXA1 as no changes in FOXA2 binding were observed (Figure 5E) despite the two factors having similar binding motifs and some overlapping functions (33). Moreover, CTCF/RAD21 depletion led to a significant increase in CDX2 binding at a cis-regulatory element in intron 10, the strong, intestinal-selective enhancer in intron 11, and the insulator at +15.6 kb (Figure 5F, marked by */**).
Contribution of architectural proteins to nuclear positioning of the CFTR locus
The increase in CFTR mRNA and CFTR protein, coupled with the changes in chromatin structure and TF occupancy upon CTCF/RAD21 depletion, led us to evaluate the positioning of the CFTR alleles within the nucleus. A previous study demonstrated that the perinuclear positioning of the inactive CFTR locus was dependent on type A lamins, CTCF and a histone deacetylase (HDAC) (34). FISH followed by 2D analysis revealed that treatment of cells with a CTCF siRNA significantly decreased the percentage of alleles at the nuclear periphery in two cell types (34). To determine the effect of CTCF and RAD21 depletion on nuclear positioning of the CFTR alleles we performed 3D FISH with cells grown on glass coverslips. Caco2 cells, which are highly aneuploid, generally have three copies of chromosome 7 per cell, as evidenced by the three distinct FISH signals in each nucleus in negative control, CTCF, or RAD21 siRNA-treated cells (Figure 6A–C). Quantification of CFTR nuclear positioning after CTCF or RAD21 depletion showed a significant increase in the average minimum distance to the nuclear periphery relative to non-targeting siRNA treated cells (Figure 6D). Hence, depletion of both architectural proteins is associated with internalization of the CFTR locus within the nucleus.
Figure 6.
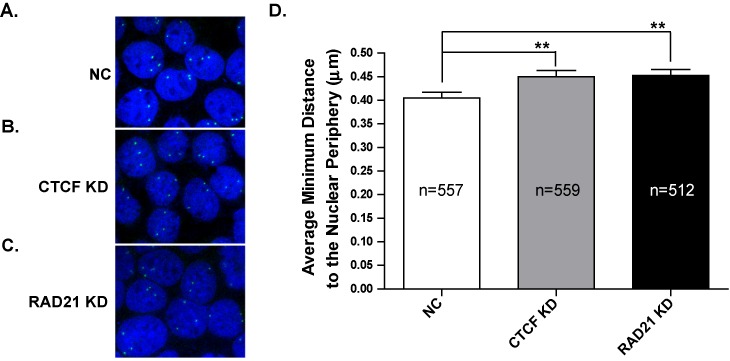
Altered nuclear positioning of CFTR alleles after loss of CTCF or RAD21. (A)–(C) Representative FISH images for CFTR in Caco2 cells treated with NC siRNA (A), CTCF siRNA (B), or RAD21 siRNA (C). Nuclei are stained with DAPI and the CFTR alleles are white dots. (D) Quantification of the minimum distance to the nuclear periphery of the CFTR alleles in all three conditions. **P < 0.01 as determined by an unpaired, two-tailed Student's t-test.
DISCUSSION
The looped structure of the active CFTR locus (1,2) led us to investigate how this 3D structure is established and maintained, and examine whether it is critical for cell-type-specific control of gene expression. We first focused on the architectural proteins, CTCF and the cohesin complex, which were shown previously to bind at sites near CFTR (1,3,4). Our aim was to determine their role, independently or in combination, at enhancer-blocking insulators flanking the locus and within neighboring genes. ChIP after knockdown of either factor showed differential loss of CTCF, RAD21 and SMC1 at each site. CTCF and RAD21 binding are co-dependent at site IV in the last intron of the CTTNBP2 gene, which is located on the 3′ side of CFTR, while CTCF has a dominant role in interactions at site I, within an intron of the ASZ1 gene, which flanks CFTR on the 5′ side and site II −20.9 kb upstream of the CFTR TSS (Figure 1B and C). Though RAD21 has a modest impact on occupancy at site V, CTCF appears to be more important in mediating CTCF and RAD21 binding at this site (Figure 1B and C). However, despite the interdependent recruitment of both CTCF and cohesin, q3C data show distinct roles for the proteins in 3D organization of the locus (Figure 2A–D).
Depletion of CTCF has a marked effect on interactions between the CFTR promoter and insulator elements flanking the gene, the insulator 5′ to the gene and both insulators and other CTCF/RAD21 sites 3′ to the locus. These data implicate a higher order looped structure between elements 5′ and 3′ to the CFTR gene, which may create a chromatin domain to isolate the locus for appropriate regulation. This is particularly relevant as the flanking genes, ASZ1 (5′) and CTTNBP2 (3′), are expressed in different cell types from CFTR. ASZ1 is primarily expressed in ovaries and testes (35), while CTTNBP2 is expressed at high levels in the brain and kidney, and at lower levels in other tissues (36). This model is consistent with our previous data, which show that two of the CTCF/cohesin binding sites flanking CFTR (−20.9 kb and +6.8 kb) have enhancer-blocking insulator activity (3,4).
Depletion of the cohesin component, RAD21, leads to lower interaction frequencies across the entire locus between the promoter, intronic enhancers and flanking CTCF/cohesin binding sites. With a bait at site II (−20.9 kb) a reciprocal decrease in interaction is seen with the promoter, though only cis-elements within CFTR exhibit a reduced interaction and CTCF/cohesin sites 5′ and 3′ to the locus remain unchanged (with the exception of site V). These data suggest that cohesin has a critical role in stabilizing and maintaining interactions between the gene promoter and intronic enhancers which drive expression of CFTR, consistent with the role of cohesin in chromatin organization (reviewed in (37,38)). For example, at the IFNG (interferon-γ) and β-globin loci, cohesin along with CTCF plays a role in maintaining cell-type selective interactions between CTCF and cohesin binding sites (39,40). Additional work on the β-globin locus showed that cohesin also mediated associations between different genes and their enhancers (41). These data are consistent with our results. However, though our q3C data show that CTCF and cohesin contribute to higher order organization of the CFTR locus, they are clearly not the only components of the complex involved in looping CFTR since >80% depletion of CTCF or RAD21 did not have proportional effects on the structure. The likelihood that other factors are involved led us to investigate whether tissue-specific TFs binding to cis-regulatory elements also have a mechanistic role. The pioneer TFs FOXA1/A2 bind to several cis-elements across CFTR and play a dominant role in driving enhancer function in intestinal cells (8). We show that siRNA-mediated depletion of FOXA1/A2 decreases looping interactions between the CFTR promoter and cis-elements in introns 10 and 11, including a strong cell-type selective enhancer ((8,27), Figure 2E and F). Hence, our data suggest two levels of chromatin organization: the first in which TFs binding to intronic cis-regulatory elements initiate looping to the promoter to drive CFTR expression and the second where the cohesin complex acts to stabilize the 3D organization. Other work published while our manuscript was in revision also suggests that CTCF and cohesin have different roles in chromatin interactions genome-wide (42) and that cohesin has a CTCF-independent role in enhancer-promoter looping (43).
Targeted depletion of both CTCF and cohesin together results in a dramatic loss of interactions across the CFTR locus as measured by q3C (Figure 3). A significant loss of associations is observed between the promoter and CTCF/cohesin binding sites I, II, IV, V (Figure 3A) and between the −20.9 kb bait and the other CTCF/cohesin binding site at +48.9 kb (Figure 3B). These data provide further evidence that a higher order looped structure is formed between CTCF/cohesin binding sites. With the bait at the CFTR promoter, a loss of interactions is also observed with cis-regulatory elements within introns of the gene, which have been shown to activate CFTR expression (Figure 3A, (1,8,27)).
The observation that despite the disruption of the higher order chromatin structure of the locus, resulting from depletion of architectural proteins, CFTR expression increases, was initially unexpected. There are several reports of decreases in gene expression after loss of chromatin looping between CTCF/cohesin binding sites or gene promoters and enhancers, where looping facilitates gene expression (40–41,44–45). However, genome-wide analyses showed that genes may be either up- or down-regulated after loss of CTCF/RAD21 (5), (46). Increased gene expression was also observed at the HOXA gene cluster through loss of barrier activity (47), consistent with our q3C data showing decreased interactions between CTCF/cohesin binding sites, some of which have enhancer-blocking insulator activity. Moreover, both CTCF and cohesin have many diverse functions and have been implicated in gene repression as well as activation (reviewed in (48,49)).
A well-established role for CTCF is positioning nucleosomes and creating distinct chromatin domains (14,50). To investigate whether the increase in CFTR expression after CTCF knockdown occurred through changes in the chromatin landscape, we evaluated three histone modifications: H3K9me3 (inactive), H3K9ac (active) and H3K27ac (active). Depletion of CTCF causes a decrease in H3K9me3 and a concurrent increase in H3K9ac and H3K27ac at specific sites across the locus, which may explain the increase in CFTR after CTCF knockdown. Moreover, the increase in H3K27ac occurs specifically at the intestinal-selective enhancers in introns 1, 10 and 11, which drive CFTR expression in Caco2 cells. These changes in histone modifications could arise from alterations in the chromatin structure that increase accessibility to cis-regulatory elements, either due to loss of insulator or barrier activity after CTCF depletion.
In addition to the alterations in chromatin structure across the locus, we observe changes in TF occupancy after CTCF and RAD21 depletion (Figure 5D–F). The increase in FOXA1 binding at the -35 kb enhancer and the CTCF/cohesin binding sites at −20.9 kb (II) and +48.9 kb (IV) (Figure 5D) suggests that the altered chromatin landscape is facilitating TF binding to other sites. These data also indicate that loss of CTCF and cohesin modifies the function of the sites as it allows other factors to associate with these regions. In silico analysis reveals that the −20.9 kb (II) site has a FOXA binding motif, which may have been previously inaccessible due to the presence of CTCF and cohesin components at that region (Figure 1B and C). Interestingly, the same effect is not observed with another FOXA family member, FOXA2 (Figure 5E), which binds together with FOXA1 at several sites in CFTR (27). These data may illustrate the emerging importance of distinct functions performed by FOXA1 and FOXA2 genome-wide. CDX2 occupancy at the locus is also significantly augmented at sites in introns 10 and 11 (Figure 5F). CDX2 was shown previously to bind to these and other sites across the locus and its reduction leads to a decrease in CFTR mRNA levels (8). These data further demonstrate that the 3D looped structure established and maintained by CTCF and cohesin may fine-tune CFTR expression by restricting TF binding to levels sufficient to drive CFTR expression, but not achieve maximal expression.
Finally, we investigated the role of CTCF and cohesin in the nuclear positioning of CFTR alleles. 3D FISH after CTCF or RAD21 depletion showed an increase in the average minimum distance to the nuclear periphery relative to negative control siRNA treated cells (Figure 6A–D). Loss of either CTCF or RAD21 alters the positioning of the CFTR alleles in Caco2 cells such that they move further into the interior of the nucleus. This is critical as tethering of genes to the nuclear periphery has been associated with silencing of genes, while movement to the interior of the nucleus is generally indicative of gene activation (51–54). These data may also partly explain the increase observed in CFTR mRNA and protein levels after CTCF and RAD21 depletion (Figure 4).
Several other mechanisms can be envisaged to account for the increase in CFTR expression after the depletion of CTCF and cohesin. One possibility involves the proposed role of CTCF and cohesin in stalling RNA polymerase II (55). The constraining higher order structure mediated by CTCF and cohesin at CFTR could stall RNA pol II, thus decreasing transcription rates, and this would be reversed after loss of CTCF or cohesin. An additional explanation could arise from data showing that CTCF physically interacts with the transcriptional repressor SIN3A and that these proteins may be in a complex at some sites in the CFTR locus (56). Loss of CTCF could then be accompanied by depletion of SIN3A, thus relieving the transcriptional repression. It is possible that several of these mechanisms and others contribute to the role of CTCF and the cohesin complex at the CFTR locus. However, our data clearly show that these architectural proteins are responsible for fine-tuning CFTR expression through the maintenance and organization of its 3D structure. Moreover, they are also involved in regulating the chromatin landscape, TF occupancy and the positioning of the CFTR alleles in the nuclear space.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Dr. S-H Leir for his contribution and Cystic Fibrosis Foundation Therapeutics (Bethesda, MD, USA) for the generous gift of anti-CFTR antibodies.
FUNDING
Cystic Fibrosis Foundation; National Institutes of Health (NIH) [NIH R01 HL094585 and HD068901 to A.H.]/NIGMS New Innovator Award [DP2 OD008717 to S.T.K.]. Funding for open access charge: Cystic Fibrosis Foundation; NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ott C.J., Blackledge N.P., Kerschner J.L., Leir S.-H., Crawford G.E., Cotton C.U., Harris A. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19934–19939. doi: 10.1073/pnas.0900946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gheldof N., Smith E.M., Tabuchi T.M., Koch C.M., Dunham I., Stamatoyannopoulos J.A., Dekker J. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010;38:4325–4336. doi: 10.1093/nar/gkq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackledge N.P., Ott C.J., Gillen A.E., Harris A. An insulator element 3′ to the CFTR gene binds CTCF and reveals an active chromatin hub in primary cells. Nucleic Acids Res. 2009;37:1086–1094. doi: 10.1093/nar/gkn1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackledge N.P., Carter E.J., Evans J.R., Lawson V., Rowntree R.K., Harris A. CTCF mediates insulator function at the CFTR locus. Biochem. J. 2007;408:267–275. doi: 10.1042/BJ20070429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 6.Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H.C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T., et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Rubio E.D., Reiss D.J., Welcsh P.L., Disteche C.M., Filippova G.N., Baliga N.S., Aebersold R., Ranish J.A., Krumm A. CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerschner J.L., Harris A. Transcriptional networks driving enhancer function in the CFTR gene. Biochem. J. 2012;446:203–212. doi: 10.1042/BJ20120693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell A.C., West A.G., Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 10.Recillas-Targa F., Bell A.C., Felsenfeld G. Positional enhancer-blocking activity of the chicken β-globin insulator in transiently transfected cells. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14354–14359. doi: 10.1073/pnas.96.25.14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kellum R., Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 12.Kim T.H., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H., Tian Y., Shu W., Bo X., Wang S. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS One. 2012;7:e41374. doi: 10.1371/journal.pone.0041374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuddapah S., Jothi R., Schones D.E., Roh T.-Y., Cui K., Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanyal A., Lajoie B.R., Jain G., Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ong C.-T., Corces V.G. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui L., Aleksandrov L., Chang X.-B., Hou Y.-X., He L., Hegedus T., Gentzsch M., Aleksandrov A., Balch W.E., Riordan J.R. Domain interdependence in the biosynthetic assembly of CFTR. J. Mol. Biol. 2007;365:981–994. doi: 10.1016/j.jmb.2006.10.086. [DOI] [PubMed] [Google Scholar]
- 19.Mouchel N., Henstra S.A., McCarthy V.A., Williams S.H., Phylactides M., Harris A. HNF1alpha is involved in tissue-specific regulation of CFTR gene expression. Biochem. J. 2004;378:909–918. doi: 10.1042/BJ20031157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagège H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forné T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat. Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 21.Cremer M., Grasser F., Lanctôt C., Müller S., Neusser M., Zinner R., Solovei I., Cremer T. Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol. Biol. Clifton NJ. 2008;463:205–239. doi: 10.1007/978-1-59745-406-3_15. [DOI] [PubMed] [Google Scholar]
- 22.Gillen A.E., Lucas C.A., Haussecker P.L., Kosak S.T., Harris A. Characterization of a large human transgene following invasin-mediated delivery in a bacterial artificial chromosome. Chromosoma. 2013;122:351–361. doi: 10.1007/s00412-013-0418-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ENCODE Project Consortium. Bernstein B.E., Birney E., Dunham I., Green E.D., Gunter C., Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith A.N., Wardle C.J., Harris A. Characterization of DNASE I hypersensitive sites in the 120kb 5′ to the CFTR gene. Biochem. Biophys. Res. Commun. 1995;211:274–281. doi: 10.1006/bbrc.1995.1807. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z., Ott C.J., Lewandowska M.A., Leir S.-H., Harris A. Molecular mechanisms controlling CFTR gene expression in the airway. J. Cell. Mol. Med. 2012;16:1321–1330. doi: 10.1111/j.1582-4934.2011.01439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z., Leir S.-H., Harris A. Immune mediators regulate CFTR expression through a bifunctional airway-selective enhancer. Mol. Cell. Biol. 2013;33:2843–2853. doi: 10.1128/MCB.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerschner J.L., Gosalia N., Leir S.-H., Harris A. Chromatin remodeling mediated by the FOXA1/A2 transcription factors activates CFTR expression in intestinal epithelial cells. Epigenetics. 2014;9:557–565. doi: 10.4161/epi.27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith D.J., Nuthall H.N., Majetti M.E., Harris A. Multiple potential intragenic regulatory elements in the CFTR gene. Genomics. 2000;64:90–96. doi: 10.1006/geno.1999.6086. [DOI] [PubMed] [Google Scholar]
- 29.Gosalia N., Leir S.-H., Harris A. Coordinate regulation of the gel-forming mucin genes at chromosome 11p15.5. J. Biol. Chem. 2013;288:6717–6725. doi: 10.1074/jbc.M112.437400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou V.W., Goren A., Bernstein B.E. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 31.Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Roh T.-Y., Cuddapah S., Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hannenhalli S., Kaestner K.H. The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muck J.S., Kandasamy K., Englmann A., Günther M., Zink D. Perinuclear positioning of the inactive human cystic fibrosis gene depends on CTCF, A-type lamins and an active histone deacetylase. J. Cell. Biochem. 2012;113:2607–2621. doi: 10.1002/jcb.24136. [DOI] [PubMed] [Google Scholar]
- 35.Yan W. Identification of gasz, an evolutionarily conserved gene expressed exclusively in germ cells and encoding a protein with four ankyrin repeats, a sterile- motif, and a basic leucine zipper. Mol. Endocrinol. 2002;16:1168–1184. doi: 10.1210/mend.16.6.0864. [DOI] [PubMed] [Google Scholar]
- 36.Cheung J. Identification of the human cortactin-binding protein-2 gene from the autism candidate region at 7q31. Genomics. 2001;78:7–11. doi: 10.1006/geno.2001.6651. [DOI] [PubMed] [Google Scholar]
- 37.Dorsett D., Merkenschlager M. Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr. Opin. Cell Biol. 2013;25:327–333. doi: 10.1016/j.ceb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merkenschlager M., Odom D.T. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 39.Hadjur S., Williams L.M., Ryan N.K., Cobb B.S., Sexton T., Fraser P., Fisher A.G., Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou C., Dale R., Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl. Acad. Sci. U.S.A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chien R., Zeng W., Kawauchi S., Bender M.A., Santos R., Gregson H.C., Schmiesing J.A., Newkirk D.A., Kong X., Ball A.R., et al. Cohesin mediates chromatin interactions that regulate mammalian β-globin expression. J. Biol. Chem. 2011;286:17870–17878. doi: 10.1074/jbc.M110.207365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuin J., Dixon J.R., van der Reijden M.I.J.A., Ye Z., Kolovos P., Brouwer R.W.W., van de Corput M.P.C., van de Werken H.J.G., Knoch T.A., van IJcken W.F.J., et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. U.S.A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He B., Chen C., Teng L., Tan K. Global view of enhancer-promoter interactome in human cells. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E2191–E2199. doi: 10.1073/pnas.1320308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Majumder P., Gomez J.A., Chadwick B.P., Boss J.M. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J. Exp. Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S., et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seitan V., Faure A., Zhan Y., McCord R., Lajoie B., Ing-Simmons E., Lenhard B., Giorgetti L., Heard E., Fisher A., et al. Cohesin-based chromatin interactions enable regulated gene expression within pre-existing architectural compartments. Genome Res. 2013;23:2066–2077. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim Y.J., Cecchini K.R., Kim T.H. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7391–7396. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips J.E., Corces V.G. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dorsett D. Cohesin: genomic insights into controlling gene transcription and development. Curr. Opin. Genet. Dev. 2011;21:199–206. doi: 10.1016/j.gde.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu Y., Sinha M., Peterson C.L., Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Croft J.A., Bridger J.M., Boyle S., Perry P., Teague P., Bickmore W.A. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sadoni N., Langer S., Fauth C., Bernardi G., Cremer T., Turner B.M., Zink D. Nuclear organization of mammalian genomes. Polar chromosome territories build up functionally distinct higher order compartments. J. Cell Biol. 1999;146:1211–1226. doi: 10.1083/jcb.146.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kosak S.T., Skok J.A., Medina K.L., Riblet R., Le Beau M.M., Fisher A.G., Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 54.Tanabe H., Müller S., Neusser M., von Hase J., Calcagno E., Cremer M., Solovei I., Cremer C., Cremer T. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc. Natl. Acad. Sci. U.S.A. 2002;99:4424–4429. doi: 10.1073/pnas.072618599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wada Y., Ohta Y., Xu M., Tsutsumi S., Minami T., Inoue K., Komura D., Kitakami J., Oshida N., Papantonis A., et al. A wave of nascent transcription on activated human genes. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18357–18361. doi: 10.1073/pnas.0902573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramachandran S., Karp P.H., Jiang P., Ostedgaard L.S., Walz A.E., Fisher J.T., Keshavjee S., Lennox K.A., Jacobi A.M., Rose S.D., et al. A microRNA network regulates expression and biosynthesis of wild-type and DeltaF508 mutant cystic fibrosis transmembrane conductance regulator. Proc. Natl. Acad. Sci. U.S.A. 2012;109:13362–13367. doi: 10.1073/pnas.1210906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


