Abstract
Telomere protection and length regulation are important processes for aging, cancer and several other diseases. At the heart of these processes lies the single-stranded DNA (ssDNA)-binding protein Pot1, a component of the telomere maintenance complex shelterin, which is present in species ranging from fission yeast to humans. Pot1 contains a dual OB-fold DNA-binding domain (DBD) that fully confers its high affinity for telomeric ssDNA. Studies of S. pombe Pot1-DBD and its individual OB-fold domains revealed a complex non-additive behavior of the two OB-folds in the context of the complete Pot1 protein. This behavior includes the use of multiple distinct binding modes and an ability to form higher order complexes. Here we use NMR and biochemical techniques to investigate the structural features of the complete Pot1-DBD. These experiments reveal one binding mode characterized by only subtle alternations to the individual OB-fold subdomain structures, resulting in an inaccessible 3′ end of the ssDNA. The second binding mode, which has equivalent affinity, interacts differently with the 3′ end, rendering it available for interaction with other proteins. These findings suggest a structural switch that contributes to telomere end-protection and length regulation.
INTRODUCTION
Linear eukaryotic chromosomes terminate in telomeres, which themselves end in a 3′ single-stranded DNA (ssDNA) overhang. This overhang must be protected against recognition by DNA damage response machinery to prevent inappropriate resection and chromosomal fusion (1). The telomeric protein complex shelterin contains an ssDNA-binding protein, Pot1, conserved from Schizosaccharomyces pombe to humans, that serves this function (2). Pot1 plays a critical role in telomere end-protection and its deletion results in activation of the ATR DNA damage response pathway and chromosomal fusion (1,2).
In addition to its role in end-protection, Pot1 limits the ability of telomerase to extend the 3′ overhang (3). Deletion of Pot1 or its DNA-binding domain results in dramatic telomere elongation in vivo (4–6). Additionally, human Pot1 (hPOT1) inhibits telomerase activity in vitro, presumably by sequestering the 3′ ssDNA overhang that telomerase requires as a substrate (7,8). Addition of the shelterin component TPP1, which also localizes Pot1 to the telomere in vivo, ameliorates this effect and significantly increases repeat addition processivity of telomerase (9–14). Further experiments revealed a physical association between TPP1 and telomerase important for telomerase recruitment in vivo (15–18). Consistent with these proposed functions of Pot1, mutations to the DNA-binding domain are found in a subset of chronic lymphocytic leukemia and melanoma patients and these mutations appear to disrupt ssDNA binding, deprotect telomeres and increase telomere length (19–21).
Despite this detailed understanding of Pot1 function, it remains unclear how telomerase accesses the 3′ end when Pot1 is bound. Forty percent of 3′ overhangs in telomerase-active human cells terminate in the sequence 5′-GGTTAC-3′ (22). Based on the crystal structure of hPOT1 bound to ssDNA, this end sequence should be bound and sequestered from telomerase (23). Additionally, hPOT1-TPP1 dissociates from ssDNA with a half-life of nearly 30 min, suggesting the need for active regulation of hPOT1 binding (12). Fewer experiments have been performed with S. pombe proteins, but many of the broad features of telomere biology are conserved. For example, S. pombe shelterin plays a role in recruiting telomerase, but does so via the Ccq1 subunit rather than Tpz1, the TPP1 homolog (11,24). Similarly, S. pombe Pot1 is somewhat structurally divergent from hPOT1, but has a similar domain topology and DNA-binding characteristics (25–27). These similarities include an ssDNA-bound half-life of ∼1 h, suggesting a shared requirement for active regulation to allow telomerase access (28).
Pot1 is a modular protein with a dual OB-fold domain (Pot1-DBD) at the N-terminus that fully recapitulates the DNA-binding affinity and specificity of the full-length protein, while the C-terminal half of the protein (also predicted to be an OB-fold) interacts with TPP1/Tpz1 (11–13,28). The structure of the tightly integrated dual OB-fold of human POT1-DBD was solved in complex with a minimal 10-nucleotide telomeric ssDNA sequence, which explained its inhibitory effect on telomerase, but revealed no obvious mechanism for regulation of 3′ access (23). Unlike human POT1-DBD, S. pombe Pot1-DBD has an extended linker between OB-folds, which allows each OB-fold subdomain, Pot1pN and Pot1pC, to be individually expressed and characterized (2,26,29–30). Biochemical studies revealed a division of function between the two domains, with Pot1pN specifically binding telomeric ssDNA while Pot1pC binds with little to no sequence specificity (28). The crystal structures of these individual OB-folds were solved in complex with their minimal 6- and 9-nt ssDNA ligands, respectively, providing a structural basis for the observed sequence specificities (27,31). Pot1pN conveys sequence specificity by binding a core GTT structure via extensive hydrogen-bonding and stacking interactions, while Pot1pC combines several structural features to create a malleable and sequence non-specific ssDNA-binding surface.
Sequence and structural divergence between Pot1pC and the second human OB-fold (hOB2) precludes accurate modeling of the complete S. pombe Pot1-DBD on the hPOT1-DBD structure. Residues at the hOB1-hOB2 interface in hPOT1 have only 18% identity and 31% similarity in S. pombe Pot1, suggesting the lack of a strongly conserved interdomain interface. Thus, it remains unclear how Pot1pN and Pot1pC come together in the context of Pot1-DBD and if these subdomains structurally and biochemically recapitulate Pot1-DBD. Alternatively, Pot1pN and Pot1pC could interact in a fashion that modulates their activities in isolation. Pot1-DBD binds a 15-nt ssDNA ligand—d(GGTTACGGTTACGGT) (15mer)—that is the combination of the minimal sequences bound by its individual subdomains, and it does so with an impressive 2 pM affinity (28,30). However, Pot1-DBD also binds a shorter 12-nt ligand—d(GGTTACGGTTAC) (12mer)—that lacks the three 3′ nucleotides of 15mer, with an affinity equal to that of 15mer (25). Interestingly, these two ligands bind with distinct biochemical activities such that the three additional 3′ nucleotides present in 15mer do not appear to simply protrude out of the binding pocket in a non-productive fashion (28). Further complicating matters, 12mer and 15mer contain multiple Pot1pN-binding sites and Pot1-DBD appears to dimerize on 12mer (32). These data suggest that Pot1-DBD binds oligonucleotides in a complex manner that is not entirely recapitulated by the subdomain activity.
Here we describe our use of solution-based nuclear magnetic resonance (NMR) methods to elucidate what features of the individual DNA-binding subdomains are maintained or altered when tethered together in the biologically relevant Pot1-DBD construct. These studies reveal that the overall structure of the individual subdomains is maintained with the addition of a subtle interface between domains. Additionally, we find that Pot1-DBD is capable of binding DNA in at least two structurally distinct binding modes. One of these modes strictly occludes the 3′ end of the bound ssDNA, while the other does not. These findings suggest a potential mechanism for telomere length regulation by switching between telomerase-extendible and non-extendible Pot1-binding modes.
MATERIALS AND METHODS
Pot1-DBD cloning, expression and purification for binding and size-exclusion chromatography-multi-angle light scattering
The Pot1-DBD and Pot1pC Y224A mutations and the Pot1-DBD linker deletion mutant were created by site-directed mutagenesis according to the QuikChange protocol (Stratagene). The linker deletion mutant lacked amino acids 179–200. All sequences were confirmed by Sanger sequencing (Genewiz Inc.). Both wild-type and mutant Pot1-DBD were expressed and purified as described previously (28). Briefly, intein/chitin-binding domain fusion protein was expressed in Escherichia coli BL21(DE3) cells. Fusion protein was purified using chitin beads (New England BioLabs) and cleaved by static incubation with 100 mM βME for at least 20 h at 4°C. Upon elution, Pot1-DBD was further purified by size-exclusion chromatography (GE Healthcare HiLoad 16/600 Superdex 200) in 20 mM Tris–HCl, pH 8.0, 150 mM NaCl and 3 mM βME. Appropriate fractions were pooled and poured over 5 ml chitin beads to reduce contamination from chitin-binding domain. Protein was concentrated to 145 μM, snap frozen in liquid nitrogen and stored at −70°C.
Expression and purification of 2H/15N /13C labeled Pot1-DBD+15mer
Transformed BL21(DE3) cells were grown overnight on a Luria Broth (LB) agar plate containing ampicillin. A 40-ml LB + ampicillin culture was inoculated with a single colony and grown overnight at 20–25°C. Twenty milliliter of the overnight growth was pelleted by centrifugation and resuspended in 300 ml unlabeled M9/H2O (33). Cells were grown at 37°C to an OD600 of 0.6, pelleted and resuspended in 1.2 l M9/D2O, containing 15N ammonium sulfate and 2H/13C glucose. Cells were evenly divided between six 1-l flasks and grown at 37°C to an OD600 of 0.6. 467 ml 2H/13C/15N M9/D2O was added to each flask to reach a total volume of 4 l and cells were grown at 37°C to an OD600 of 0.6. Cells were placed on ice for 45 min, induced with 0.5 mM isopropyl-β-D-thio-galactopyranoside (IPTG) and incubated 20 h at 20°C with shaking. Protein was purified as described for unlabeled Pot1-DBD, except that 1 M equivalent of 15mer was added following elution from the chitin column prior to gel filtration. The final sample was 700 μM 2H/13C /15N Pot1-DBD with 1.3 M equivalent 15mer.
Expression and purification of unlabeled Pot1pC_Y224A and 15N/13C Pot1pC+9mer
Pot1pC was expressed as a C-terminal intein/chitin-binding domain fusion and unlabeled Pot1pC was purified following the same protocol used for Pot1-DBD, as described previously (27).
15N/13C Pot1pC was grown in modified M9 minimal media containing 15N ammonium sulfate as the sole nitrogen source and 1H/13C glucose as the sole carbon source (34). Purification was performed as described for unlabeled Pot1pC with the addition of 1.2 M equivalents of 9mer following elution from the chitin column.
Expression and purification of 15N/13C Pot1pN+6mer
Pot1pN was expressed from a pET11a vector as an N-terminal 6X-His fusion with a PreScission protease cleavage site, as described previously (35). Briefly, BL21(DE3) cells were grown in modified M9 minimal media (34). Protein was expressed overnight at 25°C. Cells were resuspended in lysis buffer (50 mM Tris–HCl pH 8, 50 mM potassium phosphate pH 8, 150 mM NaCl, 2 mM imidazole, 10% glycerol, 3 mM βME) and lysed by sonication. Following sonication, cells were incubated 30 min at 4°C with lysozyme, DNaseI and 1 mM MgCl2. Insoluble material was removed by centrifugation and lysate was incubated at least 1 h with 5 ml Ni-NTA agarose beads while rocking at 4°C. Beads were then washed with 50 ml buffer P9 (50 mM potassium phosphate pH 8, 50 mM Tris–HCl pH 8, 150 mM NaCl, 3 mM βME) at 1 ml/min and 50 ml buffer P9 + 10 mM imidazole at 1 ml/min. Protein was eluted with 30 ml buffer P9 + 300 mM imidazole. The concentration of the eluent was estimated by A280 using the previously calculated extinction coefficient (ϵ280 = 27,900) and 2 M equivalents of 6mer were added (36). Protein was concentrated and injected onto a G75 size exclusion column pre-equilibrated in SEC buffer (50 mM Tris–HCl pH 8, 150 mM NaCl, 3 mM βME). Appropriate fractions were pooled, concentrated and buffer exchanged into NMR buffer as described for Pot1pC and Pot1-DBD. An additional 0.5 M equivalent 6mer was added to the sample before final concentration. The final sample concentration based on A260/280 values was 1.5 mM Pot1pN with 1.7 M equivalent 6mer. This is a slightly lower protein:DNA ratio than expected, likely due to protein loss during concentration.
Resonance assignment of Pot1pN+6mer, Pot1pC+9mer and Pot1-DBD+15mer spectra
Resonance assignments for 15N/13C Pot1pN+6mer were transferred from those made at slightly different buffer conditions with the aid of HSQC and HNCACB spectra at the new buffer conditions (35,36). HSQC and HNCACB spectra of 15N/13C Pot1pN+6mer were collected at 27°C on a Varian Inova 600 spectrometer. All pulse sequences used were BioPack pulse sequences and data were processed using NMRPipe and analyzed using CCPNMR analysis (37,38).
Transverse relaxation-optimized spectroscopy (TROSY) HSQC, HNCACB and CBCA(CO)NH spectra of 15N/13C Pot1pC+9mer were collected at 27°C on a Varian Inova 600 or a VNMRS 800 MHz spectrometer, each equipped with a z-axis gradient HCN cold probe. Manually selected peaks were submitted to the PINE v2.0 server and these automated assignments were manually confirmed and edited (39).
TROSY HSQC, HNCA, HN(CA)CB, HN(CO)CA and HN(COCA)CB spectra of 2H/15N/13C Pot1-DBD+15mer were collected at 27°C on an Agilent VNMRS 800 or DD2 900 spectrometer equipped with a z-axis gradient HCN cold probe. Peak overlap and poor signal strength precluded automatic assignment using the PINE server, so Pot1-DBD was manually assigned using traditional methods as well as comparison to the Pot1pN+6mer and Pot1pC+9mer spectra to resolve ambiguities. Several sharper peaks that were unique to the Pot1-DBD spectra were assigned and found to belong to a chitin-binding domain contaminant despite no detectable presence by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and Coomassie blue staining.
Composite chemical shift values were calculated using the equation (40):
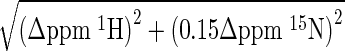 |
Electrophoretic mobility shift assay DNA binding experiments
Oligonucleotides were 5′-end-labeled with [γ-32P] adenosine triphosphate (ATP) using T4 polynucleotide kinase (New England BioLabs) according to manufacturer's specifications. Labeling reactions were quenched at 95°C and snap cooled on ice to prevent secondary structure formation. Unincorporated [γ-32P] ATP was removed using a G25 spin column (GE Healthcare) according to manufacturer's specifications. Labeled oligonucleotides were diluted to 1–2 pM in binding buffer (20 mM Tris–HCl pH 8.4 at 25°C, 50 mM NaCl, 1 mg/ml bovine serum albumin, 3 mM βME, 15% glycerol) and incubated for at least 30 min on ice.
Protein was diluted to ∼40 μM in binding buffer and the concentration was confirmed by absorbance at 280 nm using theoretical extinction coefficients (41). The protein was then serially diluted to concentrations ranging from 1 fM to 2 μM and kept on ice before addition of labeled oligonucleotide. 22.5 μl of each binding reaction was loaded onto a 1.5 mm thick native gel made from 6% acrylamide, 5% glycerol and 1X 89 mM Tris–HCl, 89 mM boric acid, 2 mM ethylenediaminetetraacetic acid disodium salt (TBE). Gels were run in running buffer (1X TBE and 5% glycerol) at 200 V for 20–25 min in a 4°C cold room with ice packs in the gel apparatuses. Gels were dried on Whatman filter paper and exposed to phosphorimaging screens (GE Healthcare) for at least 2 days before imaging on a Typhoon Imager. Data were quantified using ImageQuant version 5.1 (GE Healthcare), corrected for background and fit using Kaleidagraph version 4.1.3 (Synergy Software). Normalized free and bound values were used to calculate fraction bound and fit to a standard two-state binding model to calculate KD (28). Experiments were performed in triplicate using different protein aliquots on different days. Errors in KD were calculated as the standard error of the mean of the three calculated KD values.
Protein specific activity was measured to correct for inactive protein, errors in the extinction coefficient, and/or errors in dilution. Activity assays were performed in the same fashion as binding assays but with 250–500 pM 32P-labeled 15mer and 100 nM unlabeled 15mer to ensure concentrations well above the KD. Activity corrections were performed in parallel with every binding experiment to account for errors in dilution on that day. Active concentrations ranged from 30%-75% of the calculated concentration.
Electrophoretic mobility shift assay protein competition experiments
Oligonucleotides were 5'-end-labeled as described for electrophoretic mobility shift assays. 1–2 pM labeled oligonucleotide was preincubated with 200 pM Pot1-DBD for 30 min. Subsequently, Pot1pN was added at concentrations ranging from 0 to 4 μM and incubated at 4°C ice for 24 h. 22.5 μl of each sample was loaded onto gels as described for binding experiments and these gels were run for 1.5 h at 200 V. Gels were run in a 4°C cold room with ice packs in the gel apparatuses, which were replaced after 45 min. Gels were dried and imaged as described for binding experiments above.
Isothermal titration calorimetry
Isothermal titration calorimetry was performed as described previously with a MicroCal iTC200 (GE Healthcare Life Sciences) instrument (27). Briefly, 40 μM ssDNA was titrated into 5 μM Pot1pC in 20 mM potassium phosphate pH 8.0, 50 mM NaCl and 3 mM βME at 25°C. Experiments were done at least in triplicate and errors were calculated as the standard error of the mean. Heat of dilution experiments produced no discernible signal; thus, no correction factors were added.
Size-exclusion chromatography-multi-angle light scattering
Pot1-DBD and the Y224A mutant were buffer exchanged and concentrated in size-exclusion chromatography-multi-angle light scattering (SEC-MALS) buffer (20 mM Tris–HCl pH 8.0 and 50/400 mM NaCl) using 10 000 MWCO Vivaspin Turbo concentrators pre-rinsed with water (Fisher Scientific). Oligonucleotides were resuspended in SEC-MALS buffer and added in the ratios described. Hundred microliter of each sample was injected onto a Shodex Protein KW-802.5 column pre-equilibrated in SEC-MALS buffer. Refractive index (RI) was measured by an Optilab DSP Interferometric Refractometer (Wyatt Technologies), multi-angle scattering was measured by a Dawn EOS MALS photometer (Wyatt Technologies) and ultraviolet (UV) absorbance at 280 nm was measured by a Spectrasystem UV3000.
Scattered light was measured using 10 detectors ranging from θ = 43° to 132° and a refractive increment value (dn/dc) of 0.185 ml/g was used to calculate the molar mass of particles in solution.
RESULTS
The Pot1-DBD+15mer complex is structurally similar to its constituent subcomplexes: Pot1pN+6mer and Pot1pC+9mer
The crystal structures of the Pot1pN+6mer and Pot1pC+9mer subdomains provide some structural insight into the complete S. pome Pot1-DBD (27,31). However, these structures fail to capture the relative orientation of the two domains, the potential influence the domains may have upon one another and the role of the extended linker between domains. To answer these questions, we created a new Pot1-DBD construct using the optimized C-terminus of Pot1pC (27). This construct, spanning residues 1–339, structurally and biochemically recapitulates the original 1–389 Pot1-DBD (Supplementary Figure S1). Despite the ease of Pot1pC+9mer crystallization, the new Pot1-DBD+15mer complex, while displaying enhanced solubility properties, remained recalcitrant to crystallization. Therefore, in lieu of X-ray crystallography, we assigned and compared 15N-HSQC spectra of Pot1pN+6mer, Pot1pC+9mer and Pot1-DBD+15mer and mapped information on local chemical shift changes onto the available subdomain structures (Figure 1)(42).
Figure 1.
Comparison of subdomain and Pot1-DBD+15mer spectra shows global similarity except at a putative subdomain interface. (A) The 15N -HSQC spectrum of Pot1-DBD (red) bound to 15mer overlays well with the spectra Pot1pN (blue) and Pot1pC (green) bound to 6mer and 9mer, respectively. (B) An enlarged portion of the superpositioned spectra in (A) shows examples of residues that change chemical environment (dark red arrow) and residues that experience multiple distinct chemical environments in Pot1-DBD+15mer, resulting in peak splitting (orange arrows). (C) Altered residues cluster at a putative subdomain interface when mapped onto crystal structures of Pot1pN+6mer (PDB ID: 1QZH) and Pot1pC+9mer (PDB ID: 4HIK). Amino acids with amides shifted >0.05 ppm are shown as dark red spheres and amino acids with split peaks in Pot1-DBD are shown as orange spheres. Unshifted amino acids are colored blue and green for Pot1pN and Pot1pC, respectively. Amino acids that are unassigned in both Pot1-DBD+15mer and subcomplex spectra are colored gray and amino acids unassigned only in Pot1-DBD are colored yellow. DNA is represented by violet sticks. This panel was created using MacPyMOL (42). See supplementary Table S1 for a list of shifted amino acids.
Backbone resonances for Pot1pN+6mer had previously been assigned in different buffer conditions; thus, we collected a minimal set of spectra to transfer these assignments to buffer conditions amenable to the study of Pot1-DBD, ultimately providing assignments for 104 of the 176 observable residues (36). Pot1pC+9mer was assigned using standard three-dimensional spectroscopic techniques, which yielded backbone amide assignments for 107 of the 137 observable residues. Eleven peaks in the 15N-HSQC spectrum remained unassigned due to poor or ambiguous connectivity in the three-dimensional spectra, leaving ∼19 residues with no discernible signal. Many of these unassigned residues map to contiguous sequences in loops of the protein, suggesting their absence is due to exchange broadening. While the presence of dynamics in protein loops is unsurprising, it is interesting to note that L23 remains dynamic even while in contact with ssDNA, perhaps contributing to the plastic ssDNA-binding surface previously described (27).
Deuteration of Pot1-DBD allowed independent assignment of 198 out of 326 non-proline residues in complex with 15mer. Roughly 40 peaks are present but unassigned in the Pot1-DBD+15mer spectrum, leaving 88 amino acids with no detectible signal either due to exchange broadening or signal overlap. Almost all of these unassigned residues are also unassigned in the Pot1pN+6mer and Pot1pC+9mer spectra, but some appear to be specifically absent in the Pot1-DBD+15mer spectrum either due to exchange broadening or simply because they fall below the signal to noise threshold in the larger complex (Figure 1 and Supplementary Table S1). Overall though, the large number shared assignments act as probes that can be compared between complexes to provide structural and dynamic information.
Overall, the 15N-HSQC spectrum of Pot1-DBD+15mer largely recapitulates those of Pot1pN+6mer and Pot1pC+9mer (Figure 1A). The vast majority of assigned residues exhibit composite 15N, 1H chemical shifts of less than 0.05 ppm between the subdomain and Pot1-DBD spectra, suggesting an overall structural similarity between Pot1-DBD and its constituent subdomains (Figure 1B and C and Supplementary Table S1). This structural similarity is particularly notable at the regions of the protein that contact the 5′ and 3′ ends of the 15mer. There are, however, several peaks that clearly shift between spectra, suggesting a change in chemical environment at these sites. Five residues undergo peak splitting in the Pot1-DBD+15mer spectrum and 13 residues shift more than 0.05 ppm in composite chemical shift (Figure 1B and C and Supplementary Table S1). These split and shifted residues map to three regions: the N-terminus, the linker region and a putative interdomain interface (Figure 1C). The shifts at the N-terminus are likely due to the different tagging strategies used for Pot1pN and Pot1-DBD, while the shifts near the linker are explained by its absence in either subdomain construct. Of most significance are the shifts near the putative interdomain interface. Several of these residues contact DNA, suggesting altered nucleotide binding in this area. Indeed, this agrees with biochemical data that suggest Pot1-DBD has decreased nucleotide specificity in this region relative to the specificities observed for Pot1pN+6mer and Pot1pC+9mer (28–30).
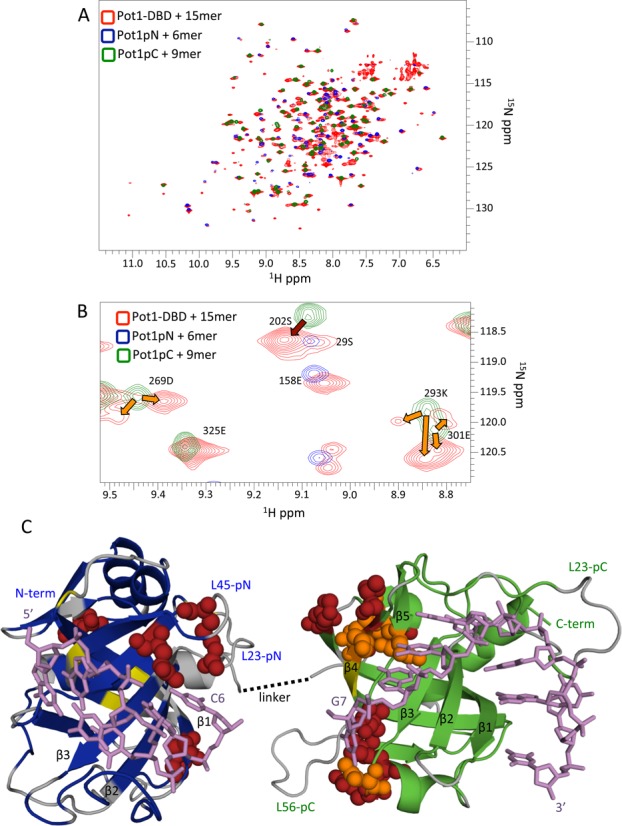
The linker between subdomains is unassigned in the Pot1-DBD+15mer spectrum due to either exchange broadening or overlap. Thus, we investigated its role in nucleic acid binding by creating a linker deletion construct. Deletion of 22 of the 25 amino acids that connect Pot1pN and Pot1pC had no negative impact upon binding affinity (Figure 2 and Supplementary Table S2)(43). In fact, affinity for both 12mer and 15mer increases slightly, likely due to a reduction in the entropic penalty of binding. These data suggest that the structurally uncharacterized linker does not play a role in nucleic acid binding, further confirming the validity of the individual subdomain structures.
Figure 2.
Deletion of 22 amino acids from the interdomain linker does not hinder nucleic acid binding. (A) The 22 amino acids deleted (shaded gray) are structurally undefined in the Pot1pN+6mer and Pot1pC+6mer crystal structures and predicted to be unstructured (43). (B) Representative binding curves for wild-type and linker deletion proteins. Linker deletion has very little effect on Pot1-DBD binding to either 15mer (blue) or 12mer (green) ligands relative to wild-type (red). The slight increase in affinity is likely due to entropic effects.
Pot1-DBD binds 12mer with variable stoichiometry dependent on concentration and buffer conditions
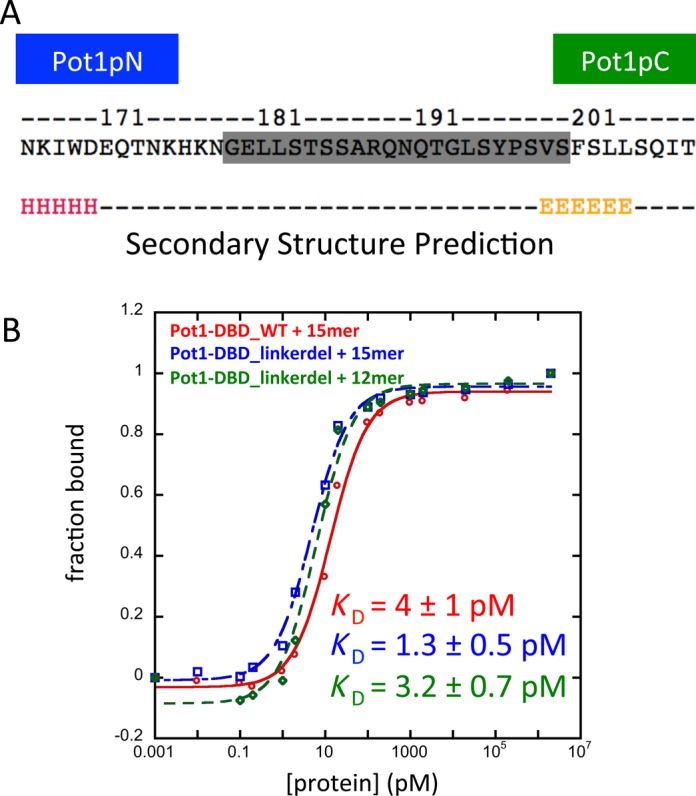
The Pot1-DBD+12mer complex has fewer nucleotides and thus cannot be represented by a simple combination of the two subdomain structures. We attempted to characterize this Pot1-DBD+12mer complex by NMR as well; however, the 15N-HSQC spectrum of an equimolar mixture of Pot1-DBD and 12mer at 900 μM shows poor peak intensity, indicative of a high molecular weight complex (data not shown). This observation is consistent with the previously reported formation of a 2:1 Pot1-DBD:12mer complex (32). To better characterize the composition of this complex, we used size-exclusion chromatography followed by multi-angle light scattering.
At lower concentrations (34 μM), Pot1-DBD+12mer and Pot1-DBD+15mer elute with molar masses of ∼45–50 kDa, similar to the predicted molecular weights of 43.2 and 44.1 kDa for the 1:1 Pot1-DBD+12mer and 1:1 Pot1-DBD+15mer complexes, respectively (Figure 3A). Pot1-DBD+15mer elutes as a uniform peak, while Pot1-DBD+12mer elutes as a broad peak, suggesting the presence of multiple conformations or complexes in the presence of 12mer. Despite this mixture of species, the 1:1 complex appears to be far more prevalent than the 2:1 complex based on the calculated molar mass.
Figure 3.
Pot1-DBD forms primarily a 1:1 complex at low concentrations and can form a larger complex at high concentrations in the presence of 12mer, but not 15mer. Amino acid mutation or base substitution can switch between these two binding modes. (A) RI traces (lines) and molar masses calculated by MALS (markers) of 34 μM Pot1-DBD+15mer and 34 μM Pot1-DBD+12mer complexes in 20 mM Tris–HCl pH 8.0, 50 mM NaCl. (B) Increasing the concentration of NaCl to 400 mM causes Pot1-DBD+12mer to elute earlier than Pot1-DBD+15mer and with a variable but larger average molar mass. Protein and DNA are both at 34 μM. (C) Increasing the concentration of Pot1-DBD to 170 μM and increasing the molar ratio to 2:1, Pot1-DBD:oligonucleotide pushes the 12mer-binding mode toward the 2:1 complex. This 12mer-binding mode can be forced to form a 1:1 complex upon 12mer_T9A substitution. Conversely, Pot1-DBD_Y224A mutation or 15mer_G13C substitution can switch the 15mer-binding mode to the 12mer-binding mode. All complexes were injected at 170 μM protein + 85 μM DNA.
To test whether Pot1-DBD+12mer is in a monomer/dimer equilibrium at these conditions, we attempted to push the equilibrium further toward the putative dimeric state. Previous experiments suggested that high salt concentrations promote dimerization on 12mer; thus, we increased the concentration of NaCl from 50 to 400 mM (25). Indeed, the higher salt concentration causes Pot1-DBD+12mer to elute earlier from the size-exclusion column relative to Pot1-DBD+15mer and with an increased molar mass, while the molar mass of Pot1-DBD+15mer is unaffected by salt concentration (Figure 3B). The molar mass value of Pot1-DBD+12mer is highly variable throughout the peak and the RI and light scattering peaks are offset, indicative of a fast-exchanging mixture of species (Supplementary Figure S2). Additionally, free Pot1-DBD elutes with a molar mass very similar to the predicted 39.5 kDa, suggesting that the higher molecular weight species is mediated by the presence of 12mer.
SEC-MALS does not provide reliable molar mass values for mixtures of species, so we increased the concentration of Pot1-DBD to 170 μM to push the equilibrium completely toward the 2:1 state. Additionally, we added only 0.5 M equivalent oligonucleotide to promote the 2:1 complex through principles of mass balance. These conditions cause Pot1-DBD+12mer to elute even earlier, and with an increased molar mass, but fast exchange between species still prevents accurate calculation of the mass (Figure 3C). In contrast, Pot1-DBD+15mer elutes as a homogenous 1:1 complex even at these conditions.
While SEC-MALS cannot be used to quantitatively measure the concentration of Pot1-DBD at which the 2:1 complex forms, these data are consistent with its formation at low μM concentrations of Pot1-DBD. This value is in agreement with EMSAs, in which a supershift begins to form near 1 μM Pot1-DBD in the presence of 12mer (Figure 4A). This supershift is absent in Pot1-DBD+15mer EMSAs, corroborating the SEC-MALS data (Figure 4B). We note that the approximate concentration at which the 2:1 complex is observed (low μM) is well above the KD (2 pM) for Pot1-DBD+12mer (28). Thus, Pot1-DBD+12mer exists predominantly as a 1:1 complex at the concentrations required for ssDNA binding.
Figure 4.
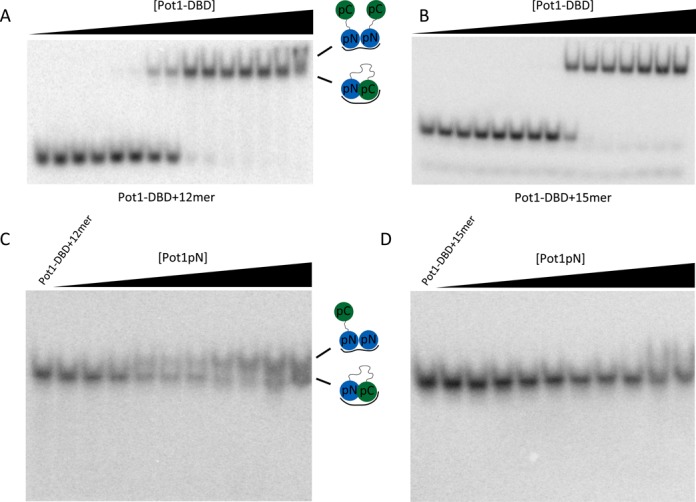
The 3′ end of 12mer, but not 15mer, is available for protein interaction when bound by Pot1-DBD. (A) Pot1-DBD begins to form a larger complex visible by EMSA in the presence of 12mer (presumably the 2:1 complex observed by SEC-MALS in Figure 3). Presumed complexes present are shown in cartoon. (B) In the presence of 15mer, however, there is no evidence of a larger, supershifted species at high concentrations of Pot1-DBD. Concentrations of Pot1-DBD are 0.34 fM, 3.4 fM, 34 fM, 68 fM, 340 fM, 680 fM, 3.4 pM, 6.8 pM, 34 pM, 68 pM, 340 pM, 680 pM, 6.8 nM, 68 nM and 680 nM. (C) Titration of Pot1pN into a solution of Pot1-DBD+12mer also forms a larger Pot1-DBD+Pot1pN+12mer complex. (D) Pot1pN, however, fails to form a larger complex with Pot1-DBD+15mer. Concentrations of Pot1pN are 0 pM, 200 pM, 400 pM, 2 nM, 4 nM, 20 nM, 40 nM, 200 nM, 400 nM, 2 μM and 4 μM.
Pot1-DBD binds 12mer in a fashion that allows proteins to bind the 3′ end of the ssDNA
The ability of DBD to dimerize on 12mer suggests a binding mode distinct from the Pot1-DBD+15mer-binding mode described above. The 2:1 Pot1-DBD+12mer complex is proposed to form via two Pot1pN-subdomain-binding events, suggesting increased access to 3′ end in the 12mer-binding mode (32). To test this model, we used mutagenesis to specifically disrupt only the 3′ Pot1pN-binding site. Complementary base substitution at position three of the six-nucleotide minimal Pot1pN-binding sequence (6mer_T3A) decreases affinity of Pot1pN by over three orders of magnitude (29). Indeed, when this substitution is engineered into the 3’ Pot1pN-binding site of 12mer (12mer_T9A), the 2:1 Pot1-DBD+12mer complex is completely disrupted, instead forming a homogenous 1:1 complex as assessed by SEC-MALS (Figure 3C). Crucially, however, this substitution does not affect the affinity of Pot1-DBD+12mer, further supporting the proposed model in which Pot1-DBD forms a high affinity 1:1 complex with 12mer independently of dimerization (28).
EMSA competition experiments further confirm the model of a second Pot1pN-binding event at high protein concentrations (Figure 4C and D). In these experiments, Pot1-DBD is first bound to 12mer or 15mer and Pot1pN is subsequently added. As expected, Pot1pN is able to bind the 3′ end of 12mer at high concentrations, resulting in a higher molecular weight Pot1-DBD+Pot1pN+12mer complex (Figure 4C). Pot1-DBD+15mer, however, does not form this higher molecular weight species (Figure 4D).
Disruption of the interaction between Pot1-DBD and the 3′ end of 15mer forces reversion to the 12mer-binding mode
The consistent 1:1 stoichiometry of Pot1-DBD+15mer and the structural similarity of the complex to the individual subdomains suggest a binding mode that fully engages the 3′ end. This occlusion of the 3′ Pot1pN-binding site prevents a second Pot1-DBD molecule from binding, unlike the 12mer-binding mode. Indeed, the crystal structure of Pot1pC+9mer reveals an extended aromatic stack involving two amino acid side chains (Trp223 and Tyr224) and two 3′ bases (G7 and T9) (27) (Figure 5A). Furthermore, based on the conserved chemical shifts in the 15N-HSQC spectra, this structural feature is maintained in the Pot1-DBD+15mer complex (Figure 1).
Figure 5.
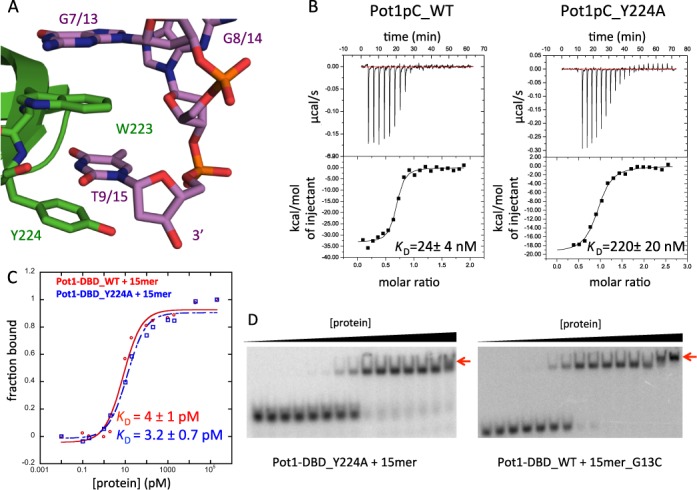
Disruption of the interaction between Pot1pC and the 3′ end of 15mer switches Pot1-DBD to the 12mer-binding mode. (A) In the Pot1pC+9mer complex, nucleotides G7 and T9 of 9mer (corresponding to G13 and T15 in 15mer) form an extended aromatic stack with Trp223 and Tyr224 of Pot1pC/Pot1DBD (PDB ID: 4HIK). Pot1pC is colored green, 9mer is colored violet and atoms are colored by element. (B) Y224A mutation disrupts binding of Pot1pC to 9mer by nearly 10-fold. (C) The same Y224A mutation in Pot1-DBD (blue) has no effect on affinity relative to wild-type Pot1-DBD (red). (D) However, Y224A mutation apparently switches to the 12mer-binding mode based on the supershift (red arrow) at high protein concentration. G13C substitution also creates a supershift indicative of the 12mer-binding mode.
As predicted based on the extensive contact between these residues, disruption of this 3′ stacking interaction, by either base substitution (G7C) or alanine mutation (Y224A), decreases affinity between Pot1pC and 9mer by 8- and 10-fold, respectively (30) (Figure 5B). Surprisingly, the same substitutions and mutations have no impact on the affinity between Pot1-DBD and 15mer (28) (Figure 5C and Supplementary Table S2). However, careful examination of these native gels reveals the formation of a larger species at high concentrations of Pot1-DBD (Figure 5D). This supershifted species is not present in the wild-type Pot1-DBD+15mer experiments, but is a hallmark of the Pot1-DBD+12mer-binding mode due to its ability to form a 2:1 complex (Figure 4A and B). Interestingly, Y224A mutation also has no effect on 12mer affinity or dimer equilibrium, likely due to the shorter length of 12mer and its inability to access this extreme 3′ binding pocket.
SEC-MALS was used to further confirm that G13C substitution and Y224A mutation force the 15mer-binding mode to switch to the dimerization-competent 12mer-binding mode (Figure 3D). Indeed, these mutants form higher molecular weight complexes at 170 μM more akin to Pot1-DBD+12mer than Pot1-DBD+15mer. Together, these data suggest that Pot1-DBD binds DNA in the 12mer-binding mode when the 15mer-binding mode is disrupted.
DISCUSSION
End-protection complexes at the telomere have the intriguing ability to orchestrate several cellular processes including protection from DNA-repair proteins, replication of telomeric DNA, and telomerase recruitment and regulation (44,45). These myriad activities require the multi-protein complex shelterin, which in turn is made up of multi-domain proteins. Here we investigate Pot1, the ssDNA-binding protein in shelterin, from S. pombe, in which individual DNA-binding subdomains have been structurally characterized, but not the complete DNA-binding domain. The NMR spectrum of Pot1-DBD+15mer largely resembles those of the individual Pot1pN+6mer and Pot1pC+9mer subcomplexes, validating the ‘divide and conquer’ approach as a tool for understanding the general structure of Pot1-DBD. Deletion of the structurally uncharacterized linker between domains has little effect on affinity, suggesting that it does not play a direct role in ligand binding and further supporting the validity of the subcomplex crystal structures. The similarity between Pot1-DBD+15mer and its subdomains is particularly notable where Pot1pC contacts the 3′ end of 15mer, implying that the crystal structure of Pot1pC+9mer provides an accurate representation of how Pot1-DBD occludes the 3′ end. hOB2 is structurally divergent from Pot1pC, but interestingly, both species have a tyrosine residue that caps the 3′ end of the terminal nucleotide (23,27). We propose that this structural feature prevents fraying that would allow inappropriate access to the 3′ end.
There are some informative perturbations to the subcomplexes in the context of the complete Pot1-DBD. NMR analysis of Pot1-DBD reveals several residues that differ in chemical environment from the subcomplexes, highlighting regions that likely undergo some type of structural perturbation relative to their isolated domain structures. Shifted residues map to complementary surfaces on Pot1pN and Pot1pC, suggesting an interdomain interface distinct from that present in human POT1, as might be expected from the structural differences between Pot1pC and hOB2 (27). As these chemical shift changes can arise from many factors, such as differences in DNA-binding, the presence of the linker, or differential tagging strategies, a precise interface cannot be mapped by NMR alone. In fact, the interface between Pot1pN and Pot1pC may simply be a transient or non-specific interaction created solely due to the proximity of DNA-binding sites. Consistent with this interpretation, docking algorithms are unable to find a high-quality interdomain interface and mutation of several interface residues had no impact on affinity for DNA (data not shown) (46–48). These data suggest a non-specific or transient interaction between domains.
The Pot1-DBD+12mer complex is more challenging to structurally characterize due to its propensity to form a higher molecular weight complex at concentrations necessary for NMR. We have found, however, that Pot1-DBD forms a high-affinity 1:1 complex with 12mer and only forms the 2:1 complex when the concentration of Pot1-DBD is sufficient to compete with the intramolecular interaction between the monomeric Pot1-DBD and the 3′ end of 12mer. This conclusion is supported by the ability of Pot1-DBD to bind 12mer-T9A with low picomolar affinity despite the inability of this oligonucleotide to promote the formation of the 2:1 complex (28). Despite difficulties in characterizing the 1:1 Pot1-DBD+12mer complex, the ability of Pot1-DBD to dimerize on 12mer suggests that the interaction with the 3′ end of 12mer differs significantly from that of 15mer. In contrast, the 5′ end of 12mer and 15mer are identical in sequence and it is likely that their interactions with Pot1pN are more similar. The salt dependence of dimerization suggests that the interaction between Pot1pC and the 3′ end of 12mer has a large electrostatic component, which is consistent with the large number of positively charged resides near the interdomain interface that may participate in binding 12mer (Figure 6A)(49–51). Together, these observations support a model in which the 3′ end of 12mer is tucked into a positively charged pocket of Pot1pC, providing the favorable energetics required for low pM binding. This interaction, however, likely allows fraying of the 3′ end or partial dissociation of Pot1pC, allowing access to the 3′ end.
Figure 6.
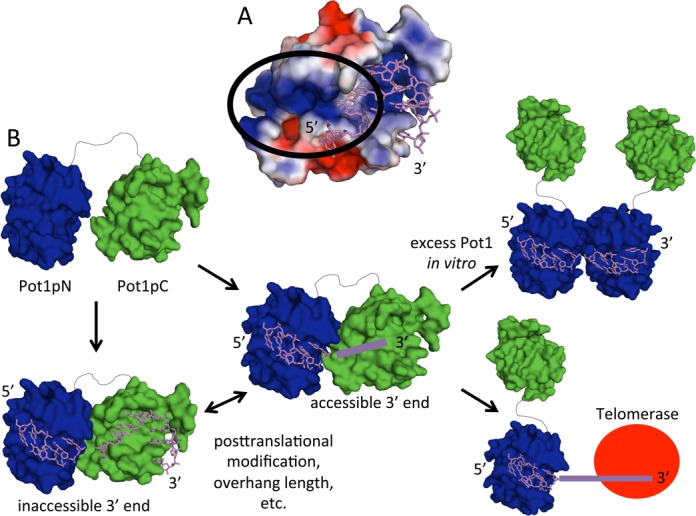
Model of how Pot1 binding modulates 3′ end accessibility. (A) The electrostatic surface representation of Pot1pC shows an electropositive patch (within the black circle) that may be involved in binding the 3′ end of 12mer (PDB ID: 4HIK). Electrostatic potentials were created using the APBS plug-in in PyMOL with electrostatic potential bounds of 2 ±kT/e (49–51). (B) Pot1-DBD can adopt multiple binding modes that may be important for function. Pot1-DBD binds 15mer as a homogeneous 1:1 complex with an occluded 3′ end. Upon disruption of this binding mode, the complex switches to the 12mer-binding mode, which allows the formation of higher order complexes in vitro and may allow telomerase access in vivo.
Despite their unique biochemical and structural features, the 12mer- and 15mer-binding modes have surprisingly similar affinities at physiological salt concentrations, suggesting that both 1:1 binding modes have to be considered when evaluating biological function. Access to the 3′ overhang at the telomere is an essential step in regulating telomerase activity. Minimally, telomerase extension is inhibited by the presence of Pot1 in vitro, presumably by physically preventing access to the 3′ overhang substrate (7). Telomerase activity is restored when the Pot1-binding site is moved away from the 3′ end, leaving eight nucleotides accessible (12). High-affinity binding of Pot1 to the 3′ overhang is crucial for the protection of telomeres from DNA damage machinery; thus, binding is likely regulated to allow telomerase access (31). The ability of a second Pot1-DBD molecule or another Pot1pN molecule to bind the 3′ end of 12mer suggests that this binding mode does not completely sequester the 3′ end and may represent an extendible telomeric state (Figure 6B). Sufficient local concentration of telomerase could be achieved via localization of telomerase via TPP1 in humans or Ccq1 in S. pombe. The dimeric Pot1-DBD complex is less likely to be biologically relevant, but the presence of multiple shelterin complexes or TPP1 dimerization may increase the local concentration of Pot1 as well (52). In opposition to the 12mer-binding mode, the tightly associated 15mer-binding mode may represent the sheltered, non-extendible telomeric state. As either single amino acid mutation (Y224A) or ligand length can trigger a switch between these two binding modes, in vivo factors, such as post-translational modification or overhang length, may trigger a similar switch that regulates telomerase access.
It is intriguing to consider whether this mechanism of regulation is conserved in human POT1. While Pot1pC and hOB2 bind ssDNA in different fashions, both subdomains exhibit reduced sequence specificity relative to Pot1pN/hOB1 and both utilize a 3′ tyrosine cap described above. Telomerase, TPP1, or other cellular factors may destabilize the interaction between hOB2 and the 3′ end of ssDNA, allowing partial dissociation of hPOT1 and access by telomerase. Indeed, the maintenance of the contact between hOB1 and ssDNA would serve to keep telomerase localized to the 3′ end via its interaction with TPP1 and would provide a model for the observed increase in repeat addition processivity in the presence of POT1/TPP1 (12). We have shown it is possible to selectively disrupt the 15mer-binding mode, suggesting potential targets for post-translational modification or allosteric structural changes that could trigger a switch between extendable and non-extendable states.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Dr Geoff Armstrong and the Rocky Mountain Regional 900-MHz NMR Facility of the University of Colorado at Denver Health Sciences Center for assistance with NMR. We thank Dr Annette Erbse for assistance with SEC-MALS and Kenneth Lyon for assistance with EMSAs. We thank all members of the Wuttke lab for review of the manuscript.
FUNDING
National Institutes of Health [GM059414 to D.S.W., T32 GM65013 to T.H.D.]; National Science Foundation [MCB1121842 to D.S.W.]. Funding for open access charge: National Science Foundation [MCB1121842 to D.S.W.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Denchi E.L., de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 2.Baumann P., Cech T.R. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 3.Baumann P., Price C. Pot1 and telomere maintenance. FEBS Lett. 2010;584:3779–3784. doi: 10.1016/j.febslet.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Churikov D., Wei C., Price C. Vertebrate POT1 restricts G-overhang length and prevents activation of a telomeric DNA damage checkpoint but is dispensable for overhang protection. Mol. Cell Biol. 2006;26:6971–6982. doi: 10.1128/MCB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loayza D., de Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 6.Bunch J.T., Bae N.S., Leonardi J., Baumann P. Distinct requirements for Pot1 in limiting telomere length and maintaining chromosome stability. Mol. Cell Biol. 2005;25:5567–5578. doi: 10.1128/MCB.25.13.5567-5578.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelleher C., Kurth I., Lingner J. Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol. Cell Biol. 2005;25:808–818. doi: 10.1128/MCB.25.2.808-818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei M., Zaug A.J., Podell E.R., Cech T.R. Switching human telomerase on and off with hPOT1 protein in vitro. J. Biol. Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- 9.Ye J.Z.-S., Hockemeyer D., Krutchinsky A.N., Loayza D., Hooper S.M., Chait B.T., de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu D., Safari A., O'Connor M.S., Chan D.W., Laegeler A., Qin J., Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 11.Miyoshi T., Kanoh J., Saito M., Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- 12.Wang F., Podell E.R., Zaug A.J., Yang Y., Baciu P., Cech T.R., Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 13.Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., O'Connor M.S., Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 14.Latrick C.M., Cech T.R. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 2010;29:924–933. doi: 10.1038/emboj.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nandakumar J., Bell C.F., Weidenfeld I., Zaug A.J., Leinwand L.A., Cech T.R. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature. 2012;492:285–289. doi: 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong F.L., Batista L.F.Z., Freund A., Pech M.F., Venteicher A.S., Artandi S.E. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell. 2012;150:481–494. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sexton A.N., Youmans D.T., Collins K. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J. Biol. Chem. 2012;287:34455–34464. doi: 10.1074/jbc.M112.394767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Chen L.-Y., Han X., Xie W., Kim H., Yang D., Liu D., Songyang Z. Phosphorylation of TPP1 regulates cell cycle-dependent telomerase recruitment. Proc. Natl Acad. Sci. U.S.A. 2013;110:5457–5462. doi: 10.1073/pnas.1217733110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramsay A.J., Quesada V., Foronda M., Conde L., Martínez-Trillos A., Villamor N., Rodríguez D., Kwarciak A., Garabaya C., Gallardo M., et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat. Genet. 2013;45:526–530. doi: 10.1038/ng.2584. [DOI] [PubMed] [Google Scholar]
- 20.Shi J., Yang X.R., Ballew B., Rotunno M., Calista D., Fargnoli M.C., Ghiorzo P., Bressac-de Paillerets B., Nagore E., Avril M.F. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat. Genet. 2014;46:482–486. doi: 10.1038/ng.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robles-Espinoza C.D., Harland M., Ramsay A.J., Aoude L.G., Quesada V., Ding Z., Pooley K.A., Pritchard A.L., Tiffen J.C., Petljak M., et al. POT1 loss-of-function variants predispose to familial melanoma. Nat. Genet. 2014;46:478–481. doi: 10.1038/ng.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sfeir A.J., Chai W., Shay J.W., Wright W.E. Telomere-end processing the terminal nucleotides of human chromosomes. Mol. Cell. 2005;18:131–138. doi: 10.1016/j.molcel.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 23.Lei M., Podell E.R., Cech T.R. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 24.Tomita K., Cooper J.P. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev. 2008;22:3461–3474. doi: 10.1101/gad.498608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croy J.E., Podell E.R., Wuttke D.S. A new model for Schizosaccharomyces pombe telomere recognition: the telomeric single-stranded DNA-binding activity of Pot11-389. J. Mol. Biol. 2006;361:80–93. doi: 10.1016/j.jmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Theobald D.L., Wuttke D.S. Prediction of multiple tandem OB-fold domains in telomere end-binding proteins Pot1 and Cdc13. Structure. 2004;12:1877–1879. doi: 10.1016/j.str.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 27.Dickey T.H., McKercher M.A., Wuttke D.S. Nonspecific recognition Is achieved in Pot1pC through the use of multiple binding modes. Structure. 2013;21:121–132. doi: 10.1016/j.str.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschuler S.E., Dickey T.H., Wuttke D.S. Schizosaccharomyces pombe protection of telomeres 1 utilizes alternate binding modes to accommodate different telomeric sequences. Biochemistry. 2011;50:7503–7513. doi: 10.1021/bi200826a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei M., Baumann P., Cech T.R. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry. 2002;41:14560–14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- 30.Croy J.E., Altschuler S.E., Grimm N.E., Wuttke D.S. Nonadditivity in the recognition of single-stranded DNA by the Schizosaccharomyces pombe protection of telomeres 1 DNA-binding domain, Pot1-DBD. Biochemistry. 2009;48:6864–6875. doi: 10.1021/bi900307x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lei M., Podell E.R., Baumann P., Cech T.R. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 32.Nandakumar J., Cech T.R. DNA-induced dimerization of the single-stranded DNA binding telomeric protein Pot1 from Schizosaccharomyces pombe. Nucleic Acids Res. 2012;40:235–244. doi: 10.1093/nar/gkr721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardner K.H., Kay L.E. The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu. Rev. Biophys. Biomol. Struct. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]
- 34.Anderson E.M., Halsey W.A., Wuttke D.S. Delineation of the high-affinity single-stranded telomeric DNA-binding domain of Saccharomyces cerevisiae Cdc13. Nucleic Acids Res. 2002;30:4305–4313. doi: 10.1093/nar/gkf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Croy J.E., Wuttke D.S. Insights into the dynamics of specific telomeric single-stranded DNA recognition by Pot1pN. J. Mol. Biol. 2009;387:935–948. doi: 10.1016/j.jmb.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croy J.E., Fast J.L., Grimm N.E., Wuttke D.S. Deciphering the mechanism of thermodynamic accommodation of telomeric oligonucleotide sequences by the Schizosaccharomyces pombe protection of telomeres 1 (Pot1pN) protein. Biochemistry. 2008;47:4345–4358. doi: 10.1021/bi701778x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 38.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 39.Bahrami A., Assadi A.H., Markley J.L., Eghbalnia H.R. Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput. Biol. 2009;5:e1000307. doi: 10.1371/journal.pcbi.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mulder F.A., Schipper D., Bott R., Boelens R. Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol. 1999;292:111–123. doi: 10.1006/jmbi.1999.3034. [DOI] [PubMed] [Google Scholar]
- 41.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schrodinger, L.L.C. The PyMOL Molecular Graphics System. 2010. Version 1.5.0.4. [Google Scholar]
- 43.Cole C., Barber J.D., Barton G.J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis K.A., Wuttke D.S. Telomerase and telomere-associated proteins: structural insights into mechanism and evolution. Structure. 2012;20:28–39. doi: 10.1016/j.str.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palm W., de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 46.Lyskov S., Gray J.J. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008;36:W233–W238. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macindoe G., Mavridis L., Venkatraman V., Devignes M.-D., Ritchie D.W. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 2010;38:W445–W449. doi: 10.1093/nar/gkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Comeau S.R., Gatchell D.W., Vajda S., Camacho C.J. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004;32:W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. U.S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dolinsky T.J., Czodrowski P., Li H., Nielsen J.E., Jensen J.H., Klebe G., Baker N.A. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dolinsky T.J., Nielsen J.E., McCammon J.A., Baker N.A. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han X., Liu D., Zhang Y., Li Y., Lu W., Chen J., Songyang Z. Akt regulates TPP1 homodimerization and telomere protection. Aging Cell. 2013;12:1091–1099. doi: 10.1111/acel.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


