Abstract
Replication Protein A (RPA) is a eukaryotic single stranded (ss) DNA binding protein that plays critical roles in most aspects of genome maintenance, including replication, recombination and repair. RPA binds ssDNA with high affinity, destabilizes DNA secondary structure and facilitates binding of other proteins to ssDNA. However, RPA must be removed from or redistributed along ssDNA during these processes. To probe the dynamics of RPA-DNA interactions, we combined ensemble and single molecule fluorescence approaches to examine human RPA diffusion along ssDNA and find that an hRPA hetero-trimer can diffuse rapidly along ssDNA. Diffusion of hRPA is functional in that it provides the mechanism by which hRPA can transiently disrupt DNA hairpins by diffusing in from ssDNA regions adjacent to the DNA hairpin. hRPA diffusion was also monitored by the fluctuations in fluorescence intensity of a Cy3 fluorophore attached to the end of ssDNA. Using a novel method to calibrate the Cy3 fluorescence intensity as a function of hRPA position on the ssDNA, we estimate a one-dimensional diffusion coefficient of hRPA on ssDNA of D1 ~5000 nucleotide2s−1 at 37°C. Diffusion of hRPA while bound to ssDNA enables it to be readily repositioned to allow other proteins access to ssDNA.
Keywords: RPA, single molecule fluorescence, FRET, dynamics, DNA hairpin melting, diffusion coefficient
Introduction
Replication protein A (RPA) is the major eukaryotic single stranded (ss) DNA binding (SSB) protein that plays a central role in genome maintenance by binding tightly to ssDNA[1–4]. Similarly to bacterial SSB proteins[5], RPA binds with high affinity to ssDNA that is formed transiently during DNA replication, recombination and repair, protecting it from nucleases, and destabilizing unwanted secondary structures (e.g., hairpins[6] and G-quadruplexes[7]). Additionally, through direct protein-protein interactions, RPA coordinates the targeting of other DNA processing proteins to their sites of action on DNA[1–3]. RPA also serves as an important intermediate in DNA damage checkpoint signaling[8]. In all eukaryotes, RPA is a hetero-trimer consisting of Rpa1 (~70 kDa), Rpa2 (~32 kDa) and Rpa3 (~14 kDa). All three subunits contain oligonucleotide/oligosaccharide binding (OB) folds[9], with Rpa1 containing 4 OB-folds (F, A, B and C), while one additional OB-fold is contained in each of Rpa2 (D) and Rpa1 (E) (see Figure 1A). RPA2 also contains a winged helix domain that is primarily involved in protein interactions[1–4]. OB-folds A, B, C and D function in ssDNA binding[3, 10] as shown in a crystal structure of Ustilago maydis RPA bound to ssDNA (dT32) (Figure 1B) in which 25 of the DNA nucleotides are observable[11]. OB-fold F is primarily involved in protein interactions[12] but it also seems to bind weakly to ssDNA[13] and affects interactions with partial duplex DNA[14]. OB-fold E shows weak affinity for telomeric DNA[15].
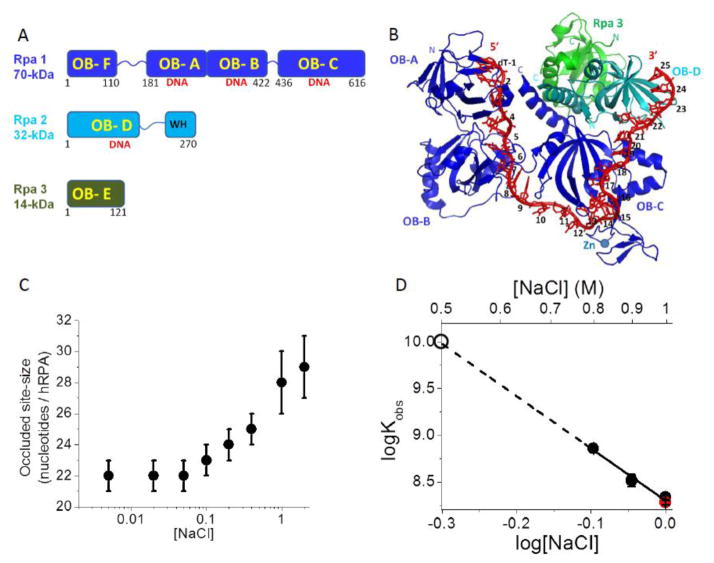
Figure 1. Binding of hRPA to ssDNA.
(A) Subunit composition of the hetero-trimeric hRPA showing the six OB-folds. (B) Structure of Ustilago maydis RPA bound to (dT)32 showing OB-folds A, B, and C of Rpa1 and OB-fold D of Rpa2 interacting with 25 nucleotides (PDB ID 4GOP)[11]. (C) The occluded site-size of hRPA bound to poly(dT) is dependent on [NaCl] (Buffer T, 25.0°C). (D) Dependence on [NaCl] of the equilibrium association constant (Kobs) for hRPA binding to (dT)30 (log-log plot) (Buffer T, 25.0°C). The dashed line shows a linear extrapolation to obtain an estimate of Kobs = 1x1010 M−1 at 0.50 M NaCl (open circle). A measurement of Kobs for hRPA binding to (dT)29-Cy3-T-3′ (red circle) is also shown at 1.0 M NaCl indicating that the presence of Cy3 on the DNA has little effect on Kobs.
Whereas RPA must bind with high affinity to ssDNA to carry out its functions, it must also be displaced from ssDNA or be redistributed along ssDNA to make room for other DNA processing proteins to carry out their functions. Yet, there is little known about the dynamics of RPA while bound to ssDNA. Using a combination of ensemble and single molecule fluorescence approaches, we show in this report that human RPA (hRPA) protein diffuses rapidly along ssDNA, while remaining bound to ssDNA. We further show that hRPA diffusion along ssDNA is functional in that it enables RPA to transiently invade and destabilize (melt) a DNA hairpin structure, which is an essential property of hRPA. We propose that this ability to diffuse on ssDNA provides RPA with a simple mechanism by which it can coordinate assembly and disassembly of other proteins during its multiple functions in genome maintenance.
Results
Equilibrium binding affinity and kinetics of hRPA binding to ssDNA
Human RPA is a hetero-trimer composed of a 70 kDa subunit, Rpa1, a 32 kDa subunit, Rpa2, and a 14 kDa subunit, Rpa3 (Figure 1A). A recent structure of a truncated version of the Ustilago maydis RPA bound to (dT)32 (Figure 1B) shows three OB-folds within Rpa1 (A, B and C) interacting with ~20 nts, while OB-folds A, B, C and D from Rpa2 interact with 25 nts[11]. Our previous studies showed that S. cerevisiae RPA (scRPA) undergoes a [NaCl]-dependent transition between two ssDNA (poly(dT)) binding modes[16]. Figure 1C indicates that hRPA displays the same behavior. At [NaCl] < 50 mM, hRPA binds poly(dT) with an occluded site size of 22±1 nucleotides (nts), whereas at [NaCl] > 1 M, hRPA binds poly(dT) with an occluded site size of 28–30 nts[16]. This suggests that below 50mM NaCl, RPA interacts with ssDNA using primarily the three OB-folds (A, B, and C) within Rpa1, whereas at higher [NaCl] RPA binds with a larger site size that involves additional interactions with OB-fold (D) within Rpa2[10, 16].
The majority of the experiments reported here were performed in Buffer T plus 500 mM NaCl at 25°C so that the DNA hairpins that we investigate have high stability in the absence of hRPA. hRPA binds to ssDNA with high affinity even at such high [NaCl] concentrations[16]. To determine the equilibrium constant, Kobs, at 500 mM NaCl for hRPA binding to (dT)30, a ssDNA long enough to maintain all contacts, we performed equilibrium titrations monitoring the quenching of hRPA tryptophan fluorescence at 1.0, 0.9 and 0.8 M NaCl. The values of Kobs, obtained from fitting the hRPA-(dT)30 binding isotherms to a 1:1 binding model are plotted as logKobs vs. log[NaCl] in Figure 1D. Linear extrapolation of these data yields Kobs ~1010 M−1 for hRPA binding to (dT)30 at 500 mM NaCl. Figure 1D also shows that Kobs at 1M NaCl for hRPA binding to 3′-Cy3-(dT)29 and (dT)30 are the same thus indicating little effect of the Cy3 fluorophore label on Kobs. We also examined the association and dissociation kinetics of hRPA binding to 3′-Cy3-(dT)30, monitoring the change in Cy3 fluorescence that accompanies binding/dissociation (data not shown). In Buffer T, 500 mM NaCl at 25.0°C, stopped-flow kinetic measurements (data not shown) indicate a bimolecular association rate constant, ka = 1.8×108 M−1 s−1, and a dissociation rate constant, kd=0.018 s−1, consistent with our estimate of Kobs. Hence, under these conditions, hRPA has an average lifetime of ~55 sec on (dT)30.
Diffusion of hRPA along ssDNA probed by single molecule fluorescence
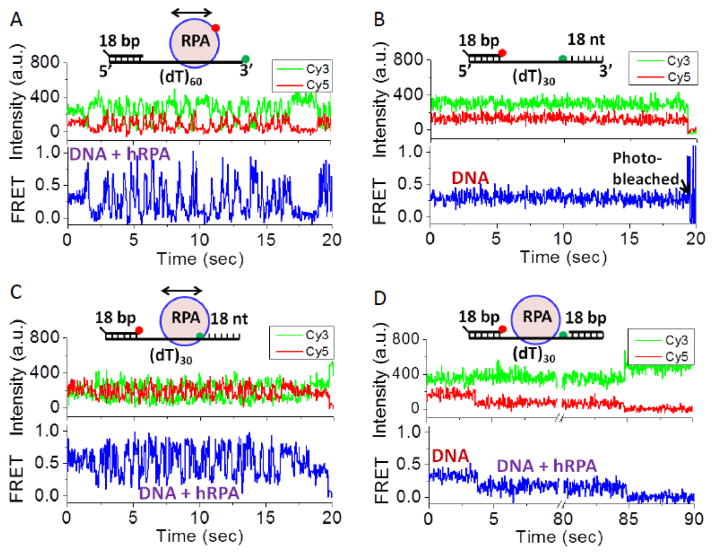
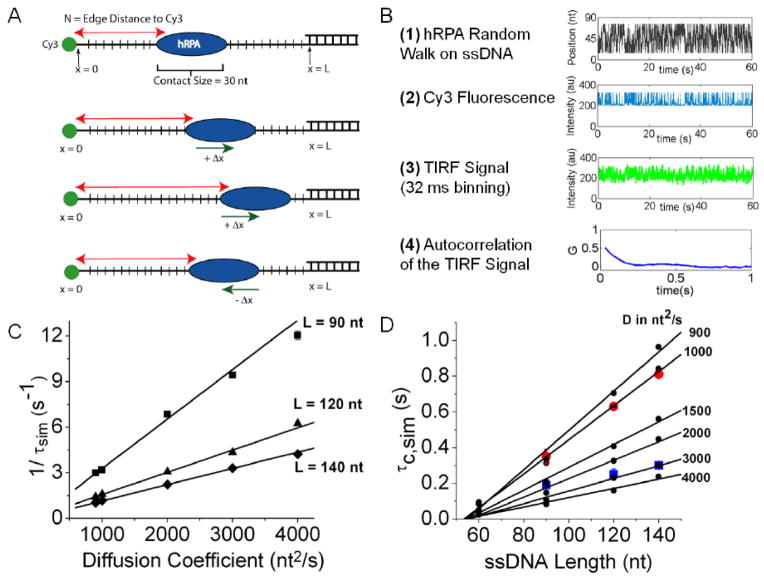
Single molecule total internal reflectance fluorescence (smTIRF) studies were carried out as described[17] and shown schematically in Figure S1. DNA with a biotinylated 18 bp duplex “handle” and a ssDNA flanking region (sequences of all oligodeoxynucleotides are given in Table S1) was immobilized on a glass slide using neutravidin-biotin chemistry as described in Methods. As an initial qualitative test of whether hRPA can diffuse along ssDNA, we performed a smTIRF experiment by binding hRPA labeled with an average of one Cy5 fluorophore (see Methods) to a surface bound 3′-Cy3-(dT)60 and monitored the FRET (Förster Resonance Energy Transfer) efficiency between the Cy3 (donor) and Cy5 (acceptor) (Figure 2A). The Cy3 and Cy5 fluorescence signals show anti-correlated fluctuations, consistent with movement of Cy5-hRPA relative to the Cy3 at the 3′ end of the ssDNA. These FRET fluctuations are not due to multiple hRPA binding and dissociation events since the average lifetime of hRPA on (dT)60 under these identical conditions is at least 55 sec (see above). An alternative is that hRPA is stably bound to a position on the DNA and that the FRET fluctuations arise due to the dynamics of the flexible Cy3-labeled ssDNA enabling contact with the acceptor labeled protein. If this were the case similar FRET fluctuations should be observed when the Cy5 acceptor is placed directly on the DNA substrate. A control experiment (Figure S2) with a DNA containing Cy3 and Cy5 separated by (dT)60 shows a stable FRET signal near 0.15, indicating that the large fluctuations observed in Figure 2A are not due solely to the conformational dynamics of the ssDNA, but require the presence of bound hRPA. These and previous experiments show that the conformational dynamics of the ssDNA are very fast and averaged out within the 32 ms time resolution of these experiments[18]. We suggest that the FRET fluctuations reflect diffusion of hRPA toward and away from the Cy3 at the 3′ end of the DNA. A second test of hRPA diffusion was performed using a DNA in which Cy5 and Cy3 are both on the DNA and separated by (dT)30, followed by an additional 18 nt of mixed sequence ssDNA containing all four bases (Figure 2B). The DNA alone shows a stable FRET signal with a value near 0.30 (Figure 2B). Upon hRPA binding, large anti-correlated Cy3 and Cy5 FRET fluctuations are observed (Figure 2C). These are expected if hRPA moves along the DNA due in part to the bending of the DNA as seen in the crystal structure (Figure 1B). However, when a complementary ssDNA is annealed to the 18 nt sequence to form a duplex, a stable FRET signal at 0.20 is observed due to confinement of hRPA to the (dT)30 ssDNA (Figure 2D). The experiments in Figure 2 provide qualitative evidence that hRPA can diffuse along ssDNA. We do not know the precise position of the Cy5 label on the hRPA used in the experiments in Figure 2A since labeling could occur at the N-termini of any of the three hRPA subunits (see Methods), hence we are unable to analyze the fluctuations observed in Figure 2A more quantitatively. An alternative method to obtain quantitative information about hRPA diffusion is discussed below.
Figure 2. hRPA can diffuse along single stranded DNA.
(A) Representative smTIRF time trace of a surface bound 3′-Cy3-(dT)60 bound with a single Cy5-labeled hRPA (250 nM, unbound RPA removed by buffer flow; n=37 complexes analyzed containing Cy3-labeled DNA and one Cy5-labeled hRPA, N=2204 DNA molecules in field). (B) smTIRF time trace of a DNA containing a 49 nt ssDNA region in which Cy5 and Cy3 are separated by (dT)30, followed by an additional 18 nt of mixed sequence ssDNA (n=70, N=251). (C) smTIRF time trace of the DNA in panel (B) bound with unlabeled hRPA showing large FRET fluctuations due to hRPA diffusion (100 nM hRPA, unbound RPA removed by buffer flow; n=63, N=637). (D) smTIRF time trace of the DNA molecule in panels (B) and (C) to which a complementary 18 nt ssDNA was annealed to the 18 nt mixed sequence region at the 3′ end ofthe DNA, thus constraining hRPA to remain bound to the (dT)30 region, thus reducing the FRET fluctuations (500 nM hRPA, unbound RPA removed by buffer flow; n=47, N=624).
hRPA can transiently melt DNA hairpins using its ability to diffuse on ssDNA
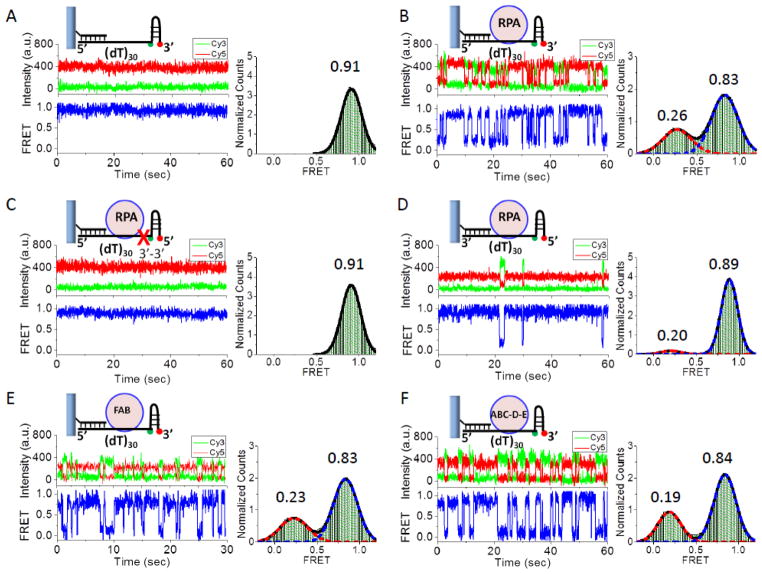
One important function of hRPA is to destabilize secondary structures, such as hairpins and G-quadruplexes, which can inhibit the functions of other proteins that act on ssDNA[1, 5, 19]. To determine if hRPA diffusion plays a role in destabilizing DNA secondary structures, we examined the ability of hRPA to destabilize a seven bp DNA hairpin possessing a stretch of (dT)30 on the 5′ side of the hairpin. Cy3 and Cy5 labels were incorporated at the base of the hairpin so that a decrease in FRET would accompany hairpin melting. The (dT)30 stretch can accommodate one hRPA. In the absence of hRPA, a stable high FRET signal (0.91) is observed, consistent with a stable, closed hairpin (Figure 3A). Upon addition of hRPA large anti-correlated Cy3/Cy5 fluctuations occur between a high FRET state (0.83) and a low FRET state (~0.26) (Figure 3B). To assess the extent to which the DNA hairpin is melted, we examined a DNA in which the hairpin was replaced by (dT)20 so that the Cy3 and Cy5 were separated by the same DNA contour length. In the absence of hRPA a stable FRET efficiency (0.66) was observed (Figure S3A), but when hRPA was added, FRET fluctuations between 0.21 and 0.66 were observed (Figure S3B). The 0.21 FRET efficiency is the same as the low FRET efficiency observed in Figure 3B and likely occurs when the hRPA moves fully toward the 3′ end. These results suggest that hRPA is able to fully melt the seven bp hairpin, albeit transiently. We also examined whether hRPA can invade a longer 18 bp hairpin on the 5′ side of a (dT)30 as monitored by Cy3/Cy5 probes placed internally in the hairpin, 9 bp from the base (Figure S3C). However, we observed no effect of hRPA on the FRET signal from these probes indicating that a single hRPA is not able to move this far into the hairpin.
Figure 3. Transient melting of a DNA hairpin by a diffusing hRPA.
(A) A 7 bp DNA hairpin with Cy3 and Cy5 at the base of the hairpin and a (dT)30 ssDNA on the 5′ side of the hairpin is stable in the absence of hRPA as indicated by the stable high FRET state (0.91) and the single Gaussian FRET probability distribution (n=64, N=795). (B) In the presence of hRPA, anti-correlated Cy3/Cy5 fluctuations are observed between high FRET (0.83 - closed hairpin) and low FRET state (0.26-melted hairpin). The FRET distribution is well described by a two Gaussian fit (n=100, N=2417). (C) hRPA melting of the DNA hairpin is blocked for a DNA in which the phosphodiester backbone polarity linking (dT)30 and the hairpin is reversed via a 3′-3′ linkage (red X) as indicated by the absence of FRET fluctuations and the stable FRET signal at 0.91 (n=41, N=424). (D) The frequency of hRPA-induced hairpin opening decreases when the (dT)30 is placed on the 3′ of the DNA hairpin, indicating a preference for hRPA melting of the hairpin when it invades from the 5′ side. The low FRET state (0.20-melted hairpin) is much less populated than the high FRET state (0.89-closed hairpin) (n=29, N=341). (E) An hRPA truncation possessing only the FAB OB-folds of the Rpa1 subunit can also melt the DNA hairpin. The FRET distribution is two state (n=143, N=3435). (F) An hRPA in which OB-fold F has been deleted can melt the DNA hairpin. The FRET distribution is two state (n=130, N=4480). The concentration of hRPA or its variants was 500 nM, and the unbound proteins were removed before data collection.
As a further test of whether hRPA melts the hairpin by diffusing in from the (dT)30 binding site, we examined a DNA containing a (dT)30 stretch and the same 7 bp DNA hairpin, but these two regions are connected by a 3′-3′ phosphodiester linkage (marked by a red X in the DNA in Figure 3C). Since the ssDNA binding site of RPA is polar[11, 20] (Figure 1B), if RPA melts the hairpin by a diffusional mechanism then both DNA segments ((dT)30 and the hairpin) must have the same continuous backbone polarity. In fact, a stable FRET signal (0.91) is observed for this 3′-3′-linked DNA in the presence of hRPA indicating that reversal of the phosphodiester backbone blocks RPA diffusion and its access to the DNA hairpin. Ensemble fluorescence experiments monitoring hRPA Trp fluorescence quenching demonstrates that hRPA can bind to this DNA. Therefore, on the normal hairpin DNA, hRPA binds to the (dT)30 region and transiently melts the hairpin DNA by diffusing in from the (dT)30 region. A control DNA containing a hairpin, but no (dT)30 loading site shows no FRET changes even in the presence of 300 nM hRPA (Figure S3D) indicating that the (dT)30 binding site is required for hRPA to melt the hairpin. The rates of hRPA-induced opening (kop) and closing (kcl) of the DNA hairpin were estimated by fitting the dwell times of the closed and open states, as identified by a Hidden Markov analysis[21], to a single exponential decay (details in Methods and Figures S4B and S5). From these we estimate kop = 3.6±0.2 s−1 and kcl = 6.1±0.5 s−1 at 500 mM NaCl (Table 1). We note that hRPA is also able to transiently melt the hairpin in the presence of Mg2+ (Buffer T containing 100 mM NaCl, 5 mM MgCl2) (Figure S4A), conditions often used in enzymatic studies of replication, recombination and repair, with similar rates (Table 1), as well as at lower [NaCl] (20 mM (Figure S4B, Table 1). hRPA is also able to melt out the hairpin with similar rates regardless of whether the Cy3 is located on the thymidine base or as part of the DNA backbone (Table 1).
Table 1.
Rates of DNA hairpin opening and closing induced by hRPA

|
|
|||||
|---|---|---|---|---|---|---|
|
| ||||||
| kopen (s−1) | kclose (s−1) | kclose/kopen | kopen (s−1) | kclose (s−1) | kclose/kopen | |
| hRPA(a) | 3.6 ± 0.2 | 6.1 ± 0.5 | 1.7 | 0.54 ± 0.08 | 6.3 ± 0.8 | 11 |
| hRPA(b) | 1.2 ± 0.4 | 3.3 ± 0.6 | 2.8 | |||
| hRPA(c) | 2.7 ± 0.1 | 1.8 ± 0.1 | 0.6 | |||
| ABC-D-E(a) | 2.0 ± 0.1 | 6.6 ± 0.3 | 3.3 | |||
| FAB(a) | 4.8 ± 0.1 | 8.2 ± 0.3 | 1.7 | |||
Unless indicated, experiments were conducted in Buffer I (20 mM Tris-HCl pH 8.1, 0.1 mM Na2EDTA, 1 mM DTT, 0.8% (w/v) dextrose, 2.5 mM Trolox, 20 units/ml glucose oxidase, 20 units/ml catalase) plus 0.5 M NaCl, 25 °C. The flow channel was washed with 200 μl of the imaging buffer to remove free protein before data collection.
Buffer I plus 0.10 M NaCl, 5 mM MgCl2, no EDTA, 25 °C.
Buffer I plus 0.020 M NaCl, 25 °C.
We next examined the ability of hRPA to melt out the same seven bp DNA hairpin, but with a (dT)30 on the 3′ side of the hairpin (Figure 3D). Although hRPA is able to melt the hairpin with this opposite orientation, it does so much less efficiently. The population of melted hairpins is very low with rates, kop = 0.54±0.8 s−1 and kcl = 6.3±0.8 s−1 at 500 mM NaCl. Interestingly, it is the rate of hairpin opening that is affected by having the (dT)30 on the 3′ side of the DNA hairpin. This suggests the interesting possibility that the hRPA does not invade the hairpin by simply capturing the ssDNA that forms transiently due to fluctuations in the hairpin, but may facilitate the melting process. Otherwise it should not matter whether the hairpin is on the 3′ or 5′ side of the (dT)30. This is a distinct possibility since the region of hRPA that faces the hairpin differs when it is bound to these two DNA molecules (see cartoon in Table 1).
We also examined two variants of hRPA in which one or more of the OB-folds were deleted. We used two truncations of the Rpa1 subunit, FAB, in which OB-fold C, Rpa2 and Rpa3 were removed, and ABC-D-E in which OB-fold F was removed. Both variants retain the ability to transiently melt the DNA hairpin possessing a (dT)30 on the 5′ side of the hairpin (Figure 3E and F). Hence, both variants must also be able to diffuse along ssDNA. The rates for hRPA-induced hairpin opening and closing (Table 1) differ somewhat from the values for the full length hRPA, again suggesting that hRPA may play some role in facilitating hairpin DNA melting. Since OB-folds A and B are sufficient for high affinity binding of RPA[22, 23], these data suggest that OB-folds A and B are sufficient to support diffusion.
Use of Cy3 fluorescence on ssDNA to monitor diffusion of unlabeled hRPA
Cy3 fluorescence is enhanced when in the vicinity of a protein as first noted in studies of UvrD[24] and this has been used to study the binding of many proteins to nucleic acids[25–36]. This is also the case for hRPA (Figure 4). Equilibrium titrations of hRPA binding to a series of DNA molecules differing in ssDNA length, L (3′-Cy3-(dT)L), were analyzed using the Binding Density Function Method[37] to determine the average Cy3 fluorescence enhancement when one hRPA is bound (Figure S6). The Cy3 fluorescence enhancement per one hRPA bound decreases with increasing ssDNA length (Figure 4E). This indicates that the average Cy3 fluorescence intensity is dependent on its distance from hRPA along the ssDNA contour length (Figure 4E). As we show quantitatively below, Cy3 fluorescence intensity is sensitive to the ssDNA contour length between the Cy3 and a single hRPA bound to the DNA. Thus, the Cy3 fluorescence can be used to monitor diffusion of hRPA on ssDNA due to the change in the DNA contour length that separates hRPA from the Cy3 end of the DNA.
Figure 4. hRPA binding to 3′-Cy3-(dT)L increases the average Cy3 fluorescence intensity and the Cy3 fluorescence fluctuations.
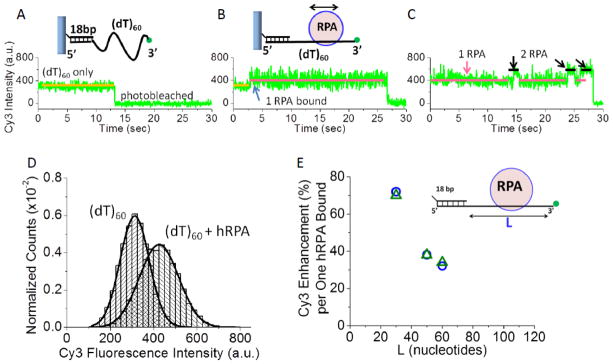
(A) Representative single molecule TIRF time trace of the Cy3 fluorescence from a single DNA molecule (3′-Cy3-(dT)60) attached to the surface via a biotin-neutravidin-biotin-18 bp handle. (B) Upon addition of hRPA (37 pM), an increase in average Cy3 fluorescence and fluorescence fluctuations occurs due to binding of a single hRPA. (C) At much higher hRPA concentrations (370 nM), transient binding of a second hRPA molecule can be observed. (D) The Cy3 fluorescence intensity distributions for (3′-Cy3-(dT)60) (n=29) and upon one hRPA bound to the same 3′-Cy3-(dT)60 molecules shows the increase in both Cy3 fluorescence and Cy3 fluorescence fluctuations. The 34% increase in Cy3 intensity indicates a single hRPA bound as shown by ensemble studies (Figure S6). (E) The average Cy3 fluorescence enhancement per one hRPA bound decreases with increasing ssDNA length, L. This was observed in both ensemble (blue circles) and smTIRF studies (green triangles).
Figure 4A shows a Cy3 smTIRF time trace for a single DNA molecule with a ssDNA region (3′-Cy3-(dT)60). Upon addition of 37 pM hRPA, the average Cy3 fluorescence intensity increases (Figure 4B). Histograms of hRPA binding events (Figure 4D) indicate that the average Cy3 fluorescence enhancement (34%) observed in the smTIRF experiment is the same as for a single hRPA binding to the same DNA in ensemble studies (Figure 4E). Notably, in addition to the increase in the average Cy3 fluorescence intensity, we also observe an increase in the fluctuations in the Cy3 fluorescence intensity (Figure 4B). At much higher hRPA concentration (370 nM), transient binding of a second hRPA to a single (dT)60 can be observed as an additional Cy3 fluorescence enhancement (60%) due to the fact that one of the hRPA molecules is forced to bind closer to the Cy3 end (Figure 4C), consistent with ensemble titrations (Figure S6). However, this second hRPA dissociates quickly, consistent with RPA lacking cooperative binding[16, 38, 39]. The binding of a second hRPA is also marked by smaller Cy3 fluorescence fluctuations (compared to 1 hRPA bound). As we show below, the increased Cy3 fluorescence fluctuations shown in Figure 4B are due to diffusion of a single hRPA hetero-trimer along the ss DNA.
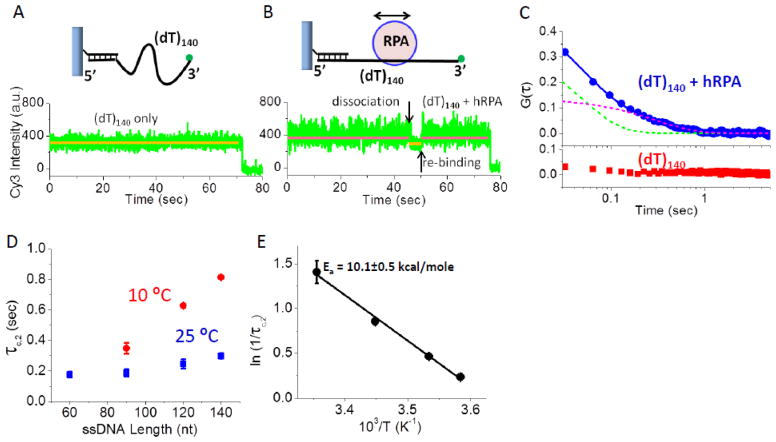
The same experiment performed for a DNA with a longer ssDNA region (3′-Cy3-(dT)140) showed a similar result (Figure 5A and B). Binding of hRPA (10 pM) results in both an increase in Cy3 fluorescence intensity and fluctuations. The time trace in Figure 5B also shows an example of hRPA dissociation and rebinding events that clearly differ from the Cy3 fluctuations observed when hRPA remains bound to the DNA. To gain insight into the increased Cy3 fluorescence intensity fluctuations due to binding of a single hRPA, we performed autocorrelation analyses on the Cy3 fluctuations associated with the DNA alone and the DNA in the presence of hRPA (10 pM). Low hRPA concentrations were used to minimize the probability that more than one hRPA is bound per DNA. For 3′-Cy3-(dT)140 alone, the average autocorrelation function, G(τ), is flat (red symbols in Figure 5C) indicating that those fluctuations are random. However, a measureable averaged auto-correlation function was obtained upon analysis of (3′-Cy3-(dT)140) bound with a single hRPA (blue symbols in Figure 5C). The auto-correlation function was best described by a two exponential decay, with τc,1 = 46±4 msec and τc,2 = 298±18 msec. We examined and performed auto-correlation analyses on data for DNA molecules with ssDNA lengths of L= 60, 90, 120 and 140 nucleotides at low [hRPA] (1–50 pM) at two temperatures (10°C and 25°C). For each DNA length, the resulting G(τ) was fit to a two exponential decay (Figure S7). The values of τc,1 ranged from 20 to 50 msec, are near the time resolution of the instrument (32 msec) and are due to photophysical or detection noise as noted previously[40] and thus were not considered further. For ssDNA molecules with a single hRPA bound, τc,2 increases with increasing ssDNA length, with a steeper increase observed for the data at 10°C (Figure 5D). As shown below, Monte Carlo simulations of a protein undergoing a one-dimensional random walk along DNA indicate that τc should increase nearly linearly with increasing DNA length and that τc varies inversely with the one-dimensional diffusion coefficient of the protein on the DNA. Figure 5E shows an Arrhenius plot of ln(1/τc,2) vs. inverse temperature for hRPA bound to 3′-Cy3-(dT)120 from which we calculate an activation energy for hRPA diffusion (Ea = 10.1±0.5 kcal/mole). The dependence of τc,2 on ssDNA length and temperature supports our conclusion that the hRPA-induced Cy3 fluorescence intensity fluctuations reflect hRPA diffusion along the ssDNA.
Figure 5. hRPA diffusion monitored by fluorescence fluctuations of Cy3-labeled ssDNA.
(A) smTIR Cy3 fluorescence intensity time trace of a single DNA 3′-Cy3-(dT)140 without hRPA. (B) An increase in Cy3 fluorescence intensity and fluctuations upon hRPA (10 pM) addition to 3′-Cy3-(dT)140 ; a dissociation/rebinding event is also shown. (C) Auto-correlation functions of the Cy3 fluorescence traces from DNA (3′-Cy3-(dT)140) alone (red squares-average of 105 molecules) and DNA with one hRPA bound (3′-Cy3-(dT)140 with one RPA bound (blue circles-average of 98 molecules). In the absence of hRPA, the Cy3 fluctuations are random whereas upon binding one hRPA a finite G(τ) is observed that can be fit by a two exponential function with τC,1 = 46±4 msec (green dashed curve) and τC,2 = 298±18 msec (red dashed curve). (D) τC,2 increases with increasing ssDNA length at 10°C and 25°C (see Figure S7). (E) Arrhenius plot of 1/τC,2 for one hRPA bound to 3′-Cy3-(dT)120 yields Ea = 10.1±0.5 kcal/mole.
Calibration of the ssDNA contour length dependence of the hRPA-induced Cy3 fluorescence enhancement
Since the values of τC in Figure 5D appear to be sensitive to hRPA diffusion along ssDNA, we sought to use these to obtain a quantitative estimate of the one-dimensional diffusion coefficient, D1, for hRPA on ssDNA. Our approach was to perform Monte Carlo simulations of a random walk of hRPA on ssDNA for an input value of D1 to simulate the time-dependent fluctuations of the position of hRPA on the DNA. These could then be analyzed in the same way that we analyzed the Cy3 fluorescence fluctuations to obtain simulated values of τC (τsim) to compare with the experimental values. However, in order to use this approach we need to know how the Cy3 fluorescence changes as a function of the DNA contour length between the hRPA and the Cy3 labeled 3′-end of a DNA (dT)L. For this we synthesized a series of chimeric DNA molecules containing two distinct single stranded segments with widely different affinities for hRPA (Figure 6A). These contain an 18 bp duplex DNA “handle” with a stretch of (dT)30 followed by an additional “N” dT nucleotides in which the phosphodiester bonds between each successive nucleotide alternate between 3′-3′ phosphodiester linkages and 5′-5′phosphodiester linkages (referred to as (dTalt)N, and represented by the red saw tooth in Figure 6A). Such a DNA molecule has a high affinity site for hRPA binding ((dT)30) that is short enough to limit hRPA diffusion, followed by a stretch of (dTalt)N to which hRPA has much lower affinity (~450-fold) as shown by the equilibrium isotherms for hRPA binding to (dT)30 vs. (dTalt)30 (Figure 6C). This affinity difference ensures that in a 1:1 hRPA complex with a chimeric (dT)30-(dTalt)30 more than 99% of the hRPA will be bound to the (dT)30 stretch. Importantly, the alternating ssDNA (dTalt)N has the same flexibility as normal (dT)N since the same average FRET efficiency value is observed for (Cy5-(dT)60-Cy3) and (Cy5-(dT)30-(dTalt)30-Cy3) (Figure S2).
Figure 6. Calibration of the effect of ssDNA contour length on the hRPA-induced Cy3 fluorescence enhancement.
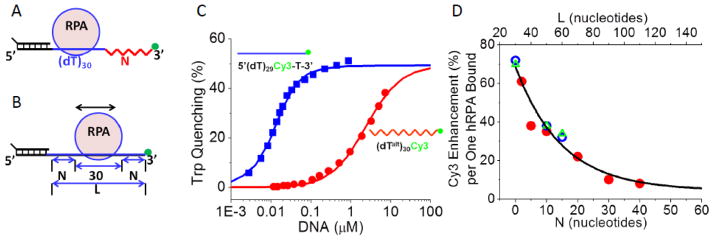
(A) Schematic of the chimeric DNA molecules containing a normal polarity (dT)30 linked to an alternating polarity (dTalt)N with a Cy3 at the 3′ end. (B) Schematic of a normal polarity DNA with ssDNA of length, L ((dT)L). If hRPA binds, on average to the middle of the ssDNA, then the relationship between L and N is given by L=30 + 2N. (C) hRPA binds with 450-fold lower affinity to (dTalt)30 in which the phosphodiester backbone polarity is reversed after each nucleotide (Kobs=(4.2±0.2)x105 M−1) than to normal polarity (dT)30 (Kobs=(1.9±0.1)x108 M−1) in (Buffer T, 1.0 M NaCl, 25°C). (D) Dependence on N of the Cy3 fluorescence enhancement induced by binding a single hRPA to a series of DNA chimeras (with varying N) obtained from analysis of ensemble equilibrium titrations of DNA with hRPA (red circles). Cy3 fluorescence enhancement for a single hRPA bound to normal polarity (dT)L from ensemble titrations (open blue circles) and from smTIRF studies (open green triangles). The data were fit to a single exponential function, (E = E0 + Emexp(−N/NC), with E0 = 4±7, Em = 65±7 and NC = 15±4) or equivalently (E = E0 + Emexp(−(L−30)/LC), with Lc =30±7).
Ensemble titrations at two different DNA concentrations were performed for the series of DNA chimeras ((dT)30-(dTalt)N–Cy3-3′), with N= 2, 5, 10, 20, 30 and 40, monitoring the enhancement of Cy3 fluorescence upon binding of hRPA. These were analyzed using the Binding Density Function Analysis (Figure S8)[37] to obtain the Cy3 fluorescence enhancement for binding one hRPA to the high affinity (dT)30 site (Figure 6D). The Cy3 fluorescence enhancement displays an exponential decrease with increasing N. For comparison, the binding of one hRPA to the (dT)30 site of (dT)30-(dTalt)30-Cy3-3′ induces only a ~10% increase in Cy3 fluorescence compared to a ~32% increase upon binding to a normal DNA of the same length ((dT)60). The lower enhancement results from the fact that in the chimeric DNA containing the (dTalt)30, hRPA binds only to the normal polarity (dT)30 site and thus is constrained to be further away from the Cy3 (in nucleotides along the contour length). If we assume the absence of end effects, then when one hRPA is bound to a (dT)L with normal polarity, its average position will be in the middle of the ssDNA region. Then the Cy3 enhancement for one hRPA bound to a chimeric DNA with N alternating polarity nucleotides ((dT)30-(dTalt)N-Cy3-3′) should be the same as the Cy3 enhancement for a normal polarity DNA ((dT)L-Cy3-3′) with L= (30+2N) (see Figure 6B). Using this relationship, Figure 6D shows that the Cy3 fluorescence enhancement from ensemble studies with (dT)30-(dTalt)N-Cy3-3′, and (dT)L-Cy3-3′, as well as single molecule experiments with normal (dT)L-Cy3-3′ (L = 30, 50 and 60) all fall on the same exponential curve given by E = E0 + Emexp(−N/NC) (where E0 = 4±7, Em = 65±7 and NC = 15±4) or equivalently (E = E0 + Emexp(−(L−30)/LC), where Lc =30±7). This indicates that hRPA binding to the normal polarity ssDNA does not show significant end effects and that the hRPA enhancement of Cy3 fluorescence upon binding to these ssDNA molecules decays exponentially with the ssDNA contour length that separates them. These results also provide further support to the conclusion that (dTalt)N has the same flexibility as normal (dT)N. Studies performed at 10°C indicate that the same dependence of Cy3 fluorescence enhancement on N applies at both 10°C and 25°C (data not shown). These results enable us to use the Cy3 fluorescence enhancement upon hRPA binding to calculate the average number of nucleotides that separate the hRPA from the Cy3-3′end for each (dT)L.
We also performed smTIRF experiments for hRPA binding to the chimeric ((dT)30-(dTalt)N-Cy3-3′) molecules to examine the Cy3 fluctuations and the auto-correlation times obtained when hRPA cannot diffuse along the ssDNA. In this case, the only mechanism to bring Cy3 close to the hRPA would be due to the flexibility of the (dTalt)N. When hRPA binds to the high affinity (dT)30 site a Cy3 fluorescence enhancement is observed, however, the Cy3 fluctuations are much smaller and an autocorrelation function with a much smaller amplitude is obtained (Figure S9). This suggests that although the increase in average Cy3 fluorescence intensity upon hRPA binding to the normal polarity Cy3-(dT)L is due to Cy3 interactions with hRPA due to DNA flexibility, the increase in non-random Cy3 fluorescence fluctuations is due to hRPA diffusion along the ssDNA.
One-dimensional diffusion coefficients of hRPA on ssDNA
We performed Monte Carlo simulations in order to estimate one-dimensional diffusion coefficients (D1) of hRPA on ssDNA based on the experimental auto-correlation times. We simulated one-dimensional random walks for a protein with a contact size on the DNA of 30 nucleotides, for different input values of D1 and ssDNA length, L, as outlined in Figure 7A and B (see Methods). We used the empirically determined relationship between Cy3 fluorescence enhancement and N (number of nucleotides along the ssDNA contour length between the Cy3 and the edge of hRPA (Figure 7C) to convert RPA position into Cy3 fluorescence intensity. Use of this calibration assumes that the conformational dynamics of the ssDNA are much more rapid than the movement of hRPA along the DNA so that the average Cy3 fluorescence equilibrates as hRPA moves from one position to the next along the ssDNA, thus providing a measure of the number of nucleotides between Cy3 and the protein edge. This assumption is consistent with smTIRF studies[18] that show no time-dependent FRET changes for the same types of ssDNA molecules used here when labeled with Cy3 and Cy5 that are separated by (dT)L (see Figure S2). We then averaged the Cy3 signal in 32 ms bins to match the time resolution of the experiment. Auto-correlation functions were then performed on 100 sets of simulated Cy3 time traces for each input value of L and D1 and averaged in the same manner as for the experimental data. These averaged auto-correlation functions were then fit to a single exponential decay function to obtain τsim. We note that it is important to know the quantitative relationship between Cy3 fluorescence intensity and hRPA position, since this will affect the quantitative results of the simulations, emphasizing the need to calibrate this effect for the particular protein and DNA under study.
Figure 7. Estimation of one-dimensional diffusion coefficient of hRPA on ssDNA.
(A) Cartoon describing the model used to simulate a one-dimensional random walk along ssDNA of hRPA with a contact size of 30 nucleotides and a 1 nt step size. (B) A simulated auto-correlation function for L= 90 nucleotides and D1 = 3000 nt2s−1, was obtained in four steps. (1)-The position (in nucleotides) of the 3′-Cy3 on the DNA relative to the edge of a diffusing hRPA was generated; (2)-The Cy3 fluorescence calibration curve was used to obtain the hRPA position dependence into a Cy3 fluorescence trace; (3)-The Cy3 signal was averaged in 32 msec bins to reflect the time resolution of the CCD camera, and (4)- Auto-correlation analyses were performed on 100 simulations and averaged to obtain G(τ). (C) 1/τsim values from single exponential fits to the simulated G(τ) obtained for hRPA diffusion along ssDNA of three lengths (L= 90 (■), 120 (▲), and 140 (◆) nucleotides) as a function of diffusion coefficient. Black lines represent linear fits to the data. (D) τsim values (●) plotted as a function of ssDNA length, L, fordifferent values of D1. Error bars are smaller than the data symbols and ranged from 1 to 4 ms. Black lines represent linear fits excluding the points for L=60 nt. Experimental values of τC for hRPA on ssDNA at 10 C (red circles) and 25 C (blue squares) are overlaid on the plot.
Figures 7C and D show the dependence of the simulated auto-correlation times, τsim on D1 for the four ssDNA lengths used in our experiments (L=60, 90, 120 and 140 nucleotides). The values of τsim are inversely proportional to D1 for each length (Figure 7C) and increase with increasing ssDNA length (Figure 7D) for a given value of D1. This is the same behavior observed for the experimental τC (Figure 5D). The simulated data in Figure 7C indicate that τsim is more sensitive to D1 for the longer lengths of ssDNA, with L=60 nts showing the least sensitivity. This suggests that the data obtained for the longer ssDNA lengths should provide a more accurate estimate of D1.
Figure 7D shows τsim as a function of ssDNA length for values of D1 from 900 to 4000 nt2s−1. The τsim values (small black circles) increase linearly with L, for L= 90 to 140 nt, but deviate from linearity for L≤ 60 nt. Both the individual τsimvalues and the slopes of the τsimvs. L plots decrease with increasing D1. The experimental τC values determined at 10°C and 25°C are also shown in Figure 7D. The τC values determined at 10°C show the predicted linear dependence on L, and agree well with the τsim for D1= 1000 nt2/s. The experimental values of τC determined at 25°C increase with L for L= 90, 120 and 140 nt, but the value for L= 60 nt is indistinguishable from the value for L=90 nt. This suggests that at 25°C, the hRPA diffusion is too fast to be accurately assessed for the shorter DNA lengths. Comparison of the experimental τC and τsim values for L=120 and 140 nt indicates a diffusion coefficient of 2800±200 nt2/s at 25°C and 1050±10 nt2/s at 10°C. Based on these estimates and the activation energy of 10.1±0.5 kcal/mol determined from τC for experiments with L= 120 nt (Figure 5E), we extrapolate to a value of D1 ~5100±400 nt2/s for hRPA at 37°C.
Discussion
Single molecule fluorescence has proved invaluable for studies of the dynamics of proteins on single stranded DNA[41]. We present evidence from single molecule TIRF experiments that a single hRPA hetero-trimer can diffuse along ssDNA, while bound with high affinity. Furthermore, this ability to diffuse along ssDNA is important functionally since it provides the mechanism by which hRPA is able to transiently melt DNA hairpins. We also designed a novel set of chimeric DNA molecules to calibrate the dependence of the Cy3 fluorescence enhancement on the distance (along the contour length) between the bound hRPA and the Cy3 fluorophore on 3′-Cy3-(dT)L. The origin of the protein-induced Cy3 fluorescence enhancement has been proposed to be due to an effect on the photo-induced cis-trans isomerization within the Cy3 fluorophore[34, 36, 42]. The ssDNA molecules used in our study are very flexible, hence transient loop formation can bring the Cy3 in direct contact with the hRPA. Our calibration studies show that the magnitude of the Cy3 fluorescence enhancement decreases exponentially with the number of nucleotides along the contour length between the edge of the hRPA and the Cy3 end of the DNA. This is consistent with the expectation for the end-to-end distance of a semi-flexible or worm-like chain[18, 43] whose flexibility can be described by a single parameter, its persistence length. Previous studies of duplex DNA binding proteins concluded that the protein-induced Cy3 enhancement effect decreases linearly with protein distance from Cy3[34]; however, those data are also well described as an exponential decrease. More studies on other systems will be needed to determine whether the particular protein under study affects the magnitude and/or distance dependence of the Cy3 fluorescence enhancement.
Although ssDNA flexibility can explain the distance dependence of the average Cy3 fluorescence enhancement, the observation that the amplitude of the Cy3 fluorescence fluctuations increases upon hRPA binding reflects movement of hRPA along the ssDNA. Our smTIRF studies with the ssDNA chimeras that restrict hRPA diffusion show that ssDNA flexibility is not responsible for the increase in Cy3 fluctuations upon hRPA binding and the finite values of τC obtained from auto-correlation analysis of those fluctuations. The conformational fluctuations of the ssDNA are fast and equilibrate within the 32 msec resolution of our CCD camera (see Figure S2 and ref[18]). Hence, the average Cy3 fluorescence intensity enhancement likely equilibrates for each position of hRPA as it diffuses along the ssDNA. Using the Cy3 fluorescence calibration data and Monte Carlo simulations, we show that the ssDNA length dependence of the experimental τC values is consistent with τC being sensitive to one-dimensional diffusion of hRPA on ssDNA. Based on these simulations, we estimate D1 for hRPA on poly(dT) at several temperatures. The agreement between simulations and experiment is excellent at 10°C, yielding D1 = 1050±10 nt2s−1. At 25°C, we estimate D1 = 2800±200 nt2s−1, although we observe deviations of the experimental values of τC from the τsim for the shorter ssDNA lengths (60 and 90 nts). Those deviations may be due to the ability of RPA to partially melt the 18 bp duplex handle, which would yield a higher than expected value for the experimental τC. Such deviations are expected to be more significant at 25°C than at 10°C and contribute more for shorter DNA lengths, as we observe. The advantage of the approach presented here to quantitatively examine diffusion of proteins bound to ssDNA is that unlabeled protein can be used and longer DNA lengths can be studied as compared to FRET-based approaches[40, 44, 45]. The extension of this method to other proteins should be straightforward, requiring only calibration of the distance dependence of the protein-induced Cy3 fluorescence enhancement.
We also demonstrate that hRPA diffusion provides the mechanism for hRPA to transiently disrupt DNA secondary structure. As shown for E. coli SSB[40], this ability to diffuse also allows hRPA to be pushed along the ssDNA, either by a directionally polymerizing protein, such as a Rad51 filament, or by a DNA polymerase. Our observation that transient melting of a DNA hairpin is more efficient when hRPA invades the hairpin from ssDNA on the 5′ side of the hairpin is intriguing and suggests a role for hRPA in facilitating the DNA hairpin disruption. Interestingly, scRPA has been shown to selectively promote the nuclease activity of Dna2 on a 5′ flap DNA substrate, but inhibit it on a 3′ flap DNA substrate[46]. It is possible that the preference that we observe here for hairpin melting may play a role in that selectivity.
Our demonstration that hRPA can diffuse on ssDNA and use this to disrupt DNA secondary structures adds these features to the growing list of similarities with the generally homo-tetrameric bacterial SSB proteins, such as E. coli SSB. E. coli SSB is a tetramer with 4 OB-folds that interact with ssDNA in its (SSB)65 mode[47, 48] and hRPA has four OB-folds that primarily interact with ssDNA[10, 49, 50]. Both display salt-dependent transitions between ssDNA binding modes that involve switches in the numbers of OB-folds used to interact with ssDNA[16, 47, 48]. Although RPA and E. coli SSB bind with high affinity with very long lifetimes, they both can undergo a reasonably rapid protein concentration dependent exchange reaction with free protein[51, 52]. RPA has yet to be shown to undergo a direct or intersegment transfer reaction between ssDNA sites as has been shown for E. coli SSB[28, 53].
Although both hRPA and E. coli SSB can diffuse along ssDNA, the diffusion coefficient on poly(dT) for an hRPA hetero-trimer at 37°C (~5,000 nt2s−1) is ~20-fold larger than that estimated for an E. coli SSB tetramer at 37°C (270 nt2s−1)[40]. An additional difference is that the activation energy for hRPA diffusion (10.1±0.5 kcal/mol) is half of that estimated for E. coli SSB (19.6±1.7 kcal/mol)[40]. These differences suggest that different mechanisms are used for diffusion of these two proteins. In its fully wrapped (SSB)65 binding mode, ~65 nucleotides of ssDNA interact with all four subunits[47, 48, 54, 55], whereas RPA interacts with only 30 nts in its largest mode[11]. Hence it is not surprising that the more extensively wrapped SSB diffuses more slowly along ssDNA than does RPA. The Monte Carlo simulations that we performed to estimate a diffusion coefficient assumes that hRPA diffuses by a sliding mechanism; however, other mechanisms are also possible. The mechanism of E. coli SSB diffusion seems to involve a reptation mechanism in which a small ssDNA bulge forms where the ssDNA enters the SSB protein[56]. In addition to this possibility, the mechanism of RPA diffusion might involve sequential local dissociation and rebinding of two or more of its OB-folds. Our observation that the hRPA truncation, FAB, can diffuse along ssDNA indicates that diffusion requires a minimum of only two DNA binding OB-folds. Recent experiments have shown that RPA bound to ssDNA can undergo rapid exchange with unbound RPA and the rate of exchange increases with increasing free RPA concentration[51]. This implies a transient intermediate in which both RPA molecules undergoing the exchange are bound simultaneously to the ssDNA. The ability of RPA to diffuse along ssDNA would facilitate this exchange.
There are now several examples of proteins that can diffuse along double stranded (ds) DNA (see [57] for a review). The TRBP family of RNA binding proteins have also been shown to diffuse along dsRNA[45]. Estimates of one-dimensional diffusion coefficients have been made for some of the dsDNA binding proteins, starting with the classic lac repressor (D ~ 106 bp2s−1)[58–61], DNA glycosylase 1 (D = 4.8±1.1x106 bp2s−1)[62], p53 tumor suppressor protein (D = (2.60 ± 2.17)×106 bp2/s)[63], Msh2-Msh6 DNA repair complex (D = 2.2×105 bp2s−1)[64], T. aquaticus MutS (D = 3.0 to 5.0×105 bp2s−1)[65], and a Type III restriction enzyme (D = 8.0±0.5×106 bp2s−1) [66]. These are considerably larger than the diffusion coefficient that we estimate for hRPA on ssDNA and much larger than the estimate for E. coli SSB on ssDNA[40], suggesting different mechanisms of diffusion for these two classes of proteins. One difference between the dsDNA and ssDNA binding proteins is that the affinities of E. coli SSB and hRPA are much higher than the affinities of the dsDNA binding proteins in their non-specific DNA binding modes. The function of diffusion for proteins on dsDNA is generally thought to increase the rate of location of a target[67]; fast diffusion in this case would be essential. However, for hRPA and SSB, the rate of diffusion may be less important than the fact that they can diffuse at all and in doing so destabilize hairpins and be moved to avoid inhibiting other proteins. Ensemble kinetic studies also inferred that the phage T4 gene 32 protein can diffuse along ssDNA[68]. Two other E. coli-like SSB proteins from Plasmodium falciparum[17] and T. thermophiles[69] can also diffuse on ssDNA, as can cytidine deaminase[70], although no estimates of diffusion coefficients have been made. Finally, the POT1-TPP1 complex has also been shown to diffuse on a telomeric ssDNA overhang, although POT1, which contains only two OB-folds does not show this ability[44], indicating that the ability to diffuse along ssDNA is not a general property of all ssDNA binding proteins.
Materials and Methods
Buffers
Buffer T is 10 mM Tris, pH 8.1, 0.1 mM Na2EDTA, 1 mM 2-mercaptoethanol. For experiments at different temperatures, the buffer was prepared so that pH 8.1 was maintained at each temperature. Buffer I[71] is 20 mM Tris-HCl pH 8.1, 0.1 mM Na2EDTA, 1 mM DTT, 0.8% (w/v) dextrose, 2.5 mM Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid, Sigma Aldrich, MO), 20 units/ml glucose oxidase and 20 units/ml catalase with 0.5 M NaCl.
Proteins
hRPA was expressed from plasmid p11d-tRPA in BL21(DE3) cells (Novagen) by auto-induction[72] in one liter of ZYP5052 medium shaken at 300 rpm at 37°C for 24h. RPA was purified as described[73, 74] with modifications (see SI Materials and Methods). The hRPA concentrations were determined by absorbance at 277 nm using an extinction coefficient of 8.57×104 M−1 cm−1 determined from the hRPA amino acid sequence and the absorbance spectrum of hRPA denatured in 6 M guanidinium HCl[75, 76]. The hRPA variants, ABC-D-E and FAB, were purified as described[77, 78]. The amino termini of the three subunits of hRPA were targeted for labeling with Cy5 as described[79] (for details see SI Materials and Methods). The hRPA was 92% labeled (overall efficiency for the heterotrimer) with a 4:3:1 labeling ratio for the 70:32:14 kDa subunits (as determined by denaturing PAGE).
DNA
Poly(dT) was from Midland Certified Reagent Co. (Midland, TX) and had an average length of 850 nucleotides and its concentration was determined using, ε260 = 8140 M−1 cm−1 (nucleotides)[80]. Oligodeoxynucleotides were either synthesized using a Mermaid 4 synthesizer (Plano, TX) with reagents from Glen Research (Sterling, VA) or purchased from Integrated DNA Technologies (IDT), Inc. (Coralville, IA). After purification by polyacrylamide gel electrophoresis[81], the oligodeoxynucleotides were further purified by reverse phase HPLC using an Xterra MS C18 Column (Waters, Milford, MA). The oligodeoxynucleotides used in this study are given in Table S1. DNA and proteins were dialyzed into the indicated buffers. Extinction coefficients for the oligodeoxynucleotides were calculated using the nearest neighbor assumption[82].
Ensemble Fluorescence experiments
Fluorescence titrations were conducted in Buffer T using a PTI QM-4 fluorometer (Photon Technology International, Birmingham, NJ, USA) as described[16] (for details see SI Materials and Methods). Titrations of Cy3 labeled DNA with hRPA monitored the Cy3 fluorescence enhancement (λex = 515 nm, λem= 570 nm). Excitation and emission slit widths were set at 0.50 mm (2 nm bandpass).
Occluded site sizes for hRPA binding to poly(dT) and equilibrium constants, Kobs, for hRPA binding to (dT)30 and (dT)30-Cy3 were measured in Buffer T, pH 8.1, 25.0°C at different [NaCl] as described[16]. The average Cy3 fluorescence enhancements upon binding one hRPA molecule to the series of (dT)L-3′-Cy3 and the chimeric, (dT)L-(dTalt)N,-3′-Cy3 were obtained by analysis of two titrations performed at two [DNA] (10–40 nM) in Buffer T, pH 8.1, 0.5 M NaCl, monitoring Cy3 fluorescence using the binding density function method[37] (for details see SI Materials and Methods).
Single molecule total internal reflectance fluorescence (smTIRF)
The smTIRF experiments were performed with an objective type total internal reflectance microscope (Olympus IX71, model IX2_MPITIRTL) equipped with a 3-laser system (488nm, 532nm and 635nm) and an oil-immersed, high numerical aperture TIRFM objective (PlanApo N, 60X/1.45 N.A., Olympus) as described[17] (see Figure S1). The slide holder was temperature controlled by a BC-110 Bionomic controller (20/20 Technology Inc., Wilmington, NC) and the objective was also connected to an objective heater (Bioptechs Inc., Butler, PA). Data were collected and analyzed using software packages generously provided by Taekjip Ha (University of Illinois, Urbana) (for details see SI Materials and Methods)..
Random Walk Simulations
Monte Carlo simulations of a random walk of a protein, with contact size of 30 nucleotides along a ssDNA of length, L, were performed using MatLab (Mathworks, Natick, MA) as depicted in Figure 7A (for details see SI Materials and Methods).
Supplementary Material
Highlights.
RPA binds single stranded DNA tightly and is essential for genome maintenance
human RPA can diffuse along single stranded DNA
hRPA can transiently melt DNA hairpins by diffusing in from adjacent ssDNA
Hairpin melting is more efficient if RPA diffuses in from the 5′ side of a hairpin
We estimate an hRPA diffusion coefficient (5000 nt2/s at 37°C) using a novel method
Acknowledgments
We thank Thang Ho for oligodeoxynucleotide synthesis and purification, Yanfei Jiang for discussions of the Monte Carlo simulations, Taekjip Ha for advice and generously supplying the software for analysis of the smTIRF time traces and Hidden Markov analysis and Vince Waldman and Rohit Pappu for comments. This work was supported in part by NIH grants GM030498 (T.M.L.), GM044721 (M.S.W.), GM098509 (R.G.) and HL109505 (E.L.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wold MS. Replication Protein A: A Heterotrimeric, Single-stranded DNA-Binding Protein Required for Eukaryotic DNA Metabolism. AnnRevBiochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 2.Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. Journal of cellular physiology. 2006;208:267–73. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–37. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oakley GG, Patrick SM. Replication protein A: directing traffic at the intersection of replication and repair. Front Biosci (Landmark Ed) 2010;15:883–900. doi: 10.2741/3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H, Lisby M, Symington LS. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol Cell. 2013;50:589–600. doi: 10.1016/j.molcel.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salas TR, Petruseva I, Lavrik O, Bourdoncle A, Mergny JL, Favre A, et al. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006;34:4857–65. doi: 10.1093/nar/gkl564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi JH, Lindsey-Boltz LA, Kemp M, Mason AC, Wold MS, Sancar A. Reconstitution of RPA-covered single-stranded DNA-activated ATR-Chk1 signaling. Proc Natl Acad Sci U S A. 2010;107:13660–5. doi: 10.1073/pnas.1007856107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–7. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bastin-Shanower SA, Brill SJ. Functional analysis of the four DNA binding domains of replication protein A. The role of RPA2 in ssDNA binding. J Biol Chem. 2001;276:36446–53. doi: 10.1074/jbc.M104386200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan J, Pavletich NP. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012;26:2337–47. doi: 10.1101/gad.194787.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X, Vaithiyalingam S, Glick GG, Mordes DA, Chazin WJ, Cortez D. The basic cleft of RPA70N binds multiple checkpoint proteins, including RAD9, to regulate ATR signaling. Mol Cell Biol. 2008;28:7345–53. doi: 10.1128/MCB.01079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daughdrill GW, Ackerman J, Isern NG, Botuyan MV, Arrowsmith C, Wold MS, et al. The weak interdomain coupling observed in the 70 kDa subunit of human replication protein A is unaffected by ssDNA binding. Nucleic Acids Res. 2001;29:3270–6. doi: 10.1093/nar/29.15.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lao Y, Lee CG, Wold MS. Replication protein A interactions with DNA. 2. Characterization of double-stranded DNA-binding/helix-destabilization activities and the role of the zinc-finger domain in DNA interactions. Biochemistry. 1999;38:3974–84. doi: 10.1021/bi982371m. [DOI] [PubMed] [Google Scholar]
- 15.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–14. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 16.Kumaran S, Kozlov AG, Lohman TM. Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes. Biochemistry. 2006;45:11958–73. doi: 10.1021/bi060994r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antony E, Kozlov AG, Nguyen B, Lohman TM. Plasmodium falciparum SSB tetramer binds single-stranded DNA only in a fully wrapped mode. J Mol Biol. 2012;420:284–95. doi: 10.1016/j.jmb.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys J. 2004;86:2530–7. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu Rev Biochem. 1994;63:527–70. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 20.Kolpashchikov DM, Khodyreva SN, Khlimankov DY, Wold MS, Favre A, Lavrik OI. Polarity of human replication protein A binding to DNA. Nucleic Acids Res. 2001;29:373–9. doi: 10.1093/nar/29.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinney SA, Joo C, Ha T. Analysis of Single-molecule FRET Trajectories Using Hidden Markov Modeling. Biophys J. 2006 doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfuetzner RA, Bochkarev A, Frappier L, Edwards AM. Replication protein A. Characterization and crystallization of the DNA binding domain. J Biol Chem. 1997;272:430–4. doi: 10.1074/jbc.272.1.430. [DOI] [PubMed] [Google Scholar]
- 23.Walther AP, Gomes XV, Lao Y, Lee CG, Wold MS. Replication protein A interactions with DNA. 1. Functions of the DNA-binding and zinc-finger domains of the 70-kDa subunit. Biochemistry. 1999;38:3963–73. doi: 10.1021/bi982370u. [DOI] [PubMed] [Google Scholar]
- 24.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Wong CJ, Lucius AL, Lohman TM. Energetics of DNA end binding by E.coli RecBC and RecBCD helicases indicate loop formation in the 3′-single-stranded DNA tail. J Mol Biol. 2005;352:765–82. doi: 10.1016/j.jmb.2005.07.056. [DOI] [PubMed] [Google Scholar]
- 26.Brendza KM, Cheng W, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci U S A. 2005;102:10076–81. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. Bacillus stearothermophilus PcrA Monomer Is a Single-stranded DNA Translocase but Not a Processive Helicase in Vitro. J Biol Chem. 2007;282:27076–85. doi: 10.1074/jbc.M704399200. [DOI] [PubMed] [Google Scholar]
- 28.Kozlov AG, Lohman TM. Kinetic mechanism of direct transfer of Escherichia coli SSB tetramers between single-stranded DNA molecules. Biochemistry. 2002;41:11611–27. doi: 10.1021/bi020361m. [DOI] [PubMed] [Google Scholar]
- 29.Wu CG, Bradford C, Lohman TM. Escherichia coli RecBC helicase has two translocase activities controlled by a single ATPase motor. Nat Struct Mol Biol. 2010;17:1210–7. doi: 10.1038/nsmb.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, et al. PcrA Helicase Dismantles RecA Filaments by Reeling in DNA in Uniform Steps. Cell. 2010;142:544–55. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomko EJ, Fischer CJ, Lohman TM. Ensemble methods for monitoring enzyme translocation along single stranded nucleic acids. Methods. 2010;51:269–76. doi: 10.1016/j.ymeth.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer CJ, Tomko EJ, Wu CG, Lohman TM. Fluorescence methods to study DNA translocation and unwinding kinetics by nucleic acid motors. Methods Mol Biol. 2012;875:85–104. doi: 10.1007/978-1-61779-806-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, et al. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–4. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci U S A. 2011;108:7414–8. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galletto R, Tomko EJ. Translocation of Saccharomyces cerevisiae Pif1 helicase monomers on single-stranded DNA. Nucleic Acids Res. 2013;41:4613–27. doi: 10.1093/nar/gkt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang H, Myong S. Protein induced fluorescence enhancement (PIFE) for probing protein-nucleic acid interactions. Chemical Society reviews. 2014;43:1221–9. doi: 10.1039/c3cs60201j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lohman TM, Bujalowski W. Thermodynamic methods for model-independent determination of equilibrium binding isotherms for protein-DNA interactions: spectroscopic approaches to monitor binding. Methods Enzymol. 1991;208:258–90. doi: 10.1016/0076-6879(91)08017-c. [DOI] [PubMed] [Google Scholar]
- 38.Kim C, Paulus BF, Wold MS. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994;33:14197–206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- 39.Kim C, Wold MS. Recombinant Human Replication Protein A Binds to Polynucleotides with Low Cooperativity. Biochemistry. 1995;34:2058–64. doi: 10.1021/bi00006a028. [DOI] [PubMed] [Google Scholar]
- 40.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–7. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ha T, Kozlov AG, Lohman TM. Single-Molecule Views of Protein Movement on Single-Stranded DNA. Annu Rev Biophys. 2012 doi: 10.1146/annurev-biophys-042910-155351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stennett EM, Ciuba MA, Levitus M. Photophysical processes in single molecule organic fluorescent probes. Chemical Society reviews. 2014;43:1057–75. doi: 10.1039/c3cs60211g. [DOI] [PubMed] [Google Scholar]
- 43.Thirumalai D, Ha BY. Statistical mechanics of Semi-flexible chains. In: Grosberg A, editor. Theoretical and Mathematical Models in Polymer Research. Academic Press; San Diego, CA: 1998. pp. 1–35. [Google Scholar]
- 44.Hwang H, Buncher N, Opresko PL, Myong S. POT1-TPP1 regulates telomeric overhang structural dynamics. Structure. 2012;20:1872–80. doi: 10.1016/j.str.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koh HR, Kidwell MA, Ragunathan K, Doudna JA, Myong S. ATP-independent diffusion of double-stranded RNA binding proteins. Proc Natl Acad Sci U S A. 2013;110:151–6. doi: 10.1073/pnas.1212917110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–6. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J Biol Chem. 1985;260:3594–603. [PubMed] [Google Scholar]
- 48.Bujalowski W, Lohman TM. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986;25:7799–802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 49.Philipova D, Mullen JR, Maniar HS, Lu J, Gu C, Brill SJ. A hierarchy of SSB protomers in replication protein A. Genes Dev. 1996;10:2222–33. doi: 10.1101/gad.10.17.2222. [DOI] [PubMed] [Google Scholar]
- 50.Brill SJ, Bastin-Shanower S. Identification and characterization of the fourth single-stranded-DNA binding domain of replication protein A. Mol Cell Biol. 1998;18:7225–34. doi: 10.1128/mcb.18.12.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gibb B, Ye LF, Gergoudis SC, Kwon Y, Niu H, Sung P, et al. Concentration-dependent exchange of replication protein a on single-stranded DNA revealed by single-molecule imaging. PLoS One. 2014;9:e87922. doi: 10.1371/journal.pone.0087922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunzelmann S, Morris C, Chavda AP, Eccleston JF, Webb MR. Mechanism of interaction between single-stranded DNA binding protein and DNA. Biochemistry. 2010;49:843–52. doi: 10.1021/bi901743k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider RJ, Wetmur JG. Kinetics of transfer of E. coli single strand deoxyribonucleic acid binding protein between single-stranded DNA molecules. Biochem. 1982;21:608–15. doi: 10.1021/bi00533a002. [DOI] [PubMed] [Google Scholar]
- 54.Bujalowski W, Lohman TM. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. II. Salt, temperature and oligonucleotide length effects. J Mol Biol. 1989;207:269–88. doi: 10.1016/0022-2836(89)90455-5. [DOI] [PubMed] [Google Scholar]
- 55.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E-coli SSB bound to ssDNA. Nature structural biology. 2000;7:648–52. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 56.Zhou R, Kozlov AG, Roy R, Zhang J, Korolev S, Lohman TM, et al. SSB functions as a sliding platform that migrates on DNA via reptation. Cell. 2011;146:222–32. doi: 10.1016/j.cell.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gorman J, Greene EC. Visualizing one-dimensional diffusion of proteins along DNA. Nat Struct Mol Biol. 2008;15:768–74. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- 58.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein trans-location on nucleic acids. I. Models and theory. Biochem. 1981;20:6929–48. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 59.Winter RB, Berg OG, von Hippel PH. Diffusion driven mechanisms of protein translocation on nucleic acids. III. The E. coli lac repressor-operator interaction: kinetic measurements and conclusions. Biochem. 1981;20:6961. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 60.Winter RB, von Hippel PH. Diffusion driven mechanisms of protein translocation on nucleic acids. II. The E. coli repressor-operator interaction: equilibrium measurements. Biochem. 1981;20:6948. doi: 10.1021/bi00527a029. [DOI] [PubMed] [Google Scholar]
- 61.Wang YM, Austin RH, Cox EC. Single molecule measurements of repressor protein 1D diffusion on DNA. Phys Rev Lett. 2006;97:048302. doi: 10.1103/PhysRevLett.97.048302. [DOI] [PubMed] [Google Scholar]
- 62.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci U S A. 2006;103:5752–7. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tafvizi A, Huang F, Leith JS, Fersht AR, Mirny LA, van Oijen AM. Tumor suppressor p53 slides on DNA with low friction and high stability. Biophys J. 2008;95:L01–3. doi: 10.1529/biophysj.108.134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat Struct Mol Biol. 2010;17:932–8. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cho WK, Jeong C, Kim D, Chang M, Song KM, Hanne J, et al. ATP alters the diffusion mechanics of MutS on mismatched DNA. Structure. 2012;20:1264–74. doi: 10.1016/j.str.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwarz FW, Toth J, van Aelst K, Cui G, Clausing S, Szczelkun MD, et al. The helicase-like domains of type III restriction enzymes trigger long-range diffusion along DNA. Science. 2013;340:353–6. doi: 10.1126/science.1231122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.von Hippel PH, Berg OG. Facilitated target location in biological systems. JBiolChem. 1989;264:675–8. [PubMed] [Google Scholar]
- 68.Lohman TM, Kowalczykowski SC. Kinetics and mechanism of the association of the bacteriophage T4 gene and 32 (helix destabilizing) protein with single-stranded nucleic acids. Evidence for protein translocation. JMolBiol. 1981;152:67–109. doi: 10.1016/0022-2836(81)90096-6. [DOI] [PubMed] [Google Scholar]
- 69.Zhang J, Zhou R, Inoue J, Mikawa T, Ha T. Single molecule analysis of Thermus thermophilus SSB protein dynamics on single-stranded DNA. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Senavirathne G, Jaszczur M, Auerbach PA, Upton TG, Chelico L, Goodman MF, et al. Single-stranded DNA scanning and deamination by APOBEC3G cytidine deaminase at single molecule resolution. J Biol Chem. 2012;287:15826–35. doi: 10.1074/jbc.M112.342790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nature methods. 2006;3:891–3. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 72.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein expression and purification. 2005;41:207–34. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 73.Wold MS, Kelly T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci U S A. 1988;85:2523–7. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Binz SK, Dickson AM, Haring SJ, Wold MS. Functional Assays for Replication Protein A (RPA) Methods Enzymol. 2006;409:11–38. doi: 10.1016/S0076-6879(05)09002-6. [DOI] [PubMed] [Google Scholar]
- 75.Lohman TM, Chao K, Green JM, Sage S, Runyon G. Large-scale purification and characterization of the Escherichia coli rep gene product. JBiolChem. 1989;264:10139–47. [PubMed] [Google Scholar]
- 76.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein science : a publication of the Protein Society. 1995;4:2411–23. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gomes XV, Wold MS. Functional domains of the 70-kilodalton subunit of human replication protein A. Biochemistry. 1996;35:10558–68. doi: 10.1021/bi9607517. [DOI] [PubMed] [Google Scholar]
- 78.Gomes XV, Wold MS. Structural analysis of human replication protein A. Mapping functional domains of the 70-kDa subunit. J Biol Chem. 1995;270:4534–43. doi: 10.1074/jbc.270.9.4534. [DOI] [PubMed] [Google Scholar]
- 79.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–8. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 80.Kowalczykowski SC, Lonberg N, Newport JW, von Hippel PH. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. I. Characterization of the binding interactions. JMolBiol. 1981;145:75–104. doi: 10.1016/0022-2836(81)90335-1. [DOI] [PubMed] [Google Scholar]
- 81.Ferrari ME, Bujalowski W, Lohman TM. Co-operative binding of Escherichia coli SSB tetramers to single-stranded DNA in the (SSB)35 binding mode. J Mol Biol. 1994;236:106–23. doi: 10.1006/jmbi.1994.1122. [DOI] [PubMed] [Google Scholar]
- 82.Fasman GD. Handbook of Biochemistry and Molecular Biology. 1975. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.