Abstract
To investigate the origin and evolutionary history of the spread of HIV-1 subtype B in China, a total of 409 sequences of pol gene sampled from 1994 to 2012 in 29 provinces across China was subjected to phylogenetic and Bayesian molecular clock analyses. The study reveals that subtype B strains in China are genetically diverse and can be classified into four distinct subgroups, namely B′ (Thai-B), BJ-B (Beijing-B), Pan-B (Pandemic-B), and TW-B (Taiwan-B), according to the origin of the sequences. The BJ-B and TW-B are reported for the first time. Phylogeographic analysis reveals that B′ exhibits a nationwide, transprovincial distribution, and is found in 21 provinces in China in this study, whereas the Pan-B, BJ-B, and TW-B lineages are restricted to particular regions. From the same common ancestor of B′, there arise two subclusters in which sequences from Yunnan occupy the basal position. The times of the most recent common ancestors (tMRCAs) of B′ and BJ-B are estimated to be 1983.6 (1975.9–1990.3) and 1995.3 (1989.6–2000.3), respectively. The skyline plot profile reveals an exponential decrease in median number of effective infections of subtype B in China from 1994 to 2009. The existence of four types of B clades also indicates distinct transmission networks of subtype B, originating from different introduction events at different time points. The data presented here offer a new perspective on the epidemic of HIV-1 subtype B in China.
China is the world's most populous country in which the human immunodeficiency virus type 1 (HIV-1) epidemic still prevails. Since the first HIV/AIDS case in China was reported in 1985,1 an estimated 780,000 individuals by the end of 2011 had contracted HIV in China. It is estimated that there were 48,000 new cases of HIV in 2011, and that the prevalence rate among the population as a whole was 0.057%. Sexual transmission continues to be the primary mode of transmission, and homosexual transmission is also increasing rapidly. Of the 780,000 people estimated to have been living with HIV in 2011, the percentage of cases infected through sexual transmission was 63.9%, of which 46.5% were infected through heterosexual transmission and 17.4% through homosexual transmission. The percentage of infected cases through intravenous drug use (IDU) was 28.4%. In the past 25 years, the main drivers of China's HIV epidemic have shifted considerably, from blood transmission to sexual transmission. The proportions of IDUs, heterosexuals, and men who have sex with men (MSM) among HIV-positive persons have changed from 44.2%, 11.3%, and 0.3% in 1985–2005 to 18.0%, 52.2%, and 29.4% in 2011, respectively.2
The HIV-1 epidemic in China is primarily driven by four major strains, including CRF07_BC, CRF01_AE, CRF08_BC, and B′ (Thailand variant of subtype B; also referred to as Thai-B), which account for 35.5%, 27.5%, 20.2%, and 9.6% of all infections, respectively. Among these HIV-1 strains, B′ plays a unique role in the genesis of the HIV-1 epidemic in China. HIV-1B′ was the most widespread HIV-1 strain in mainland China in terms of geographic reach and was identified in 28 provinces (except Ningxia and Xizang). While subtype B (circulating in the United States and Europe, also referred to as Pandemic-B or Pan-B), which constituted only 1.05% in China, was the minor HIV-1 genotype.3–6 Subtypes B and B′ were first identified almost concurrently among the IDUs in Yunnan province China in 1989. Over time, B′ overtook the prototypical B subtype in frequency, increasing from 20% of all subtype B in 1990 to 90% in 1996.7–9 Later, the B subtype disappeared without a trace and B′ constituted only 2.3% in Yunnan in 2009.10 Subtype B isolates were also identified in MSM in Beijing,5 and were the dominant strains to the extent that in 1993–2001, 100% of MSM infections in the city were caused by subtype B.11
The emergence and dispersal patterns of HIV-1B′ have been studied extensively in China. Discoveries made independently by three groups have provided similar answers recently to its temporal and geographic distribution patterns, including its origin and dissemination.12–14 However, the spatial diffusion of the epidemic from the perspective of Pan-B has not previously been traced. In this study, we used phylogenetic analyses and a Bayesian coalescent-based approach to investigate the temporal and spatial dynamics of HIV-1 subtype B transmission including B′ and Pan-B together in order to provide a greater understanding of the epidemic growth model of the HIV-1B virus.
We initially downloaded all published B subtype sequences for which province of origin and sampling year were known in China, covering the protease (PR) and partial reverse transcriptase (RT) gene regions from the Los Alamos HIV database (www.hiv.lanl.gov/content/hiv-db). A total of 1,202 nucleotide sequences labeled as subtype B pol sequences were retrieved, of which 926 were from China Mainland, 3 from Hong Kong, and 273 from Taiwan. We then subjected the resulting sequence set to strict quality control measures to remove (1) incomplete sequences and (2) multiple sequences from the same patient, resulting in a set of 212 pol sequences. Other subtype B sequences that prevailed globally were selected according to the study of Li et al.12 The subtype C sequences (86ETH2220, 95IN21068, and 92BR025d) comprised the outgroup. Sequences were also collected from molecular epidemiology studies conducted from 2006 to 2012 mainly in Beijing, the capital city of China.
The epidemiological information on the patients including age, gender, ethnicity, place of birth, route of infection, and CD4 cell count was also collected. The subtyping process was performed by phylogenetic analysis. We eventually obtained 197 sequences and aligned them with the 212 sequences retrieved from the Los Alamos HIV database; the GenBank accession numbers of the 197 B pol gene sequences from our laboratory are KM011653–KM011849. To reduce excessive computational load, closely related sequences from the same province were manually removed without compromising the genetic or geographic heterogeneity of each alignment, resulting in a down-sampling of the B data sets to 116 representative sequences for Bayesian phylogeographic analysis. The 409 local B pol sequences were aligned with 60 HIV-1B reference sequences isolated, of which 20 were from the United States/Europe, 20 from Thailand, and 20 from other countries using Sequencher 5.1. Multiple alignments were performed automatically by BioEdit 7.9 with minor manual adjustments.
A phylogenetic tree based on the pol sequence was constructed with the Kimura two-parameter model and neighbor-joining (NJ) method, using MEGA 4. The bootstrap test was performed with 1,000 replications.15,16 The evolution rate, time of the most recent common ancestor (tMRCA), and demographic history of the B strains circulating in mainland China were inferred using the Bayesian Markov chain Monte Carlo (MCMC) method in BEAST 1.7.5. The Relaxed Clock:Uncorrelated Exponential model was tested in combination with four different coalescent prior trees (“Constant Size,” “Exponential Growth,” “Logistic Growth,” and “Bayesian Skyline”) under an HKY nucleotide substitution model with heterogeneity among sites modeled with a gamma distribution and invariant sites.
For each model, the MCMC chain was run for 20,000,000 steps and sampled every 2,000 steps. The first 2,000,000 steps of each run were discarded as burn-in. The resulting log-files were analyzed in Tracer v.1.6.2 and the Bayes Factor was calculated to compare molecular clock models, using marginal likelihood as implemented in Tracer v.1.6.2. The maximum clade credibility with time scale (MCC) tree was obtained by TreeAnnotator v1.7.5 with a burn-in of the first hundred trees.17–19 A total of 409 subtype B pol gene (2263–3325 nt, HXB2) sequences isolated from 29 provinces in China from the years 1994 to 2012 were used in this study. Of the cases 197 were newly characterized from ongoing molecular epidemiological studies mainly in Beijing, the capital city of China, and 212 cases were obtained from the Los Alamos HIV sequence database (www.hiv.lanl.gov) from previously published reports.
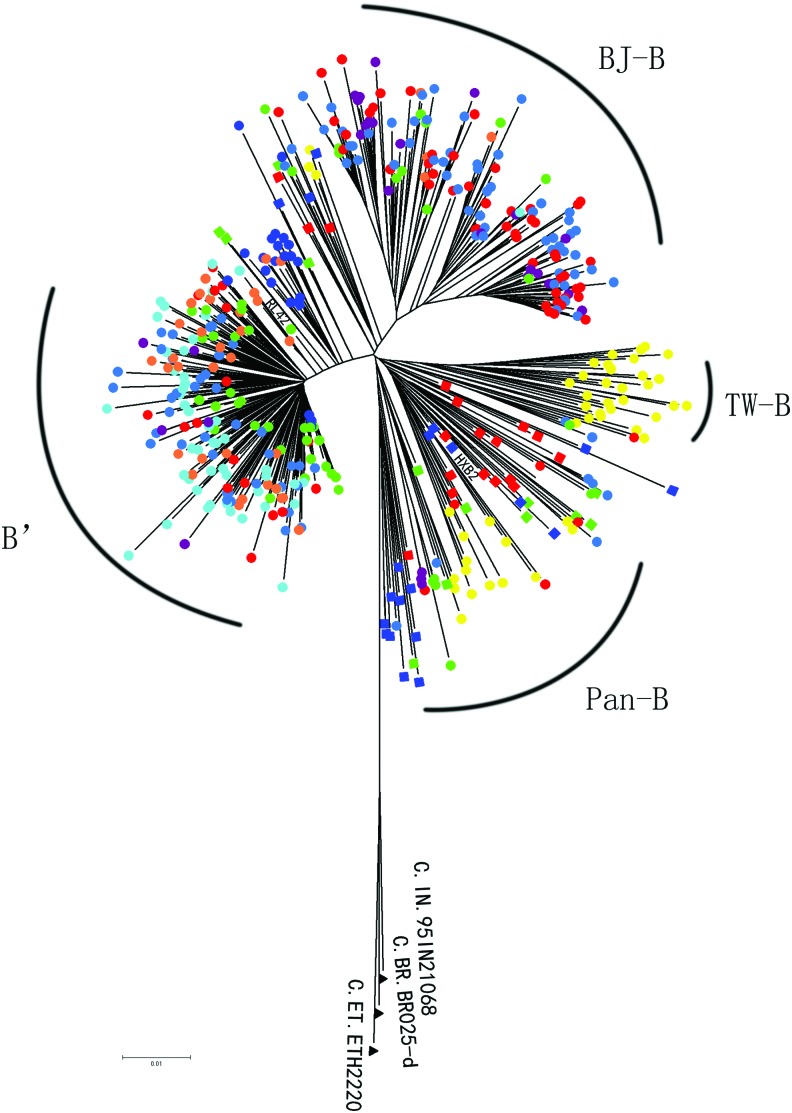
The clinical and demographic characteristics of the 409 HIV-1B infectors are listed in Table 1. A phylogenetic tree was reconstructed using the NJ method with 469 pol sequences, consisting of 409 sequences from China and 60 B reference sequences isolated in the United States/Europe (20), Thailand (20), and other countries (20). The tree demonstrated 13 out of 409 sequences from China (3.2%), which were scattered among the other sequences, while the other 386 sequences were segregated into four major distinct clusters with bootstrap values above 70% and more than 20 members (Fig. 1). A monophyletic subtype B subcluster (designated Pan-B) included 38 sequences mainly from Taiwan (21), Hong Kong (3), Beijing (6), and Liaoning (3) and was found to cluster with the reference sequences isolated from the HIV-1 infector in the United States/Europe. The information on nine individuals is as follows: the median age range was 32.6 (22–63) years and eight were males and one was female. Four belonged to the MSM risk group, whereas five were infected through heterosexual contacts.
Table 1.
The Distribution of the Geographic Source, Sampling Year, and Risk Factor for HIV-1 Subtype B Strains Used in the Study
| Risk groupb | ||||||||
|---|---|---|---|---|---|---|---|---|
| Geographic source | Sampling year | na | Hetero | MSM | IDU | FPD | MTC | Unkown |
| Beijing | 2006–2012 | 73 (18) | 18 | 32 | 1 | 22 (18) | ||
| Hebei | 2006–2012 | 22 | 8 | 12 | 2 | |||
| Liaoning | 2000–2010 | 49 (40) | 2 | 7 | 40 (40) | |||
| Henan | 2001–2012 | 37 (5) | 18 | 4 | 6 | 1 | 8 (5) | |
| Hubei | 2005–2012 | 48 (43) | 2 | 1 | 1 | 1 | 43 (43) | |
| Yunnan | 1994–2010 | 22 (19) | 2 | 1 | 19 (19) | |||
| Unknown | 2007–2012 | 20 | 4 | 12 | 1 | 3 | ||
| Taiwan | 1994–2009 | 48 (48) | 48 (48) | |||||
| Hong Kong | 2006 | 3 (3) | 3 (3) | |||||
Numbers in parenthesis indicate sequences downloaded from the Los Alamos HIV sequence database (www.hiv.lanl.gov).
Risk groups: FPDs, former plasma donors; Hetero, heterosexuals; IDUs, injecting drug users; MTC, mother-to-child transmission.
FIG. 1.
Phylogenetic tree analysis of HIV-1B pol gene sequences in China. The phylogenetic tree was constructed using neighbor-joining methods (Mega 4.0) for the pol region. The solid squares indicate reference sequences from the Los Alamos HIV sequence database. Colors indicate the geographic location of sampling as follow: United States in red, Thailand in green, other countries in blue. Samples from Beijing, Liaoning, Yunnan, Hebei, Taiwan, Hubei, Henan, and other provinces are shown by red, green, blue, purple, yellow, cyan, orange, and acid blue solid circles in the tree. Color images available online at www.liebertpub.com/aid
The second subcluster (designated B′) contained 193 B′ sequences sampled from 22 provinces in China and was found to cluster with sequences RL42 isolated from the HIV-1 infector in Yunnan. Information on the epidemic for 60 individuals indicated that the median age range was 36.1 (4–68) years and 34 were males and 26 were females. Heterosexual transmission (60.0%, 36 of 60) and transmission by former plasma donors (FPDs) (15.0%, 9 of 60) accounted for the major transmission routes for these isolates. MSM and mother-to-child transmission were also responsible for 6.7% (4 of 60) and 5.0% (3 of 60), respectively.
The third subcluster (designated BJ-B) featured the 139 subtype B strains mainly circulating among MSMs in Beijing and Hebei Province. The information on the epidemic reported for 121 of these patients revealed that the median age range was 30.8 (19–61) years and that 120 were males and one was female; the majority of patients (71.9%, 87 of 121) were MSM. Heterosexual (26) and IDUs (3) also accounted for 21.5% and 2.5%, respectively.
The forth subcluster (designated TW-B) featured the 26 subtype B strains circulating in Taiwan province. This cluster was pure Taiwan B clusters, which do not contain any foreign strains.
Phylogenetic analyses of the viral pol genes showed that B′ in China was closely related to the strains identified in Thailand, suggesting that the two clusters have close genetic relationships with these strains. In the pol phylogeny, the B′ sequences are most closely related to the isolate 92FR-BX08, which was identified in France in 1992. Marked differences were found among the 29 provinces in China (Table 2). As in Beijing and Hebei province, B infections were caused by BJ-B (67.1%, 49 of 73 and 71.4%, 20 of 28), with smaller proportions of infections caused by B′ (23.3%, 17 of 73, 28.6%, 8 of 28), and Pan-B (8.2%, 6 of 73, 0%, 0 of 28) strains. Meanwhile, in Liaoning, Henan, Hubei, and Yunnan, the dominant B strains were B′, which were found to be responsible for 63.3% (31 of 49), 88.9% (32 of 36), 98.0% (47 of 48), and 95.5% (21 of 22) of all infections, respectively. In Liaoning, BJ-B also constituted 20.4% (10 of 49). In Taiwan, the most prevalent B strains were TW-B (54.2%, 26 of 48) and Pan-B (43.8%, 21 of 48).
Table 2.
Distribution of B Isolates in Different Province of China
| Province | Overall | Pan-B | B′ | BJ-B | TW-B | Other |
|---|---|---|---|---|---|---|
| Beijing | 73 | 6 | 17 | 49 | 1 | |
| Hebei | 28 | 5 | 20 | 3 | ||
| Liaoning | 49 | 3 | 31 | 10 | 5 | |
| Henan | 36 | 32 | 4 | |||
| Hubei | 48 | 47 | 1 | |||
| Yunnan | 22 | 1 | 21 | |||
| Taiwan | 48 | 21 | 26 | 1 | ||
| Hong Kong | 3 | 3 | ||||
| Others 21 | 82 | 4 | 36 | 39 | 3 | |
| Province unknown | 20 | 4 | 16 | |||
| Total | 409 | 38 | 193 | 139 | 26 | 13 |
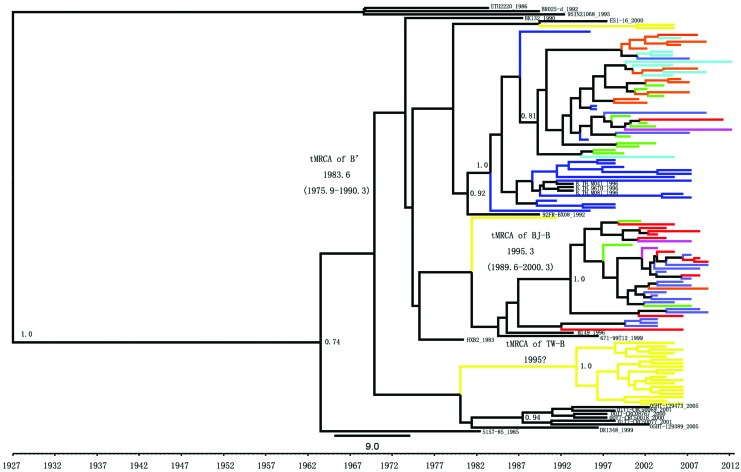
To explore these relationships in greater depth, we conducted a detailed Bayesian MCMC phylogenetic analysis using 106 representative China sequences, representing 25.9% of the total alignment, and a set of 17 reference sequences from France (n=2), Haiti (n=2), Japan (n=1), Netherlands (n=1), Thailand (n=5), Trinidad and Tobago (n=4), and the United States (n=2). The resulting tree predicts with high support that the C subtype group is located outside the B subtype, and the Haitian sequences occupy the basal position of the B subtype and can be regarded as being closest to the common B subtype ancestor. The phylogenetic and evolutionary relationships of subtype B inferred by BEAST were similar to that inferred by Mega 4. As opposed to the NJ trees, the MCC tree shows separate clades for three types of subtype B strains. As shown in the MCC tree, a total of 51 sequences from China grouped together with three Thailand sequences (96TH, M041, and M081) to form a single monophyletic clade, while 12 of 17 Yunnan sequences grouped basally with respect to this B′ clade, and the 39 isolates from other provinces including five isolates from Yunnan occupied more derived positions.
Twenty-nine BJ-B sequences from China formed a single monophyletic cluster with no related isolate sequence. It was also noted that a single B subtype sequence named M149 sampled in Thailand in 1996 appeared to be more closely associated with the BJ-B group; the relationship was not supported by high clade credibility (0.12), which might provide some insight into the origin of the BJ-B subtype, but there is little available information about this patient.
Clade BJ-B is mainly composed of isolates from Beijing's permanent resident and floating population (93.1%, 27 of 29), especially MSM (51.7%, 15 of 29). Within this clade, the sequences positioned nearest the tree root were also derived from Beijing, suggesting that this clade may originate in that region. However, 19 Taiwan sequences formed a single well-supported group displaying short branch lengths suggestive of a local transmission chain. There was no other related sequence in this cluster either. Although one sequence (DR1348) from Japan appeared to be more closely associated with the strongly clustered TW-B sequences, the relationship was not supported by high clade credibility (0.12), thus we were unable to clearly identify a sister cluster in the B subtype. The grouping of subtype B sequences into B′, BJ-B, and TW-B clusters was supported by high posterior probabilities (1.0) in each case (Fig. 2). No significant cluster of Pan-B was observed; they are all interspersed with strains from different geographic origins mainly in the United States and Europe.
FIG. 2.
Maximum clade credibility trees of the HIV-1B pol gene sequences in China. The maximum clade credibility (MCC) trees of pol gene sequences obtained by Bayesian Markov chain Monte Carlo (MCMC) analyses with BEAST 1.7.5. HIV-1 subtype C sequences (86ETH2220, 95IN21068, and 92BR025d) were used as outgroups. Colors indicate the geographic location of sampling as follow: Beijing in red, Liaoning in green, Yunnan in blue, Hebei in purple, Taiwan in yellow, Hubei in cyan, Henan in orange, and other provinces in cobalt blue. The time of most common ancestor (tMRCA) means and 95% highest posterior density (HPD) regions for the key nodes are indicated. Posterior probability values greater than 0.70 are noted above well-supported nodes. Color images available online at www.liebertpub.com/aid
To access the possible timeline of subtype B expansion in China, we estimated the tMRCAs of the respective subtype B clusters, using the relaxed molecular clock approach implemented in BEAST v1.7. 5. Different models were compared by calculating the Bayes factor, which is the ratio of the marginal likelihoods of the two models being compared. The results showed that the Bayesian skyline plot model performed better than the other three demographic models. As summarized in Tables 3 and 4, the estimated evolutionary rates were 1.25 (0.84–1.67)×103 substitutions per site per year for HKY models with a skyline coalescent model and the estimated tMRCAs of B′ were 1983.6 [95% credible region: highest posterior density (HPD): 1975.9–1990.3]. This is in good agreement with previous estimates by Li et al.12 and Deng et al.14 The rate of evolution for BJ-B was estimated to be 2.43 (1.51–3.32)×103 substitutions per site per year, and the tMRCA of HIV-1 BJ-B dated to 1995.3 (95% credible region: 1989.6–2000.3), which is in agreement with previous epidemiological investigations involving the first report of subtype B in the middle of 1980 in China. The likely year of the origin of subtype TW-B was estimated to be 1999 in the MCC tree (Fig. 2), but the conclusion remains questionable because the time span was too short.
Table 3.
Estimated Substitution Rates and Time of the Most Recent Common Ancestors for HIV-1 Subtype B′ Lineages
| Modela | Bayes factor (log10)b | Rate of evolutionc | tMRCA (year)d |
|---|---|---|---|
| Constant | — | 1.47 (1.05, 1.92) | 1979.4 (1968.3, 1988.4) |
| Exponential | 0.18 | 0.86 (0.41, 1.34) | 1971.7 (1952.4, 1986.4) |
| Logistic | −0.51 | 0.68 (0.22, 1.16) | 1959.9 (1917.6, 1984.9) |
| Skyline | 2.14 | 1.25 (0.84, 1.67) | 1983.6 (1975.9, 1990.3) |
Based on BEAST analysis under a relaxed molecular clock with a HKY substitution model and a constant population, exponential growth, logistic growth, or skyline plot model.
The Bayes factor (log10) reflects the difference between the constant population model and the other three models.
Estimates of the mean evolutionary rate (μ:×103 nucleotide substitutions/site/year) for subtype B′ with the range of 95% highest posterior density in parentheses.
The time of the most recent common ancestor (tMRCA: year) for subtype B′ with the range of 95% highest posterior density in parentheses.
Table 4.
Estimated Substitution Rates and Time of the Most Recent Common Ancestors for HIV-1 Subtype BJ-B Lineages
| Modela | Bayes factor (log10)b | Rate of evolutionc | tMRCA (year)d |
|---|---|---|---|
| Constant | — | 2.50 (1.75, 3.21) | 1994.1 (1988.8, 1998.7) |
| Exponential | −0.13 | 2.20 (1.35, 3.06) | 1992.8 (1985.5, 1998.4) |
| Logistic | −0.04 | 2.17 (1.29, 3.15) | 1988.1 (1984.3, 1999.0) |
| Skyline | 0.58 | 2.43 (1.51, 3.32) | 1995.3 (1989.6, 2000.3) |
Based on BEAST analysis under a relaxed molecular clock with a HKY substitution model and a constant population, exponential growth, logistic growth, or skyline plot model.
The Bayes factor (log10) reflects the difference between the constant population model and the other three models.
Estimates of the mean evolutionary rate (μ:×103 nucleotide substitutions/site/year) for subtype BJ-B with the range of 95% highest posterior density in parentheses.
The time of the most recent common ancestor (tMRCA: year) for subtype BJ-B with the range of 95% highest posterior density in parentheses.
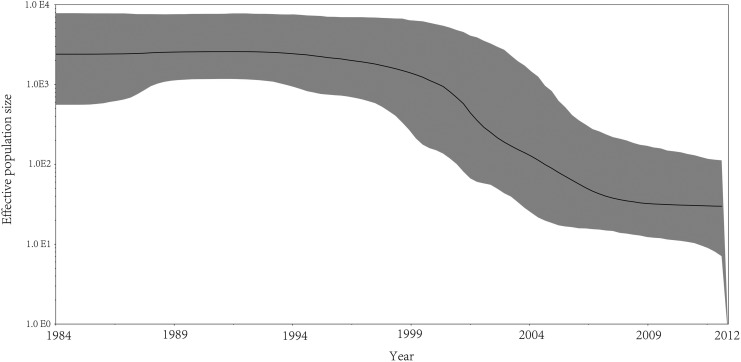
We also investigated the history of the epidemic involving subtype B in China by estimating a Bayesian skyline plot (BSP) for our subtype B pol sequences dataset, which depicts the change in effective population size over time. The demographic history from the B pol BSP identified three growth phases of the epidemic (Fig. 3), an initial stationary phase until the year 1994, followed by an exponential decreased phase until 2009, followed by another stationary phase approaching 2012. The most notable feature of Fig. 3 is the change at the onset of 1998 from a high and relatively constant effective population size to low epidemic growth and an effective population size decrease of approximately 100-fold during the period. We also reconstruct the demographic history of B′ and BJ-B in China, respectively. Our reconstruction of the demographic history of HIV-1B′ in China identified an initial, stable phase until 2001 followed by a period of quick exponential-like decrease from 2001 to 2012 when the growth of the epidemic slowed (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/aid). The demographic analysis of BJ-B in China showed a trend similar to that of the B′ epidemic, with a decrease in the median number of effective infections that occurred between 2006 and 2012 (Supplementary Fig. S2).
FIG. 3.
Demographic history of the expansion of HIV-1 subtype B lineages in China. Bayesian skyline plot was estimated to reconstruct the demographic history of subtype B in China using BEAST 1.4. The x-axis is the time in units of years and the y-axis is equal to the effective population size. The thick solid line is the mean estimates; the 95% HPD credible region is shown by shaded areas.
In this study, we investigated the origin and evolutionary history of HIV-1 subtype B in China using the phylogenetic, geographic, and coalescent method. For this purpose, we determined a total of 409 subtype B pol sequences sampled between 1994 and 2012 from various risk populations, including FPDs, heterosexuals, IDUs, MSM, and mother-to-child transmission (MCT), in 29 provinces across China (Table 1). Our results demonstrate that there exist four types of B subtypes in China, namely B′, BJ-B, Pan-B, and TW-B, according to the origin of the sequences, with the BJ-B and TW-B reported for the first time. B′ was the main subtype B strain in China, constituting 47.2% (193 of 409). This virus was distributed nationwide and was identified in 22 provinces of China; it was mainly transmitted by FPD and heterosexually. The B′ strains in China can be classified into two distinct subgroups: (1) B′ strains circulating among IDUs in Yunnan (B′YN) that occupy the most basal position of the B′ clade in China and (2) a monophyletic B′ cluster (B′CN) consisting of B′ sequences mainly from FPDs and heterosexuals across China. As subtype B′CN is paraphyletic with respect to B′YN, B′CN is likely a descendant lineage of subtype B′YN. It is thus tempting to speculate that a founder strain of the B′CN cluster originated from one of the B′ lineages circulated in Yunnan and subsequently transferred across China.
Pan-B constituted a relatively small proportion in China, and was mainly found in Taiwan and Beijing, and the transmission methods were MSM and heterosexually. The 38 Pan-B isolates form a cluster in the NJ tree, but are genetically diverse in the MCC tree. Within the Pan-B cluster, there exist many strains from the United States and Europe indicating that the origins of the Pan-B are complicated. BJ-B strains that were first identified in this study constituted 34.0% in China and were prevalent strains in Beijing (67.1%) and Hebei (71.4%) province.
The BJ-B strains are obviously phylogenetically distinct in the trees, representing novel B HIV-1 diversity, and show no clear relationship to any previously reported lineage. The MSM undoubtedly played an important role in the HIV-1 subtype BJ-B epidemic. Of the 116 BJ-B infectors of which the risk method was available, 87 (75.0%) were MSM. The overland migration of subtype BJ-B strains is most likely related to migration movements of MSMs. There is one well-supported cluster of Taiwan sequences having 54.2% (26 of 48) of all TW-B strains. Phylogenetic analyses of the viral pol genes showed that TW-B strains were closely related to a strain identified in Japan (DR1348), suggesting that TW-B has a close genetic relationship with the strains. The Taiwan clusters can be described as “local epidemic” strains, which transmitted in specific locations but did not spread nationwide in the manner of the “national epidemic” B′. Local epidemic B strains have previously been noted in Korea,20 and our results indicate that local epidemic subtypes also exist in Taiwan.
Using an evolutionary time scale spanning 26 years, the Bayesian analysis indicates that the HIV-1B′ epidemic in China evolved from a common ancestor introduced around 1983.6 (95% credible region: HPD: 1975.9–1990.3) from Thailand. In contrast, the estimated date of origin of the BJ-B clade was more recent, at 1995.3 (95% credible region: 1989.6–2000.3). Our evolutionary analysis of HIV-1B′ agrees with other studies in the timing of its introduction into China.12–14
Perhaps the number of HIV-1 sequences recovered from early victims of AIDS was too small; in the BSP analysis, we did not observe an increase in the period of the B subtype on the whole, as others reported. Demographic reconstruction of the entire B subtype showed an exponential decrease in the median number of effective infections from 1994 to 2009, followed by an asymptotic phase afterward. A decreasing trend in the epidemic involving B′ and BJ-B was also observed. This change coincides with the time blood donations were forbidden in 1995; as a result, the proportion of HIV-1B subtypes has greatly decreased in MSM.
BJ-B has been described for the first time in this study. As shown, this strain had been circulating in China for approximately 17 years. Why was this strain not identified before? We think the main reason was that BJ-B was mainly prevalent in MSM in Beijing, Hebei, and Liaoning province and the number of nucleotide sequences submitted to the Los Alamos database was too small; of the 139 BJ-B identified in this study, only 18 were from the Los Alamos database. After removing the sequences from our laboratory, we could no longer identify the BJ-B cluster.
Taken together, we identified four distinct subclades of subtype B circulating in China. Our analyses are based on a 1.3-kb pol nucleotide sequence dataset and provide the first thorough investigation of the subtype B epidemiology in China. Our analyses suggest that B′ is one of four variants or subclades of subtype B circulating in China.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundational of China (81302440 and 81273136), the China megaprojects in infectious diseases (2012ZX10001-005), and the Beijing Postdoctoral Research Foundation. We thank all the laboratory members of the Beijing Center for Disease Prevention and Control, College of Life Science and Bio-engineering, Beijing University of Technology, and the National Institute for Viral Disease Control and Prevention of China for their support in carrying out this project.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Zeng Y, Fan J, Zhang Q, et al. : Detection of antibody to LAV/HTLV-III in sera from hemophiliacs in China. AIDS Res 1986;2(Suppl 1):S147–149 [PubMed] [Google Scholar]
- 2.Ministry of Health of the People's Republic of China, UNAIDS WHO: China 2011 AIDS Epidemic Report. Chin J AIDS STD 2012;18:1–5(in Chinese). [Google Scholar]
- 3.He X, Xing H, Ruan Y, et al. : A comprehensive mapping of HIV-1 genotypes in various risk groups and regions across China based on a nationwide molecular epidemiologic survey. PLoS One 2012;7:e47289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xin R, He X, Xing H, et al. : Genetic and temporal dynamics of human immunodeficiency virus type 1 CRF07_BC in Xinjiang, China. J Gen Virol 2009;90:1757–1761 [DOI] [PubMed] [Google Scholar]
- 5.Ye JR, Yu SQ, Lu HY, Wang WS, Xin RL, and Zeng Y: Genetic diversity of HIV type 1 isolated from newly diagnosed subjects (2006–2007) in Beijing, China. AIDS Res Hum Retroviruses 2012;28:119–123 [DOI] [PubMed] [Google Scholar]
- 6.Meng Z, Xin R, Zhong P, et al. : A new migration map of HIV-1 CRF07_BC in China: Analysis of sequences from 12 provinces over a decade. PLoS One 2012;7:e52373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Y, Li Z, and Zhao SD: HIV infected people were first identified in intravenous drug users in China. Chin J Epidemiol 1990;11:184–185 [Google Scholar]
- 8.Shao Y, Zhao Q, Wang B, et al. : Sequence analysis of HIV env genes among HIV infected drug injecting users in Dehong epidemic area of Yunnan province, China. Chin J Virol 1994;10:291–299 [Google Scholar]
- 9.Graf M, Shao Y, Zhao Q, et al. : Cloning and characterization of a virtually full-length HIV type 1 genome from a subtype B′-Thai strain representing the most prevalent B-clade isolate in China. AIDS Res Hum Retroviruses 1998;14:285–288 [DOI] [PubMed] [Google Scholar]
- 10.Chen M, Yang L, Ma Y, et al. : Emerging variability in HIV-1 genetics among recently infected individuals in Yunnan, China. PLoS One 2013;8:e60101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao J, Zhang FJ, He ZP, et al. : Subtype and sequence analysis of the C2-V3 region of env gene among HIV-1 infected homosexual men in Beijing. Chin J STD/AIDS Prev Cont 2002;8:131–133(in Chinese). [Google Scholar]
- 12.Li Z, He X, Wang Z, et al. : Tracing the origin and history of HIV-1 subtype B' epidemic by near full-length genome analyses. AIDS 2012;26:877–884 [DOI] [PubMed] [Google Scholar]
- 13.Deng X, Liu H, Shao Y, Rayner S, and Yang R: The epidemic origin and molecular properties of B′: A founder strain of the HIV-1 transmission in Asia. AIDS 2008;22:1851–1858 [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Uenishi R, Hase S, et al. : Explosive HIV-1 subtype B' epidemics in Asia driven by geographic and risk group founder events. Virology 2010;402:223–237 [DOI] [PubMed] [Google Scholar]
- 15.Saitou N. and Nei M: The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4:406–425 [DOI] [PubMed] [Google Scholar]
- 16.Tamura K, Dudley J, Nei M, and Kumar S: MEGA4: Molecular evolutionary genetics analysis (MEGA) software, version 4.0. Mol Biol Evol 2007;24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 17.Lemey P, Rambaut A, Drummond AJ, and Suchard MA: Bayesian phylogeography finds its roots. PLoS Comput Biol 2009;5:e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drummond AJ, Suchard MA, Xie D, and Rambaut A: Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 2012;29:1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drummond AJ, Ho SY, Phillips MJ, and Rambaut A: Relaxed phylogenetics and dating with confidence. PLoS Biol 2006;4:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim MS, Jang SY, Park CS, Lee KM, Lee DH, and Lee CH: Timing and evolution of the most recent common ancestor of the Korean clade HIV subtype B based on nef and vif sequences. J Microbiol 2009;47:85–90 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.