Abstract
The last decade has seen an incredible breakthrough in technologies that allow histones, transcription factors (TFs), and RNA polymerases to be precisely mapped throughout the genome. From this research, it is clear that there is a complex interaction between the chromatin landscape and the general transcriptional machinery and that the dynamic control of this interface is central to gene regulation. However, the chromatin remodeling enzymes and general TFs cannot, on their own, recognize and stably bind to promoter or enhancer regions. Rather, they are recruited to cis regulatory regions through interaction with site-specific DNA binding TFs and/or proteins that recognize epigenetic marks such as methylated cytosines or specifically modified amino acids in histones. These “recruitment” factors are modular in structure, reflecting their ability to interact with the genome via one region of the protein and to simultaneously bind to other regulatory proteins via “effector” domains. In this chapter, we provide examples of common effector domains that can function in transcriptional regulation via their ability to (a) interact with the basal transcriptional machinery and general co-activators, (b) interact with other TFs to allow cooperative binding, and (c) directly or indirectly recruit histone and chromatin modifying enzymes.
12.1 Introduction
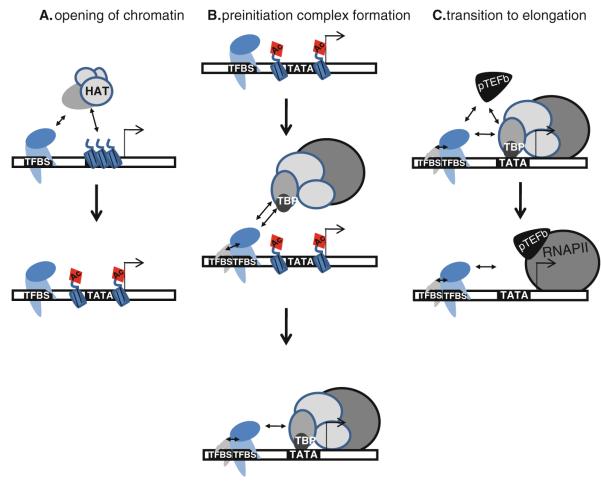
Transcriptional activation is a stepwise process that requires (a) creating and maintaining an open chromatin structure, (b) assembly of the preinitiation complex, and (c) transition to productive elongation (Fig. 12.1). Successful completion of each of these steps involves a diverse group of proteins, some of which function in a relatively promoter-specific manner whereas others regulate large sets of genes. Recent advances in molecular and computational biology allow histone and DNA modifications, TFs, and RNA polymerases to be precisely mapped throughout the genome, relative to active or silent promoters (see [1–3] for reviews). From this research, it is becoming clear that there is a complex interaction between the chromatin landscape and the transcriptional machinery and that the dynamic relationship of this interface is central to biological control over gene expression [4]. It is now recognized that regulatory factors can exert their influence on transcriptional activation either via co-localization with other proteins that are bound at or near core promoter regions or they can be recruited to distal enhancer regions and interact with promoter-bound proteins via looping mechanisms. However, generally speaking, the chromatin remodeling enzymes and the general transcription factors involved in initiation and elongation cannot, on their own, recognize and stably bind to the promoter or enhancer regions.
Fig. 12.1.
Regulation of transcription. Shown is a schematic representing the three steps needed for productive transcription, including Step 1: the creation of open chromatin, which involves interactions between DNA-bound proteins and histone modifying enzymes (e.g. a HAT which can create an acetylated (Ac) histone); Step 2: assembly of the preinitiation complex, which can involve interactions between different DNA binding proteins and between DNA-bound proteins and general factors (such as TBP which binds to the TATA box); and Step 3: transition to productive elongation, which involves interaction between DNA-bound proteins and enzymes such as the pTEFb kinase. Although in this schematic the TFs are shown binding to transcription factor binding sites (TFBS) proximal to the transcription start site (indicated by the bent arrow), many transcriptional regulators can also bind to sites quite far from the core promoter regions
One way in which chromatin remodeling enzymes and general transcription factors are recruited to cis-regulatory regions is through interaction with site-specific DNA binding TFs (Fig. 12.2a). The three largest classes of site-specific DNA binding proteins in mammals contact the genome via conserved DNA binding domains called zinc fingers, homeodomains, and helix–loop–helix domains [5] (Chapter 3 of this volume provides a catalog of eukaryotic DNA binding domains, and Chapters 4 and 5 specifically review C2H2 zinc fingers and homeodomains). Each of these classes of site-specific DNA binding factors contains many different proteins; for example, in humans there are over 650 zinc finger proteins, ~ 250 homeodomain proteins, and ~80 helix-loop-helix proteins [5]. Within each class, individual TFs can bind to and regulate hundreds to thousands of different genes. Site-specific TFs are modular in their structure reflecting their ability to bind to DNA via their DNA binding domains and simultaneously bind to other transcriptional regulatory proteins via so-called effector domains. The modular nature of site-specific TFs has been repeatedly demonstrated using in vitro and in vivo reporter assays. In these experiments, effector domains are separated from their natural DNA binding domains and then engineered to be part of a fusion protein having a heterologous DNA binding domain. Numerous studies have shown that simply bringing such effector domains to promoter regions can modulate transcription [6–8].
Fig. 12.2.
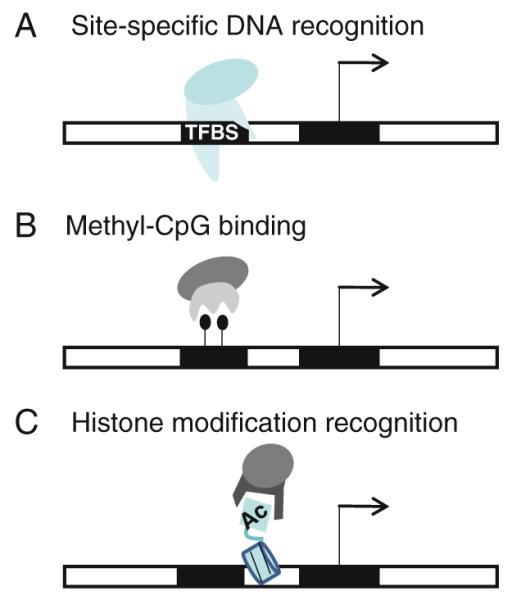
Modular structure of effector domain-containing proteins. Effector domains can be recruited to specific genomic regions via a DNA binding domains that recognize short DNA sequence motifs (TFBS), b recognition of a methylated cytosine (shown as a black ball), and c recognition of a modified amino acid of a histone (e.g. an acetylated histone, shown as Ac)
Another way in which chromatin remodeling enzymes and general transcription factors can be brought to the genome is via effector domains that reside in proteins that can recognize epigenomic marks. Similar to recognition of a short nucleotide motif by a DNA binding protein, other proteins can distinguish distinctively modified DNA and histone protein “motifs”. For example, methylated cytosine in the 5′-CpG-3′ dinucleotide sequence is specifically recognized by members of a family of proteins containing a conserved methyl-CpG binding domain (MBD). MBD-containing proteins, which include MeCP2, MBD1, MBD2 and MBD4, bind specifically to methyl-CpG motifs located throughout the genome [9]; see Fig. 12.2b. MBD-containing proteins function by recruiting various co-regulators to methyl-CpG sites. For example, MeCP2 simultaneously binds promoter regions containing methyl-CpG motifs and the Sin3-containing histone deacetylase complex via a transcriptional repression domain (TRD), resulting in histone deacetylation and transcriptional silencing [10, 11]. Likewise, MBD1 and MBD2 copurify with distinct cellular complexes which link DNA methylation with chromatin modification and transcriptional repression. Similarly, posttranslational modifications of the amino termini of core histones are correlated to transcriptional states and are recognized by relevant chromatin-associated proteins (Fig. 12.2c). Several different histone modifications have been identified, including acetylation, phosphorylation, and methylation, and specific protein domains have evolved to recognize several of these different modifications. For example, different methylation states of histone H3 at lysine 4 can be recognized by tudor, chromo, and plant homeodomains (PHD), by malignant brain tumor (MBT) domains, and by WD40 repeat domains (many of these domains are structurally related and are collectively referred to as the “royal family” [12], reviewed [13, 14]). Other examples of this family include the chromodomain of HP1, which interacts with lower (mono- and di-) methylation states of lysine 9 of histone H3 but preferentially binds to the trimethylated state [15, 16] and the tudor domain of 53BP1, which can discriminate between the diand tri-methyl state of H4K20, preferring the dimethyl form [17, 18]. Acetylated lysine is also recognized by specific protein modules called the bromodomain [19], which is found in many chromatin-associated proteins and in nearly all known nuclear histone acetyltransferases (HATs). Of course, epigenetic marks such as DNA methylation and histone modifications are located at specific genomic regions (which can vary in different cell types), indicating that DNA methylases and histone modifying enzymes must be recruited to the genome by sequence-specific mechanisms such as site-specific TFs or RNAs. For example, KRAB-ZNFs can recruit the KAP1/SETDB1 histone methylating complex and long non-coding RNAs can recruit the PRC2 histone methylation complex [20–23].
The focus of this chapter is on the effector domains that are brought to specific sites of the genome by DNA binding proteins, methyl-CpG binding proteins, or histone binding proteins. (The interaction of TFs with chromatin more generally is discussed in Chapter 11). We provide examples of common effector domains that can function in transcriptional regulation via their ability to influence each of the steps outlined in Fig. 12.1. Specifically, we discuss effector domains that can: (a) interact with the basal transcriptional machinery and general co-activators, (b) interact with other TFs to allow cooperative binding, and (c) directly or indirectly recruit histone and chromatin modifying enzymes. It is important to understand that a given effector domain does not have a one-to-one interaction with only one type of regulatory partner. Rather, some effector domains can interact with the general transcriptional machinery, with various co-activator complexes, and with chromatin remodeling proteins. To provide a specific example, nuclear receptors (NRs) are very specialized ligand-dependent TFs that regulate cellular gene expression programs in response to a variety of small molecules, including endocrine hormones, fatty acids, and lipid metabolites (discussed in detail in Chapter 6). NR transactivation domains (also referred to as Activating Function, or AF, domains) have the capacity to drastically alter transcriptional activities in a context-dependent manner by recruiting many different types of multi-protein co-regulatory complexes, often referred to as co-activators and co-repressors. For instance, the AF-1 and AF-2 terminal regions of the human glucocorticoid receptor alpha (hGRα) can link the receptor with different complexes depending on cellular signals. In the presence of glucocorticoids, the AF domains of hGRα interact with transcription-activating factors including basal TFs (e.g. RNA polymerase II, TATA-binding protein (TBP) and a host of TBP-associated proteins (TAFIIs)), coactivators such as p300/CBP and members of the p160/SRC family, site-specific factors including AP-1 and NFκB, and chromatin modulators such as the SWI/SNF and SAGA (Spt-Ada-Gcn5 acetyltransferase) complexes (reviewed in [24]). On the other hand, when bound to different gene regulatory regions, the same AF domains can, in response to glucocorticoid signals, recruit transcription repression complexes including corepressors, histone deacetylases (HDACs) and chromatin remodelers to down regulate transcriptional activity [25, 26]; see Fig. 12.3. Therefore, in addition to directing the TF to a specific genomic target, the precise nucleotide sequence of a regulatory factor binding site may also specify the mode of transcriptional regulation by directing the assembly of distinct regulatory complexes. The ability of a given DNA binding site to differentially affect hGRα activity has recently been investigated with much attention. Structural studies indicate that interactions between the hGRα DBD and different DNA elements can allosterically modulate interdomain interactions and thereby expose different surfaces for the recruitment of specific coregulator molecules [27].
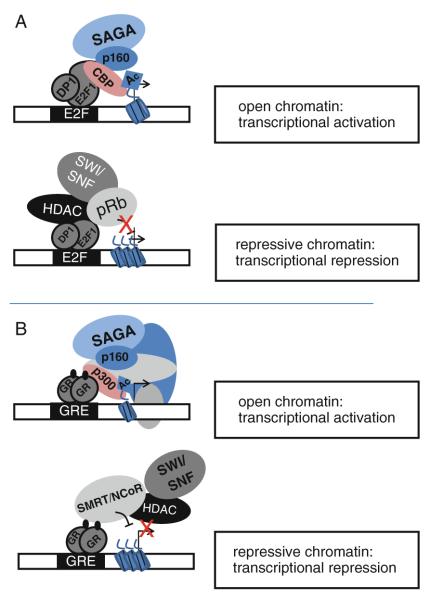
Fig. 12.
Effector domains interact with different types of proteins to confer transcriptional regulation. The effector domains of both ubiquitously expressed factors such as E2F1 (panel a) and cell type-specific factors such as the glucocorticoid receptor (panel b) can interact with many different proteins, resulting in either transcriptional activation or transcriptional repression (see text for references and descriptions of the various proteins; see also [118] for a review on context-dependent transcriptional regulation)
12.2 Effector Domains Can Interact with the Basal Transcription Machinery
Sequence-specific transcriptional activators play an important role in transcription initiation by mediating the interaction of components of the transcriptional machinery with the DNA (see [28, 29] for a review of the eukaryotic basal transcriptional machinery). The domains that stimulate transcriptional activation through contacts with general TFs are called transactivation domains (TADs). Specifically, TADs interact with components of the preinitiation complex (PIC) to enhance recruitment and stabilization of the general factors at target promoters. The TADs from many regulatory TFs, such as E2F1, have been shown to make direct contacts with general TFs, including TATA-binding protein (TBP), TBP-associated factors (TAFs), TFIIA, TFIIB, and TFIIH from sites located both near to and far from core promoters [30–39]. TADs can also interact with and recruit components of the mediator protein complex, a multi-protein complex involved in activating a large number of genes [29].
Eukaryotic transactivation domains are typically classified with respect to their amino acid composition. TADs can be rich in acidic amino acid residues (e.g. E2F1 and p53), in glutamine residues (e.g. Oct1, Oct2, Sp1) or in proline residues (e.g. AP-2 and CTF/NF1). Each of these classes of transactivation domains has been shown to interact with various components of the basal transcriptional machinery, such as TFIIB and certain of the TBP-associated factors (TAFs) [40–42]. For example, the glutamine-rich transactivation domain of the site-specific DNA binding factor Sp1 can interact directly with a specific subunit of TFIID (TAFII 130) and point mutations within the transactivation domain inhibit binding of TFIID and reduce activation of transcription [32, 33, 43]. The discovery that the glutamine-rich domains of Sp1 interact with TAFII 130, whereas several acidic and proline-rich transactivation domains do not interact with TAFII 130, provides support for the association of specific transactivation domains with specific general coactivators [33, 43, 44]. However, it should be noted that not all glutamine-rich domains interact with TAFII 130, highlighting the limitations in understanding TAD function that arise by grouping activation domains by their most common amino acids [43, 44]. Along these lines, mutational analysis of the glutamine-rich TAD of SP1 and the acidic-rich TADs of p53 and RelA revealed that the ability of these TADs to stimulate transcription is more sensitive to mutation of bulky hydrophobic amino acids than to the mutation of the glutamine or acidic amino acids that broadly define them [32, 45, 46]. Thus, the pattern of bulky hydrophobic residues may be more important than the more obvious features used to distinguish the classes of different activation domains.
More appropriately, eukaryotic TADs have been functionally grouped into those that stimulate initiation versus those that stimulate elongation, based on the different contacts they make with general transcription factors [47]. Prevailing models suggest that many activators act primarily at the level of transcription initiation [48]. However, contact between a TAD and the general transcriptional machinery can stimulate transcription not only by stabilizing the preinitiation complex (PIC), but can also facilitate promoter clearance and enhance the rate of elongation [45, 47, 49–55]. For example, in addition to stimulating transcriptional initiation, the activation domain of c-Myc also promotes transcription elongation through the recruitment of the RNA polymerase II Ser2 C-terminal domain (CTD) kinase called P-TEFb (positive transcription elongation factor b, which is composed of CycT1 and Cdk9) [55]. The c-Myc activation domain interacts directly with CycT1. Interestingly, the c-Myc transactivation domain can also increase mRNA cap maturation, polysome loading, and the rate of translation, processes that result from c-Myc-mediated phosphorylation of the RNA polymerase II CTD [56].
Structural studies of transactivation domains have revealed that many TADs are largely unstructured in solution [57]. For example, NMR studies have shown a lack of structure in the N-terminal region of p53 containing its acidic TAD [58, 59]. Further analysis revealed that specific motifs in the TAD fold into an α-helix upon binding to either the transcription initiation complex or to the p53 transcriptional attenuator Mdm2. Such studies propose that subdomains within the TADs become conformationally constrained upon interaction with a target protein [60–62]. Additionally, much evidence supports a structural and functional mechanism for the AF of hormone nuclear receptors that involves induced folding into an α-helical structure in response to protein–protein interactions and exposure to certain solutes [63–66]. These findings suggest that the target (i.e. a general transcription factor) is a template for the shaping of an unstructured TAD, allowing TADs to interact with numerous different components of the general transcriptional machinery. This mechanism creates a situation in which there is not a restricted relationship between certain general factors and specific types of TADs. Rather, TADs have evolved “flexible” ways to contact multiple components of the general machinery to activate transcription.
12.3 Effector Domains Can Interact with Other Site-Specific TFs
Another important type of effector domain that specifies TF functions is one that mediates direct interaction with other site-specific factors. Cooperative interactions between unrelated TFs expand the possibilities for extending DNA sequence recognition, perhaps allowing binding of a site-specific factor to a sequence not quite matching the preferred consensus motif. Additionally, the physical association of TFs at enhancers or promoters not only stabilizes weak protein–DNA interactions of one factor to the genome but also allows combinatorial regulation, an important mechanism that enables integration of different signaling pathways [67].
One type of protein–protein interaction between site-specific DNA binding factors is the obligate hetero-or homodimer. bZIP, bHLH, and certain nuclear hormone receptors are examples of TFs that form dimers at their target genes. In such cases, the protein–protein interactions generally form in solution with the dimeric complex binding to DNA as a preassembled unit. The members of the E2F family of TFs, which are involved in the regulation of the cell cycle and many other cellular processes, also function via heterodimerization. These factors possess a centrally-located DNA binding domain immediately followed by a dimerization domain, which allows interactions with an obligate dimerization partner (DP) protein that contains similar DNA binding and heterodimerization domains. Dimerization between DP and E2F is required for high-affinity, sequence-specific DNA binding [68]. Thus, the ability of E2F TFs to form dimers can determine the strength of the resultant protein–DNA interactions as well as confer an ability to regulate a variety of different target genes [69]. In contrast to the E2F/DP dimeric complexes that are mediated by similar domains in each partner, other heterodimeric TF complexes dimerize using two dissimilar domains. In such cases, a heterodimeric TF complex might then preferably recognize half-binding sites arranged in a head-to-tail configuration. Examples of such an arrangement have been shown to occur in vivo for heterodimers of the retinoic acid receptor with vitamin D3 receptors, peroxisome proliferator-activated receptors or thyroid hormone receptors (reviewed in [70]).
Other effector domains mediate the interaction of one site-specific factor with another site-specific factor only subsequent to a DNA binding event. This DNA-dependent mode of association suggests that the individual proteins are unable to interact in solution, perhaps due to a relatively low dimerization constant. It is thought that the binding of a site-specific factor to DNA may induce an allosteric change in the protein structure, which in turn increases its affinity for another site-specific factor. This has been shown to be the case for binding of hGRα to different DNA motifs that differ by as little as a single base pair and for the DNA–dependent interaction of specific thyroid hormone receptor isoforms with the retinoid X receptor at specific DNA motifs, each of which can differentially affect the conformation and activity of the factors in response to hormone [27, 71–75]. Therefore DNA can be a sequence-specific allosteric ligand that modifies the activity of a site-specific factor at certain target genes. An important example of DNA–mediated protein interaction comes from studies of the Oct4 and Sox2 proteins, which are critical TFs involved in regulating embryonic stem cell (ESC) self-renewal and pluripotency. Oct4 co-localizes with different sets of TFs at many genomic sites, including promoters and enhancers [76–79]. The Oct4 binding sites co-occupied by Sox2 correlate with the ESC-specific expression of the nearby genes. Oct4 and Sox2 have low affinity for each other in solution, yet this affinity is critical for the cooperative binding of Oct4 and Sox2 proteins to adjacent sites on DNA. Electrophoretic mobility shift assays indicate that the Sox2-Oct4 heterodimer forms more efficiently on specific composite elements than do the single proteins [80]. Thus, the effector domains in Oct4 and Sox2 that mediate this specific protein–protein interaction play crucial roles in ESC-specific transcriptional regulation.
12.4 Effector Domains Can Recruit Chromatin-Modifying Enzymes
In addition to general and site-specific TFs, there are other types of regulatory proteins recruited by TFs to target genes. Many of these so-called transcriptional co-regulators harbor enzymatic activities that assist in gene regulation through post-translational histone modification. Numerous different histone-modifying enzymes have been identified; in particular, HATs and histone methyltransferases (HMTs) are critically involved in setting up active chromatin regions. Protein–protein interactions between TFs and histone modifying enzymes appear to play a dominant role in eukaryotic gene regulation and may ultimately determine the transcriptional output of a given promoter. For example, histone acetylation is associated with open chromatin and gene activation whereas histone methylation can be associated with both activation (e.g. methylation of lysine 4 of histone H3) and repression (e.g. methylation of lysine 9 or lysine 27 of histone H3). Although some subunits of the basal transcriptional machinery encode HAT functions (e.g. TAF1), in many cases the histone modifying enzyme is a component of a large multi-protein complex (see [29, 81] for reviews).
Many different TFs co-purify with histone modifying enzymes, including ubiquitous factors such as E2F family members and cell type-specific nuclear receptors. E2F family members possess domains that mediate interactions with histone modifying complexes that confer either activation or repression. For example, E2F family members can interact directly with the histone acetyltransferases p300/CBP [82, 83] via their C terminal transactivation domain. The transcriptional coactivators p300 and CBP (CREB binding protein) are versatile transcriptional regulator proteins that are highly related in primary structure and have many overlapping functions (thus they are referred to as p300/CBP). p300/CBP is a promiscuous acetyltransferase in that it catalyzes the acetylation of lysines on all four core histones, as well as acetylating more than 70 non-histone proteins, including itself. p300/CBP proteins have multiple protein interaction domains as well as a bromodomain, which recognizes acetylated lysines, thus providing extra contacts to specific “active” regions of the genome. E2F family members also interact with repressive histone-modifying complexes. For example, E2F6 copurifies in a repression complex with euchromatic HMTases called GLP and G9a, both of which are implicated in methylation of lysine 9 of histone H3 [84]. E2F6 has also been shown to interact with polycomb group protein complexes that contain H3K27me3-specific histone methyltransferases [85, 86]. These studies suggest that E2F6 may function to silence E2F-responsive genes via formation of heterochromatin. However, other studies [87, 88] have shown that E2F6 can also repress transcription via mechanisms other than lysine 9 or lysine 27 methylation, indicating that E2F6 must also be involved in other types of repressive complexes. Other E2F family members (i.e. E2F1-5) can interact with repressive chromatin complexes through interaction of their transactivation domains with members of the retinoblastoma (Rb) tumor suppressor protein family. Rb proteins serve as a bridge between E2Fs and histone methyltransferases that target H4K20, DNA methyltransferases, histone deacetylases, and chromatin compaction complexes [89–96]. Thus, the same effector domain in an E2F protein can mediate both activation and repression; see Fig. 12.3.
Nuclear receptors can also recruit various sets of co-regulators and histone modifying enzymes to their DNA binding sites (also called hormone response elements or HREs) to modulate target gene transcription [97]. For example, liganded nuclear receptors recruit the p160/SRC family of proteins that, in turn, provide a scaffold for the recruitment of HATs, such as p300/CBP, HMTs, and histone arginine methyltransferases such as CARM1 and PRMT1. These enzymes covalently modify histone and non-histone proteins to permit changes in the chromatin architecture and to alter the assembly of transcriptional complexes (for reviews see [98, 99]). The p160/SRC proteins have been shown to interact directly with the AF2 activation domain of NRs via conserved LxxLL motifs (where L stands for leucine and × is any other amino acid) [97]. Other transcriptional regulatory proteins have also been shown to associate with p160/SRC intermediaries, including AP-1, Smad3, NF-κB, E2F1, Rb, and p53 [100–105], demonstrating the widespread use of a p160/SRC scaffold to build transcription complexes. Additionally, ATP-dependent chromatin remodeling complexes, including the SWI/SNF, ISWI, and WINAC complexes, are recruited to HREs through direct interactions with NRs in a hormone-dependent manner, where they play critical roles in regulating transcriptional activation through remodeling chromatin structure [106]. In addition to NRs, other sequence-specific activators, including AP-1, ELKF, C/EBPβ and c-Myc can interact with ATP-dependent chromatin remodeling complexes. Interestingly, histone acetylation has been shown to stabilize SWI/SNF binding to nucleosomes (several SWI/SNF subunits, including BRG1, BAF250, BAF60a, and BAF57, contain bromodomains which are known to bind acetylated histone tails). Thus, multiple interactions are likely involved in both the recruitment and stabilization of SWI/SNF to activator-bound target genes. Once these multi-subunit complexes are recruited by effector domains to the regulatory regions of target genes, nucleosomal rearrangement and further chromatin modifications such as histone acetylation occur, allowing the Mediator complex, the general TFs, and the RNAPII machinery access to the promoter region to activate transcription.
A subset of nuclear receptors, including Thyroid hormone Receptor (TR), Retinoic Acid Receptor (RAR), and the Vitamin D Receptor (VDR), can repress transcription in the absence of their ligands [107]. Repression mediated by NRs involves the direct association of specific corepressor complexes containing the NCoR and SMRT corepressors. Analogous to coactivator recruitment, corepressors interact with nuclear receptors via effector domains and assemble in large multi-protein complexes that possess distinct enzymatic activities. However, rather than facilitating an open chromatin structure, corepressors generate repressive chromatin through the actions of HMTs, HDACs, histone demethylases, and specific chromatin remodeling complexes, including NURD [97]. In general, the specific mechanisms that lead to chromatin compaction and transcriptional repression are not well understood. Interestingly, many of these repressive chromatin complexes are shared for a number of site-specific TFs involved in transcriptional repression. For example, the BTB/POZ (Broad complex, Tramtrack, Bric-a-brac/POxvirus and Zinc finger) effector domain, a highly conserved protein–protein interaction domain, has been shown to interact with NCoR and SMRT corepressors [108–111]. There are approximately 80 different human BTB/POZ-containing proteins, including PLZF, HIC-1, BCL-6, Kaiso, FAZF and LRF, suggesting that this effector domain may be widely utilized for transcriptional repression [112]. Similarly, the extremely large family of TFs containing the Krüppel associated box (KRAB) domain, of which there are over 300 different members, have been suggested to repress the transcription of specific genes via an interaction with the KAP1 corepressor protein [113]. In turn, the KAP1 corepressor functions as a scaffold to recruit heterochromatin protein 1 (HP1) isoforms, histone deacetylases, and SETDB1, a SET-domain histone methyltransferase that methylates histone H3 at lysine 9 [23, 114, 115]. This modification is associated with closed chromatin and therefore KRAB effector domains of KRAB-zinc finger proteins link the KAP1 corepressor complex to specific genomic sites and silence gene expression by forming a facultative heterochromatin environment [116]. Due to the extremely large number of KRAB- zinc finger proteins, the KRAB domain may turn out to be one of the most commonly used effector domains involved in repression.
12.5 Summary
The molecular framework involved in transcription initiation consists of a multitude of cellular factors. A deep understanding of transcriptional regulation requires a detailed knowledge of the structural lattice in which TFs and co-regulators build hierarchical protein assemblies that provide control and specificity to transcriptional programs. The laths that link transcriptional regulators to their ultimate genomic targets are composed of a series of protein–protein interactions that recruit and confine transcriptional proteins to an appropriate regulatory location. Thus, knowledge of protein domains that serve as the biological effectors to recruit chromatin-modifying and nucleotide-synthesizing enzymes is critical for understanding how a cell type-specific transcriptome is established. A comprehensive cataloging of effector domains encoded in the human genome is beyond the scope of this review. However, we have provided examples of common effector domains utilized in eukaryotic transcriptional regulation. We suggest that researchers query the pfam website to identify conserved domains in specific TFs (http://pfam.janelia.org/; see also [117]). Although there are as many ways to regulate transcription as there are genes, several unifying themes can be derived from the many years of study of transcriptional regulation. These include:
Effector domains can mediate gene activation or repression by promoting the formation of active or repressed chromatin, by interacting with domains in other factors to form “platforms” for recruitment of co-regulatory proteins, or by stimulating or inhibiting preinitiation complex formation or productive elongation (Fig. 12.1).
Effector domains can be brought to DNA in multiple ways, including as a modular domain of a site-specific DNA binding factor or as a domain or interacting partner with a protein that binds to methylated DNAs or modified histones (Fig. 12.2).
There is not a one-to-one relationship between an effector domain and a specific co-regulatory protein; rather, many effector domains can interact with the same general factor, coregulator, or histone modifying complex and a single effector domain can interact with multiple other proteins, including proteins involved in both activation and repression (Fig. 12.3).
References
- 1.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nature Rev Genet. 2009;(10):669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laird PW, Jaenisch R. DNA methylation and cancer. Human Molecular Genetics. 1994;3:1487–1495. doi: 10.1093/hmg/3.suppl_1.1487. [DOI] [PubMed] [Google Scholar]
- 3.Farnham PJ. Insights from genomic profiling of transcription factors. Nature Rev Genet. 2009;(10):605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Reviews Genetics. 2009;10(4):252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 6.Keegan L, Gill G, Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- 7.Lin YS, Carey MF, Ptashne M, Green MR. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988;54(5):659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- 8.Brent R, Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985;43(3 Pt 2):729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- 9.Sasai N, Defossez PA. Many paths to one goal? The proteins that recognize methylated DNA in eukaryotes. Int J Dev Biol. 2009;53(2–3):323–334. doi: 10.1387/ijdb.082652ns. [DOI] [PubMed] [Google Scholar]
- 10.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylases to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 11.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393(6683):386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 12.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28(2):69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 13.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25(1):15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 14.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14(11):1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410(6824):120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 16.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410(6824):116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 17.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127(7):1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7(4):397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26(37):5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- 20.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frietze S, O’Geen H, Blahnik KR, Jin VX, Farnham PJ. ZNF274 recruits the histone methyltransferase SETDB1 to the 3′ ends of ZNF genes. PLoS One. 2010;5(12):e15082. doi: 10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang X-P, Neilson EG, et al. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes & Dev. 1996;10(16):2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 23.Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol. 2006;26(22):8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavery DN, McEwan IJ. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem J. 2005;391(Pt 3):449–464. doi: 10.1042/BJ20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falkner KC, Pinaire JA, Xiao GH, Geoghegan TE, Prough RA. Regulation of the rat glutathione S-transferase A2 gene by glucocorticoids: involvement of both the glucocorticoid and pregnane X receptors. Mol Pharmacol. 2001;60(3):611–619. [PubMed] [Google Scholar]
- 26.Szapary D, Huang Y, Simons SS., Jr. Opposing effects of corepressor and coactivators in determining the dose-response curve of agonists, and residual agonist activity of antagonists, for glucocorticoid receptor-regulated gene expression. Mol Endocrinol. 1999;13(12):2108–2121. doi: 10.1210/mend.13.12.0384. [DOI] [PubMed] [Google Scholar]
- 27.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324(5925):407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41(3):105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 29.Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 30.Cujec TP, Cho H, Maldonado E, Meyer J, Reinberg D, Peterlin BM. The human immunodeficiency virus transactivator Tat interacts with the RNA polymerase II holoenzyme. Mol Cell Biol. 1997;17(4):1817–1823. doi: 10.1128/mcb.17.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fry CJ, Slansky JE, Farnham PJ. Position-dependent transcriptional regulation of the murine dihydrofolate reductase promoter by the E2F transactivation domain. Mol Cell Biol. 1997;17(4):1966–1976. doi: 10.1128/mcb.17.4.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gill G, Pascal E, Tseng ZH, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA. 1994;91(1):192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodrich JA, Hoey T, Thut C, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75(3):519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 34.Horikoshi M, Hai T, Lin YS, Green MR, Roeder RG. Transcription factor ATF interacts with the TATA factor to facilitate establishment of a preinitiation complex. Cell. 1988;54(7):1033–1042. doi: 10.1016/0092-8674(88)90118-3. [DOI] [PubMed] [Google Scholar]
- 35.Kashanchi F, Piras G, Radonovich MF, Duvall JF, Fattaey A, Chiang CM, et al. Direct interaction of human TFIID with the HIV-1 transactivator tat. Nature. 1994;367(6460):295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- 36.Lin Y-S, Ha I, Maldonado E, Reinberg D, Green MR. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991;353(6344):569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 37.Roberts SG, Choy B, Walker SS, Lin YS, Green MR. A role for activator-mediated TFIIB recruitment in diverse aspects of transcriptional regulation. Curr Biol. 1995;5(5):508–516. doi: 10.1016/s0960-9822(95)00103-5. [DOI] [PubMed] [Google Scholar]
- 38.Stringer KF, Ingles CJ, Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990;345(6278):783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 39.Zhu H, Joliot V, Prywes R. Role of transcription factor TFIIF in serum response factor-activated transcription. J Biol Chem. 1994;269(5):3489–3497. [PubMed] [Google Scholar]
- 40.Kim TK, Roeder RG. Proline-rich activator CTF1 targets the TFIIB assembly step during transcriptional activation. Proc Natl Acad Sci U S A. 1994;91(10):4170–4174. doi: 10.1073/pnas.91.10.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiang C-M, Roeder RG. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267(5197):531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 42.Tanese N, Pugh BF, Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991;5:2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- 43.Hoey T, Weinzierl RO, Gill G, Chen JL, Dynlacht BD, Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72(2):247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 44.Chen J-L, Attardi DL, Verrijzer CP, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79(1):93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 45.Blair WS, Bogerd HP, Madore SJ, Cullen BR. Mutational analysis of the transcription activation domain of RelA: identification of a highly synergistic minimal acidic activation module. Mol Cell Biol. 1994;14(11):7226–7234. doi: 10.1128/mcb.14.11.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes & Dev. 1994;8(10):1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 47.Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domains. Mol Cell Biol. 1996;16(5):2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choy B, Green MR. Eukaryotic activators function during multiple steps of preinitiation complex assembly. Nature. 1993;366(6455):531–536. doi: 10.1038/366531a0. [DOI] [PubMed] [Google Scholar]
- 49.Krumm A, Hickey LB, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 1995;9(5):559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 50.Mahanta SK, Scholl T, Yang FC, Strominger JL. Transactivation by CIITA, the type II bare lymphocyte syndrome-associated factor, requires participation of multiple regions of the TATA box binding protein. Proc Natl Acad Sci U S A. 1997;94(12):6324–6329. doi: 10.1073/pnas.94.12.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yankulov K, Blau J, Purton T, Roberts S, Bentley DL. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77(5):749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 52.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461(7261):186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selth LA, Sigurdsson S, Svejstrup JQ. Transcript Elongation by RNA Polymerase II. Annu Rev Biochem. 2010;79:271–293. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- 55.Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J Biol Chem. 2002;277(42):40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- 56.Cowling VH, Cole MD. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol Cell Biol. 2007;27(6):2059–2073. doi: 10.1128/MCB.01828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J Mol Biol. 2006;359(4):1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 58.Ayed A, Mulder FA, Yi GS, Lu Y, Kay LE, Arrowsmith CH. Latent and active p53 are identical in conformation. Nat Struct Biol. 2001;8(9):756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- 59.Dawson R, Muller L, Dehner A, Klein C, Kessler H, Buchner J. The N-terminal domain of p53 is natively unfolded. J Mol Biol. 2003;332(5):1131–1141. doi: 10.1016/j.jmb.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 60.Chi SW, Lee SH, Kim DH, Ahn MJ, Kim JS, Woo JY, et al. Structural details on mdm2-p53 interaction. J Biol Chem. 2005;280(46):38795–38802. doi: 10.1074/jbc.M508578200. [DOI] [PubMed] [Google Scholar]
- 61.Uesugi M, Verdine GL. The alpha-helical FXXPhiPhi motif in p53: TAF interaction and discrimination by MDM2. Proc Natl Acad Sci U S A. 1999;96(26):14801–14806. doi: 10.1073/pnas.96.26.14801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garza AS, Ahmad N, Kumar R. Role of intrinsically disordered protein regions/domains in transcriptional regulation. Life Sci. 2009;84(7–8):189–193. doi: 10.1016/j.lfs.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 63.Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389(6652):753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 64.Kumar R, Betney R, Li J, Thompson EB, McEwan IJ. Induced alpha-helix structure in AF1 of the androgen receptor upon binding transcription factor TFIIF. Biochemistry. 2004;43(11):3008–3013. doi: 10.1021/bi035934p. [DOI] [PubMed] [Google Scholar]
- 65.Pike AC, Brzozowski AM, Hubbard RE, Bonn T, Thorsell AG, Engstrom O, et al. Structure of the ligand-binding domain of oestrogen receptor beta in the presence of a partial agonist and a full antagonist. EMBO J. 1999;18(17):4608–4618. doi: 10.1093/emboj/18.17.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar R, Litwack G. Structural and functional relationships of the steroid hormone receptors’ N-terminal transactivation domain. Steroids. 2009;74(12):877–883. doi: 10.1016/j.steroids.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panne D. The enhanceosome. Curr Opin Structural Biol. 2008;18(2):236–242. doi: 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 68.Huber HE, Edwards G, Goodhart PJ, Patrick DR, Huang PS, Ivey-Hoyle M, et al. Transcription factor E2F binds as a heterodimer. Proc Natl Acad Sci USA. 1993;90(8):3525–3529. doi: 10.1073/pnas.90.8.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Helin K, Wu C-L, Fattaey AR, Lees JA, Dynlacht BD, Ngwu C, et al. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 1993;7(10):1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- 70.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83(6):841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 71.Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17(16):2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Remenyi A, Tomilin A, Scholer HR, Wilmanns M. Differential activity by DNA-induced quarternary structures of POU transcription factors. Biochem Pharmacol. 2002;64(5-6):979–984. doi: 10.1016/s0006-2952(02)01164-4. [DOI] [PubMed] [Google Scholar]
- 73.Kitayner M, Rozenberg H, Rohs R, Suad O, Rabinovich D, Honig B, et al. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat Struct Mol Biol. 2010;17(4):423–429. doi: 10.1038/nsmb.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leng X, Blanco J, Tsai SY, Ozato K, O’Malley BW, Tsai MJ. Mechanisms for synergistic activation of thyroid hormone receptor and retinoid X receptor on different response elements. J Biol Chem. 1994;269(50):31436–31442. [PubMed] [Google Scholar]
- 75.Reginato MJ, Zhang J, Lazar MA. DNA-independent and DNA-dependent mechanisms regulate the differential heterodimerization of the isoforms of the thyroid hormone receptor with retinoid X receptor. J Biol Chem. 1996;271(45):28199–28205. doi: 10.1074/jbc.271.45.28199. [DOI] [PubMed] [Google Scholar]
- 76.Squazzo SL, Komashko VM, O’Geen H, Krig S, Jin VX, Jang S-W, et al. Suz12 silences large regions of the genome in a cell type-specific manner. Genome Research. 2006;16(7):890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jin VX, O’Geen H, Iyengar S, Green R, Farnham PJ. Identification of an OCT4 and SRY regulatory module using integrated computational and experimental genomics approaches. Genome Research. 2007;17(6):807–817. doi: 10.1101/gr.6006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mathur D, Danford TW, Boyer LA, Young RA, Gifford DK, Jaenisch R. Analysis of the mouse embryonic stem cell regulatory networks obtained by ChIP-chip and ChIP-PET. Genome Biol. 2008;9(8):R126. doi: 10.1186/gb-2008-9-8-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133(6):1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 80.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, et al. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 2005;280(26):24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 81.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 82.Trouche D, Cook A, Kouzarides T. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res. 1996;24(21):4139–4145. doi: 10.1093/nar/24.21.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fry CJ, Pearson A, Malinowski E, Bartley SM, Greenblatt J, Farnham PJ. Activation of the murine dihydrofolate reductase promoter by E2F1: A requirement for CBP recruitment. J Biol Chem. 1999;274(22):15883–15891. doi: 10.1074/jbc.274.22.15883. [DOI] [PubMed] [Google Scholar]
- 84.Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296(5570):1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 85.Trimarchi JM, Fairchild B, Wen J, Lees JA. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc Natl Acad Sci U S A. 2001;95(6):2850–2855. doi: 10.1073/pnas.041597698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Attwooll C, Oddi S, Cartwright P, Prosperini E, Agger K, Steensgaard P, et al. A novel repressive E2F6 complex containing the polycomb group protein, EPC1, that interacts with EZH2 in a proliferation-specific manner. J Biol Chem. 2005;280(2):1199–1208. doi: 10.1074/jbc.M412509200. [DOI] [PubMed] [Google Scholar]
- 87.Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, et al. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals iterchangeable roles of E2F family members. Genome Res. 2007;17(11):1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oberley MJ, Inman D, Farnham PJ. E2F6 negatively regulates BRCA1 in human cancer cells without methylation of histone H3 on lysine 9. J Biol Chem. 2003;278(43):42466–42476. doi: 10.1074/jbc.M307733200. [DOI] [PubMed] [Google Scholar]
- 89.Vandel L, Nicolas E, Vaute O, Ferreira R, Ait-si-ali S, Trouche D. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol Cell Biol. 2001;21(19):6484–6494. doi: 10.1128/MCB.21.19.6484-6494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7(4):420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- 91.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. nature genetics. 2000;25(3):338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 92.Pradhan S, Kim G-D. The retinoblastoma gene product interacts with maintenance human DNA (cytosine-5) methyltransferase and modulates its activity. EMBO J. 2002;21:779–788. doi: 10.1093/emboj/21.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Trojer P, Li G, Sims RJ, 3rd, Vaquero A, Kalakonda N, Boccuni P, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129(5):915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 94.Trojer P, Reinberg D. Beyond histone methyl-lysine binding: how malignant brain tumor (MBT) protein L3MBTL1 impacts chromatin structure. Cell Cycle. 2008;7(5):578–585. doi: 10.4161/cc.7.5.5544. [DOI] [PubMed] [Google Scholar]
- 95.Trojer P, Zhang J, Yonezawa M, Schmidt A, Zheng H, Jenuwein T, et al. Dynamic Histone H1 Isotype 4 Methylation and Demethylation by Histone Lysine Methyltransferase G9a/KMT1C and the Jumonji Domain-containing JMJD2/KDM4 Proteins. J Biol Chem. 2009;284(13):8395–8405. doi: 10.1074/jbc.M807818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Longworth MS, Dyson NJ. pRb, a local chromatin organizer with global possibilities. Chromosoma. 2010;119(1):1–11. doi: 10.1007/s00412-009-0238-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20(11):1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 98.Lee YH, Stallcup MR. Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol. 2009;23(4):425–433. doi: 10.1210/me.2008-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41(1):185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 100.Lee SK, Kim HJ, Na SY, Kim TS, Choi HS, Im SY, et al. Steroid receptor coactivator-1 coactivates activating protein-1-mediated transactivations through interaction with the c-Jun and c-Fos subunits. J Biol Chem. 1998;273(27):16651–16654. doi: 10.1074/jbc.273.27.16651. [DOI] [PubMed] [Google Scholar]
- 101.Li G, Heaton JH, Gelehrter TD. Role of steroid receptor coactivators in glucocorticoid and transforming growth factor beta regulation of plasminogen activator inhibitor gene expression. Mol Endocrinol. 2006;20(5):1025–1034. doi: 10.1210/me.2005-0145. [DOI] [PubMed] [Google Scholar]
- 102.Gao Z, Chiao P, Zhang X, Lazar MA, Seto E, Young HA, et al. Coactivators and corepressors of NF-kappaB in IkappaB alpha gene promoter. J Biol Chem. 2005;280(22):21091–21098. doi: 10.1074/jbc.M500754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Louie MC, Zou JX, Rabinovich A, Chen HW. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol. 2004;24(12):5157–5171. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Batsche E, Desroches J, Bilodeau S, Gauthier Y, Drouin J. Rb enhances p160/SRC coactivator-dependent activity of nuclear receptors and hormone responsiveness. J Biol Chem. 2005;280(20):19746–19756. doi: 10.1074/jbc.M413428200. [DOI] [PubMed] [Google Scholar]
- 105.Lee SK, Kim HJ, Kim JW, Lee JW. Steroid receptor coactivator-1 and its family members differentially regulate transactivation by the tumor suppressor protein p53. Mol Endocrinol. 1999;13(11):1924–1933. doi: 10.1210/mend.13.11.0365. [DOI] [PubMed] [Google Scholar]
- 106.Belandia B, Parker MG. Nuclear receptors: a rendezvous for chromatin remodeling factors. Cell. 2003;114(3):277–280. doi: 10.1016/s0092-8674(03)00599-3. [DOI] [PubMed] [Google Scholar]
- 107.Privalsky ML. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- 108.Melnick A, Carlile G, Ahmad KF, Kiang CL, Corcoran C, Bardwell V, et al. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol Cell Biol. 2002;22(6):1804–1818. doi: 10.1128/MCB.22.6.1804-1818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell. 2003;12(3):723–734. doi: 10.1016/j.molcel.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 110.Muto A, Hoshino H, Madisen L, Yanai N, Obinata M, Karasuyama H, et al. Identification of Bach2 as a B-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3′ enhancer. EMBO J. 1998;17(19):5734–5743. doi: 10.1093/emboj/17.19.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dhordain P, Albagli O, Lin RJ, Ansieau S, Quief S, Leutz A, et al. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc Natl Acad Sci U S A. 1997;94(20):10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kelly KF, Daniel JM. POZ for effect–POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16(11):578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 113.Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4(10):231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16(8):919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schultz DC, Friedman JR, Rauscher F., Jr Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PhD and bromodomains of KAP-1 form a coopeative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15(4):428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Denervaud N, et al. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6(3):e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fry CJ, Farnham PJ. Context-dependent transcriptional regulation. J Biol Chem. 1999;274(42):29583–29586. doi: 10.1074/jbc.274.42.29583. [DOI] [PubMed] [Google Scholar]