Abstract
Optogenetics allows the manipulation of neural activity in freely moving animals with millisecond precision, but its application in Drosophila has been limited. Here we show that a recently described Red activatable Channelrhodopsin (ReaChR) permits control of complex behavior in freely moving adult flies, at wavelengths that are not thought to interfere with normal visual function. This tool affords the opportunity to control neural activity over a broad dynamic range of stimulation intensities. Using time-resolved activation, we show that the neural control of male courtship song can be separated into probabilistic, persistent and deterministic, command-like components. The former, but not the latter, neurons are subject to functional modulation by social experience, supporting the idea that they constitute a locus of state-dependent influence. This separation is not evident using thermogenetic tools, underscoring the importance of temporally precise control of neuronal activation in the functional dissection of neural circuits in Drosophila.
Drosophila melanogaster is one of the most powerful model organisms available for the genetic dissection of neural circuit function1,2. Likewise, the use of light-sensitive microbial opsins, such as channelrhodopsin, has revolutionized the functional dissection of neural circuits in behaving animals3,4. Unfortunately, with the exception of larval neurons and peripheral sensory neurons in adults5-13 this powerful technology and model organism have been largely incompatible in the case of adult flies (but see refs10,13). Therefore Drosophila researchers have, to a large extent, been unable to exploit the rapidly expanding optogenetic toolkit for neural circuit manipulation. Although P2X2, an ionotropic purinergic receptor, has been used as an optogenetic tool in adult Drosophila, this technique requires injection of caged ATP into the brains of individual anesthetized flies14. This relatively invasive technology is sub-optimal for many applications, especially large-scale, high-throughput screening.
In the absence of facile optogenetic manipulation, dTRPA1, a thermosensitive cation channel, has been the preferred method for neuronal activation in freely behaving adult flies1,15. Since this method depends on changes in temperature to control neuronal activity, however, it lacks precision in both the temporal and intensity domains and suffers from potentially confounding influence of temperature changes on behavior.
Here, we demonstrate that expression of ReaChR in adult central nervous system (CNS) neurons enables rapid and temporally precise control of behavior in freely moving adult Drosophila. Using this optogenetic method, we have separated the control of wing extension, a male-specific courtship behavior, into probabilistic, state-dependent and deterministic, command-like components. Moreover, by combining ReaChR activation with functional calcium imaging, we have also identified a neural correlate of the influence of social experience on male courtship behavior.
Results
Optogenetic vs. thermogenetic control of gustatory neurons
We reasoned that previously described ChR2 variants do not work well in adult Drosophila due, at least in part, to low penetrance of blue light through the cuticle. Indeed, direct measurements in vivo indicated that the penetrance of blue light through the cuticle is much weaker (c.a. 1 %) than that of longer wavelengths such as green or red light (5–10 %) (Fig. 1a). Therefore, we created transgenic flies that express the recently developed red-shifted channelrhodopsins, C1V1(T/T)16 and ReaChR17 under the control of the Gal4-UAS system, to test whether red shifted light can penetrate the cuticle sufficiently to activate neurons expressing these channels (see Supplementary Table 1 for a listing of all transgenic fly strains created).
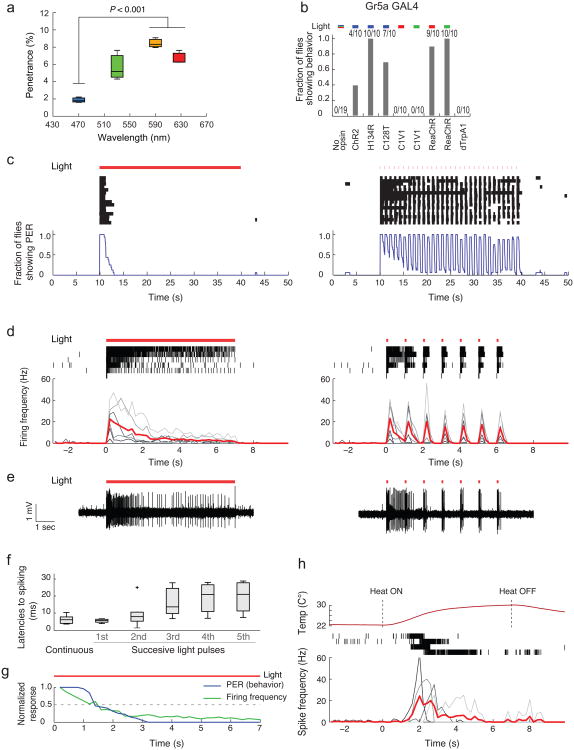
Figure 1. Optogenetic vs. thermogenetic control of Gr5a GRNs.
(a) Penetrance of light through the adult cuticle. n = 3. P-value represents one-way ANOVA (P = 0.0046) followed by t-test with Bonferroni correction (b) Fraction of flies showing PER triggered by different opsins expressed in Gr5a GRNs. Fractions at top indicate number of responders/number of flies tested. Activation wavelengths are represented as blue (470 nm), green (530 nm), red (627 nm) bars. For no opsin control, all the wavelengths were tested. (c) Behavioral and (d, e) electrophysiological responses of flies expressing ReaChR in Gr5a GRNs. Red lines (c–e): photostimulation pattern (627 nm, 1.1 mW/mm2); pulsed photostimulation (right) was delivered at 1 Hz (100 msec pulse width). Raster plots (c): PER bouts. Blue curves (c): Fraction of flies showing PER (time bins: 1sec; n = 16). Raster plots (d): Gr5a GRNs spikes. Lower plots: Average spiking rate (red lines); spiking rates for individual flies are overlaid (gray lines; time bins: 200 msec; n = 6). (e) Sample traces. (f) Latencies to first spike following photostimulation onset, from (d). Boxplots: whiskers represent 1.5 inter-quartile range of the lower and upper quartiles; boxes indicate lower quartile, median, and upper quartile, from bottom to top. (g) Overlay of normalized PER and firing frequencies during continuous photostimulation based on (c, d). (h) Top row: Measured temperature change caused by a heat source placed near the labellum. Middle row: Raster plots representing spikes in Gr5a GRNs expressing dTrpA1. Bottom row: spiking responses plotted as in (d). n = 4.
We first compared the efficacy of different opsins to activate sugar-sensing gustatory receptor neurons (GRNs) that express the receptor Gr5a18. Optogenetic activation of Gr5a neurons using channelrhodopsin-2 (ChR2) has previously been shown to trigger the proboscis extension reflex (PER) in Drosophila6,12 (Supplementary Video 1). All of the blue light-sensitive opsin variants tested (ChR219,20, H134R21 and C128T22) induced PER behavior in response to photostimulation at 470 nm, although only H134R yielded responses in 100 % of flies (Fig. 1b). Flies expressing ReaChR in Gr5a GRNs yielded robust PER responses to both red (627 nm) and green (530 nm) light. In contrast, flies expressing C1V1(T/T) did not exhibit PERs in response to either red or green light (Fig. 1b). Instead, they moved their proboscis slightly, albeit in a manner time-locked to photostimulation, suggesting that C1V1(T/T) has only a weak ability to activate Gr5a GRNs.
Surprisingly, in flies expressing dTrpA1 in Gr5a GRNs, we did not observe any behavioral response at an ambient temperature known to activate the ion channel (32 °C)8,15 (Fig. 1b), or during gradual ramping to this temperature from 22 °C (data not shown). Interestingly, activation of dTrpA1 in Gr5a GRNs using heat pulses from an IR laser23 has been reported to induce a PER. Upon continuous current injection some neurons develop a depolarization block24. We reasoned that if Gr5a neurons are continuously or gradually activated via TrpA1, they may undergo a rapid depolarization block that prevents PER behavior. Consistent with this idea, continuous illumination of Gr5a-ReaChR flies produced only a transient PER reaction (half-time for decay = 1.5 sec), while pulsatile illumination (1 Hz, 100 msec pulse duration) evoked a train of PERs time-locked to each light pulse (Fig. 1c).
Consistent with this, electrophysiological recording of Gr5a GRNs revealed that pulsed light caused continuous bursts of spiking throughout the stimulation period (Fig. 1d, e) with short latencies (Fig. 1f). In contrast, spiking activity decayed exponentially during continuous light stimulation (half-time for decay, ∼1.5 sec; Fig. 1d, e). The rapid decay of both spiking and PER behavior during continuous activation of ReaChR (Fig. 1g; Pearson's correlation coefficient: r = 0.96), suggests that the former likely accounts for the latter.
Similar to the results obtained using continuous ReaChR activation, TrpA1 activation triggered only transient spiking in Gr5a GRNs, with a strong decay after several seconds (Fig. 1h). Together, these data may explain why PER responses were not induced by constitutive or gradual thermal activation in Gr5a-TrpA1-expressing flies (Fig. 1b). They also reconfirm the importance of pulsed activation of neurons to avoid depolarization block, as reported previously in other systems4 (but note that depolarization block does not occur in all neuronal subtypes8).
Activation of CNS neurons with ReaChR
Only a few studies have reported successful elicitation of behavior in adult Drosophila by activating CNS neurons expressing blue light-sensitive opsins10,13. To determine whether activation using ReaChR would be more effective, we directly compared the behavioral responses of flies expressing blue light- vs. red light- sensitive opsins in GAL4 lines driving expression in different populations of CNS neurons. These lines included: HB9-GAL425 whose activation induces side walking (Supplementary Video 2) and, at higher intensities, paralysis (loss of postural control and immobility); Corazonin (Crz)-GAL4 whose activation induces abdominal bending and ejaculation26 (Supplementary Video 3); Fru-GAL427, which labels ∼1,500 neurons throughout the brain and whose activation in males induces mating behavior including wing extension28, and abdominal bending; at higher intensities, paralysis is observed (Supplementary Video 4); and “P1-GAL4,” a split-GAL429,30 driver generated from parental GAL4 lines31 identified in a behavioral screen (E.D.H. and D.J.A., unpublished), that is specifically expressed in ∼16–20 male-specific P1 neurons, activation of which elicits wing extension in males in the absence of females28,32. To facilitate the control and monitoring of light-induced behaviors in freely moving adult flies in a high-throughput, cost-effective and flexible manner, we developed a high power LED-based activation system (Fig. 2a–c; Supplementary Fig. 1a–e, Table 2, Software and Online Methods).
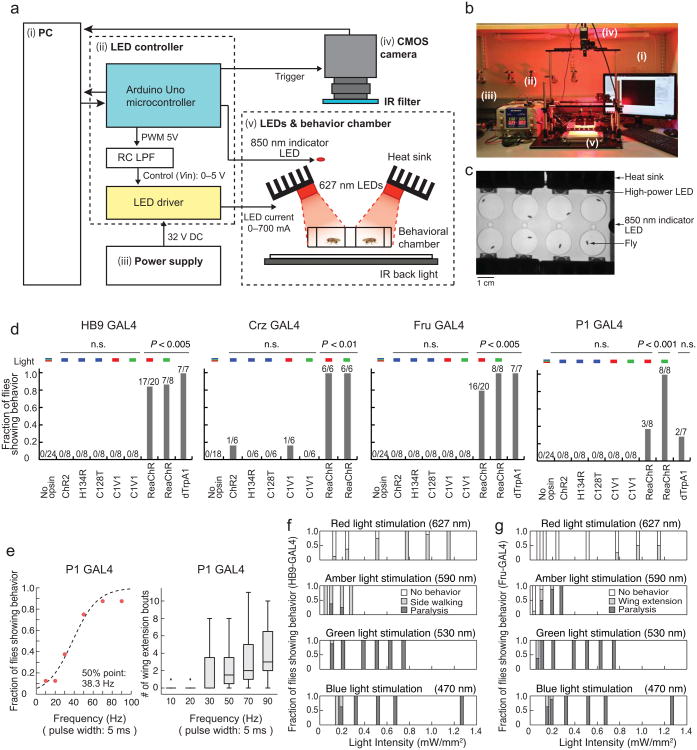
Figure 2. ReaChR enables light-dependent activation of the CNS neurons in Drosophila.
(a-b) Experimental setup for high power LED-based activation system. Each number in the diagram (a) corresponds to a number in the photograph (b). See Supplementary Fig.1 and Table 2 for detail. (c) View from the CMOS camera. (d) Comparison of behavioral responses of flies expressing different channelrhodopsin variants in distinct CNS subpopulations. Plot properties as in Fig. 1b. “Fraction of flies showing behavior” indicates: side walking or knock-out phenotype (HB9 GAL4); ejaculation (Crz GAL4); wing extension or knock-out (Fru GAL4); or wing extension (P1 GAL4). No opsin: empty promoter GAL4 (BFP-GAL4. See Online Methods) crossed with UAS-ReaChR. Flies showing any of the characteristic behaviors during 1 min of continuous photostimulation were scored as responders. P values represent Fisher's exact test with Bonferroni correction (comparing No-opsin vs. each column). (e) ReaChR-mediated activation of P1 neurons using different frequencies of red light pulses (627 nm, 1.1 mW/mm2, 1 min) (P1-GAL4; UAS-ReaChR (attP40)). The fraction of flies showing wing extension during 1 min photostimulation trials was fitted by a sigmoidal function to calculate the 50% point. n = 8. (f–g) Fraction of flies exhibiting characteristic behaviors at different photostimulation intensities and wavelengths, in animals expressing HB9 GAL4 (f) or Fru GAL4 (g), and UAS-ReaChR. n = 8.
Strikingly, among all 5 opsins tested using these CNS drivers, ReaChR was the only one whose activation yielded robust behavioral phenotypes in a light-dependent manner (Fig. 2d). The evoked behaviors were not due to innate responses to light, because control flies lacking UAS-ReaChR did not exhibit them in response to all the wavelengths tested (Fig. 2d). The fact that blue-light activated opsins yielded a behavioral response (PER) when expressed in GRNs, but not in the CNS neurons tested here, likely reflects the fact that the peripheral GRNs are located close to the cuticle, where blue light may penetrate more easily. Analysis of C1V1(T/T) expression in CNS neurons revealed that this opsin is expressed weakly in cell somata and not trafficked to arborizations, while ReaChR is strongly expressed in somata and trafficked to arborizations as well (Supplementary Fig. 2a). This difference likely accounts for the different efficacies of the two red-shifted opsins in this system.
The peak of the ReaChR action spectrum (measured in cultured hippocampal neurons) is ∼590 nm17. The efficacy of ReaChR activation by different wavelengths in freely behaving flies will, however, reflect a combination of factors including cuticular penetration and intensity, as well as proximity to peak sensitivity. To empirically determine the optimal wavelength of light for behavioral assays, therefore, we compared the ability of blue (470 nm), green (530 nm), amber (590 nm) and red (627 nm) light to induce behavior in flies expressing ReaChR under the control of different CNS GAL4 drivers. When not normalized for intensity, green LEDs had the strongest capacity to elicit ReaChR-dependent behaviors (Fig. 2d, f, g). In some cases (pIP10 neurons; see below), robust behavioral responses were detected only using green light, and hardly at all using other wavelengths. Although amber light is closest to the peak of the ReaChR action spectrum, commercial LEDs of this wavelength are dimmer than the others and therefore did not elicit strong behavioral responses (Fig. 2f, g).
Although TrpA1-mediated activation of P1 neurons can elicit wing extension28,32, in our direct comparison the fraction of solitary male flies showing a wing extension phenotype was much higher using ReaChR and green light, than using TrpA1 (Fig. 2d). This suggests that the intensity of activation obtained using ReaChR (and green light) can be substantially stronger than that achieved using dTrpA1, without subjecting flies to the high temperatures necessary to activate the latter. Nevertheless, although green LEDs elicited the strongest behavioral responses, flies can see this wavelength, whereas their sensitivity to wavelengths > 620 nm is much lower 33,34 (see, however, Hanai (2008)35). Therefore we used red LEDs whenever possible to avoid behavioral artifacts caused by strong visual stimulation.
To investigate whether the strength of a given ReaChR-dependent behavioral phenotype can be quantitatively tuned, we tested multiple frequencies and intensities of light pulses using the P1-GAL4 driver (pulse width: 5 msec). There was a frequency-dependent increase in the fraction of flies showing wing extension, as well as in the average number and duration of wing extension bouts per fly (Fig. 2e and Supplementary Fig. 2e), even when correcting for the total duration of illumination (Supplementary Fig. 2f). The HB9 and Fru-GAL4 drivers also yielded an increase in the fraction of flies showing the respective behavioral responses as the intensity was increased, albeit over different ranges (Fig. 2f, g). Together, these data indicate that ReaChR can be used to tune behavioral phenotypes by varying the light intensity and/or pulse frequency, over a relatively broad dynamic range.
Probabilistic vs. deterministic control of wing extension
Previous studies of the neural circuitry underlying male courtship behavior in Drosophila have used neuronal activation methods, including P2X2 and TrpA1, to identify different neuronal subclasses that control courtship song28,32,36,37. In particular, studies using TrpA1 have described two neuronal classes in the central brain controlling this behavior: one, called P1 or pMP4, constitutes a population of interneurons28,32,37, while the other, called pIP10, constitutes a small group of descending neurons that project to the ventral nerve cord (VNC)28 (Fig. 3b). The pre-synaptic terminals of P1 neurons overlap the dendrites of pIP10 neurons, suggesting that they may be synaptic partners28; however the difference, if any, between the roles of these neurons in controlling courtship song has not been apparent, as similar behaviors are evoked by TrpA1-mediated stimulation of both classes28.
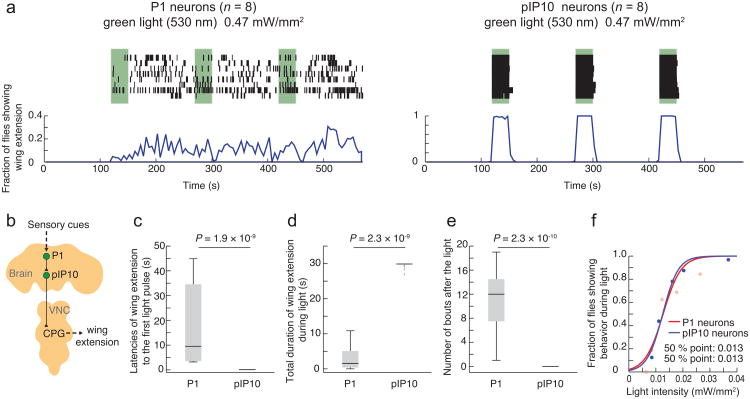
Figure 3. Probabilistic vs. deterministic optogenetic control of courtship song.
(a) Activation of P1 neurons (P1-Gal4; UAS-ReaChR(VK5)) (left) and pIP10 neurons (VT40556/UAS>stop>ReaChR (attP40); fru-FLP) (right) with green light (530 nm, 0.47 mW/mm2). Top: Raster plot representing wing extension bouts (n = 8 flies per genotype). Green bars represent 30 sec continuous photostimulation trials with 120 sec inter-trial intervals. Bottom: Fraction of flies showing wing extension (time bins: 5 sec). Note different y-axis scales in (a, left) and (a, right). P1 responses during trials 2 and 3 are more clearly phased to the onset of photostimulation at lower light intensities (Supplementary Fig. 3a). (b) Schematic illustrating neuronal circuit control of courtship song, simplified from ref28. (c) Latency to first wing extension after onset of the first photostimulation. (d) Total duration of wing extension during photostimulation. (e) Number of wing extension bouts during 30 sec following photostimulation offset. Plots in (c–e) are based on data in (a). P-values represent Mann-Whitney U tests. (f) Fraction of flies showing wing extension during a single photostimulation trial as a function of light intensity (green light: 530 nm, continuous, 30 sec). The data were fitted by a sigmoidal function to calculate the 50% point. n = 32 for both P1 neurons, and pIP10 neurons.
We exploited the time-resolved control of neuronal activation afforded by ReaChR to compare the temporal patterns of stimulation-evoked behavioral responses in P1 vs. pIP10 neurons. To express ReaChR in the latter cells, we used an intersectional strategy combining a specific GAL4 line (VT4055628) with fru-FLP38 and a UAS>STOP>ReaChR transgene (where “>” denotes FRT sites, the target of FLP recombinase; see Supplementary Fig. 2b, c and Supplementary Table 1). Anatomical analysis using a citrine reporter fused to the C-terminus of ReaChR confirmed the restricted expression of ReaChR in flies of the appropriate intersectional genotype (Supplementary Fig. 2d).
Surprisingly, we found that the temporal dynamics of wing extension evoked by activation of P1 vs. pIP10 neurons were strikingly different. ReaChR-mediated activation of P1 neurons evoked wing extension in a probabilistic or stochastic manner: the initiation of wing extension was not time-locked to the onset of illumination, but rather occurred with variable latencies throughout the stimulation period (17.7 ± 27.5 sec) (Fig. 3a, c). The average duration of each bout was short (0.99 ± 0.48 sec) relative to the duration of photostimulation (30 sec). Finally, the offset of the behavior was not time-locked to the offset of stimulation; rather, we observed persistent wing extension bouts in the intervals between photostimulation trials (Fig. 3a, e: Pearson's correlation coefficient between stimulation pattern and behavioral response: r = 0.004).
In contrast to the results observed with P1 neurons, activation of pIP10 neurons triggered robust wing extension in a deterministic manner (Fig. 3a). The onset of the behavior was strongly time-locked to the onset of stimulation, with a very short latency (0.08 ± 0.04 sec) (Fig. 3a, c). Once initiated, wing extension continued throughout the photostimulation period, and co-terminated, with few exceptions, with the offset of photostimulation (Fig. 3a, d: Pearson's correlation coefficient between stimulation pattern and behavioral response: r = 0.993). With weaker intensities of illumination close to threshold (≤ 0.012 mW/mm2), wing extension responses were less efficiently evoked, but responses were still restricted to the photostimulation period and no persistent behavior between trials was observed (Supplementary Fig. 3a).
These differences between P1 and pIP10 neurons in the temporal dynamics of ReaChR activation-evoked wing extensions do not reflect a higher sensitivity of pIP10 than P1 neurons, because the intensity dependence of pIP10-evoked wing extension by green light was almost identical to that of P1 neurons (Fig. 3f). Moreover, these properties were largely independent of illumination intensity (Figs. 4a–c and 5a–c).
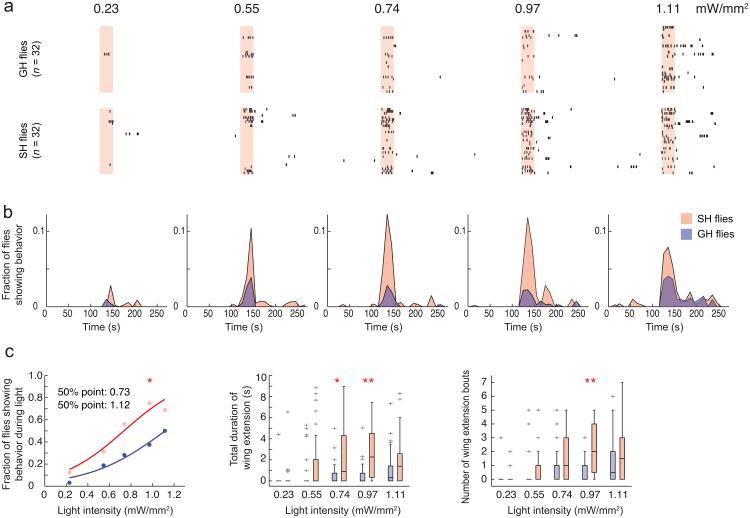
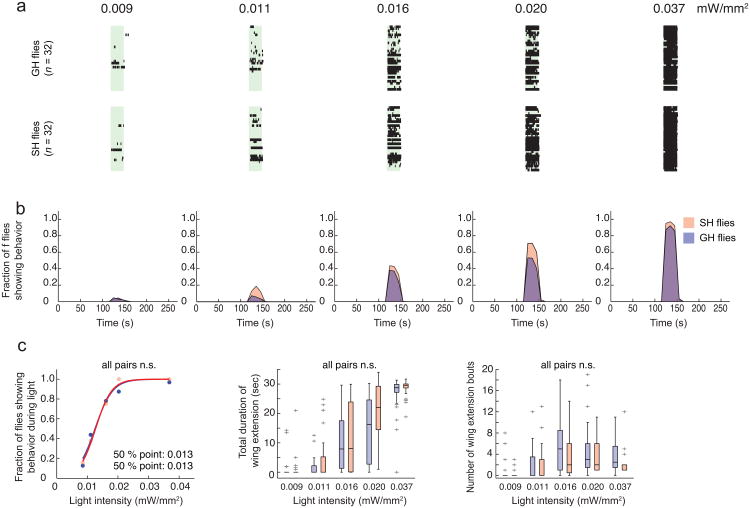
Figure 4. Social isolation lowers the threshold for ReaChR-activated male courtship behavior.
(a) Raster plots representing wing extension bouts from group-housed (GH: top row) or single-housed (SH: bottom row) flies expressing ReaChR in P1 neurons (P1-GAL4/UAS-ReaChR (VK5)). Flies were activated with different intensities of red light (627 nm). Light red bars in raster plots indicate photostimulation trials (30 sec continuous light), with different intensities indicated above the bars. n = 32 flies per intensity. (b) Fraction of flies showing wing extension based on the raster plots in (a). Data was binned every 10 sec. Time scale is the same in (a) and (b). In this and in Fig. 5, red or blue points/traces/boxplots represent data from single-housed or group-housed flies, respectively. (c) Different parameters were extracted from the raster plots in (a). Properties of boxplots in this and in Fig. 5 are as in Fig. 1f; “+” indicates outlier data bigger than the upper whisker. P-values in (c, left) represent Friedman's test comparing SH vs. GH (P = 6.3 × 10-28) followed by Fisher's exact test with Bonferroni correction comparing SH vs. GH at each intensity of light (*: P =0.01). P-values in (c, middle and right) represent Krusukal-Wallis one-way ANOVA followed by Mann-Whitney U tests with Bonferroni correction (*: P =0.027 ; **: P < 0.005). P-values for Krusukal-Wallis one-way ANOVA: (c, middle) P = 6.4 × 10-11; (c, right) P = 8.2 × 10-12.
Figure 5. Optogenetic activation of pIP10 neurons is not modulated by social isolation.
(a) Raster plot representing wing extension bouts from group-housed (GH: top row) or single-housed (SH: bottom row) flies expressing ReaChR in pIP10 neurons (VT40556-GAL424/UAS>stop>ReaChR(attP40); fru-FLP). Flies were activated with different intensities of green light (530 nm). Green bars indicate photostimulation trials (30sec continuous light), with different intensities indicated above the bars. n = 32 flies per intensity. (b) Fraction of flies showing wing extension based on the raster plot in (a). Time scale is the same in (a) and (b). (c) Different parameters extracted from the raster plots in (a). The GH data in (c, left) (blue points) are the same as those used in Fig. 3f, and are replotted here for purposes of comparison. (c, left) Friedman's test comparing SH vs. GH followed by Fisher's exact test with Bonferroni correction comparing SH vs. GH at each light intensity. (c, middle and right) Krusukal-Wallis one-way ANOVA followed by Mann-Whitney U tests with Bonferroni correction. All statistical tests yielded P-values > 0.05.
Social isolation modulates ReaChR-activated wing extension
The probabilistic or biasing nature of the wing extension responses elicited by ReaChR-mediated activation of P1 neurons suggested that these neurons might encode, or be modified by, state-dependent influences on male courtship behavior. Interestingly, social isolation of male flies for more than several days enhances courtship behavior, including singing, towards females39. To investigate whether P1 neurons might be modulated by such experience, we first determined whether social isolation lowers the threshold for eliciting wing extension using ReaChR-mediated stimulation of these neurons. Indeed, the intensity of red light that evoked wing extension in 50 % of flies expressing ReaChR in P1 neurons was lower in males that were socially isolated for 7 days, than in group-housed males (Fig. 4a–c). A similar effect was observed using green light as well (Supplementary Fig. 3b). For each of 3 different parameters measured, socially isolated flies exhibited significantly higher values than group housed flies (Fig. 4c). Thus, social isolation effectively “tuned” the response to ReaChR activation of P1 neurons, such that the probability of a wing extension response was increased. These data suggest that the increased sensitivity to ReaChR activation of wing extension occurs in P1 neurons themselves, or in a functionally downstream population.
Because pIP10 neurons are thought to be functionally “downstream” of P1 neurons28 (Fig. 3b), we investigated whether ReaChR activation of wing extension via these descending neurons was also sensitive to social experience. Because red light was not strong enough to activate wing extension in male flies expressing ReaChR in pIP10 neurons, we used green light to trigger wing extension. Activation of pIP10 neurons using ReaChR did not reveal any differences between single vs. group-housed flies in the efficiency with which photostimulation evoked wing extension behavior, even at lower intensities that evoked responses in only a subset of flies (Fig. 5a–c). These data indicate that the enhanced sensitivity of ReaChR-evoked wing extension in single-housed flies using the P1-GAL4 driver is likely to occur in P1 neurons themselves (or in other downstream neurons), rather than in pIP10 neurons. They also indicate that the sensitization of the P1 response by social isolation does not reflect a general increase in sensitivity among all neurons involved in wing extension behavior.
Functional calcium imaging combined with ReaChR activation
To examine directly whether social isolation enhances the sensitivity of P1 neurons to ReaChR activation, we performed calcium imaging experiments using laser-scanning 2-photon microscopy, taking advantage of the relative separation of the action spectrum peaks for ReaChR and GCaMP3.040 (Fig. 6a). Importantly, co-expression of GCaMP3.0 in P1 neurons together with ReaChR did not diminish the ability of the latter to mediate light-evoked wing extension in freely moving flies, indicating that the calcium buffering effect of GCaMP3.0 does not interfere with this behavior (data not shown).
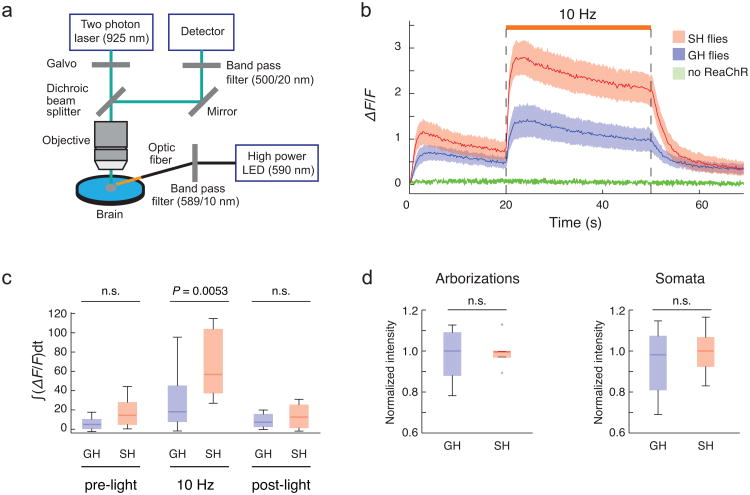
Figure 6. Functional calcium imaging of P1 neurons.
(a) Experimental setup for calcium imaging (see online Methods for detail). (b) Responses of P1 neurons (ΔF/F) to ReaChR activation were monitored using 2-photon LSM. Flies expressing both ReaChR and GCaMP3 in P1 neurons (P1-GAL4/UAS-ReaChR(attP40); UAS-GCaMP3(VK5)) were single-housed (SH: red line) or group-housed (GH: blue line) and imaged (n>10 brains for each curve). Amber light (590 nm, 1.7 mw/mm2) with a 5 msec pulse-width was delivered at 10 Hz for 30 sec (orange line above traces). GCaMP3.0 emissions were monitored in the arborizations of P1 neurons. Flies expressing GCaMP3.0 but not ReaChR in P1 neurons (P1-GAL4; UAS-GCaMP3(VK5)) were used as negative controls (green line) (n = 3). Solid red and blue lines represent average traces, and envelopes indicate SEMs. (c) Quantification of fluorescent changes. ∫ΔF/Fdt, integrated ΔF/F during 30 sec of light activation. Data were analyzed from (b). (d) Expression level of ReaChR at the arborizations and somata of P1 neurons were quantified using a citrine tag fused to the C-terminus of ReaChR. P-values represent Mann-Whitney U tests with Bonferroni correction.
An amber LED (590 nm) was used for photostimulation during imaging experiments in order to maximize overlap with the peak of the ReaChR action spectrum. Excitation scanning caused an initial increase in baseline GCaMP3.0 fluorescence in flies co-expressing ReaChR in P1 neurons, even in the absence of amber light excitation of ReaChR (Fig. 6b). These increases were not observed in flies lacking UAS-ReaChR (Fig. 6b) implying that they reflect cross-activation of ReaChR by the GCaMP3.0 excitation beam (925 nm). Nevertheless, amber light still evoked a clear increase in the strength of GCaMP3.0 emissions over this background (Fig. 6b).
Using these conditions, we compared the GCaMP3.0 response of P1 neurons to ReaChR activation of these same neurons, between single-housed (SH) vs. grouped-housed (GH) flies. P1 neurons in SH flies showed larger ReaChR-evoked calcium influxes than did those in GH flies (Fig. 6b, c). Quantitative analysis of ReaChR-citrine expression in these cells indicated that this difference was not due to higher levels of P1-GAL4 expression in SH vs. GH flies (Fig. 6d). Together, these combined behavioral and imaging experiments suggest that the excitability of P1 neurons can be modulated by prior social experience. Attempts to monitor calcium transients in pIP10 neurons were precluded by the complex expression pattern and low level of GCaMP3 expression driven by this GAL4 line.
Discussion
Here we describe a system for optogenetic activation of behavior in freely moving adult flies using ReaChR, a newly described red-shifted opsin17 (see Supplementary Note for discussion why ReaChR is more effective than other channelrhodopsins tested). The strength of activation obtained using ReaChR, and the broad dynamic range of intensities and frequencies over which stimulation can be delivered, offer a more quantitative and temporally controlled approach to investigating the neuronal control of behavior than is provided by available thermogenetic tools (but see ref23). The use of ReaChR with red light also reduces the confounding influence of strong visual stimulation that occurs when using blue light-activated opsins or the temperature increases required by thermogenetic effectors. Finally, the ability to control activation using LEDs, rather than lasers14,23, permits a relatively inexpensive approach to large-scale, high-throughput screening of behaviors.
Using ReaChR to monitor both behavioral sensitivity and neuronal activation, we discovered that 1) P1 and pIP10 neurons control male courtship song in a state-like (probabilistic and persistent) vs. command-like (deterministic and time-locked) manner, respectively; and 2) the effect of social isolation to increase male courtship behavior is mediated, at least in part, through an increase in the excitability of P1 neurons (see Supplementary Table 3). It has been proposed, based on anatomical data, that P1 neurons are part of a circuit integrating multimodal sensory cues that control courtship behavior38. Our observations suggest that P1 neurons also integrate this information with the flies' history of social experience, in a manner that influences the probability that the flies will exhibit courtship behavior. To our knowledge, this represents the first observation of a neural correlate of social experience in Drosophila. Interestingly, we did not observe any evidence of persistent calcium transients in P1 neurons after photoactivation (Fig. 6b), implying that the persistent wing extension triggered by P1 activation is mediated by other neurons. The mechanisms underlying the influence of social state on P1 excitability, and persistent activity, are interesting questions for future investigation.
While ReaChR-based activation of behavior was effective in all the GAL4 lines tested, the optogenetic toolkit in Drosophila could benefit from further engineering of red-shifted opsins with a narrower action spectrum and faster kinetics, and also by development of red-shifted variants of inhibitory opsins. Together, such tools would further enhance the applicability of optogenetics to neural circuit dissection in Drosophila.
Online Methods
Construction of transgenic animals
Plasmids were constructed by standard DNA cloning and PCR methods. All PCR reactions were performed using PrimeStar® HS DNA polymerase (Takara). Following amplification all sequences were verified by DNA sequencing.
UAS-ChR2(H134R)∷EYFP-2A-ChR2(H134R)∷EYFP
A DNA fragment containing the ChR2(H134R) coding sequence kindly provided by Dr. Karl Deisseroth and an intervening F2A sequence12,41 were amplified by PCR using primers, 5F-EcoRI-chr2 (CGGAATTCACCatggactatggcggcgctttg), 3R-2a-YFP (ctccagaacctgatctcttagcccgcttgtacagctcgtccatgccg), 5F-2a-Chr2 (ggagtccaacccagggcccatggactatggcggcgctttg), 3R-Xba-YFP (GCTCTAGAttacttgtacagctcgtccatgccg), 5F-2a (cgggctaagagatcaggttctggagcaccagtgaaacagactttgaattttgaccttctcaagttggcaggagacgtggagtccaacccagggccc) and 3R-2a (gggccctgggttggactccacgtctcctgccaacttgagaaggtcaaaattcaaagtctgtttcactggtgctccagaacctgatctcttagcccg), and subcloned into pUAST vector in a tandem manner using restriction enzymes. Several transgenic flies were created with different insertion sites. We picked the line that exhibited the strongest induction of PER when crossed to Gr5a-GAL4.
UAS-C1V1(T/T)
A DNA fragment containing the coding sequence of C1V1(E122T/E162T)-TS-eYFP kindly provided by Dr. Karl Deisseroth was amplified by PCR using primers, C1V1-f (TCTTATCCTTTACTTCAGGCCAAAATGTCGCGGAGGCCATGGCTTCTTGCCCTA) and C1V1-EGFP-r (GGTTCCTTCACAAAGATCCTTCTCGGCATGGACGAGCTGTACAAGTGA). This PCR product was subcloned into the vector pJFRC230 using SLIC cloning42. This vector was injected and integrated into attP40 and VK5 sites30.
UAS-ReaChR, LexAop-ReaChR, UAS-frt-mCherry-frt-ReaChR, and LexAop-frt-mCherry-frt-ReaChR
A DNA fragment containing the ReaChR∷citrine coding sequence was amplified by PCR using primers, ReaChR-f (TCTTATCCTTTACTTCAGGCCAAAATGGTGAGCAGAAGACCCTG) and ReaChR-citrine-r (GGTTCCTTCACAAAGATCCTCTAGACTACTTGTACAGCTCGTCCATGC). This PCR product was subcloned into pJFRC2 and pJFRC1930 using SLIC cloning42 for UAS-, and LexAop-driven versions, respectively. For the version containing an frt-mcherry-stop-frt cassette, the frt sequences (GAAGTTCCTATTCTCTAGAAAGTATAGGAACTTC) and ReaChR DNA fragments were subcloned together into pJFRC2 and pJFRC19 using SLIC cloning42. These vectors were injected and integrated into attP40, attP5 and VK5 sites30.
Fly strains
UAS-ChR25, UAS-dTrpA115, UAS-GCaMP3.040, Gr5a-GAL418, and BDP-GAL443 (empty promoter GAL4: an enhancer-less GAL4 containing a Drosophila basal promoter) were generously provided by Dr. André Fiala, Dr. Paul A. Garrity, Dr. Loren L. Looger, Dr. Kristin Scott and Dr. Gerald M. Rubin, respectively. Fru-GAL427, Fru-FLP38 and VT40556 GAL428 were kindly provided by Dr. Barry J. Dickson. HB9-GAL4 was obtained from Bloomington Stock Center (BL #32555). Crz-GAL426 and UAS-C128T12 were previously created in the lab. All the transgenic flies created for this paper are summarized in Supplementary Table 1. These flies are available on request.
All experimental flies were maintained on a 12/12 hour day-night cycle. Newly eclosed male flies were CO2 anesthetized and allowed to recover for more than 3-7 days prior to behavioral tests at 25 °C. For dTrpA1 experiments, flies were raised at 18 °C. For experiments with Gr5a-GAL4, female flies were used, and for all the other experiments male flies were used.
Feeding of retinal
All trans-retinal powder (Sigma) was stored in -20 °C as a 40 mM stock solution dissolved in DMSO (×100). 400 μl of sugar-retinal solution (400 μM all trans-retinal diluted in 89 mM sucrose) was directly added to surface of solid food in food vials when larvae were at the first or second instar stage. After collecting newly eclosed flies, they were transferred into a vial containing food with 400 μM all trans-retinal (food was heated and liquefied to mix the retinal evenly in the food). We found that larval feeding is not necessary, but it was performed for all the experiments in this paper to be consistent.
Behavioral setup
See Supplementary Table 2 for a list of components used to assemble the behavioral setup. See Supplementary Fig. 1 for details of the setup and the behavioral chamber. In brief, high power LEDs mounted on heat sinks were placed above the behavioral chamber to provide an illumination source (Fig. 2a and Supplementary Fig. 1a, b). The range of available light intensities in our setup is approximately 0.001 -1 mW/mm2 (note that intensity ranges are different for different LEDs; see Supplementary Fig. 1d). LED units were designed to be switchable to facilitate testing of different photostimulation wavelengths. The LEDs were controlled by an externally dimmable LED driver (700 mA; Externally dimmable, Buckpuck DC driver with leads) and its output was adjusted using custom software controlling an Arduino UNO board (Smart Projects, Italy). The Arduino digital PWM output was converted into analogue voltage using an RC-filter (electronic low-pass filter composed of resistor and capacitor. RC LPF in Fig. 2a) containing a 200 Ω resistor and 1 μF capacitor to control the output current of the LED driver. Fly behavior was monitored using a CMOS camera equipped with an IR long-pass filter to avoid detection of light from the high power LEDs. IR back-light was used to visualize the behaving flies. Video capture and LED control were time-locked using the Arduino UNO board. To time-stamp photostimulation trials in the videos, we placed an IR indicator LED, whose illumination was synchronized to that of the photostimulation LEDs, in the field of view of the camera. The temperature inside the behavioral chamber was minimally affected by the high intensity photostimulation. After illumination using the highest available intensities of blue, green or red LEDs (1.1, 0.67 and 1.27 mW/mm2, respectively) for 1 minutes, the biggest change in ambient temperature, detected using a thermocouple inserted into the chamber, was 0.7 °C.
Behavioral experiments and quantification of behaviors
For experiments to activate Gr5a-GRNs, non-starved flies were mounted into 200 μl pipetman tips as described previously12. Mounted flies were placed beneath high power LEDs and PERs were monitored using a videocamera. Mounted flies were not placed in the behavioral chamber but placed at the same location as the wells of behavioral chamber in Supplementary Fig. 1b. Bouts of PER were counted manually. Definition of bouts: a bout starts when flies start extending their proboscis, and ends when they retract the proboscis. Incomplete proboscis extensions were not counted. LEDs were used at maximum intensities in Figs. 1b, c and 2d, e (Red: 1.1 mW/mm2, Amber: 0.22 mW/mm2, Green: 0.67 mW/mm2, and Blue: 1.27 mW/mm2). For Fig. 1b, 100msec photostimulation trials (1 Hz) were delivered (3 trials) and flies showing more than one PER during this activation period were counted as responders. Fly genotype: w-;Gr5a-GAL4(II);GR5a-GAL4(III)/UAS-ReaChR(VK5) (Fig. 1b–g); w-;Gr5a-GAL4(II)/UAS-dTrpA1(II);GR5a(III)-GAL4/UAS-dTrpA1 (III) (Fig. 1h).
To activate Crz neurons (Fig. 2 d), males expressing each opsin in crz-GAL4 neurons were mounted dorsal side down on a glass slide as previously described26. Flies were illuminated using the maximum available intensity of light for each type of LED, continuously for 1 minute, while monitoring them from the ventral side using a video camera. The number of flies exhibiting ejaculation during light stimulation was manually counted.
For all other behavioral experiments, we used acrylic behavioral chambers (16mm diameter) in a 2 × 4 array (Fig. 2 and Supplementary Fig. S1) to monitor fly behavior. Unless otherwise indicated, chambers were photostimulated using the maximum intensity available for each LED, for 1 minute using continuous illumination, while monitoring them with the camera from above. The number of flies showing continuous side-walking during stimulation using the HB9-GAL4 driver was manually counted (Fig. 2 d, f). Fru-GAL4 neurons were activated in the same manner, and flies showing wing extension or paralysis phenotypes were counted manually (Fig. 2 d, g). Paralysis was defined as the cessation of locomotion and loss of postural control. Flies that showed a weaker behavioral phenotypes (HB9, side walk; Fru, wing extension) at the onset of photostimulation, but that were paralyzed before the 1 minute stimulation was terminated, were counted as paralysis (Fig. 2f, g).
Wing extension evoked by activation of P1 or pIP10 neurons were tested in solitary males in the absence of female flies. The wing extension was manually scored (Figs. 2–5). Grooming (rapid wing movements while touching with hind leg) was excluded. Definition of bouts: a bout starts when flies begin to increase the wing angle, and ends when they stop decreasing it.
In order to fit the data into a sigmoidal curve, sigmoid interpolation was performed. The sigmoid curves were defined as follows:
Where
Fbehav : Fraction of flies showing the behavior
X : Light intensity (Figs. 3f, 4c and 5c) or frequency (Fig. 2e)
X50: Light intensity (Figs. 3f, 4c and 5c) or frequency (Fig. 2e) where 50% of flies show the behavuor
α : slope of the sigmoid curve
Based on the experimentally measured quantities (X and Fbehav), X50 and α were chosen to best fit the data. For all experimental data, polynomial curve fitting, which finds the coefficients that fit the data by the least squares method, was calculated with Matlab (MathWorks). Goodness-of-fit was tested by two-way ANOVA between the sigmoidal curve and the actual PER response curve, which indicated a good fit for all cases (P < 0.05. two-way ANOVA). The X50 is shown as 50 % point in the figures (Figs. 2e, 3f, 4c and 5c).”
Measurement of light intensity
A photodiode power sensor (S130VC, Thorlab) was placed at the location of the behavioral chamber, but in the absence of the chamber. The peak wavelength of each LED (Red: 627 nm, Amber: 590 nm, Green: 530 nm, Blue: 470 nm) was measured at different voltage inputs. Measurements were repeated 4 times and averaged. The baseline intensity of each wavelength before LED illumination was subtracted. Note that light intensity can drop during stimulation at high input voltages. In this study, intensity after 10 sec of stimulation was measured.
Measurement of penetrance of different wavelengths of light through the fly cuticle
Proboscis of female adult fly is removed and a 10 μm multimode optic fiber (N.A. 0.1. Thorlab) was inserted into the brain through the window. The amount of light entering the optic fiber inside or outside the fly was measured using a power meter (Model 1931, New port). Penetrance was calculated as the amount of light that entered the optic fiber inside the fly, divided by the amount of light measured outside the fly. The long axis of the optic fiber was always aligned with the light source. Different wavelengths of high power LEDs (470 nm, 530 nm, 590 m, 627 nm) were used as light sources.
Fly histology
All fixation and staining procedures were performed at 4 °C in PBS, unless otherwise specified. Dissected brains were fixed in 4 % formaldehyde in PEM (0.1 M PIPES, pH 6.95, 2 mM EGTA, 1 mM MgSO4) for 2 hours. After three 15 min rinses with PBS, brains were incubated with primary antibodies overnight. Following three 15 min rinses with PBS, brains were incubated with secondary antibody overnight. Following three 15 min rinses, brains were incubated in 50 % glycerol in PBS for 2 hours and cleared with VECTASHIELD® (VECTA). All procedures were performed at 4 °C. A Fluoview™ FV1000 Confocal laser scanning biological microscope (Olympus) with a 30×, 1.05 N.A. silicone oil objective (Olympus) was used to obtain confocal serial optical sections. The antibodies used for Supplementary Fig. 2a, d were: anti-GFP, rabbit polyclonal antibody unconjugated (A11122, invitrogen) and Alexa Fluor® 488 donkey anti Rabbit IgG(H+L) (A11008, invitrogen). Both of the antibodies were diluted to 1/300. Expression of mCherry in Supplemental Fig. 2d was monitored using native fluorescence without antibody staining.
Fluorender software44 (http://www.sci.utah.edu/software/13-software/127-fluorender.html) was used to make 3D image reconstructions. To measure the expression levels of ReaChR∷citrine in P1 neurons in Fig. 6d, the native fluorescence of citrine in different specimens was monitored using the same intensity of laser power (470 nm) and PMT voltage. Signal intensity was quantified in imageJ (http://rsbweb.nih.gov/ij/).
Calcium imaging
Two-photon imaging was performed on an Ultima two-photon laser scanning microscope (Prairie Technology) with an imaging wavelength of 925 nm (Fig. 6). To filter out auto-fluorescence of the brain and light from the amber stimulation LED (for ReaChR activation), we used a 500/20 nm (center wavelength/band width) band-pass filter (Chroma) in the emission pathway to detect the GCaMP3 fluorescence. Using this laser and filter setting, fluorescent emissions from the Citrine tag (on ReaChR) were not detectable by our PMT. This was confirmed by examination of P1-GAL4;UAS-ReaChR∷Citrine flies (the flies without GCaMP3.0), which exhibited no fluorescent signal under our imaging conditions. Therefore the detected fluorescence signals are purely from GCaMP3.0.The scanning resolution was 128 × 128 pixels, dwell time per pixel was 8 μsec, and the optical zoom was ×4. The scanning speed was ca.10 Hz. The excitation intensity of the 2-photon laser was varied among samples depending on the level of GCaMP3.
In both cases, a 40×, 0.80 N.A. water-immersion objective (Olympus) was used for imaging. A high power amber LED (590 nm) collimated with an optic fiber (M590F1, Thorlab) was used as a light source to activate ReaChR. To narrow the band width of the LED output, we connected the optic fiber to a fiber optic filter holder (World Precision Instruments) equipped with 589/10 nm (center wavelength/band width) bandpass filter (Edmund optics). A 200 μm core multimode optic fiber (N.A. 0.39) (FT200EMT, Thorlab) was used to deliver the light from the fiber optic holder to the brain. One side of the optic fiber was custom-made to be a bare tip (Thorlab) and was dipped into the saline imaging bath and placed 430 μm away from the brain. A 10×, 0.30 N.A. water-immersion objective (Olympus) was used to locate the brain and align the optic fiber. The distance between brain and the fiber was measured with an objective micrometer (Olympus). We set the light intensity to be 170 μW at the tip of optic fiber. Thus, at a distance of 430 μm from the tip of a 0.39 N.A. optic fiber, the light power is calculated to be approximately 1.7 mW/mm2 at the brain surface (the size of light spot should be approximately 0.10 mm2 at the brain). In addition to the PMT used to monitor GCaMP emissions, we used another PMT to monitor the 590 nm ReaChR activation light. This was to ensure that the intensities of 590 nm light were comparable between samples. To prepare the brain for imaging, an ex-vivo prep was used. After a brief anesthesia on ice, the brain was dissected out using a sharp forceps into a 35 mm plastic petri dish (35 3001, FALCON®) containing Drosophila imaging saline (108 mM NaCl, 5 mM KCl, 2 mM CaCl2, 8.2 mM MgCl2, 4 mM NaHCO3, 1 mM NaH2PO4, 5 mM Trehalose, 10 mM Sucrose, 5 mM HEPES, pH 7.5)45. The fat body, air sacs, and esophagus were gently removed to give a clear view of the brain and to minimize its movement. The brains were attached to the bottom of the plate by static. The saline was changed once after dissection to remove debris. Calcium imaging was performed within 10–15 minutes after the dissection to ensure that the brains were healthy.
Electrophysiology
The tip recording method was used to record the electrophysiological responses of labellar taste neurons46. Briefly, the fly was mounted and immobilized for recording by inserting a pulled glass capillary (BF150-86-10, Sutter instruments) from the dorsal surface of the thorax to the tip of the labellum, passing through the cervical connective and the head. The mounting glass capillary was filled with recording solution (7.5 g/L NaCl, 0.35 g/L KCl, 0.279 g/L CaCl2·2H2O and 11.915 g/L HEPES (Sigma-Aldrich)) and served as a ground electrode. Another glass capillary, pulled to a tip diameter of 10 to 20 micrometers and filled with 30 mM tri-choline chloride (TCC; Sigma-Aldrich), as an electrolyte, was used for recording the electrophysiological responses of the gustatory neurons innervating this sensillum. All the recordings were obtained from L7 sensilla. The recordings were made using a MultiClamp 700B amplifier and Digidata 1440A A/D converter (Molecular Devices). The recorded data was sampled at a rate of 10 kHz, filtered (band pass filter between 100 Hz and 3 kHz) and stored on a PC hard drive using Clampex 10 software (Molecular Devices). The data were analyzed by sorting the action potentials and measuring their frequency within the indicated time windows using Clampfit software (Molecular Devices).
For PER activation experiments, a high power amber LED (590 nm) collimated with an optic fiber (M590F1, Thorlab) was used as a light source to activate ReaChR. To deliver light to the labellum a 200 μm core multimode optic fiber with bare end (N.A. 0.39) (Thorlab) was used. The distance of optic fiber from the labellum was set to be 540 μm using a micrometer. The estimated light intensity at the labellum was approximately 1.0 mW/mm2.
To activate TrpA1 (Fig. 1h), a custom-made heat source was used. In brief, the heat source is a small piece of thermistor (2K Bead Thermistor, Fenwal), which emits heat in proportion to the electrical current passed through it. The distance of the heat source from the labellum was set to be 540 μm using micrometer. The temperature at this distance was measured using a thermocouple (Omega) (top panel in Fig. 1h).
Supplementary Material
Acknowledgments
We thank K. Deisseroth (Stanford University), and B. Pfeiffer (Janelia Farm Research Campus) for plasmids. Fly stocks were generously provided by the Bloomington Stock Center, A. Fiala (Georg-August-Universität Göttingen), G. M. Rubin, L. L Looger, B.J. Dickson (Janelia Farm Research Campus), and P. A. Garrity (Brandeis University). We also thank members of the Anderson lab for helpful discussion and sharing of flies. H.K.I. was supported by the Nakajima Foundation. J.Y.L. was funded by Foundation of Research, Science and Technology New Zealand. The project was supported by grants from the National Institutes of Health to R.Y.T. (NS027177) and to D.J.A (R01DA031389-03). D.J.A. and R.Y.T are investigators of the Howard Hughes Medical Institute.
Footnotes
Author Contribution: H.K.I. and D.J.A. designed the experiments. H.K.I., Y.J., and N.M. performed behavioral experiments. H.K.I. and A.M.W. created the transgenic flies. H.K.I performed physiological experiments. E.D.H. provided P1-GAL4. J.Y.L. and R.Y.T. provided the ReaChR reagent and advices on its biophysical properties. H.K.I. and Y.J. performed the data analysis. H.K.I. and D.J.A. prepared the figures and wrote the paper.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Venken KJ, Simpson JH, Bellen HJ. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011;72:202–30. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–60. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annual review of neuroscience. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Schroll C, et al. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Current biology : CB. 2006;16:1741–7. doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 6.Zhang W, Ge W, Wang Z. A toolbox for light control of Drosophila behaviors through Channelrhodopsin 2-mediated photoactivation of targeted neurons. The European journal of neuroscience. 2007;26:2405–16. doi: 10.1111/j.1460-9568.2007.05862.x. [DOI] [PubMed] [Google Scholar]
- 7.Suh GS, et al. Light activation of an innate olfactory avoidance response in Drosophila. Current biology : CB. 2007;17:905–8. doi: 10.1016/j.cub.2007.04.046. [DOI] [PubMed] [Google Scholar]
- 8.Pulver SR, Pashkovski SL, Hornstein NJ, Garrity PA, Griffith LC. Temporal dynamics of neuronal activation by Channelrhodopsin-2 and TRPA1 determine behavioral output in Drosophila larvae. Journal of neurophysiology. 2009;101:3075–88. doi: 10.1152/jn.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61:373–84. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmermann G, et al. Manipulation of an innate escape response in Drosophila: photoexcitation of acj6 neurons induces the escape response. PloS one. 2009;4:e5100. doi: 10.1371/journal.pone.0005100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellmann D, et al. Optogenetically Induced Olfactory Stimulation in Drosophila Larvae Reveals the Neuronal Basis of Odor-Aversion behavior. Frontiers in behavioral neuroscience. 2010;4:27. doi: 10.3389/fnbeh.2010.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inagaki HK, et al. Visualizing neuromodulation in vivo: TANGO-mapping of dopamine signaling reveals appetite control of sugar sensing. Cell. 2012;148:583–95. doi: 10.1016/j.cell.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Vries SE, Clandinin T. Optogenetic stimulation of escape behavior in Drosophila melanogaster. Journal of visualized experiments : JoVE. 2013;71:e50192. doi: 10.3791/50192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lima SQ, Miesenbock G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–52. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Hamada FN, et al. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–20. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yizhar O, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–8. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nature neuroscience. 2013;16:1499–508. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott K, et al. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell. 2001;104:661–73. doi: 10.1016/s0092-8674(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 19.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nature neuroscience. 2005;8:1263–8. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 20.Nagel G, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13940–5. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagel G, et al. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Current biology : CB. 2005;15:2279–84. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 22.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nature neuroscience. 2009;12:229–34. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 23.Keene AC, Masek P. Optogenetic induction of aversive taste memory. Neuroscience. 2012;222:173–80. doi: 10.1016/j.neuroscience.2012.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bianchi D, et al. On the mechanisms underlying the depolarization block in the spiking dynamics of CA1 pyramidal neurons. Journal of computational neuroscience. 2012;33:207–25. doi: 10.1007/s10827-012-0383-y. [DOI] [PubMed] [Google Scholar]
- 25.Odden JP, Holbrook S, Doe CQ. Drosophila HB9 is expressed in a subset of motoneurons and interneurons, where it regulates gene expression and axon pathfinding. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2002;22:9143–9. doi: 10.1523/JNEUROSCI.22-21-09143.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tayler TD, Pacheco DA, Hergarden AC, Murthy M, Anderson DJ. A neuropeptide circuit that coordinates sperm transfer and copulation duration in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20697–702. doi: 10.1073/pnas.1218246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stockinger P, Kvitsiani D, Rotkopf S, Tirian L, Dickson BJ. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 28.von Philipsborn AC, et al. Neuronal control of Drosophila courtship song. Neuron. 2011;69:509–22. doi: 10.1016/j.neuron.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 29.Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–36. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–55. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenett A, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell reports. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan Y, Meissner GW, Baker BS. Joint control of Drosophila male courtship behavior by motion cues and activation of male-specific P1 neurons. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:10065–70. doi: 10.1073/pnas.1207107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi S, Desplan C, Heisenberg M. Contribution of photoreceptor subtypes to spectral wavelength preference in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5634–9. doi: 10.1073/pnas.0809398107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stavenga DG. Colour in the eyes of insects. Journal of comparative physiology A, Neuroethology, sensory, neural, and behavioral physiology. 2002;188:337–48. doi: 10.1007/s00359-002-0307-9. [DOI] [PubMed] [Google Scholar]
- 35.Hanai S, Hamasaka Y, Ishida N. Circadian entrainment to red light in Drosophila: requirement of Rhodopsin 1 and Rhodopsin 6. Neuroreport. 2008;19:1441–4. doi: 10.1097/WNR.0b013e32830e4961. [DOI] [PubMed] [Google Scholar]
- 36.Clyne JD, Miesenbock G. Sex-specific control and tuning of the pattern generator for courtship song in Drosophila. Cell. 2008;133:354–63. doi: 10.1016/j.cell.2008.01.050. [DOI] [PubMed] [Google Scholar]
- 37.Kohatsu S, Koganezawa M, Yamamoto D. Female contact activates male-specific interneurons that trigger stereotypic courtship behavior in Drosophila. Neuron. 2011;69:498–508. doi: 10.1016/j.neuron.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 38.Yu JY, Kanai MI, Demir E, Jefferis GS, Dickson BJ. Cellular organization of the neural circuit that drives Drosophila courtship behavior. Current biology : CB. 2010;20:1602–14. doi: 10.1016/j.cub.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 39.Dankert H, Wang L, Hoopfer ED, Anderson DJ, Perona P. Automated monitoring and analysis of social behavior in Drosophila. Nature methods. 2009;6:297–303. doi: 10.1038/nmeth.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nature methods. 2009;6:875–81. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donnelly ML, et al. The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. The Journal of general virology. 2001;82:1027–41. doi: 10.1099/0022-1317-82-5-1027. [DOI] [PubMed] [Google Scholar]
- 42.Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nature methods. 2007;4:251–6. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 43.Pfeiffer BD, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9715–20. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan Y, Otsuna H, Chien CB, Hansen C. An Interactive Visualization Tool for Multi-channel Confocal Microscopy Data in Neurobiology Research. Ieee Transactions on Visualization and Computer Graphics. 2009;15:1489–1496. doi: 10.1109/TVCG.2009.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong AM, Wang JW, Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109:229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- 46.Hodgson ES, Lettvin JY, Roeder KD. Physiology of a primary chemoreceptor unit. Science. 1955;122:417–8. doi: 10.1126/science.122.3166.417-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.