Significance
Genomic prediction is a new field of quantitative genetics. Individual performance can be predicted using genome-wide markers before the phenotype is measured. Genomic prediction for hybrid performance is even more promising because genotype of a hybrid is predetermined by the parents. We propose a genomic best linear unbiased prediction to predict hybrid performance of rice, and incorporate dominance and epistasis into the prediction model. Simulation studies showed that predictability can be further improved after incorporating dominance and epistasis into the model. The new strategy of marker-guided hybrid prediction is called genomic hybrid breeding, and it represents a new technology that may potentially revolutionize hybrid breeding in agriculture.
Keywords: hybrid rice, IMF2, mixed model, restricted maximum likelihood, variance component analysis
Abstract
Genomic selection is an upgrading form of marker-assisted selection for quantitative traits, and it differs from the traditional marker-assisted selection in that markers in the entire genome are used to predict genetic values and the QTL detection step is skipped. Genomic selection holds the promise to be more efficient than the traditional marker-assisted selection for traits controlled by polygenes. Genomic selection for pure breed improvement is based on marker information and thus leads to cost-saving due to early selection before phenotypes are measured. When applied to hybrid breeding, genomic selection is anticipated to be even more efficient because genotypes of hybrids are predetermined by their inbred parents. Hybrid breeding has been an important tool to increase crop productivity. Here we proposed and applied an advanced method to predict hybrid performance, in which a subset of all potential hybrids is used as a training sample to predict trait values of all potential hybrids. The method is called genomic best linear unbiased prediction. The technology applied to hybrids is called genomic hybrid breeding. We used 278 randomly selected hybrids derived from 210 recombinant inbred lines of rice as a training sample and predicted all 21,945 potential hybrids. The average yield of top 100 selection shows a 16% increase compared with the average yield of all potential hybrids. The new strategy of marker-guided prediction of hybrid yields serves as a proof of concept for a new technology that may potentially revolutionize hybrid breeding.
The mission of plant breeding is to develop high-yield varieties to increase crop productivity to meet the need of human population. Hybrid breeding has been proved to be an important tool to improve yield. The most successful examples are hybrid maize and rice, which have greatly increased the global food security. Despite the successes of hybrid breeding programs, selection of desirable hybrids has largely been a practice of trial and error in the past. It takes much luck to find desired matches between selected parents. The biggest challenge in hybrid breeding is how to predict the performance of future crosses based on existing data. Large efforts have been made in the past to develop methods for hybrid prediction, with the goal to facilitate hybrid breeding by obtaining better hybrids with fewer crosses. A common approach in the early days was to find correlations between marker polymorphisms and hybrid performance in crosses involving diverse germplasms. Extensive studies in corn and rice using this approach have produced variable results depending on the germplasms used in the studies (1).
Bernardo (2) applied best linear unbiased prediction technology to predict hybrid corn. He used existing hybrids and the pedigree relationship between them and untested hybrids to make prediction. Recent development in genomic research has greatly increased the availability of molecular makers to easily cover the entire genome, which are used to calculate the relationship matrix, leading to a method called genomic best linear unbiased prediction (GBLUP) (3). This method has been used to predict heterotic traits in maize hybrids (4). However, no attempt has been made to incorporate nonadditive effects into the prediction models.
Genomic selection aims to use whole-genome markers to predict future individuals, and it differs from traditional predictions in that the marker-detection step is skipped; instead, all markers are used to predict genomic values. Theoretically, when the number of markers is larger than the sample size, there is no unique estimation of effects, but the total genomic value remains estimable. Therefore, genomic prediction does not require accurate estimates of effects; it concerns the predictability resulted from the combination of all markers and their collective effects. Such genomic prediction has been applied to many agricultural species, including dairy cattle (5), crops (6), mice (7), and even humans (8). However, this approach has not been used to predict hybrid performance, an unexplored area that has a great potential to significantly improve efficiency of hybrid breeding.
Current methods for genomic prediction include Bayes B (9), empirical Bayes (10), least absolute shrinkage selection operator (LASSO) (11), and GBLUP (3). The first three procedures are classified into a category called selective shrinkage. Although simulation studies repeatedly showed that selective shrinkage is superior over GBLUP (12), experimental studies using cross-validation often showed similar performance (13), and GBLUP can be superior if a trait is controlled largely by polygenes, which implies that GBLUP may be more robust than the selective shrinkage methods. These genomic selection tools are mainly based on the additive model. Incorporating nonadditive variances is the next step in genomic prediction, but little study has been done so far. Incorporating dominance has been shown to be effective (14), but the benefit of incorporating epistasis has never been correctly demonstrated. One goal of this study is to investigate the effect of nonadditive variances on the efficiency of genomic prediction for hybrid performance.
Results
Predicting Hybrid Performance Using GBLUP.
Results of the restricted maximum likelihood (REML) analysis under the additive model are summarized in Table 1. The narrow sense heritability, defined as the ratio of the additive variance to the phenotypic variance, ranges from 0.38 for yield to 0.84 for 1,000 grain weight (KGW). The goodness of fit (squared correlation between observed and predicted trait values) varies from 0.51 for yield to 0.89 for KGW, which are all relatively high. The predictability drawn from fivefold cross-validation is much lower than the goodness of fit. Yield has the lowest predictability (0.13), and KGW has the highest predictability (0.68). The other two traits have predictabilities between the two. This analysis shows that genomic selection will be effective for all traits, especially for KGW. The difference between goodness of fit and predictability will be explained elsewhere in the text.
Table 1.
Parameters estimated using REML method for four traits in rice under the additive model
| Parameter | YIELD | TILLER | GRAIN | KGW |
| Additive variance | 14.4912 | 1.3879 | 254.6365 | 2.8200 |
| Residual variance | 23.3308 | 1.3998 | 124.1658 | 0.5472 |
| Heritability | 0.3831 | 0.4979 | 0.6722 | 0.8375 |
| Goodness of fit | 0.5148 | 0.6052 | 0.7280 | 0.8980 |
| Predictability | 0.1269 | 0.2259 | 0.3471 | 0.6797 |
Comparison of GBLUP with LASSO and SSVS.
The estimated marker effects (including the intercepts) of GBLUP for all traits are given in Dataset S1, where the results of the two competing methods are stored in separate sheets named LASSO and stochastic search variable selection (SSVS), respectively. The predictabilities drawn from fivefold cross-validation are listed in Table 2 along with the predictability from GBLUP. Results of the fivefold cross-validation depend on the partitioning of the sample. We randomly partitioned the sample into five parts of equal size and repeated the partitioning 20 times. Table 2 gives the average predictability for each trait under each method. The LASSO method barely outperformed GBLUP. The SSVS method is the worst one, especially for trait yield, which is 0.0943, compared with 0.1264 for GBLUP and 0.1601 for LASSO. In general, the three methods produced similar results for three of the four traits.
Table 2.
Comparison of the predictability for three methods drawn from fivefold cross-validation analysis
| Trait | GBLUP | LASSO | SSVS |
| YIELD | 0.1264 | 0.1601 | 0.0943 |
| TILLER | 0.2259 | 0.2046 | 0.2115 |
| GRAIN | 0.3471 | 0.3706 | 0.3527 |
| KGW | 0.6797 | 0.6868 | 0.6720 |
GBLUP Incorporating Epistasis.
The immortalized F2 (IMF2) population is unique in the sense that we can incorporate dominance and all four types of epistatic variances into the model. There are six genetic variance components, which are additive (a), dominance (d), additive by additive (aa), dominance by dominance (dd), additive by dominance (ad), and dominance by additive (da) variance components. The estimated variance components are given in Table 3, where ad and da are combined into a single composite term named (ad). Yield (YIELD) and tiller number (TILLER) are largely controlled by the (ad) interactions. Additive variance plays a major role for grain number (GRAIN) and KGW. None of the traits are controlled by the dominance variance. We also evaluated six different models, designated models 1–6, where the model number also represents the model size (number of genetic variances included in the model). For example, model 4 means that the model contains four genetic variance components, a, d, aa, and dd. Model 6 means that all six genetic variances are included (see Table S1 for model definitions).
Table 3.
Estimated variances and proportions (in parentheses) of phenotypic variance contributed by the variances
| Trait | a | aa | dd | (ad)* | e† |
| YIELD | 0.00 (0.00) | 7.01 (0.18) | 5.27 (0.14) | 23.83 (0.63) | 1.96 (0.05) |
| TILLER | 0.45 (0.17) | 0.59 (0.22) | 0.00 (0.00) | 1.25 (0.47) | 0.37 (0.14) |
| GRAIN | 150.91 (0.42) | 66.84 (0.19) | 6.58 (0.02) | 110.18 (0.31) | 21.51 (0.06) |
| KGW | 2.27 (0.73) | 0.31 (0.10) | 0.23 (0.07) | 0.19 (0.06) | 0.11 (0.04) |
Composite additive × dominance interaction that represents the sum of ad and da.
Residual variance. The dominance variance (d) is zero for all traits.
Fig. 1 shows the goodness of fit (Upper) and the predictability (Lower) plotted against the model size. Both goodness of fit and predictability were expressed as the squared correlation between observed and predicted phenotypes, but the predicted phenotypes were calculated using different approaches. For the goodness of fit, individuals predicted were also used to estimate parameters. For the predictability, the predicted values were drawn from fivefold cross-validation where individuals predicted did not contribute to parameter estimation. As the model size grows, the goodness of fit also grows until it reaches perfect fit when all six genetic variances are included in the model. To our surprise, the predictability does not show noticeable change as the model grows. The conclusion is that adding dominance and epistatic variances did not help genomic prediction. The estimated variance components for all traits are given in Table S2 for all six models. The lack of improvement is due to the large SEs of the estimated variances (Figs. S1 and S2 and Table S3) and the high correlation between different estimated variance components (Table S4). Large sample sizes are required to demonstrate the benefit of adding epistatic variances.
Fig. 1.
Goodness of fit (Upper) and predictability (Lower) of four traits plotted against model size, where the model size is determined by the number of variance components.
Simulation Study on Prediction Under Epistasis.
We performed a simulation study to demonstrate the effects of sample size and model size on model predictability. A hypothetic trait was simulated with equal values for all variance components (six genetic variances and a residual variance). We took the genotypes of n randomly selected hybrids (of 21,945 potential hybrids) as the true genotypes, where n ranged from 200 to 1,000 incremented by 100. The results are illustrated in Fig. 2. The goodness of fit started at ∼60% (additive variance only) and reached 100% for the full model (all six variance components) under all sample sizes. Small samples sizes tend to have a higher goodness of fit (Fig. 2, Upper). The predictability (Fig. 2, Lower) shows that adding dominance has increased the predictability under all sample sizes, but no further improvement are observed when the sample sizes are below n = 500. When n > 500, the predictability has progressively increased as the model size grows. For large sample sizes, there is benefit in prediction by including epistatic variances in the model. Our simulation experiments demonstrated the benefit of adding dominance for all sample sizes, but the real data analysis did not support this because we actually simulated dominance in the experiment. However, in the real experiment, dominance may be absent or very small. Further simulation study showed that it is safe to include dominance and epistatic variances in the model, even if the trait is only controlled by additive variance.
Fig. 2.
Effects of model size and sample size on the goodness of fit (Upper) and the predictability (Lower) of genomic prediction.
Prediction of Genomic Values for Future Crosses.
The 278 hybrids analyzed in this experiment are a random sample of all potential crosses, where 210 is the number of recombinant inbred lines (RILs) that initiated the current hybrid population. From the available RILs, we deduced the genotypes of all potential hybrids. We now try to predict the phenotypic values of the 21,667 remaining hybrids using the genetic parameters given in Table 1 under the additive model. The extended kinship matrix for all of the 21,945 hybrids is partitioned as follows:
| [1] |
where is the kinship matrix () for the current sample, is the kinship matrix () for the future hybrids, and is the relationship matrix () between the future hybrids and the current hybrids. The predicted phenotypic values for all of the crosses are given in Dataset S2. We also predicted the genomic values using the LASSO and SSVS methods; their predicted genomic values are also given in Dataset S2. The Spearman rank correlation coefficients of the predicted genomic values between the three methods are given in Table S5. The correlations are all high except the correlation between GBLUP and SSVS for yield, which is 0.65. The highest correlation occurs between GBLUP and LASSO for KGW (0.98).
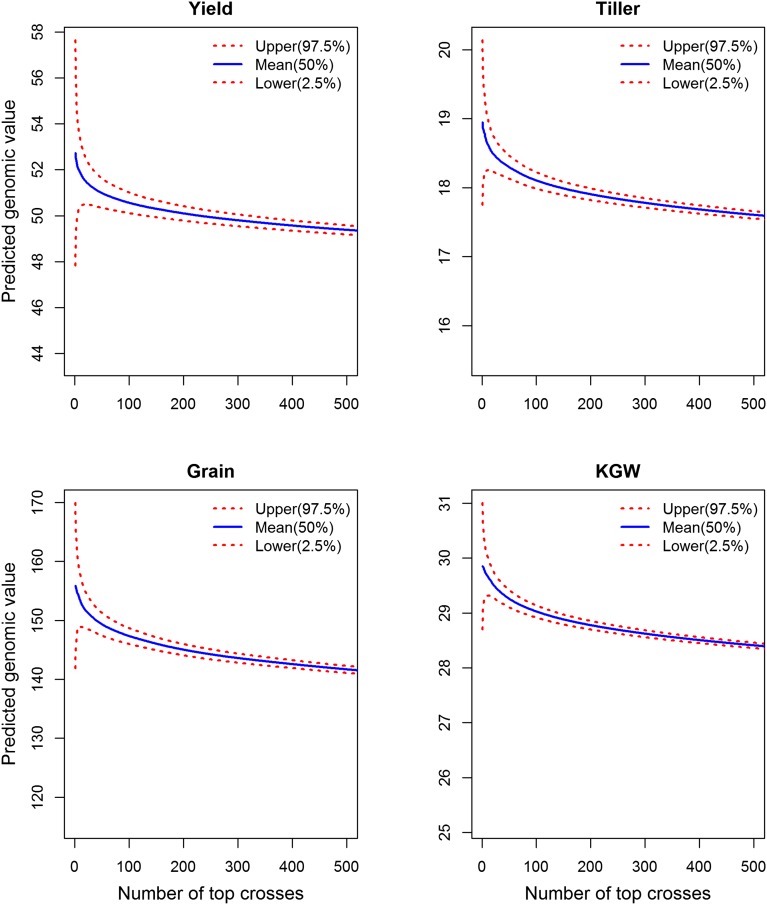
We then sorted the predicted phenotypic values in descending order and calculated the running average. For example, if we choose the top 100 crosses, the mean predicted breeding value of the top 100 crosses will be 50.5589 ± 0.23034 for yield. The average predicted genomic value of the entire hybrid population for yield is 43.6152. With genomic selection of the top 100 crosses, we expect to gain g in yield. Breeders can actually produce these 100 crosses based on the result of this study. Fig. 3 shows the average predicted genomic value for each trait against the top crosses selected for breeding (figure only shows the plot up to the top 500 crosses).
Fig. 3.
Average predicted genomic value of selected top crosses plotted against the number of crosses selected. The two dotted curves define the 95% confidence intervals of the mean predicted genomic value. The minimum value of the y axis for each trait is the average predicted genomic value for that trait. The plot is truncated at 500, and the total number of top crosses can run to 21,945 (all potential crosses).
Among the 21,667 potential crosses, we field evaluated 105 crosses in year 2012. These crosses were not included in the training sample, but their trait values have been predicted from the training sample. We calculated the squared correlation between the predicted and the observed trait values for the 105 crosses. This squared correlation is the actual predictability of our model under the assumption of no G×E interaction. The predictabilities are given in Table 4 for the four traits using the three competing methods. YIELD and TILLER have lost their predictability due to G×E interaction. The predictabilities for GRAIN and KGW remain relatively high, although both are lower than the cross-validation generated predictabilities due to possible G×E interaction. Recall that our training sample was collected in years 1998 and 1999, but the 105 additional crosses were collected in year 2012, which experienced an unusually high temperature. For traits with strong G×E interaction, such as YIELD and TILLER, our model can fail to predict the genomic values. However, heritable traits with less G×E interaction, such as GRAIN and KGW, are highly predictable.
Table 4.
Predictability (squared correlation between predicted and observed trait values) for 105 additional crosses evaluated in year 2012 using three competing methods
| Trait | GBLUP | LASSO | SSVS |
| YIELD* | 0.0053 | 0.0014 | 0.0076 |
| TILLER | 0.0727 | 0.0566 | 0.0773 |
| GRAIN | 0.2685 | 0.2473 | 0.2862 |
| KGW | 0.6107 | 0.6397 | 0.6378 |
Predictabilities for YIELD are not significantly different from zero. All other predictabilities listed are significant.
Discussion
The study demonstrated the application of advanced technologies, including genotype by sequencing and new statistical models to hybrid prediction. We used 278 existing hybrids derived from 210 RIL parents to predict the genomic values of all 21,945 potential hybrids for yield component traits in rice. The predicted best-performing hybrids can be generated from the original RIL parents. Genotypes of the future hybrids are not measured but determined from their inbred parents. The top crosses can be used immediately and converted to high-performing hybrids. What is the optimal proportion of the top crosses that should be selected for hybrid breeding? Two factors should be considered: one is the estimation error of the average performance of the selected top crosses. From Fig. 3, it is obvious that we should not select only the top five crosses because the average predicted value has a large prediction error. We need to keep at least 10 top crosses to reduce the prediction error. The other point to consider is that the genetic diversity of the top crosses tends to be narrow relative to that of the entire hybrid population. To maintain a high genetic diversity, one should select as many top crosses as possible, but the average predicted genomic value should remain high. For example, if we select the top 100 crosses (of 21,945 hybrids) for yield, the gain would be g per plant. This number represents a substantial gain in yield. Similar gains would also be obtained with other traits, which would potentially bring a substantial improvement of future hybrids. The predictability for yield (∼0.1) appears to be low; yet the gain obtained from selection of the top 100 hybrids would mean a significant achievement. How can we get the high gain with the low predictability? The key relies on the high selection intensity, represented by the small proportion selected (100/21,945 = 0.004556). Recall that selection response equals the product of the heritability and the selection differential, called the breeder’s equation. The predictability drawn from cross-validation is analogous to the heritability, and the proportion selected is inversely related to the selection differential. Therefore, the marker-guided selection of hybrids may achieve high gain largely through the high selection intensity.
One special feature of this experiment was that all of the RILs are from the same two parents, and this will limit the inference space of the results, i.e., the estimated parameters cannot be used to predict crosses of RILs that are not derived from the two parents. However, these types of crosses (IMF2) represent the best scenario to capture genetic variation of the parents because the F2-like genotypes provide the largest possible genetic variation. In addition, the two inbred lines that initiated the IMF2 were carefully selected to represent the best germplasms in hybrid rice breeding; they are the parents of Shanyou 63, the most popular rice hybrid in China and other Asian countries. Moreover, many widely used hybrids have one or the other line as their parents. Furthermore, most of the parental lines now commonly used in hybrid rice breeding programs have parentage of the two lines. Therefore, the result obtained here is important in its own rights.
Our cross-validation analysis showed that incorporating nonadditive variances did not show improvement in prediction, which may give the impression that we are not taking advantage of special combining ability and using only the general combining ability to predict heterosis. In fact, we are predicting hybrid performance, not heterosis, which is defined as the difference of the hybrid performance from the midparent performance. It is also important to emphasize that the six genetic kinship matrices are highly correlated. Therefore, the additive kinship matrix may already capture much information about the other kinship matrices. We need a very large sample size to well-separate the six variance components due to the multicollinearity of the kinship matrices.
For general application to a broader range of germplasms, imagine that if a half-diallel cross is to be conducted from 1,000 varieties, there would be possible hybrids. If the top 100 hybrids are selected, the proportion selected would be ; even a low predictability would be translated into a huge gain. However, it is practically impossible to conduct a half-diallel cross in such a large scale. An experimental design involving a subset of the crosses has to be used. Such a design is called partial diallel cross. For example, an experiment with 500 crosses is certainly realistic. Parameters estimated from the 500 hybrids can be used to predict all of the 499,500 potential hybrids, provided that the 500 crosses are selected in such a way that their genome composition represents the parental genomes as uniformly as possible. Further study on the optimal design should be the first priority of genomic hybrid breeding. As discussed earlier, IMF2 represents the best scenario for genetic analysis aiming to detect the difference between the two parents. Crosses from randomly selected inbred lines (not derived from two parents) do not share the same features as the IMF2 crosses. Therefore, detecting nonadditive variances, especially the epistatic variances, can be difficult. This argument is true for detecting individual pairwise epistatic variance. In our prediction model, we used genome-wide epistasis for prediction. Although, each pair of loci may be hard to detect because of the rare combination of some genotypes, all these rare events are given the same variance and thus all are combined together. Therefore, the genome-wide epistatic variances may not be as hard to detect as pairwise epistatic effects. It is the multicollinearity of the kinship matrices that causes the difficulty of separation. Therefore, predicting hybrid performance using a large number of inbred lines may be as efficient as IMF2 crosses and certainly better in terms of the broader inference space.
We also compared the performance of three different statistical methods to ensure that there are no artifacts caused by human errors in any single method. Three methods produced very similar results. Theoretically, selective shrinkage methods (LASSO and SSVS) perform better for traits controlled by a few large QTL, whereas GBLUP performs better for traits controlled under the infinitesimal model. GBLUP is more robust than the other methods because it does not depend on estimated marker effects, and has an additional advantage of being able to incorporate epistatic variances. In real-life experiments, any one of the three methods may be used under the additive and dominance model. If software packages are available, all methods should be tried to cross-confirm the results.
We reported the results of hybrid prediction under the main effect model, models incorporating dominance and epistatic effects were also investigated (see SI Text for methods and results incorporating epistasis). We originally hoped to demonstrate some improvement of the epistatic model over the additive model. To our surprise, there was no noticeable improvement; this does not disqualify the epistatic model because our training sample size is not large enough. The fact that the simple additive model performed equally well as the nonadditive models does not mean that nonadditive variances are not important to these traits. These additional variances are mostly captured by the additive variance because of the high correlations among the different types of kinship matrices (Table S4). Increasing sample size may not necessarily help to decrease the correlations of the kinship matrices but will reduce the estimation errors of different variance components, which in turn will improve prediction.
Materials and Methods
The rice population was constructed by randomly dividing 240 RILs derived from a cross between two indica rice, Zhenshan 97 and Minghui 63, into two groups and pairing lines in the two groups at random to create 120 crosses (15). Two additional rounds of crossing resulted in 360 crosses, IMF2. The two inbred lines that initiated the IMF2 were carefully selected to represent the best germplasms in hybrid rice breeding.
Field Data and Genotyping of the Population.
Field data of yield (YIELD), number of tillers per plant (TILLER), number of grains per panicle (GRAIN), and 1,000 grain weight (KGW) for the IMF2 population and the RILS were collected in the 1998 and 1999 rice-growing seasons from replicated field trials on the experimental farm of Huazhong Agricultural University (15). The RILs were genotyped using next-generation sequencing (16). More than 250,000 high-density SNP markers were obtained to infer recombination breakpoints (crossovers) and then construct bins. The 1,619 bins were treated as markers, and the genotypes of the hybrids in the IMF2 were deduced based on the bin genotypes of the RILs. Only 278 of the 360 crosses were available in both phenotypes and bin genotypes. For each trait, there were two temporal replications (years 1998 and 1999). The phenotypic values of the two replicates were pooled for each cross after removing the year effect using , where and are the mean values of the trait measured in 1998 and 1999, respectively. This pooled trait value was treated as the actual phenotypic value for analysis. Apparently, we ignored G×E effects, if there were any.
Statistical Methods.
Three methods were used to predict hybrid performance: GBLUP (3), LASSO (11), and SSVS (17). The LASSO and SSVS methods are well known in statistics and in the genomic selection community, and the GBLUP method is not as familiar to the plant-breeding community. In addition, we adopted an efficient algorithm to perform variance component analysis for GBLUP. Therefore, GBLUP will be described in more detail than the other two methods.
Mixed model.
Let y be an vector for the phenotypic values of a quantitative trait measured from individuals of a diploid population with genotypes denoted by , , and , respectively. We first numerically coded the genotype of individual j at locus k by a variable Z with for , for and for . The linear model that includes all m markers is
| [2] |
where is an design matrix, is a vector of nongenetic effects (called fixed effects), is an vector for the genotype indicator variable of all n individuals for marker k, is the effect of marker k, and is a vector of residual errors with an assumed distribution. The residual variance is an unknown parameter. Assume that all marker effects follow a normal distribution with mean zero and a common variance, i.e., , where is called the polygenic variance. The expectation of y is and the variance matrix is , where is the variance ratio and is a marker-generated kinship matrix, which measures the genetic similarity of all individuals in the sample. The REML method was used to estimate the variance ratio. The log likelihood function is
| [3] |
where is substituted by and by . Therefore, the likelihood function only involves . Such a likelihood function is called the profiled likelihood function. When is very large, the eigen-decomposition algorithm can be used to estimate , which is briefly described as follows. The eigen-decomposition is , where is the eigenvector (an matrix) and is the eigenvalue (a diagonal matrix). The two matrices, U and D, are obtained using marker information only before the REML analysis. With this algorithm, the variance matrix is rewritten as . The inverse and determinant of are calculated using and , respectively. Because matrix is diagonal, the inverse and determinant can be computed instantly using and . Any numerical optimization algorithm can be used to search for the REML estimate of , e.g., the Newton’s iterative method.
The model goodness of fit is expressed as the squared correlation coefficient between the observed (y) and the predicted () phenotypic values, where the latter were calculated using where . The model goodness of fit is not the same as the model predictability because individuals predicted also contribute to parameter estimation. The predictability should be obtained using an independent validation sample or via cross-validation where individuals predicted should not contribute to parameter estimation.
Genomic best linear unbiased prediction.
Let us define as the polygenic effect (the sum of all marker effects) and rewrite Eq. 2 by . Denote the polygenic covariance matrix by and the residual covariance matrix by . Suppose that we have two independent samples collected from the same population. One sample contains individuals with both phenotypes and genotypes, denoted by sample one. The other sample contains individuals with genotypes only, denoted by sample two. The models for the phenotypic values of the two samples are written together as
| [4] |
where is a vector of length for the observed phenotypic values from sample 1 and is a vector of length for the unobserved phenotypes for individuals from sample 2. The purpose of genomic selection is to predict the polygenic values for individuals in sample 2 using observed phenotypes for individuals in sample 1. The GBLUP estimate of is interpreted as the conditional expectation of given , denoted by . The predicted phenotypic values for individuals of sample 2 are
| [5] |
In the future when is measured, we will be able to calculate the predictability using the . The GBLUP method of genomic prediction does not require estimation of marker effects. The information to predict the genomic values of sample 2 comes from the genomic covariance between the unobserved and the observed individuals, . We predict the phenotypic value of a new individual in sample 2 by comparing marker genotypes of the new individual with the genotypes of individuals in sample 1, which is analogous to comparing DNA sample of a suspect to the DNA database to determine the criminal status of the suspect. In other words, genomic selection using GBLUP requires a database (the training sample) containing both the phenotypes of traits and the genotypes of markers.
Alternatively, estimated polygenic effects can be converted into estimated marker effects using the following equation, , which are then used to predict the genomic values of future individuals. This approach of genomic selection (by estimating ) does not need to store the marker genotypes and the trait phenotypes for the training sample. Information of those types has already been incorporated into the estimated marker effects. Details about estimation of marker effects using GBLUP are given in SI Text.
LASSO and SSVS.
Two additional models were compared with the GBLUP method, which are the LASSO method and the SSVS method; the latter is also called Bayes B (9) and is the very first method for genomic selection. Both methods use the model given in Eq. 2, i.e., they directly estimate marker effects in the training sample and predict the genomic values for individuals in the testing sample. The LASSO method minimizes a penalized sum of squares and was implemented using the GlmNet/R program (18) in this study. The SSVS method is a Markov chain Monte Carlo (MCMC) sampling-based method; it assumes that each marker effect has a mixture of two normal distributions, described as , where and are preset by the investigator. The mixing label is a binary variable to indicate whether is from or distribution. The situation of from distribution is equivalent to the effect being included in the model. Finally, is a beta variable representing the proportion of the effects included in the model relative to the total number of markers in the dataset. All parameters were sampled via the MCMC algorithm. The SSVS algorithm was implemented using a SAS/IML program written by S.X. (10).
The three methods (GBLUP, LASSO, and SSVS) were compared using the predictability drawn from fivefold cross-validation, in which four parts of the sample were used to estimate parameters for prediction of the phenotypic values in the remaining part of the sample. Eventually, each individual was predicted once and used four times to estimate parameters. The squared Pearson correlation coefficient between the observed and the predicted phenotypic values is a measure of the predictability.
Incorporation of nonadditive variances.
In addition to , here we define a dominance genotype indicator variable with for heterozygote and for homozygotes. Let be an vector for all hybrids at locus k. The polygenic effect is now partitioned into six polygenic components,
| [6] |
where and are the polygenic additive and dominance effects, respectively, and the remaining terms are the polygenic epistatic effects,
| [7] |
Note that represents element-wise vector multiplication, and and are the additive and dominance effects. The four terms, , , , and , are the additive × additive, dominance × dominance, additive × dominance, and dominance × additive effects, respectively, between markers k and for . These four terms are called the epistatic effects. By treating each genetic effect as a randomly distributed normal variable with mean zero and a common variance across all markers or marker pairs, the model becomes a mixed model. Let , , , , , and be the variance components of the six types of genetic effects. The expectation of is and the variance matrix of y is , where is the residual error covariance matrix and
| [8] |
is the genetic covariance matrix, in which the s are marker-generated kinship matrices developed by Xu (19). Given these marker-generated kinship matrices, the variance components were estimated using the standard mixed-model procedure. We used the REML method to estimate the parameters, a vector denoted by . The fixed effects were expressed as a function of the six variance components using the generalized least-squares equation. The REML likelihood function is defined as
| [9] |
where . The Newton–Raphson iteration was used to find the solutions of the parameters. The variance matrix of the REML estimated variance components was approximated by the negative inverse of the hessian matrix, which is a covariance matrix; square roots of its diagonal elements are the SEs of the estimated parameters. The model with six genetic variance components is the full model. Various reduced models were also evaluated. For example, if only the additive variance is included, the model is called the additive model or model 1 with a model size 1. The dominance model includes both the additive and dominance variances and is thus called model 2. The full model is called model 6. The model number represents the model size. Table S1 lists all six models evaluated in this study. Finally, the GBLUP analysis was performed using the mixed procedure in SAS. The SAS code is provided in Dataset S3.
Supplementary Material
Acknowledgments
This project was supported by US Department of Agriculture National Institute of Food and Agriculture Grant 2007-02784 (to S.X.), National Natural Science Foundation Grant 31330039, and 111 Project of China Grant B07041 (to Q.Z.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413750111/-/DCSupplemental.
References
- 1.Zhang Q, et al. Relationship between molecular marker polymorphism and hybrid performance in rice. In: Khush GS, editor. Rice Genetics III: Proceedings of the Third International Rice Genetics Symposium. Los Baños, Laguna, Philippines: Intl Rice Res Inst; 1995. pp. 317–326. [Google Scholar]
- 2.Bernardo R. Testcross additive and dominance effects in best linear unbiased prediction of maize single-cross performance. Theor Appl Genet. 1996;93(7):1098–1102. doi: 10.1007/BF00230131. [DOI] [PubMed] [Google Scholar]
- 3.VanRaden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91(11):4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- 4.Riedelsheimer C, et al. Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat Genet. 2012;44(2):217–220. doi: 10.1038/ng.1033. [DOI] [PubMed] [Google Scholar]
- 5.Hayes BJ, Bowman PJ, Chamberlain AJ, Goddard ME. Invited review: Genomic selection in dairy cattle: Progress and challenges. J Dairy Sci. 2009;92(2):433–443. doi: 10.3168/jds.2008-1646. [DOI] [PubMed] [Google Scholar]
- 6.Heffner EL, Sorrells ME, Jannink J-L. Genomic selection for crop improvement. Crop Sci. 2009;49(1):1–12. [Google Scholar]
- 7.Legarra A, Robert-Granié C, Manfredi E, Elsen JM. Performance of genomic selection in mice. Genetics. 2008;180(1):611–618. doi: 10.1534/genetics.108.088575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meuwissen THE, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157(4):1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu S. An empirical Bayes method for estimating epistatic effects of quantitative trait loci. Biometrics. 2007;63(2):513–521. doi: 10.1111/j.1541-0420.2006.00711.x. [DOI] [PubMed] [Google Scholar]
- 11.Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc, B. 1996;58:267–288. [Google Scholar]
- 12.Usai MG, Goddard ME, Hayes BJ. LASSO with cross-validation for genomic selection. Genet Res. 2009;91(6):427–436. doi: 10.1017/S0016672309990334. [DOI] [PubMed] [Google Scholar]
- 13.Resende MF, Jr, et al. Accuracy of genomic selection methods in a standard data set of loblolly pine (Pinus taeda L.) Genetics. 2012;190(4):1503–1510. doi: 10.1534/genetics.111.137026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vitezica ZG, Varona L, Legarra A. On the additive and dominant variance and covariance of individuals within the genomic selection scope. Genetics. 2013;195(4):1223–1230. doi: 10.1534/genetics.113.155176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hua J, et al. Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA. 2003;100(5):2574–2579. doi: 10.1073/pnas.0437907100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie W, et al. Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc Natl Acad Sci USA. 2010;107(23):10578–10583. doi: 10.1073/pnas.1005931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi N, George V, Allison DB. Stochastic search variable selection for identifying multiple quantitative trait loci. Genetics. 2003;164(3):1129–1138. doi: 10.1093/genetics/164.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 19.Xu S. Mapping quantitative trait loci by controlling polygenic background effects. Genetics. 2013;195(4):1209–1222. doi: 10.1534/genetics.113.157032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.