Abstract
Homo sapiens are genetically diverse, but dramatic demographic and socioeconomic changes during the past century have created further diversification with respect to age, nutritional status, and the incidence of associated chronic inflammatory disorders and chronic infections. These shifting demographics pose new challenges for vaccination, as emerging evidence suggests that age, the metabolic state, and chronic infections can exert major influences on the immune system. Thus, a key public health challenge is learning how to reprogram suboptimal immune systems to induce effective vaccine immunity. Recent advances have applied systems biological analysis to define molecular signatures induced early after vaccination that correlate with and predict the later adaptive immune responses in humans. Such “systems vaccinology” approaches offer an integrated picture of the molecular networks driving vaccine immunity, and are beginning to yield novel insights about the immune system. Here we discuss the promise of systems vaccinology in probing humanity’s diverse immune systems, and in delineating the impact of genes, the environment, and the microbiome on protective immunity induced by vaccination. Such insights will be critical in reengineering suboptimal immune systems in immunocompromised populations.
Vaccinating Obstinately Diverse Populations
Vaccines represent one of the greatest public health achievements of the 20th century. Vaccination programs during the last century have resulted in the global eradication of smallpox and the near eradication of polio, and exerted a dramatic impact on reducing the morbidity and mortality caused by infectious diseases, such as measles, rubella, tetanus, diphtheria, and Haemophilus influenzae type b (1). Furthermore, vaccination—through the control of infectious diseases—has contributed to a striking increase in life expectancy in many countries from about 40 y in 1900 to more than 80 y today (2). Although these impressive achievements will no doubt continue in the present century, vaccinologists face several new challenges. First, as reviewed extensively recently, the development of vaccines against global pandemics, such as HIV, malaria, tuberculosis, and dengue has been stymied by the unique set of challenges posed by these pathogens (3, 4). These challenges require a deeper understanding of the interaction between the pathogen and the host immune system, and a clearer definition of types of immune responses necessary to confer protective immunity.
Second, a major emerging challenge for vaccinologists is learning how to vaccinate what Sir Peter Medawar once termed “obstinately diverse populations” (5). Medawar was referring to genetic diversity, but dramatic demographic and socioeconomic changes during the past century have diversified the human species now, more so than ever before. Thus, the world is polarized with respect to several parameters, such as the nutritional status and age of its citizens, the incidence of chronic inflammatory disorders, and chronic infectious diseases. One in six children in developing countries is underweight, and up to one in three are stunted because of inadequate nutrition (6). In contrast, the success of vaccines in improving morbidity and mortality has allowed more children to grow to adulthood, with the potential for development of chronic diseases and conditions. For example, there has been a drastic increase in obesity and an array of associated disorders, such as insulin resistance type 2 diabetes, cardiovascular diseases, as well as diseases associated with low-grade chronic inflammation such as cancer (7, 8). Indeed, according to estimates from the World Health Organization (WHO), more than 1 billion adults worldwide are overweight and some 300 million people are clinically obese (8). The percentage of children aged 6–11 y in the United States who were obese increased from 7% in 1980 to nearly 18% in 2012 (9). Furthermore, there has been a major shift in the age distribution of populations. The WHO estimates that the percentage of individuals over 60, which was only a few percent in 1900, will reach nearly 25% by 2050 (www.who.int/ageing/about/facts/en/). These shifting demographics in the age distribution and the rise in the incidence of obesity and metabolic disorders pose new challenges for vaccination, as emerging evidence suggests that age (10) and the metabolic state of individuals can exert major influences on the immune system (8, 11, 12). This diversity in the physiological states of the human species is further enhanced by the increasing incidence of allergic disorders, such as asthma and allergic rhinitis, food allergies, and eczema (13, 14), as well as autoimmune diseases (14).
Emerging evidence suggests that vaccines induce suboptimal immunity in some special populations; therefore, a major challenge is learning how to design vaccines that induce protective immunity in populations with diverse immune systems. Here it is important to note that host genetics, as well as the human microbiome and other environmental influences, may impact immunity (15–17). Indeed, genes, the microbiome, and environmental cues (such as diet, allergies, and psychological stress) represent interacting variables that impinge on the physiology of the host, including immunity to infections and vaccination. These challenges demand a deeper understanding of the molecular networks that drive immunity to vaccination in diverse human populations. More recently, systems biological approaches have been developed that allow exploration of the complex, interconnected networks involved in immune responses to vaccines (3, 18–21). Specifically, systems approaches have been used to predict vaccine immunogenicity and are beginning to offer novel insights into the mechanisms of action of vaccines (22, 23). This emerging science of “systems vaccinology” has the potential to transform the field in enabling the identification of the multifactorial signatures associated with immunological protection and providing clues to protective mechanisms in diverse populations (Fig. 1). In this review, I discuss recent advances in systems vaccinology, and then discuss its potential application in identifying signatures of vaccine efficacy in clinical trials and in enabling rational vaccine design.
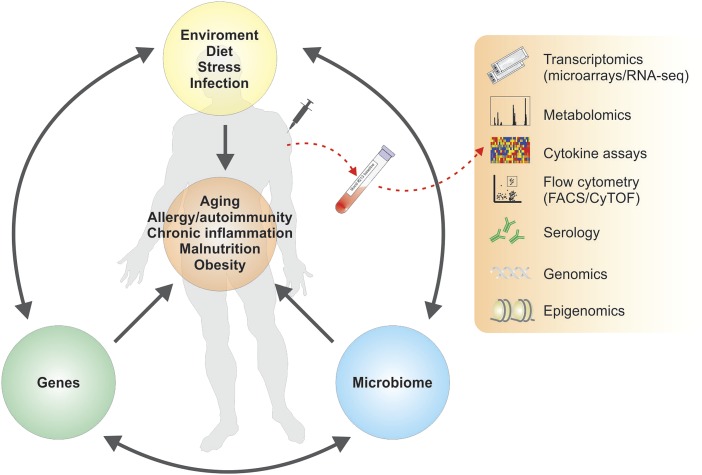
Fig. 1.
Systems vaccinology approaches to probe humanity’s immune systems. Genes, the environment, and the microbiome are three interdependent determinants of human physiology. Variations in each of these three determinants impact fundamental aspects of physiology, such as aging, nutritional status, and susceptibility to chronic infections and autoimmunities and allergies, and result in a staggering diversity of human physiologies. Recent studies have applied systems biological approaches to understand the molecular networks driving immune responses in humans. Such systems vaccinology approaches will be of great value in probing humanity’s diverse immune systems, and in enabling the rational design of vaccines that can induce effective immunity in special immunocompromised populations.
Systems Vaccinology: Predicting Vaccine Efficacy
Vaccines represent a diverse range of immunologic stimuli (live viruses, bacteria, carbohydrates, recombinant proteins) and are administered to millions of people every year. Vaccination results in a precisely synchronized perturbation of the immune system, and the ability to obtain blood samples from a vaccine at intervals provides an opportunity to study the ensuing immune response from the earliest few minutes to several decades after vaccination. Therefore, vaccines offer an efficient means to probe the immune system in humans. Recent advances have used the tools of systems biology to probe the immune response to vaccination in humans (22–32). Novel systems biology techniques allow integration of hierarchical levels of information, leading to a deconvolution of the complexity of biological systems (33). These approaches include “omics” measurements, such as genomic, transcriptomic, proteomic, metabolomic, and lipidomic technologies. Immunologists have begun to apply such “omic” technologies to make systems-wide measurements of immune responses, and use computational approaches to identify molecular signatures (e.g., patterns of gene expression induced after vaccination) that correlate with and predict subsequent adaptive immune responses (22–24). The ability to predict vaccine immunity offers a solution to a major challenge in vaccinology: to prospectively determine vaccine efficacy. This ability is of considerable public health importance in identifying “nonresponders” in special populations in which a vaccine may induce suboptimal immunity. In addition, it would be great value in clinical trials, in providing a rapid means to assess vaccine efficacy, without the need to assess the incidence of the target disease in vaccinated versus unvaccinated populations or subgroups of vaccinated populations.
The first examples of the application of systems biology to understanding vaccine-induced immune responses came from studies with the yellow fever live-attenuated vaccine YF-17D, one of the most successful vaccines ever developed (22, 24). YF-17D is a live-attenuated virus, derived several decades ago from the pathogenic strain of yellow fever (19). A single immunization with YF-17D stimulates robust antigen-specific CD8+ T cells and neutralizing antibody responses that persist for several decades (19, 34). Thus, YF-17D represents a “gold standard” vaccine. We (22) and Sékaly and colleagues (24) did independent studies in which we performed transcriptomic analysis of peripheral blood mononuclear cells (PBMCs) isolated 3 to 7 d after vaccination of healthy healthy young adults with YF-17D. Both studies revealed a pattern of gene expression profile consisting of genes encoding proteins involved in antiviral sensing and viral immunity, including the type I IFN pathway. This finding is consistent with the fact that vaccination with YF-17D causes an acute viral infection.
Using computational analysis we could identify signatures of gene expression, induced 3 or 7 d after vaccination, which correlated with the magnitude of the later antigen-specific CD8+ T-cell and neutralizing antibody responses (22). We subsequently used machine-learning techniques to validate the predictive capacity of such signatures by assessing their ability to predict the magnitude of the CD8+ T-cell and neutralizing antibody response in an independent clinical study with subjects vaccinated with YF-17D. Importantly, our recent work demonstrates the functional relevance of one of the genes contained within the predictive signatures, eukaryotic initiation factor-α kinase 4 (EIF2AK4), in programming dendritic cells to stimulate CD8+ T-cell responses (22). In the case of neutralizing antibody responses, a different gene signature was found to be predictive. One of the genes within this signature was TNFRSF17, which encodes the receptor for the B-cell growth factor BLyS-BAFF, which is known to play a key role in the differentiation of plasma cells (35).
This study provides proof-of-concept evidence that systems approaches could indeed be used to identify early “signatures” that could predict the later immunogenicity of the vaccine. At the time, this was a surprising result, as it was unclear whether there would be any detectable signature in the blood, in response to a vaccine that was administered subcutaneously. In retrospect however, the identification of molecular signatures in the blood, capable of predicting immunity to YF-17D, was perhaps not unexpected because vaccination results in an acute viral infection in which blood cells come into direct contact with the virus. This finding raised the question of whether this approach could be used to predict vaccine immunity in other vaccines, particularly inactivated vaccines. We and others therefore extended this approach to studying immunity to the trivalent inactivated seasonal influenza vaccine (TIV) (23, 26, 28) and the live-attenuated seasonal influenza vaccine (LAIV) (23). TIV is an inactivated nonreplicating vaccine administered intramuscularly and LAIV is a live-attenuated vaccine administered intranasally; it was thus of interest to determine whether signatures could be detected in the blood in response to such vaccines and, if so, whether such signatures were similar to that induced by YF-17D and capable or predicting the subsequent immune response. To address these issues, we performed a systems analysis of responses to TIV and LAIV in young healthy adults over three consecutive influenza seasons (23). TIV induced higher antibody titers and more plasmablasts than LAIV did. Consistent with the fact that LAIV results in a viral infection in mucosal tissues, vaccination of humans with LAIV induces a robust type I IFN antiviral transcriptomic signature. Vaccination with TIV also induced some genes encoding type I IFNs and related proteins, as well as genes encoding proinflammatory mediators (23).
In subjects vaccinated with TIV, consistent with studies by Bucasas et al. (26) and Obermoser et al. (29), there was enhanced expression of genes involved innate sensing of viruses and antiviral responses within 1–3 d after vaccination (23). This was followed 3–7 d later by the expression of genes known to be involved in the differentiation of plasmablasts (e.g., TNRSF17, genes such as XBP-1, which regulate the unfolded protein response) was highly correlated with—and predictive of—the magnitude of the hemagglutin titers after 28 d, in independent studies done in previous or subsequent years (23). Of particular interest was the expression of the gene encoding calmodulin-dependent protein kinase IV (CaMKIV) at day 3, which was inversely correlated with later antibody titers. Vaccination of CaMKIV-deficient mice with TIV induced enhanced antigen-specific antibody titers, which demonstrated an unappreciated role for CaMKIV in the regulation of antibody responses (23). This “plasmablast signature” and its capacity to predict antibody titers has been confirmed by several groups (29, 31, 36).
Recent studies have attempted to identify baseline signatures that can predict vaccine immunogenicity. Tsang et al. (36) analyzed PBMC transcriptomes and FACS analysis of cell subpopulation frequencies in 63 individuals before and after vaccination with TIV, and strikingly they could identify a baseline signature constructed with data from FACS analysis alone, which was capable of predicting the magnitude of the later antibody response to the vaccine. This result raises the prospect of predicting the outcome of vaccination, before immunization.
Finally, a key question is whether there are “universal signatures” of immunity to vaccines. For example, since in the case of antibody responses, certain aspects of the sequelae of immunological events (e.g., innate sensing of vaccine by dendritic cells and other innate cells, T-cell expansion, B-cell expansion, generation of plasmablasts) that lead to antibody production may be conserved between different vaccines, it could be argued that there may be overlapping signatures of antibody responses to different vaccines, that are detectable in the blood. However, because different vaccines trigger distinct innate receptors [e.g., YF-17D activates the Toll-like receptors (TLR) 2, 3, 7, 8, 9, and the RNA helicases RIG-I and MDA-5 (22, 37); LAIV triggers TLR7 (38, 39) and vaccines containing bacterial carbohydrates trigger TLR4 (27)], there may be expected to be different signatures of innate activation. To address this issue we performed a comparative systems analysis of signatures induced by different types of vaccines [YF-17D, TIV, the carbohydrate meningococcal vaccine (Menimmune), and the conjugate meningococcal vaccine (Menectra)] to see whether there are common predictors of antibody responses (27). A striking observation was that in contrast to what was observed with YF-17D, LAIV, and TIV, only a small number of genes was found to be differentially expressed 3 d after vaccination for Menimmune and Menectra, and at 7 d for Menimmune. To address this issue, we did a large-scale network integration of publicly available human blood transcriptomes and systems-scale databases in specific biological contexts and deduced a set of blood transcription modules (BTMs). Those modules revealed distinct transcriptional signatures of antibody responses to different classes of vaccines. Thus, recall antibody responses to inactivated vaccines (e.g., TIV and diphtheria toxoid component of Menectra) were highly correlated to BTMs induced at day 3 or day 7, which contained genes associated with plasmablast differentiation. In contrast, antibody responses to a live virus such as YF-17D were highly correlated BTMs associated with antiviral and type I IFN responses. Furthermore, the antibody responses to the carbohydrate components of Menimmune or Menectra were highly correlated with BTMs containing genes associated with inflammatory responses. These results highlight the fact that there are unlikely to be “universal signatures” of antibody responses to vaccines, but rather that different types of vaccines may have distinct signatures. Clearly further work with many more vaccines and greater numbers of subjects is necessary to address this issue definitively.
Application of Systems Approaches in Clinical Trials
To what extent systems vaccinology approaches are likely to be of value in defining correlates of protection in vaccine development has received much attention lately (18, 40, 41). Although the magnitude of the antibody response has been considered to be the correlate of protection for many vaccines, for diseases such as HIV, malaria, and tuberculosis we lack any real correlates. In the case of many vaccines, humoral immunity may not be the only or even the relevant correlate of protection. For example, for varicella vaccines, although efficacy is usually determined by measuring antibody titers, persistent varicella-specific T cells have been suggested as possible additional or alternative correlates of protection in children and the elderly (42, 43). Furthermore, in the case of seasonal influenza vaccines, although a serum HAI antibody titer of 1:40 is now widely accepted as a correlate of protection for immunity induced by inactivated influenza vaccines (44), no such immune correlate of protection has been identified for LAIV. Furthermore, such serological correlates are measured several weeks or months after vaccination, and efficacy studies for vaccines with unknown correlates can take years. Thus, the identification of correlates induced within a few days after vaccination or even at baseline will be of great value in accelerating vaccine testing in clinical trials. Therefore, the integration of systems approaches into clinical trials maybe useful in helping identify early predictors of efficacy, and accelerate the vaccine testing pipeline by allowing larger numbers of vaccine candidates to be screened more rapidly. Here it is worth considering different scenarios concerning vaccines with or without known correlates of protection and for which challenge models are available.
Vaccines with Known Correlates of Protection.
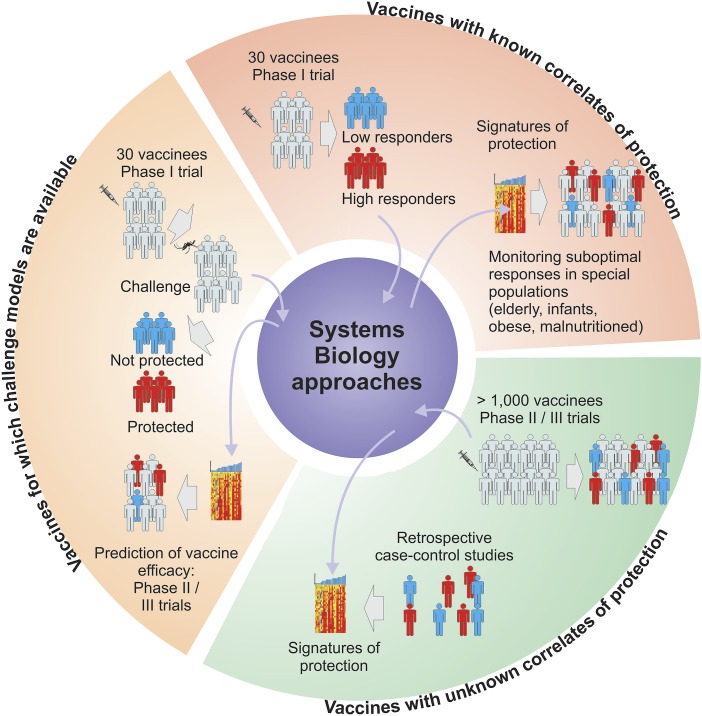
For vaccines where the correlate of protection is established, such as TIV (44), systems approaches can be used to identify signatures induced rapidly after vaccination, which predict the later immune response, similar to what has been done with the recent studies with YF-17D and TIV (Fig. 2, Upper Right). This would facilitate the rapid screening of nonresponders in special populations.
Fig. 2.
The application of systems vaccinology in clinical trials. Systems vaccinology approaches hold promise for predicting vaccine efficacy in clinical trials. (Upper Right) For vaccines with a known correlate of protection, systems approaches could be used to identify signatures induced 1 d or 3 after vaccination that is capable of predicting the later immunogenicity. Such signatures can be used to rapidly identify nonresponders in special populations. (Left) For vaccines for which challenge with the pathogen is feasible, systems approaches permit a direct identification of the correlates of protection. (Lower Right) For vaccines for which correlates are unknown, systems approaches can be applied to a retrospective nested case control study to identify novel correlates.
Vaccines for Which Challenge Models Are Available.
In some rare cases, as is the case with the malaria RTS,S vaccine, small phase 1 clinical trials can be conducted in which vaccinees can be challenged with the pathogen to assess efficacy (25). Such studies allow a careful dissection of the correlates of protection. Indeed, recent work has begun to use systems approaches to define the correlates of protection against RTS,S (25). In such cases, it is important to confirm the predictive capacity of such signatures using machine learning techniques, in repeated and iterative small phase 1 trials (Fig. 2, Left). Once a correlate has been thus rigorously defined in these phase 1 trials, then the signature can be used to rapidly predict vaccine efficacy in larger phase II and III trials.
Vaccines for Which There Are No Known Correlates.
This is the most challenging scenario, but retrospective nested case-control studies can be done to decipher correlates. For example, in the recent RV144 HIV 1 vaccine efficacy trial, the estimated efficacy of a vaccine against acquisition of HIV 1 was 31.2%. Haynes et al. performed a nested case-control analysis to identify correlates of risk from infection (45). Their analysis revealed that the titer of the IgG binding antibodies against the variable regions 1 and 2 (V1V2) of HIV-1 envelope at week 26 after vaccination, was inversely correlated with the rate of HIV 1 infection. Conversely, the titer of IgA binding antibodies in the plasma was directly correlated with rate of infection. Although these results need to be confirmed in additional studies, they provide a starting point for systems analysis of correlates. For example, are there signatures induced within a few days after vaccination that correlate with the magnitude of the binding antibody response to V1V1 of Env? If so, then the efficacy may be predicted within a few days of vaccination, not 26 wk later (Fig. 2, Lower Right). To what extent such approaches will be successful in predicting vaccine efficacy in clinical trials remains to be tested.
Gnostic Predictors: From Data ➔ Knowledge ➔ Understanding
A major challenge in systems vaccinology, and indeed for systems biology in general, is learning how to extract knowledge and eventually understanding from a “sea of data” (46). The identification of signatures that predict vaccine efficacy does not necessarily provide any mechanistic insights of how that vaccine stimulates protective immunity. This issue of developing knowledge-based gene-expression predictors [gnostic predictors (47)] has been recently discussed. One approach to this problem is to use analytical approaches to identify predictors that consist, not of sets of single genes that reach a certain statistical cut off, but rather a group of biologically related genes that are coordinately regulated in response to vaccination. Examples of such approaches include gene-set enrichment analysis (48), which has been widely used to reveal biologically meaningful differences in response a given stimulus, based on the coordinated regulation of biologically related genes. Such efforts have been enhanced by the development of tools for analysis of pathways. These tools include pathway annotations, such as the Kyoto Encyclopedia of Genes and Genomes (49), the National Cancer Institute-Pathway Interaction Database (50), the Molegular Signatures Database (MSigDB) (48), and Reactome (51). Efforts of manual curation of immune pathways are ongoing at the Innate Database and MSigDB (52). An alternative approach is to learn immune responses from existing data in the form of gene modules (27, 53).
Although such tools are of great value, ultimately it is human knowledge and intuition together with experimental validation, which are perhaps the most effective means of extracting biological insights from gene signatures. Thus, the creation of novel hypotheses based on the data, and experimental validation of such hypotheses is of paramount importance, and is already beginning to yield new insights. For example, in our study with the YF-17D (22), vaccine induced expression of EIF2AK4 [eukaryotic initiation factor 2 α-kinase 4, also known as general control nonderepressible 2 kinase (GCN2)] in PBMCs was found to correlate strongly and be predictive of the later CD8+ T-cell response. GCN2 is a sensor of amino acid starvation in mammals and is a key mediator of the “integrated stress response” (54, 55). GCN2 is activated by conditions of amino acid starvation and regulates protein synthesis through phosphorylation of eukaryotic translation initiation factor 2α (eIF2α). The phosphorylation of eIF2α shuts down translation of mRNA and polysome formation and induces the formation of stress granules, which are small cytoplasmic vesicles that contain untranslated mRNA (56). Despite its key role in nutritional sensing and the stress response, GCN2’s role in mediating immune response is poorly understood. The fact that its early expression was a strong predictor of immunity to vaccination raised the possibility that it may play a role in innate programming of adaptive immunity. Subsequent work in mice revealed a fundamental role for virus induced GCN2 activation in programming dendritic cells to initiate autophagy and enhanced antigen presentation to CD8+ T cells (57). These results reveal an unappreciated link between activation of a nutritional sensing pathway in dendritic cells and the adaptive immune response. A key question concerns the evolutionary significance of coupling a nutritional sensing pathway with innate control of adaptive immunity. One possibility is that such a mechanism evolved to sense the “footprints of infection”: nutritional depletion in the local microenvironments where pathogen replication occurred. Alternatively, such a mechanism may have evolved to amino acid depletion in sites of tumor replication, resulting in antitumor immunity.
Another unexpected insight emerged from our studies with TIV in which we observed an intriguing correlation between the expression of TLR5 in PBMCs isolated 3–7 d after vaccination, and the magnitude of the antibody responses at day 28 (23). This finding was curious because TLR5 is a sensor of bacterial flagellin and is not known to play any role in viral immunity. Our initial hypothesis was that the commercially available TIV contained contaminants that activated TLR5. However, this was experimentally shown not to be the case. Nevertheless, vaccination of mice deficient in TLR5 with TIV resulted in a markedly reduced antibody response relative to control mice. We investigated whether the intestinal microflora may signal through TLR5 to enhance antibody responses to vaccination. Consistent with this finding, antibiotic treatment of mice before vaccination or vaccination of germ-free mice resulted in greatly impaired antibody responses to TIV. These results reveal a striking role of the intestinal microbiota in regulating immunity to vaccination.
In addition to these results, several other interesting insights about the molecular circuitry of the immune responses to vaccination are emerging. For example, the studies with TIV have revealed an inverse correlation between the early expression of the gene encoding CaMKIV and hemagglutinin titers at day 28 (23). CaMKIV is a calcium/calmodulin-dependent serine/threonine kinase. Some members in this family, such as CaMKI and CaMKII, are ubiquitously expressed, but CaMKIV is chiefly expressed in cells of the nervous system and the immune system (58, 59). CaMKIV has been known to be involved in neuronal memory, but its impact on immune responses is poorly understood. The inverse correlation between CaMKIV expression and antibody responses led to the hypothesis that CaMKIV may suppress the antibody response to influenza vaccination. Vaccination of Camk4−/− mice resulted in enhanced antibody responses to TIV (23). The precise mechanism of this is under investigation. These observations highlight the utility of systems biological approaches in stimulating novel insights about the workings of the immune system.
This type of gene-by-gene analysis of function is beginning to provide many additional insights but a major challenge is the relatively large number of genes contained within predictive signatures. The approach of validating the function of genes one at a time using knockout mouse models is not optimally suited to validate large numbers of genes. However, the recent development of highly parallel assessment of gene function, such as shRNA vectors or pools of siRNA, allow a more rapid functional validation of larger gene sets in vitro, and the consequent delineation of gene regulatory circuits (60–62). Such an approach is likely to yield a broad and integrated view of the complex processes set in motion by injection of a vaccine. Here the challenge is a relevant surrogate in vitro read out for in vivo vaccine immunity.
Confounding Variables: Genes, Microbiota, and the Environment
The physiological states of humans are determined by the complex interplay between genes and environment. The evidence for the influence of host genetics on vaccine immunity comes from studies of identical twins. In one such study, immune responses to bacillus Calmette–Guérin, polio, hepatitis B, diphtheria, pertussis, and tetanus vaccines were measured at 5 mo of age in 207 pairs of Gambians recruited at birth. Both monozygotic and dizygotic twins were studied to delineate the genetic versus environmental components. There was a high heritability for antibody responses to hepatitis B (77%), oral polio (60%), tetanus (44%), and diphtheria (49%) (63). Other similar studies have assessed the influence of genetics on vaccine induced immunity and found these to be variable, ranging from 39–89% heritability (64–67). However, the interpretation of such data is confounded by several variables, such as age, sex, pre-exposure to the vaccine or pathogen, and other environmental factors. Furthermore, the identities of the particular genes that mediate heritability to a given vaccine are largely unknown. There have been several studies that have assessed the influence of candidate gene polymorphisms on immune responses to vaccination, and recently studies are beginning to use the genome-wide association studies approach, but such studies are still in their infancy (reviewed in ref. 67). Replication of results in independent studies is needed and importantly the functional relevance of genes identified in such studies to vaccine immunity, remain largely unexplored.
Emerging evidence also points to an important role for the host microbiome in exerting a major influence on the metabolic state, and contributing to anomalies such as obesity (16, 68). The dynamic interplay between host genetics, the environment and the microbiome can generate a staggering diversity of metabolic states in humans, manifest in conditions, such as aging, obesity, starvation, and susceptibility to allergies and infections (16). These three variables are also interdependent: genes may control susceptibility to environmental stimulants, such as high fat diet and allergens, and the expression of genes can be influenced by the environment and microbiome. The composition of the microbiome can be controlled by environmental conditions, such as diet, and it in turn may influence susceptibilities to environmental triggers, such as allergens and diet (16). Therefore, it is the intricate interplay between these variables that results in a particular metabolic state, and a key question is how immunity to vaccination is affected by such metabolic states. There are several studies documenting impaired immune responses associated with obesity (69, 70) and undernutrition (71–73). Systems vaccinology offers a promising way to dissect the molecular mechanisms that result in impaired vaccine immunity in such abnormal conditions. This issue is of importance now, more so than ever before, because of dramatic increases in conditions such as obesity and its associated metabolic disorders.
Recent evidence suggests a very robust connectivity between the metabolic and immune systems (8, 11). The metabolic system represents one of the most ancient homeostatic systems that are capable of responding to the most primal of environmental perturbations: changes in nutrient or oxygen conditions or other such perturbations. Mammals have evolved sensors of various such environmental stress signals—for example GCN2, protein kinase R-like endoplasmic reticulum kinase, and heme-regulated inhibitor kinase—that can sense such changes in environmental conditions and orchestrate responses like the integrated stress response (56). Subsequently, in evolution when cells evolved pathogen-sensing mechanisms, it is possible that some of the sensors and mechanisms that had evolved to detect metabolic changes were co-opted into the pathogen-sensing mechanism. Our recent finding that GCN2, a sensor for amino acid starvation, is also capable of orchestrating immunity to viral vaccines is one example of this concept (57). In addition, emerging evidence suggests that obesity results in chronic low grade inflammation leading to metabolic disorders. Excess nutrients induce stressed protein assembly pathways in the endoplasmic reticulum, leading to secretion of proinflammatory cytokines by adipocytes (8, 11, 12). This process results in the recruitment of inflammatory cells, such as proinflammatory macrophages leading to an inflammatory cascade. In the central nervous system, inflammatory cytokines inhibit leptin signaling and perturb energy balance. The impact of any of these pathways on the development of vaccine immunity and immunological memory is poorly understood and should be a focus of systems vaccinology studies.
A critical factor in the relationship between obesity and inflammation is the intestinal microbiome. It is known that the intestinal microflora of obese individuals differs from that of healthy individuals (16, 68), and consistent with this mice deficient in TLR5 contain a distinct composition of microflora that sensitizes them to obesity and metabolic syndrome (74). Furthermore, our recent data demonstrate a critical role for intestinal microflora in promoting immunity to influenza vaccine immunity. This finding may be of particular relevance in developing countries, where the efficacy of oral vaccines against polio, rotavirus, and cholera have showed a lower immunogenicity relative to efficacy in developed countries (75–78). In a study testing a live cholera oral vaccine, Lagos et al. (79) demonstrated that excessive bacterial growth in the small intestine of children in less-developed countries might contribute to the low antibody response to the vaccine. Therefore, future systems vaccinology studies should integrate metagenomics approaches to determine the composition of microflora that correlates with a particular profile of vaccine immunity. Subsequent studies should aim to experimentally perturb the microflora composition with antibiotics or probiotics and assess the impact on vaccine immunity.
In summary, recent efforts at applying systems-level approaches to vaccine development show promise, particularly in defining molecular signatures induced early after vaccination that correlate with and predict the later adaptive immune responses in humans, and are beginning to provide new insights into the immune system. Such an approach will be transformative in understanding the molecular networks underlying the immune response to vaccination in diverse human populations and will enable strategies to re-engineer these networks to generate protective immunity. Such studies should aim to obtain an integrated high-resolution picture of the dynamic changes in the expression of genes, proteins, metabolites, and cellular composition in response to vaccination, as well as the composition of the microflora before, during, and after vaccination, in diverse populations. Such analyses will undoubtedly generate a sea of data, but the meaningful analyses of large sets of data in a multitude of formats also require substantial computational technologies to facilitate data visualization, model building, and ultimately the capacity to simulate the necessary immune responses in silico. Therefore, deployment of systems biology approaches in vaccine research requires the integrated efforts of vaccinologists, clinicians, immunologists, systems biologists, and computational specialists.
Supplementary Material
Acknowledgments
The author thanks Walt Orenstein for valuable comments on the paper, and Helder Nakaya, Dmitri Kazmin, and Mario Cortese for artwork in Figs. 1 and 2. This work was supported by National Institutes of Health Grants U19AI090023, U54AI057157, R37AI48638, R37DK057665, U19AI057266, and AI100663-02 (to B.P.), and a grant from the Bill and Melinda Gates Foundation (to B.P.)
Footnotes
The author declares no conflict of interest.
This article is a PNAS Direct Submission.
This article is part of the special series of PNAS 100th Anniversary articles to commemorate exceptional research published in PNAS over the last century.
References
- 1.D’Argenio DA, Wilson CB. A decade of vaccines: Integrating immunology and vaccinology for rational vaccine design. Immunity. 2010;33(4):437–440. doi: 10.1016/j.immuni.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Rappuoli R, Mandl CW, Black S, De Gregorio E. Vaccines for the twenty-first century society. Nat Rev Immunol. 2011;11(12):865–872. doi: 10.1038/nri3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rappuoli R, Aderem A. A 2020 vision for vaccines against HIV, tuberculosis and malaria. Nature. 2011;473(7348):463–469. doi: 10.1038/nature10124. [DOI] [PubMed] [Google Scholar]
- 4.Koff WC, et al. Accelerating next-generation vaccine development for global disease prevention. Science. 2013;340(6136):1232910. doi: 10.1126/science.1232910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medawar P. 1959. The Future of Man. The Reith Lectures, 1959. BBC Radio 4. Available at http://downloads.bbc.co.uk/rmhttp/radio4/transcripts/1959_reith4.pdf. Accessed June 4, 2014.
- 6.World Food Programme 2014. Hunger Statistics. Available at www.wfp.org/hunger/stats03/10/2014. Accessed June 4, 2014.
- 7.Lumeng CN, Saltiel AR. Inflammatory links between obesity and metabolic disease. J Clin Invest. 2011;121(6):2111–2117. doi: 10.1172/JCI57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 9. Center for Disease Control and Prevention (2014) Childhood Obesity Facts. Available at www.cdc.gov/healthyyouth/obesity/facts.htm. Accessed June 4, 2014.
- 10.Duraisingham SS, et al. Systems biology of vaccination in the elderly. Curr Top Microbiol Immunol. 2013;363:117–142. doi: 10.1007/82_2012_250. [DOI] [PubMed] [Google Scholar]
- 11.Ouchi N, Parker JL, Lugus JJ, Walsh K. Adipokines in inflammation and metabolic disease. Nat Rev Immunol. 2011;11(2):85–97. doi: 10.1038/nri2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donath MY, Shoelson SE. Type 2 diabetes as an inflammatory disease. Nat Rev Immunol. 2011;11(2):98–107. doi: 10.1038/nri2925. [DOI] [PubMed] [Google Scholar]
- 13.Pawankar R, Canonica GW, Holgate ST, Lockey RF. Allergic diseases and asthma: A major global health concern. Curr Opin Allergy Clin Immunol. 2012;12(1):39–41. doi: 10.1097/ACI.0b013e32834ec13b. [DOI] [PubMed] [Google Scholar]
- 14.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347(12):911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 15.Casanova J-L, Abel L. The genetic theory of infectious diseases: A brief history and selected illustrations. Annu Rev Genomics Hum Genet. 2013;14:215–243. doi: 10.1146/annurev-genom-091212-153448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474(7351):327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamada N, Seo S-U, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 18.Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity. 2010;33(4):516–529. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pulendran B. Learning immunology from the yellow fever vaccine: Innate immunity to systems vaccinology. Nat Rev Immunol. 2009;9(10):741–747. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- 20.Buonaguro L, Pulendran B. Immunogenomics and systems biology of vaccines. Immunol Rev. 2011;239(1):197–208. doi: 10.1111/j.1600-065X.2010.00971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mooney M, McWeeney S, Canderan G, Sékaly R-P. A systems framework for vaccine design. Curr Opin Immunol. 2013;25(5):551–555. doi: 10.1016/j.coi.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Querec TD, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10(1):116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakaya HI, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12(8):786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaucher D, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205(13):3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vahey MT, et al. Expression of genes associated with immunoproteasome processing of major histocompatibility complex peptides is indicative of protection with adjuvanted RTS,S malaria vaccine. J Infect Dis. 2010;201(4):580–589. doi: 10.1086/650310. [DOI] [PubMed] [Google Scholar]
- 26.Bucasas KL, et al. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J Infect Dis. 2011;203(7):921–929. doi: 10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol. 2014;15(2):195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franco LM, et al. Integrative genomic analysis of the human immune response to influenza vaccination. eLife. 2013;2:e00299. doi: 10.7554/eLife.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obermoser G, et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38(4):831–844. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zak DE, et al. Merck Ad5/HIV induces broad innate immune activation that predicts CD8⁺ T-cell responses but is attenuated by preexisting Ad5 immunity. Proc Natl Acad Sci USA. 2012;109(50):E3503–E3512. doi: 10.1073/pnas.1208972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furman D, et al. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol Syst Biol. 2013;9(1):659. doi: 10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Germain RN. Vaccines and the future of human immunology. Immunity. 2010;33(4):441–450. doi: 10.1016/j.immuni.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 33.Diercks A, Aderem A. Systems approaches to dissecting immunity. Curr Top Microbiol Immunol. 2013;363:1–19. doi: 10.1007/82_2012_246. [DOI] [PubMed] [Google Scholar]
- 34.Pulendran B, Oh JZ, Nakaya HI, Ravindran R, Kazmin DA. Immunity to viruses: Learning from successful human vaccines. Immunol Rev. 2013;255(1):243–255. doi: 10.1111/imr.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vincent FB, Saulep-Easton D, Figgett WA, Fairfax KA, Mackay F. The BAFF/APRIL system: Emerging functions beyond B cell biology and autoimmunity. Cytokine Growth Factor Rev. 2013;24(3):203–215. doi: 10.1016/j.cytogfr.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsang JS, et al. Baylor HIPC Center; CHI Consortium Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157(2):499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Querec T, et al. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J Exp Med. 2006;203(2):413–424. doi: 10.1084/jem.20051720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geeraedts F, et al. Superior immunogenicity of inactivated whole virus H5N1 influenza vaccine is primarily controlled by Toll-like receptor signalling. PLoS Pathog. 2008;4(8):e1000138. doi: 10.1371/journal.ppat.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koyama S, et al. Plasmacytoid dendritic cells delineate immunogenicity of influenza vaccine subtypes. Sci Transl Med. 2010;2(25):25ra24–25ra24. doi: 10.1126/scitranslmed.3000759. [DOI] [PubMed] [Google Scholar]
- 40.Nakaya HI, Li S, Pulendran B. Systems vaccinology: Learning to compute the behavior of vaccine induced immunity. Wiley Interdiscip Rev Syst Biol Med. 2012;4(2):193–205. doi: 10.1002/wsbm.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakaya HI, Pulendran B. Systems vaccinology: Its promise and challenge for HIV vaccine development. Curr Opin HIV AIDS. 2012;7(1):24–31. doi: 10.1097/COH.0b013e32834dc37b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arvin AM. Humoral and cellular immunity to varicella-zoster virus: An overview. J Infect Dis. 2008;197(Suppl 2):S58–S60. doi: 10.1086/522123. [DOI] [PubMed] [Google Scholar]
- 43.Levin MJ, et al. Veterans Affairs Cooperative Studies Program Shingles Prevention Study Investigators Varicella-zoster virus-specific immune responses in elderly recipients of a herpes zoster vaccine. J Infect Dis. 2008;197(6):825–835. doi: 10.1086/528696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohmit SE, Petrie JG, Cross RT, Johnson E, Monto AS. Influenza hemagglutination-inhibition antibody titer as a correlate of vaccine-induced protection. J Infect Dis. 2011;204(12):1879–1885. doi: 10.1093/infdis/jir661. [DOI] [PubMed] [Google Scholar]
- 45.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366(14):1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brenner S. Sequences and consequences. Philos Trans R Soc Lond B Biol Sci. 2010;365(1537):207–212. doi: 10.1098/rstb.2009.0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haining WN, Pulendran B. Identifying gnostic predictors of the vaccine response. Curr Opin Immunol. 2012;24(3):332–336. doi: 10.1016/j.coi.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanehisa M, et al. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006;34(Database issue) suppl 1:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaefer CF, et al. PID: The pathway interaction database. Nucleic Acids Res. 2009;37(Database issue) suppl 1:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vastrik I, et al. Reactome: A knowledge base of biologic pathways and processes. Genome Biol. 2007;8(3) doi: 10.1186/gb-2007-8-3-r39. R39; corrected Genome Biol (2009) 10(2):402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaussabel D, et al. A modular analysis framework for blood genomics studies: Application to systemic lupus erythematosus. Immunity. 2008;29(1):150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hinnebusch AG. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci USA. 1984;81(20):6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sood R, Porter AC, Olsen DA, Cavener DR, Wek RC. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2α. Genetics. 2000;154(2):787–801. doi: 10.1093/genetics/154.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anderson P, Kedersha N. Visibly stressed: The role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7(2):213–221. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ravindran R, et al. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science. 2014;343(6168):313–317. doi: 10.1126/science.1246829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ribar TJ, et al. Cerebellar defects in Ca2+/calmodulin kinase IV-deficient mice. J Neurosci. 2000;20(22):RC107. doi: 10.1523/JNEUROSCI.20-22-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wayman GA, Tokumitsu H, Davare MA, Soderling TR. Analysis of CaM-kinase signaling in cells. Cell Calcium. 2011;50(1):1–8. doi: 10.1016/j.ceca.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amit I, Regev A, Hacohen N. Strategies to discover regulatory circuits of the mammalian immune system. Nat Rev Immunol. 2011;11(12):873–880. doi: 10.1038/nri3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chevrier N, et al. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell. 2011;147(4):853–867. doi: 10.1016/j.cell.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yosef N, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496(7446):461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newport MJ, et al. MRC Gambia Twin Study Group Genetic regulation of immune responses to vaccines in early life. Genes Immun. 2004;5(2):122–129. doi: 10.1038/sj.gene.6364051. [DOI] [PubMed] [Google Scholar]
- 64.Höhler T, et al. Differential genetic determination of immune responsiveness to hepatitis B surface antigen and to hepatitis A virus: A vaccination study in twins. Lancet. 2002;360(9338):991–995. doi: 10.1016/S0140-6736(02)11083-X. [DOI] [PubMed] [Google Scholar]
- 65.Lee YC, et al. MRC Twin Study Group Influence of genetic and environmental factors on the immunogenicity of Hib vaccine in Gambian twins. Vaccine. 2006;24(25):5335–5340. doi: 10.1016/j.vaccine.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 66.Konradsen HB, Henrichsen J, Wachmann H, Holm N. The influence of genetic factors on the immune response as judged by pneumococcal vaccination of mono- and dizygotic Caucasian twins. Clin Exp Immunol. 1993;92(3):532–536. doi: 10.1111/j.1365-2249.1993.tb03433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O’Connor D, Pollard AJ. Characterizing vaccine responses using host genomic and transcriptomic analysis. Clin Infect Dis. 2013;57(6):860–869. doi: 10.1093/cid/cit373. [DOI] [PubMed] [Google Scholar]
- 68.Le Chatelier E, et al. MetaHIT consortium Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 69.Lamas O, Marti A, Martínez JA. Obesity and immunocompetence. Eur J Clin Nutr. 2002;56(Suppl 3):S42–S45. doi: 10.1038/sj.ejcn.1601484. [DOI] [PubMed] [Google Scholar]
- 70.Bandaru P, Rajkumar H, Nappanveettil G. The impact of obesity on immune response to infection and vaccine: An insight into plausible mechanisms. Endocrinol Metab Synd. 2013;2:113. [Google Scholar]
- 71.Savy M, et al. Landscape analysis of interactions between nutrition and vaccine responses in children. J Nutr. 2009;139(11):2154S–2218S. doi: 10.3945/jn.109.105312. [DOI] [PubMed] [Google Scholar]
- 72.Haque R, et al. Oral polio vaccine response in breast fed infants with malnutrition and diarrhea. Vaccine. 2014;32(4):478–482. doi: 10.1016/j.vaccine.2013.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qadri F, Bhuiyan TR, Sack DA, Svennerholm A-M. Immune responses and protection in children in developing countries induced by oral vaccines. Vaccine. 2013;31(3):452–460. doi: 10.1016/j.vaccine.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 74.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328(5975):228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ferreira RB, Antunes LCM, Finlay BB. Should the human microbiome be considered when developing vaccines? PLoS Pathog. 2010;6(11):e1001190. doi: 10.1371/journal.ppat.1001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.John TJ. Experience with poliovaccines in the control of poliomyelitis in India. Public Health Rev. 1993-1994-1994;21(1-2):83–90. [PubMed] [Google Scholar]
- 77.Patriarca PA, Wright PF, John TJ. Factors affecting the immunogenicity of oral poliovirus vaccine in developing countries: Review. Rev Infect Dis. 1991;13(5):926–939. doi: 10.1093/clinids/13.5.926. [DOI] [PubMed] [Google Scholar]
- 78.Hanlon P, et al. Trial of an attenuated bovine rotavirus vaccine (RIT 4237) in Gambian infants. Lancet. 1987;1(8546):1342–1345. doi: 10.1016/s0140-6736(87)90649-0. [DOI] [PubMed] [Google Scholar]
- 79.Lagos R, et al. Effect of small bowel bacterial overgrowth on the immunogenicity of single-dose live oral cholera vaccine CVD 103-HgR. J Infect Dis. 1999;180(5):1709–1712. doi: 10.1086/315051. [DOI] [PubMed] [Google Scholar]