Abstract
In Escherichia coli, small RNAs GlmY and GlmZ feedback control synthesis of glucosamine-6-phosphate (GlcN6P) synthase GlmS, a key enzyme required for synthesis of the cell envelope. Both small RNAs are highly similar, but only GlmZ is able to activate the glmS mRNA by base-pairing. Abundance of GlmZ is controlled at the level of decay by RNase adaptor protein RapZ. RapZ binds and targets GlmZ to degradation by RNase E via protein–protein interaction. GlmY activates glmS indirectly by protecting GlmZ from degradation. Upon GlcN6P depletion, GlmY accumulates and sequesters RapZ in an RNA mimicry mechanism, thus acting as an anti-adaptor. As a result, this regulatory circuit adjusts synthesis of GlmS to the level of its enzymatic product, thereby mediating GlcN6P homeostasis. The interplay of RNase adaptor proteins and anti-adaptors provides an elegant means how globally acting RNases can be re-programmed to cleave a specific transcript in response to a cognate stimulus.
Keywords: small RNA GlmZ, glucosamine-6-phosphate synthase GlmS, RNase E adaptor protein RapZ (YhbJ), decoy small RNA GlmY, RNA mimicry, two-component system GlrK/GlrR (QseE/QseF)
Introduction
Small trans-encoded RNAs present an eminent class of ribo-regulatory molecules found in all three domains of life. In bacteria, sRNAs engage in regulatory circuits in virtually all physiological processes.1,2 Nutrient intake, metabolism, and metabolite fluxes are intricately controlled by small RNAs, as exemplified by sRNA Spot42 participating in the regulation of substrate transport and carbon catabolite repression, sRNA GlmZ mediating glucosamine-6-phosphate (GlcN6P) homeostasis, and the dual-function sRNA SgrS counteracting sugar phosphate stress.3-5 Many sRNAs regulate multiple transcripts by imperfect base-pairing, thereby altering translation efficiency and/or stability of the paired RNA. In Gram-negative bacteria, most base-pairing sRNAs require the Sm-like RNA chaperon Hfq for functionality.6 Hfq stabilizes sRNAs and stimulates formation of cognate sRNA/mRNA duplexes. In contrast, few sRNAs act by protein-binding to alter the activity of their cognate binding partners.7,8
Due to their pivotal regulatory roles, the abundance of small RNAs must be firmly controlled. Often, sRNA genes are elaborately regulated at the level of transcription.9,10 Many global regulators, such as alternative sigma factors and two-component systems (TCS), expand and invert their regulatory repertoires by integrating sRNAs into their regulons.11-13 Regulatory proteins and sRNAs often form various network motifs, resulting in feedback- and feed-forward loops or more elaborate regulatory circuits to coordinate complex physiological responses and social behavior.10,12,14 As opposed to biogenesis, sRNA decay and how it might be regulated is barely understood. Endoribonucleases RNase E and RNase III and the 3′→5′ exoribonuclease PNPase are key factors responsible for sRNA degradation in Gram-negative bacteria.6,15 RNase E often promotes the coupled degradation of sRNAs paired to their target transcripts.16-18 However, RNase E may also cleave unpaired sRNAs, either initiating their further decay or leading to variants with distinct regulatory properties.19-22
Research on the GlmY/GlmZ system in E. coli has revealed novel principles in how sRNA activities can be controlled at the level of transcription as well as decay. The homologous sRNAs GlmY and GlmZ feedback regulate expression of the key enzyme for cell wall biosynthesis, glucosamine-6-phosphate synthase (GlmS), in a hierarchical manner. In contrast, other homologous small RNAs act redundantly and/or additively on their targets. GlmZ activates translation of the glmS mRNA by base-pairing. Abundance of GlmZ is governed at the level of degradation catalyzed by RNase E. GlmZ is not recognized by its decay machinery. Rather, degradation of GlmZ requires the specific RNase adaptor protein RapZ, which binds GlmZ and targets it to decay by a mechanism involving physical interaction with RNase E. This process can be specifically countered by anti-adaptor GlmY, which functions as a decoy sRNA and sequesters RapZ when GlcN6P is limiting. The GlmY/GlmZ regulatory cascade therefore features a unique mechanism to specifically control sRNA decay in response to physiological cues by employing a dedicated RNase adaptor protein.
The Central Role of Enzyme GlmS for Synthesis of Bacterial Cell Envelope Precursors
GlmS is the key enzyme in the hexosamine biosynthesis pathway, which generates precursors for synthesis of important structural macromolecules in bacteria and eukaryotes (Fig. 1A). The product of this pathway is UDP-N-acetyl D-glucosamine (UDP-GlcNAc), an essential building block of peptidoglycan in bacteria. In Gram-negative bacteria it is additionally required for biosynthesis of lipopolysaccharides of the outer membrane. GlmS converts fructose-6-phosphate (Fru6P) into GlcN6P, which provides the first and rate-limiting step in this pathway.23,24 Subsequently, GlcN6P is converted to UDP-GlcNAc in three reactions involving enzyme GlmM and the bi-functional enzyme GlmU (Fig. 1A). GlmM and GlmU are essential under all conditions. GlmS is also essential, unless amino sugars are available in the environment. These sugars can be taken up and converted to GlcN6P, thereby bypassing the reaction catalyzed by GlmS.25 Gram-negative bacteria even possess a sophisticated system for recycling of GlcN6P from degradation of peptidoglycan, highlighting the importance of this metabolite for bacterial growth (Fig. 1A).26 Some bacteria naturally produce inhibitors of GlmS exhibiting bactericidal or fungicidal properties, thus indicating that GlmS is an important target in microbial warfare.27 However, bacteria also possess powerful mechanisms allowing them to overcome GlmS inhibition by instant and drastic overproduction of this enzyme.28 This response acts at the post-transcriptional level and involves sophisticated mechanisms of riboregulation.
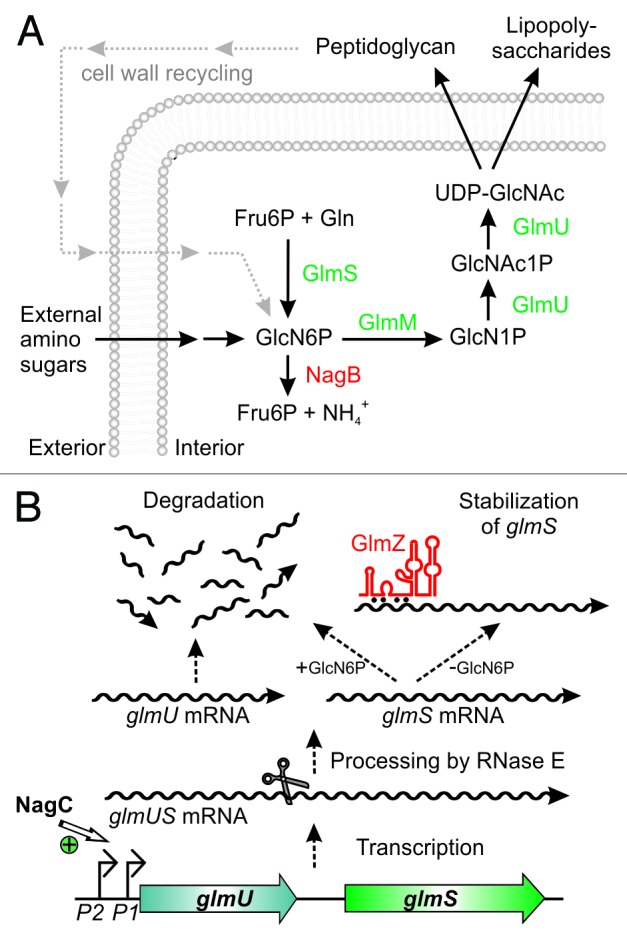
Figure 1. Key role of glucosamine-6-phosphate synthase GlmS for bacterial cell envelope synthesis. (A) The hexosamine pathway in Enterobacteriaceae. This pathway generates UDP–GlcNAc from Fru6P and glutamine (Gln) in four sequential reactions catalyzed by enzymes GlmS, GlmM, and GlmU. UDP–GlcNAc is the dedicated precursor for biosynthesis of peptidoglycan and lipopolysaccharides. GlmS catalyzes synthesis of GlcN6P, which is the key reaction. If available, various amino sugars can be taken up and converted to GlcN6P, bypassing the need for GlmS. GlcN6P can also be recycled from degradation of peptidoglycan. Degradation by enzyme NagB allows utilization of GlcN6P as nitrogen and carbon source. (B) Origin and fate of the glmUS transcript in E. coli. Genes glmU and glmS are co-transcribed from two promoters. In the absence of external amino sugars, promoter P1 is activated by transcriptional regulator NagC increasing transcription rates 3-fold. The glmUS co-transcript is processed by RNase E generating monocistronic mRNAs that are usually rapidly degraded. Upon GlcN6P depletion, the glmS mRNA can be stabilized by base-pairing with sRNA GlmZ enhancing GlmS synthesis.
Regulation of GlmS Synthesis by a GlcN6P Responsive Ribozyme in Gram-Positive Bacteria
In order to achieve metabolite homeostasis in the hexosamine pathway, activity of GlmS must be tightly controlled. Although very different in Gram-positive and Gram-negative bacteria, post-transcriptional mechanisms of glmS regulation perform the same physiological task: they mediate feedback inhibition of GlmS synthesis by its product GlcN6P, thereby regulating flux through the hexosamine pathway. Gram-positive bacteria use a metabolite responsive ribozyme to adjust GlmS enzymatic activity to the requirements of the cell.29 This cis-regulatory RNA element resides in the 5′ UTR of the glmS transcript and is inactive at low GlcN6P levels. However, at high concentrations, GlcN6P binds the glmS ribozyme and activates self-cleavage. This activity generates a 5′-hydroxylated glmS transcript that is specifically recognized and rapidly degraded by RNase J1.30 The glmS ribozyme is unique among riboswitches because ligand binding does not provoke any structural rearrangements in the RNA. In contrast, GlcN6P acts as co-enzyme and participates in the transesterification reaction leading to cleavage of the phosphodiester bond.31
The Gram-Negative Silver Bullet: sRNA GlmZ Mediates Intra-Operonic Regulation of glmS
In the Gram-negative bacterium E. coli and related species, the genes encoding GlmU and GlmS are present in one operon (Fig. 1B). The two genes are separated by an intercistronic region of 161 nt (Fig. 2A), but a ribozyme is not detectable.32 Transcription initiation at the glmUS operon is modulated 3-fold by the transcriptional regulator NagC in response to external amino sugars (Fig. 1B).33 Since GlmS is dispensable in the presence of exogenous amino sugars, this modulation of transcription frequency cannot account for the required differential expression of both enzymes.
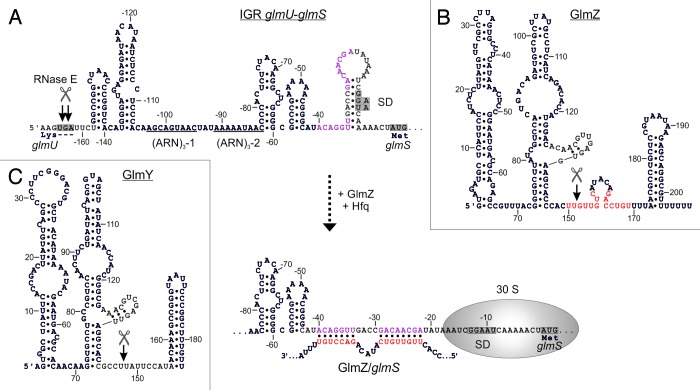
Figure 2. Activation of glmS by base-pairing with sRNA GlmZ. (A) Secondary structure of the glmUS intercistronic region and mechanism of activation of glmS translation by GlmZ. The glmUS co-transcript is processed by RNase E at the glmU stop codon (top). The two adjacent stem-loops might contribute to recognition by RNase E. A stem loop masks the SD in the glmS mRNA, thereby limiting translation initiation (top). Assisted by Hfq, full-length GlmZ base-pairs with the left half-site of this stem loop, opening the structure and providing access to ribosomes (bottom). The 5′ UTR of glmS contains two (ARN)3 motifs, providing binding sites for Hfq. Base-pairing nucleotides in glmS and GlmZ are shown in purple and red, respectively. (B and C) Secondary structures of homologous sRNAs GlmZ and GlmY. N.B., the glmS base-pairing site in GlmZ (marked red) is absent in GlmY. Processing sites are labeled by scissors.
This paradox was solved when sRNA GlmZ was found to mediate differential regulation within the glmUS operon. GlmZ (formerly RyiA or SraJ) is encoded in an intergenic region opposite to the adjacent genes aslA and hemY (Fig. 4A).34,35 As a prerequisite for regulation, the glmUS co-transcript undergoes rapid and seemingly unregulated cleavage by RNase E at the glmU stop codon, generating monocistronic glmU and glmS mRNAs (Figs. 1B and 2A).28,36 The resulting glmU mRNA lacks a stop codon and is rapidly degraded, indicating that protein GlmU is synthesized from the primary co-transcript.28,36,37 The glmS mRNA is also unstable, unless it becomes activated by base-pairing with GlmZ (Fig. 1B).28,38 Thus, GlmZ selectively activates a downstream cistron within an operon. In silico analysis predicted interaction of GlmZ with nucleotides located between positions -41 and -19 upstream of the glmS start codon (Fig. 2A).28 Indeed, activation of glmS was abolished by mutations in this region and could be rescued through compensatory base mutations in GlmZ, demonstrating direct interaction of both molecules.38 The base-pairing site in GlmZ is composed of 15 nt located in the single-stranded region between stem loops 2 and 3 (Fig. 2B).
Figure 4. Control of GlmY and GlmZ expression in E. coli. (A) Genomic context of genes glmY and glmZ and role of the TCS GlrK/GlrR (QseE/QseF) for glmY transcription. GlmY can be transcribed from overlapping σ70/σ54 promoters. Transcription of glmY by σ54 RNA-polymerase relies on activator protein GlrR and integration host factor (IHF). GlrR is a response regulator and requires phosphorylation by histidine kinase GlrK for increased DNA binding activity. The TCS GlrK/GlrR is encoded downstream of glmY within the glrK-yfhG-glrR locus, suggesting a functional connection with outer membrane protein YfhG. In E. coli, glmZ is transcribed from a constitutively active σ70 promoter. (B) The glmY promoter in E. coli K12. The sequences of the overlapping σ70/σ54 promoters are boxed in purple and yellow, respectively. The GlrR and IHF binding sites are depicted in green and orange. A putative binding site for response regulator QseB is marked in purple. Formation of the open complex by σ54 RNA-polymerase requires interaction of σ54 with a GlrR hexamer. This process is facilitated by the DNA bending activity of IHF (bottom).
GlcN6P-Regulated Decay of GlmZ Triggers Feedback Control of GlmS Synthesis
Primarily, GlmZ activates translation of glmS through an anti-antisense mechanism similar to the few other base-pairing sRNAs known to stimulate translation.39 Base-pairing with GlmZ disrupts an inhibitory stem-loop structure that sequesters the Shine-Dalgarno sequence (SD) of glmS (Fig. 2A).28,38 However, at least in a fraction of glmS mRNAs the SD might be accessible. That is, the basal level of glmS translation in absence of GlmZ is sufficient to allow growth under standard laboratory conditions. In addition, base-pairing with GlmZ also stabilizes the glmS mRNA,28 possibly resulting from protection against nucleolytic attack by increased translation.40
As expected for a base-pairing sRNA, Hfq is essential for activation of glmS by GlmZ.28,38 GlmZ strongly associates with Hfq in vivo and in vitro.22,35,41 This interaction also contributes to stability of GlmZ as observed for many other base-pairing sRNAs.6,22 Consistently, Hfq also binds with high affinity to the 5′ UTR of glmS.42 Two tripartite ARN repeats (i.e., [ARN]3 motifs, where R denotes a purine and N any nucleotide) are detectable in this region (Fig. 2A). (ARN)X motifs are believed to mediate binding to the distal surface of Hfq.20 In agreement, the (ARN)3-2 motif is essential for regulation of glmS by GlmZ.43 In conclusion, Hfq facilitates base-pairing of GlmZ and glmS similar to many other sRNA/target RNA interactions.6
Small RNA GlmZ is an exceptional case as its activity is controlled at the level of decay rather than expression. There are two versions of GlmZ: the primary GlmZ transcript, which is 207 nt long, and a shorter variant of ∼151 nt resulting from processing.22,35 Cleavage removes the base-pairing nucleotides generating a species that is unable to activate glmS (Fig. 2B). Intriguingly, processing of GlmZ is not a constant process, but is modulated by GlcN6P.28,44 Decreasing cellular concentrations of GlcN6P incrementally inhibit processing of GlmZ. Accordingly, full-length GlmZ accumulates and activates synthesis of GlmS, which replenishes GlcN6P. Thus, GlcN6P homeostasis is established at the level of GlmZ decay.
It Takes Two to Tango: RapZ is an Adaptor Protein Targeting GlmZ to Cleavage by RNase E
A search for the corresponding RNase catalyzing cleavage of GlmZ in vivo indicated involvement of RNase E. Surprisingly, in a pure in vitro system RNase E alone is insufficient to cleave GlmZ, indicating requirement for an additional factor. It was fortuitously observed that mutants lacking protein RapZ (formerly YhbJ) accumulate enormous amounts of GlmS.28 Subsequent studies established that RapZ exerts its effect on GlmS synthesis via sRNA GlmZ. Indeed, in rapZ mutants, processing of GlmZ is abolished resulting in chronic activation of glmS expression.28,38 Vice versa, overproduction of RapZ increases GlmZ cleavage rates beyond wild-type levels suggesting that RapZ is a limiting factor for processing. However, RapZ is not a ribonuclease as it lacks nucleolytic activity. In fact, cleavage of GlmZ requires the simultaneous presence of both proteins, RNase E and RapZ. In vitro, RapZ triggers correct processing of GlmZ by RNase E in a concentration-dependent manner.22
In E. coli, RapZ is encoded in the rpoN (Sigma 54) operon. Although located in different genetic contexts, homologs of RapZ are present in a wide range of bacteria indicating an important function.22,45,46 Apart from a Walker A/Walker B motif,45 RapZ does not exhibit any extended homology to other proteins. However, a C-terminal RNA binding domain was predicted for RapZ in Enterobacteriaceae.22 Notably, occurrence of this domain coincides with the presence of GlmZ (and GlmY; see below), suggesting a functional connection. Indeed, RapZ specifically binds GlmZ in vivo and in vitro with high affinity and this interaction is a prerequisite for proper processing of the sRNA.22 Intriguingly, processing of GlmZ by the concerted action of RapZ and RNase E also involves physical interaction between these proteins.22 Initial experiments suggest that RapZ forms a homotrimer and might associate with RNase E in a 3:1 stoichiometry.22,47 In bacteria that do not possess sRNAs GlmZ (and GlmY), the roles of RapZ homologs remain elusive. However, at least for the Bacillus subtilis homolog, a function in regulation of late competence genes has been described.45
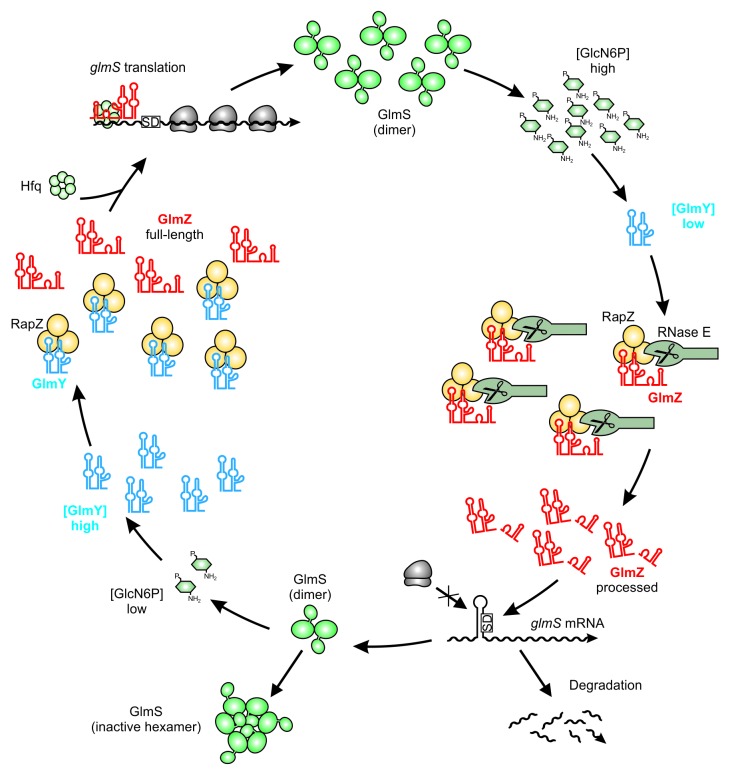
In conclusion, GlmZ can meet two fates: at limiting GlcN6P concentrations GlmZ remains unprocessed and binds Hfq to activate glmS through base-pairing. In contrast, GlmZ is preferably bound by RapZ, and consequently, degraded at high GlcN6P concentrations (Fig. 3). Hence, RapZ acts as an adaptor protein specifically directing cleavage of a sRNA by a globally acting RNase.
Figure 3. Maintenance of GlcN6P homeostasis by the regulatory GlmY/GlmZ/RapZ circuit. Under ample GlcN6P supply, sRNA GlmY is present in low amounts. Therefore, adaptor protein RapZ recruits the homologous sRNA GlmZ for cleavage by RNase E in a process that involves physical interaction of both proteins. Processed GlmZ lacks complementarity to glmS and is unable to activate glmS expression. Consequently, the glmS SD is not accessible to ribosomes, leading to low translation rates and rapid degradation of the mRNA. In addition, high GlcN6P concentrations trigger conversion of preexisting GlmS dimers to enzymatically inactive hexamers, providing feedback regulation at the protein level.82 Upon GlcN6P limitation, the processed variant of GlmY accumulates and sequesters RapZ by an RNA mimicry mechanism. As a result, GlmZ cannot be cleaved by RNase E. Consequently, unprocessed GlmZ accumulates and base-pairs with the glmS mRNA in an Hfq-dependent manner. Base-pairing disrupts the inhibitory stem loop occluding the SD, thereby allowing translation of glmS, which concomitantly stabilizes the transcript. The newly synthesized GlmS replenishes GlcN6P.
Reprogramming RNase E Activity by Association with Accessory Proteins
RNase E consists of an N-terminal catalytic domain and an unstructured C-terminal scaffolding domain, which binds RNA substrates and provides interaction sites for RNA helicase RhlB, the glycolytic enzyme enolase, and PNPase.48-50 The resulting complex, designated RNA degradosome, is required for degradation of bulk RNA. However, RNase E can associate with additional proteins leading to formation of alternative degradosomes that may serve specialized functions. For instance, helicase RhlB can be replaced by other helicases under specific conditions such as cold shock.51,52 Additional association of the degradosome with ribosomal protein L4 may selectively inhibit degradation of stress-related transcripts.53 Proteins RraA and RraB are able to change the activity and/or composition of the degradosome upon binding, rerouting cleavage activity.54,55 Hfq may replace helicase RhlB in the degradosome, thereby recruiting RNase E for degradation of sRNA-targeted transcripts.2,56
Although the various canonical and alternative degradosome components impact RNA decay by different mechanisms, they have two features in common: they simultaneously influence a multitude of transcripts and they all bind to the scaffolding domain of RNase E. In contrast, RapZ targets a single RNA molecule and interacts with the N-terminal catalytic domain of RNase E.22 This raises the possibility that the role of the catalytic domain as potential hub for interacting proteins has been underappreciated. Targeting the N terminus could provide a means for direct regulation of the nucleolytic activity of RNase E. At least for a sRNA, such a direct mode has recently been demonstrated: sRNA MicC allosterically activates RNase E through interaction with its 5′-monophosphate to trigger cleavage of its target mRNA ompD.17 The discovery of RapZ implies that more adaptors exist, which could confer substrate specificity to general ribonucleases such as RNase E and RNase III. This might provide a mechanistic basis for how globally acting RNases can be redirected to cleave specific transcripts in a controlled manner.
The Homologous Decoy sRNA GlmY Indirectly Activates glmS by Sequestration of Adaptor RapZ
RapZ targets GlmZ to cleavage by RNase E. Yet, how is this process controlled by GlcN6P? The homologous sRNA GlmY acts as a molecular mimic for GlmZ (Fig. 2B and C). When GlcN6P is limiting, GlmY accumulates and sequesters RapZ. As a consequence, GlmZ remains unprocessed and associates with Hfq to activate synthesis of GlmS (Fig. 3).22 GlmY (formerly SroF or Tke1)57,58 is a 184 nt long sRNA that undergoes rapid and apparently unregulated processing by a yet unknown enzyme at its 3′-end. The resulting 148 nt variant represents the molecule responsible for regulation in vivo. Strikingly, GlmY and GlmZ are highly similar in structure and sequence (Fig. 2B and C).22,38,44 Both sRNAs are conserved in Enterobacteriaceae. Multiple sequence alignments show that homology does not extend beyond the central stem loop structures. Thus, GlmY lacks complementarity to the glmS mRNA (Fig. 2C). Nonetheless, GlmY mediates discoordinated expression within the glmUS operon similar to GlmZ.37 Subsequent studies revealed that GlmY and GlmZ operate in a hierarchical manner to jointly attune synthesis of GlmS to the cellular GlcN6P concentration. Essentially, GlmY controls GlmS levels indirectly by antagonizing processing of GlmZ.38,44 Limiting GlcN6P concentrations induce accumulation of the processed form of GlmY by a yet unknown mechanism, ultimately leading to activation of glmS. In conclusion, GlmY and GlmZ represent a unique mechanism employed by homologous sRNAs (Fig. 3). Regulation of gene expression by redundant or additive action of homologous sRNAs is widespread in bacteria. In contrast, a hierarchical mode of action has so far only been observed for GlmY and GlmZ.12
Unlike GlmZ, GlmY is not bound by Hfq with high affinity and does not require Hfq for stability, indicating a protein-binding rather than base-pairing function.22 Indeed, RapZ binds GlmY with a slightly higher affinity as compared with GlmZ. In a ligand-fishing experiment using RapZ as bait, GlmY and GlmZ were highly enriched and collectively accounted for 80% of the co-purifying RNA, emphasizing that RapZ is highly specific for both sRNAs. RapZ interacts with the sRNAs’ central stem loop, which is a structure shared by both molecules. Consequently, GlmY and GlmZ compete for binding to RapZ. When GlmY accumulates in the cell as a consequence of GlcN6P deprivation, it sequesters RapZ and precludes GlmZ from binding. As a result, RNase E cannot be recruited to cleave GlmZ and glmS is activated (Fig. 3). This regulation could even be reconstituted in vitro: Presence of GlmY strongly inhibits processing of GlmZ by the concerted action of RNase E and RapZ. Thus, GlmY is the first example for a sRNA that regulates another sRNA through molecular mimicry.22
RNA Mimicry—A Hot Topic in Post-Transcriptional Regulation
As exemplified by the role of GlmY as mimic for GlmZ, RNA mimicry becomes an increasingly recognized mechanism governing RNA activity through titration. A paradigm is provided by the carbon storage regulatory Csr system in γ-Proteobacteria. Protein CsrA regulates translation and/or stability of target RNAs by direct binding.7,59 The cognate sRNAs CsrB and CsrC antagonize CsrA. Both sRNAs are enriched in GGA-motifs that function as CsrA-recognition sequences and are therefore capable of sequestering multiple CsrA proteins.7 Further, CsrA can even be counteracted through sequestration by an mRNA, as demonstrated for regulation of fimbriae gene expression in Salmonella.60 Another example found in the chitosugar catabolism highlights the importance of decoy RNAs for regulation of interaction between RNA molecules themselves.61,62 Presence of substrate induces the chb operon required for utilization of chitosugars. Synthesis of the separately encoded chitooligosaccharide-specific outer membrane porin ChiP is repressed by sRNA ChiX. ChiX also base-pairs with the chb mRNA. Interestingly, this interaction functions as an RNA trap that relieves chiP from repression by ChiX.
These findings may just be scratches at the surface: for eukaryotes, evidence is accumulating that transcripts may cross-regulate one another via competition for shared microRNAs.63 Similarly, bacterial RNAs may communicate with each other by trapping sRNA regulators, or acting as sponges for global RNA-binding proteins, such as CsrA or Hfq.7,61,64 Using this mechanism, untranslated regions of RNAs may also communicate with other transcripts as opposed to solely controlling stability and expression of the cognate RNA molecule. In sum, competition between RNAs for binding of shared regulators emerges as a widespread mechanism adopted for post-transcriptional regulation in all living organisms.
Polyadenylation Impacts on glmS Expression by Targeting GlmY Stability
GlmY was also the first sRNA reported to influence gene expression dependent on the poly(A) status of its 3′−end. Initially, it was observed that absence of poly(A) polymerase I (PAP I) causes accumulation of GlmS, reminiscent of the phenotype of a rapZ mutant.36 Generally, polyadenylation by PAP I facilitates the degradation of transcripts in bacteria.65 However, rather than being directly targeted by PAP I, the glmS transcript is indirectly controlled via polyadenylation of GlmY.38,44 Absence of PAP I leads to stabilization of GlmY, and consequently, of GlmZ and glmS. That is, the processed form of GlmY requires polyadenylation at its 3′-end for efficient decay. The poly(A) tail presumably provides a toehold allowing PNPase to overcome the extensive stem loop structure in GlmY.44,66 Similarly, sRNAs MicA and SraL are polyadenylated by PAP I to facilitate their degradation by PNPase.67,68 Finally, stability of antisense RNAs maintaining plasmid copy numbers are controlled by PAP I-dependent polyadenylation.65 Thus, it is possible that many of the effects exerted by PAP I on gene expression and bacterial physiology are the indirect consequence of differentially polyadenylated sRNA regulators.69
Exceptional Promoter Architectures Control Expression of GlmY and GlmZ
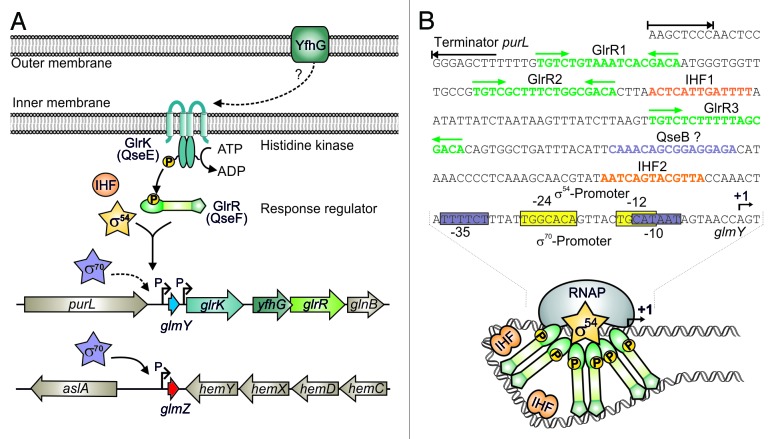
GlmY was the first sRNA shown to be controlled by sigma factor 54 in Enterobacteriaceae.37,70 Response regulator GlrR activates transcription initiation at the σ54 promoter directing expression of glmY.70 The cognate TCS GlrK/GlrR (formerly YfhK/YfhA) is encoded downstream of glmY and transcribed from an independent promoter (Fig. 4A). Sensor histidine kinase GlrK activates GlrR by phosphorylation. Phosphorylated GlrR binds to three conserved TGTCN10GACA motifs located more than 100 bp upstream of the glmY promoter (Fig. 4B).70,71 Multiple binding sites may facilitate formation of GlrR hexamers. Generally, activator proteins assemble in hexamers to catalyze open complex formation at σ54 promoters.72 In addition, binding of integration host factor IHF to two distinct sites may facilitate GlrR–RNA polymerase contacts through DNA looping (Fig. 4B).71 Gene yfhG encoding an outer membrane protein, co-localizes with the genes encoding GlrK and GlrR, suggesting a functional connection (Fig. 4A).
Surprisingly, expression of glmY is not abolished in mutants lacking σ54. This is explained by an overlapping σ70 promoter (Fig. 4B), an arrangement that is also observed in other Enterobacteriaceae.70,71 Intriguingly, both promoters start transcription at the same nucleotide, thus preventing the generation of GlmY species with altered 5′−ends,70 which may lead to functionally different variants as observed for sRNAs IstR1 and IstR2.73 Although such an overlapping σ54/σ70 promoter architecture was never observed before, more recent studies indicate that it may also apply to other genes.74 The σ70 promoter only marginally contributes to glmY transcription, suggesting that its activity could be increased under specific conditions.70
The TCS GlrK/GlrR and small RNAs GlmY and GlmZ are highly conserved among Enterobacteriaceae and their occurrence strictly coincides.71 Strikingly, in most species, glmZ is also transcribed from σ54 promoters controlled by GlrK and GlrR. Again, overlapping σ54/σ70 promoters are present upstream of glmZ in a subset of species. In contrast, in Escherichia species, glmZ is transcribed exclusively from an apparently unregulated σ70 promoter (Fig. 4A). Hence, glmY and glmZ compose a regulon controlled by GlrK/GlrR and σ54 in most Enterobacteriaceae. However, in a subset of species including E. coli, this regulon is apparently in evolutionary transition to a σ70-dependent system for reasons that remain elusive.71
A Second Function for GlmY and GlmZ in Interaction with Host Cells?
The TCS GlrK/GlrR also plays a role in virulence of various enterobacterial pathogens. In Salmonella, GlrK is required for an undisturbed expression of virulence genes and glrK mutants are impaired in invasion of epithelial cells, survival within macrophages, and in vivo colonization of liver and spleen in mice.75 In Yersinia pseudotuberculosis, glrR mutants are significantly less virulent than the wild-type as assessed in a mouse model.76 In enterohemorrhagic E. coli (EHEC), the orthologs of GlrK/GlrR are named QseE/QseF for quorum-sensing regulators E and F. Together, with the second TCS QseB/QseC, QseE/QseF controls virulence functions presumably in response to autoinducer-3 (AI-3), which is a quorum-sensing signal produced by the intestinal microbiota, and to host signals epinephrine/norepinephrine. Thus, these TCSs may function in inter-kingdom signaling and virulence regulation during host colonization.77 Both TCSs coordinate expression of espFU, which encodes an effector protein translocated to host cells, and genes located within the locus of enterocyte effacement (LEE). The LEE genes are required for adhesion of EHEC to epithelial cells and for effacement of the colonic epithelium, which includes actin rearrangement within host cells.78 However, as direct regulation of virulence gene expression by QseF could not be demonstrated, GlmY is a likely candidate linking TCS QseE/QseF to pathogenesis. Indeed, attachment of EHEC to host cells and remodeling of the host cytoskeleton by bacterial effector proteins was recently shown to rely on GlmY and GlmZ, providing the first example for sRNA-mediated virulence gene expression in EHEC.78 The sRNAs promote expression of espFU and selectively downregulate genes within the LEE4 and LEE5 loci by so far unknown mechanisms. These antagonistic regulatory effects on the expression of virulence genes seem to be confounding. However, GlmY and GlmZ may contribute to proper timing, precise modulation, and rapid adaptation of virulence gene expression during host infection. Interestingly, response regulator QseB was shown to modulate glmY expression in EHEC 2-fold. QseB apparently binds to the glmY promoter in vitro and a corresponding binding site has also been suggested in E. coli K-12 (Fig. 4B).78 Hence, both TCSs may employ GlmY and GlmZ for regulation of virulence functions. In conclusion, GlmY and GlmZ provide a further example for core-genome encoded sRNAs that were coopted for regulation of horizontally acquired genes within pathogenicity islands.79
Open Questions and Perspectives
The enterobacterial GlmYZ system represents a novel mechanism in sRNA-based regulation of unusual complexity: In response to a specific stimulus the regulatory output of a base-pairing sRNA (GlmZ) is determined by its programmed decay, which involves an adaptor protein (RapZ) for the degrading RNase, and a decoy sRNA (GlmY) that functions as an anti-adaptor (Fig. 3). So far, regulation of sRNAs has mainly been studied at the level of biogenesis revealing sophisticated and extensive control of transcription (e.g., Fig. 4). In contrast, the regulatory potential of programmed degradation of sRNAs has long been neglected. Interestingly, degradation of sRNAs CsrB and CsrC by RNase E also relies on an additional protein designated CsrD.80 The discovery of RapZ and CsrD opens the intriguing possibility that selective targeting of sRNAs to degradation by dedicated adaptor proteins might provide a ubiquitous mechanism to control sRNAs. Switching sRNA activities by regulated decay may allow cells to adapt instantly to changing physiological conditions. RapZ physically interacts with the N-terminal catalytic domain of RNase E. Hence, RapZ could serve as a co-factor to activate RNase E allosterically. Alternatively, RapZ could deliver GlmZ to membrane-bound RNase E increasing its local concentration or remodel the structure of GlmZ to a substrate that is recognized by RNase E. In conclusion, GlmYZ may represent a model system for similar mechanisms of programmed decay of sRNA regulators, not only in bacteria but perhaps even in eukaryotes.
Another elusive question concerns the mechanism of GlcN6P sensing by the GlmYZ cascade. GlmY accumulates upon GlcN6P depletion and counteracts processing of GlmZ (Fig. 3). In a glmY mutant, GlcN6P has no effect on GlmZ, emphasizing that GlmY is essential for perception of this metabolite.44 However, the TCS GlrK/GlrR, which controls glmY transcription, does not sense GlcN6P, and consequently, activity of the dual glmY promoter is not affected by GlcN6P. Therefore, GlcN6P acts post-transcriptionally.70 Does GlcN6P facilitate decay of GlmY or does it act by preventing its association with RapZ?
The unusually complex GlmY/GlmZ sRNA circuit provides a potential hub for interconnection with additional processes and regulatory pathways in the cell. Recent findings suggest that sRNAs GlmY and GlmZ have been recruited for regulation of virulence functions in EHEC and perhaps in other pathogens.78 How GlmY and GlmZ cooperate in fine-tuning of virulence gene expression and whether RapZ also plays a role in this process remains elusive. GlmY might serve additional regulatory functions even in non-pathogenic Enterobacteriaceae as it strongly accumulates at the onset of stationary phase, when GlmS synthesis is dispensable.70 The crucial role of GlmS attracts much interest to target this enzyme for antimicrobial chemotherapy.24,81 However, inhibitors of GlmS enzymatic activity are only marginally effective against Enterobacteriaceae. Activation of the GlmY/GlmZ cascade triggers overproduction of GlmS, which overcomes inhibition.28 Consequently, co-administration of compounds that prevent activation of GlmY/GlmZ is expected to potentiate the antimicrobial activity of GlmS inhibitors. The recently discovered involvement in bacterial virulence even emphasizes suitability of GlmY/GlmZ as target for antimicrobial chemotherapy.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Petra Dersch, Ralf Ficner, Eliane Hajnsdorf, Slawomir Milewski, and Jörg Vogel for fruitful collaborations and all members of the DFG priority program SPP1258 for inspiring discussions. Our research on GlmY and GlmZ was supported by grants in the framework of the German DFG priority program SPP1258 “Sensory and Regulatory RNAs in Prokaryotes.”
References
- 1.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–91. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beisel CL, Storz G. The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell. 2011;41:286–97. doi: 10.1016/j.molcel.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Görke B, Vogel J. Noncoding RNA control of the making and breaking of sugars. Genes Dev. 2008;22:2914–25. doi: 10.1101/gad.1717808. [DOI] [PubMed] [Google Scholar]
- 5.Bobrovskyy M, Vanderpool CK. Regulation of bacterial metabolism by small RNAs using diverse mechanisms. Annu Rev Genet. 2013;47:209–32. doi: 10.1146/annurev-genet-111212-133445. [DOI] [PubMed] [Google Scholar]
- 6.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–89. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romeo T, Vakulskas CA, Babitzke P. Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ Microbiol. 2013;15:313–24. doi: 10.1111/j.1462-2920.2012.02794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldelari I, Chao Y, Romby P, Vogel J. RNA-mediated regulation in pathogenic bacteria. Cold Spring Harb Perspect Med. 2013;3:a010298. doi: 10.1101/cshperspect.a010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3:a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandin P, Guillier M. Expanding control in bacteria: interplay between small RNAs and transcriptional regulators to control gene expression. Curr Opin Microbiol. 2013;16:125–32. doi: 10.1016/j.mib.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Gogol EB, Rhodius VA, Papenfort K, Vogel J, Gross CA. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc Natl Acad Sci U S A. 2011;108:12875–80. doi: 10.1073/pnas.1109379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Göpel Y, Görke B. Rewiring two-component signal transduction with small RNAs. Curr Opin Microbiol. 2012;15:132–9. doi: 10.1016/j.mib.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Fröhlich KS, Papenfort K, Berger AA, Vogel J. A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res. 2012;40:3623–40. doi: 10.1093/nar/gkr1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol Rev. 2010;34:866–82. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev. 2010;34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 16.Caron MP, Lafontaine DA, Massé E. Small RNA-mediated regulation at the level of transcript stability. RNA Biol. 2010;7:140–4. doi: 10.4161/rna.7.2.11056. [DOI] [PubMed] [Google Scholar]
- 17.Bandyra KJ, Said N, Pfeiffer V, Górna MW, Vogel J, Luisi BF. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell. 2012;47:943–53. doi: 10.1016/j.molcel.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lalaouna D, Simoneau-Roy M, Lafontaine D, Massé E. Regulatory RNAs and target mRNA decay in prokaryotes. Biochim Biophys Acta. 2013;1829:742–7. doi: 10.1016/j.bbagrm.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Davis BM, Waldor MK. RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol Microbiol. 2007;65:373–85. doi: 10.1111/j.1365-2958.2007.05796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci U S A. 2010;107:9602–7. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viegas SC, Silva IJ, Saramago M, Domingues S, Arraiano CM. Regulation of the small regulatory RNA MicA by ribonuclease III: a target-dependent pathway. Nucleic Acids Res. 2011;39:2918–30. doi: 10.1093/nar/gkq1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Göpel Y, Papenfort K, Reichenbach B, Vogel J, Görke B. Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti-adaptor RNA. Genes Dev. 2013;27:552–64. doi: 10.1101/gad.210112.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durand P, Golinelli-Pimpaneau B, Mouilleron S, Badet B, Badet-Denisot MA. Highlights of glucosamine-6P synthase catalysis. Arch Biochem Biophys. 2008;474:302–17. doi: 10.1016/j.abb.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 24.Milewski S. Glucosamine-6-phosphate synthase--the multi-facets enzyme. Biochim Biophys Acta. 2002;1597:173–92. doi: 10.1016/S0167-4838(02)00318-7. [DOI] [PubMed] [Google Scholar]
- 25.Plumbridge J, Vimr E. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J Bacteriol. 1999;181:47–54. doi: 10.1128/jb.181.1.47-54.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park JT, Uehara T. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan) Microbiol Mol Biol Rev. 2008;72:211–27. doi: 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mora I, Cabrefiga J, Montesinos E. Antimicrobial peptide genes in Bacillus strains from plant environments. Int Microbiol. 2011;14:213–23. doi: 10.2436/20.1501.01.151. [DOI] [PubMed] [Google Scholar]
- 28.Kalamorz F, Reichenbach B, März W, Rak B, Görke B. Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol Microbiol. 2007;65:1518–33. doi: 10.1111/j.1365-2958.2007.05888.x. [DOI] [PubMed] [Google Scholar]
- 29.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–6. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 30.Collins JA, Irnov I, Baker S, Winkler WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 2007;21:3356–68. doi: 10.1101/gad.1605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferré-D’Amaré AR. The glmS ribozyme: use of a small molecule coenzyme by a gene-regulatory RNA. Q Rev Biophys. 2010;43:423–47. doi: 10.1017/S0033583510000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plumbridge J. Co-ordinated regulation of amino sugar biosynthesis and degradation: the NagC repressor acts as both an activator and a repressor for the transcription of the glmUS operon and requires two separated NagC binding sites. EMBO J. 1995;14:3958–65. doi: 10.1002/j.1460-2075.1995.tb00067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–50. doi: 10.1016/S0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 35.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–51. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joanny G, Le Derout J, Bréchemier-Baey D, Labas V, Vinh J, Régnier P, Hajnsdorf E. Polyadenylation of a functional mRNA controls gene expression in Escherichia coli. Nucleic Acids Res. 2007;35:2494–502. doi: 10.1093/nar/gkm120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urban JH, Papenfort K, Thomsen J, Schmitz RA, Vogel J. A conserved small RNA promotes discoordinate expression of the glmUS operon mRNA to activate GlmS synthesis. J Mol Biol. 2007;373:521–8. doi: 10.1016/j.jmb.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 38.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fröhlich KS, Vogel J. Activation of gene expression by small RNA. Curr Opin Microbiol. 2009;12:674–82. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 40.Deana A, Belasco JG. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 2005;19:2526–33. doi: 10.1101/gad.1348805. [DOI] [PubMed] [Google Scholar]
- 41.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–19. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salim NN, Faner MA, Philip JA, Feig AL. Requirement of upstream Hfq-binding (ARN)x elements in glmS and the Hfq C-terminal region for GlmS upregulation by sRNAs GlmZ and GlmY. Nucleic Acids Res. 2012;40:8021–32. doi: 10.1093/nar/gks392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reichenbach B, Maes A, Kalamorz F, Hajnsdorf E, Görke B. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res. 2008;36:2570–80. doi: 10.1093/nar/gkn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luciano J, Foulquier E, Fantino JR, Galinier A, Pompeo F. Characterization of YvcJ, a conserved P-loop-containing protein, and its implication in competence in Bacillus subtilis. J Bacteriol. 2009;191:1556–64. doi: 10.1128/JB.01493-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boël G, Mijakovic I, Mazé A, Poncet S, Taha MK, Larribe M, Darbon E, Khemiri A, Galinier A, Deutscher J. Transcription regulators potentially controlled by HPr kinase/phosphorylase in Gram-negative bacteria. J Mol Microbiol Biotechnol. 2003;5:206–15. doi: 10.1159/000071072. [DOI] [PubMed] [Google Scholar]
- 47.Resch M, Göpel Y, Görke B, Ficner R. Crystallization and preliminary X-ray diffraction analysis of YhbJ from Escherichia coli, a key protein involved in the GlmYZ sRNA regulatory cascade. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69:109–14. doi: 10.1107/S1744309112048622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Górna MW, Carpousis AJ, Luisi BF. From conformational chaos to robust regulation: the structure and function of the multi-enzyme RNA degradosome. Q Rev Biophys. 2012;45:105–45. doi: 10.1017/S003358351100014X. [DOI] [PubMed] [Google Scholar]
- 49.Mackie GA. RNase E: at the interface of bacterial RNA processing and decay. Nat Rev Microbiol. 2013;11:45–57. doi: 10.1038/nrmicro2930. [DOI] [PubMed] [Google Scholar]
- 50.Domínguez-Malfavón L, Islas LD, Luisi BF, García-Villegas R, García-Mena J. The assembly and distribution in vivo of the Escherichia coli RNA degradosome. Biochimie. 2013;95:2034–41. doi: 10.1016/j.biochi.2013.07.022. [DOI] [PubMed] [Google Scholar]
- 51.Prud’homme-Généreux A, Beran RK, Iost I, Ramey CS, Mackie GA, Simons RW. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome’. Mol Microbiol. 2004;54:1409–21. doi: 10.1111/j.1365-2958.2004.04360.x. [DOI] [PubMed] [Google Scholar]
- 52.Khemici V, Toesca I, Poljak L, Vanzo NF, Carpousis AJ. The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: functional replacement of RhlB by RhlE. Mol Microbiol. 2004;54:1422–30. doi: 10.1111/j.1365-2958.2004.04361.x. [DOI] [PubMed] [Google Scholar]
- 53.Singh D, Chang SJ, Lin PH, Averina OV, Kaberdin VR, Lin-Chao S. Regulation of ribonuclease E activity by the L4 ribosomal protein of Escherichia coli. Proc Natl Acad Sci U S A. 2009;106:864–9. doi: 10.1073/pnas.0810205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Górna MW, Pietras Z, Tsai YC, Callaghan AJ, Hernández H, Robinson CV, Luisi BF. The regulatory protein RraA modulates RNA-binding and helicase activities of the E. coli RNA degradosome. RNA. 2010;16:553–62. doi: 10.1261/rna.1858010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao J, Lee K, Zhao M, Qiu J, Zhan X, Saxena A, Moore CJ, Cohen SN, Georgiou G. Differential modulation of E. coli mRNA abundance by inhibitory proteins that alter the composition of the degradosome. Mol Microbiol. 2006;61:394–406. doi: 10.1111/j.1365-2958.2006.05246.x. [DOI] [PubMed] [Google Scholar]
- 56.Ikeda Y, Yagi M, Morita T, Aiba H. Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol Microbiol. 2011;79:419–32. doi: 10.1111/j.1365-2958.2010.07454.x. [DOI] [PubMed] [Google Scholar]
- 57.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jäger JG, Hüttenhofer A, Wagner EG. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–43. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rivas E, Klein RJ, Jones TA, Eddy SR. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr Biol. 2001;11:1369–73. doi: 10.1016/S0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- 59.Yakhnin AV, Baker CS, Vakulskas CA, Yakhnin H, Berezin I, Romeo T, Babitzke P. CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol Microbiol. 2013;87:851–66. doi: 10.1111/mmi.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sterzenbach T, Nguyen KT, Nuccio SP, Winter MG, Vakulskas CA, Clegg S, Romeo T, Bäumler AJ. A novel CsrA titration mechanism regulates fimbrial gene expression in Salmonella typhimurium. EMBO J. 2013;32:2872–83. doi: 10.1038/emboj.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009;23:2004–15. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Overgaard M, Johansen J, Møller-Jensen J, Valentin-Hansen P. Switching off small RNA regulation with trap-mRNA. Mol Microbiol. 2009;73:790–800. doi: 10.1111/j.1365-2958.2009.06807.x. [DOI] [PubMed] [Google Scholar]
- 63.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol. 2011;82:1545–62. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Régnier P, Hajnsdorf E. Poly(A)-assisted RNA decay and modulators of RNA stability. Prog Mol Biol Transl Sci. 2009;85:137–85. doi: 10.1016/S0079-6603(08)00804-0. [DOI] [PubMed] [Google Scholar]
- 66.Andrade JM, Pobre V, Matos AM, Arraiano CM. The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA. 2012;18:844–55. doi: 10.1261/rna.029413.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J, Arraiano CM. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007;35:7651–64. doi: 10.1093/nar/gkm916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andrade JM, Arraiano CM. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA. 2008;14:543–51. doi: 10.1261/rna.683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maes A, Gracia C, Bréchemier D, Hamman P, Chatre E, Lemelle L, Bertin PN, Hajnsdorf E. Role of polyadenylation in regulation of the flagella cascade and motility in Escherichia coli. Biochimie. 2013;95:410–8. doi: 10.1016/j.biochi.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 70.Reichenbach B, Göpel Y, Görke B. Dual control by perfectly overlapping sigma 54- and sigma 70- promoters adjusts small RNA GlmY expression to different environmental signals. Mol Microbiol. 2009;74:1054–70. doi: 10.1111/j.1365-2958.2009.06918.x. [DOI] [PubMed] [Google Scholar]
- 71.Göpel Y, Lüttmann D, Heroven AK, Reichenbach B, Dersch P, Görke B. Common and divergent features in transcriptional control of the homologous small RNAs GlmY and GlmZ in Enterobacteriaceae. Nucleic Acids Res. 2011;39:1294–309. doi: 10.1093/nar/gkq986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bush M, Dixon R. The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol Mol Biol Rev. 2012;76:497–529. doi: 10.1128/MMBR.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wagner EG, Unoson C. The toxin-antitoxin system tisB-istR1: Expression, regulation, and biological role in persister phenotypes. RNA Biol. 2012;9:1513–9. doi: 10.4161/rna.22578. [DOI] [PubMed] [Google Scholar]
- 74.Zhao K, Liu M, Burgess RR. Promoter and regulon analysis of nitrogen assimilation factor, sigma54, reveal alternative strategy for E. coli MG1655 flagellar biosynthesis. Nucleic Acids Res. 2010;38:1273–83. doi: 10.1093/nar/gkp1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moreira CG, Sperandio V. Interplay between the QseC and QseE bacterial adrenergic sensor kinases in Salmonella enterica serovar Typhimurium pathogenesis. Infect Immun. 2012;80:4344–53. doi: 10.1128/IAI.00803-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Flamez C, Ricard I, Arafah S, Simonet M, Marceau M. Phenotypic analysis of Yersinia pseudotuberculosis 32777 response regulator mutants: new insights into two-component system regulon plasticity in bacteria. Int J Med Microbiol. 2008;298:193–207. doi: 10.1016/j.ijmm.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 77.Njoroge J, Sperandio V. Enterohemorrhagic Escherichia coli virulence regulation by two bacterial adrenergic kinases, QseC and QseE. Infect Immun. 2012;80:688–703. doi: 10.1128/IAI.05921-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gruber CC, Sperandio V. Posttranscriptional control of microbe-induced rearrangement of host cell actin. MBio. 2014;5:e01025–13. doi: 10.1128/mBio.01025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vogel J. A rough guide to the non-coding RNA world of Salmonella. Mol Microbiol. 2009;71:1–11. doi: 10.1111/j.1365-2958.2008.06505.x. [DOI] [PubMed] [Google Scholar]
- 80.Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006;20:2605–17. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wojciechowski M, Milewski S, Mazerski J, Borowski E. Glucosamine-6-phosphate synthase, a novel target for antifungal agents. Molecular modelling studies in drug design. Acta Biochim Pol. 2005;52:647–53. [PubMed] [Google Scholar]
- 82.Mouilleron S, Badet-Denisot MA, Pecqueur L, Madiona K, Assrir N, Badet B, Golinelli-Pimpaneau B. Structural basis for morpheein-type allosteric regulation of Escherichia coli glucosamine-6-phosphate synthase: equilibrium between inactive hexamer and active dimer. J Biol Chem. 2012;287:34533–46. doi: 10.1074/jbc.M112.380378. [DOI] [PMC free article] [PubMed] [Google Scholar]