Abstract
Wor1 (white-opaque switching regulator 1) is a master regulator of the white-opaque switching in Candida albicans, an opportunistic human fungal pathogen, and is associated with its pathogenicity and commensality. Wor1 contains a conserved DNA-binding region at the N-terminus, consisting of two conserved segments (WOPRa and WOPRb) connected by a non-conserved linker that can bind to specific DNA sequences of the promoter regions and then regulates the transcription. Here, we report the crystal structure of the C. albicans Wor1 WOPR segments in complex with a double-stranded DNA corresponding to one promoter region of WOR1. The sequentially separated WOPRa and WOPRb are structurally interwound together to form a compact globular domain that we term the WOPR domain. The WOPR domain represents a new conserved fungal-specific DNA-binding domain which uses primarily a conserved loop to recognize and interact specifically with a conserved 6-bp motif of the DNA in both minor and major grooves. The protein-DNA interactions are essential for WOR1 transcriptional regulation and white-to-opaque switching. The structural and biological data together reveal the molecular basis for the recognition and binding specificity of the WOPR domain with its specific DNA sequences and the function of Wor1 in the activation of transcription.
Keywords: crystal structure, Wor1, WOPR domain, white-opaque switching, protein-DNA recognition, transcriptional regulation
Introduction
The yeast Candida albicans is an invasive pathogen of humans but can also exist as a normal commensal in human gastrointestinal microbiome1,2. Important to its pathogenesis and commensalism is its ability to switch between different morphological forms2,3,4. C. albicans can switch between two distinct cell types, white and opaque, which have different properties including cell shape, colonial morphology, metabolic preference, mating ability, gene expression pattern, and host tissue preference5,6,7,8,9. White-opaque switching is controlled through expression of a master regulator, Wor1 (white-opaque switching regulator 1), which is highly upregulated in opaque cells and is required for both the transition to and maintenance of the opaque cell type10,11,12,13. Wor1 can genetically interact with the other five key regulators, forming a network of positive and negative feedback loops to control white-opaque switching14,15. Recently, Wor1 is also found to regulate a phenotypic switch that promotes commensalism when passing through the mammalian gut in MTLa/α cell types2.
Homologs of Wor1 have been found in every sequenced fungal genome. Besides the C. albicans Wor1 (CaWor1), all the identified Wor1 homologs are also shown to play critical roles in regulation of key developmental processes. Ryp1 from human pathogen Histoplasma capsulatum is a master regulator of yeast-mycelia transition and affects transcription of hundreds of genes16. Sge1 from plant pathogen Fusarium oxysporum is required for parasitic growth and is involved in regulation of the expression of effector genes17. Mit1 governs pseudohyphal growth in Saccharomyces cerevisiae18. Gti1 plays a role in regulation of gluconate uptake in Schizosaccharomyces pombe19. The Wor1 homologs from several necrotrophic plant pathogens are all important for pathogenesis and toxin synthesis20,21,22.
As a transcriptional regulator, Wor1 can bind to more than 100 target promoters as well as its own upstream region (nearly 200 sites in the C. albicans genome)15. CaWor1 is a transcription regulator of 785 residues, consisting of a conserved N-terminal region (residues 1-325) termed WOPR box and a non-conserved and functionally and structurally undefined C-terminal region23. The conserved WOPR box is required for binding specific DNA sequences from different promoter regions and the binding is sufficient for the Wor1-dependent transcriptional activation23. It is predicted to consist of two globular domains (WOPRa and WOPRb), dissimilar to each other, but well-conserved across the fungal lineage, connected by a low-complexity insertion23,24. The WOPR box binds DNA as a monomer and Wor1 binding requires the presence of both WOPRa and WOPRb23. However, how WOPRa and WOPRb interact with each other and bind to the specific DNA in a concerted way is unknown.
In this report, we analyzed the crystal structure of the C. albicans Wor1 WOPR box in complex with a double-stranded DNA (dsDNA) corresponding to a Wor1-binding sequence in the WOR1 promoter. Our findings reveal that WOPRa and WOPRb are structurally interwound together to form a compact globular domain which we term the WOPR domain. A conserved loop in the WOPR domain and a conserved 6-bp motif of the DNA are important for Wor1-DNA specific recognition and binding. The protein-DNA interactions are validated by both in vitro and in vivo functional assays. The structural and biological data together show that the WOPR domain of CaWor1 utilizes a combination of the base readout mechanism and the local shape readout mechanism to recognize and bind its specific DNA, which has important implications for the function of CaWor1 in the activation of white-to-opaque switching.
Results
Crystal structure of the CaWor1 WOPR-dsDNA complex
Previous biochemical data showed that the N-terminal region of CaWor1 exhibits sequence-specific DNA-binding ability and can recognize and bind specifically to six different DNA sequences corresponding to the promoter regions of three genes23. To carry out the structural studies of CaWor1 in complex with its specific DNA, we first constructed the entire WOPR box of CaWor1 (residues 1-325) into the pET28a expression plasmid with the two CUG codons (residues Ser60 and Ser200) converted to UCG to compensate for the alternative genetic code in C. albicans25 and expressed it in E. coli. The purified protein of this fragment was unstable and easily degraded into several small fragments as detected by SDS-PAGE (data not shown). Addition of various protease inhibitors including the cocktail (Roche) and PMSF in the purification process could not prevent the degradation of the protein. Considering that the low-complexity insertion is predicted to be largely unstructured and thus could be prone to proteolysis, we then made a construct consisting of the conserved WOPRa (residues 5-101) and WOPRb (residues 196-321) domains without the linker into the pETDuet expression plasmid and expressed it in E. coli. The purified protein of this construct was stable and could bind tightly to the DNA substrates, and the protein-DNA complex could be crystallized but the crystals diffracted very poorly. Sequence alignment of the WOPRb domain from several species showed that the C-terminal region of the WOPRb domain is not conserved (Supplementary information, Figure S1). Thus, we made a series of the WOPR constructs with the linker deletion and different truncated forms of the C-terminal region of the WOPRb domain. Eventually, the construct containing two fragments of WOPRa (residues 5-93) and WOPRb (residues 201-273) yielded a stable protein which could also bind effectively to the six DNA fragments (Supplementary information, Figure S2), and this protein-DNA complex led to the successful crystallization and structure determination of the WOPR-DNA complex. However, neither WOPRa nor WOPRb could be expressed and purified individually as soluble proteins (data not shown). To prepare the WOPR-dsDNA complex, we choose the 20-bp DNA sequence 5′-AAGAAGTTAAACTTTTTTGA-3′ corresponding to site 2 (−5 992 to −5 973) of the CaWOR1 promoter region as the template strand of the dsDNA.
Crystallization experiments of CaWor1 WOPR in complex with dsDNA of different lengths (13, 15, 17, and 20 bp) all yielded crystals with yet varied diffraction qualities. Finally, the crystal structure of CaWor1 WOPR in complex with a 17-bp dsDNA (5′-AAGTTAAACTTTTTTGA-3′) (WOPR-17bp dsDNA) was determined to 3.0 Å resolution, and the crystal structure of CaWor1 WOPR in complex with a 13-bp dsDNA (5′-AAGTTAAACTTTT-3′) (WOPR-13bp dsDNA) to 2.1 Å resolution (Table 1). In the structure of the WOPR-13bp dsDNA complex, there is one complex molecule in the asymmetric unit, and residues 5-90 of WOPRa and residues 203-270 of WOPRb and all nucleotides 1-13 of both DNA strands are well defined (Figure 1). In the structure of the WOPR-17bp dsDNA complex, there are two complex molecules in the asymmetric unit which are related by a non-crystallographic two-fold symmetry and have almost identical overall structure (a root-mean-square deviation (RMSD) of 0.4 Å for all Cα atoms of WOPR). Residues 6-86 of WOPRa and residues 203-266 of WOPRb and nucleotides 1-17 of both DNA strands are well defined (Supplementary information, Figure S3). Structural comparison shows that the overall structures of CaWor1 WOPR in the two complexes are very similar with an RMSD of 0.7 Å for all Cα atoms even though they have different space groups and crystal packing modes (Supplementary information, Figure S4). The only notable conformational difference occurs in the region of residues 216-226 which forms a long loop connecting the β5 and β6 strands in the 17-bp dsDNA complex but a short helix α4 and a short loop in the 13-bp dsDNA complex. This region is not involved in crystal packing in the WOPR-17bp dsDNA complex, but interacts with the C-terminus of WOPRa of a symmetry-related complex in the WOPR-13bp dsDNA complex. In addition, this region does not participate in the DNA binding in both complexes. As the structure of the WOPR-13bp dsDNA complex has a higher resolution and a better quality, we choose it as the representative in the description and analysis hereafter.
Table 1. Diffraction data and structure refinement statistics of WOPR-dsDNA complex.
| WOPR-13bp dsDNA | WOPR-17bp dsDNA (Se) | |
|---|---|---|
| Diffraction data | ||
| Wavelength (Å) | 0.9791 | 0.9793 |
| Space group | P3221 | P21 |
| Cell parameters | ||
| a, b, c (Å) | 46.6, 46.6, 215.5 | 75.7, 49.9, 84.3 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 98.0, 90.0 |
| Resolution (Å)a | 50.0-2.1 (2.2-2.1) | 50.0-3.0 (3.1-3.0) |
| Observed reflections | 121 309 | 90 116 |
| Unique reflections (I/σ(I) > 0) | 16 046 | 12 474 |
| Average redundancy | 7.6 (7.8) | 7.2 (7.4) |
| Average I/σ(I) | 31.5 (5.0) | 24.0 (6.2) |
| Completeness (%) | 95.2 (94.0) | 97.7 (98.5) |
| Rmerge (%)b | 5.7 (44.6) | 10.7 (45.6) |
| Refinement and structure model | ||
| No. of reflections (Fo > 0σ(Fo)) | ||
| Working set | 15 975 | 11 844 |
| Free R set | 810 | 609 |
| R factor (%)c | 20.3 | 22.0 |
| Free R factor (%) | 24.6 | 27.1 |
| No. of atoms | ||
| Overall | 1 916 | 3 738 |
| Protein | 1 244 | 2 351 |
| DNA | 527 | 1 382 |
| Water | 120 | 5 |
| Average B factor (Å2) | ||
| Overall | 59.5 | 84.5 |
| Protein | 61.9 | 84.1 |
| DNA | 53.9 | 85.6 |
| Water | 58.6 | 54.8 |
| RMS deviations | ||
| Bond lengths (Å) | 0.007 | 0.007 |
| Bond angles (°) | 1.3 | 1.0 |
| Ramachandran plot (%) | ||
| Favored | 98.7 | 95.8 |
| Allowed | 1.3 | 4.2 |
aNumbers in parentheses represent the highest-resolution shell.
bRmerge = ∑hkl∑i|Ii(hkl)i−<I(hkl)>|/∑hkl∑iIi(hkl)
cR = ∑hkl ||Fo|−|Fc||/∑hkl|Fo|
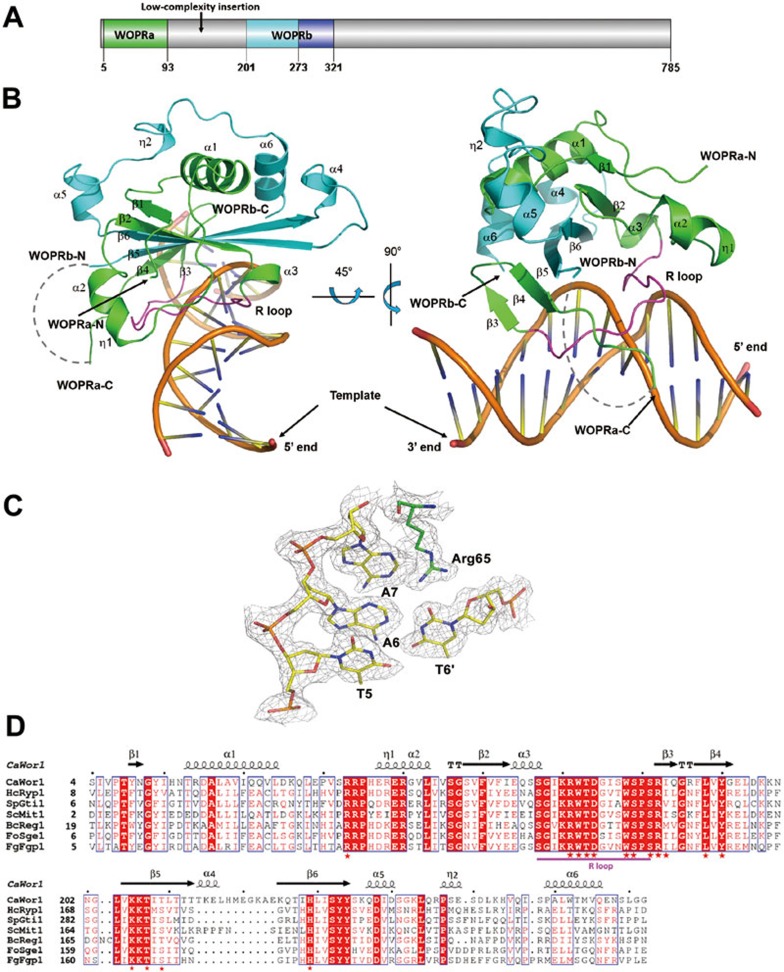
Figure 1.
Crystal structure of the Wor1 WOPR-dsDNA complex. (A) A schematic representation of the full-length CaWor1. The conserved WOPRa (residues 5-93) and WOPRb (residues 201-273) segments of CaWor1 are colored in green and cyan, respectively, the C-terminal region of WOPRb (residues 274-321) in blue, and the other regions in gray. (B) Overall structure of the WOPR-13bp dsDNA complex in two different views. The WOPR domain is shown with a ribbon model with the WOPRa and WOPRb segments colored in green and cyan, respectively, and the recognition loop is highlighted in magenta. The missing linker region between the WOPRa and WOPRb domains is indicated by a gray dash line. The dsDNA is shown with a coil model in yellow. (C) Representative simulated annealing composite omit map of the WOPR-13bp dsDNA complex. The map is contoured at 1.0σ level with the final structure shown in stick model and colored as in B. (D) Sequence alignment of the WOPRa and WOPRb segments of Wor1 from different fungal species. CaWor1, C. albicans Wor1; BcReg1, Botrysis cinerea Reg1; FgFgp1, Fusarium graminearum Fgp1; HcRyp1, Histoplasma capsulatum Ryp1; FoSge1, Fusarium oxysporum Sge1; SpGti1, Schizosaccharomyces pombe Gti1; ScMit1, Saccharomyces cerevisiae Mit1. The secondary structures of CaWor1 WOPR are placed on the top of the alignment. Strictly conserved residues are highlighted in shaded red boxes and conserved residues in open red boxes. The recognition loop is underlined in magenta. The residues involved in the interactions with dsDNA are marked with asterisks.
In the crystal structure of the WOPR-dsDNA complex, WOPRa and WOPRb, the two previously defined separate domains, are interwound with each other to form a compact globular domain which we term the WOPR domain (Figure 1B). The core structure of the WOPR domain consists of an antiparallel six-strand β-sheet (β1-β6), of which the β1-β4 strands belong to WOPRa and the β5-β6 strands belong to WOPRb. The central β-sheet is flanked by two α-helices (α2 and α3 of WOPRa) on one side and four α-helices (α1 of WOPRa and α4, α5, and α6 of WOPRb) on the other side. The WOPR domain represents a new conserved fungal DNA-binding domain (see Discussion later). The bound dsDNA assumes a largely B-form conformation with 11 base pairs per turn and a pitch of about 35 Å.
Interactions between the WOPR domain and the DNA
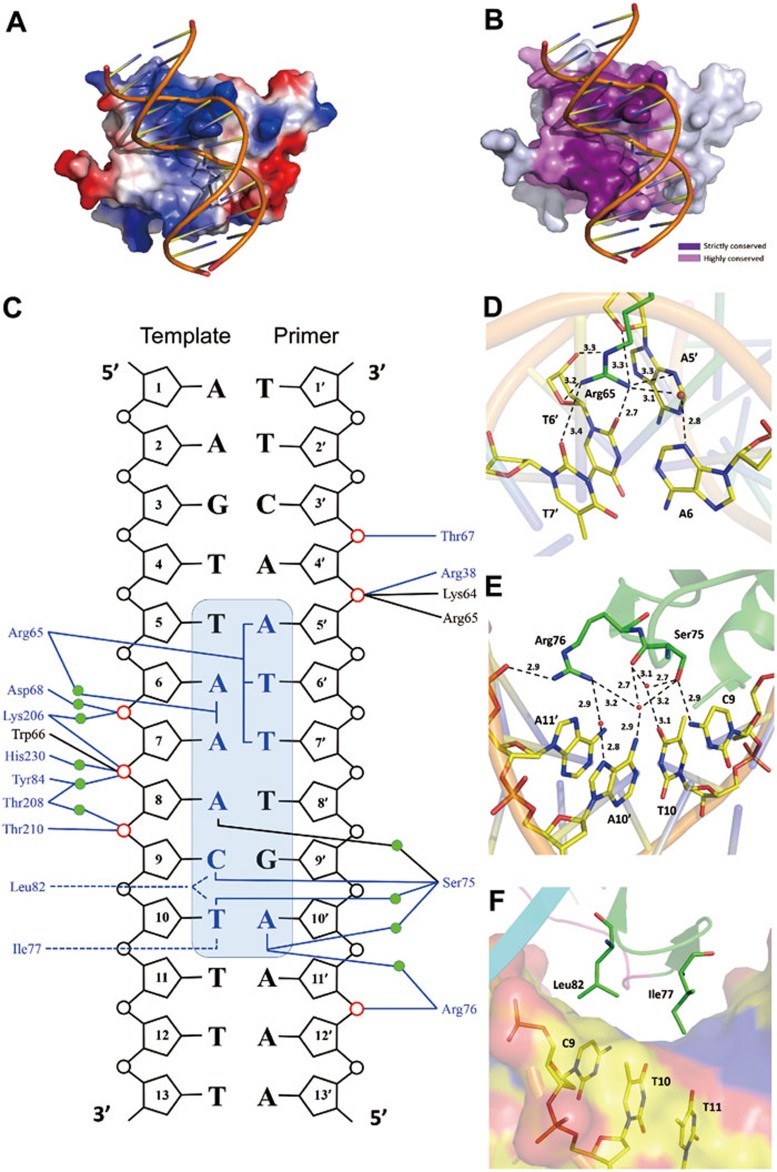
In the WOPR-dsDNA complex, the WOPR domain uses mainly the α3-β3 loop and the β3-β6 strands to interact with both the minor and major grooves of the dsDNA (Figure 1B). The interaction buries a total solvent-accessible surface area of 1 520 Å2, and the interaction interface of the WOPR domain is composed of highly conserved residues and exhibits a largely positively-charged electrostatic surface in complementary to the negatively-charged electrostatic surface of the DNA (Figure 2A and 2B). The α3-β3 loop spans from the minor groove to the major groove and several residues of this loop have hydrophilic interactions with the bases of a 6-bp core motif either directly or indirectly via water molecules (Figure 2C and Supplementary information, Table S1). Specifically, the side chain of Arg65 inserts into the minor groove and recognizes the bases of nucleotides A5′, A6, T6′, A7 and T7′, and the side chains of Ser75 and Arg76 are embedded into the major groove and recognize the bases of C9, A10′ and T10 (Figure 2D and 2E). In addition, a number of residues of this loop including Lys64, Arg65, Trp66, Thr67, Asp68, and Arg76 make hydrophilic interactions with several phosphates either directly or indirectly via water molecules. On the other hand, one edge of the β3-β6 sheet lies in the major groove and several residues including Tyr84 (β4), Lys206 (β5), Thr208 (β5), Thr210 (β5), and His230 (β6) make hydrophilic interactions with a number of phosphates mainly via water molecules (Figure 2C and Supplementary information, Table S1). In addition, the side chains of Ile77 (β3) and Leu82 (β4) make hydrophobic contacts with the bases of C9 and T10 in the major groove (Figure 2F). Sequence comparison indicates that most of the interacting residues of WOPR are strictly or highly conserved (Figure 1D). Particularly, the α3-β3 loop contains two strictly conserved sequence motifs KRWTD and WSPSR of which several residues play critical roles in the recognition and interaction of the bases of a 6-bp core motif of the DNA, and thus we term it the recognition loop (R loop).
Figure 2.
Interactions between the WOPR domain and the dsDNA. (A) A surface representation of the WOPR-dsDNA complex showing the electrostatic surface of the WOPR domain. The interaction interface of the WOPR domain exhibits a largely positively-charged electrostatic surface which is complementary to the negatively-charged electrostatic surface of the DNA. The bound dsDNA is shown with a coil model in yellow. (B) A surface representation of the WOPR-dsDNA complex showing the residue conservation of the WOPR domain. The residues of the WOPR at the interaction interface are either strictly or highly conserved. (C) A schematic representation of the interactions between the WOPR domain and the dsDNA. Hydrophilic interactions are indicated by blue solid lines and hydrophobic contacts by blue dashed lines. The core motif of the dsDNA is highlighted in a shaded blue box and the bases involved in the interactions are colored in blue. The phosphates involved in interactions with the protein are highlighted in red. The residues interacting with the nucleotides via the side chains and the main chains are colored in blue and black, respectively. Water molecules are indicated by green circles. (D) Hydrophilic interactions between the side chain of Arg65 and the nucleotides A5′, A6, T6′, and T7′ in the minor groove. Water molecules are shown as red spheres. The hydrophilic interactions are shown with dashed lines and distances. (E) Hydrophilic interactions between the side chains of Ser75 and Arg76 and the nucleotides C9, T10, A10′, and A11′ in the major groove. (F) Hydrophobic contacts between the side chains of Ile77 and Leu82 and the nucleotides C9 and T10 in the major groove. The protein is shown in ribbon and the dsDNA in van der Waals surface. Surface of the DNA is colored according to the atom types: carbon in yellow, oxygen in red, and nitrogen in blue, respectively.
Binding of the specific DNA is essential for the function of CaWor1
To test whether deletion of the linker between the WOPRa and WOPRb domains has any effect on the DNA binding, we performed in vitro quantitative electrophoretic mobility shift assay (EMSA). Our result shows that the linker deletion has no significant effect on the DNA binding (Supplementary information, Figure S2B). This is consistent with the structural data showing that in the structure of the WOPR-DNA complex, the C-terminal end of WOPRa and the N-terminal end of WOPRb are both surface exposed and point away from the DNA (Figure 1B), suggesting that the linker is likely surface exposed and is not involved in DNA binding. Similarly, our in vitro quantitative EMSA result also shows that truncation of the C-terminal region of the WOPRb domain has no significant effect on the DNA binding (Supplementary information, Figure S2B).
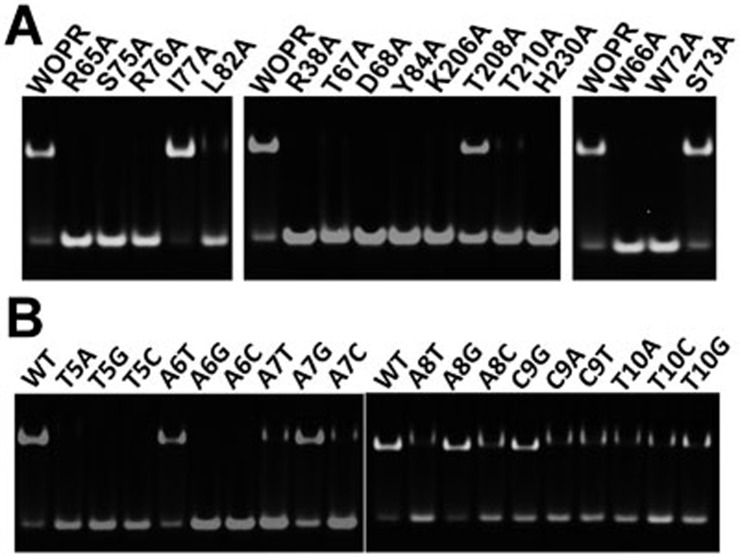
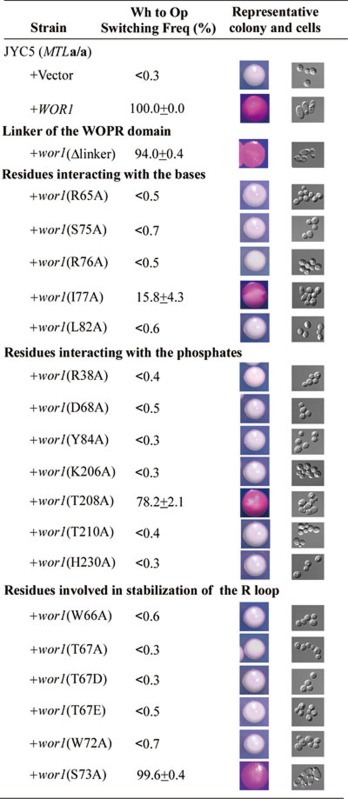
To validate the biological relevance of the WOPR-DNA interaction, we mutated the interacting residues of WOPR to Ala and tested their effects on the DNA-binding ability using in vitro qualitative and quantitative EMSA assays. Our results show that among the five residues that have interactions with the bases, mutations of four of them (R65A, S75A, R76A, and L82A) completely or severely disrupt the DNA binding (Figure 3A and Supplementary information, Table S2). However, mutation I77A has no effect on the DNA binding probably because this residue has only weak hydrophobic contact with the T10 base (Supplementary information, Table S1). Among the eight residues that have interactions with the phosphates via their side chains, mutations of seven of them (R38A, T67A, D68A, Y84A, K206A, T210A, and H230A) also completely or severely abolish the DNA-binding ability and mutation of the other (T208A) significantly impairs the DNA-binding ability (Figure 3A and Supplementary information, Table S2). These biochemical data indicate that the WOPR-DNA interaction is biologically relevant and the interactions of WOPR with the bases and phosphates of DNA are equally important.
Figure 3.
Mutational analyses of the WOPR-dsDNA interaction in vitro. (A) Mutations of the residues of WOPR involved in interactions with dsDNA and stabilization of the R loop. Left panel, mutations of the residues interacting with the bases of the dsDNA. Middle panel, mutations of the residues interacting with the phosphates of the dsDNA. Right panel, mutations of the residues involved in stabilization of the R loop. (B) Mutations of the DNA core motif recognized by the WOPR domain. The protein concentration is 10 μM and the dsDNA concentration is 5 μM (A, B).
It was shown previously that binding of the specific DNA sequence by CaWor1 is essential for the Wor1-dependent activation of transcription. To verify the functional role of the WOPR-DNA interaction in vivo, we performed morphological white-to-opaque switching assays to analyze whether CaWor1 with the linker deletion and CaWor1 mutants could activate the expression of endogenous CaWor1 and then initiate the white-to-opaque switching. Our in vivo assay result shows that the linker deletion has no significant effect on the proper functions of CaWor1 in the activation of transcription and the regulation of white-to-opaque switching (Table 2). Among the five residues that interact with the bases, mutations R65A, S75A, R76A, and L82A completely abolish the function of CaWor1, and mutation I77A significantly impairs the function (Table 2). Among the eight residues that have interactions with the phosphates via side chains, mutations of seven of them (R38A, T67A, D68A, Y84A, K206A, T210A, and H230A) also completely abolish the function of CaWor1 and mutation T208A impairs the function of CaWor1 (Table 2). The results are also consistent with the in vitro DNA binding assay results and indicate that the binding of the specific sites of the promoter region by the CaWor1 WOPR domain is essential for the proper functions of CaWor1. The discrepancy between the in vitro and in vivo assay results for mutation I77A might be due to the binding of the CaWor1 WOPR domain to other sites of the promoter region in vivo.
Table 2. White-to-opaque switching of C. albicans strains harboring wild-type WOR1 or wor1 mutants.
 |
Stabilization of the R loop is critical for DNA binding
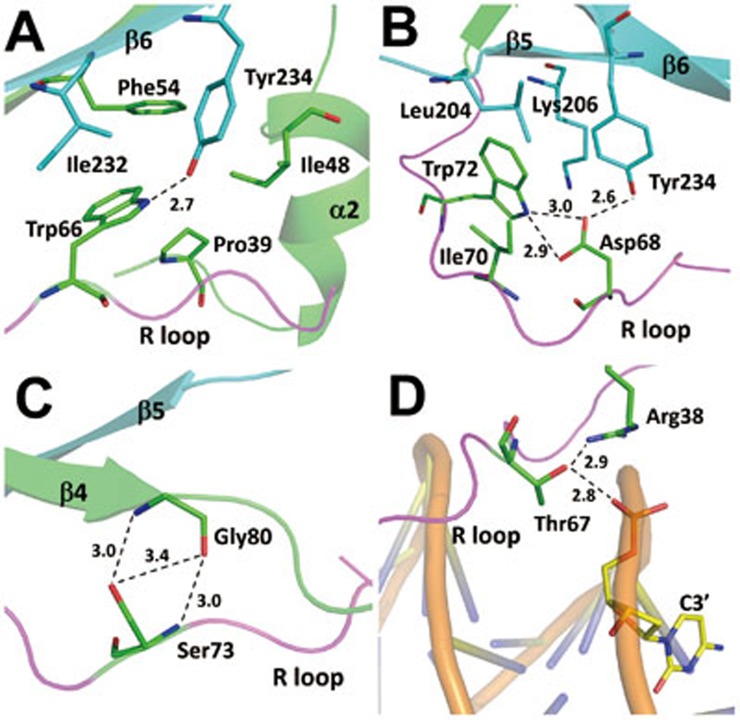
The structural and biochemical data show that the R loop plays an important role in the recognition and interaction of the DNA. A long loop usually has high conformational flexibility and needs to be stabilized in a specific conformation to recognize and interact with its partner. Structural analysis of the WOPR-dsDNA complex reveals that several strictly conserved residues in the R loop form interactions with other residues of this loop or other structural elements of WOPR which appear to play critical roles in stabilizing the conformation of the R loop. In particular, Trp66 of the R loop forms a hydrophobic cluster with Pro39 (the α1-η1 loop), Ile48 (the α2-β2 loop), Phe54 (β2), Ile232 and Tyr234 (β6), and the side chain of Trp66 also forms a hydrogen bond with the side chain of Tyr234 (Figure 4A). In addition, Trp72 of the R loop makes extensive hydrophobic interactions with Ile70 (the R loop), Leu204, and Lys206 (β5), and the side chain of Trp72 makes two hydrogen bonds with the side chain of Asp68 of the R loop (Figure 4B). Furthermore, the side chain of Ser73 of the R loop forms two hydrogen bonds with the main chain of Gly80 (β4) (Figure 4C). Our biochemical and cell biological data also show that mutations W66A and W72A completely disrupt the DNA binding in vitro (Figure 3A) and impair the proper functions of CaWor1 in the activation of transcription and the regulation of white-to-opaque switching in vivo (Table 2). However, mutation S73A has no significant effects on the DNA binding in vitro and the proper functions of CaWor1 in vivo (Figure 3A, Table 2 and Supplementary information, Table S2). These results together indicate that stabilization of the conformation of the R loop is critical for DNA binding.
Figure 4.
Stabilization of the R loop. (A) Interactions of Trp66 of the R loop with the surrounding residues. (B) Interactions of Asp68 and Trp72 of the R loop with the surrounding residues. (C) Interactions of Ser73 of the R loop with the surrounding residues. (D) Interactions of Thr67 of the R loop with the surrounding residue and nucleotide. WOPRa and WOPRb are shown as ribbon models and colored in green and cyan, respectively. The R loop is highlighted in magenta. The dsDNA is shown as a coil model and colored in yellow. Hydrogen bonds are shown with dashed lines and distances.
DNA core motif recognized by the WOPR domain
In the WOPR-dsDNA complex, the WOPR domain recognizes and interacts with the bases of the DNA sequence motif TAAACT as well as several phosphates of the backbone (Figure 2C and Supplementary information, Table S1). Our biochemical data show that the WOPR domain can bind all the six 20-bp DNA fragments (Supplementary information, Figure S2C). As the TAAACT motif is not strictly conserved among them except for site 2 of the orf19.4394 promoter (Supplementary information, Figure S2D), the biochemical results suggest that the core sequence motif of the DNA recognized by the WOPR domain can tolerate some variations. To determine the conservation of the key bases of the DNA, we designed a series of 20-bp DNA fragments of the WOR1 promoter site 2 that contain single base substitution in the core motif TAAACT (Supplementary information, Table S3) and tested their effects on the binding ability with the WOPR domain (Figure 3B). The EMSA results show that mutations T5A (which denotes mutation of T5 to A in the template strand), T5G, T5C, A6G, and A6C completely abolish the binding ability with the WOPR domain, mutations A7T, A7C, A8T, A8C, C9A, C9T, T10A, T10C, and T10G severely impair the binding ability, and mutations A6T, A7G, A8G, and C9G slightly impair the binding ability. In other words, the base at position 1 of the core motif has to be strictly a T, the base at position 2 can be either an A or T but not C and G, the bases at positions 3 and 4 prefer a purine (A or G) than pyrimidine (C or T), the base at position 5 prefers a C or G than T and A, and the base at position 6 strongly prefers a T than other bases. These results can be well explained by the structural data as these nucleotides are recognized by WOPR via hydrophilic and hydrophobic interactions and most of the substitutions would either disrupt favorable interactions or cause unfavorable steric hindrances with the protein. The structural and biochemical data together indicate that the WOPR domain can recognize and bind a dsDNA containing the characteristic motif TAAACT with a strictly conserved T at position 1 and a highly conserved T at position 6 but some variations at the other positions with base preferences, which are largely in agreement with the biochemical results by Lohse et al.23. This finding may explain why there are so many different CaWor1-binding sites (nearly 200 sites) in the C. albicans genome15. Based on these results, we predict the specific binding sites of the other four DNA fragments as TAAGGT for the MDR1 promoter, TAAAGT for the orf19.4394 promoter site 1, TAAAAA for the WOR1 promoter site 1, and TAGAGT for the WOR1 promoter site 3 (Supplementary information, Figure S2D). It should be noted that in the previous biochemical study, Lohse et al.23 identified a 9-bp core motif of the DNA substrates recognized by the full-length WOPR domain. In the structure of the WOPR-DNA complex, the C-terminal end of WOPRb points toward the major groove of the DNA (Figure 1B). Thus, it is possible that the C-terminal region of the WOPRb domain might be involved in interactions with the 3′-end nucleotides of the DNA substrate, particularly the last two thymines of the highly conserved TTT repeats of the core motif identified by Lohse et al.
Discussion
In this study, we determined the crystal structure of the WOPR domain in complex with a dsDNA of a specific sequence and carried out in vitro and in vivo functional assays to validate the protein-DNA interaction and their functional roles in the Wor1-dependent transcriptional activation and white-to-opaque switching. The structural, biochemical, and cell biological data together reveal the molecular basis for the recognition mechanism of the WOPR domain with its specific DNA. In addition, the structure of the WOPR-dsDNA complex has important implications in understanding the functions of Wor1 and other members of this protein family.
The WOPR domain represents a new conserved fungal-specific DNA-binding domain
In the structure of the WOPR-dsDNA complex, WOPRa and WOPRb, the two previously defined separate domains in sequence are interwound with each other to form a compact globular domain to bind the DNA. This explains why neither WOPRa nor WOPRb could be expressed and purified alone (this work) and exhibited DNA-binding ability23. A structural similarity search of the WOPR domain using the Dali server reveals no obvious similar structure fold in the Protein Data Bank (PDB)26. The most similar protein fold is the NAC domain (PDB: 3SWP) with a score of 3.9, a RMSD of 3.7 Å for the Cα atoms of 86 aligned residues, and 10.5% sequence identity (Supplementary information, Figure S5). The NAC domain is mainly consisted of a 5-strand β-sheet flanked by one α-helix on each side and the lengths of the β-strands are relatively longer than those in the WOPR domain. In addition, the NAC domain functions as a dimer and each monomer inserts one edge of the β-sheet into the major groove of dsDNA to interact with the phosphates and bases27, which is different from the recognition mode of the WOPR domain with its specific DNA. Thus, the WOPR domain represents a new conserved fungal-specific DNA-binding domain.
The WOPR domain utilizes both the base readout and the shape readout mechanisms to recognize and bind its specific DNA
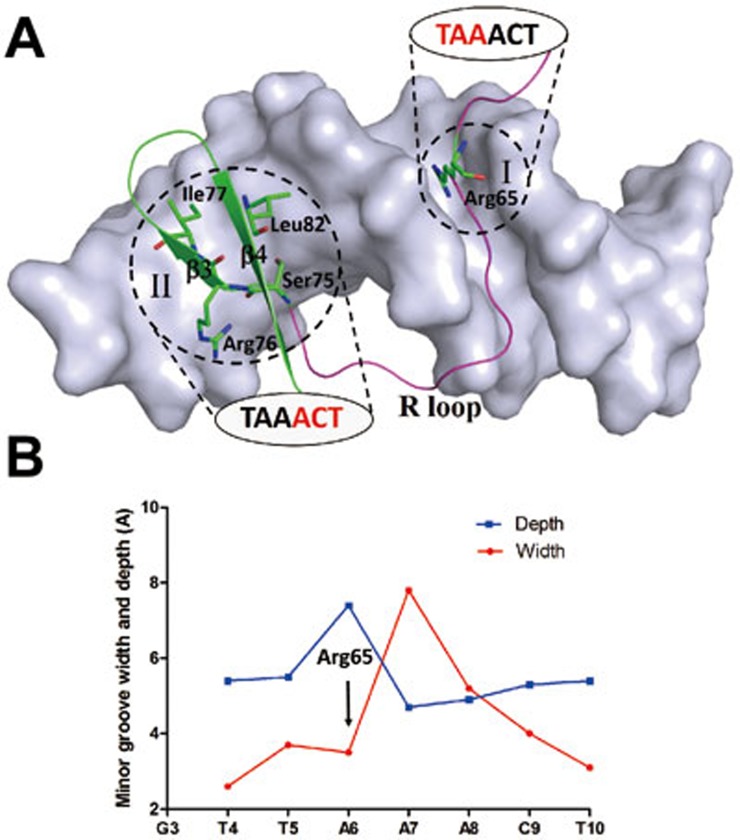
Interactions of proteins with their specific DNAs are fundamental to many biological processes. In the protein-DNA complexes, the protein can specifically recognize the DNA via two types of mechanisms: the base readout mechanism involving formation of hydrogen bonds or hydrophobic contacts with the specific bases of DNA primarily in the major groove and the shape readout mechanism involving sequence-dependent deformation of the DNA in the narrow minor groove28. In the structure of the WOPR-dsDNA complex, the WOPR domain uses mainly the R loop and the β3-β6 strands of WOPR to interact with the bases of a 6-bp core motif of the dsDNA. Several strictly conserved residues of the R loop recognize the bases of the core motif in both the minor and major grooves of the DNA, and those of the β3-β6 strands make a few hydrophobic interactions with the bases of the core motif in the major groove, indicating that the WOPR domain utilizes a typical base readout mechanism to recognize the DNA (Figure 5A). However, a detailed analysis of the WOPR-dsDNA interaction shows that Arg65 of the R loop inserts into the minor groove and specifically recognizes the first three base pairs of the DNA core motif, and compared with the ideal B-DNA, the width of the minor groove at this binding site is narrowed to about 3.5 Å and the depth is deepened to about 7.4 Å (Figure 5B). This conforms to a typical local shape readout mechanism in the narrow minor groove28. In other words, the CaWor1 WOPR domain uses both the base readout mechanism in the major groove and the local shape readout mechanism in the minor groove to recognize the core motif of the sequence-specific DNA. As CaWor1 can bind to multiple promoter regions of different genes to regulate the transcription, this combined recognition mechanism might be advantageous for CaWor1 to achieve high and distinct DNA-binding specificity. Since Wor1 is well conserved across the fungal kingdom, we suggest that the recognition mechanism of the CaWor1 WOPR domain with its specific DNA is also applicable to members of this protein family in other fungal species.
Figure 5.
Recognition mechanism of the WOPR domain with its specific DNA. (A) DNA is shown with a surface model in light blue. The R loop is colored in magenta and strands β3 and β4 in green. The five key residues of the WOPR domain (Arg65, Ser75, Arg76, Ile77, and Leu82) involved in the recognition of the DNA are shown with side chains. The specific interactions between WOPR and DNA can be divided into two regions. In region I, the side chain of Arg65 of the R loop is inserted into the narrow minor groove and recognizes the first three bases of the core motif via the local shape readout mechanism. In region II, the last three bases of the core motif in the major groove are recognized by the side chains of Ser75 and Arg76 of the R loop and Ile77 and Leu82 of the β-sheet via the base readout mechanism. (B) Graphs showing the width and depth of the minor groove of the dsDNA. The binding site of Arg65 in the minor groove is indicated. The geometrical parameters of the DNA were calculated using Curves+38.
CaPth2 might recognize the dsDNA in a similar manner
In many fungi species, there are two distinct sets of Wor1 homologous proteins23. The related family is even less studied than the Wor1 family. In C. albicans, Pth2 is a short homolog of Wor1 with yet unknown function, which consists of the two conserved WOPRa and WOPRb segments connected by a much shorter linker. Previous biochemical study showed that CaPth2 can also recognize the core DNA motif23. Sequence alignment shows that the key residues of CaWor1 involved in interactions with the dsDNA and in stabilization of the R loop are strictly or highly conserved in CaPth2 (Supplementary information, Figure S6). Among the five residues interacting with the bases, three residues (Arg65, Ser75, and Leu82) are strictly conserved, Ile77 is changed to Val and Arg76 is changed to Lys. Among the eight residues interacting with the phosphates by side chains, six residues (Arg38, Thr67, Asp68, Tyr84, Lys206, and His230) are strictly conserved, and Thr208 and Thr210 are substituted with Ser. The three residues involved in stabilization of the R loop (Trp66, Trp72, and Ser73) are also strictly conserved. Thus, we predict that the CaPth2 WOPR domain would recognize and bind the dsDNA in a similar manner to the CaWor1 WOPR domain. It is plausible that CaPth2 might be able to compete with CaWor1 to bind to the same site(s) of the promoter regions of the genes to regulate the transcription of these genes in different cellular processes.
Thr67 of the R loop is unlikely a potential phosphorylation site
It was predicted previously that Thr67 in the strictly conserved motif KRWTD of the WOPR domain is a putative Pka1 phosphorylation site and mutation of this residue could severely impair the function of Wor129 and its S. pombe homolog Gti119. However, there is no direct evidence so far showing that this residue could be phosphorylated. In the structure of the WOPR-dsDNA complex, Thr67 of the R loop is positioned on the edge of the minor groove and forms two hydrogen bonds with the side chain of Arg38 and a phosphate group (C3′) of the DNA (Figure 4D). These interactions play a critical role in the stabilization of the R loop conformation near the minor groove and thus are critical for the specific recognition of Arg65 with the narrow minor groove. Phosphorylation of Thr67 would disrupt its interaction with the DNA due to electrostatic repulsion. Indeed, our in vitro and in vivo functional assay results show that mutation of Thr67 to Ala completely abolishes the DNA binding and the proper function of CaWOR1 (Figure 3A and Table 2). Moreover, our cell biological data also show that the CaWor1 mutants containing mutations T67D and T67E which mimic the potential phosphorylation state of Thr67 cannot activate the expression of endogenous CaWor1 and then initiate the white-to-opaque switching (Table 2). These results indicate that Thr67 is unlikely a potential phosphorylation site and it plays an important role in the stabilization of the R loop to assist the specific recognition of Arg65 with the minor groove.
Materials and Methods
Cloning, expression, and purification of proteins and preparation of dsDNAs
The full-length CaWOR1 gene was amplified from the C. albicans genomic DNA and inserted into the pET-28a expression plasmid (Novagen) which attaches an N-terminal His6 tag. To express the protein properly in E. coli, the six CUG codons in CaWOR1 were mutated to UCG by several rounds of PCR25. For structural and in vitro functional studies, the cDNAs corresponding to WOPRa (residues 5-93) and WOPRb (residues 201-273) were inserted into two cloning sites of the pETDuet expression plasmid (Novagen) with a His6 tag at the C-terminus of WOPRb. Mutants of the WOPR domain were constructed using the QuikChange® Site-Directed Mutagenesis kit (Strategene).
The reconstructed expression plasmids were transformed into E. coli BL21 (DE3) Codon-Plus strain (Novagen). The transformed cells were grown in 2× YT medium at 37 °C containing 0.05 mg/ml ampicillin until OD600 reached 0.8 and then induced with 0.1 mM IPTG at 16 °C for 24 h. The cells were harvested by centrifugation and lysed by sonication in lysis buffer (20 mM Tris-HCl, 300 mM NaCl, 1 mM MgCl2, and 1 mM PMSF). The target proteins were purified by affinity chromatography using a Ni-NTA column (Qiagen) and then gel filtration chromatography using a Superdex 75 16/60 (preparative grade) column (GE Healthcare), and stored in a buffer containing 20 mM Tris-HCl, pH 8.0, 300 mM NaCl, and 1 mM MgCl2. For the Se-Met derivative protein, the expression and purification procedures were the same as the native protein, except that the cells were grown in M9 medium supplemented with amino acids Lys, Thr, Phe, Leu, Ile, Val, Se-Met, and 1% lactose. The purified proteins were of high purity (> 95%) as determined by SDS-PAGE.
The single-stranded oligonucleotides of different lengths (13, 15, 17, and 20 bases) corresponding to the template strands and their complementary strands were synthesized by Sangon Biotech (Shanghai). The dsDNAs were prepared by annealing of the template and complementary strands from 95 °C to 22 °C over a period of 6 h in the same buffer as for the protein.
Crystallization, data collection, structure determination, and refinement
To obtain the WOPR-dsDNA complexes, the dsDNA was incubated with the WOPR protein at a molar ratio of 1.5:1 at 4 °C overnight. The final concentration of the complexes for crystallization was about 20 mg/ml. Crystallization was performed using the hanging drop vapor diffusion method. Crystals were grown at 16 °C from the drop containing equal volumes (1 μl) of the complex solution and the reservoir solution (0.1 M Bis-Tris, pH 5.7, 0.2 M ammonium sulfate, and 20% PEG3350). For diffraction data collection, the crystals were cryoprotected using the reservoir solution supplemented with 40% PEG3350 and then flash-cooled into liquid nitrogen. Diffraction data were collected at 100 K at BL17U of Shanghai Synchrotron Radiation Facility, and processed, integrated, and scaled together using HKL200030.
Structure of the WOPR-17bp dsDNA complex was solved using the single wavelength anomalous dispersion (SAD) method as implemented in Phenix31 and structure of the WOPR-13bp dsDNA complex was solved using the molecular replacement (MR) method as implemented in Phenix using the WOPR-17bp dsDNA complex as the search model. Structure refinement was carried out using Phenix and Refmac532,33 and model building was performed using Coot34. Stereochemistry of the structure models was analyzed using Procheck35. Structural analyses were carried out using the CCP4 suite33 and the PISA server36. The figures were generated using Pymol37. The statistics of the structure refinement and the final structure models are summarized in Table 1.
EMSA
EMSA was performed to analyze the DNA-binding ability of the wild-type and mutant WOPR domain with 20-bp dsDNA fragments of different sequences. Specifically, 20 μl of the protein-dsDNA mixture consisting of 2 μl 20-bp dsDNA (1 μmol), 10 μl WOPR (0.5, 1, and 2 μmol), 5 μl loading buffer (50% glycerol and 0.02% bromophenol blue), and 3 μl buffer (30 mM Tris, pH 8.0, and 30 mM NaCl) were incubated in 4 °C for 30 min, and then run on gel containing 10% acrylamide, 0.5× TBE (45 mM Tris-borate, pH 8.0, and 1 mM EDTA), and 2.5% glycerol at 100 V for 120 min. The gels were stained with GelGreen (Biotium). Quantitative fluorescent EMSA is described in Supplementary information, Data S1.
White-to-opaque switching assay
To verify the functional role of the WOPR-DNA interaction in vivo, we examined the activation of wild-type Wor1 or Wor1 mutants in C. albicans white-to-opaque switching using a modified method reported previously13. White state cells or colonies were selected on SCD (synthetic complete medium with 2% glucose) plates at 37 °C for 3 days in air. For white-to-opaque switching, the white state cells were cultured in SCD and spread onto SCD (pH 6.8) plates, and incubated at 22 °C for 5-10 days in air. SCD medium was supplemented with 5 μg/ml phloxine B for detecting the opaque state. The percentage of opaque colonies or white colonies with opaque regions was calculated as the white-to-opaque switching frequency based on the method of Miller and Johnson9. The number of assessed colonies ranged between 1 000 and 2 000. The pACT1-WOR1 mutant plasmid for the expression of Wor1 mutant in C. albicans was constructed by PCR-based mutagenesis. The pACT1-WOR1 or pACT1-WOR1 mutant plasmids were linearized with AscI and introduced into MTLa/a strain JYC5 at ADE2 locus13. The ectopically expressed wild-type Wor1 from ACT1 promoter can bind to endogenous WOR1 promoter to induce its expression, and the induced endogenous Wor1 can in turn promote white-to-opaque switching. The binding affinity between ectopic Wor1 and endogenous WOR1 promoter is correlated with the activation of white-to-opaque switching.
Accession codes
The crystal structures of the WOPR-13bp dsDNA and WOPR-17bp dsDNA complexes have been deposited in PDB under accession codes 4QTJ and 4QTK, respectively.
Acknowledgments
We thank the staff members at BL17U of Shanghai Synchrotron Radiation Facility (SSRF), China, for technical support in diffraction data collection and other members of our groups for helpful discussion. This work was supported by the National Natural Science Foundation of China (31370105 and 31230017), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08010300), and the Ministry of Science and Technology of China (2011CB966301 and 2010CB912103).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
Sequence comparison of the WOPRb domain from different fungal species.
Binding of the Wor1 WOPR domain with dsDNA of different sequences.
Crystal structure of the WOPR-17bp dsDNA complex.
Comparison of the WOPR-13bp dsDNA complex and the WOPR-17bp dsDNA complex.
Comparison of the CaWor1 WOPR domain with the NAC domain.
Sequence comparison of the WOPR domain between CaWor1 and CaPth2.
Interactions between theWOPR domain and the dsDNA
Binding affinity of WOPR mutations with dsDNA
dsDNA used in the EMSA
Materials and Methods
References
- Odds FC.Candida and Candidosis, A Review and Bibliography. 2nd Edition, London: WB Saunders, 1988
- Pande K, Chen C, Noble SM. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet. 2013;45:1088–1091. doi: 10.1038/ng.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow NA, van de Veerdonk FL, Brown AJ, Netea MG. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol. 2011;10:112–122. doi: 10.1038/nrmicro2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M, Bachewich C. Morphogenesis in Candida albicans. Annu Rev Microbiol. 2007;61:529–553. doi: 10.1146/annurev.micro.61.080706.093341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger J, Wessels D, Lockhart SR, Soll DR. Release of a potent polymorphonuclear leukocyte chemoattractant is regulated by white-opaque switching in Candida albicans. Infect Immun. 2004;72:667–677. doi: 10.1128/IAI.72.2.667-677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvaal CA, Srikantha T, Soll DR. Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect Immun. 1997;65:4468–4475. doi: 10.1128/iai.65.11.4468-4475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan CY, Newport G, Murillo LA, et al. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc Natl Acad Sci USA. 2002;99:14907–14912. doi: 10.1073/pnas.232566499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- Xie J, Tao L, Nobile CJ, et al. White-opaque switching in natural MTLa/α isolates of Candida albicans: evolutionary implications for roles in host adaptation, pathogenesis, and sex. PLoS Biol. 2013;11:e1001525. doi: 10.1371/journal.pbio.1001525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci USA. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha T, Borneman AR, Daniels KJ, et al. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell. 2006;5:1674–1687. doi: 10.1128/EC.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci USA. 2006;103:12813–12818. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernday AD, Lohse MB, Fordyce PM, Nobile CJ, DeRisi JL, Johnson AD. Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol Microbiol. 2013;90:22–35. doi: 10.1111/mmi.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VQ, Sil A. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc Natl Acad Sci USA. 2008;105:4880–4885. doi: 10.1073/pnas.0710448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse CB, van Wijk R, Reijnen L, et al. The nuclear protein Sge1 of Fusarium oxysporum is required for parasitic growth. PLoS Pathog. 2009;5:e1000637. doi: 10.1371/journal.ppat.1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain CW, Lohse MB, Homann OR, Sil A, Johnson AD. A conserved transcriptional regulator governs fungal morphology in widely diverged species. Genetics. 2012;190:511–521. doi: 10.1534/genetics.111.134080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspari T. Onset of gluconate-H+ symport in Schizosaccharomyces pombe is regulated by the kinases Wis1 and Pka1, and requires the gti1+ gene product. J Cell Sci. 1997;110 (Pt 20):2599–2608. doi: 10.1242/jcs.110.20.2599. [DOI] [PubMed] [Google Scholar]
- Michielse CB, Becker M, Heller J, Moraga J, Collado IG, Tudzynski P. The Botrytis cinerea Reg1 protein, a putative transcriptional regulator, is required for pathogenicity, conidiogenesis, and the production of secondary metabolites. Mol Plant Microbe Interact. 2011;24:1074–1085. doi: 10.1094/MPMI-01-11-0007. [DOI] [PubMed] [Google Scholar]
- Okmen B, Collemare J, Griffiths S, van der Burgt A, Cox R, de Wit PJ. Functional analysis of the conserved transcriptional regulator CfWor1 in Cladosporium fulvum reveals diverse roles in the virulence of plant pathogenic fungi. Mol Microbiol. 2014;92:10–27. doi: 10.1111/mmi.12535. [DOI] [PubMed] [Google Scholar]
- Jonkers W, Dong Y, Broz K, Kistler HC. The Wor1-like protein Fgp1 regulates pathogenicity, toxin synthesis and reproduction in the phytopathogenic fungus Fusarium graminearum. PLoS Pathog. 2012;8:e1002724. doi: 10.1371/journal.ppat.1002724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Zordan RE, Cain CW, Johnson AD. Distinct class of DNA-binding domains is exemplified by a master regulator of phenotypic switching in Candida albicans. Proc Natl Acad Sci USA. 2010;107:14105–14110. doi: 10.1073/pnas.1005911107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkel H, Michael S, Weatheritt RJ, et al. ELM — the database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40:D242–D251. doi: 10.1093/nar/gkr1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MA, Tuite MF. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 1995;23:1481–1486. doi: 10.1093/nar/23.9.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welner DH, Lindemose S, Grossmann JG, et al. DNA binding by the plant-specific NAC transcription factors in crystal and solution: a firm link to WRKY and GCM transcription factors. Biochem J. 2012;444:395–404. doi: 10.1042/BJ20111742. [DOI] [PubMed] [Google Scholar]
- Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, Soll DR. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 2010;6:e1000806. doi: 10.1371/journal.ppat.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W.Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW, Sweet RM, eds. Macromolecular Crystallography: Part A. New York: Academic Press1997307–326. [DOI] [PubMed]
- Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Schrodinger LLC.The PyMOL Molecular Graphics System, , http://www.pymol.org , Version 1.3r1. 2010
- Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence comparison of the WOPRb domain from different fungal species.
Binding of the Wor1 WOPR domain with dsDNA of different sequences.
Crystal structure of the WOPR-17bp dsDNA complex.
Comparison of the WOPR-13bp dsDNA complex and the WOPR-17bp dsDNA complex.
Comparison of the CaWor1 WOPR domain with the NAC domain.
Sequence comparison of the WOPR domain between CaWor1 and CaPth2.
Interactions between theWOPR domain and the dsDNA
Binding affinity of WOPR mutations with dsDNA
dsDNA used in the EMSA
Materials and Methods