Abstract
The majority of hepatitis C virus (HCV) infection develops chronic infection, which causes steatosis, cirrhosis and hepatocellular carcinoma. However, understanding HCV chronicity and pathogenesis is hampered by its narrow host range, mostly restricted to human and chimpanzee. Recent endeavour to infect a variety of humanized mice has not been able to achieve persistent HCV infection unless the essential innate immune responsive genes are knocked out. Nevertheless, such immune-compromised humanized mice still lacked HCV infection-induced hepatopathogenesis. Here we report that transgenic mice in ICR background harboring both human CD81 and occludin genes (C/OTg) are permissive to HCV infection at a chronicity rate comparable to humans. In this mouse model, HCV accomplishes its replication cycle, leading to sustained viremia and infectivity for more than 12 months post infection with expected fibrotic and cirrhotic progression. Host factors favorable for HCV replication, and inadequate innate immune-response may contribute to the persistence. Lastly, NS3/4 protease inhibitor telaprevir can effectively inhibit de novo RNA synthesis and acute HCV infection of C/OTg mice. Thus, chronic HCV infection with complete replication cycle and hepatopathologic manifestations is recapitulated, for the first time, in immune-competent mice. This model will open a new venue to study the mechanisms of chronic hepatitis C and develop better treatments.
Keywords: HCV, animal model, humanized mice, immune-competent
Introduction
The hepatitis C pandemic has been severely impacting the healthcare system, with estimated over 185 million individuals infected with hepatitis C virus (HCV) worldwide1. Spontaneous clearance of the virus occurs in only 15% - 20% of the infected, while remainders develop chronic infection2. Chronic HCV infection becomes one of the most common causes of chronic hepatitis, and other severe liver diseases including steatosis, cirrhosis and hepatocellular carcinoma (HCC)3. Much progress has been made on HCV biology4, and how immune evasion leads to HCV persistent infection5,6. However, understanding of HCV infection and pathogenesis is still hampered by its narrow host range, mostly restricted to humans and chimpanzees. Chimpanzees are by far the best available animal model of HCV infection7, but they are unsuitable to study chronic hepatitis and associated HCC6,8, and restricted from medical research supported by the NIH or USA funding as an endangered species. Several sophisticated approaches, represented by SCID/Alb-uPA9 and Fah−/−Rag2−/−Il2rg−/− models10, have revealed how HCV infects and replicates in vivo. These models, however, are limited by lack of adaptive immune system and liver pathology, among other technical hurdles in the transplants. Two additional humanized mouse models have been developed. AFC8-huHSC/hep model engrafts both human hematopoietic stem cells and hepatocyte progenitor cells to partially reconstitute human innate immune system11, where HCV genome in liver, virus-specific T cell response and hepatic fibrosis can be detected. Rosa26-Fluc model ectopically expresses human receptor CD81 and Occludin (OCLN) carried by an adenovirus vector, enables HCV entry and transient replication in mouse hepatocytes in an immune-competent background12. However, both models suffer from hardly detectable viremia and infectious HCV particles in the periphery. Recently, a 4hEF mouse model transgenized with HCV entry receptors in C57BL/6 background show sustained serum viremia only after certain essential interferon-stimulated genes (ISGs) were knocked out13. Nevertheless, these mice show no liver immunopathology. Hence, a more reliable small animal model of HCV persistent infection with sustained viremia and immunopathological manifestations is still in the hunt.
In this work, we have managed to generate transgenic ICR mice with human CD81 and OCLN specifically expressed in hepatocytes. The humanized mice can support persistent infection by HCV particles derived from either cell cultures or chronically infected patients. HCV can accomplish the entire replication cycle in liver, and the released viral particles are fully infectious and transmissible. HCV infection of the transgenic mice showed similar rates of persistent infection to those of humans, with due manifestations of chronic hepatitis C. Induction of a unique expression profile of certain host factors and ISGs upon HCV infection may provide a clue of persistent infection. In all, we have established for the first time a valid model of HCV persistent infection in an immune-competent mouse stock.
Results
HCV persistent infection of C/OTg mice
Transgenic mice in ICR background harboring both human CD81 and OCLN genes (C/OTg) driven by a minimal mouse albumin promoter and mouse alpha fetoprotein enhancer II were generated (Supplementary information, Figure S1A). Offsprings were verified for genomic integration of the transgenes and expression of cognate receptor proteins (Supplementary information, Figure S1B and S1C). Furthermore, both human CD81 and OCLN had a dominant hepatic expression with expected hepatic cell surface localization (Supplementary information, Figure S1D and S1E). All transgenic mice were fertile and healthy (Supplementary information, Figure S2), without apparent hepatopathological alteration (Supplementary information, Figure S5B and S5E; see Supplementary information, Table S1 for baseline levels of liver function parameters and proinflammatory cytokines and chemokines in serum).
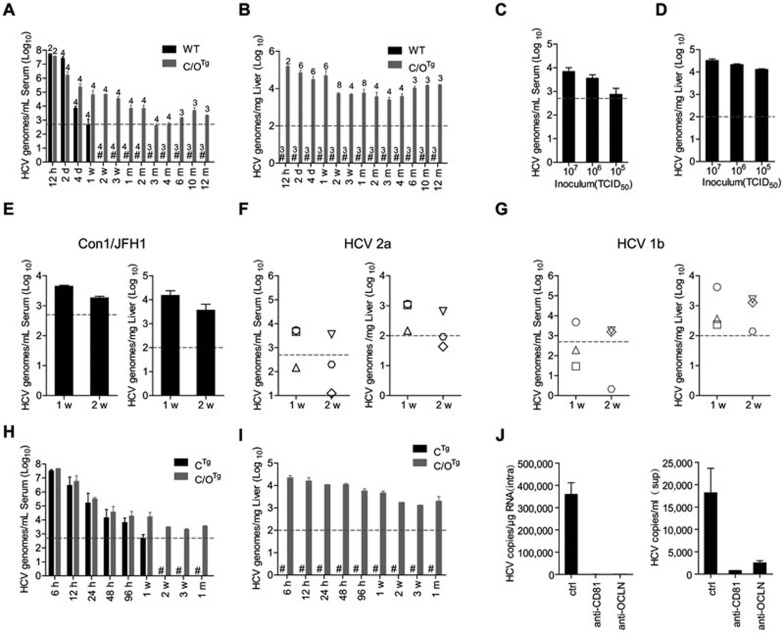
The first set of C/OTg (n = 84 in total) or wild-type (WT) (n = 44 in total) ICR mice were tail-vein injected (1 ml in 1-2 min to avoid liver damage) with HCV J399EM (TCID50 = 1 × 108/ml), a highly replicative strain derived from HCV JFH114. Most WT mice remained healthy after inoculation (Supplementary information, Table S2 and Figure S2). Quantitative RT-PCR analyses showed that HCV RNA levels declined rapidly in the blood of WT mice in the first 7 days post injection (dpi), and became undetectable afterwards (Figure 1A; see Supplementary information, Table S3 for mouse allocation and Supplementary information, Table S4 for genome copy numbers). Moreover, no HCV RNA was detected in WT liver throughout the course of study (Figure 1B). Thus, HCV merely passed through blood in these naïve mice. In contrast to the abortive infection in WT mice, HCV could infect C/OTg mice (n = 67) with sustained viral RNA levels (see Supplementary information, Tables S3 and S4 for mouse allocation and genome copy numbers, respectively) for over 12 months post infection (mpi), in both serum (Figure 1A) and liver (Figure 1B). Three C/OTg mice were succumbed to fulminant hepatitis, and 14 showed no detectable HCV RNA (due to either spontaneous clearance or unknown reasons) in either blood or liver when measured at 1 month post infection (mpi). These mice were excluded from the following prospective studies on persistent infection. HCV replicated and spreaded efficiently in C/OTg liver, with the viral loads remaining steadily above 103−4 copies/mg two weeks post infection (wpi) till the end of the study. Viral loads decreased to the lowest point at 3 mpi and started to increase at 4 mpi (Figure 1B), concomitant with the elevated hepatic inflammation and liver damage (see below). All chronic C/OTg mice exhibited no fulminant hepatitis or jaundice during the course of studies. Male but not female C/OTg mice showed a slower gain of body weight than WT (Supplementary information, Figure S2), albeit both genders had a similar degree of reduced food consumption (Supplementary information, Table S2). Abortive infection of WT or mock infection of C/OTg mice yielded no difference in body weights. Therefore, factors other than appetite change induced by HCV infection might contribute to the preferential loss of body weight in HCV-infected male C/OTg mice. These results suggested that HCV established persistent infection in immune-competent C/OTg mice, with an apparent chronicity rate (67/84 = 79.8%) similar to that in human15.
Figure 1.
HCV persistent infection of C/OTg mice. HCV RNA in serum and liver after WT and C/OTg (A-B), or CTg and C/OTg (H-I) mice were infected with HCV for the indicated time. (C, D) HCV RNA in serum (C) and liver (D) after ICR-C/OTg mice were infected with the indicated dose of HCV J399EM for 2 weeks. (E-G) HCV RNA in serum and liver of C/OTg mice after infection with Con1/JFH1 (E, TCID50 = 5 × 106/ml), or patient sera positive for HCV2a (F, 1 × 105 IU) or HCV1b (G, 1.4 × 105 IU). Each symbol in F-G indicates HCV RNA copies in individual C/OTg mouse. (J) PHTTg cells (1 × 106) were infected with HCV (MOI = 1) for 72 h, in the presence of blocking antibodies to CD81 (2 μg, sc70804, Santa Cruz), OCLN (2 μg, ab64482, Abcam) or isotype control, respectively. HCV RNA levels in cells (left) and supernatants (right) were determined. Unless indicated, 1 ml HCV J399EM (TCID50 = 1 × 108/ml) was used and HCV copies were expressed in log scales. Three mice at each time point or different treatment were used unless indicated above the bar. All data are shown as mean ± SD of at least three independent experiments. #, not detectable. Dashed grey lines indicate limit of detection, 100 copies/mg for liver tissue detection and 500 copies/ml for serum detection.
To exclude the possibility that the persistent infection in C/OTg mice was caused by a high dose of HCV, we infected C/OTg mice (n = 3 each group) with reduced inocula (1 ml, TCID50 = 107, 106 and 105/ml, respectively). The virus productively replicated in C/OTg mice with HCV RNA detectable in both blood (Figure 1C) and liver (Figure 1D) at 2 wpi, albeit hepatopathological onset was delayed with reduced doses (data not shown). C/OTg mice were permissive not only to infection by HCV J399EM in 2a genotype, but also to infection by HCV1b/2a chimeric virus Con1/JFH1 (Figure 1E; TCID50 = 5 × 106/ml, n = 6; 5 out of 6 were infected). Besides these cell culture-derived HCV (HCVcc) particles, C/OTg mice (n = 3 each group) were also able to be infected by patient sera (patient information was in Supplementary information, Table S5) positive for HCV2a (Figure 1F; 1 × 105 IU) and HCV1b (Figure 1G; 1.4 × 105 IU), which showed considerable viremia at 2 wpi (n = 1 for HCV2a and n = 2 for HCV1b, judged by serum viral loads). Furthermore, infection in C/OTg mice apparently depended on the presence of both CD81 and OCLN, because transgenic mice with CD81 alone (CTg, n = 3 for each time point) completely failed to support HCV J399EM replication (Figure 1H and 1I). This was further confirmed by in vitro studies that HCV J399EM infection of primary hepatocytes isolated from C/OTg mice (PHTTg) was efficiently blocked by antibodies to either CD81 or OCLN (Figure 1J). Therefore, C/OTg mice are permissive to infection by both HCVcc of different genotypes and clinical isolates.
Complete viral life cycle
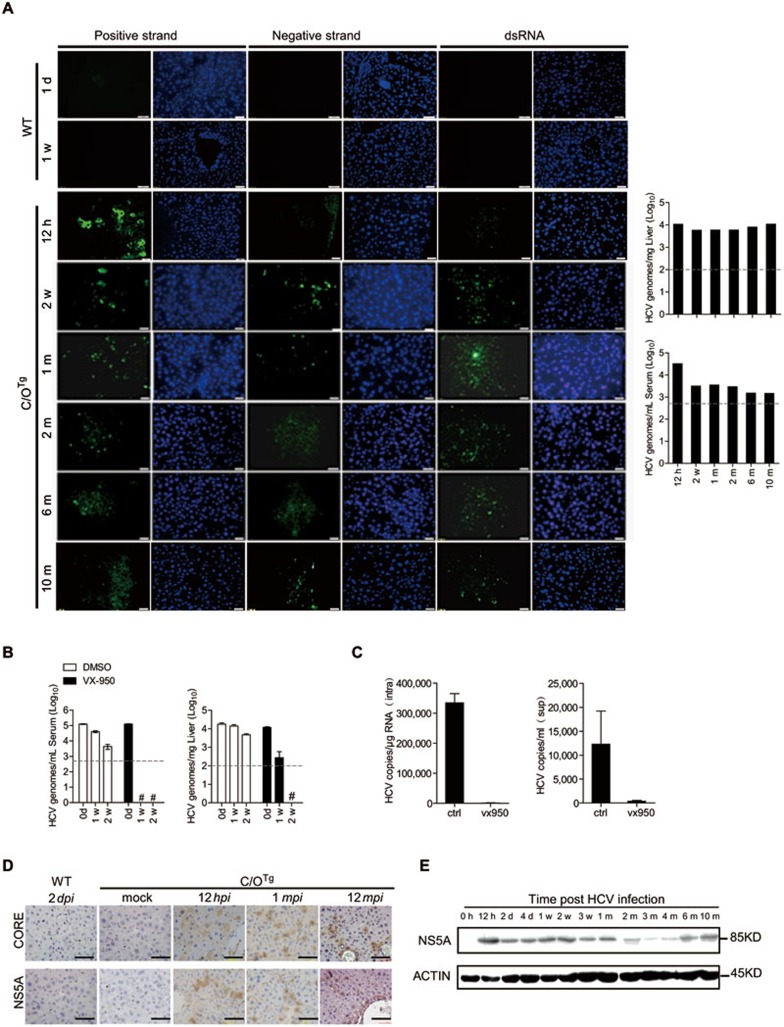
Sustained viremia in C/OTg mice suggests that HCV might have completed its replication cycle in vivo. To test whether C/OTg mice fully supported HCV RNA replication, we performed in situ hybridization of HCV ssRNA and immunofluorescent staining of dsRNA (Supplementary information, Table S6 for strand-specific probes) in the liver tissues of C/OTg mice after HCV infection. As J399EM expresses EGFP fused to NS5A, we used JFH1 (TCID50 = 5 × 106/ml) to infect C/OTg mice in this particular set of experiments to avoid fluorescent interference (n = 1 for examination at each time point; at least 5 out of 6 mice were persistently infected). Like J399EM, stable levels of JFH1 RNA (Figure 2A, bar graphs) could be detected during the entire infection (12 hpi - 10 mpi). The results of HCV ssRNA in situ hybridization and dsRNA immunofluorescent staining showed that HCV genome started to replicate as early as 12 hpi, and accomplished its genome replication steps steadily throughout the course of infection (Figure 2A). Negative-stranded RNA synthesis was confirmed by nested RT-PCR16 with an add-on primer for specific amplification of negative strands (Supplementary information, Figure S3A). To verify that the viral genome detected in liver and blood was de novo synthesized, NS3/4 protease inhibitor telaprevir (VX-950) was administered (200 mg/kg, i.p., daily for 2 weeks) in C/OTg mice (n = 6 for each time point) one week after HCV infection. Viral loads in both liver and serum were significantly reduced after VX-950 treatment for 1-2 weeks (Figure 2B). HCV replication (MOI = 1) in PHTTg could also be inhibited by VX-950 (Figure 2C). Lastly, effective genome replication led to efficient expression of HCV structural (Core) and non-structrual (NS5A) proteins in C/OTg liver as early as 12 hpi (Figure 2D and Supplementary information, Figure S3B), as depicted by immunohistochemistry (IHC) and immunoblotting (Figure 2E for NS5A). IHC scan analysis of Core protein confirmed the spread of infection from central veins to hepatocytes of hepatic lobules (Supplementary information, Figure S3C). It is of interest to point out that detection of viral antigens and HCV RNA, using conventional IHC methods and fluorescent microscopy, respectively, is technically challenging and controversial17,18,19. It has, however, become a routine and reproducible practice in our laboratories and for the contracted commercial service (see Materials and Methods) to perform these analyses on C/OTg and patient (data not shown) liver tissues.
Figure 2.
Completion of viral life cycle. (A) Complete genome replication. C/OTg (n = 6) and WT (n = 2) mice were injected with 1 ml JFH1 (TCID50 = 5 × 106/ml) for the indicated time (12 hpi-10 mpi). HCV positive and negative strands were determined in situ by molecular beacons probes (green), and dsRNA by J2 antibody (green) in liver biopsies (left). HCV RNA in liver and serum was quantified in parallel at the indicated time (right). Scale bars, 100 μm. (B) de novo RNA synthesis in vivo. C/OTg mice (n = 3 for each time point) were infected with 1 ml HCV J399EM (TCID50 = 108/ml) for 1 week before administration of VX-950 (200 mg/kg) or DMSO. HCV copies in serum and liver were measured at the indicated time of VX-950 treatment. (C) HCV RNA copies in cells (left) and supernatant (right) were determined, after PHTTg cells (1 × 106) were infected with HCV (MOI = 1) for 72 h, in the presence of VX-950 (100 μM) or DMSO control. (D) Representative IHC on Core and NS5A in liver sections (n = 2 for each time point) from infected C/OTg mice in Figure 1A. Scale bars, 100 μm. (E) Liver homogenates (100 μg total proteins) from mice in Figure 1A were immunoblotted for NS5A after C/OTg mice were infected for the indicated time. β-actin is used as loading control. Dashed grey lines indicate limit of detection.
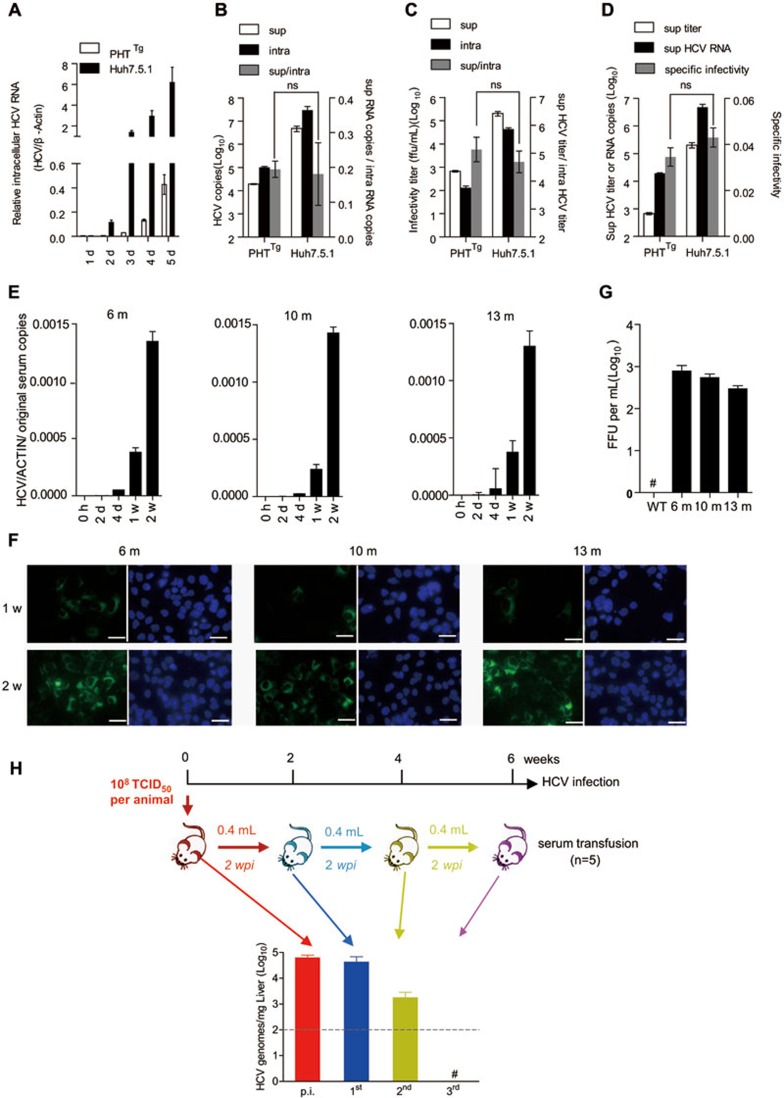
We then went on to test whether other essential viral replication steps were accomplished in C/OTg liver. Quantitative RT-PCR analysis of intracellular HCV RNA showed that HCV efficiently replicated in PHTTg cells (Figure 3A), although less robustly than in Huh7.5.1 cells, which are defective in interferon production owing to RIG-I mutation. The maturation processes of HCV, including assembly (Figure 3B for ratios of HCV RNA copies in supernatants and cells) and budding (Figure 3C for ratios of virus titers in supernatants and cells), were equally efficient in both cells. The progeny viruses released from PHTTg had the same infectivity20 as those from Huh7.5.1 cells (Figure 3D for ratios of HCV titer and HCV copies in supernatants). More importantly, the progeny virions produced in C/OTg mice were infectious. Sera prepared from HCV-infected C/OTg mice at 6 (1 896 copies/ml), 10 (6 950 copies/ml) and 13 mpi (1 745 copies/ml) effectively infected Huh7.5.1 cells (Figure 3E and 3F), with invariable infectivity (Figure 3G). Sequential passage experiments further indicated that HCV particles in circulation were fully transmissible to naïve C/OTg mice (Figure 3H). Of course, whether adaptive and/or reverse mutations are selected requires further analysis of quasispecies during the course of persistent infection. These results thus provided solid evidence that HCV had completed its life cycle and produced fully infectious virions in C/OTg mice, in good agreement with previous in vitro studies21.
Figure 3.
HCV in the C/OTg serum was infectious and transmissible. (A) Intracellular HCV RNA copies were determined after PHTTg and Huh7.5.1 cells (both 1 × 106) were infected with J399EM (MOI = 1) for the indicated time. Similar in vitro infections for 96 h were performed to measure intracellular (intra) and supernatant (sup) HCV RNA copies and titers. The assembly efficiency (B) was then plotted as the ratios of HCV RNA copies in sup over intra (sup/intra), budding efficiency (C) as ratios of sup/intra, and specific infectivity (D) as the ratios of sup HCV titer/sup HCV copies in log scales. Data were average of three independent experiments, and analyzed by student's t-test. NS, no significant difference. (E, F) Huh7.5.1 (1 × 106) cells were infected with 100 μl sera collected from C/OTg mice persistently infected by HCV at 6 mpi (1 896 copies/ml), 10 mpi (6 950 copies/ml) and 13 mpi (1 745 copies/ml). Intracellular HCV RNA levels after incubation for the indicated time were determined by quantitative RT-PCR (E), and NS5A expression (green) by immunofluorescent microscopy (F). Scale bars, 10 μm. (G) HCV FFU was assayed using sera collected at different time points from WT and C/OTg mice in E. (H) Experimental scheme of the sequential transmission (3 rounds, n = 5 for each round). C/OTg mice were infected with HCV J339EM (TCID50 = 1 × 108 ml) for 2 weeks (red). Infected mice were then sacrificed to collect liver tissue and serum. The serum (0.4 ml) was transfused to the next group of naïve C/OTg mice (n = 5) as the first round of transmission (blue mice and bars). The second (green) and the third (purple) rounds of transmission were carried out similarly. HCV RNA levels in liver tissue were analyzed by quantitative RT-PCR. Data were mean ± SD. #, not detectable. Dashed grey lines indicate limit of detection.
Histopathological manifestations
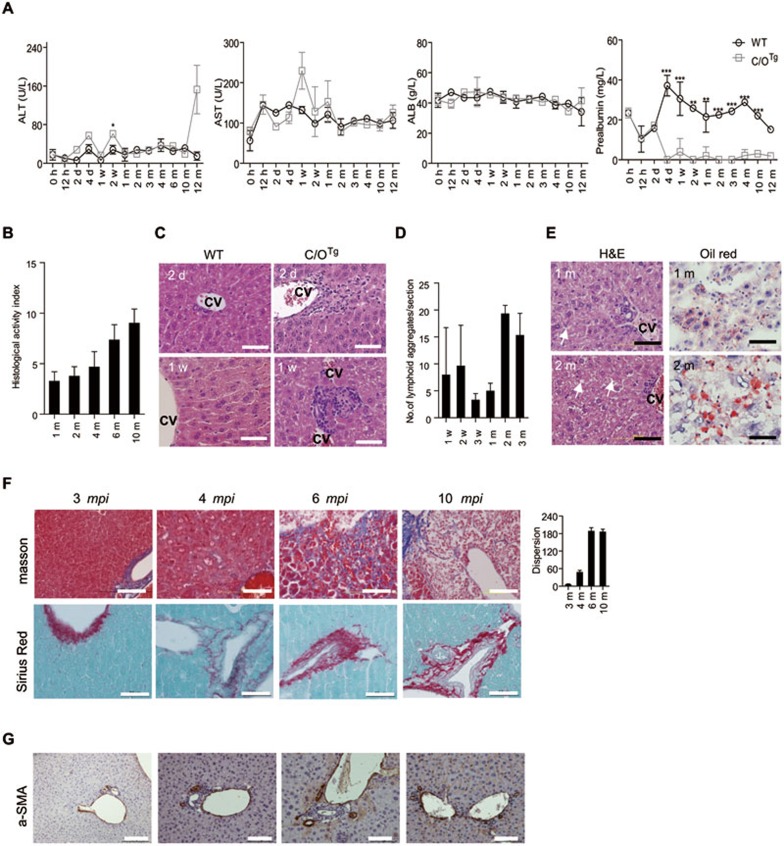
Acute HCV infection is often asymptomatic, whereas chronic manifestations progress slowly from an asymptomatic state to steatosis, fibrosis and cirrhosis in patients3. We tentatively chose 2 wpi to divide acute and persistent infection in C/OTg mice, because it was the earliest time point when the sharp reduction of viral load slowed down and leukocyte infiltration, especially NK cells (Supplementary information, Figure S4), came to a halt in liver, suggesting that homeostasis of viral replication and immune response might have reached. Similar to abortive infection in WT mice (n = 44), persistently infected C/OTg mice (n = 67) manifested normal liver function during either acute or persistent phase of infection, as demonstrated by the baseline fluctuation of ALT, AST and ALB levels (Figure 4A). ALT level was slightly higher (∼2-fold), if any, than that in WT controls at 2 wpi (n = 3 for each time point). Of note, severe reduction of pre-albumin (PA), possibly a biomarker of persistent HCV infection22, was observed in C/OTg but not WT mice, after 4 dpi (Figure 4A). Reduction of PA is seen in patients with chronic and serious health conditions, especially malnutrition and liver fibrosis23. Of note, PA has a faster turnover rate than albumin, thus representing a more sensitive biomarker of liver function. For example, albumin remains normal in about 30% patients with various liver diseases, in which PA level significantly reduces. Whether PA could serve as a valid diagnostic marker to segregate acute from chronic hepatitis C remains to be determined.
Figure 4.
Pathological manifestations of chronic hepatitis. (A) Serum ALT, AST, ALB and pre-albumin levels were measured in WT and C/OTg mice (n = 3) from Figure 1A. (B) Histological activities (Knodell index) were scored on liver sections from Figure 1A with H&E staining at the indicated time. (C) Representative H&E staining of liver sections of C/OTg(n = 4) or WT (n = 3) mice showed leukocyte infiltration (2 dpi) and lymph nodules (1 wpi). (D) Average number of lymphoid aggregates shown by histogram after C/OTg mice were infected for the indicated time. (E) Vesicular steatosis was shown by H&E and Oil Red O staining in representative C/OTg liver sections after HCV infection for 1 m and 2 m. (F, G) Representative Masson trichrome and Sirius red stainings (F), and IHC of α-SMA (G) showed fibrotic lobules in persistently infected C/OTg liver (n = 2, 3 mpi; n = 3, 4 mpi; n = 4, 6 mpi; n = 4, 10 mpi). The fibrotic dispersity (fibrosis area/fibrosis area quantity) was histogramed. cv, central vein. Scale bars, 100 μm except for 50 μm in Oil Red O and Sirius red staining. All data are shown as mean ± SD of three independent experiments. Data were analyzed by ANOVA test, *P < 0.05; **P < 0.01; ***P < 0.001.
Abortive infection in WT (Supplementary information, Figure S5A) or mock infection in C/OTg mice (Supplementary information, Figure S5B) yielded no obvious histopathological alteration in the liver. Albeit liver function remained normal, moderate hepatic inflammation occurred in persistently infected C/OTg mice (n = 4 for each time point), as scored by Knodell histological activities in H&E staining (Figure 4B). This was correlated with leukocyte infiltration, which expanded gradually from central veins (Supplementary information, Figure S5C, 12 hpi) to lobules (Figure 4C, 2 dpi). More profoundly, intrahepatic lymphoid follicles or aggregates formed around central veins, small veins and within lobules at as early as 1 wpi (Figure 4C; 4 of 8 mice) and elaborated at 2 wpi (Supplementary information, Figure S5C; 6 of 8 mice). The number of lymphnodules increased during the course of persistent infection (Figure 4D), agreeing with that in chronic hepatitis C patients24. Moreover, both H&E and Oil Red staining showed that micro- or macro-vesicular steatotic areas increased from ∼15% (1 mpi, 2 of 6 C/OTg mice) to ∼40% (2 mpi, 3 of 5 C/OTg mice) of C/OTg liver tissues (Figure 4E). Such hepatic inflammation would lead to fibrosis and cirrhosis. Indeed, serum hyaluronic acid and laminin, the direct measures of fibrinogenesis or fibrinolysis, became pronounced in persistently infected C/OTg mice (n = 4) at 4 mpi (Supplementary information, Figure S5D). Masson's and Sirius Red staining (Figure 4F) confirmed progressive fibrogenesis in C/OTg liver (3 out of 8 mice; see Supplementary information, Figure S5E and S5F for control stains), which correlated with activation of hepatic stellate cells as evidenced by α-smooth muscle actin (α-SMA) expression (Figure 4G). Fibrosis became more evident surrounding regenerative nodules at 6 (1 of 3 mice) and 10 mpi (2 of 3 mice) as revealed by liver ultrasonography (Supplementary information, Figure S6A) and non-enhanced computed tomography (CT; Supplementary information, Figure S6B; see Supplementary information, Tables S7 and S8 for fibrotic/cirrhotic classifications). Enhanced CT analysis of C/OTg liver at 6, 10 and 13 mpi (Supplementary information, Figure S6C, n = 4 for each time point) further depicted heterogeneity in morphology typified in severe fibrosis and cirrhosis25,26. None of these mice showed ascites except much leaner, but some exhibited apparent splenomegaly, a typical sign of cirrhosis due to portal hypertension (data not shown). Atypical amyloidosis around lobules (Supplementary information, Figure S5C) occurred at 3 mpi, and enlarged at 4 and 6 mpi, which potentially led to hepatic infarction (10 mpi). Finally, using aforementioned histological and non-invasive imaging classifications, we defined the time frame of severe hepatitis (Supplementary information, Figure S7), which identified 5 of 11 persistently infected mice with steatosis (1-3 mpi), 3 out of 8 showing fibrosis (6-10 mpi) and 1 out of 4 exhibiting early stage of cirrhosis (13 mpi). Compared to the rest of persistent C/OTg mice in the same category, no apparent association was observed between viral loads and liver injury, fibrosis or cirrhosis, except that hepatic viral load was correlated with steatosis. One mouse showed higher ALT level than non-cirrhotic group (Figure 4A and Supplementary information, Figure S7). In conclusion, HCV infection in our transgenic mice recapitulated the natural history of both acute2 and chronic3,27 hepatitis C, from asymptomatic in acute infection to progressive liver injury in chronic infection.
Potential mechanisms for permissiveness to HCV infection in ICR-C/OTg mice
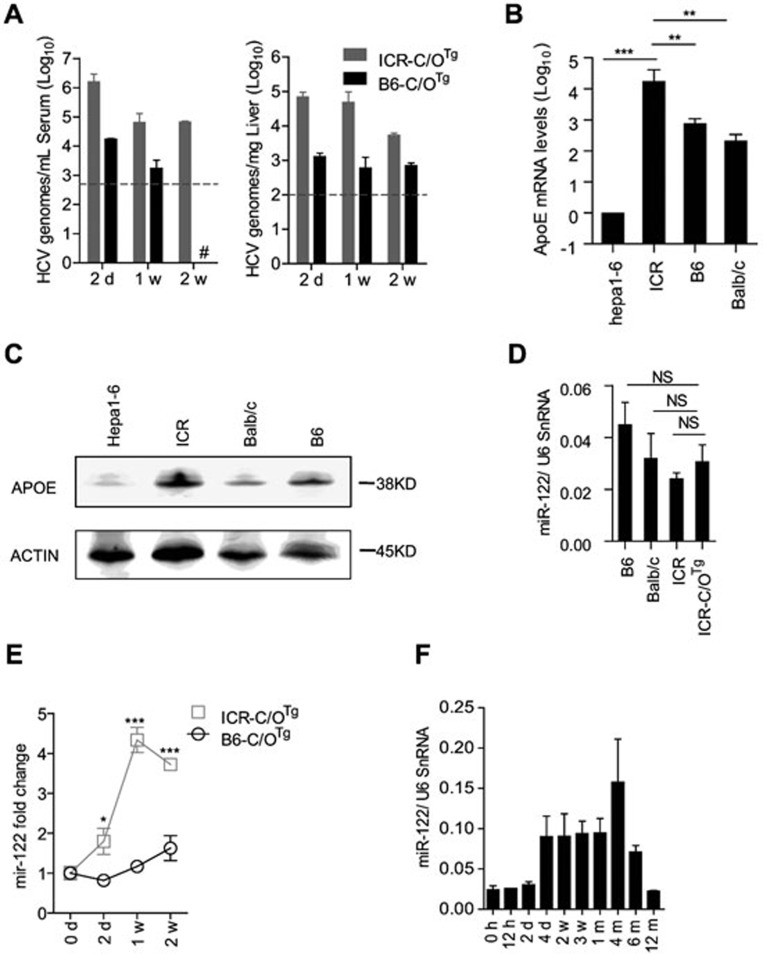
It is rather striking that HCV can establish persistent infection in immune-competent ICR-C/OTg mice, as a similar transgenic approach using 4hEF mouse model in C57BL/6 background fails to show sustained HCV replication13. This promoted us to first assess whether any known host factors essential for HCV replication and infection may differ between ICR and C57BL/6 strains. To address this issue, we managed to backcross C/OTg to C57BL/6 background (B6-C/OTg). Strikingly, HCV RNA copy number was significantly lower in serum or liver of B6-C/OTg mice at 2 wpi, compared to that in ICR-C/OTg mice (Figure 5A). Cyclophilin A (CypA), required for sustainable HCV replication in 4hEF Stat1−/− mice13, was induced to the similar level by HCV infection in both strains (data not shown). CypA thus is unlikely a determinant factor for HCV persistant infection in ICR-C/OTg mice. Surprisingly, ICR hepatocytes expressed much higher levels of apoE mRNA (Figure 5B) and protein (Figure 5C) than those of C57BL/6. This is consistent with a previous report that ectopic expression of apoE in mouse hepatocytes, which are poor producer of infectious progeny HCV particles28,29, can significantly improve HCV production30. Whether the higher level of apoE could facilitate efficient spread of HCV in hepatocytes of ICR-C/OTg mice will be a subject of future studies. Additionally, miR-122 is a liver-specific facilitator for HCV replication31, which expressed at a similar basal level in ICR and C57BL/6 hepatocytes (Figure 5D). Interestingly, miR-122 was significantly induced during HCV acute infection (Figure 5E) and kept increasing through the persistent infection (Figure 5F) in ICR-C/OTg mice, while its expression level barely changed upon HCV infection in B6-C/OTg mice (Figure 5E). Therefore, unique hepatic components critical for HCV replication and assembly in ICR-C/OTg mice might help explain HCV persistent infection. The roles of apoE and miR-122 in assistance of HCV infection in ICR-C/OTg mice is worth of further investigation.
Figure 5.
ICR-C/OTg mice showed unique host factor response to HCV infection. (A) Measurement of HCV RNA in serum and liver after HCV infection of ICR-C/OTg or B6-C/OTg mice for the indicated time. (B, C) Comparison of apoE mRNA (B) and protein (C) levels in different mouse strains. Hepa1-6 cell line was used as a control. (D) Quantitation of hepatic miR-122 levels in various mouse strains. U6 SnRNA as an internal control. (E) Comparison of hepatic miR-122 levels after HCV infection of ICR-C/OTg and B6-C/OTg mice for the indicated time. (F) Hepatic miR-122 response to HCV persistent infection of ICR-C/OTg mice. Unless indicated, three mice were used for infection with 1 ml HCV J399EM (TCID50 = 1 × 108/ml) at each time point. Data were analyzed by ANOVA test and shown as mean ± SD, *P < 0.05; **P < 0.01; ***P < 0.001. NS, no significant difference. Dashed grey lines indicate limit of detection.
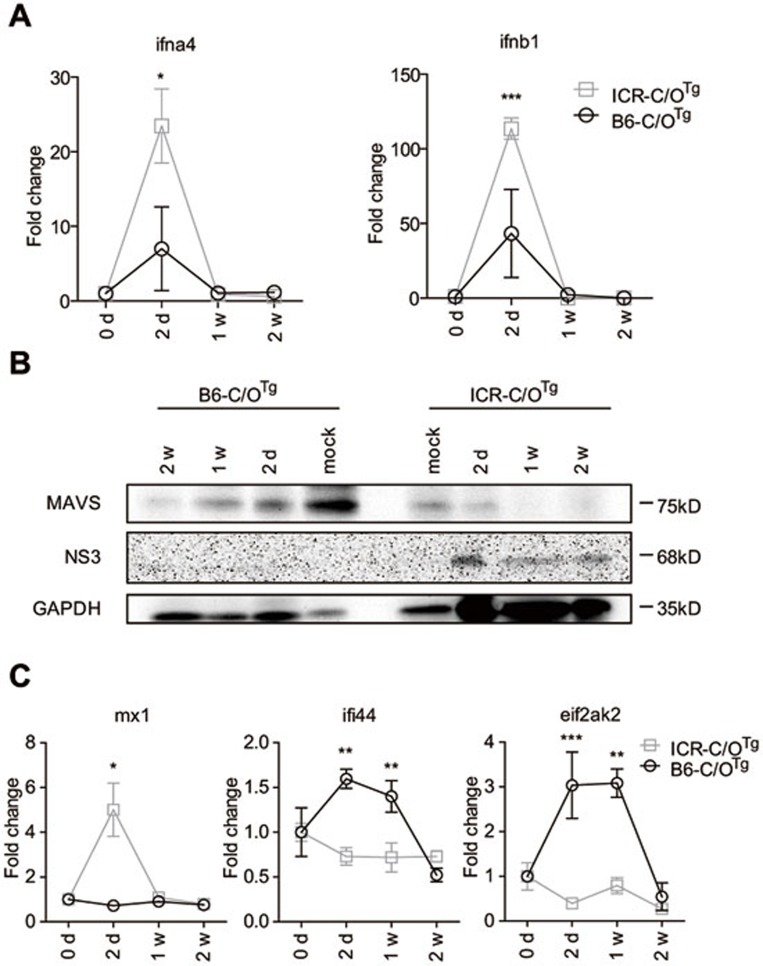
Despite these host factor response favorable for HCV replication in ICR hepatocytes, HCV still has to overcome the next block(s) of host immunity for efficient infection and spread32. Therefore, HCV persistent infection in ICR-C/OTg mice suggests either a specific defect in host immune response or an effective immune evasion of HCV. ICR mice are innate immune-competent and have been widely used in studying the innate immunity33. When NF-κB and IRF3 signaling was compared using peritoneal macrophages isolated from ICR- and B6-C/OTg mice, both mouse models showed intact and indistinguishable response to polyI:C (via TLR3, Supplementary information, Figure S8A) and LPS (via TLR4, Supplementary information, Figure S8B). Moreover, ICR-C/OTg mice exhibited stronger type I IFN response to VSV infection (via RIG-I) than B6- C/OTg mice (Supplementary information, Figure S8C). Therefore, CD81/OCLN transgenes in ICR mice would unlikely perturb the due innate immune response to HCV. Indeed, quantitative RT-PCR measurement showed that hepatic IFNα4 and IFN1 were transiently induced by HCV infection in both ICR- and B6-C/OTg mice (Figure 6A), albeit the overall level of IFNβ in serum was low throughout the course of infection in ICR-C/OTg mice (Supplementary information, Figure S8D). Why more efficient HCV replication in ICR-C/OTg liver did not lead to stronger IFN responses than in B6-C/OTg mice remains to be determined. Intriguingly, IFN was only transiently induced within 1 wpi, which might be partly due to MAVS proteolysis by HCV NS3 in both mouse strains, albeit NS3 was barely detectable in B6-C/OTg liver (Figure 6B). Even if HCV can evade antiviral IFN induction by MAVS cleavage, it still has to overcome the other blocks of ISGs for efficient replication. This is also suggested by recent work that HCV cannot establish persistent infection in 4hEF mice unless essential genes downstream of IFN signaling (Ifnar1, Stat1, Irf1, Irf7 and other ISGs) but not those upstream of IFN induction (Mavs and Irf3) are removed13. When the induction of ISGs known as antagonists of HCV persistent infection was compared, Ifi44 and Eif2ak2/PKR were specifically upregulated in B6- but not ICR-C/OTg mice, whereas Mx-1 induction was opposite (Figure 6C; other ISG genes, including Viperin, Oas1a and IP-10, were induced similarly in both strains, Supplementary information, Figure S9A; null response by Ifnl3 and Ifi27, data not shown). Failure to induce Ifi44 and Eif2ak2/PKR was not due to any intrinsically genetic defect in ICR-C/OTg mice as both genes could be activated by polyI:C in peritoneal macrophages (Supplementary information, Figure S9B) and by HCV infection in ICR-PHTTg cells (Supplementary information, Figure S9C). Ifi44 and Eif2ak2/PKR are known as strong anti-HCV ISGs, while Mx-1 possesses trivial anti-HCV activity32,34. Whether such a specific suppression of Eif2ak2/PKR and Ifi44 expression contributed to HCV persistent infection of ICR-C/OTg mice will be determined by future studies. Of note, previous overexpression experiments have shown that HCV NS5A35,36 or E237 can also target PKR for evasion of type I IFN response in vitro.
Figure 6.
ISG response to HCV differed between ICR- and B6-C/OTg mice. (A) Induction of hepatic type I IFN mRNA (ifna4 and ifnb1) during HCV acute infection of ICR-C/OTg and B6-C/OTg mice. (B) Proteolysis of MAVS during acute infection. ICR- and B6-C/OTg hepatocytes after HCV infection were immunoblotted for MAVS and HCV NS3. PBS was used in mock infection. GAPDH was used as loading control. (C) Induction of hepatic mx1, ifi44 and eif2ak2 mRNA in mice used in A. Three mice were used for infection with 1 ml HCV J399EM (TCID50 = 1 × 108/ml) at each time point. Data were analyzed by ANOVA test and shown as mean ± SD, P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
This work has shown for the first time that HCV can persistently infect an immune-competent mouse model with completion of viral life cycle. It is particularly important that C/OTg mouse model supports persistent levels of viremia for HCVcc of different genotypes and primary isolates from chronic hepatitis C patients. One would argue that ICR mouse is an outbred stock whose genetic background is ill-defined38. It would be ideal that ICR-C/OTg mice backcross to an inbred mouse strain (e.g., C57BL/6), to study the function of the transgenes. However, we found that B6-C/OTg mice show limited susceptibility to HCV infection, as also demonstrated in the recently developed 4hEF model13. It therefore calls for more comprehensive analyses of the determinants of persistent HCV infection in this ICR transgenic model. Preliminary assessment by this work has shown that the overall induction of antiviral interferons and pro-inflammatory cytokine responses are similar between ICR and C57BL/6 transgenic mice. However, a unique induction of host factors and ISGs favorable for HCV replication occurred in ICR-C/OTg mice, which may contribute to premissiveness to HCV infection of ICR-C/OTg mice. Another feasible approach is to identify genetic traits and selective immune response that would affect the course of viral infectivity and chronic hepatitis, especially given that humanized ICR, C57BL/6 and their backcrossed derivatives have now become available. In this sense, outbred stocks of ICR-C/OTg mice provide a better phenotype and genotype pool than C57BL/6 strain to study specific traits of interest.
It has to be cautious to interpret serum HCV loads (103−4 copies/ml) in ICR-C/OTg mouse model, which are relatively lower than those in chronic patients (103−6 copies/ml) or 4hEF Stat1−/− immune-compromised mice (104−5 copies/ml). Lower serum viral load does not undermine efficient HCV replication and spread in persistently infected ICR-C/OTg mice. The reasons include: first, the proportion of hepatocytes infected with HCV is about 1% (by IHC detection of antigens) to 10% (by in situ hybridization of HCV RNA) of ICR-C/OTg liver tissue, depending on the sensitivity of different detection methods. This is in close proximity to that (7% - 20% of hepatocytes infected with plasma HCV RNA loads of 105IU/ml) measured by highly sensitive two-photon method19. The infected ICR-C/OTg hepatocytes are present in clusters, in agreement with those in chronic patients19,39; second, 106−7 copies of HCV RNA are present per gram of C/OTg liver tissue. Although this amount is less than that in chronically infected patients and chimpanzees40,41 (∼108-11 HCV genomes per gram of liver tissue), we estimate that ICR-C/OTg hepatocyte contains about 1-10 genome copies of HCV RNA (assuming that 1% of liver tissues are infected), which is comparable to that in chronic patients42; third, and most importantly, circulating HCV particles derived from ICR-C/OTg mice are fully infectious and transmissible, which makes C/OTg mice a very reliable small animal model to study HCV persistent infection.
This work has further shown that due manifestations of chronic viral hepatitis, from steatosis to cirrhosis, can be impressively developed in C/OTg mice within 12 months post HCV infection. Therefore, C/OTg transgenic model would provide a new tool for studies on mechanisms of viral evasion and immune tolerance involved in HCV persistent infection. Unexpectedly, we fail to observe HCC development in persistently infected C/OTg mice even at 20 mpi, either by blood biochemical indices or non-invasive imaging analysis (data not shown). Whether different prognosis of HCC in mice and human reflects the species difference, or simply more infected mice and/or longer incubation time are needed to score HCC in the mice, remain to be determined.
The other puzzling phenomenon of the current study is a greater loss of body weights in male C/OTg mice than in females in response to HCV infection. HCV infection usually disproportionately affects men more than women43. Apetite hormone ghrelin is reduced in chronic hepatitis C patients44. Because both male and female mice have a similar degree of reduced food intake, greater reduction of body weight in males may not be caused by gender-specific loss of apetite. Women are more likely to clear the virus spontaneously and have slower rates of liver disease progression than men if they become chronically infected43. Overall, both genders of mice showed similar rates of persistent infection, but liver pathology progresses slightly faster, if any, in males (Supplementary information, Table S9). Whether sex hormones45, IL-28B polymorphism46, and natural killer p46 expression47, as suggested in the gender differences in humans, play a role in this mouse model remains to be investigated.
Given the urgent need for a robust immunocompetent small animal model to investigate HCV pathogenesis and immune control, this work certainly addresses a very important research area. Therefore, C/OTg transgenic mouse model would provide a new tool to study the mechanisms of adaptive immune evasion and tolerance involved in HCV persistent infection, and develop therapeutics and vaccines against HCV infection.
Materials and Methods
Animal study design
We used 8-12 weeks of age- and gender-matched mice for in vivo experiments. Unless specified, WT, CTg and C/OTg mice were in ICR background. In brief, C/OTg mice (n = 84) and ICR WT (n = 44) mice were divided in 3 batches and injected with HCV J399EM (Batch 1-3, Supplementary information, Table S3). Mice were randomly withdrawn from each batch at the indicated time points and sacrificed for varies analyses, including viral loads, H&E, Oil Red O and Sirius Red staining, and IHC of HCV proteins and α-SMA. Where indicated, ultrasonograph and CT were performed before the mice were euthanized. In analogy, different batches of mice were allocated (Supplementary information, Table S3) for various experiments: 12 C/OTg mice for HCV patient sera, 6 C/OTg mice for Con1/JFH1 (Batch 4); 9 C/OTg mice for HCV of different doses (Batch 5); 18 C/OTg mice for VX950 treatment (Batch 6); 9 C/OTg and 9 B6-C/OTg mice for comparison of innate response to HCV (Batch 7); 27 C/OTg and 27 CTg mice for assays of entry receptor requirement (Batch 8). Six C/OTg mice for JFH1 infection (Batch 9). Infection experiments were performd in a non-blind manner. Uneless otherwise specified, genders of mice used in different assays were random.
CD1 (ICR), C57BL/6 and Balb/C mice were purchased fromVital River Laboratory Animal Technology Co. (Beijing). Animal experiments were performed in accordance with institutional and NIH guidelines and were approved by the Animal Care and Use Committees of Institute of Biophysics, Chinese Academy of Sciences.
Viruses
HCV J399EM infectious clone (pJ399EM) was derived from pJFH-1 (HCV type 2a) by insertion of EGFP gene into NS5A region14. Plasmids (1 μg) of pJ399EM, pJFH-1 or pCon1/JFH148 (genotype 1b/2a mosaic virus, a gift from Pro Zhong Jin) were then in vitro transcribed (Ambion, USA) and RNA (10 μg) was electroporated into 1 × 107 naïve Huh7.5.1 cells, as previously described49. Progeny viruses were collected to infect a larger quantity of naive Huh7.5.1 cells (1 × 108) for virus production and ultracentrifugal purification14. Virus titers in supernatants and cell lysates were measured for 50% tissue culture infective dose (TCID50) by endpoint dilution assays (EPDA) as previously described50. Expansion and titration of Vesicular stomatitis virus (VSV) was performed as previously described51.
Sera positive for HCV1b and 2a were obtained from 4 chronic patients before IFN therapy administered (Supplementary information, Table S5). None of the patients had virological evidence of coexisting HBV infection at time of study (serological positive for HBV infection; one patient does not have records of HBV tests). All participants provided written informed consent and protocols were approved by the ethical commission of the Department of Hepatology, The First Hospital of Jilin University, China and in accordance with the Helsinki Declaration.
The uses of HCVcc and derivatives, HCV-positive sera, and VSV in tissue culture and animals were approved by the Institutional Biosafety Committee at Institute of Biophysics and Wuhan Institute of Virology, Chinese Academy of Sciences.
Generation of transgenic mice
Human CD81 and OCLN cDNA fragments were PCR amplified and cloned into pLIVE vector (Mirus, WI, USA) in between XhoI and BamHI, or SalI and XhoI restriction sites, respectively. DNA fragments encompassing hCD81 or hOCLN, and mouse AFP enhancer II and mouse minimal albumin promoter were excised by BglII and NdeI, or XbaI and NdeI, and electrophoretically purified by Elutip-D columns (Whatman, MA, USA). Transgenic procedures were adopted from a previous method52. In brief, each DNA fragment (1 ng/μl) was microinjected into CD1 (ICR) zygotes (n = 200-300) by a DMI300B micro-injector (Leica, Germany). Eggs were then transplanted to pseudopregnant CD1 (ICR) mothers (n = 10) to generate founders of transgenic mice. Transgenic mice heterozygous for hCD81 and hOCLN were backcrossed to generate and routinely breed double transgenic mice of hCD81 and hOCLN (C/OTg). All offspring transgenic mice were genotyped by PCR with human transgene-specific primers (Supplementary information, Table S6). Transgenic hCD81 (CTg) and hOCLN (OTg) mice in ICR background were backcrossed 10 generations to C57BL/6 strains. The derived C57BL/6 mice carrying CD81 or OCLN were further crossed for breeding of double transgenic mice (B6-C/OTg). All offsprings were genotyped by PCR using the method as for ICR-C/OTg.
HCV infection, transmission and treatments
For infection in vivo, mice (WT or transgenic in ICR or C57BL/6 background, where indicated) were tail-vein injected with 1 ml HCV (HCV J399EM, Con1/JFH1 or JFH-1) at indicated doses, in 60∼120 s to avoid liver injury. For infection with patients sera, C/OTg mice were i.v. injected with 200 μl of serum positive for HCV1b (pooled serum of patient #840 and #858, final HCV RNA = 7 × 105IU/ml) or 2a (pooled serum of patient #5331 and #900, final HCV RNA = 5 × 105IU/ml). Mouse blood and liver tissues were harvested at the indicated time, for measurements of HCV genomic RNA by quantitative RT-PCR. Subtype-specific primers were shown in Supplementary information, Table S6.
Three consecutive rounds of transmission experiments were performed. In breif, C/OTg mice (n = 5) were tail-vein injected with HCV J399EM (TCID50 = 1 × 108/ml) and liver tissues and sera were collected 2 wpi. The sera (in 400 μl for each mouse) were transfused to naïve C/OTg mice (n = 5) via tail vein. Two weeks later, mice were sacrificed, and liver tissues were used for HCV genome measurements, and sera for the next round of transfusion. HCV copy numbers in liver were monitored for each round.
For monotherapy, VX-950 (200 mg/kg, NDA 201917, Vertex Pharma, MA) or DMSO were i.p. administered daily at 1 week post HCV infection (TCID50 = 1 × 108/ml). Liver tissues and sera were collected at the indicated time for measurement of HCV RNA.
For infection in vitro, PHTTg or Huh7.5.1 cells (1 × 106) were infected with J399EM (MOI = 1) for the indicated time. Intercellular (inter), intracellular (intra) and supernatant (sup) HCV RNAs were determined by quantitative RT-PCR, and viral titers were measured by FFU. The assembly efficiency was plotted in log scales as the ratios of HCV RNA copies in sup over intra (sup/intra), budding efficiency as ratios of sup titer/intra titer, and infectivity as the ratios of sup HCV titer/sup HCV copies after viral infection for 96 h.
To measure the infectivity of circulating HCV, sera (100 μl) collected from HCV persistently infected C/OTg mice at 6, 10 and 13 mpi were used to infect Huh7.5.1 cells (1 × 106). Intracellular HCV RNA levels at the indicated incubation time were determined by quantitative RT-PCR. Viral titers in supernatants and cell lysates were measured by FFU50.
For receptor blocking and VX-950 treatment in primary hepatocytes, anti-CD81 (2 μg, sc70804, Santa Cruz), anti-OCLN (2 μg, ab64482, Abcam) or VX-950 (100 μM) was added to the medium. HCV RNAs of the intercellular and supernatant were extracted at 72 hpi and determined by quantitative RT-PCR.
Isolation of primary hepatocytes (PHT) and peritoneal macrophages
PHT cells were isolated from WT or C/OTg mice by a 2-step collagenase perfusion protocol. The hepatic portal vein was ligated and perfused with Ca2+/Mg2+-free HBSS (Beyotime, China) with 5 mM EDTA, then 0.025% type IV collagenase (Sigma, St. Louis, USA) in HBSS. After perfusion, liver was excised and hepatocytes were suspended in serum-free DMEM (Gibco, MD) and passed through 100-mm strainer. The filtrate was centrifuged at 50× g for 3 min at 4 °C. The pellet was re-suspended in 10% FBS containing William's Essential Medium (Gibco). The viability (> 85%) was determined by trypan blue exclusion.
To isolate macrophages, ICR- or B6-C/OTg mice were injected i.p. with 2 ml of 4% thioglycollate broth to elicit peritoneal macrophages. Purified macrophages were stimulated with 100 μg/ml polyI:C (Sigma) or 200 ng/ml LPS (Sigma) in a 96-well plate for 16 h, and supernatants were then collected for cytokine analyses.
RT-PCR
Total RNA was extracted from different tissues (0.1 g liver or tissues indicated in Supplementary information, Figure S1D), cells (1 × 106) or serum (0.1 ml) using Trizol or Trizol LS reagent (Invitrogen). Quantitative RT-PCR analyses with One-step QuantiTect SYBR Green Kit (Qiagen, CA, USA) of the indicated genes were performed as previously described14 in an ABI 7500 (ABI, CA, USA).The internal standard HCV RNA was synthesized by in vitro transcription (MEGAscript T7, Ambion, TX, USA) from pJFH-1 plasmid and quantified by a commercial kit (path-HCV, PrimerDesign, UK) at 1.1 × 1010 copies/ml. Serial dilutions of the internal standard HCV RNAs were spiked in ICR sera and liver homogenates to determine the linear range of detection and the limit of detection, using One-step QuantiTect SYBR Green Kit. The limit of detection of HCV RNA by this method is defined as the lowest concentration at which 95% of HCV RNA are detected53, i.e., 500 copies/ml (serum) and 100 copies/mg (liver), respectively. Nested PCR was performed to detect HCV negative strand16 (see Supplementary information, Table S6 for primers). Quantitative RT-PCR analyses were carried out similarly for mRNA levels of IFNα4, IFNβ1, ISGs, miR-122, apoE, CypA and IL-28 (see Supplementary information, Table S6 for primers). Data were analyzed with ABI software version 2.0.3 (Applied Biosystems).
Fluorescent in situ hybridization
WT (n = 2) or C/OTg mice (n = 6) were chronically infected with HCV JFH-1 (TCID50 = 5 × 106). The frozen sections of liver tissues at the indicated time post infection were fixed in 4% paraformaldehyde and hybridized with HCV positive or negative strand-specific molecular beacons probe (400 nM, see Supplementary information, Table S6 for sequences). Images were captured with a Olympus FV1000 confocal microscope and analyzed with Olympus FluoView FV1000 software. The nuclei were counterstained with DAPI, λEx = 488 nm.
Pathology, immunofluorescence and immunohistochemistry analyses
A single-blind test was performed for histochemistry and immunohistochemistry. H&E, Masson's trichrome and Sirius red staining of formalin-fixed paraffin-embedded liver tissues (C/OTg, n = 4 for each time point, 3 sections per mouse, 4 views per section; WT, n = 3 for each time point, 3 sections per mouse, 4 views per section) was performed according to manufacturer's instructions (Leica, Germany). Oil red O staining was carried out using standard protocol on cryostat sectioned tissues that had been embedded in OCT. All images were acquired with a Leica DS-Ri1 microscope.
For IHC staining, the right lobes of livers embedded at the indicated time were serially sliced (thickness 5 μm) and processed on an automated Tissue Processor (Shandon Excelsior, Thermo China). The antigens were repaired by high pressure cooking and tryptic digestion. The sections were incubated with primary antibodies to α-SMA (1:200, SPM332, Abcam), NS5A (1:20, 9e10) and Core (1:50, C7-50, ab2740, Abcam) overnight. A three-step indirect peroxidase-labeled antibody method was used (Guge Biotech, Wuhan, China). Following incubation overnight with the antibodies, a peroxidase-labeled rabbit anti-mouse IgG was applied, followed by multiple peroxidase-labeled goat anti-rabbit IgG, using tyramide signal amplification kit (Invitrogen). Nuclei were stained with Mayer's hematoxylin. Images were acquired by Leica SCN400 and analyzed with QWin V3 imaging software (Leica).
Lymphoid infiltration and aggregation, Core and NS5A expression were quantified by software IPP6.0 (Media Cybernetics, MD, USA) after determining the average optical density (AOD) of positive stains (magnification 400×, 10 random viewfields). The percentage of lymphoid aggregates or Core/NS5A positive cells/areas of entire viewfield were plotted after deduction of the background stained cells in the control groups at the same time point. Two blinded pathologists independently scored each liver section, and the data represented the average scores using the modified Knodell scoring system. The representative images were shown in the indicated figures.
Western blotting
Proteins were routinely prepared from cells or tissues lysed with ice-cold RIPA buffer (25 mM Tris/HCl, pH 7.6, 150 mM NaCl, 1% Nonidet P40, 1% sodium deoxycholate, 0.1% SDS) containing phosphatase and protease inhibitors (Cat# 04906837001 and Cat# 11836170001, Roche). The proteins were resolved in appropriate Bis/Tris NuPage polyacrylamide gels (Invitrogen). The following antibodies were used: NS5A (1:500, 9e10), NS3 (1:1,000, 8G2, ab65407, Abcam), CD81 (1:200, sc70804, Santa Cruz), OCLN (1:1 000, ab64482, Abcam), MAVS (1:1 000, Cat# 4983, CST), APOE (1:1 000, D6E10, ab1906, Abcam), GAPDH (1:1 000, #5174, CST) and β-actin (1:10 000, A2066, Sigma). Proteins were visualized with HRP-conjugated IgG (1:100 000, JacksonImmunol) using Super Signal West Pico (Thermo Scientific), and analyzed by MiniChemi Chemiluminescence Imaging System with Lane 1D software (SageCreation, Beijing).
Serum chemistry and serology
Mice were routinely retro-orbital bled at the indicated time post infection. About 40 μl plasma were collected to measure enzymes and proteins (pre-albumin, albumin, alanine aminotransferase, aspartate aminotransferase and alkaline phosphatase) using automated Blood Chemistry Analyzer (ZS-200B, BioSino Biotech, China).
ELISA
IFN-α and IFN-β in the sera were determined by Mouse IFN-alpha and IFN-beta ELISA Kits (PBL Interferon Source, NJ, USA). IL-28 was determined by ELISA Kits (Sino-American Biotech, China). The intensities were quantitated in a Plate CHAMELEON V reader (Hidex, Finland, λ = 450 nm) with Mikrowin 2000 software (Berthold Technologies, Germany).
Flow cytometry
Intrahepatic leukocytes were isolated at the indicated time post HCV infection as previously described with modifications54. Multi-color flow cytometry was performed in LSRFortessa (BD Biosciences, NJ, USA) after leukocytes (1 × 106) were incubated with mAb (10 μg/ml each) in PBS containing 2% FBS. The following antibodies were from BioLegend: PerCP-Cy5.5-CD4 (GK1.5), FITC-NK1.1 (PK136), BV605-CD8α (53-6.7), PE-Cy7-CD3ɛ (145-2C11), PE-Ly6C (HK1.4), FITC-Ly6G (1A8) and APC-Cy7-CD11b (OX-42). Data were analyzed with FlowJo Software (Tree Star Inc.) and represented three independent experiments (mean ± SD).
Serum cytokine and chemokine detection
FLEXMAP 3D quantification (Luminex, TX, USA) of concentrations of cytokines and chemokines was performed on sera (10 μl) stored from various HCV-infected WT or C/OTg mice. Three Milliplex MAP kits (Millipore, MA, USA) were combined for measurement of IL1α, IL1β, IL2, IL-3, IL-4, IL-5, IL-6, IL-10, IL-12p40, IL-12p70, IL-13, IL-17A, IL-17F, IL-21, TNF-α, MCP-1, IP-10, KC, IFN-, GM-CSF, TGF-α. Data were analyzed by xPONENT software (Luminex). Data were average of at least two independent measurements (mean ± SD).
Micro-ultrasonography
C/OTg mice at different time post infection were starved overnight before anesthetized with 4% isoflurane. The transabdominal ultrasound images (axial resolution of 40 mm with a 14.6-mm field of view) were acquired by Vevo770 high resolution micro-imaging ultrasound system (VisualSonics, Canada). In brief, mice were positioned in dorsal recumbence on a rectangular table, and liver was assessed by placing the RMV704 scan head (40 MHz, 6 mm focal depth) just distal to the last right costal cartilages and angling its beam cranially, obtaining multiple transversal and longitudinal scans. The hepatic artery and portal vein were scanned in the longitudinal views. Histological stages were determined by the scoring system as previously described55 using the VisualSonics Vevo770 software.
CT scan analysis of hepatic fibrosis and cirrhosis
Micro CT scans from the diaphragm to the pelvis of the dorsal right lobe of liver (2 mm intervals) were obtained with (for enhanced CT) or without iobitridol emulsion in a helical CT apparatus (LaTheta LCT-200, Hitachi-Aloka Medical, Japan) after 2% isoflurane anesthesia. CT images were sent to a referral center for blind analysis after an expert radiologist determined the ROI (liver parenchyma). Histological algorithm was deduced with fast Fourier transform to the different gray shades in each square. Enhanced CT was used to diagnose cirrhosis. Histological stages were determined by the scoring system as previously described25, and quantified with LaTheta software (version 3.00).
Statistical analysis
Power In compliance with ethical guidelines to minimize the number of animals used, we ususally used a minimum of 3 mice for each data point (except indicated in figure legend) to ensure statistical power. The sample size is thus determined according to the duration of experiments and number of data points pre-determined. Batches of infection were carried out to ensure accuracy, repeatability and enough animals for each data point.
Randomization Mice are grouped with the matched age, gender, body weight and timing of experiments, between two cohorts. To form the cohort, each batch of infection, contained the number of mice more than the number of data points, to ensure the randomization and accidental exclusion of animals. The cohort was then grouped using randomly selected animals from the batches of infection (Supplementary information, Table S3). In vitro analyses (HCV copies, chemistry, cytokines and chemokines) were usually performed on specimen (sera, liver) from animals at each data point to ensure a minimal 3 biological replicates. Other in vitro assays (western blotting, quantitative RT-PCR and Luminex assays) were carried out non-randomly, except for loading samples to multi-well device.
Sample or animal exclusion The pre-established criteria to exclude animals from entire study are the mice negative for viral load in liver at the time of assay, or death of fulminant hepatitis or unkonwn reasons. Animals with insufficent sera are excluded from serum viral titer analysis. Serum insufficiency is usually caused by the comsuption of the same serum for other assays.
Statistics Routinely, data collection and data analysis were performed by differrent persons, to blind the potential bias. All measurement data are expressed as mean ± SD to maximally show deriviations, unless otherwise specified. Differences between two groups were assessed using unpaired two-tailed student's t-test. For time-series data, the two-way by ANOVA procedure was applied. Data involving more than two groups were assessed ANOVA. Statistical tests are justified as appropriate. Data were in normal distribution, after variation within each group of data was estimated. The variance usually was within the range of statistic similarity. P values < 0.05 were considered significant. Statistical analyses were performed using GraphPad Prism.
Acknowledgments
We are grateful to Drs T Wakita (National Institute of Infectious Diseases, Japan) for pJFH-1 clone, F Chisari (The Scripps Research Institute) for Huh7.5.1 cell line, L Su (University of North Carolina) for cDNA clones of human CD81 and Occludin, and C Rice (The Rockefeller University) for NS5A antibody. We thank Drs Y Tian and S Meng for making a series of transgenic mice; C-X Wei for the genotyping of transgenic mice. We thank Drs L Su, Y-X Fu (The University of Chicago), R Sun (University of California, Los Angeles) and P Hertzog (Monash University, Australia) for their critical reading and comments on the manuscript. This work was supported by grants from the National Basic Research Program of China (2009CB522506 and 2011CB946104 to HT, 2009CB118903 to XC, and 2010CB530100 to LP), the National Natural Science Foundation of China (31200135 to XC and 31030031 to HT), Knowledge Innovation Program Projects of Chinese Academy of Science (2010-Biols-CAS-0201to HT, and KSCX1-YW-10 to XC and HT).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Transgene constructs of C/OTg mice.
Gender difference in gain of body weight after HCV J399EM infection (TCID50 = 1×108/ml).
(A) Negative strand HCV RNA after ICR-C/OTg mice (n = 3) infected with HCV for indicated time was detected by nested PCR.
Lymphocyte infiltration in C/OTg liver after HCV J339EM (TCID50 = 1×108/ml) infection.
Representative H&E of liver sections from (A) abortive infection of wt mice (n = 3), (B) PBS mock infection of ICR-C/OTg mice (n = 4), and (C) HCV J399EM infection of ICR-C/OTg (n = 3) at indicated time.
Fibrotic alteration in chronic C/OTg liver.
Persistently infected C/OTg mice at steatotic (11 mice within 1-3 mpi), fibrotic (8 mice within 3-10 mpi) and cirrhotic (4 mice within 10-14 mpi) stages of infection were randomly selected to compare the viral loads in serum and liver, and ALT levels.
Intact innate immune response of ICR C/OTg mice.
(A) Induction of hepatic IP-10, Viperin and Oas1a mRNA from mice in Figure 5A.
Baselines of serum chemistry profiles, cytokines and chemokines in C/OTg mice.
Health conditions of chronic hepatitis C mice
Mice allocation in HCV infection cohort$
HCV genome copies of the cohorts in Fig. 1A-B
Patients' information
Primes and probes.
Ultrasonography assessment of liver fibrosis
Computed tomography assessment of liver cirrhosis
Gender-specific response to HCV J399EM infection
References
- Thomas DL. Global control of hepatitis C: where challenge meets opportunity. Nat Med. 2013;19:850–858. doi: 10.1038/nm.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari A, Ray S, Thuluvath PJ. Acute hepatitis C. Lancet. 2008;372:321–332. doi: 10.1016/S0140-6736(08)61116-2. [DOI] [PubMed] [Google Scholar]
- Chisari FV. Unscrambling hepatitis C virus-host interactions. Nature. 2005;436:930–932. doi: 10.1038/nature04076. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- Sklan EH, Charuworn P, Pang PS, Glenn JS. Mechanisms of HCV survival in the host. Nat Rev Gastroenterol Hepatol. 2009;6:217–227. doi: 10.1038/nrgastro.2009.32. [DOI] [PubMed] [Google Scholar]
- Gale M, Jr, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939–945. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- Bukh J. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology. 2012;142:1279–1287. doi: 10.1053/j.gastro.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Walker CM. Comparative features of hepatitis C virus infection in humans and chimpanzees. Springer Semin Immunopathol. 1997;19:85–98. doi: 10.1007/BF00945027. [DOI] [PubMed] [Google Scholar]
- Mercer DF, Schiller DE, Elliott JF, et al. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- Bissig KD, Wieland SF, Tran P, et al. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J Clin Invest. 2010;120:924–930. doi: 10.1172/JCI40094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn ML, Bility MT, Zhang L, et al. A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease. Gastroenterology. 2011;140:1334–1344. doi: 10.1053/j.gastro.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner M, Horwitz JA, Robbins JB, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner M, Horwitz JA, Donovan BM, et al. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature. 2013;501:237–241. doi: 10.1038/nature12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q, Xu C, Wu C, Zhu W, Yang R, Chen X. Compensatory mutations in NS3 and NS5A proteins enhance the virus production capability of hepatitis C reporter virus. Virus research. 2009;145:63–73. doi: 10.1016/j.virusres.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat H, Berby F, Trabaud MA, et al. Specific detection of hepatitis C virus minus strand RNA in hematopoietic cells. J Clin Invest. 1996;97:845–851. doi: 10.1172/JCI118485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczynski K, Beach MJ, Bradley DW, et al. Hepatitis C virus antigen in hepatocytes: immunomorphologic detection and identification. Gastroenterology. 1992;103:622–629. doi: 10.1016/0016-5085(92)90856-t. [DOI] [PubMed] [Google Scholar]
- Sansonno D, Lauletta G, Dammacco F. Detection and quantitation of HCV core protein in single hepatocytes by means of laser capture microdissection and enzyme-linked immunosorbent assay. J Viral Hepat. 2004;11:27–32. doi: 10.1046/j.1365-2893.2003.00474.x. [DOI] [PubMed] [Google Scholar]
- Liang Y, Shilagard T, Xiao SY, et al. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology. 2009;137:1448–1458. doi: 10.1053/j.gastro.2009.07.050. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Uprichard SL, Chung J, Chisari FV, Wakita T. Replication of a hepatitis C virus replicon clone in mouse cells. Virol J. 2006;3:89. doi: 10.1186/1743-422X-3-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasmin MY, Aziz B, Nazim M, Madhavan RK. Prealbumin rather than albumin is a more sensitive indicator of acute liver disease. Malays J Pathol. 1993;15:147–150. [PubMed] [Google Scholar]
- Kawamoto M, Mizuguchi T, Katsuramaki T, et al. Assessment of liver fibrosis by a noninvasive method of transient elastography and biochemical markers. World J Gastroenterol. 2006;12:4325–4330. doi: 10.3748/wjg.v12.i27.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloisi F, Pujol-Borrell R. Lymphoid neogenesis in chronic inflammatory diseases. Nat Rev Immunol. 2006;6:205–217. doi: 10.1038/nri1786. [DOI] [PubMed] [Google Scholar]
- Brancatelli G, Federle MP, Ambrosini R, et al. Cirrhosis: CT and MR imaging evaluation. Eur J Radiol. 2007;61:57–69. doi: 10.1016/j.ejrad.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Ito K, Mitchell DG, Gabata T, Hussain SM. Expanded gallbladder fossa: simple MR imaging sign of cirrhosis. Radiology. 1999;211:723–726. doi: 10.1148/radiology.211.3.r99ma31723. [DOI] [PubMed] [Google Scholar]
- Carithers RL, Jr, Sugano D, Bayliss M. Health assessment for chronic HCV infection: results of quality of life. Dig Dis Sci. 1996;41:75S–80S. doi: 10.1007/BF02087879. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Guo JT, Seeger C. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J Virol. 2003;77:9204–9210. doi: 10.1128/JVI.77.17.9204-9210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey AP, Ohashi K, Meuse L, et al. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol Ther. 2002;5:676–684. doi: 10.1006/mthe.2002.0600. [DOI] [PubMed] [Google Scholar]
- Long G, Hiet MS, Windisch MP, Lee JY, Lohmann V, Bartenschlager R. Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology. 2011;141:1057–1066. doi: 10.1053/j.gastro.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013;10:542–552. doi: 10.1038/nrgastro.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner SM, Gale M., Jr Regulation of hepatic innate immunity by hepatitis C virus. Nat Med. 2013;19:879–888. doi: 10.1038/nm.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KS, Cai Z, Zhang C, Sen GC, Williams BR, Luo G. Replication of hepatitis C virus (HCV) RNA in mouse embryonic fibroblasts: protein kinase R (PKR)-dependent and PKR-independent mechanisms for controlling HCV RNA replication and mediating interferon activities. Journal of virology. 2006;80:7364–7374. doi: 10.1128/JVI.00586-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugheber J, Fredericksen B, Sumpter R, Jr, et al. Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc Natl Acad Sci USA. 2002;99:4650–4655. doi: 10.1073/pnas.062055699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- Chia R, Achilli F, Festing MF, Fisher EM. The origins and uses of mouse outbred stocks. Nat Genet. 2005;37:1181–1186. doi: 10.1038/ng1665. [DOI] [PubMed] [Google Scholar]
- Lau DT, Fish PM, Sinha M, Owen DM, Lemon SM, Gale M., Jr Interferon regulatory factor-3 activation, hepatic interferon-stimulated gene expression, and immune cell infiltration in hepatitis C virus patients. Hepatology. 2008;47:799–809. doi: 10.1002/hep.22076. [DOI] [PubMed] [Google Scholar]
- Sugano M, Hayashi Y, Yoon S, et al. Quantitation of hepatitis C viral RNA in liver and serum samples using competitive polymerase chain reaction. J Clin Pathol. 1995;48:820–825. doi: 10.1136/jcp.48.9.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger CB, Guerra B, Brasky KM, et al. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J Virol. 2004;78:13779–13792. doi: 10.1128/JVI.78.24.13779-13792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier C, Champenois K, Gervais A, et al. Predictive value of liver enzymes and inflammatory biomarkers for the severity of liver fibrosis stage in HIV/HCV co-infected patients. PloS One. 2013;8:e 59205. doi: 10.1371/journal.pone.0059205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden R, Rockstroh JK, Buti M. Natural history and management of hepatitis C: does sex play a role. J Infect Dis. 2014;209 Suppl 3:S81–S85. doi: 10.1093/infdis/jiu057. [DOI] [PubMed] [Google Scholar]
- Moreno M, Chaves JF, Sancho-Bru P, et al. Ghrelin attenuates hepatocellular injury and liver fibrogenesis in rodents and influences fibrosis progression in humans. Hepatology. 2010;51:974–985. doi: 10.1002/hep.23421. [DOI] [PubMed] [Google Scholar]
- Mekky RY, Abdelaziz AI. Sex hormones and HCV: an unresolved mystery. Expert Rev Gastroenterol Hepatol. 2013;7:69–75. doi: 10.1586/egh.12.69. [DOI] [PubMed] [Google Scholar]
- van den Berg CH, Grady BP, Schinkel J, et al. Female sex and IL28B, a synergism for spontaneous viral clearance in hepatitis C virus (HCV) seroconverters from a community-based cohort. PloS One. 2011;6:e27555. doi: 10.1371/journal.pone.0027555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden-Mason L, Stone AE, Bambha KM, Cheng L, Rosen HR. Race- and gender-related variation in natural killer p46 expression associated with differential anti-hepatitis C virus immunity. Hepatology. 2012;56:1214–1222. doi: 10.1002/hep.25771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Weady P, Duggal R, Hao W. Novel chimeric genotype 1b/2a hepatitis C virus suitable for high-throughput screening. Antimicrob Agents Chemother. 2008;52:666–674. doi: 10.1128/AAC.01133-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Chung J, et al. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J Virol. 2006;80:11082–11093. doi: 10.1128/JVI.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Chen G, Guo B, Cheng G, Tang H. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 2008;18:1105–1113. doi: 10.1038/cr.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittner LM, Gotz J. Pronuclear injection for the production of transgenic mice. Nat Protoc. 2007;2:1206–1215. doi: 10.1038/nprot.2007.145. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Ohtsuka K, Kimura M, et al. Details of an isolation method for hepatic lymphocytes in mice. J Immunol Methods. 1992;146:145–154. doi: 10.1016/0022-1759(92)90223-g. [DOI] [PubMed] [Google Scholar]
- Macias Rodriguez MA, Rendon Unceta P, Navas Relinque C, Tejada Cabrera M, Infantes Hernandez JM, Martin Herrera L. Ultrasonography in patients with chronic liver disease: its usefulness in the diagnosis of cirrhosis. Rev Esp Enferm Dig. 2003;95:258–264. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transgene constructs of C/OTg mice.
Gender difference in gain of body weight after HCV J399EM infection (TCID50 = 1×108/ml).
(A) Negative strand HCV RNA after ICR-C/OTg mice (n = 3) infected with HCV for indicated time was detected by nested PCR.
Lymphocyte infiltration in C/OTg liver after HCV J339EM (TCID50 = 1×108/ml) infection.
Representative H&E of liver sections from (A) abortive infection of wt mice (n = 3), (B) PBS mock infection of ICR-C/OTg mice (n = 4), and (C) HCV J399EM infection of ICR-C/OTg (n = 3) at indicated time.
Fibrotic alteration in chronic C/OTg liver.
Persistently infected C/OTg mice at steatotic (11 mice within 1-3 mpi), fibrotic (8 mice within 3-10 mpi) and cirrhotic (4 mice within 10-14 mpi) stages of infection were randomly selected to compare the viral loads in serum and liver, and ALT levels.
Intact innate immune response of ICR C/OTg mice.
(A) Induction of hepatic IP-10, Viperin and Oas1a mRNA from mice in Figure 5A.
Baselines of serum chemistry profiles, cytokines and chemokines in C/OTg mice.
Health conditions of chronic hepatitis C mice
Mice allocation in HCV infection cohort$
HCV genome copies of the cohorts in Fig. 1A-B
Patients' information
Primes and probes.
Ultrasonography assessment of liver fibrosis
Computed tomography assessment of liver cirrhosis
Gender-specific response to HCV J399EM infection