Summary
The bacterium Xenorhabdus nematophila is a mutualist of entomopathogenic Steinernema carpocapsae nematodes and facilitates infection of insect hosts. X. nematophila colonizes the intestine of S. carpocapsae which carries it between insects. In the X. nematophila colonization-defective mutant nilD6::Tn5, the transposon is inserted in a region lacking obvious coding potential. We demonstrate that the transposon disrupts expression of a single CRISPR RNA, NilD RNA. A variant NilD RNA also is expressed by X. nematophila strains from S. anatoliense and S. websteri nematodes. Only nilD from the S. carpocapsae strain of X. nematophila rescued the colonization defect of the nilD6::Tn5 mutant, and this mutant was defective in colonizing all three nematode host species. NilD expression depends on the presence of the associated Cas6e but not Cas3, components of the Type I-E CRISPR-associated machinery. While cas6e deletion in the complemented strain abolished nematode colonization, its disruption in the wild-type parent did not. Likewise, nilD deletion in the parental strain did not impact colonization of the nematode, revealing that the requirement for NilD is evident only in certain genetic backgrounds. Our data demonstrate that NilD RNA is conditionally necessary for mutualistic host colonization and suggest that it functions to regulate endogenous gene expression.
Keywords: small non-coding RNA, mutualism, symbiosis, entomopathogen
Introduction
Entomopathogenic Steinernema spp. nematodes are mutualistically associated with bacteria of the genus Xenorhabdus (Stock and Goodrich-Blair, 2008). Together, these symbiont pairs infect, kill, and reproduce within insect hosts. A specialized infective juvenile (IJ) stage of the Steinernema nematode transmits bacterial symbionts between insects, ensuring maintenance of the symbiosis through generations. The association between S. carpocapsae and its symbiont X. nematophila has been well studied with regard to cellular and molecular aspects of symbiosis, particularly with respect to the mechanisms by which the IJ is colonized (Goodrich-Blair, 2007; Herbert and Goodrich-Blair, 2007; Chaston et al., 2013). The bacteria occupy a region known as the receptacle in the anterior portion of the IJ intestine (Poinar, 1966; Wouts, 1980; Bird and Akhurst, 1983; Martens et al., 2003; Flores-Lara et al., 2007; Snyder et al., 2007). Although the processes by which X. nematophila bacteria are selected and gain entry to the receptacle remain obscure, only one or two individual clones are founders for the final population (~30–200 CFU/IJ) that ultimately fills the space (Martens et al., 2003; Chaston et al., 2013).
To better understand the bacterial molecular factors necessary during colonization of the IJ nematode, a signature tagged mutagenesis screen to identify X. nematophila mutants defective in this process was conducted in the S. carpocapsae nematode-associated strain X. nematophila HGB081 AN6/1 (hereafter referred to as XnSc 081) (Heungens et al., 2002). In one of the mutants identified in this screen, nilD6::Tn5, the transposon had inserted into a region of the genome lacking obvious coding potential. Complementation studies then confirmed the nilD region was necessary for nematode colonization but was dispensable for virulence in an insect model of infection (Heungens et al., 2002). Bioinformatic analyses have since indicated that the nilD locus encodes a single, free-standing CRISPR (clustered regularly interspaced short palindromic repeats) element comprising one spacer and two repeats, which was disrupted by the transposon insertion (Fig. 1).
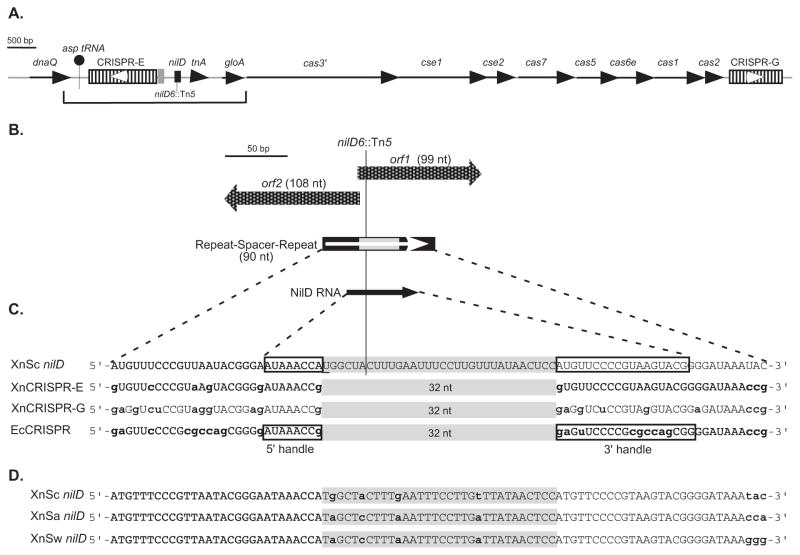
Figure 1.
Schematic representation of the nilD CRISPR locus.
A. Schematic diagram of the X. nematophila genomic regions containing CRISPR loci, cas/cse genes, and nilD. The bracket indicates the 3240-bp region previously sequenced in the HGB081 (XnSc 081) strain background (AY077466) (Heungens et al., 2002), which is identical in the sequenced ATCC 19061 (HGB800/XnSc 800) genome (NC_014228.1). Line arrows represent open reading frames, with gene names indicated above each. CRISPR loci are represented by hatched rectangles and are named CRISPR-E and CRISPR-G according to their position on the X. nematophila genome, with nilD, shown as a black rectangle, considered CRISPR-F. The location of the nilD6::Tn5 transposon insertion within nilD is indicated. The gray shaded box represents the 135-bp leader sequence of CRISPR-E. White arrowheads indicate the predicted orientations of CRISPR-E and -G transcription, based on comparison to E. coli CRISPR transcription.
B. Detail of the nilD locus, showing the two small overlapping open reading frames (orf1 and orf2) represented by checkered block arrows. The positions of nilD locus repeats and spacer are indicated by black and gray rectangles, respectively. The white arrow indicates the predicted orientation of transcription based on comparison to E. coli CRISPR transcription. The black arrow represents the position of the small RNA transcript encoded by the nilD locus. The position of the nilD6::Tn5 insertion site is indicated by a line.
C. Sequence of NilD RNA aligned with CRISPR small RNAs predicted to be encoded by X. nematophila (XnCRISPR-E and -G) and CRISPR RNAs expressed in E. coli (EcCRISPR). Spacer regions are shaded in gray. Lower case, bold nucleotides indicate those that differ from NilD RNA. The 5′ and 3′ handles as described by Brouns et al. (2009), and experimentally determined for NilD RNA are boxed. The underlined “U” in the nilD spacer sequence is necessary for colonization (Heungens et al., 2002).
D. Alignment of nilD locus repeat-spacer-repeat sequences of X. nematophila (carpocapsae) (XnSc nilD), X. nematophila (anatoliense) (XnSa nilD), and X. nematophila (websteri) (XnSw nilD). Lower case bold letters indicate nucleotides that differ among the three sequences.
CRISPRs are genetic elements broadly distributed among bacteria and archaea and can provide resistance to foreign nucleotide sequences (Barrangou and Marraffini, 2014). CRISPRs comprise a series of short repeat sequences that are separated by variable regions, called spacers. Many CRISPR spacers exhibit identity to sequences, termed proto-spacers, within bacteriophage genomes or other mobile DNA elements (Bolotin et al., 2005; Mojica et al., 2005; Pourcel et al., 2005; Deveau et al., 2008). This observation led to the discovery that CRISPR elements encode a rapidly evolving acquired immune defense system against incoming phages and plasmids (reviewed in (Sorek et al., 2013; Barrangou and Marraffini, 2014)). CRISPR arrays are transcribed as single RNA molecules that are then processed by components of the Cas (CRISPR associated sequences) machinery into individual CRISPR RNA (crRNA) molecules (Barrangou et al., 2007; Brouns et al., 2008; Carte et al., 2008). There are three major classes of CRISPR systems (Types I–III). These three types are further subdivided into several subtypes, differentiated by criteria including the phylogeny of the cas genes and the sequence of the CRISPR repeats (Makarova et al., 2011a; Makarova et al., 2011b). In E. coli, a type I-E system, five proteins, Cse1 (Cas subtype E. coli), Cse2, Cas7, Cas5, and Cas6e (previously named CasA, B, C, D, and E, respectively) are associated in a complex termed “Cascade” and Cas6e, a putative RNA binding protein, is the subunit responsible for RNA processing (Brouns et al., 2008; Carte et al., 2008; Jore et al., 2011). Processed crRNAs target and interact with proto-spacer encoding DNA or RNA molecules, resulting in gene silencing and/or degradation (Gasiunas et al., 2014). A 6–8 nt seed sequence within the crRNA exhibits 100% identity to the target and is predicted to guide the interaction between the crRNA and the proto-spacer (Semenova et al., 2011; Wiedenheft et al., 2011). CRISPR targeting and silencing also require the presence of a short, proto-spacer adjacent motif (PAM) within the exogenous target sequence, located directly upstream of the seed sequence. The PAM provides a mechanism by which the system differentiates between target and non-target (e.g. endogenous) sequences, thereby preventing potentially lethal auto-immunity due to targeting of chromosomal “self” sequences (Mojica et al., 2009; Westra et al., 2013). The hybridization of the crRNA molecule to the PAM-encoding DNA sequence results in the formation of an R-Loop within the crRNA that acts as a binding site for another member of the Cas protein family, Cas3. Cas3 contains helicase and nuclease activities that are responsible for degradation of the target molecule (Sinkunas et al., 2011; Westra et al., 2012; Sinkunas et al., 2013). Evolution of resistance to new challenges occurs by the addition of spacers to the promoter-proximal end of the CRISPR repeat array, in a Cas1 and Cas2-dependent process (Barrangou and Marraffini, 2014).
In addition to providing resistance to exogenous sequences, E. coli CRISPRs have activity against lysogeny and induction of temperate bacteriophages (Edgar and Qimron, 2010). Induction of the CRISPR-Cas system results in E. coli cell death if crRNA targets are present on the chromosome, but the system also selects for bacterial populations that have lost prophages. Other functions attributed to CRISPR-Cas systems include modulation of bacterial community behaviors, gene expression, and DNA repair (Methe et al., 2005; Viswanathan et al., 2007; Zegans et al., 2009; Aklujkar and Lovley, 2010; Babu et al., 2010). Lastly, recent studies have implicated or established a role for CRISPR-Cas systems in promoting virulence of several pathogens including Legionella pneumophila, Campylobacter jejuni, and Francisella novicida (Gunderson and Cianciotto, 2013; Louwen et al., 2013; Sampson et al., 2013; Louwen et al., 2014). Thus, the repertoire of cellular activities impacted by CRISPR-Cas appears to be diverse and much remains to be learned about these versatile elements particularly with regard to their influence on host interactions.
The work presented here was undertaken to determine the relationship of nilD to the CRISPR-Cas system and its function in mutualistic colonization of host nematodes. We demonstrate that the nilD locus expresses a CRISPR RNA molecule that contributes, in a Cas6e-dependent manner, to colonization of three distinct nematode species. Our data are consistent with a model that NilD functions to regulate endogenous bacterial sequences in a way that requires neither Cas3 nor perfect sequence identity to the target.
Results
The nilD locus is encoded within a CRISPR-Cas region
Heungens et al. (2002) previously reported a 3185-bp sequence (AY077466) of XnSc 081containing the nilD locus required for association with S. carpocapsae nematodes. Further sequence analysis revealed this locus encodes a CRISPR repeat element, comprising one spacer and two repeats, which is disrupted by the transposon insertion in the colonization deficient strain nilD6::Tn5 (Fig. 1) (Heungens et al., 2002). Additional CRISPR repeat sequences were noted upstream of nilD (bracketed region in Fig. 1A). These data indicate that X. nematophila encodes multiple CRISPR loci and that disruption of one of these, nilD, can inhibit nematode colonization.
To gain further information on the chromosomal context of nilD and to identify other CRISPR loci, the genome of XnSc 081 was sequenced and compared to that of the published sequence of X. nematophila strain HGB800 (NC_014228; ATCC 19061, hereafter referred to as XnSc 800) (Table S1). In both genomes there are six CRISPR elements (Table S2), labeled alphabetically in order of their occurrence in the chromosome (A–E and G) in addition to nilD (CRISPR-F). In both genomes, the nilD locus is located approximately 250-nt downstream of CRISPR-E. The nilD CRISPR is most similar to loci C and E: Each of these three loci (nilD, CRISPR-C, and CRISPR-E) encodes 32-nt spacer sequences and 29-nt repeats that are similar in sequence to those of E. coli K12 (Heungens et al., 2002) (Fig. 1C). In turn, the nilD repeat sequences are similar, but not identical, to those of the loci C and E. In the 29 nt comprising each repeat, 6 and 4 differences occurred in the left and right repeats of nilD respectively, relative to the CRISPR-E repeats (Fig. 1C), indicating the nilD locus has diverged from the other CRISPR loci in the genome.
Encoded downstream of nilD is the previously sequenced gloA gene, as well as cas genes and CRISPR locus G (Fig. 1A). The sequences of each of these regions are identical between XnSc 081 and XnSc 800. CRISPR loci are preceded by A/T rich leader sequences containing promoters driving CRISPR transcription (Pul et al., 2010). These leaders can be necessary for CRISPR function (Marraffini and Sontheimer, 2008; Sorek et al., 2008) and their sequence tends to be conserved within, but not across, species (Jansen et al., 1999; Lillestol et al., 2006). In both XnSc 800 and XnSc 081, a 99-nt sequence adjacent to CRISPR-E (Fig. 1A) is 93% identical to that found upstream of CRISPR-C, and is likely the leader sequence. This sequence is not found adjacent to any other CRISPR locus (A, B, D, or G), nor is it found adjacent to nilD, suggesting that these loci may be regulated in a manner distinct from CRISPR-C and -E.
The X. nematophila cas genes are of the Type I-E subset and include the broadly conserved cas1, cas2, and cas3 genes, as well as cse1 (casA), cse2 (casB), cas7 (casC), cas5 (casD), and cas6e (casE) (Fig. 1A) (Haft et al., 2005; Makarova et al., 2006; Chakraborty et al., 2010; Makarova et al., 2011b). Based on comparisons to E. coli and other systems (Brouns et al., 2008; Sinkunas et al., 2011; Westra et al., 2012; Sinkunas et al., 2013), we predict cas3 encodes a protein with nuclease and helicase activity necessary for mediating degradation of crRNA-DNA hybrids, while the other five genes encode components of the ribonucleoprotein Cascade complex necessary for CRISPR RNA processing and target DNA degradation. In other systems cas1 and cas2 genes are not necessary for CRISPR RNA processing or activity, but rather encode nucleases that form a complex necessary for acquisition of new spacers (Fineran and Charpentier, 2012; Nuñez et al., 2014). In addition, cas1 is involved in DNA repair and chromosome segregation (Babu et al., 2010), while cas3 promotes plasmid replication in E. coli (Ivancic-Bace et al., 2013).
Genomic analyses indicate that the spacer composition of the E. coli Type I-E system diversifies more slowly than would be expected if the CRISPRs were primarily involved in immunity (Touchon et al., 2011). To address if X. nematophila spacer content diversifies during association with nematode and insect hosts we isolated DNA from 10 individual colonies of X. nematophila from our laboratory stock population of S. carpocapsae IJ nematodes that had been maintained for ~15 years by repeated (~monthly) passage through Galleria mellonella insect larvae. These “evolved” X. nematophila are the result of >1500 rounds of the natural life cycle, comprising persistence in non-feeding IJ nematodes during storage in water, infection and growth within insect larvae (including exposure to insect-associated microbiota), and colonization of nematode IJs (Richards and Goodrich-Blair, 2009). In contrast, XnSc 800 and XnSc 081 stocks have been stored for a similar period frozen in glycerol without propagation. There were no spacer sequence differences in CRISPR loci C, E or nilD in the 10 isolated colonies relative to each other or to the frozen stock strains (data not shown), indicating that these loci are not evolving during laboratory passage through nematodes and insects. While we did not examine spacer content of the other 4 CRISPR loci in the evolved strain, the absence of spacer content changes in CRISPR loci C and E after more than 1500 passages through insects supports the idea that the CRISPR-Cas machinery in X. nematophila may function in a role outside of the canonical immunity against exogenous nucleic acids (Takeuchi et al., 2012). Overall, the genomic analyses described above indicate that the nilD locus, which is necessary for X. nematophila to colonize S. carpocapsae nematodes, encodes a non-canonical CRISPR element.
The nilD CRISPR sequence is sufficient to promote nematode colonization
We previously reported that the colonization defect of the nilD6::Tn5 mutation was partially rescued by introduction of a plasmid (pSR2-312, Table 1) carrying a 312-bp fragment of wild type nilD-containing DNA, confirming this region is necessary for colonization (Heungens et al., 2002). Likewise, insertion of a 387-bp fragment encoding the nilD locus (pEVS107-nilD, Table 1) in single copy at the attTn7 insertion site of the XnSc 081 nilD6::Tn5 genome (referred to hereafter as nilD6::Tn5 + nilD) was sufficient to restore nematode colonization, in this case to wild type levels (Fig. 2B). The DNA surrounding the transposon insertion encodes two putative divergent and overlapping small open reading frames, orf1 and orf2 that encompass the CRISPR element (Heungens et al., 2002) (Fig. 1B) and may encode small peptides that could be involved in colonization. A plasmid carrying the nilD sequence with a mutation at the common “T” of the start codons of these two ORFs did not rescue the colonization defect of the nilD6::Tn5 mutant (Heungens et al., 2002). However, since this nucleotide is also the first within the 32-nt spacer (see underlined nucleotide in Fig. 1C), these previously reported data did not clarify if orf1, orf2, or the CRISPR-like element is involved in colonization. To help address this question, we transformed the nilD6::Tn5 mutant with derivatives of plasmid pSR2-312 (Heungens et al., 2002), containing the 312-bp fragment sufficient to rescue colonization. Deletions were made in the pSR2-312 backbone such that the 5′ (ΔL) and/or the 3′ (ΔR) ends of the 312-bp nilD region were truncated (Table 1, Fig. S1). These constructs were transformed into either the nilD6::Tn5 mutant or its wild-type parent XnSc 081 and transformants were tested for the ability to colonize IJ nematodes (Table S3). Constructs lacking substantial regions of orf1 or orf2 were able to rescue the colonization defect of the nilD6::Tn5 mutant, indicating that neither ORF is required in its entirety to encode the colonization determinant. In contrast, the deletion constructs in which portions of the CRISPR repeat-spacer sequence are truncated were unable to rescue the colonization defect of the nilD6::Tn5 mutant. Furthermore, a plasmid (pSR2-ΔP90/ΔL84, Table 1) carrying a 137-bp central fragment containing the 90-nt nilD CRISPR-like region did rescue the colonization defect of the nilD6::Tn5 mutant. These data support the hypothesis that within the nilD locus, the CRISPR-like sequences, not the short coding regions are necessary for colonization.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant Characteristics | Source or Reference |
|---|---|---|
| E. coli | ||
| DH5α | General cloning host | (Sambrook et al., 1989) |
| DH5α (λpir) | General cloning strain for maintenance of oriR6K plasmids | |
| S17-1 (λpir) | E. coli donor strain for conjugations | |
| X. nematophila | ||
| HGB081 (XnSc 081) | Rifampicin resistant derivative of wild-type X. nematophila AN6/1 (carpocapsae) | S. Forst |
| HGB151 | X. nematophila ATCC 19061 rpoS1::kan | (Vivas and Goodrich- Blair, 2001) |
| HGB315 | HGB081 nilD6::Tn5 | (Heungens et al., 2002) |
| HGB829 | HGB315 nilD6::Tn5 pECM20-gfp | (Martens et al., 2003) |
| HGB1186 | HGB315 nilD6::Tn5 pECM20-gfp sup-1 | This study |
| HGB1578 | HGB081 Δcas3-3::kan | This study |
| HGB1695 | HGB081 ΔcasE4::kan (cas6e mutant) | This study |
| HGB1418 (XnSa 1418) | X. nematophila (anatoliense) isolated from S. anatoliense nematodes | This study |
| HGB1419 (XnSw 1419) | X. nematophila (websteri) isolated from S. websteri nematodes | This study |
| HGB1421 | X. nematophila strain of unknown origin | S. P. Stock |
| HGB007 (XnSc 007) | Wild-type X. nematophila (carpocapsae) ATCC 19061, acquired in 1995 | ATCC |
| HGB800 (XnSc 800) | Wild-type X. nematophila (carpocapsae) ATCC 19061, acquired in 2003 | ATCC |
| HGB1764 | XnSc 081 nilD56::kan | This study |
| HGB1756 | XnSc 800 nilD56::kan | This study |
| HGB1940 | HGB315 nilD6::Tn5 attTn7::Tn7/nilD | This study |
| HGB1986 | HGB315 nilD6::Tn5 attTn7::Tn7/nilD-SDM | This study |
| HGB1901 | XnSc 081 Δcas3-5::strep | This study |
| HGB1909 | XnSc 081 ΔcasE6::strep (cas6e mutant) | This study |
| HGB1907 | HGB1940 Δcas3-5::strep | This study |
| HGB1915 | HGB1940 ΔcasE6::strep (cas6e mutant) | This study |
| X. bovienii | ||
| HGB1166 | ATCC 35271 X. bovienii attTn7::miniTn7 | |
| HGB1167 | ATCC 35271 X. bovienii attTn7::miniTn7/SR1 (containing nilA, nilB, and nilC) | (Cowles and Goodrich- Blair, 2004) |
| HGB1649 | HGB1166 pECMXb-Empty | This study |
| HGB1651 | HGB1166 pECMXb-SR2; nilD+ | This study |
| HGB1653 | HGB1167 pECMXb-Empty; nilABC+ | This study |
| HGB1655 | HGB1167 pECMXb-SR2; nilD+ nilABC+ | This study |
| Plasmids | ||
| pBCSK+ | General cloning vector, CmR, | Stratagene |
| pCR2.1®-TOPO | General cloning vector, AmpR, KanR | Invitrogen, Carlsbad, CA |
| pCR2.1-TOPOmini | General cloning vector, AmpR | This study |
| pTopoSR2-2 | 312-bp XnSc 007 nilD region amplified with KPH62 and KPH63 primers and cloned into pCR2.1®-TOPO | (Heungens et al., 2002) |
| pSR2-312 | ApaI-SacI fragment from pTopoSR2-2 subcloned into pBCSK+, formerly named pBCSR2-2 | (Heungens et al., 2002) |
| pAWA1 | 137-bp XnSc 007 nilD region PCR amplified from HGB007 chromosomal DNA with primers KPH57 and KPH58 and cloned into pCR®II-TOPO | This study |
| pCR2.1-TOPO-nilD-XnSa | pCR2.1-TOPO-nilD modified by site directed mutagenesis to match the nilD spacer sequence of XnSa and XnSw. Use for RPA analysis. | This study |
| pEVS107 | Source of Kanr cassette for cas3 and cas6e mutations | (McCann et al., 2003) |
| pEVS107-nilD | KanR; vector for insertion of 387-bp nilD fragment at attTn7 site of XnSc 081 | This study |
| pEVS107-nilD-SDM | KanR; pEVS107-nilD altered by site directed mutagenesis to change NilD RNA spacer sequence | This study |
| pKNG101 | SmR; oriR6K suicide vector | (Kaniga et al., 1991) |
| pECM20 | pECM2 containing a 614-bp chromosomal insert from XnSc 007 | (Martens et al., 2003) |
| pECMXb-Empty | pECM20 with X. bovienii sequence replacing the X. nematophila insertion sequence | This study |
| pECMXb-SR2 | pECMXb-Empty with 312bp nilD region from pTopoSR2-2 in the XbaI site | This study |
| pKNG cas3-5::strep | SmR; pKNG101 with Δcas3::strep for replacing cas3 gene with SmR cassette | This study |
| pKNG casE6::strep | SmR; pKNG101 with ΔcasE:::strep for replacing cas6e gene with SmR cassette | This study |
| pKR100 | CmR, oriR6K suicide vector | |
| pKR100 nilD56::kan | KanR, CmR; pKNG101 with Δcas-3::kan for replacing nilD encoding region with KanR cassette. | This study |
| pBS-5S | AmpR | (Trotochaud and Wassarman, 2005) |
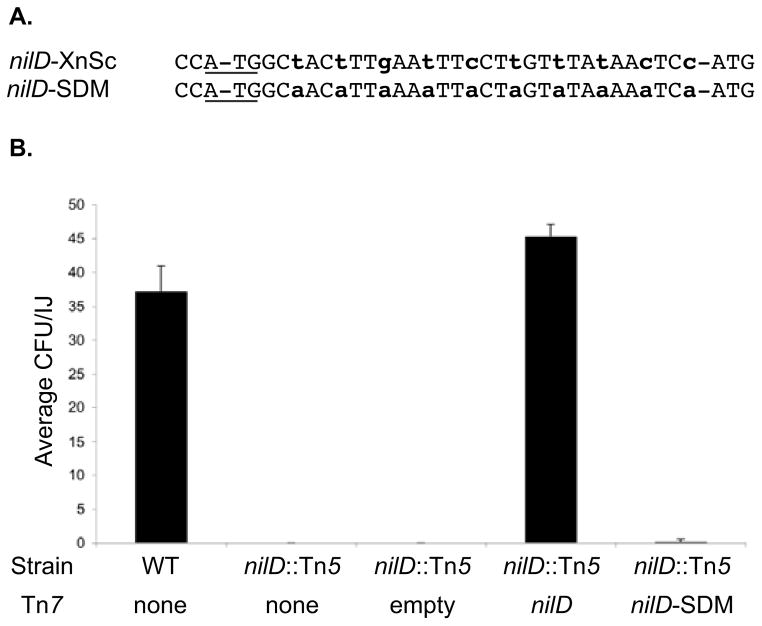
Figure 2.
Wild type, but not mutant nilD provided in trans complements the colonization defect of the nilD6::Tn5 mutant
A. Alignment of the spacer sequences of X. nematophila (from S. carpocapsae) wild type nilD (nilD-XnSc) and the mutated allele (nilD-SDM).
B. XnSc 081, XnSc 081 nilD::Tn5, and complemented strains were co-cultivated with axenic S. carpocapsae nematodes. The average colony forming units (CFU) colonizing the resulting progeny infective juveniles was measured by sonication and dilution plating.
The role of the predicted NilD RNA in colonization was investigated further by introducing base substitutions that would alter the putative NilD RNA sequence but not the Orf1 or Orf2 peptide coding sequences (Fig. 2A). This construct (pEVS107-nilD-SDM, Table 1) was introduced in single copy in the attTn7 site on the XnSc 081 nilD6::Tn5 genome and the colonization phenotypes of the resulting strains were examined. Complementation with this construct failed to restore nematode colonization, indicating that NilD RNA rather than either Orf peptide is essential for nematode colonization and further supporting the predicted function of NilD as a crRNA (Fig. 2B). Consistent with this conclusion, attempts to detect expression of the Orf1 and Orf2 peptides by immunoblot and assaying lacZ translational fusions were unsuccessful (data not shown), indicating that these factors may not be stably expressed.
nilD encodes a small RNA transcript expressed during growth in lab culture and colonization of nematodes
E. coli CRISPR repeats are transcribed and processed into small RNAs of ~57 nt (Brouns et al., 2008). We sought to determine if the nilD locus similarly expresses a small RNA. Northern blots were insufficiently sensitive to detect an RNA transcript from the nilD region (data not shown). We therefore performed an RNase protection assay (RPA) using probes containing nilD sequence specific for either sense or anti-sense RNAs (relative to the transcript orientation of the CRISPR-like element predicted by comparison to E. coli (Brouns et al., 2008)). No protected signal was observed in any reactions specific for anti-sense RNA (data not shown). In contrast, RNA harvested from wild type cultures, but not from the nilD6::Tn5 mutant, protected a fragment of approximately 58-nt when probes specific for the sense strand were used (Fig. 3A), indicating the nilD region encodes a 58-nt RNA that is expressed under in vitro growth conditions. Similar results were observed when RNA was extracted from wild type bacteria harvested from IJ stage S. carpocapsae nematodes (Fig. 3B), demonstrating that the NilD RNA is also expressed during mutualistic interactions with its nematode host. Additionally, RPA analysis of RNA isolated from X. nematophila wild type cells grown under various in vitro conditions indicate that NilD RNA levels are elevated in nutrient-limited or aged cells (Fig. S2A). Higher NilD RNA levels were detected after growth in LB supplemented with 2, 2-dipyridil (an Fe(II) chelator) relative to LB alone or with additional supplementation with exogenous Fe2SO3, indicating NilD RNA levels may increase after iron limitation (Fig. S2B).
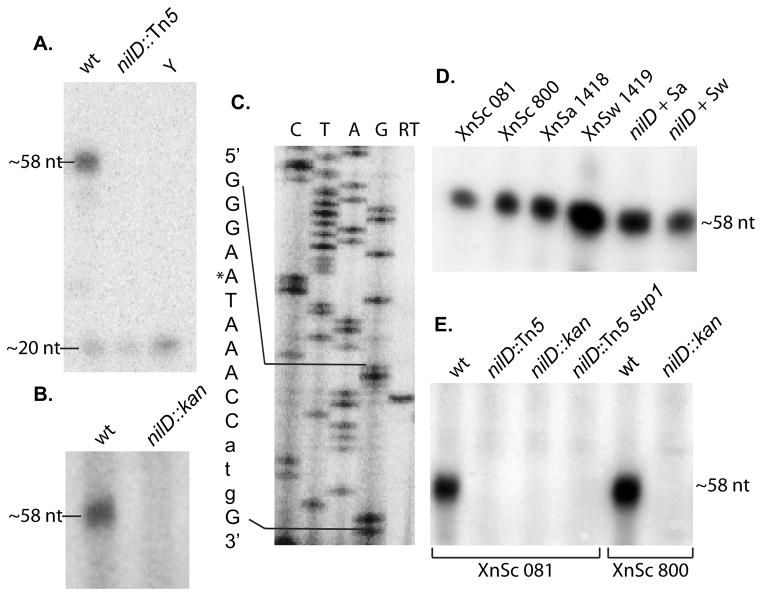
Figure 3.
The nilD locus of X. nematophila encodes a small RNA
RNase protection analysis, using a radiolabeled NilD RNA-specific probe, was used to detect expression of the NilD RNA in RNA harvested from X. nematophila strains during both in vitro growth (panels A, C, and E) and mutualistic interactions with the nematode host (panel B). Primer extension analysis was performed on RNA isolated from laboratory grown X. nematophila cultures to map the 5′ end of NilD RNA (panel C). A. RNA was harvested from stationary phase cultures of X. nematophila Sc 081, and X. nematophila Sc 081 nilD6::Tn5. The probe was also incubated with yeast RNA (Y) as a negative control. Radioactive markers (not shown) were used to estimate the sizes of the labeled fragments as indicated on the left. The 20-nt band represents the smallest nuclease-resistant single stranded RNA product of RNase cleavage. B. To assay for in vivo expression of NilD, RNA was extracted from Sc 081 and Sc 081 nilD56::kan harvested from infective-juvenile stage S. carpocapsae nematodes after co-cultivation. C. RNA isolated from wild type X. nematophila HGB800 (ATCC19061) after overnight growth in LB was used as a template for reverse transcriptase extension (RT) from the radioactively labeled primer AAP2. The resulting products were loaded on each gel adjacent to a sequencing ladder (C, T, A, and G lanes) derived from the same primer on pAWA1 template DNA. The relevant sequence is indicated to the left of the panel. The asterisk represents the starting nucleotide of the observed product. The lower case atg in the sequence to the left of the panel indicates the predicted start codon of orf1. D. To determine if the NilD RNA is expressed in distinct X. nematophila strains, RNA was harvested from the indicated strains: XnSc 081, XnSc 800 and strains harvested from S. anatoliense and S. websteri nematodes (XnSa 1418 and XnSw 1419, respectively). Likewise, RNA was harvested from nilD6::Tn5 strains complemented with the nilD locus derived from XnSa 1418 and XnSw 1419 (nilD + Sa and nilD + Sw, respectively). E. RNA was isolated from wild type X. nematophila strains (XnSc 081 and XnSc 800), nilD mutant strains (nilD::Tn5 or nilD::kan), and the nilD6::Tn5 suppressor strain (XnSc 081 nilD::Tn5 sup1). For each panel, each image was processed in its entirety with Adobe Photoshop CS3 to optimize visibility of bands by adjusting brightness and contrast.
Primer extension was used to determine the 5′ end of the ~58-nt NilD RNA (Fig. 3C). The run-off fragment indicates that the 5′ end of NilD RNA is an adenine (designated +1 and indicated by an asterisk in Fig. 3C) seven nucleotides upstream of the spacer region. In addition to this 5′ end, we occasionally observed run off fragments consistent with the +2U as the 5′ end (data not shown), which may indicate flexibility in processing or transcription initiation. The mapped 5′ end of NilD RNA and the predicted 5′ end match the 5′ and 3′ handles identified for E. coli crRNAs (Brouns et al., 2008) (Fig. 1C). These results confirm that the nilD locus encodes a small CRISPR RNA (genome coordinates: 3579434…3579491), hereafter referred to as NilD RNA (Fig. 1C).
NilD RNA expression requires Cas6e
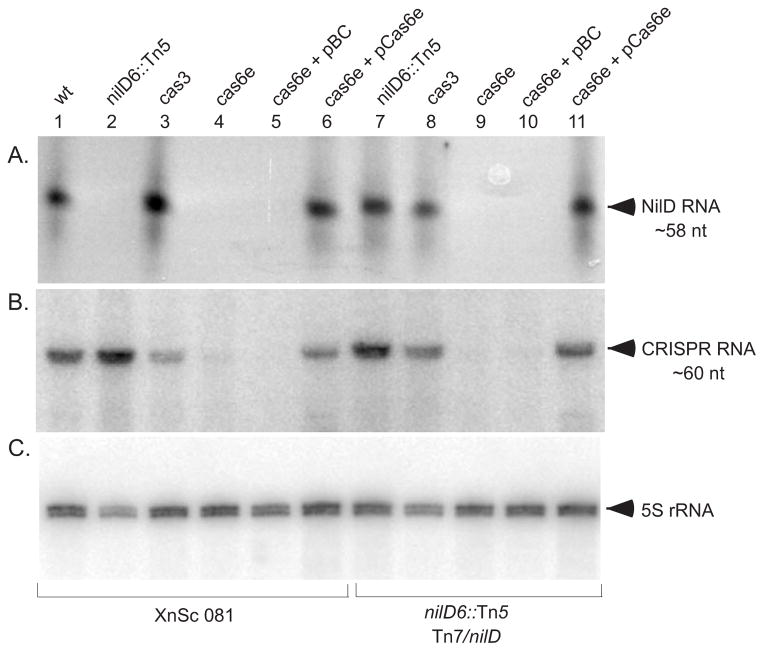
To investigate the role of the Cas machinery in NilD RNA processing and nematode colonization, we used allelic exchange to generate mutations in cas6e, a gene predicted to encode an endoribonuclease responsible for processing of CRISPR transcripts into small RNAs, and in cas3, which is predicted to encode a helicase/nuclease required for CRISPR-mediated resistance to infection (Brouns et al., 2008; Sinkunas et al., 2011). RPA was used to detect X. nematophila NilD RNA and northern hybridization, using a general crRNA probe, was used to detect total CRISPR RNA in wild type and cas mutants (Fig. 4). In the XnSc 081 background, CRISPR small RNAs were absent in the cas6e mutant, but were present in the cas6e mutant complemented with the cas6e gene on a plasmid (compare Fig. 4B lanes 4 and 5 with lane 6). CRISPR RNAs were also apparent in the cas3 mutant (Fig. 4B, lane 3). These data indicate that, as in E. coli, cas6e but not cas3 is necessary for normal processing of crRNAs (Brouns et al., 2008). Furthermore, crRNAs were expressed in the nilD6::Tn5 mutant (Fig. 4B, lane 2), suggesting that the nematode colonization defect of this mutant is not due to general disruption of crRNA expression. RPA analysis revealed NilD RNA, like other crRNAs, is apparent in the cas3-deficient (Fig. 4A, lanes 3 and 8) but not the cas6e-deficient strains (Fig. 4A, lanes 4, 5, 9, and 10), when expressed from its native locus (in the HGB081 background) or from the attTn7 locus (in the nilD6::Tn5 + nilD background). Furthermore, providing a wild-type copy of the cas6e gene on a plasmid restored NilD processing (Fig. 4A, lanes 6 and 11). These results establish that the expression of the 58-nt NilD RNA depends on a component of the Cas machinery, further supporting its identity as a CRISPR RNA.
Figure 4.
cas6e is necessary for presence of CRISPR RNA, but not for NilD RNA or colonization of S. carpocapsae nematodes. RNA was isolated from XnSc 081 wild type (wt), nilD6::Tn5, Δcas-3::strep (cas3), and ΔcasE::strep (cas6e) with or without the empty vector control (pBC) or the cas6e complement plasmid (pCas6e). All cells were grown to stationary phase in LB at 30°C. RNAse protection assays using radiolabeled AAP2 primer (A) were used to detect NilD RNA while radiolabeled primers AAP1 (B) or Xn 5S RNA (C) were used in northern blots to detect CRISPR RNA and 5S RNA respectively. CRISPR RNA is detected as a band of ~60 nt. 5S RNA was detected as a band of ~113 nt.
nilD and casE are only necessary for colonization in a specific genetic background
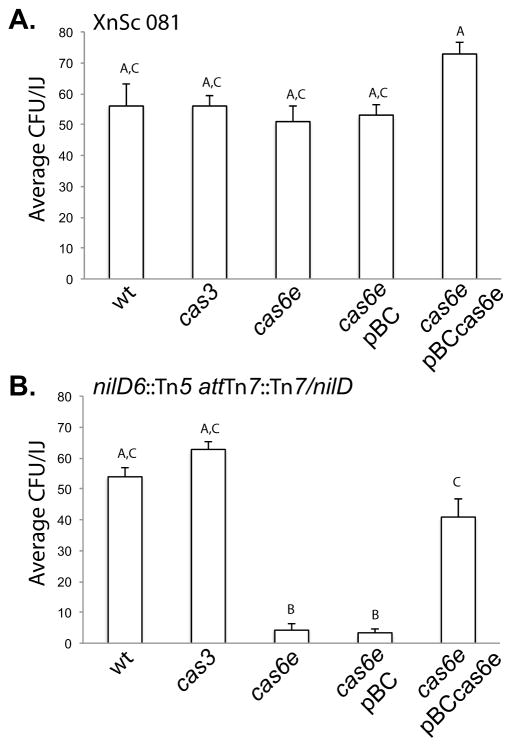
Our data indicate that NilD RNA is not processed in the absence of cas6e. Furthermore, in E. coli, Cas3 is predicted to be required for processed crRNA function. We therefore predicted that in X. nematophila cas6e and cas3, like nilD, would be required for nematode colonization. We first tested this by replacing the cas6e and cas3 genes with a streptomycin resistance cassette in XnSc 081 and the nilD6::Tn5 mutant with (Tn7-nilD) or without (eTn7) nilD, generating a panel of cas6e::strep and cas3::strep strains. As predicted, disruption of cas6e in the nilD6::Tn5 + nilD strain resulted in a significant colonization defect, which was rescued by providing a wild type copy of cas6e on a plasmid (Fig. 5B). These data are consistent with the model that the function of Cas6e in colonization is to process NilD RNA into a crRNA, although we have not ruled out the possibility that it has a NilD-independent function in colonization. Contrary to our prediction, deletion of cas3 resulted in no significant effects on nematode colonization. Therefore, in contrast to crRNAs in other systems, NilD activity is independent of Cas3.
Figure 5.
cas6e is necessary for colonization in the nilD6::Tn5 attTn7::Tn7/nilD background.Colonization ability was measured for (A) XnSc 081 and (B) nilD6::Tn5 attTn7::Tn7/nilD (the nilD mutant with wild type nilD expressed in trans at the attTn7 site). Each strain carried wild type cas genes (wt) or cas3 and cas6e mutations. In each background, the colonization phenotypes of the cas6e mutant carrying the control vector pBC or wild type cas6e (pBCcas6e) was also assessed. Each strain was co-cultivated with axenic S. carpocapsae nematodes and colony forming units (CFU) within the resulting progeny infective juveniles (IJ) was measured by sonication and dilution plating. The average CFU of strain/IJ ± standard error (n ≥ 5) is shown. In this experiment the uncomplemented nilD6::Tn5 strain colonized at 0.22 ± 0.11 CFU/IJ Different letters indicate significant differences in colonization levels between bacterial strains: p<0.0001 One-way ANOVA with Tukey’s post-test.
Surprisingly, neither the cas6e::strep nor the cas3::strep mutations caused a colonization defect in the XnSc 081 strain background (the parent of the nilD6::Tn5 mutant) (Fig. 5A), raising the possibility that the requirement for NilD RNA is specific to the nilD6::Tn5 background. To further explore this hypothesis, nilD was replaced by allelic exchange with a kanamycin resistance cassette in the XnSc 081 and XnSc 800 wild type backgrounds. Like XnSc 081 cas6E::strep, the resulting strains, XnSc 081 nilD56::kan and XnSc 800 nilD56::kan mutants colonized S. carpocapsae to wild type levels (Table 2), despite a lack of NilD RNA detected by RPA (Fig. 3E and Fig. 4A).
Table 2.
A nilD::kan mutation does not confer a colonization defect in the HGB081 and HGB800 backgrounds.
| Relevant Alleles | Average CFU of strain/IJ ± standard error (n ≥ 3)a | |
|---|---|---|
| XnSc 081 | HGB800 | |
|
|
||
| none | 66.36 ± 2.57 | 50.03 ± 4.02 |
| nilD::kan | 60.03 ± 3.86 | 61.73 ± 0.96 |
| nilD::kan cas3::str | 60.52 ± 5.97 | 60.05 ± 3.17 |
| nilD::kan cas6e::str | 58.83 ± 2.76 | 58.72 ± 2.80 |
In this experiment the nilD6::Tn5 strain colonized at 0.10 ± 0.01 CFU/IJ. None of the values shown in the table were significantly different from each other, but all were significantly different from the nilD6::Tn5 strain (p<0.001 using one-way ANOVA with Tukey’s post-test).
These data verify that NilD and Cas6e are required for mutualism, but only within a specific genetic background of X. nematophila, suggesting a synthetic allele arose during the transposon mutagenesis process that gave rise to nilD6::Tn5. To examine possible synthetic mutations present in this background, we sequenced and compared the nilD6::Tn5 draft genome to that of its parent XnSc 081 and found only one difference, a single nucleotide variation (SNV) located within the intergenic region of genes XNC1_3346 and XNC1_3347 (T-3271541-C). Gene XNC1_3346 is predicted to encode a P4-like DNA primase while XNC1_3347 is a small hypothetical gene in the DUF1795 superfamily that is followed immediately by XNC1_3348, predicted to encode an Rhs-like, YD-repeat-containing protein of unknown function (Fig. S3). Overlapping XNC1_3346 is an 1147-bp repeat sequence that occurs with 80–87% identity in two other locations of the genome (one full length copy and one truncated copy), also overlapping with genes with homology to those encoding P4-like DNA primases (Fig. S3). The SNV occurs within this repetitive region and an alignment of the three repeat regions shows variability in the nucleotides around the SNV (Fig. S3D). That the SNV associated with the nilD6::Tn5 strain background is located within a phage-encoding region is consistent with it being involved in the activity of the CRISPR-Cas system, potentially as a target for NilD RNA. However, no obvious regions of sequence identity or complementarity were observed between NilD RNA (repeats or spacer) and the region surrounding the SNV.
A suppressor of the nilD6::Tn5 colonization phenotype
The low level of colonization observed for the nilD6::Tn5 mutant (~0.1 CFU/IJ; see for example Table 3) could reflect a majority of nematodes colonized by few bacterial cells, or full colonization (~50 bacteria) in one of many nematodes. The former phenotype might indicate the nilD6::Tn5 mutant has a defect in outgrowth (Martens et al., 2003), whereas the latter phenotype might indicate the nilD6::Tn5 mutant has a defect in initiation of colonization which can be occasionally suppressed. To distinguish between these possibilities we examined by epifluorescence microscopy the frequency of colonized nematodes after cultivation on a GFP-expressing nilD6::Tn5 strain (XnSc 829; Table 1). This analysis showed that the majority of individual nematodes were uncolonized and that approximately 1 in 600 nematodes were fully colonized (data not shown). To determine if rare colonization events were due to genetic or epi-genetic suppression of the nilD6::Tn5 mutation we examined the colonization phenotype of a colony isolate derived from nilD6::Tn5-colonized nematodes. Upon re-cultivation with nematodes, this colony isolate, XnSc 1186 (sup-1) exhibited significantly higher levels of colonization than its nilD6::Tn5 parent (5.48 ± 0.57 vs. 0.15 ± 0.47 Avg. CFU/IJ respectively, n≥6, p<0.001 by unpaired Student’s t-test), despite the absence of detectable NilD RNA by RPA (Fig. 3E). These data indicate that the phenotype caused by the nilD6::Tn5 mutation can be suppressed, presumably by nucleotide or epigenetic changes elsewhere on the chromosome. The latter seems most likely, as sequencing of the XnSc 1186 sup-1 genome did not reveal mutations that could explain the suppression phenotype (data not shown).
Table 3.
nilD is required for X. nematophila colonization of S. anatoliense and S. websteri nematodes.
| Bacterial Strain | attTn7 locus insertiona | Average CFU/IJ ± standard deviation (n=3)
|
||
|---|---|---|---|---|
| S. carpocapsae | S. anatoliense | S. websteri | ||
| XnSc 081 | none | 42.07 ± 11.01A | 34.40 ± 4.12A | 26.31 ± 6.20A |
| XnSc 081 nilD6::Tn5 | none | 0.10 ± 0.04B | 0.36 ± 0.06B | 0.12 ± 0.15B |
| XnSc 081 nilD6::Tn5 | nilD-Sc | 54.40 ± 7.95A | 38.60 ± 4.77A,C | 44.10 ± 7.03A,C |
| XnSc 081 nilD6::Tn5 | nilD-Sa | 1.09 ± 0.24B | 1.08 ± 0.40B | 0.45 ± 0.23B |
| XnSc 081 nilD6::Tn5 | nilD-Sw | 0.03 ± 0.02B | 0.25 ± 0.17B | 0.02 ± 0.14B |
| XnSa | none | 77.90 ± 16.58A,C | 59.40 ± 4.13C | 55.50 ± 16.61C,D |
| XnSw | none | 63.55 ± 12.97A,C | 28.45 ± 6.66A | 74.57 ± 4.74D |
A Tn7 transposon carrying nilD loci from XnSc (nilD-Sc), XnSa (nilD-Sa), or XnSw (nilD-Sw) was integrated at the attTn7 locus. Different letters indicate significant differences between bacterial strains for colonization within each nematode species: p<0.05 using one-way repeated measures ANOVA (Prism v2.0, GraphPad, La Jolla, CA) with Tukey’s post-test. Colonization levels achieved by each bacterial strain in the three nematode species were not significantly different except XnSw for which colonization of S. anatoliense nematodes was significantly lower than those of the other two nematode species (not shown).
The nilD locus contributes to XnSc 081 association with different nematode species
In addition to S. carpocapsae, X. nematophila associates with two other nematode species, S. websteri and S. anatoliense (Lee and Stock, 2010). To examine if NilD RNA is required for colonization of these other nematode hosts, the colonization phenotypes of the nilD6::Tn5 mutant and XnSc 081 in S. anatoliense and S. websteri were assessed. Similar to the phenotypes observed in S. carpocapsae colonization assays, XnSc 081 colonized S. anatoliense and S. websteri while the nilD6::Tn5 mutant displayed a marked defect in its ability to associate with these host nematode species (Table 3). In all three nematode hosts the colonization defect of the nilD6::Tn5 mutant was rescued by insertion of the XnSc 081 nilD sequence at the attTn7 insertion site (Table 3).
Strain specific NilD RNA variants are expressed in the S. anatoliense and S. websteri symbionts but do not rescue the nilD6::Tn5 colonization defect
To determine if the nilD locus is present in all X. nematophila strains regardless of the identity of their natural nematode host, we searched for it in X. nematophila strains from S. anatoliense and X. websteri (XnSa 1418 and XnSw 1419 respectively). Oligonucleotides (NilD 5′ ApaI and NilD 3′ KpnI, Table S4) complementary to flanking regions around the nilD region of XnSc 081 successfully amplified products from XnSa 1418 and XnSw 1419 genomic DNA. In each case a product was obtained of similar size to that amplified from XnSc 081 and XnSc 800 (data not shown). The products amplified from XnSa 1418 and XnSw 1419 genomic DNA were cloned and sequenced. Relative to the S. carpocapsae-derived X. nematophila strains, the XnSa 1418 and XnSw 1419 nilD regions encode an identical left repeat, 4-nt differences within the 32-nt spacer, and an identical right repeat except for the last 3 nt (Fig. 1D). Also, unlike the XnSc 081 nilD sequence, the nilD regions of XnSa 1418 and XnSw 1419 are not predicted to encode small overlapping open reading frames (data not shown).
To assess if the nilD loci of XnSa 1418 and XnSw 1419 encode a NilD RNA molecule, we conducted RNase protection assays (RPA) using probes that match the XnSa 1418 nilD sequence. RPA using this probe detected protected fragments in both the XnSa 1418 and XnSw 1419 samples that were similar in size to those that were detected (using XnSc specific probe) in the XnSc 081 and XnSc 800 samples (Fig. 3D). These data indicate that NilD RNA is expressed and processed similarly in all four Xn strains.
Given the divergence of the XnSa 1418 and XnSw 1419 nilD spacers from those of Sc X. nematophila strains, we examined if the former could rescue the colonization defect of the nilD6::Tn5 mutant. The XnSa 1418 and XnSw 1419 nilD sequences were cloned and inserted at the attTn7 site of the XnSc 081 nilD6::Tn5 mutant, generating strains nilD6::Tn5 + nilD-XnSa (nilD + Sa) and nilD6::Tn5 + nilD-XnSw (nilD + Sw). RPA analysis was used to verify expression of the NilD RNA (Fig. 3D) while the colonization proficiency of the complemented strains was assessed using Sc nematodes (Table 3). As expected, insertion of the XnSc nilD sequence (strain nilD6::Tn5 + nilD) was sufficient to restore wild type levels of colonization. In contrast, providing the XnSa 1418 and XnSw 1419 sequences failed to restore levels of colonization above the level of the nilD6::Tn5 mutant strain (Table 3). These results are consistent with our findings described above that alteration of the NilD RNA sequence was sufficient to disrupt the activity of this molecule. nilD6::Tn5 + nilD-XnSa and nilD6::Tn5 + nilD-XnSw were also deficient for colonization of S. anatoliense and S. websteri nematodes (Table 3), indicating that distinct nilD sequences do not confer specificity for these nematode species.
To further explore the possible role of nilD in host range specificity, we introduced it into X. bovienii (the symbiont of the nematode S. jollieti) that naturally lacks nilD (Chaston et al., 2011; Sugar et al., 2012). X. bovienii is unable to colonize S. carpocapsae unless it expresses the host-range specificity determinants nilA, B, and C (Cowles and Goodrich-Blair, 2008; Chaston et al., 2013). We therefore expressed nilD in X. bovienii with and without the nilA, B, and C genes. The presence of nilD did not impact the colonization levels of X. bovienii: colonization of S. carpocapsae nematodes was below the level of detection (0.005 CFU/IJ) when nilD was present without nilA, B, and C, and colonization levels of X. bovienii carrying nilA, B, and C were not increased by the presence of nilD (data not shown).
Discussion
The goal of this study was to elucidate the mechanistic role of the X. nematophila nilD locus during colonization of S. carpocapsae host nematodes. Our work demonstrates that nilD encodes a small RNA and that expression of this molecule is sufficient to rescue the colonization defect of the nilD6::Tn5 strain. Bioinformatic predictions indicated that NilD RNA is a member of the CRISPR RNA family. Consistent with this, we present experimental evidence that NilD RNA processing and colonization function requires cas6e, encoding a homolog of the E. coli CRISPR RNA processing Cascade complex endonuclease (Cas6e) (Westra et al., 2012). However, unlike the CRISPR-Cas systems of other bacteria, NilD RNA function does not appear to require the helicase-nuclease Cas3 (Sinkunas et al., 2011), since a cas3 mutant does not display a colonization defect (Fig. 5). This may indicate that the function of NilD RNA diverges from that of other crRNAs, and that its requirement in colonization does not include Cas3-mediated target degradation.
Several lines of evidence argue against the idea that the colonization function of NilD RNA is to restrict entry of exogenous DNA (plasmids and lytic bacteriophages), the initial primary function ascribed to crRNAs (Brouns et al., 2008; Marraffini and Sontheimer, 2008). Instead, our data support the model that NilD RNA regulates endogenous sequences, as has been observed or suggested in other CRISPR-Cas systems (Zegans et al., 2009; Aklujkar and Lovley, 2010; Cady and O’Toole, 2011; Sampson et al., 2013). First, the nilD6::Tn5 colonization defect is apparent in a closed system consisting only of a bacterial clonal population and the nematode host. Therefore, the only source of potentially toxic foreign DNA is the nematode. However, nematode lysates do not inhibit growth of Xenorhabdus strains in liquid culture and do not form plaques on bacterial lawns (data not shown). Further, BLASTn analyses (Altschul et al., 1997) against the NCBI GenBank sequence database and to 13 other Xenorhabdus bacterial genomes (H. Goodrich-Blair, unpublished) failed to identify putative proto-spacers with identity to NilD (data not shown). While not conclusive, this fact is contrary to the idea that the NilD RNA targets a mobile genetic element present in other Xenorhabdus spp. Second, differences in the endogenous chromosome can bypass or cause the need for NilD RNA. The nilD6::Tn5 colonization defect is only apparent in a specific genomic background (Fig. 5) in which suppressor alleles (e.g. sup-1) can arise that are able to colonize despite the absence of nilD expression (Fig. 3). Our genomic analysis indicates the strain background associated with the requirement for nilD in colonization has a single distinguishing SNV, a T to C change at nt 3271541 in the intergenic region between a phage P4 primase and a region predicted to encode Rhs-like and associated elements (Fig. S3), while sequencing of the sup-1 strain failed to identify any genetic alterations that could account for the suppression phenotype (data not shown). These findings suggest that a single nucleotide change in the bacterial genome may confer dependence upon nilD for colonization, whereas suppression may result from epigenetic or phase variability.
An alternative explanation for the role of NilD RNA in colonization is that it is required to control expression of an endogenous genetic element that is detrimental for host interactions, with the NilD RNA 32-nt spacer region conferring specificity for this element. Similar to the E. coli CRISPR system, NilD may control expression of its targets (Edgar and Qimron, 2010). Two models of CRISPR-Cas-mediated gene regulation are that the Cascade complex, including the crRNA binds to target mRNA to block translation or to promote Cas-3-mediated cleavage, or the Cascade complex binds to the DNA target and prevents transcription. Since a cas-3 mutant does not display the same colonization defects as the nilD6::Tn5 mutant (Fig. 5), our data are most consistent with the idea that the NilD RNA-Cascade complex blocks either transcription or translation, rather than triggering target degradation. Further insights into the mechanism of NilD RNA function await identification of its target(s). No proto-spacer with 100% identity is apparent in the genomes of XnSc 800 or XnSc 081, suggesting that low levels of similarity may be sufficient for NilD targeting. Conversely, complementation experiments using mutagenized XnSc 081 nilD and the divergent nilD loci of XnSa 1418 and XnSw 1419 failed to restore the colonization defect of nilD6::Tn5 (Table 3), revealing that some sequence integrity is essential. Furthermore, these data may indicate that the NilD RNA targets in strains XnSa 1418 and XnSw 1419 have diverged in sequence, and that the nilD loci in those strains have co-evolved to maintain sequence identity. If true, further comparative sequence analysis of these strains could yield putative targets.
This report extends the limited number of studies demonstrating an impact of CRISPR-Cas systems on host-microbe interactions. The cas2 gene of Legionella pneumophila is required for intracellular growth within host amoebae (Gunderson and Cianciotto, 2013). Similar to our work, these experiments were conducted in the absence of exogenous DNA or phage, suggesting that the requirement for cas2 in L. pneumophila was independent of any interference-related functions. Likewise, a cas2 mutant was not more sensitive upon exposure to UV light or to treatment with mitomycin C or nalidixic acid, indicating that the virulence defect was not due to the a potential role for Cas2 in DNA repair (data not shown). A recent study demonstrated F. novicida Cas9-mediated negative regulation of an endogenous gene encoding a lipoprotein. In the absence of cas9, aberrant expression of the lipoprotein triggered host immunity and reduced virulence of the pathogen (Sampson et al., 2013). Together these and other studies have established a role for CRISPR-Cas machinery in facilitating pathogen virulence (Louwen et al., 2014). The work presented here demonstrates these systems can also function in mutualistic associations. Though still enigmatic, the role of NilD RNA and its synthetic allele in nematode colonization should be clarified by identifying its molecular target, and in turn the function of this target in either promoting or inhibiting the colonization process.
Experimental Procedures
Organisms and growth conditions
Strains used in this study are listed in Table 1. Unless otherwise noted, cultures were grown at 30°C in LB broth (Miller, 1972). X. nematophila growth media were stored in the dark or supplemented with 0.1% pyruvate (Xu and Hurlbert, 1990). Media were supplemented with kanamycin (Km, 50 μg/ml), rifampicin (Rif, 100 μg/ml), ampicillin (Amp, 150 μg/ml), streptomycin (Sm, 25 μg/ml), or chloramphenicol (Cm, 30 μg/ml) where appropriate. For indicated RNA isolations, cultures were supplemented with 500 μM Fe2SO3, 100 μM 2,2-dipyridyl or 1 μM deferoxamine (Sigma-Aldrich, St. Louis, MO). The nematodes Steinernema carpocapsae (Weiser) All, obtained from Harry Kaya, and S. anatoliense (Al-Jubiha Jordan) and S. websteri (Peru), obtained from S. Patricia Stock, were reared in Galleria mellonella wax moth larvae (Kaya and Stock, 1997). For in vitro co-cultures nematodes were grown at room temperature (20–26°C) on lipid agar (LA) plates with lawns of test X. nematophila strains as previously described (Vivas and Goodrich-Blair, 2001). Defined medium was as previously described (Orchard and Goodrich-Blair, 2004) except that glutamate was added at 100 mg l−1 and Bacto agar (Sigma-Aldrich, St. Louis, MO) was used instead of noble agar. X. nematophila strains from S. anatoliense and S. websteri were isolated by surface sterilization of 1000–10000 IJ nematodes for 3 min in 0.5% NaOCl as described previously (Heungens et al., 2002). Surface-sterilized nematode were sonicated for 1 min (Cowles and Goodrich-Blair, 2004) and dilution plated onto LB + 0.1% pyruvate agar. Individual colonies were streaked for isolation, cultured overnight at 30°C, and frozen in glycerol stocks. Bacterial identity was verified by Sanger sequencing of the 16S gene using primers 27F and 1492R (Table S4) (Lane, 1991).
DNA manipulations and biochemical assays
Plasmids used in this study are listed in Table 1. To create pCR2.1-TOPOmini, which lacks the KanR cassette, primers TOPO2.1mini_Fwd_NcoI and TOPO2.1mini_Rev_NcoI (Table S4) were used to amplify the backbone of the plasmid pCR2.1-TOPO. The amplified product was cut with NcoI and self-ligated. Standard protocols were used for the isolation of chromosomal DNA, DNA digestion, electrophoresis, and electroporation (Sambrook et al., 1989). Enzymes for DNA manipulations were obtained from Promega (Fitchburg, WI), New England Biosciences (Ipswich, MA) or Fermentas (Pittsburg, PA). Plasmid isolations and gel purifications were performed using appropriate kits (Qiagen, Germantown, MD). PCR amplification was performed using ExTaq polymerase, Primestar polymerase (Takara Shuzo, Kyoto, Japan) or PFU Ultra (Agilent Technologies, Madison, WI) and appropriate buffers on 100 ng Xenorhabdus chromosomal template-DNA, 0.2 μM each primer, 0.4 mM dNTPs, and 2.5 U of polymerase. After 2 min incubation at 95°C, 30 cycles of 20 s at 95°C, 30 s at an annealing temperature appropriate for each primer pair, and 60-s kb−1 at 72°C, were conducted, followed by 7 min incubation at 72°C.
pECMXb-Empty and pECMXb-SR2 construction, conjugation into X. bovienii
The pECMXb-Empty vector was constructed from pECM20 (Martens et al., 2003) by replacing the X. nematophila insertion sequence with a 588-bp fragment of X. bovienii intergenic genomic DNA (coordinates 390649-391236 of X. bovienii SS-2004; NC_013892.1) to facilitate homologous recombination of the plasmid into the X. bovienii genome. The pECM20 plasmid was divergently amplified on either side of the X. nematophila insert site to replace the insertion with the restriction sites for ApaI and KpnI. Primers used were pECM20_Xb_F and pECM20_Xb_R (Table S4). A predicted intergenic region of X. bovienii was amplified using primers pECMXb_insert_F and pECMXb_insert_R, and the sequence was inserted into pECMXb using ApaI and KpnI. The insert was confirmed by sequencing using primers pECMXb_seq_F and pECMXb_seq_R (Table S4). For construction of pECMXb-SR2, the SR2 region was sub-cloned from pTopoSR2-2 into the pECMXb XbaI site using XbaI and SpeI.
The pECMXb-Empty and pECMXb-SR2 plasmids were conjugated in X. bovienii 1166 and 1167 using previously described methods (Murfin et al., 2012). Ex-conjugants were grown on LB media supplemented with 15 μg/ml of chloramphenicol to select for insertion of the plasmid. Integration of pECMXb-Empty and pECMXb-SR2 into the genome was confirmed by positive PCR results using primers pECMXb_integratation_F and pECMXb_integration_R (Table S4).
Isolation of X. nematophila cas and nilD mutants
The 4,857 bp DNA fragment containing 2,751 bp cas3 gene and its up-stream (1,213 bp) and down-stream (893 bp) sequences were amplified from HGB800 chromosomal DNA using Pfu DNA polymerase (Stratagene, Santa Clara, CA) and primers cas3UpFwd_SpeI and cas3DownRev_XbaI. Likewise, the 2,553 bp DNA fragment containing 678-bp cas6e (termed casE in strain designations) and its up-stream (1,233 bp) and down-stream (642 bp) sequences were amplified using primers casEUpF_SpeI and casEDownR_ XbaI, respectively (Table S4). The resulting fragments were digested with XbaI and SpeI and cloned into plasmid pCR2.1-TOPOmini between XbaI and SpeI sites. The ahp kanamycin resistance cassette and its promoter region were amplified from plasmid pEVS107 using primers Kan-CleanRev_EcoRV_NEW and Kan-FullFwd_NheI_NEW (Table S4) digested with NheI and EcoRV, and used to replace the 2,362 bp NheI-EcoRV region (89-2451 bp) within the cas3 gene and the 26-321 bp region of the cas6e gene, generating pCR2.1 mini Δcas3::kan and pCR2.1 mini ΔcasE4::kan.
To create Δcas6e::strep and cas3::strep insertion constructs used in this study, the KanR cassettes in pCR2.1 mini ΔcasE4::kan (HGB1692) and pCR2.1 mini Δcas3-3::kan were removed by cutting with EcoRV and NheI. The remaining backbone of the plasmid was ligated to EcoRV/SpeI fragment, containing the SmR cassette from pKNG101, to form pCR2.1 mini ΔcasE6::strep and pCR2.1 mini Δcas3-5::strep. The ΔcasE6::strep and Δcas3-5::strep fragments were cut from the pCR2.1 mini backbone using SpeI and XbaI and cloned into the SpeI site of the mobilizable suicide plasmid pKNG101, generating pKNGDcasE6::strep and pKNGDcas3-5::strep. The resulting constructs were maintained by electroporating into E. coli SM10 (λpir) cells and then introduced into HGB081, HGB800 and HGB1940 (Table 1) by conjugation, as described previously (Forst and Tabatabai, 1997). Ex-conjugants were grown on LB agar containing 25 μg/ml streptomycin overnight and subsequently grown on LB agar plus 5% sucrose to select for sucrose-resistant ex-conjugants that had excised the vector. The SmR phenotype was verified, and deletion of the cas6e or cas3 gene fragments was confirmed by PCR amplification.
For deletion of the nilD region, a 1,542 bp fragment upstream of nilD was amplified from the XnSc 081 chromosome using PFU Ultra polymerase and the dNilD Up 5′ SalI and dNilD Up 3′ ApaI primers. Likewise, a 1,082 bp fragment downstream of nilD was amplified using the dNilD Dwn 5′ ApaI and dNilD 3′ SacI primers while the KanR cassette was amplified from plasmid pEVS107 using primers Kan 5′ ApaI and Kan 3′ ApaI (Table S4). PCR fragments were digested using appropriate restriction enzymes and then ligated into pKR100 plasmid, linearized with SalI and SacI enzymes, generating pKR100-nilD56::kan. The resulting construct was maintained by electroporation of E. coli S17.1 (λpir) and introduced into HGB081 and HGB800 by conjugation. Ex-conjugants were grown on LB agar containing 50 μg/ml kanamycin overnight and then screened for loss of resistance to chloramphenicol. The deletion of the nilD locus was confirmed by PCR and RPA was utilized to confirm NilD RNA was not being produced.
Complementation studies
To generate nilD truncation constructs, portions of the nilD locus were amplified (using primers indicated in parentheses) and cloned into pCR2.1®-TOPO to create pTopoSR2-ΔR90 (KHP62 and KHP58), -ΔR126 (KHP62 and KHP64), -ΔL84 (KHP57 and KHP63), -ΔL132 (KHP55 and KHP63), -ΔL161 (KHP36N and KHP63), and –ΔR90/ΔL84 (KHP57 and KHP58) (Table S4). Once constructed, all fragments were subcloned from pCR2.1®-TOPO into pBCSK+ using ApaI and SacI to create pSR2-ΔR90, pSR2-ΔR126, etc. To generate a stable nilD complemented strain, a 387-nt fragment encoding the nilD crRNA region and approximately 175-nt upstream, was amplified using Primestar polymerase (Takara Shuzo, Kyoto, Japan) and primers nilD 5′ ApaI and nilD 3′ KpnI (Table S4). The nilD PCR fragment and the Tn7-insertion vector, pEVS107 (Table 1), were digested with ApaI and KpnI restriction enzymes and ligated using T4 DNA ligase (New England Biosciences). The resulting vector, pEVS107-nilD, was maintained by electroporating into competent S17.1 λpir E. coli cells followed by selection on LB plates supplemented with kanamycin.
To complement the nilD-deficient strain (nilD6::Tn5) with a nilD region encoding synonymous base mutations within the spacer sequence, pEVS107-nilD was subjected to a series of site directed mutagenesis reactions. The pEVS107-nilD vector was first amplified using the NilD SDM set 1F and 1R primers (Table S4) to generate base substitutions within codons 2 and 3 in the nilD spacer region. The resulting construct was then further mutated using NilD SDM sets 2 through 5 (Table S4) to generate synonymous base substitutions within codons 4–11, forming pEVS107-nilD-SDM (Table 1). For site directed mutagenesis reactions, approximately 3 μg of pEVS107-nilD DNA were mixed with 15 pmol of each primer, 4 μl of DMSO, 50 μmol dNTP mix, 5 μl PFU Ultra buffer and 1 μl PFU ultra polymerase (2.5 units/μl) (Agilent Technologies) in 50 μl of total volume. After 1 minute at 95°C, the resulting mixtures were incubated at 95°C for 1 min, 56°C for 50 s, and 72°C for 10 min for 25 cycles, followed by 10 additional min at 72°C. Following amplification, template DNA was digested by incubation with 10 units of ApaI restriction enzyme at 37°C for 1 hr and the resulting PCR product was maintained by electroporation into S17.1 λpir E. coli cells and selection on LB supplemented with kan. All plasmids were sequenced to ensure that the appropriate mutations were present and that no additional mutations had occurred within the nilD region.
To generate the casE complementation construct, the cas6e gene was amplified using the CasE 5′ XbaI and CasE 3′ EcoRV primers (Table S4) and Primestar polymerase (Takara Shuzo, Kyoto, Japan). The PCR product and pBluescript (pBCSK+) vector (Stratagene, La Jolla, CA) were subjected to restriction digestion with XbaI and EcoRV enzymes and ligated using T4 ligase. The resulting vector, pBC-casE (Table 1), was introduced into Top 10 E. coli cells (Invitrogen, Carlsbad, CA) and maintained by selection on LB plates supplemented with chloramphenicol.
Complementation of Xenorhabdus strains was performed as previously described (Bao et al., 1991; Forst and Tabatabai, 1997). Briefly, for complementation of the nilD mutation, overnight cultures of nilD6::Tn5, S17.1 + pEVS107-Tn7/nilD and the transposition helper strain S17.1 + pUX-BF13 (Bao et al., 1991), were diluted 1:100 in LB and incubated for 3 h at 30°C. After incubation, 900 μl of X. nematophila culture was mixed with 600 μl of S17.1 + pEVS107-Tn7/nilD, and 500 μl S17.1 + pUX-BF13. The mixture was then pelleted by centrifugation, resuspended in 30 μl of LB and spotted onto LB plates supplemented with 0.1% pyruvate. Approximately 18 h after plating, cells were scraped into 1 ml of LB and 50 μl were plated onto LB supplemented with 0.1% pyruvate, and containing ampicillin and erythromycin for selection. Resistant colonies were screened for proper insertion at the Tn7 site by PCR analysis. For complementation with nilD truncation or cas6e constructs, chemically competent X. nematophila strains were generated as previously described (Xu et al., 1989) and transformed with 200 ng of individual vector constructs. Transformants were plated on LB supplemented with 0.1% pyruvate and chloramphenicol.
RNA isolations and analyses
For RNA isolation from cultured bacteria, X. nematophila strains were grown for 18 h in either liquid LB or LB containing 100 μM dipyridyl (to promote optimal NilD expression) and 0.1% pyruvate. Cultures were then diluted and cells were harvested during stationary phase or during late log phase (OD600 of 1.0). Alternatively, for analysis of gene expression on solid media, cells were harvested from LA agar plates after 1 or 8 d of incubation, or from defined medium plates after 1 d. All strains were grown at either 20° or 30°C. RNA for RPA was isolated from individual strains using Trizol Reagent (Life Technologies, Madison, WI), as previously described (Wassarman and Storz, 2000). The presence of approximately equal amounts of RNA between treatments was confirmed by agarose gel-electrophoresis (data not shown). Small RNA for northern analysis of CRISPR RNAs was isolated using mirVana kits (Applied Biosystems, Life Technologies, Madison, WI) according to manufacturer methods except for modifications to facilitate cell lysis as previously described (Cavanagh et al., 2012).
For isolation of RNA from symbiotic bacteria, approximately 100,000 nematodes were harvested from lipid agar plates, suspended in LB, and lysed by sonication for 1 minute in a water bath sonicator. Xenorhabdus bacteria were harvested by centrifugation and RNA was isolated using Trizol Reagent (Life Technologies, Madison, WI), as described above.
Primer extension experiments
For primer extension, PAGE purified primers AAP1 and AAP2 (Table S4) were radioactively labeled using T4 Polynucleotide Kinase and γ32P-ATP (Perkin Elmer, Waltham, MA) for 30 min at 37°C. Excess nucleotides were removed using a QIAquick ® (Nucleotide Removal kit, Qiagen, Valencia, CA). Labeled primer was hybridized to 5 μg of total cell RNA in Avian Myeloblastosis Virus Reverse Transcriptase (AMV-RT) buffer (Promega, Madison, WI) by heating to 80°C for 10 min and slow cooling to 37°C. The primer was extended by AMV-RT enzyme at 37°C, precipitated, and resuspended in gel loading buffer (Promega, Madison, WI). A plasmid-based copy of X. nematophila SR2-2 region was cycle sequenced using fmol® DNA Cycle Sequencing System kit (Promega, Madison, WI) and the labeled primer. The sequencing reaction was stopped using fmol® sequencing stop solution (Promega, Madison, WI). Samples were electrophoresed on a 12% polyacrylamide gel (National Diagnostics, Charlotte, NC) that was then dried and imaged on a Storm860 phosphorimager (Molecular Dynamics, Sunnyvale, CA).
RNase protection assays
RPA analyses were performed using the Ambion RPA III kit™ method (Ambion Inc., Austin, TX), following manufacturer recommendations. Probes for XnSc nilD were generated by transcription from pAWA1 (Table 1) template containing a 137 bp fragment containing the nilD locus of XnSc. For generation of a probe complementary to the nilD region of XnSa and XnSw, site directed mutagenesis was utilized to alter the spacer region of nilD within TOPOSR2-2. SDM was performed as described above using PFU Ultra polymerase and primers RPA SDM 5′ and 3′ (Table S4) to generate pTOPO-nilD-Sa (Table 1). Complementary transcripts were amplified and radioactively labeled with α32P-UTP (PerkinElmer, Waltham, MA) using the MAXIscript®-T7 reaction (Ambion, Austin, TX). Labeled transcripts were separated on a 10% polyacrylamide gel (National Diagnostics Inc, Charlotte, NC) and probes were gel purified. 10 μg of sample RNA was then hybridized to 1–2 x 105 cpm of purified probe and RNase treated using a1:100 dilution of RNase A/T1 enzyme (Ambion, Austin, TX). The RNase treated samples were electrophoresed on 10% polyacrylamide gels and visualized using XAR ECL- film (Kodak, Rochester, NY). A no-RNase control was also run to confirm probe integrity.
Northerns
Total small RNA was separated on 12% denaturing polyacrylamide MOPS gels, transferred to uncharged nylon membrane, and probed first for crRNAs with pAAP1 oligonucleotide (which anneals to the conserved 3′ repeat of all crRNAs; Table S4) using methods previously described for LNA probes except that 2.5 μg small RNA was loaded per lane and hybridization was done at 50°C (Cavanagh et al., 2012). Membranes were then reprobed with a full length RNA probe to E. coli 5S RNA (generated from pBS-5S) as previously described (Wassarman and Storz, 2000).
Nematode colonization assays
For colonization assays, lawns of individual bacterial strains were grown on lipid agar for 48 hours. Aposymbiotic infective juvenile-stage nematodes were generated by cultivation on a non-colonizing rpoS mutant, then surface sterilized, using a diluted bleach solution, and applied to the bacterial lawns (Vivas and Goodrich-Blair, 2001). In each experiment, two independent cultures of each strain were used as replicates, and 3 plates per replicate were seeded. Approximately one week post-inoculation, plates were water trapped for harvesting of IJs. Roughly 104 progeny IJs were then harvested from each plate (Vivas and Goodrich-Blair, 2001), surface sterilized and disrupted by sonication. The macerates were dilution plated on selective media to quantify CFU/IJ as previously described (Heungens et al., 2002). For nilD deletion analyses, XnSc 081 and nilD6::Tn5 were transformed (Xu et al., 1989) with the plasmids indicated for each experiment and transformants were co-cultivated with nematodes on LA plates containing rifampicin and chloramphenicol. For casE complementation analyses, strains containing pBC-casE were grown on lipid agar plates supplemented with 10 μg/ml Cm and 1 mM IPTG.
Sequencing of X. nematophila strains
The genomes of X. nematophila strains XnSc 081, nilD6::Tn5, and nilD6::Tn5 sup-1 were sequenced using Illumina paired-end libraries (mean insert length = 300bp) yielding approximately 20–30 million 75 base-pair, paired-end reads for each strain. The resulting reads were trimmed for quality and then used in a reference alignment with respect to XnSc 800 using CLC Genomics Workbench 5.1(CLC Bio). Assembled genomes were then analyzed for deletions, insertions and single nucleotide variations using CLC Genomics Workbench 5.1. SNVs were manually inspected for verification. Regions where low sequence coverage was obtained (fewer than 8 reads of coverage) were amplified and cloned from individual genomes and then sequenced. Because of significant genomic differences between XnSc 800, XnSa 1418 and XnSw 1419, performing reference alignments was not feasible. As a result, CLC Genomics Workbench 5.1 software was utilized to generate de novo genome assemblies using the reads from XnSa 1418 and XnSw 1419.
Supplementary Material
Acknowledgments
We wish to thank Amy Cavanagh (UW-Madison) for technical support on northerns and RPAs, and Jonathan Klassen (UConn-Storrs) and the Magnifying Genome team for their assistance in genome analysis and annotation. We gratefully acknowledge former and current members of the Goodrich-Blair lab: Dr. Kurt Heungens for preliminary data on the nilD locus, James Weger and Nick Feirer for contributions to cas mutant phenotypic analyses, and Ángel Casanova-Torres for quantitative reverse transcriptase PCR analysis of putative target genes. We thank Dr. Brian Tjaden (Wellesley College) for providing the Target RNA program for X. nematophila, S. Patricia Stock and S. Forst for collaborative work on S. anatoliense and S. websteri, and Caitilyn Allen and Katrina Forest (UW-Madison) for helpful discussions. Work in the Goodrich-Blair lab was funded by a grant from the National Science Foundation IOS-0950873. XL was supported by the UW-Madison Graduate School research funds. JMC and KEM were supported by a National Institutes of Health (NIH) National Research Service Award T32 (AI55397) ‘Microbes in Health and Disease’. J.M.C. was also supported by a National Science Foundation (NSF) Graduate Research Fellowship. EAH was supported by the National Institutes of Health grant 1F32AI084441. ARD was supported by a United States Public Health Service Training Grant (T32GM07616), and the Howard Hughes Medical Institute, with which PWS is an investigator.
Footnotes
Accession numbers: Genomes have been submitted and accession numbers are pending. The project accession number for HGB081 X. nematophila AN6/1 genome (XNC2) is PRJEB5061 while the project accession numbers for X. nematophila anatoliense (XNA1) and X. nematophila websteri (XNW1) are ERS451357 and ERS451358, respectively.
The authors have no conflict of interest to declare.
References
- Aklujkar M, Lovley DR. Interference with histidyl-tRNA synthetase by a CRISPR spacer sequence as a factor in the evolution of Pelobacter carbinolicus. BMC Evol Biol. 2010;10:230. doi: 10.1186/1471-2148-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, et al. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol Microbiol. 2010;79:484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Lies DP, Fu H, Roberts GP. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of Gram-negative bacteria. Gene. 1991;109:167–168. doi: 10.1016/0378-1119(91)90604-a. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AF, Akhurst RJ. The nature of the intestinal vesicle in nematodes of the family Steinernematidae. Int J Parasitol. 1983;13:599–606. [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiol. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady KC, O’Toole GA. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J Bacteriol. 2011;193:3433–3445. doi: 10.1128/JB.01411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh AT, Sperger JM, Wassarman KM. Regulation of 6S RNA by pRNA synthesis is required for efficient recovery from stationary phase in E. coli and B. subtilis. Nucleic Acids Res. 2012;40:2234–2246. doi: 10.1093/nar/gkr1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Snijders AP, Chakravorty R, Ahmed M, Tarek AM, Hossain MA. Comparative network clustering of direct repeats (DRs) and cas genes confirms the possibility of the horizontal transfer of CRISPR locus among bacteria. Mol Phylogenet Evol. 2010;56:878–887. doi: 10.1016/j.ympev.2010.05.020. [DOI] [PubMed] [Google Scholar]
- Chaston JM, Murfin KE, Heath-Heckman EA, Goodrich-Blair H. Previously unrecognized stages of species-specific colonization in the mutualism between Xenorhabdus bacteria and Steinernema nematodes. Cell Microbiol. 2013;15:1545–1559. doi: 10.1111/cmi.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston JM, Suen G, Tucker SL, Andersen AW, Bhasin A, Bode E, et al. The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: convergent lifestyles from divergent genomes. PLoS ONE. 2011;6:e27909. doi: 10.1371/journal.pone.0027909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CE, Goodrich-Blair H. Characterization of a lipoprotein, NilC, required by Xenorhabdus nematophila for mutualism with its nematode host. Mol Microbiol. 2004;54:464–477. doi: 10.1111/j.1365-2958.2004.04271.x. [DOI] [PubMed] [Google Scholar]
- Cowles CE, Goodrich-Blair H. The Xenorhabdus nematophila nilABC genes confer the ability of Xenorhabdus spp. to colonize Steinernema carpocapsae nematodes. J Bacteriol. 2008;190:4121–4128. doi: 10.1128/JB.00123-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Qimron U. The Escherichia coli CRISPR system protects from lambda lysogenization, lysogens, and prophage induction. J Bacteriol. 2010;192:6291–6294. doi: 10.1128/JB.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virol. 2012;434:202–209. doi: 10.1016/j.virol.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Flores-Lara Y, Renneckar D, Forst S, Goodrich-Blair H, Stock P. Influence of nematode age and culture conditions on morphological and physiological parameters in the bacterial vesicle of Steinernema carpocapsae (Nematoda: Steinernematidae) J Invertebr Pathol. 2007;95:110–118. doi: 10.1016/j.jip.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Forst SA, Tabatabai N. Role of the histidine kinase, EnvZ, in the production of outer membrane proteins in the symbiotic-pathogenic bacterium Xenorhabdus nematophilus. Appl Environ Microbiol. 1997;63:962–968. doi: 10.1128/aem.63.3.962-968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Sinkunas T, Siksnys V. Molecular mechanisms of CRISPR-mediated microbial immunity. Cell Mol Life Sci. 2014;71:449–465. doi: 10.1007/s00018-013-1438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich-Blair H. They’ve got a ticket to ride: Xenorhabdus nematophila-Steinernema carpocapsae symbiosis. Curr Op Microbiol. 2007;10:225–230. doi: 10.1016/j.mib.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Gunderson FF, Cianciotto NP. The CRISPR-associated gene cas2 of Legionella pneumophila is required for intracellular infection of amoebae. mBio. 2013:4. doi: 10.1128/mBio.00074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert EE, Goodrich-Blair H. Friend and foe: the two faces of Xenorhabdus nematophila. Nat Rev Microbiol. 2007;5:634–646. doi: 10.1038/nrmicro1706. [DOI] [PubMed] [Google Scholar]
- Heungens K, Cowles CE, Goodrich-Blair H. Identification of Xenorhabdus nematophila genes required for mutualistic colonization of Steinernema carpocapsae nematodes. Mol Microbiol. 2002;45:1337–1353. doi: 10.1046/j.1365-2958.2002.03100.x. [DOI] [PubMed] [Google Scholar]
- Ivancic-Bace I, Radovcic M, Bockor L, Howard JL, Bolt EL. Cas3 stimulates runaway replication of a ColE1 plasmid in Escherichia coli and antagonises RNaseHI. RNA Biol. 2013:10. doi: 10.4161/rna.23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G, Thijssen KL, Werner P, van der Horst M, Hazendonk E, Plasterk RHA. The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat Genet. 1999;21:414–419. doi: 10.1038/7753. [DOI] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Kaniga K, Delor I, Cornelis GR. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- Kaya HK, Stock SP. Techniques in insect nematology. In: Lacey LA, editor. Manual of Techniques in Insect Pathology. London: Academic Press; 1997. pp. 281–324. [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Oxford: Blackwell; 1991. pp. 115–175. [Google Scholar]
- Lee MM, Stock SP. A multilocus approach to assessing co-evolutionary relationships between Steinernema spp. (Nematoda: Steinernematidae) and their bacterial symbionts Xenorhabdus spp. (gamma-Proteobacteria: Enterobacteriaceae) Syst Parasitol. 2010;77:1–12. doi: 10.1007/s11230-010-9256-9. [DOI] [PubMed] [Google Scholar]
- Lillestol RK, Redder P, Garrett RA, Brugger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwen R, Horst-Kreft D, de Boer A, van der Graaf L, de Knegt G, Hamersma M, et al. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barré syndrome. Eur J Clin Microbiol Infect Dis. 2013;32:207–226. doi: 10.1007/s10096-012-1733-4. [DOI] [PubMed] [Google Scholar]
- Louwen R, Staals R, Endtz H, van Baarlen P, van der Oost J. The role of CRISPR-Cas systems in virulence of pathogenic bacteria. Microbiol Molec Biol Rev. 2014;78:74–88. doi: 10.1128/MMBR.00039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011a;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011b;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Heungens K, Goodrich-Blair H. Early colonization events in the mutualistic association between Steinernema carpocapsae nematodes and Xenorhabdus nematophila bacteria. J Bacteriol. 2003;185:3147–3154. doi: 10.1128/JB.185.10.3147-3154.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann J, Stabb EV, Millikan DS, Ruby EG. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl Environ Microbiol. 2003;69:5928–5934. doi: 10.1128/AEM.69.10.5928-5934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methe BA, Webster J, Nevin K, Butler J, Lovley DR. DNA microarray analysis of nitrogen fixation and Fe(III) reduction in Geobacter sulfurreducens. Appl Environ Microbiol. 2005;71:2530–2538. doi: 10.1128/AEM.71.5.2530-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N. Y: 1972. p. 466. [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiol. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Murfin KE, Chaston J, Goodrich-Blair H. Visualizing bacteria in nematodes using fluorescence microscopy. J Vis Exp. 2012;68:e4298. doi: 10.3791/4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard SS, Goodrich-Blair H. Identification and functional characterization of a Xenorhabdus nematophila oligopeptide permease. Appl Environ Microbiol. 2004;70:5621–5627. doi: 10.1128/AEM.70.9.5621-5627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinar GO. The presence of Achromobacter nematophilus in the infective stage of a Neoaplectana sp. (Steinernematidae: Nematoda) Nematologica. 1966;12:105–108. [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiol. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Pul U, Wurm R, Arslan Z, Geissen R, Hofmann N, Wagner R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol. 2010;75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- Richards GR, Goodrich-Blair H. Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cell Microbiol. 2009;11:1025–1033. doi: 10.1111/j.1462-5822.2009.01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, Siksnys V. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder HA, Stock SP, Kim SK, Flores-Lara Y, Forst S. New insights into the colonization and release process of Xenorhabdus nematophila and the morphology and ultrastructure of the bacterial receptacle of its nematode host, Steinernema carpocapsae. Appl Environ Microbiol. 2007;73:5338–5346. doi: 10.1128/AEM.02947-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- Stock SP, Goodrich-Blair H. Entomopathogenic nematodes and their bacterial symbionts: The inside out of a mutualistic association. Symbiosis. 2008;46:65–76. [Google Scholar]
- Sugar DR, Murfin KE, Chaston JM, Andersen AW, Richards GR, Deleon L, et al. Phenotypic variation and host interactions of Xenorhabdus bovienii SS-2004, the entomopathogenic symbiont of Steinernema jollieti nematodes. Env Microbiol. 2012;14:924–939. doi: 10.1111/j.1462-2920.2011.02663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi N, Wolf YI, Makarova KS, Koonin EV. Nature and intensity of selection pressure on CRISPR-associated genes. J Bacteriol. 2012;194:1216–1225. doi: 10.1128/JB.06521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Charpentier S, Clermont O, Rocha EP, Denamur E, Branger C. CRISPR distribution within the Escherichia coli species is not suggestive of immunity-associated diversifying selection. J Bacteriol. 2011;193:2460–2467. doi: 10.1128/JB.01307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud AE, Wassarman KM. A highly conserved 6S RNA structure is required for regulation of transcription. Nat Struct Mol Biol. 2005;12:313–319. doi: 10.1038/nsmb917. [DOI] [PubMed] [Google Scholar]
- Viswanathan P, Murphy K, Julien B, Garza AG, Kroos L. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J Bacteriol. 2007;189:3738–3750. doi: 10.1128/JB.00187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivas EI, Goodrich-Blair H. Xenorhabdus nematophilus as a model for host-bacterium interactions: rpoS is necessary for mutualism with nematodes. J Bacteriol. 2001;183:4687–4693. doi: 10.1128/JB.183.16.4687-4693.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- Westra ER, Semenova E, Datsenko KA, Jackson RN, Wiedenheft B, Severinov K, Brouns SJ. Type I-E CRISPR-Cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet. 2013;9:e1003742. doi: 10.1371/journal.pgen.1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, van Erp PB, Kunne T, Wong SP, Staals RH, Seegers CL, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouts WM. Biology, life cycle, and redescription of Neoaplectana bibionis Bovien, 1937 Nematoda: Steinernematidae. J Nematol. 1980;12:62–72. [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hurlbert RE. Toxicity of irradiated media for Xenorhabdus spp. Appl Environ Microbiol. 1990;56:815–818. doi: 10.1128/aem.56.3.815-818.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lohrke S, Hurlbert IM, Hurlbert RE. Transformation of Xenorhabdus nematophilus. Appl Environ Microbiol. 1989;55:806–812. doi: 10.1128/aem.55.4.806-812.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O’Toole GA. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol. 2009;191:210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.