Abstract
Study Objectives:
MicroRNAs (miRNAs) have been implicated in the pathogenesis of human diseases including neurological disorders. The aim is to address the involvement of miRNAs in the pathophysiology of central hypersomnias including autoimmune narcolepsy with cataplexy and hypocretin deficiency (type 1 narcolepsy), narcolepsy without cataplexy (type 2 narcolepsy), and idiopathic hypersomnia.
Design:
We conducted high-throughput analysis of miRNA in plasma from three groups of patients—with type 1 narcolepsy, type 2 narcolepsy, and idiopathic hypersomnia, respectively—in comparison with healthy controls using quantitative real-time polymerase chain reaction (qPCR) panels.
Setting:
University hospital based sleep clinic and research laboratories.
Patients:
Twelve patients with type 1 narcolepsy, 12 patients with type 2 narcolepsy, 12 patients with idiopathic hypersomnia, and 12 healthy controls.
Measurements and Results:
By analyzing miRNA in plasma with qPCR we identified 50, 24, and 6 miRNAs that were different in patients with type 1 narcolepsy, type 2 narcolepsy, and idiopathic hypersomnia, respectively, compared with healthy controls. Twenty miRNA candidates who fulfilled the criteria of at least two-fold difference and p-value < 0.05 were selected to validate the miRNA changes in an independent cohort of patients. Four miRNAs differed significantly between type 1 narcolepsy patients and healthy controls. Levels of miR-30c, let-7f, and miR-26a were higher, whereas the level of miR-130a was lower in type 1 narcolepsy than healthy controls. The miRNA differences were not specific for type 1 narcolepsy, since the levels of the four miRNAs were also altered in patients with type 2 narcolepsy and idiopathic hypersomnia compared with healthy controls.
Conclusion:
The levels of four miRNAs differed in plasma from patients with type 1 narcolepsy, type 2 narcolepsy and idiopathic hypersomnia suggesting that alterations of miRNAs may be involved in the pathophysiology of central hypersomnias.
Citation:
Holm A, Bang-Berthelsen CH, Knudsen S, Kornum BR, Modvig S, Jennum P, Gammeltoft S. miRNA profiles in plasma from patients with sleep disorders reveal dysregulation of miRNAs in narcolepsy and other central hypersomnias. SLEEP 2014;37(9):1525-1533.
Keywords: miRNA, hypocretin, narcolepsy, cataplexy, hypersomnia
INTRODUCTION
Hypersomnias of central origin include autoimmune narcolepsy with cataplexy and hypocretin deficiency (type 1 narcolepsy), narcolepsy without cataplexy (type 2 narcolepsy), idiopathic hypersomnia, and recurrent and secondary hypersomnias.1,2 Type 1 narcolepsy is the most common of the central hypersomnias, affecting one out of 3,000 individuals, and is the most thoroughly described.3
Type 1 narcolepsy is characterized by excessive daytime sleepiness and abnormal REM sleep manifestations including sleep paralysis, hypnagogic hallucinations, and REM periods at sleep onset.3,4 Type 1 narcolepsy is caused by a loss of hypocretin neurons in the lateral hypothalamus after an auto-immune attack, which is reflected by low hypocretin-1 levels in the cerebrospinal fluid (CSF) and a strong correlation with HLA-DQB1*06:02.5,6 Type 2 narcolepsy seems to have a more heterogeneous origin. Type 2 narcolepsy shows hypocretin-1 deficiency in only 25% of cases and positive HLA-DQB1*06:02 in 41% of cases (of Caucasian ethnicity) and the etiology of type 2 narcolepsy is not known.7,8
Idiopathic hypersomnia is less common and less well defined than type 1 narcolepsy. Patients are characterized by excessive daytime sleepiness despite long sleeping hours.2 The diagnosis is based on clinical observations, documentation of long sleep duration and otherwise normal overnight polysomnography, short sleep latencies in a multiple sleep latency test (MSLT); but no laboratory tests are available and no clear genetic predisposition has been identified. The cause is complex and only 18% of idiopathic hypersomnia patients (of Caucasian ethnicity) are HLA-DQB1*06:02-positive.9
MicroRNAs (miRNAs) have recently emerged as an important class of small RNAs, about 22 nucleotides long, that act as post-transcriptional regulators of gene expression by base-pairing with their target messengerRNAs (mRNAs).10,11 In the nervous system they regulate neuronal processes such as brain morphogenesis, neuronal cell differentiation, and transcription of neuronal-specific genes.12 Several studies have linked miRNAs to human diseases, including neurological disorders. Specific miRNAs have been identified in postmortem brain tissue from patients with neurodegenerative diseases like Parkinson and Alzheimer diseases.13 Changes in miRNA levels have been described in plasma, peripheral blood mono-nuclear cells, and CSF from patients with neurodegenerative diseases.14–16
The role of miRNAs in sleep regulation has been studied in experimental animal models. Sleep deprivation in rats results in significant changes of miRNAs in the brain and adipose tissue.17,18 Furthermore, sleep deprivation of mice resulted in changes of four miRNAs that are independent of elevated corticosterone levels.19 Finally, intraventricular and cortical injection into rat brain of miRNA as well as specific inhibitors (anti-miR) to miRNA alter sleep and electroencephalographic (EEG) slow wave activity.20,21 So far the role of miRNAs in human sleep regulation and sleep disturbances has not been studied.
In the present study we aimed to determine differences between miRNAs in central hypersomnias that may be involved in the pathophysiology of these sleep disorders. In a preliminary attempt we analyzed the miRNAs in CSF of hypersomnia patients. However, miRNA levels were borderline, making it impossible to validate significant miRNA differences between hypersomnia patients and controls (A. Holm, unpublished observation). Instead, plasma was analyzed instead of CSF because of the higher circulating miRNA levels in plasma.22 Here we used quantitative PCR (qPCR) to compare the miRNA profiles in plasma samples of hypersomnia patients with those of healthy controls. We identified four miRNAs with significantly different levels in plasma between patients with type 1 narcolepsy, type 2 narcolepsy, and idiopathic hypersomnia relative to healthy controls. The abnormal miRNA levels may give insight into the pathophysiological mechanisms responsible for the symptoms of these sleep disorders.
METHODS
Ethics Statement
After approval (KA03119) from the Danish Health Science Ethical Committee (Capital Region) and obtaining written informed consent, plasma was collected from Danish Caucasian patients with various hypersomnias and normal controls seen at the Danish Center for Sleep Medicine and the Department of Neurology, Glostrup Hospital, Denmark.
Patients
Thirty-six patients diagnosed with central hypersomnia were recruited for the initial screening of miRNAs in plasma, including 12 patients each with type 1 narcolepsy, type 2 narcolepsy, and idiopathic hypersomnia. For the independent validation of selected miRNAs in plasma, a new cohort of 12 type 1 narcolepsy patients, 6 type 2 narcolepsy patients, and 6 idiopathic hypersomnia patients were recruited. All patients were examined at the Danish Center for Sleep Medicine, Glostrup Hospital, Denmark. Exclusion criteria were the presence of neurological, psychiatric or medical disorders. All patients were free of antidepressants and stimulants 7-14 days before inclusion. The patients were evaluated by neurological examination, polysomnography, MSLT, CSF hypocretin-1 levels, and HLA-typing of the DQB1*06:02 locus. Their hypersomnia history was obtained by a semi-structured interview based on the Stanford Sleep Questionnaire.
Type 1 narcolepsy diagnosis was based on the following criteria: (1) excessive daytime sleepiness, (2) mean sleep latency < 8 min, (3) cataplexy, (4) HLA-DQB1*06:02, and (5) CSF hypocretin-1 < 110 pg/mL.23 The criteria for the diagnosis of type 2 narcolepsy were (1) excessive daytime sleepiness, (2) mean sleep latency during MSLT < 8 min, (3) absence of cataplexy, and (4) HLA-typing with approximately 20% HLA-DQB1*06:02.8 For the diagnosis of idiopathic hypersomnia, the following criteria were met: (1) complaints of excessive daytime sleepiness occurring daily for ≥ 3 months; (2) mean sleep latency during MSLT < 8 min; (3) no features to suggest cataplexy; and (4) apnea-hypopnea index < 10.24
Twelve healthy controls without medical, neurological, or sleep abnormalities were recruited for the initial screening of miRNA by advertising for normal volunteers. To validate the miRNAs, 12 healthy controls without medical, neurological, or sleep disorders were recruited by the Department of Neurology at Glostrup Hospital.
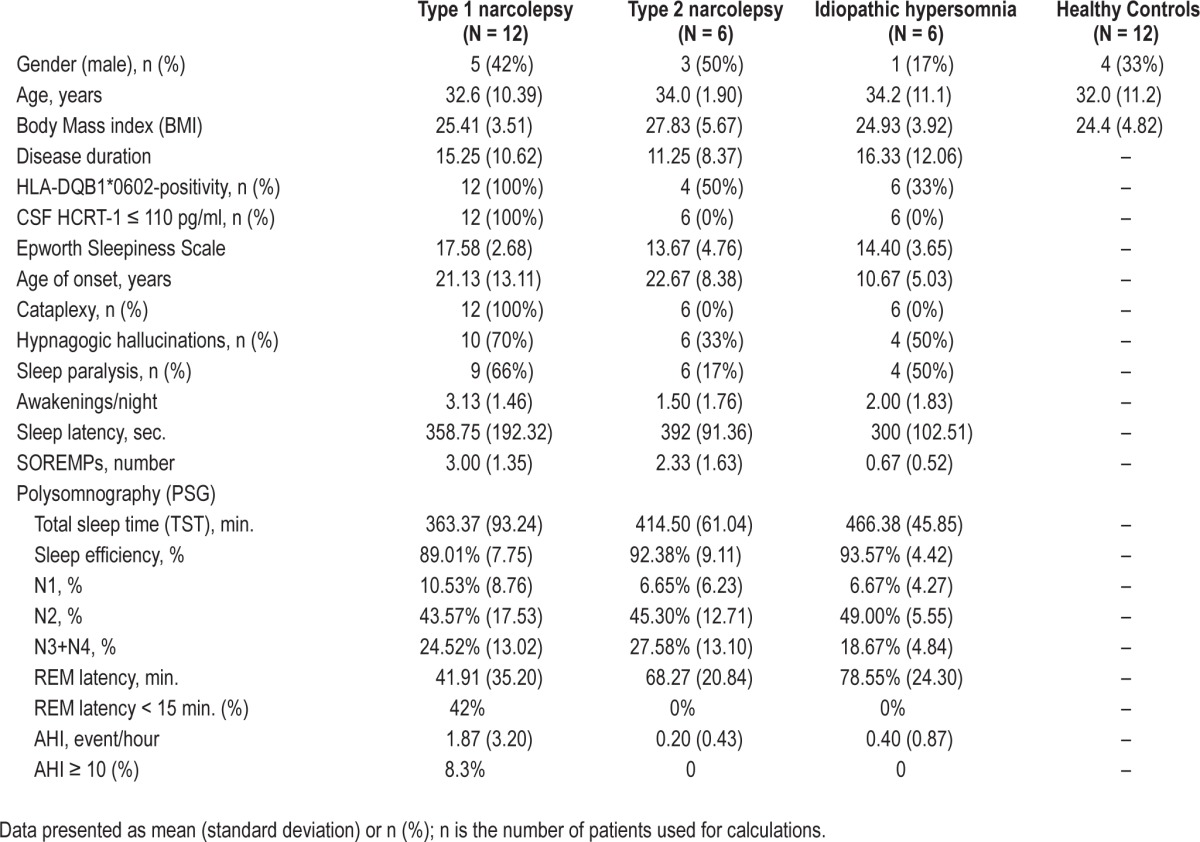
The demographic, clinical, sleep, and biological characteristics of the two cohorts of patients with hypersomnias and the healthy controls recruited for the miRNA screen and validation of miRNA candidates are presented in Tables 1 and 2. The mean age of the patient groups ranged between 27.8 to 34.2 years. Patients and controls were selected to have a mean BMI in the range of 22.1-27.8 in order to minimize confounding effects. All type 1 narcolepsy patients were HLADQB1*06:02-positive and had CSF hypocretin-1 < 110 pg/mL. Patients with type 2 narcolepsy and idiopathic hypersomnia had CSF hypocretin-1 > 200 pg/mL. The average number of HLADQB1*06:02-positive patients with type 2 narcolepsy and idiopathic hypersomnia in the screening and validation cohorts was 19.4%. All patients included in the study were Caucasian.
Table 1.
Demographic, clinical, sleep, and biological data of central hypersomnia patients and healthy controls in miRNA screen study.
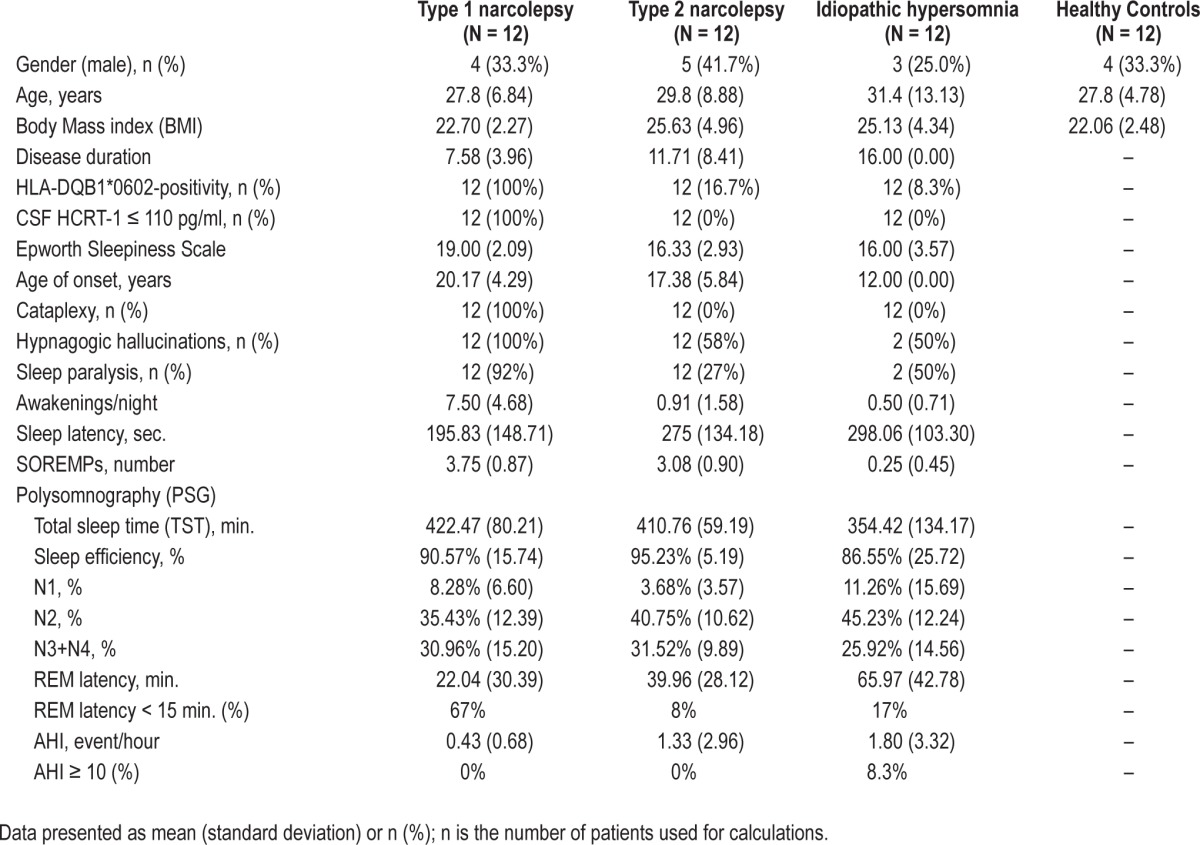
Table 2.
Demographic, clinical, sleep and biological data of central hypersomnia patients and healthy controls in miRNA validation study.
Measurement of CSF Hypocretin-1
CSF (10 mL) was collected between 08:00 am and 12:00, cooled on ice and stored within 30 min at −80°C until analysis. Hypocretin-1 was analyzed in crude CSF by radioimmuno-assay from Phoenix Pharmaceuticals (Belmont, CA, USA) as previously described.25 All samples were blindly measured in duplicate and the means of the results were calculated. If the intra-assay variability was > 5%, the sample was reanalyzed. The standard curve range was 10-1280 pg/mL. Assay quality was controlled by the positive control sample included in the assay kit. Furthermore, one external reference sample of pooled CSF from healthy individuals with an HCRT-1 concentration of 163 pg/mL was included in each assay to determine the inter-assay variability, i.e. the precision of results between the assays. The CSF samples were analyzed monthly and the inter-assay coefficient of variation was determined as 9.9%. Finally, a sample from the previous assay was included in each current assay to estimate the day-to-day variability. The mean deviation between two repeated measurements was 6.6%. The external reference CSF sample was originally donated by Dr. E. Mignot, Stanford Center for Sleep Sciences and Medicine, Stanford University, USA. Accordingly, the CSF concentration of HCRT-1 in all measurements was adjusted to the level of CSF HCRT-1 defined at Stanford University. We applied previously used groupings of CSF intervals for HCRT-1 concentrations: low (≤ 110 pg/ ml), intermediate (> 110 - ≤ 200 pg/ml), and normal (> 200 pg/ ml). 110 pg/ml and 200 pg/ml are the optimal cutoffs for type 1 narcolepsy and type 2 narcolepsy, respectively.6–8
HLA Typing
The HLA-DQB1*06:02 locus was typed in different laboratories over several years. Some type 1 narcolepsy samples were typed at Stanford University using either Innotype reverse dot-blot kits following the manufacturer's recommendations or a sequence-specific oligonucleotide probe-enzyme-linked immunosorbent assay technique. High resolution HLA typing of DQB1 was performed at Stanford University. Some samples were HLA typed at Glostrup Hospital by the PCR oligotyping technique using the primers listed in the Table 3. The PCR program was initiated for 15 min at 94°C, followed by 35 cycles of 94°C for 30 sec, 63°C for 30 sec and 72°C for 1 min, and terminated for 10 min at 72°C.
Table 3.
HLA typing of DQB1*06:02 of hypersomnia patients at Glostrup Hospital performed by the PCR oligotyping technique using two sets of primers.

Plasma Sample Handling and miRNA Isolation
Blood samples were drawn from all patients at the same time as CSF collection, i.e., between 08:00 and 12:00. This was strictly enforced to avoid possible diurnal oscillations of levels of miRNAs in plasma as reported in mice.26 Nine milliliters of blood was collected per participant by venipuncture into a BD Vacutainer CPT glass tube with EDTA (Becton-Dickinson). Within 30 min of blood collection, plasma was isolated from the tube by centrifugation (10 min, 1600 g, 4°C), and 3×1 mL aliquots were frozen at −80°C.
For the miRNA screening analysis the plasma samples were pooled in order to minimize labor and cost. The number of samples was reduced from 48 to 12 for the large screening protocol, which was performed on 3 pools of 4 plasma samples from each group of patients and controls, respectively. We were aware that pooling could prevent the characterization of individual variability and ultimately bias the analysis of the 4 groups towards inclusivity. Consequently, the validation of miRNA candidates in plasma found in the initial screen was performed on individual samples from a new cohort of 24 hypersomnia patients and 12 healthy controls. miRNA was isolated from 200 μL plasma using the NucleoSpin miRNA plasma mini spin columns from Macherey-Nagel (Düren, Germany) following the manufacturer protocol. The miRNAs were eluted in a total volume of 30 μL. NucleoSpin only isolates small RNAs, so the amount and quality of miRNA could not be evaluated. However, we tested the performance of NucleoSpin isolation by measuring the Ct values of the RNA spike-in and hsa-miR-140-3p, concluding that NucleoSpin gave a high miRNA yield.
cDNA Synthesis and miRCURY LNA Universal RT MicroRNA PCR System
cDNA was synthesized from purified miRNA using the Universal cDNA synthesis kit from Exiqon (Vedbaek, Denmark) according to the manufacturer's instructions. The cDNA product was diluted 50x in nuclease-free water and mixed 1:1 with 2× SYBR Green master mix. Ten μL of SYBR Green master mix:cDNA mix was added to each well on the Ready-to-use microRNA PCR Human panels (I+II). The qRTPCRs were run on the CFX384 Real Time System from BioRad (Hercules, CA, USA). In total, 24 human panels I and II were used to identify miRNAs in plasma. A negative control plate for each panel was performed. Negative control plates contained nuclease-free water purified on similar columns as plasma, cDNA reaction performed in similar way as with the patient samples and the qPCR were run on the CFX384 Real Time System. If there was no back-group measurement on the negative control plate, we accepted a Ct value of 38. The selected miRNAs were validated on a pick-and-mix panel performed by Exiqon (Vedbaek, Denmark).
Hemolysis in Plasma
Hemolysis is known to inhibit the expression of some miRNAs.27 We tested the samples for hemolysis by analyzing the plasma level of miR-451-5p, which is present in high concentration in erythrocytes. However, miR-451-5p can also be elevated in samples from patients as a result of disease. Therefore, the miR-451-5p level was normalized with respect to miR-23a, which was stable among the samples. Three pools of 4 plasma samples from each group of 12 patients with type 1 narcolepsy, type 2 narcolepsy, and idiopathic hypersomnia, respectively, and the 12 healthy controls were analyzed. As shown in Figure S1 (supplemental material) the Ct value of miR-451-5p minus Ct value of miR-23a was less than 7, indicating that hemolysis was not present in the samples as described in https://www.exiqon.com/ls/Documents/Scientific/microRNA-serum-plasma-guidelines.pdf. Additionally, we analyzed oxyhemoglobin absorbance at 414 nm in the plasma samples, but no peak absorbance was observed (data not shown). Based on this we concluded that our plasma samples from the initial screen and the validation cohort did not show hemolysis.
qPCR Analysis
qPCR data were analyzed with GenEx software developed by MultiD Analyses AB (Gothenburg, Sweden).28 We used a version of the GenEx qRT-PCR data analysis software specially adapted for the microRNA qRT-PCR platform from Exiqon (Vedbaek, Denmark). The analysis was performed using the step-by-step analysis user guide http://www.exiqon.com/ls/Documents/Scientific/Exiqon-data-analysis-guide.pdf. In summary, data obtained from the negative control plate were subtracted from the data from the miRNA PCR panels. The data were normalized to a global mean. Furthermore, the top candidates for normalization were identified using two algorithms: NormFinder and Gene-Norm. Normalization genes, miR-378 and miR-186, in plasma were used as normalization genes in the validation study. The fold change and statistical significance were calculated using the delta-delta Ct method and Student's t-test, respectively. A Dunn-Bonferroni analysis was used to correct for multiple testing. Data are presented as means and standard deviations. Principal component analysis and hierarchical clustering analysis were also performed using GenEx software.28
Analysis of miRNA Tissue Distribution
The distribution of the 4 miRNAs in human tissues was analyzed using the miRNA Expression Atlas in Normal Tissues (http://www.mirnabodymap.org).29
Pathway Analysis
The enrichment of the biological pathways regulated by the differentially expressed miRNAs was investigated by a pathway analysis in which the 4 aberrant miRNAs (miR-130a, miR-30c, miR-26a, and let-7f) were combined. The pathway analysis tool DIANA-mirPath Software (http://diana.cslab.ece.ntua.gr/pathways/) was used, with default settings.30,31 The miRNA database (http://www.mirbase.org)32 and the Kyoto Encyclopedia of Genes and Genomes (KEEG) (http://www.genome.jp/kegg)/)33 were applied in the DIANA pathway analysis.
RESULTS
miRNA Screening of Plasma from Central Hypersomnia Patients
The miRNA screening was performed on 3 pools of 4 plasma samples from each group of patients and controls, respectively, using qPCR on miRNA PCR panels as described in Methods. After data analysis miRNA candidates were selected by fulfillment of the criteria of a significant (P < 0.05) two-fold or more difference between type 1 narcolepsy, type 2 narcolepsy, and idiopathic hypersomnia patients compared with healthy controls.
Based on these criteria, we identified 50 miRNA differences between type 1 narcolepsy patients and healthy controls, 24 miRNA differences between type 2 narcolepsy patients and healthy controls, and 6 miRNA differences between idiopathic hypersomnia patients and healthy controls (Table S1, supplemental material). Five miRNAs (miR-100, miR-323-3p, miR-337-5p, miR-361-5p, and miR-610) were different in both type 1 narcolepsy and type 2 narcolepsy patients compared to healthy controls. Furthermore, 2 miRNAs (miR-224* and miR-643) were different in both type 2 narcolepsy and idiopathic hypersomnia. This suggests that the miRNA differences are not specific to the 3 central hypersomnias. Dunn Bonferroni correction for multiple testing confirmed the significant differences of the 3 miRNAs between type 1 narcolepsy patients and healthy controls and of one miRNA between type 2 narcolepsy patients and healthy controls (Table S2, supplemental material).
Validation of miRNA Changes in Central Hypersomnia Patients
The validation of altered miRNA candidates in plasma found in the initial screen was performed on individual samples from the new cohort of 24 hypersomnia patients comprising 12 type 1 narcolepsy, 6 type 2 narcolepsy, and 6 idiopathic hypersomnia patients compared with 12 healthy controls, as described in the Methods. Since type 1 narcolepsy is more homogenous than the other hypersomnias, the validation analysis focused on the miRNAs that were different in the type 1 narcolepsy group compared with the healthy controls. Twenty miRNA candidates that fulfilled the criteria of two-fold or more difference and a value of P < 0.05 were selected from the initial screen of type 1 narcolepsy patients versus healthy controls. Furthermore, 2 normalization genes, miR-378 and miR-186 in plasma, were identified by calculations involving the screening data using NormFinder and GeneNorm.
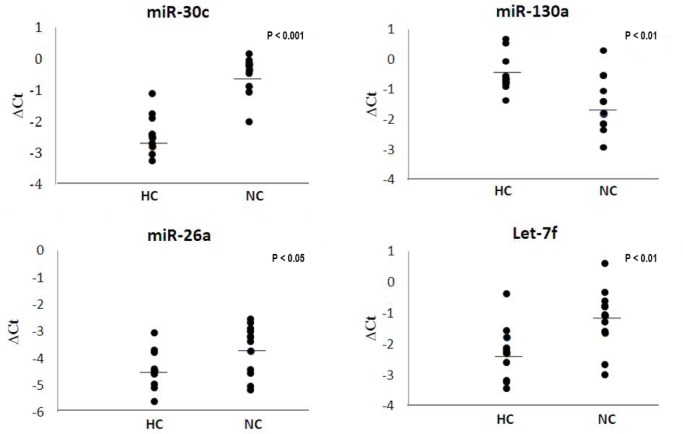
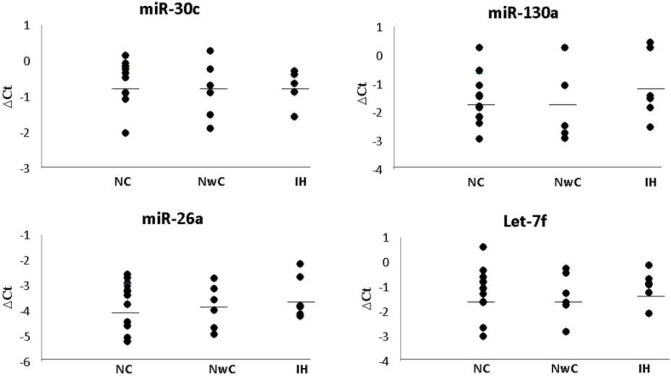
Four miRNAs in plasma were significantly different between type 1 narcolepsy and healthy controls. miR-30c, let-7f, and miR-26a were all found at a higher level in type 1 narcolepsy patients than in healthy controls; the level of miR-130a was lower in type 1 narcolepsy patients than in healthy controls (Figure 1). After Dunn Bonferroni correction for multiple testing, the level of miR-30c proved to be significantly higher (3.67-fold) in type 1 narcolepsy patients than in healthy controls (P-value = 2.0*10E-6). The miRNA changes were not specific to type 1 narcolepsy, since the 4 miRNAs were significantly different in type 2 narcolepsy and idiopathic hypersomnia compared with healthy controls (Figure 2).
Figure 1.
Levels of four miRNAs in plasma from type 1 narcolepsy patients and healthy controls. The 4 panels show levels of 4 miRNAs: miR-30c, miR-130a, miR-26a, and let-7f detected in plasma from 12 patients with type 1 narcolepsy (NC) and 12 healthy controls (HC). ΔCt = delta cycle threshold. The bars represent means of ΔCt.
Figure 2.
Levels of 4 miRNAs in plasma from patients with type 1 narcolepsy, type 2 narcolepsy and idiopathic hypersomnia. The 4 panels show levels of 4 miRNAs: miR-30c, miR-130a, miR-26a, and let-7f in plasma from patients with type 1 narcolepsy (NC), type 2 narcolepsy (NwC), and idiopathic hypersomnia (IH). ΔCt = delta cycle threshold. The bars represent means of ΔCt.
A principal component analysis (PCA) was performed on the miRNA results from the analyzed samples to see how the 4 validated miRNAs were distributed in patients with type 1 narcolepsy, type 2 narcolepsy, idiopathic hypersomnia, and in healthy controls. This clearly separated the 3 central hypersomnias from healthy controls, whereas type 1 narcolepsy, type 2 narcolepsy, and idiopathic hypersomnia could not be separated. This suggests that the observed miRNA differences are related to the pathophysiology of central hypersomnia rather than to the individual disorders (Figure 3).
Figure 3.
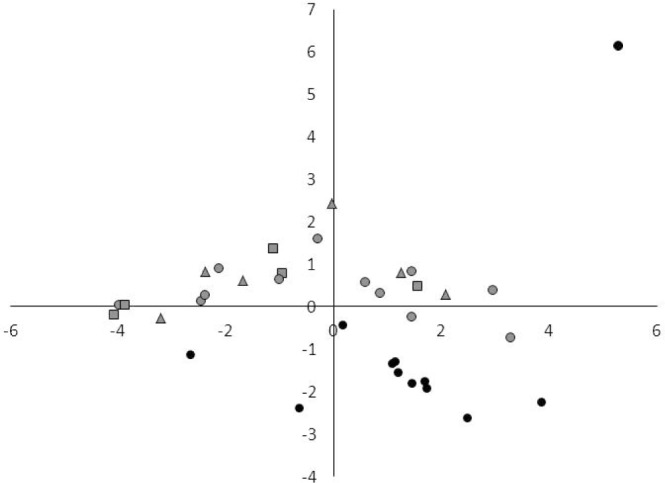
Principal component analysis of miRNAs that were detected in all samples from patients with type 1 narcolepsy, type 2 narcolepsy, and idiopathic hypersomnia, and healthy controls. Normalized values were used for PCA. Gray circles: type 1 narcolepsy; gray triangles: type 2 narcolepsy; gray squares: idiopathic hypersomnia; black circles: healthy controls.
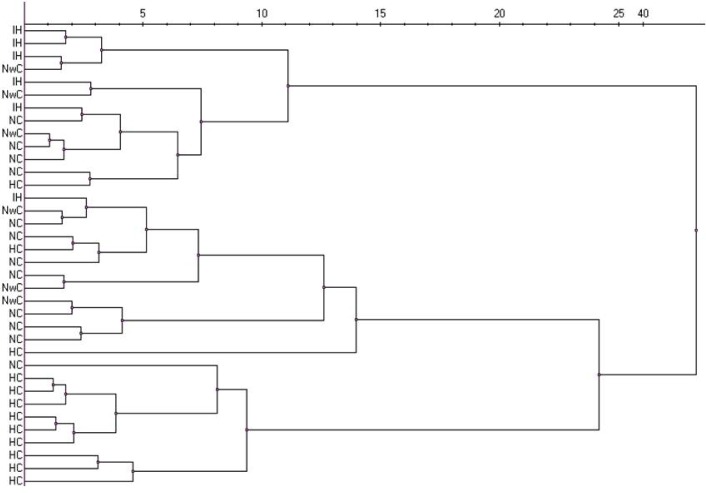
Hierarchical clustering analysis showed that healthy controls were clustered in a separate group as with the PCA. Furthermore, type 1 narcolepsy and idiopathic hypersomnia were clustered in 2 separate groups, whereas type 2 narcolepsy patients were located in both groups. This suggests that the phenotype of type 2 narcolepsy is heterogeneous and has similarities with both type 1 narcolepsy and idiopathic hypersomnia (Figure 4).
Figure 4.
Hierarchical clustering analysis miRNA pattern of all samples from patients with type 1 narcolepsy (NC), type 2 narcolepsy (NwC), and idiopathic hypersomnia (IH), and healthy controls (HC). Clustering method: Ward algorithm. Distance measure: Euclidean.
DISCUSSION
Previous reports have described the detection of miRNAs in plasma from healthy individuals as well as patients with a variety of medical disorders including neurodegenerative diseases. This study shows for the first time that it is possible to detect differences in miRNA levels in plasma from patients with sleep disorders. miRNA qPCR analysis of plasma samples from patients with type 1 narcolepsy, type 2 narcolepsy, and idiopathic hypersomnia revealed significant differences in the plasma levels of four miRNAs, including miR-130a, miR-26a, miR-30c, and let-7f. Our findings could be important in understanding the pathophysiology of central hypersomnias. We used publicly available software to analyze three aspects of the biological significance of the four aberrant miRNAs in central hypersomnias: the tissue distribution of the miRNAs, their involvement in cellular pathways, and their role in regulating mRNA and protein expression.
The distribution in human tissues of the dysregulated miRNAs was analyzed using the miRNA Expression Atlas in Normal Tissues.29 The four miRNAs show varying expression in human tissues (Figure S2, supplemental material). The levels of miR-26a, miR-30c, and let-7f are relatively high in the brain, whereas miR-130a expression is very low. However, miR-130a is expressed in the heart, lung, kidney, jejunum, colon, thyroid gland, and the reproductive system. The expression of miR-30c is particularly high in the heart, thyroid gland, blood cells, skeletal muscle, kidney and lung, but relatively low in other tissues; miR-26a is expressed in the heart, thyroid, skeletal muscle, colon, and reproductive system; and let-7f is expressed in the heart, thyroid, blood cells, skeletal muscle, and reproductive system. In general, the expression of these four miRNAs is low in adipose tissue, liver, pancreas, adrenal gland, spleen, and digestive tract.
The tissue expression pattern of the four dysregulated miRNAs implies that central hypersomnias are accompanied by differences in the brain activity. Brain-specific miRNAs have been detected in body fluids, indicating that miRNAs may cross the blood-brain barrier, possibly in exosomes or other lipid vesicles.34 Furthermore, miRNA can be delivered to the mouse brain by systemic injection of targeted exosomes.34
The different level of miRNAs in plasma that characterize central hypersomnias may reflect abnormal function of peripheral organs in the human body. This is in agreement with observations that type 1 narcolepsy is associated with abnormal cardiovascular control like altered blood pressure regulation and heart rate response during sleep, muscular symptoms in cataplexy, metabolic differences with lower energy expenditure leading to weight gain in spite of reduced appetite, and endocrine dysfunction with precocious puberty and obesity in childhood.35–40
miRNAs are key regulators of diverse biological processes and their functional analysis is crucial to our understanding of their specific effects and role in diseases. We performed pathway analysis using DIANA-miRPath v2.0, which offers tools specifically focused on miRNA-targeted genes and pathways.31 We analyzed the four distinct miRNAs together to identify how their target genes and pathways are enriched when they are combined. The analysis predicted a number of cellular processes, signaling pathways and pathological conditions, where miR-130a, miR-26a, miR-30c, and let-7f are combined in regulation of enriched sets of target genes in the relevant pathways (Table S3, supplemental material). Overall the predictions suggested that the miRNAs have broad tissue distribution, multiple functions, and a large number of putative targets. The statistically significant (P < 0.05) predictions included cellular processes of possible relevance to sleep disorders such as axon guidance, focal adhesion, circadian rhythm, and adherens junction. The predicted signaling pathways (MAPK, TGF-beta, Wnt, calcium, ErbB, mTOR, T-cell receptor, adipocytokine, and phosphatidylinositol signaling) may be involved in cellular dysfunction accompanying hypersomnias.
Finally, pathological conditions with combined alterations of miR-130a, miR-26a, miR-30c, and let-7f include several cancers as well as amyotrophic lateral sclerosis, prion disease, Huntington disease, and type 2 diabetes mellitus.
Literature search on the role of miR-130a, miR-26a, miR-30c, and let-7f in specific cell types and tissues as well as human disorders showed that the miRNAs are involved in regulating a variety of functions. We focused on targets, which may be related to the pathology of type 1 narcolepsy. miR-130a targets substance P mRNA and inhibition of miR-130a in neuronal cells resulted in substance P synthesis and release.41 The CSF concentration of substance P was significantly lower in type 1 narcolepsy patients and the decreased levels were correlated with the duration of disease, severity of cataplectic symptoms, intensity of sleepiness, and frequency of daytime sleep attacks.42 However, a relationship between CSF substance P levels and miR-130a in plasma of type 1 narcolepsy patients remains unclear.
miR-30c is expressed in primary cardiomyocytes and induces cellular hypertrophy.43 Patients with type 1 narcolepsy show altered night-time blood pressure regulation and heart rate response that may be associated with increased cardiovascular risk.35,36 Furthermore, transgenic narcoleptic mice lacking hypocretin neurons showed significant alterations in cardiovascular control during sleep.37
miR-26a as well as miR-26b downregulates brain-derived neurotrophic factor (BDNF). However, the inhibition is allele specific since there is a SNP located in the miRNA target site of BDNF 3'UTR, although its biological significance is not clear.44 Serum BDNF levels were significantly higher in type 1 narcolepsy patients than in healthy controls (64.2 ± 3.9 ng/mL vs. 47.3 ± 2.6 ng/mL, P < 0.01), while there were no significant difference in nerve growth factor levels. The increased serum BDNF levels were not correlated with disease duration and sleep parameters in type 1 narcolepsy patients.45 The mechanisms responsible for the markedly higher BDNF levels in type 1 narcolepsy patients and the role of miR-26a remain unknown.
Our miRNA target analysis further predicted that miR-26a targets tribbles homolog 2 (TRIB2) mRNA. Autoantibodies against TRIB2 have been detected by screening of sera in 14% of type 1 narcolepsy patients versus 5% of controls.46 TRIB2 is not only expressed by hypocretin neurons, but is also present in many other cell populations in the brain and in the periphery, including immune cells.47 This suggests that miR-26a may play a role in the cellular changes following the loss of hypocretin neurons in type 1 narcolepsy.
Let-7f is expressed in naïve, effector, and memory T cells,48 and let-7f inhibits IL-23 receptor expression in memory T cells, which may influence T-cell proliferation.49 There is a SNP (rs10889677) located in the 3'UTR of the IL-23 receptor, which disrupts let-7f binding leading to increased IL-23 receptor expression,50 but the SNP in IL-23 receptor is not associated with type 1 narcolepsy.51 The present study has several limitations and strengths. The limitations include the relatively small number of patients, the unknown significance of plasma miRNA levels in brain disorders, and the correlational nature of the findings. It is nonetheless the first study to report miRNA changes in plasma from patients with central hypersomnias. First, the number of patients and controls was limited to 48 individuals in the miRNA screen and 36 individuals in the miRNA validation in order to analyze the three groups of central hypersomnia patients and search for differences and similarities between the sleep disorders in comparison to controls. In spite of the small number of patients we were able to identify statistically significant changes in four miRNAs in plasma. Second, the role of circulating miRNA in pathophysiology of sleep disorders is difficult to ascertain given their unknown origin, function, or targets, i.e., each miRNA has thousands of putative targets.22 We have compensated for this by examining the expression characteristics of selected miRNAs from a tissue databank. Furthermore, we performed pathway analysis of the four altered miRNAs together to identify how their target genes and pathways are enriched. Finally, literature search of the four miRNAs identified possible targets relevant to the nervous system and hypersomnia. The predicted targets of the altered miRNAs are speculative and additional analysis of the specific functions of the four miRNAs in hypersomnia is needed. Third, the miRNAs changes in plasma from hypersomnia patients are correlational. At present we cannot link the miRNAs to the pathophysiology of sleep disorders, but they give clues about abnormalities in peripheral organs as well as the nervous system.
In conclusion, we have identified four aberrant miRNAs in plasma from patients with central hypersomnias. The biological significance of miR-130a, miR26a, miR-30c, and let-7f in the three hypersomnias—type 1 narcolepsy, type 2 narcolepsy, and idiopathic hypersomnia—remains unclear. In silico analysis of their tissue distribution, cellular pathways and biological functions suggests that they reflect changes in the brain as well as peripheral organs. We propose that looking at the targets of these miRNAs will increase our understanding of the pathogenesis of central hypersomnias. Our findings thus lend a new perspective on investigating sleep disorders, and offer a new approach to identifying relevant pathways in the pathogenesis of central hypersomnias. Further studies are required to clarify the importance of miRNAs in central hypersomnias.
DISCLOSURE STATEMENT
This study was funded by the Lundbeck Foundation. The authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
The authors are especially grateful to Birte Kofoed for skillful technical assistance. Dr. E. Mignot, Stanford Center for Sleep Sciences and Medicine, Stanford University, USA is acknowledged for the HLA analysis and the external reference CSF sample. Dr. Knudsen's present address is: Norwegian Resource Center for ADHD, TS and Narcolepsy, Oslo University Hospital, Ullevål, Norway.
SUPPLEMENTAL MATERIAL
Hemolysis test. The levels of miR-451-5p and miR-23a were analyzed by qPCR of 3 pools of 4 plasma samples from each group of healthy controls (HC), and type 1 narcolepsy (NC), type 2 narcolepsy (NwC), and idiopathic hypersomnia (IH) patients. The Ct value of miR-451-5p minus Ct value of miR-23a was plotted for each pool.
miRNA expression data in normal human tissues. Heat map illustrating the tissue expression of 4 miRNAs: miR-30c, miR-130a, miR-26a, and let-7f. White, blue, rose, and red bars respectively indicate absent, low, intermediate, and high levels of high expression.
miRNAs in plasma from type 1 narcolepsy, type 2 narcolepsy, and idiopathic hypersomnia patients with two-fold or more difference with respect to healthy controls and P < 0.05.
microRNA candidates in plasma from patients with type 1 narcolepsy, type 2 narcolepsy, and healthy controls after Dunn Bonferroni correction with P < 0.00087.
Analysis of miRNA-targeted genes and pathways using DIANA-miRPath v2.0.
REFERENCES
- 1.American Academy of Sleep Medicine. International Classification of Sleep Disorders. 3rd ed. Darien, IL: American Academy of Sleep Medicine; 2014. [Google Scholar]
- 2.Young TJ, Silber MH. Hypersomnias of central origin. Chest. 2006;130:913–20. doi: 10.1378/chest.130.3.913. [DOI] [PubMed] [Google Scholar]
- 3.Mignot E. Genetic and familial aspects of narcolepsy. Neurology. 1998;50:S16–22. doi: 10.1212/wnl.50.2_suppl_1.s16. [DOI] [PubMed] [Google Scholar]
- 4.Dauvilliers Y, Arnulf I, Mignot E. Narcolepsy with cataplexy. Lancet. 2007;369:499–511. doi: 10.1016/S0140-6736(07)60237-2. [DOI] [PubMed] [Google Scholar]
- 5.Nishino S, Ripley B, Overeem S, Lammers GJ, Mignot E. Hypocretin(orexin) deficiency in human narcolepsy. Lancet. 2000;355:39–40. doi: 10.1016/S0140-6736(99)05582-8. [DOI] [PubMed] [Google Scholar]
- 6.Mignot E, Hayduk R, Black J, Grumet FC, Guilleminault C. HLA DQB1*0602 is associated with cataplexy in 509 narcoleptic patients. Sleep. 1997;20:1012–20. [PubMed] [Google Scholar]
- 7.Oka Y, Inoue Y, Kanbayashi T, et al. Narcolepsy without cataplexy: 2 subtypes based on CSF hypocretin-1/orexin-A findings. Sleep. 2006;29:1439–43. doi: 10.1093/sleep/29.11.1439. [DOI] [PubMed] [Google Scholar]
- 8.Andlauer O, Moore H, Hong SC, et al. Predictors of hypocretin (orexin) deficiency in narcolepsy without cataplexy. Sleep. 2012;35:1247–55. doi: 10.5665/sleep.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson KN, Pilsworth S, Sharples LD, Smith IE, Shneerson JM. Idiopathic hypersomnia: a study of 77 cases. Sleep. 2007;30:1274–81. doi: 10.1093/sleep/30.10.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNeill E, Van Vactor D. MicroRNAs shape the neuronal landscape. Neuron. 2012;75:363–79. doi: 10.1016/j.neuron.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J, Inoue K, Ishii J, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–4. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martins M, Rosa A, Guedes LC, et al. Convergence of miRNA expression profiling, a-synuclein interaction and GWAS in Parkinson's disease. PLoS One. 2011;6:e25443. doi: 10.1371/journal.pone.0025443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin XF, Wu N, Wang L, Li J. Circulating microRNAs: a novel class of potential biomarkers for diagnosing and prognosing central nervous system diseases. Cell Mol Neurobiol. 2013;33:601–13. doi: 10.1007/s10571-013-9940-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cogswell J P, Ward J, Taylor IA, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 17.Davis CJ, Bohnet SG, Meyerson JM, Krueger JM. Sleep loss changes microRNA levels in the brain: a possible mechanism for state-dependent translational regulation. Neurosci Lett. 2007;422:68–73. doi: 10.1016/j.neulet.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gharib SA, Khalyfa A, Abdelkarim A, Bhushan B, Gozal D. Integrative miRNA-mRNA profiling of adipose tissue unravels transcriptional circuits induced by sleep fragmentation. PloS One. 2012;7:e37669. doi: 10.1371/journal.pone.0037669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mongrain V, Hernandez SA, Pradervand S, et al. Separating the contribution of glucocorticoids and wakefulness to the molecular and electrophysiological correlates of sleep homeostasis. Sleep. 2010;33:1147–57. doi: 10.1093/sleep/33.9.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis CJ, Clinton JM, Taishi P, Bohnet SG, Honn KA, Krueger JM. MicroRNA 132 alters sleep and varies with time in brain. J Appl Physiol. 2011;111:665–72. doi: 10.1152/japplphysiol.00517.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis CJ, Clinton JM, Krueger JM. MicroRNA 138, let-7b, and 125a inhibitors differentially alter sleep and EEG delta-wave activity in rats. J Appl Physiol. 2012;113:1756–62. doi: 10.1152/japplphysiol.00940.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Etheridge A, Gomes CP, Pereira RW, Galas D, Wang K. The complexity, function and applications of RNA in circulation. Front Genet. 2013;4:115. doi: 10.3389/fgene.2013.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overeem S, Mignot E, van Dijk JG, Lammers GJ. Narcolepsy: clinical features, new pathophysiologic insights, and future perspectives. J Clin Neurophysiol. 2001;18:78–105. doi: 10.1097/00004691-200103000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Vernet C, Arnulf I. Idiopathic hypersomnia with and without long sleep time: a controlled series of 75 patients. Sleep. 2009;32:753–9. doi: 10.1093/sleep/32.6.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knudsen S, Jennum PJ, Alving J, Sheikh SP, Gammeltoft S. Validation of the ICSD-2 criteria for CSF hypocretin-1 measurements in the diagnosis of narcolepsy in the Danish population. Sleep. 2010;33:169–76. doi: 10.1093/sleep/33.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shende VR, Goldrick MM, Ramani S, Earnest DJ. Expression and rhythmic modulation of circulating microRNAs targeting the clock gene Bmal1 in mice. PLoS One. 2011;6:e22586. doi: 10.1371/journal.pone.0022586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirschner MB, Kao SC, Edelman JJ, et al. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One. 2011;6:e24145. doi: 10.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergkvist A, Rusnakova V, Sindelka R, et al. Gene expression profiling-Clusters of possibilities. Methods. 2010;50:323–35. doi: 10.1016/j.ymeth.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 29.Mestdagh P, Van Vlierberghe P, De Weer A, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25:1991–3. doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- 31.Vlachos IS, Kostoulas N, Vergoulis T, et al. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40:W498–504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(suppl 1):D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular datasets. Nucleic Acids Res. 2012;40:D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–5. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 35.Grimaldi D, Calandra-Buonaura G, Provini F, et al. Abnormal sleepcardiovascular system interaction in narcolepsy with cataplexy: effects of hypocretin deficiency in humans. Sleep. 2012;35:519–28. doi: 10.5665/sleep.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorensen GL, Knudsen S, Petersen ER, et al. Attenuated heart rate response is associated with hypocretin deficiency in patients with narcolepsy. Sleep. 2013;36:91–8. doi: 10.5665/sleep.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silvani A, Bastianini S, Berteotti C, et al. Sleep and cardiovascular phenotype in middle-aged hypocretin-deficient narcoleptic mice. J Sleep Res. 2014;23:98–106. doi: 10.1111/jsr.12081. [DOI] [PubMed] [Google Scholar]
- 38.Burgess CR, Scammell TE. Narcolepsy: neural mechanisms of sleepiness and cataplexy. J Neurosci. 2012;32:12305–11. doi: 10.1523/JNEUROSCI.2630-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plazzi G, Parmeggiani A, Mignot E, et al. Narcolepsy-cataplexy associated with precocious puberty. Neurology. 2006;66:1577–9. doi: 10.1212/01.wnl.0000216142.21375.71. [DOI] [PubMed] [Google Scholar]
- 40.Kok SW, Roelfsema F, Overeem S, et al. Altered setting of the pituitary-thyroid ensemble in hypocretin-deficient narcoleptic men. Am J Physiol Endocrinol Metab. 2005;288:E892–9. doi: 10.1152/ajpendo.00327.2004. [DOI] [PubMed] [Google Scholar]
- 41.Greco SJ, Rameshwar P. MicroRNAs regulate synthesis of the neurotransmitter substance P in human mesenchymal stem cell-derived neuronal cells. Proc Natl Acad Sci U S A. 2007;104:15484–9. doi: 10.1073/pnas.0703037104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strittmatter M, Isenberg E, Grauer MT, Hamann G, Schimrigk K. CSF substance P somatostatin and monoaminergic transmitter metabolites in patients with narcolepsy. Neurosci Lett. 1996;218:99–102. doi: 10.1016/s0304-3940(96)13125-6. [DOI] [PubMed] [Google Scholar]
- 43.Jentzsch C, Leierseder S, Loyer X, et al. A phenotypic screen to identify hypertrophy-modulating microRNAs in primary cardiomyocytes. J Mol Cell Cardiol. 2012;52:13–20. doi: 10.1016/j.yjmcc.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 44.Caputo V, Sinibaldi L, Fiorentino A, et al. Brain derived neurotrophic factor (BDNF) expression is regulated by microRNAs miR-26a and miR-26b allele-specific binding. PLoS One. 2011;6:e28656. doi: 10.1371/journal.pone.0028656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klein AB, Jennum P, Knudsen S, Gammeltoft S, Mikkelsen JD. Increased serum brain-derived neurotrophic factor (BDNF) levels in patients with narcolepsy. Neurosci Lett. 2013;544:31–5. doi: 10.1016/j.neulet.2013.03.031. [DOI] [PubMed] [Google Scholar]
- 46.Cvetkovic-Lopes V, Bayer L, Dorsaz S, et al. Elevated Tribbles homolog 2-specific antibody levels in narcolepsy patients. J Clin Invest. 2010;120:713–9. doi: 10.1172/JCI41366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dobens LL, Jr, Bouyain S. Developmental roles of tribbles protein family members. Dev Dyn. 2012;241:1239–48. doi: 10.1002/dvdy.23822. [DOI] [PubMed] [Google Scholar]
- 48.Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA profiling of naïve, effector and memory CD8 T cells. PLoS One. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Z, Wu F, Brant SR, Kwon JH. IL-23 receptor regulation by Let-7f in human CD4+ memory T cells. J Immunol. 2011;186:6182–90. doi: 10.4049/jimmunol.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng J, Jiang L, Zhang L, et al. Functional genetic variations in the IL-23 receptor gene are associated with risk of breast, lung and nasopharyngeal cancer in Chinese populations. Carcinogenesis. 2012;33:2409–16. doi: 10.1093/carcin/bgs307. [DOI] [PubMed] [Google Scholar]
- 51.Faraco J, Lin L, Kornum BR, et al. ImmunoChip study implicates antigen presentation to T cells in narcolepsy. PLoS Genet. 2013;9:e1003270. doi: 10.1371/journal.pgen.1003270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hemolysis test. The levels of miR-451-5p and miR-23a were analyzed by qPCR of 3 pools of 4 plasma samples from each group of healthy controls (HC), and type 1 narcolepsy (NC), type 2 narcolepsy (NwC), and idiopathic hypersomnia (IH) patients. The Ct value of miR-451-5p minus Ct value of miR-23a was plotted for each pool.
miRNA expression data in normal human tissues. Heat map illustrating the tissue expression of 4 miRNAs: miR-30c, miR-130a, miR-26a, and let-7f. White, blue, rose, and red bars respectively indicate absent, low, intermediate, and high levels of high expression.
miRNAs in plasma from type 1 narcolepsy, type 2 narcolepsy, and idiopathic hypersomnia patients with two-fold or more difference with respect to healthy controls and P < 0.05.
microRNA candidates in plasma from patients with type 1 narcolepsy, type 2 narcolepsy, and healthy controls after Dunn Bonferroni correction with P < 0.00087.
Analysis of miRNA-targeted genes and pathways using DIANA-miRPath v2.0.