Abstract
Purpose
To investigate the possible association of oxidative stress with keratoconus (KC), we estimated the changes in relative mitochondrial DNA (mtDNA) content.
Methods
The study included 119 patients with KC and 208 controls matched for gender, ethnicity, and systemic disease status. We selected controls who were older than the patients since the mtDNA copy number tends to increase with age. The age mean (standard deviation) was 26.4(7.6) and 54.5(14.4) years for the patients and controls, respectively. The relative mtDNA copy number was estimated with the real-time quantitative PCR (qPCR) method using ND1 as the mtDNA gene and human globulin (HGB; also known as the cytoglobin gene, CYGB) as the reference single-copy nuclear gene.
Results
The mean relative mtDNA content was significantly higher in patients with KC (1.20±0.45) than in the normal control subjects (1.04±0.36; p = 0.0004). Subjects with high mtDNA content (>1.259, i.e., greater than 75th percentile) were at an increased risk of the disease (odds ratio = 2.62, 95% confidence interval = 1.40 to 4.89; p =0.0025).
Conclusions
Increased mtDNA content in patients with KC may indicate mitochondrial respiratory chain defects and thus mitochondrial-abnormality involvement.
Introduction
The number of mitochondria in a cell and the amount of mitochondrial DNA (mtDNA) per mitochondrion together constitute the mtDNA content of the cell. Cells carefully regulate the mtDNA content [1], but they also seem able to adjust relative (to nuclear DNA) mtDNA content upward by duplication of mitochondria or proliferation of mtDNA as compensation for reduced ATP synthesis [2,3]. For example, relative mtDNA content increases in certain tissues as respiratory function declines with age [4] and oxidative stress [5]. Keratoconus (KC) is a complex condition of multifactorial etiology, and to date, the genetic cause has not been identified. Previous studies have demonstrated evidence of mitochondrial abnormalities in KC [6-8]. We recently showed that mtDNA mutation(s) are present in a group of patients with KC, thus implying an oxidative stress mechanism indirectly influences the development and/or progression of KC [9]. We also showed that individuals with mitochondrial haplogroup H and R are at increased risk of developing KC [10]. These findings by our group and others suggest increasing evidence of mitochondrial abnormalities associated with KC. In line with this view, we investigated another oxidative stress marker associated with mtDNA copy number in a group of patients with KC.
Methods
Patients and control subjects
Patients with KC (n = 119) were selected from the anterior segment clinic at King Abdulaziz University Hospital (KAUH) from May 2009 through December 2013. Among the patient group (n= 119), there were 60 males and 55 females with a mean age of 26.4 years. Among the control group (n= 208), there were 138 males and 70 females with a mean age of 54.5 years. The study adhered to the tenets of the Declaration of Helsinki, and all participants provided Signed informed consent. The study was approved by the College of Medicine (King Saud University, Riyadh, Saudi Arabia) ethical committee (proposal number # 09–659). The study adhered to the tenets of the Declaration of Helsinki, and all participants provided signed informed consent. The study was approved by the College of Medicine (King Saud University, Riyadh, Saudi Arabia) ethical committee (proposal number # 09–659). Patients with KC (n = 119) were selected from the anterior segment clinic at King Abdulaziz University Hospital (KAUH) from May 2009 through December 2013. Patients were diagnosed with KC if the Schimpff flow-based elevation map showed posterior corneal elevation within the central 5 mm ≥+20 µm, inferior-superior dioptric asymmetry (I-S value) >1.2 diopters (D), and the steepest keratometry >47 D. Patients were considered sporadic cases after immediate family members were examined and the patient was identified as an isolated case of KC. The exclusion criteria were based on the presence of post-laser-assisted in situ keratomileusis (LASIK) ectasia and refusal to participate. All KC cases secondary to causes such as trauma, surgery, Ehlers Danlos syndrome, osteogenesis imperfecta, and pellucid marginal degeneration were excluded from the study. Controls (n = 208) were recruited from the general ophthalmology clinic and had no ocular disease(s) or previous ophthalmic surgeries. Their slit-lamp exam showed a clear cornea, and their Schimpff flow-based elevation map was within normal limits.
Sample collection and relative mtDNA copy number assay
Blood samples (5 mL) were collected in EDTA tubes from all participating individuals. The tubes were centrifuged at 5,500 ×g for 5 min. DNA was extracted from the buffy layer using the illustra blood genomicPrep Mini Spin kit (GE Healthcare, Buckinghamshire, UK) and stored at –20 °C in aliquots until further use. Relative mtDNA copy number was determined with the real-time quantitative PCR (qPCR) method as described previously [11]. The assay measures relative mtDNA copy number by determining the ratio of mitochondrial gene copy number to single-copy nuclear gene copy number based on the cycle thresholds (Cts) derived from a standard curve. In this study, ND1(Gene ID 4535; OMIM 516000) represents the mtDNA copy number and human globulin (HGB; also known as the cytoglobin gene, CYGB; Gene ID 114757; OMIM 608759) was the reference single-copy nuclear gene. Briefly, two pairs of primers were designed. One pair was used to amplify the MT-ND1 gene in mtDNA, and the second pair was used to amplify the endogenous reference single-copy nuclear gene human globulin (HGB; also known as cytoglobin gene, CYGB). The primer sequences were as follows: forward primer (ND1-F) 5′-CCC TAA AAC CCG CCA CAT CT-3′ and reverse primer (ND1-R) 5′-GAG CGA TGG TGA GAG CTA AGG T-3′ for ND1; and forward primer (HGB-F) 5′-GTG CAC CTG ACT CCT GAG GAG A-3′ and reverse primer (HGB-R) 5′-CCT TGA TAC CAA CCT GCC CAG-3′ for HGB. The PCR was performed on a 96-well plate with the 7500 Sequence Detection System (Applied Biosystems; Foster City, CA) in a 20 μl reaction mixture containing 1× SYBR Green Mastermix (Applied Biosystems), 215 nM ND1-F (or HGB-F) primer, 215 nM ND1-R (or HGB-R) primer, and 10 ng of genomic DNA. The amplification conditions for the mtDNA (MT-ND1 gene) were 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min; and for the HGB amplification were 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, and 56 °C for 1 min. Melting curve analysis was included at the end of each run to validate the specificity of the PCR products. Standard curves were plotted using a reference genomic DNA sample by a 1:2 serial dilution to produce a six-point curve between 1.25 and 40 ng of DNA. The ratio of the mtDNA copy number to the HGB (nuclear) copy number for each sample was determined using the calibration curves representing the mtDNA copy number in each cell. The ratio for each sample was then normalized to a calibrator DNA to standardize variations between different runs. The calibrator DNA was a genomic DNA sample from a healthy control subject. A standard curve of a serially diluted reference DNA, one negative control, and one calibrator DNA were included in each run in duplicate. PCR efficiency (E) for each assay were calculated using the equation, E (%) = [10(−1/slope) −1] × 100.
Statistical analysis
The mean, median, and standard deviation (SD) were calculated. Differences between the mean was determined using Student’s unpaired t test, and for the median, the Mann–Whitney U test was applied. Pearson’s chi-square test was used to test differences in gender distribution and the association between the mtDNA copy number and the disease. The odds ratio (OR) and 95% confidence interval (CI) were calculated. Analyses were performed using Stat View software version 5.0 (SAS Institute, Cary, NC). All statistical tests were two-sided, and a p value <0.05 was considered statistically significant.
Results
Standard curves derived for mtDNA ND1 and the HGB gene showed good linearity for all separate PCR runs. The correlation coefficient (R2) for each standard curve was greater than 0.98. The mean PCR amplification efficiency for the ND1 gene was 101% and that of HGB was 98% with a coefficient variation (CV) of 0.06 for both genes.
Table 1 shows various characteristics of the patients and the controls, including age, gender, diabetes mellitus status, hypertension, and mtDNA copy number. The results presented in Table 1 indicate that the patient group was significantly younger than the control group with age ranging from 12 to 50 years and 19 to 96 years, respectively. In addition, there were significantly fewer women in the control group (p = 0.012). The mean relative mtDNA content was significantly higher in the patients with KC (1.20±0.45) than the normal control subjects (1.04±0.36; mean difference = 0.164, 95% CI = 0.07–0.25; p = 0.0004). The median values, for relative mtDNA content, among the cases and the control subjects were 1.12 (range = 0.61–3.82) and 1.02 (range = 0.39–3.82) respectively and were also significantly different (p = 0.0004) (Figure 1).
Table 1. Characteristics of subjects included in the study and their mtDNA copy number.
| Variables | Controls (n=208) | Patients (n=119) | P value |
|---|---|---|---|
| Age, mean (SD) |
54.5 (14.4) |
26.4 (7.6) |
<0.0001 |
| Range, years |
19 – 96 |
12 – 50 |
|
| Male [No. (%)] |
138 (66) |
60 (52) |
- |
| Female [No. (%)] |
70 (34) |
55 (48) |
0.012 |
| Diabetes Mellitus [No. (%)] |
45 (22) |
22 (18.5) |
0.590 |
| Hypertension [No. (%)] |
27 (13) |
11 (9.2) |
0.402 |
| mtDNA content, copy number [Mean (SD)] |
1.04 (0.36) |
1.20 (0.45) |
0.0004 |
| mtDNA content, copy number [Median (Range)] | 1.02 (0.39 – 3.82) | 1.12 (0.61 – 3.82) | 0.0004 |
Figure 1.
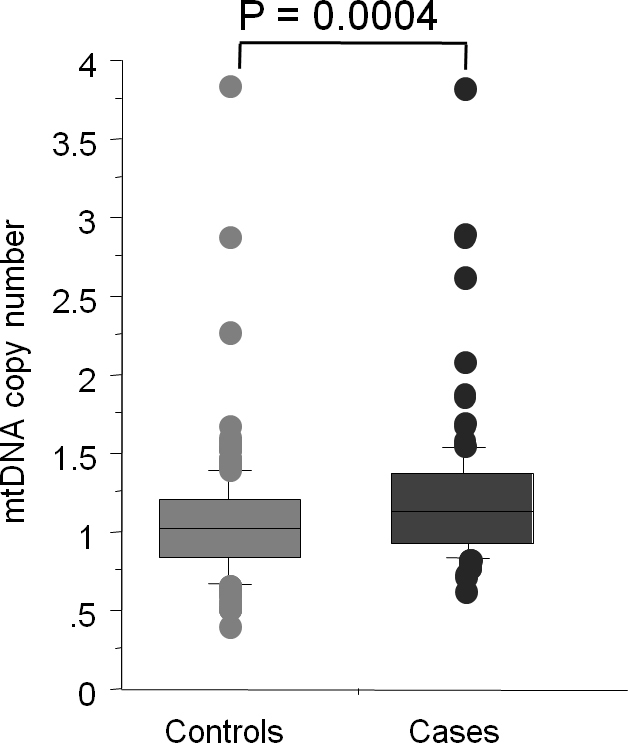
Box plot showing distribution of the mtDNA copy number in the cases and control groups.
To assess the risk of disease, the levels of mtDNA content among the control and patient subjects were initially dichotomized (uncategorized) at the 50th percentile (or median) value. However, individuals with mtDNA content ≥1.032 were not at a statistically significant risk (OR = 1.4, 95% CI = 0.89 to 2.21; p = 0.14; Table 2). The data were then further dichotomized (uncategorized) into quartiles (to assess any dose–response trend), and two cutoff points were identified: 0.817 (first quartile or the 25th percentile) and 1.259 (third quartile or the 75th percentile). Subjects were then categorized into three groups by using these two cutoff points. Overall, the dose–response trend was statistically significant (χ2 = 10.912, df = 2; p = 0.0043). Compared to individuals with the lowest mtDNA content (<0.817), individuals in the medium category (0.817–1.259; inter-quartile) did not show significant association; however, individuals in the highest category (>1.259) were at an increased risk of the disease (OR = 2.62, 95% CI = 1.40 to 4.89; p = 0.0025; Table 2).
Table 2. Risk of Keratoconus as estimated by mtDNA content.
| mtDNA copy number | Controls (n=208) No. (%) | Cases (n=119) No. (%) | OR† | 95% CI | P value |
|---|---|---|---|---|---|
|
By median |
|||||
|
<1.032 |
105 (50.5) |
50 (42) |
1 |
reference |
- |
|
≥1.032 |
103 (49.5) |
69 (58) |
1.40 |
0.89 – 2.21 |
0.14 |
|
By quartiles‡ |
|||||
|
<0.871 |
64 (30.7) |
25 (21) |
1 |
reference |
- |
|
0.871–1.259 |
101 (48.5) |
50 (42) |
1.26 |
0.71 – 2.24 |
0.41 |
| >1.259 | 43 (20.7) | 44 (37) | 2.62 | 1.40 – 4.89 | 0.0025 |
† OR – unadjusted odds ratio; CI – confidence interval. ‡ first quartile (<25th percentile); inter-quartile (25th – 75th percentile); third quartile (>75th percentile).
Discussion
KC is a complex condition of multifactorial etiology. Genetic and environmental factors are associated with KC. Evidence of genetic etiology includes the conditions of familial inheritance, discordance between dizygotic twins, and its association with other known genetic disorders. Environmental factors include contact lens wear, chronic eye rubbing, and atopy of the eye. Although numerous studies of KC have been reported, there are still problems with identifying genetic factors [12]. Phenomena such as gene–gene interactions, genetic heterogeneity, reduced penetrance, or phenocopy are frequently observed in complex diseases and significantly influence genetic factor identification process.
In Saudi Arabia, the prevalence of KC is not known. The prevalence of KC has been reported to vary in different studies, from 8.8 to 54.4 per 100,000 [13-15]. The variation is in part due to the different diagnostic criteria used in each study. KC is known to affect all ethnicities. An incidence of KC of 25 per 100,000 (1 in 4,000) per year for Asians compared with 3.3 per 100,000 (1 in 30,000) per year for Caucasians (p<0.001) has been reported [16]. Previously, we investigated our patients with KC for mutations in the visual system homebox 1 (VSX1) gene (Gene ID 30813; OMIM 605020) where mutations associated with KC cases have been found in different studies [17,18]. Sequencing the VSX1 gene in a group of Saudi patients with KC did not reveal any mutation(s) [19]. We also investigated the possible presence of chromosomal copy number variants (CNVs) in patients with isolated non-familial KC using high-resolution array comparative genomic hybridization (array CGH) technology. We did not detect any chromosomal abnormalities in this group of patients thus eliminating CNVs as a possible cause of keratoconus [20].
mtDNA copy number had been linked to many diseases and conditions such as diabetic retinopathy [21], autism [22], various human cancers [23] including breast cancer [24], renal cell carcinoma [11], Leber hereditary optic neuropathy [25], non-arteretic ischemic optic neuropathy [26], and various mitochondrial disorders [27]. Elevated mtDNA copy number is thought to be a response to oxidative stress status [28,29]. Our group recently showed that mtDNA mutations [9] and mitochondrial haplogroup H and R [10] are present in patients with KC. Based on these findings, we suggested that an oxidative stress mechanism could be contributing indirectly to the development and/or progression of KC. Other investigators have also reported evidence of oxidative stress and its markers in KC. Under the transmission electron microscope, swelling of the mitochondria were observed in KC corneal tissues [6]. KC corneas exhibited more mtDNA damage than normal corneas [7]. KC fibroblasts had increased basal generation of reactive oxygen species and were more susceptible to stressful challenges (low-pH and/or H2O2 conditions) than normal fibroblasts [8]. Additionally, cultured KC fibroblasts have an inherent, hypersensitive response to oxidative stress that involves mitochondrial dysfunction and mtDNA damage. As a result, it was suggested that KC fibroblast hypersensitivity may play a role in the development and progression of KC [30]. We therefore investigated a group of patients with KC for mtDNA content and compared their results with normal controls. Our results indicate that the patients with KC had higher relative mtDNA content than the controls although our KC patient group was younger than the controls. One would expect normal older healthy individuals would have higher mtDNA content than younger individuals. This fact strengthens our findings. We report a fairly small group of patients from a restricted ethnic population, and this type of evaluation must be repeated at other centers. If these results can be replicated, then mtDNA content (mtDNA copy number) may be considered as a genetic risk factor contributing indirectly to KC pathogenesis.
Acknowledgments
This research is supported by the Glaucoma Research Chair (GRC); Grant ID GRC-6787 at Department of Ophthalmology, College of Medicine, King Saud University, Riyadh, Saudi Arabia.
References
- 1.Robin ED, Wong R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Physiol. 1988;136:507–13. doi: 10.1002/jcp.1041360316. [DOI] [PubMed] [Google Scholar]
- 2.Wei YH. Mitochondrial DNA mutations and oxidative damage in aging and diseases: an emerging paradigm of gerontology and medicine. Proc Natl Sci Counc Repub China B. 1998;22:55–67. [PubMed] [Google Scholar]
- 3.Wei YH, Lee HC. Oxidative stress, mitochondrial DNA mutation, and impairment of antioxidant enzymes in aging. Exp Biol Med (Maywood) 2002;227:671–82. doi: 10.1177/153537020222700901. [DOI] [PubMed] [Google Scholar]
- 4.Lee HC, Lu CY, Fahn HJ, Wei YH. Aging- and smoking-associated alteration in the relative content of mitochondrial DNA in human lung. FEBS Lett. 1998;441:292–6. doi: 10.1016/s0014-5793(98)01564-6. [DOI] [PubMed] [Google Scholar]
- 5.Lee HC, Wei YH. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol. 2005;37:822–34. doi: 10.1016/j.biocel.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Aktekin M, Sargon MF, Cakar P, Celik HH, Firat E. Ultrastructure of the cornea epithelium in keratoconus. Okajimas Folia Anat Jpn. 1998;75:45–53. doi: 10.2535/ofaj1936.75.1_45. [DOI] [PubMed] [Google Scholar]
- 7.Atilano SR, Coskun P, Chwa M, Jordan N, Reddy V, Le K, Wallace DC, Kenney MC. Accumulation of mitochondrial DNA damage in keratoconus corneas. Invest Ophthalmol Vis Sci. 2005;46:1256–63. doi: 10.1167/iovs.04-1395. [DOI] [PubMed] [Google Scholar]
- 8.Chwa M, Atilano SR, Reddy V, Jordan N, Kim DW, Kenney MC. Increased stress-induced generation of reactive oxygen species and apoptosis in human keratoconus fibroblasts. Invest Ophthalmol Vis Sci. 2006;47:1902–10. doi: 10.1167/iovs.05-0828. [DOI] [PubMed] [Google Scholar]
- 9.Abu-Amero KK, Azad TA, Kalantan H, Sultan T, Al-Muammar AM. Mitochondrial sequence changes in keratoconus patients. Invest Ophthalmol Vis Sci. 2014;55:1706–10. doi: 10.1167/iovs.14-13938. [DOI] [PubMed] [Google Scholar]
- 10.Abu-Amero KK, Azad TA, Sultan T, Kalantan H, Kondkar AA, Al-Muammar AM. Association of Mitochondrial Haplogroups H and R with Keratoconus in Saudi Arabian Patients. Invest Ophthalmol Vis Sci. 2014;55:2827. doi: 10.1167/iovs.14-14300. : - [DOI] [PubMed] [Google Scholar]
- 11.Xing J, Chen M, Wood CG, Lin J, Spitz MR, Ma J, Amos CI, Shields PG, Benowitz NL, Gu J, de Andrade M, Swan GE, Wu X. Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst. 2008;100:1104–12. doi: 10.1093/jnci/djn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang HY, Chodosh J. The genetics of keratoconus. Semin Ophthalmol. 2013;28:275–80. doi: 10.3109/08820538.2013.825295. [DOI] [PubMed] [Google Scholar]
- 13.Ihalainen A. Clinical and epidemiological features of keratoconus genetic and external factors in the pathogenesis of the disease. Acta Ophthalmol Suppl. 1986;178:1–64. [PubMed] [Google Scholar]
- 14.Kennedy RH, Bourne WM, Dyer JA. A 48-year clinical and epidemiologic study of keratoconus. Am J Ophthalmol. 1986;101:267–73. doi: 10.1016/0002-9394(86)90817-2. [DOI] [PubMed] [Google Scholar]
- 15.Tanabe U, Fujiki K, Ogawa A, Ueda S, Kanai A. Prevalence of keratoconus patients in Japan. Nippon Ganka Gakkai Zasshi. 1985;89:407–11. [PubMed] [Google Scholar]
- 16.Georgiou T, Funnell CL, Cassels-Brown A, O'Conor R. Influence of ethnic origin on the incidence of keratoconus and associated atopic disease in Asians and white patients. Eye (Lond) 2004;18:379–83. doi: 10.1038/sj.eye.6700652. [DOI] [PubMed] [Google Scholar]
- 17.Karamichos D, Guo XQ, Hutcheon AE, Zieske JD. Human corneal fibrosis: an in vitro model. Invest Ophthalmol Vis Sci. 2010;51:1382–8. doi: 10.1167/iovs.09-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engler C, Chakravarti S, Doyle J, Eberhart CG, Meng H, Stark WJ, Kelliher C, Jun AS. Transforming growth factor-beta signaling pathway activation in Keratoconus. Am J Ophthalmol. 2011;151:752–9. doi: 10.1016/j.ajo.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abu-Amero KK, Kalantan H, Al-Muammar AM. Analysis of the VSX1 gene in keratoconus patients from Saudi Arabia. Mol Vis. 2011;17:667–72. [PMC free article] [PubMed] [Google Scholar]
- 20.Abu-Amero KK, Hellani AM, Al Mansouri SM, Kalantan H, Al-Muammar AM. High-resolution analysis of DNA copy number alterations in patients with isolated sporadic keratoconus. Mol Vis. 2011;17:822–6. [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Kafaji G, Golbahar J. High glucose-induced oxidative stress increases the copy number of mitochondrial DNA in human mesangial cells. BioMed Research International. 2013;2013:754946. doi: 10.1155/2013/754946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu F, Chauhan V, Kaur K, Brown WT, LaFauci G, Wegiel J, Chauhan A. Alterations in mitochondrial DNA copy number and the activities of electron transport chain complexes and pyruvate dehydrogenase in the frontal cortex from subjects with autism. Transl Psychiatry. 2013;3:e299. doi: 10.1038/tp.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin PH, Lee HC, Chau GY, Wu YT, Li SH, Lui WY, Wei YH, Liu TY, Chi CW. Alteration of the copy number and deletion of mitochondrial DNA in human hepatocellular carcinoma. Br J Cancer. 2004;90:2390–6. doi: 10.1038/sj.bjc.6601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thyagarajan B, Wang R, Nelson H, Barcelo H, Koh WP, Yuan JM. Mitochondrial DNA copy number is associated with breast cancer risk. PLoS ONE. 2013;8:e65968. doi: 10.1371/journal.pone.0065968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abu-Amero KK, Bosley TM. Mitochondrial abnormalities in patients with LHON-like optic neuropathies. Invest Ophthalmol Vis Sci. 2006;47:4211–20. doi: 10.1167/iovs.06-0295. [DOI] [PubMed] [Google Scholar]
- 26.Abu-Amero KK, Bosley TM. Increased relative mitochondrial DNA content in leucocytes of patients with NAION. Br J Ophthalmol. 2006;90:823–5. doi: 10.1136/bjo.2006.090332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu CS, Tsai CS, Kuo CL, Chen HW, Lii CK, Ma YS, Wei YH. Oxidative stress-related alteration of the copy number of mitochondrial DNA in human leukocytes. Free Radic Res. 2003;37:1307–17. doi: 10.1080/10715760310001621342. [DOI] [PubMed] [Google Scholar]
- 28.Tan X, Li Y. Copy number and deletion of 4 977 bp of granular cell mitochondria DNA in patients with diminished ovarian reserve. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2010;35:879–84. doi: 10.3969/j.issn.1672-7347.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 29.Hori A, Yoshida M, Shibata T, Ling F. Reactive oxygen species regulate DNA copy number in isolated yeast mitochondria by triggering recombination-mediated replication. Nucleic Acids Res. 2009;37:749–61. doi: 10.1093/nar/gkn993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chwa M, Atilano SR, Hertzog D, Zheng H, Langberg J, Kim DW, Kenney MC. Hypersensitive response to oxidative stress in keratoconus corneal fibroblasts. Invest Ophthalmol Vis Sci. 2008;49:4361–9. doi: 10.1167/iovs.08-1969. [DOI] [PubMed] [Google Scholar]


