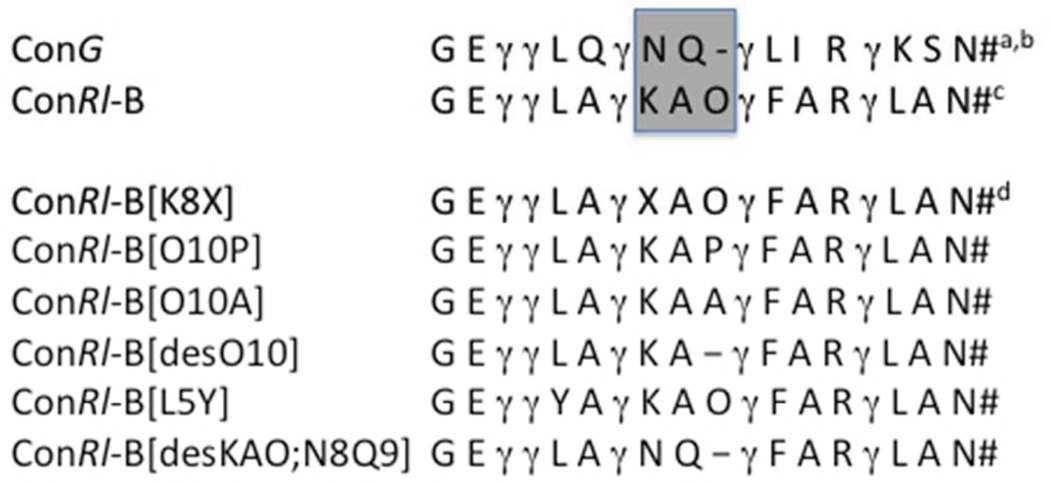Abstract
Using molecular phylogeny has accelerated the discovery of peptidic ligands targeted to ion channels and receptors. One clade of venomous cone snails, Asprella, appears to be significantly enriched in conantokins, antagonists of N-Methyl D-Asparate receptors (NMDARs). Here, we describe the characterization of two novel conantokins from Conus rolani, including conantokin conRl-B that has shown an unprecedented selectivity for blocking NMDARs that contain NR2B subunits. ConRl-B shares only some sequence similarity to the most studied NR2B-selective conantokin, conG. The divergence between conRl-B and conG in the second inter-Gla loop was used to design analogs for structure-activity studies; the presence of Pro10 was found to be key to the high potency of conRl-B for NR2B, whereas the ε-amino group of Lys8 contributed to discrimination in blocking NR2B- and NR2A-containing NMDARs. In contrast to previous findings from Tyr5 substitutions in other conantokins, conRl-B [L5Y] showed potencies on the four NR2 NMDA receptor subtypes that were similar to those of the native conRl-B. When delivered into the brain, conRl-B was active in suppressing seizures in the model of epilepsy in mice, consistent with NR2B-containing NMDA receptors being potential targets for antiepileptic drugs. Circular dichroism experiments confirmed that the helical conformation of conRl-B is stabilized by divalent metal ions. Given the clinical applications of NMDA antagonists, conRl-B provides a potentially important pharmacological tool for understanding the differential roles of NMDA receptor subtypes in the nervous system. This work shows the effectiveness of coupling molecular phylogeny, chemical synthesis and pharmacology for discovering new bioactive natural products.
Keywords: Conus peptides, conantokin, NMDA antagonist, NR2B subunits, epilepsy, anticonvulsant
N-Methyl D-Aspartate (NMDA) receptors are a major class of glutamate receptors that play critical roles in excitatory neurotransmission. These receptors have been clinically validated therapeutic drug targets, and are implicated in the synaptic plasticity in neuropathic pain, learning, mood disorders and addiction. Functional NMDARs are heterotetrameric complexes comprising two NR1 subunits and two NR2 subunits. Four genes, namely NR2A, NR2B, NR2C or NR2D, encode the NR2 subunits. NR2B targeting antagonists are being developed for the treatment of pain, epilepsy, stroke or Parkinson’s disease (1, 2). Small molecule NMDA antagonists, summarized in Table S1 have been developed that preferentially or selectively block NMDARs containing various NR2 subunits (3–14). Given the molecular complexity and importance of NMDARs, there is a constant need for novel NMDA antagonists that selectively discriminate with a wide separation in affinities among the four individual NR2 subunits. Such compounds should be useful pharmacological tools to define the role of individual NMDA receptor subtypes in the nervous system.
Conantokins are a diverse group of Conus peptides that target NMDA receptors (15, 16). Characterization using heterologous expression assays showed that conantokins act competitively at the glutamate-binding site on the NR2 subunit (17). Most conantokins have been found to preferentially target NMDA receptors containing the NR2B subunit, although the affinity for the other NR2 subunits of the NMDA receptor varies substantially (18–22). Table 1 depicts the amino acid sequences of all conantokins characterized thus far. Among the conantokins characterized, conG has demonstrated the greatest selectivity for the NR2B subunit. ConG has shown efficacy in a number of preclinical studies, including models of pain, epilepsy, and neuroprotection following ischemia (15, 21, 23–26). Based on favorable preclinical studies, ConG has reached phase I clinical trials for the treatment of epilepsy (21, 27–29).
Table 1.
Amino acid sequences of previously characterized conantokins.
| Conus Species | Conanto kin |
Amino-Acid Sequence | Ref. |
|---|---|---|---|
| C. geographus | ConG | GE γ γ LQ γ NQ γ LIR γ KSN# | (51) |
| C. tulipa | ConT | GE γ γ YQ K ML γ NLR γ AEVKKNA# | (52) |
| C. radiatus | ConR | GE γ γ VA K MAA γ LAR γ NIAKGCKVNCYP^ | (53) |
| C. lynceus | ConL | GE γ γ VA K MAA γ LAR γ DAVN# | (54) |
| C. parius | ConPr-A | GE DyYAyGIRyYQL I HGKI^ | (34) |
| C. parius | ConPr-B | DE O γ YA γ AIR γ YQL K YGKI^ | (34) |
| C. parius | ConPr-C | GE O γ VA K WA γ GLR γ KASSN# | (34) |
| C. purpurascens | ConP | GE γ γ HS KYQ γ CLR γ IRVNKVQQ γ C(^) | (55) |
| C. brettinghami | ConBr | GD γ γ YS K FI γ RER γ AGRLDLSKFP^ | (22) |
| C. rolani | ConRl-A | AD γγ YL K FI γ EQR K QGKLDPTKFP^ | (36) |
denotes amidated C-terminus, CONH2
denotes free carboxyl group on the C-terminus
Our research group has recently been using molecular phylogeny, guided discovery to facilitate the identification of novel Conus peptides targeting sodium channels, nAChRs and NMDA receptors (30–33). Several new conantokins have been discovered using this approach, each with a unique pharmacological profile (22, 34–36). Particularly noteworthy is the Asprella clade of Conus spp. that contains C. brettinghami, C. sulcatus, C. bocki, C. rolani and C. samiae, which appears to be a rich source of peptides targeted NMDA receptors. Recently two new conantokins, conantokinBr and conantokinRl-A were described from C. brettinghami, and C. rolani, respectively: despite having divergent sequences these peptides exhibited similar pharmacological properties (35, 36). Here, we describe characterization of two new conantokins from C. rolani; one of these, conantokinRl-B, has a more pronounced subtype specificity than any conantokin previously reported.
MATERIALS AND METHODS
Preparation of genomic DNA and characterization of clones encoding ConRl-B
Genomic DNA was prepared from 20 mg Conus rolani tissue using the Gentra PUREGENE DNA Isolation Kit (GentraSystems, Minneapolis, MN) according to the manufacturer’s standard protocol. 10 ng of C. rolani genomic DNA was used as a template for polymerase chain reaction (PCR) with oligonucleotides corresponding to conserved regions of the signal sequence and 3’ UTR sequences of conantokin prepropeptides, as described previously (22, 34–36). The resulting PCR product was purified using the High Pure PCR Product Purification Kit (Roche Diagnostics, Indianapolis, IN) following the manufacturer’s suggested protocol. The eluted DNA fragment was ligated to pNEB206A vector using the cloning kit (New England BioLabs, Inc., Bever1y, MA) following manufacturer’s suggested protocol and the resulting product transformed into DH5a competent E. coli cells. The nucleic acid sequences of the resulting conantokin toxin-encoding clones were determined according to the standard protocol for DNA sequencing.
Peptide Synthesis
Native peptide ConRl-B and its analogs were synthesized using an Apex 396 automated peptide synthesizer (AAPPTec, Louisville, KY) and a standard solid-phase Fmoc (9-fluorenylmethyloxycarbonyl) protocol. The peptides were assembled on preloaded Fmoc-L-Asn (Trt)-Rink Amide MBHA resin purchased from Peptides International, Inc. (Louisville, KY; substitution: 0.38 mmolg-1). All standard amino acids were purchased form AAPPTec, Fmoc-γ-carboxy-γ-(di-tert-butyl ester)-L-glutamic acid (γ-carboxyglutamic acid) from Advanced ChemTech (Louisville, KY), N-α-Fmoc-O-t-butyl-L-trans-4-hydroxyproline (Hyp) form NovaBiochem/EMD Chemicals (Gibbstown, NJ) and Fmoc-L-norleucine from ChemImpex Int. (Wood Dale, IL). Side-chain protection for the following amino acids was: Gla and Glu O-tert-butyl (OtBu); Arg 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl (Pbf) Lys: tert-butyloxycarbonyl (Boc); Hyp and Tyr: tert-butyl (tBu); Asn and Gln: trityl (Trt). Peptides were synthesized on 30 µmol scale. Coupling activation was achieved with 1 equivalent of 0.22 M benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate and 2 equivalents of 2 M N, N-diisopropylethyl amine in N-methyl-2-pyrrolidone. 10-fold excess of amino acid was used except for γ-carboxyglutamic acid for which 3-fold excess was applied. Each coupling reaction was carried out for 60 min except for γ-carboxyglutamic acid for which reaction time was 90 min. Fmoc deprotection was carried out for 20 min with 20% solution of piperidine in DMF. Each peptide was cleaved from 25 – 50 mg resin by a 3h treatment with 0.5 mL of Reagent K (trifluoroacetic acid (TFA)/water/phenol/thioanisole/ethanedithiol 82.5/5/5/5/2.5 by volume) and subsequently filtered and precipitated with cold methyl-tert-butyl ether (MTBE). The crude peptides were then collected by centrifugation at 5000g for 8 min and washed two times with cold MTBE. The washed peptide pellet was dissolved in 20% acetonitrile in 0.1% TFA and purified by reversed-phase HPLC using a preparative C18 Vydac column (218TP510, 250 mm × 10 mm, 5 µm particle size) eluted with a linear gradient ranging from 20 to 60% of solvent B in 40 min at a flow rate 4 ml/min. The HPLC solvents were 0.1% (v/v) TFA in water (solvent A) and 0.1% TFA (v/v) in 90% aqueous acetonitrile (solvent B). The eluent was monitored by measuring absorbance at 220 nm. Purity of peptides was assessed by an analytical C18 Vydac reversed-phase HPLC (218TP54, 250 mm × 4.6 mm, 5 µm particle size) using a linear gradient ranging from 20 to 55% of solvent B in 30 min (retention times and gradients specified in Table S1 of the supporting information) with a flow rate 1 ml/min. Peptides were quantified against a reference peptide using the same HPLC separation conditions. Molecular masses of all analogs were confirmed by ESI MS (Table S1 supporting information).
Heterologous expression of NMDA receptors
The rat NMDA receptor clones NR2A, NR2B, NR2C, NR2D, NR3A, NR3B, NR1–2a, NR1–2b, and NR1–4b were used (GenBank numbers AF001423, U11419, U08259, U08260, NM_001198583, NM_133308, U08262, U08264, U08268, respectively). The splice variant NR1–2b was used for all concentration-response assays, as it is widely expressed in the CNS (37, 38). To control for the possibility that exon 5 may affect NMDA receptor sensitivity to conantokins (i.e., (39, 40)), NR1–2a was separately co-expressed with all NR2 subtypes. We observed low expression levels of NR3A and NR3B when co-expressed with NR1–2b or NR1–2a; thus the NR1–4b splice variant was co-expressed with these subunits. All of the expression clones, except NR3B, were under control of a T7 promoter. A T3 promoter controlled expression of NR3B. For each clone, Ambion RNA transcription kits (Ambion, Inc.) were used to make capped RNA (cRNA) for injection into Xenopus oocytes. To express NMDA receptors, 2–5 ng of RNA encoding each subunit was injected into each oocyte. Oocytes were maintained in ND96 solution (96 mM NaCl, 2 mM KCl, 1,8 mM CaCl2, 1mM MgCl2, and 5 mM HEPES at pH 7.2–7.5) with antibiotics (Septra, Amikacin, Pen/Strep). All voltage-clamp electrophysiology was performed prior to 7 days post-injection.
Two electrode voltage-clamp electrophysiology
All oocytes were voltage clamped at −70 mV at room temperature. Oocytes were gravity-perfused with Mg2+-free ND96 buffer (96.0 mM NaCl, 2.0 mM KCl, 1.8 mM CaCl2, and 5 mM HEPES at pH 7.2 – 7.5). Mg2+ was omitted from the ND96 buffer to prevent the voltage-dependent blockade of NMDA receptors at −70mV. BSA (0.1 mg/mL) was added to reduce non-specific absorption of peptide. In an additional set of experiments, conRl-B was also assessed on oocytes in the presence of Ca2+-free, Mg2+-free ND96 substituted with barium chloride (96.0 mM NaCl, 2.0 mM KCl, 1.8 mM BaCl2, and 5 mM HEPES at pH 7.2 – 7.5); no difference in the effect of peptide was seen between oocytes tested in the presence of calcium-containing buffer or barium-containing buffer. NMDA receptor-mediated current was elicited by the administration of one-second pulses of agonist (200 uM glutamate, 20 uM glycine, in Mg+2-free ND96 for NR1/NR2 subunit combinations; 20 uM glycine, in Mg+2-free ND96 for NR1/NR3 subunit combinations). To measure the effect of conantokins and analogues on oocytes expressing NMDA receptors, the buffer flow was halted, and the peptides were applied in a static bath for duration sufficient to reach equilibrium, or a minimum of 5 minutes. A blockade of NMDA receptor-mediated current by peptides was measured by normalizing the response of the first agonist pulse following static bath to the baseline response (current in response to agonist prior to peptide application). A virtual instrument made by Dr. Doju Yoshikami at the University of Utah was used for data acquisition, and concentration-response curves were generated using Prism software (Graphpad Software, Inc.). The following equation, where nH is the Hill Coefficient, and IC50 is the concentration required to achieve half-maximal block, was used to fit concentration-response curves: % response= 100/{1+([peptide]/IC50)nH}.
Anticonvulsant assay of conRl-B
The 6 Hz partial psychomotor seizure test was performed to assess the anticonvulsant potential conRl-B as described previously (41). Adult male CF No 1 albino mice (30–35 g) obtained from Charles River, Portage, Michigan, were utilized for behavioral seizure testing in the 6 Hz model of partial psychomotor seizure activity following i.c.v. administration of conRl-B. Stock solutions of the peptide were prepared in 0.9% saline and were diluted to the required concentration prior to intracerebroventricular (i.c.v.) injections. For i.c.v. administration, the test solution was administered in a volume of 5 µL, using a Hamilton syringe (size number 701), directly through the skull into a lateral ventricle of the brain at a depth of 3 mm. A 6 Hz current of 32 mA was administered via corneal electrodes for 3 s in order to elicit a partial psychomotor seizure. Animals not displaying behavioral seizure activity, characterized by an initial momentary stun followed immediately by forelimb clonus, twitching of the vibrissae, and Straub tail, were considered “protected”.
Circular Dichroism Spectroscopy
Circular dichroism spectra were recorded on an AVIV Model 62D spectropolarimeter, using the method and parameters described in the CD studies of conRl-A (35, 36). Briefly, peptides were dissolved at 100 µM final concentration in 10 mM HEPES buffer, pH 7.0, containing with or without 2 mM CaCl2 and measurements were taken at room temperature. Subtracting the peptide CD signal with that of the buffer alone CD signal eliminated the contribution of buffer to the peptide CD signal. The spectral intensities were expressed as mean residue elipticities using the equation reported elsewhere (34) and molar ellipticity of −33530.78 degrees cm2 dmol−1 was estimated to be a perfect α-helix (100% α-helix). The percent helical conformation was calculated by assuming a linear relationship in comparison with 100% α-helix. Estimate of percent of helical conformation induced by divalent calcium to conRl-B was calculated by subtracting the percent of peptide helical content with calcium to that of peptide helical content in the absence of calcium.
RESULTS AND DISCUSSION
Molecular cloning, sequence prediction and synthesis
Two C. rolani gene sequences encoding peptide precursors with a high degree of homology to other members of the conantokin family were cloned and designated conRl-B and conRl-C. The predicted peptide precursor and mature toxin sequences corresponding to the open reading frame of conRl-B and conRl-C are shown in Fig 1 and compared to the previously elucidated sequences for conRl-A, and conG. As predicted, the propeptide regions of conRl-B and conRl-C are highly conserved with respect to other conantokin sequences (Fig. 1A).
Figure 1.
Predicted amino acid sequences of ConRl-B and conRl-C Predicted translated sequences from genomic DNA are shown for the pre/propeptide (upper panel, A) and mature toxin regions (lower panel, C) of conRl-B and ConRl-C, aligned to the sequences of conRl-A and conG for comparison. Shading indicates residues conserved among the four sequences. Two potential mature sequences predicted for conRl-B (C). The proline that may undergo post-translational modification to hydroxyproline is highlighted in bold. O denotes hydroxyproline; γ denotes gamma-carboxyglutamate, and # denotes C-terminal amidation.
Remarkably, when aligned optimally there was a high degree of similarity between the predicted mature peptide sequences of conRl-B or conRl-C and conG (65% of conG AA identical); this was in striking contrast to a comparison of conRl-B or conRl-C to conRl-A (Gowd et al., 2010) from the same species (only 17% of conRl-B and conRl-C AA identical, the majority of these being Gla residues). (Fig1B). Due to the high degree of similarity to venom-purified conG, conRl-B and conRl-C were predicted to have a similar pattern of post-translational modification: a Gla at positions 3–4 and Gla every 3–4 amino acids after, in addition to an amidated C-terminus. Interestingly, the presence of proline in position 10 in conRl-B was a novel structural feature, but given the high degree of posttranslational modifications in Conus peptides including 4-hydroxyproline (Hyp), we predicted that this proline is likely hydroxylated; conantokins from C. parius contain Hyp residues, though not at the homologous position (34).
Chemical synthesis of the predicted mature sequences of both peptides from C. rolani, was performed on a solid support as described under Materials and Methods. γ-Carboxyglutamate residues were coupled in all positions where there was a Glu codon in the corresponding mature toxin derived from the cDNA clone (except for Glu2, which is never posttranslationally modified). Given that the presence of Hyp was based on a less secure prediction, we also synthesized the conRl-B analog containing Pro10 instead of Hyp10. The HPLC elution of the purified conRl-B and conRl-C are shown in Figure S1. Mass spectrometry results, summarized in Table S2, were consistent with the predicted sequence of the synthetic peptides.
Electrophysiological characterization
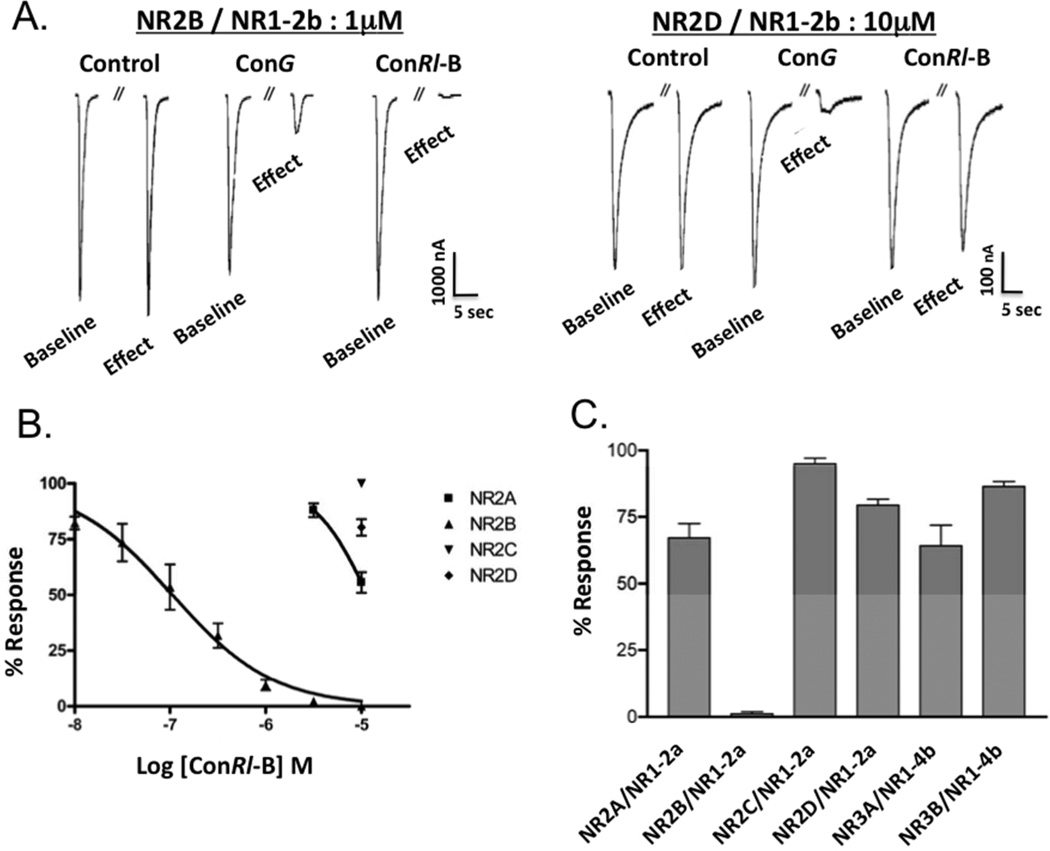
ConRl-B and conRl-C were assessed for antagonist activity on the heterologous expression of an array of NMDA receptor subtypes in Xenopus oocytes, using two-electrode voltage-clamp electrophysiology (see Methods). Figure 2A depicts agonist-elicited current traces from NMDA receptors expressing the NR2B and NR2D subunits for conRl-B. ConRl-B blocked the current in NR2B-containing NMDA receptors at 1 µM more completely than did conG (left panel). Dose-response experiments for conRl-B (Figure 2B) yielded IC50=0.1 µM for blocking NR2B. Strikingly, at the highest concentration tested (10 µM) conRl-B had little or no antagonist activity on three of the four NR2 subunits when co-expressed with NR1–2b, including NR2C and NR2D for which conG has IC50 of 1 µM (Figure 2b, right panel). Thus, conRl-B discriminated at least 100-fold between NR2B and all other NR2 subunits. As shown in Figure S2 and summarized in Table 2, ConRl-C was significantly less selective than conRl-B in blocking NMDARs containing NR2B subunits.
Figure 2.
NMDA receptor subtype selectivity of conRl-B. (A) Current traces from Xenopus oocytes expressing heterologous NR1-2b/NR2B and NR1-2b/NR2D, respectively. ConRl-B blocks most of the agonist-elicited current in oocytes expressing NR1-2b/NR2B (left) but only weakly blocks NR1-2b/NR2D (right). (B) Concentration response curves for conRl-B tested against the four NR2 NMDA receptor subtypes. Data points represent normalized peak current ± SEM from a minimum of 3 oocytes. (C) Normalized current responses of NR1-2a/NR2 and NR1-4b/NR3 subunit combinations, in response to 10 µM conRl-B.
Table 2.
IC50 values for conRl-B and its analogs determined using heterologous expression of four NMDA receptor subtypes expressed in Xenopus oocytes.
| IC50 (µM) | ||||
|---|---|---|---|---|
| Peptide | NR2A | NR2B | NR2C | NR2D |
| ConRl-B | ~10 | 0.1 | >10 | >10 |
| ConRl-B[L5Y] | >10 | 0.12 | >10 | >10 |
| ConRl-B[O10A] | >10 | 0.94 | >10 | >10 |
| ConRl-B[desO10] | >10 | 2.17 | >10 | >10 |
| ConRl-B[K8Nle] | 0.55 | 0.04 | >10 | >10 |
| ConRl-B[desKAO;N8Q9] | >10 | >10 | >10 | >10 |
| ConRl-C | 2.9 | 1.4 | >10 | >10 |
| ConGa | >10 | 0.1 | 1 | 1 |
values reported from (34)
As the potency of conantokins has been reported to vary as a function of the presence or absence of the N-terminal exon (exon 5) in the NR1 subunit (39, 40), the potency of conRl-B was also assessed using oocytes expressing the NR1–2a splice variant in combination with each of the four NR2 subunits (Fig. 2c). Similar to the effects seen on NMDA receptor subtypes expressing the exon 5-containing splice variant, NR1–2b, 10 µM conRl-B had little or no potency on any NR1–2a-containing subtypes, with the exception of NR1–2a/NR2B.
NR3 subunits have been reported to form a functional glycine receptor when expressed in Xenopus oocytes in combination with NR1 subunits (42), conRl-B was also assessed for potency on the NR1/NR3A and NR1/NR3B subtypes. As shown in Figure 2c, 10 µM conRl-B showed little or no potency on either of the NR1/NR3 subtypes tested. Thus, conRl-B is the most selective conantokin for NR2B-containing NMDA receptors characterized to date.
Anticonvulsant assay of conRl-B
Conantokins have anticonvulsant activity (reviewed in (15)); given the high subtype selectivity of conRl-B for NR2B, this peptide was assessed for activity using the 6 Hz partial psychomotor seizure test in mice. At a dose of 0.1 nmol following intracerebroventricular injection (i.c.v.) 50% of mice were protected (n=8) from seizures at time to peak effect (TPE) 1 hour, whereas no control mice (n=8) were protected (5 µl saline, i.c.v.). The rectal body temperature measured at 1 hour (TPE) did not differ between groups.
Determinants of NR2B selectivity
Comparing sequences of conRl-B and conG points to striking structural differences in the second inter-Gla fragments (Fig. 3). Indeed, the presences of either Pro10 (Hyp10) or a positively charged residue in position 8 (Lys8) are sequence features not reported for any of the conantokins characterized so far. This prompted us to examine whether the second inter-Gla loop might contain key determinants for the high subtype selectivity of conRl-B. We designed and synthesized SAR analogs in which Pro10 was either deleted (resulting in making the size of the inter-Gla loop similar to that of conG) or replaced by Ala (Fig. 3). In addition, we assessed the role of the positively charged Lys8 adjacent to Gla7 with an analog containing norleucine in this position (K8Nle). To examine the effect of Pro10 hydroxylation, we synthesized Hyp10Pro analog. Lastly, we substituted the residues found in the second inter-Gla loop of conG for those in conRl-B (desKAO; N8Q9). All analogs were chemically synthesized and tested on NMDARs containing different NR2 subunits.
Figure 3.
Sequences of native ConRl-B and its analogs. Shaded boxed region indicates region of peptide that primary sequence analysis suggests is important for the selectivity profile of conRl-B. a γ denotes gamma-carboxyglutamic acid; b # denotes C-terminal amidation; c O denotes 4-trans-hydroxyproline; X denotes L-norleucine.
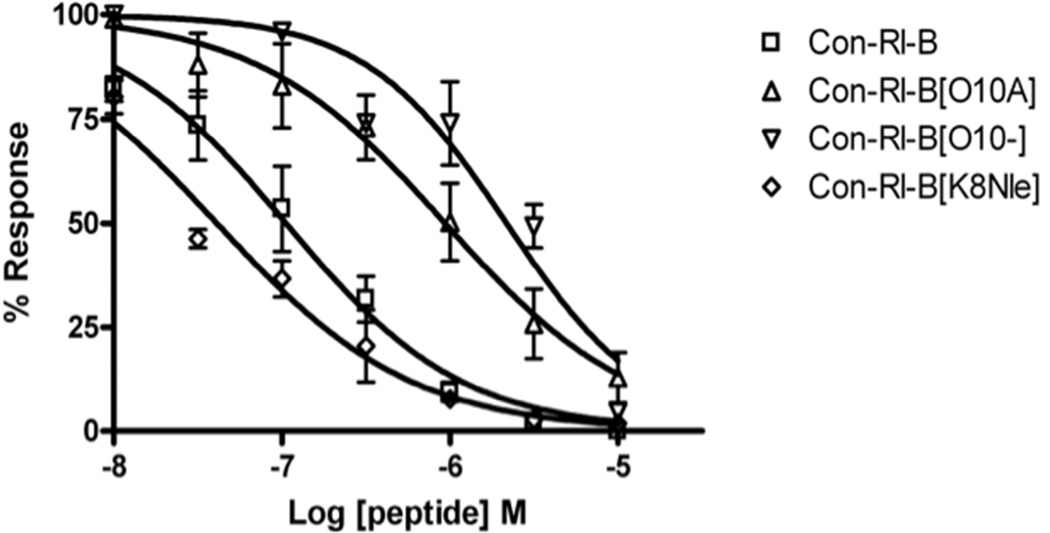
Dose-response studies for SAR analogs are summarized in Figure 4 and Table 2. The potencies of both conRl-B[O10A] and conRl-B[desO10] in blocking NR2B decreased by more than 20-fold, suggesting that this residue is an important determinant for activity. Interestingly, the Lys8Nle replacement did not affect the peptide’s ability to block NR2B, but increased the potency for NR2A-containing NMDA receptors, indicating that the ε-amino group of Lys8 is important for selectivity. No significant difference to conRl-B was observed for conRl-B containing Pro10 instead of Hyp10 (Figure S4). Surprisingly, conRl-B [desKAO; N8Q9] showed little or no activity on any of the NMDA receptor subtypes tested, further indicating that the residues found in the second inter-Gla loop are highly important for the activity of conRl-B. In addition, we also evaluated how the naturally-occurring Gla-to-Lys replacements in conantokins (see Table 1) may affect the potency of conRl-B in blocking NR2B-containing NMDA receptors. The potency of ConRl-B[γ7K] and ConRl-B[γ15K] were IC50= 0.12 µM and IC50= 0.68 µM, respectively, suggesting that this replacement has little effect. Lastly, we tested whether Leu5 is an important determinant of selectivity in conRl-B; to this end, we synthesized and tested a Tyr5 variant of conRl-B (L5Y). Interestingly, and in contrast with data from Tyr5 substitutions in other conantokins (20, 22), conRl-B [L5Y] showed potencies on the four NR2 NMDA receptor subtypes that were very similar to native conRl-B.
Figure 4.
Concentration response curves of ConRl-B analogs on NR2B/NR1-2b, compared to native ConRl-B. Potency is decreased by O10A and O10-, but not by K8Nle. Sequences of ConRl-B and variants are shown in Figure 3. Each data point represents the average peak current, normalized to baseline from a minimum of three oocytes. Error bars represent SEM.
Structural characterization of conRl-B
The characteristic structural feature of conantokins is their helical conformation. Most conantokins adopt a helical conformation in the presence of divalent cations, which aligns the Gla residues to stabilize the helical conformation. A few conantokins, such as conPr-C, conP and conRl-A, are inherently helical peptides (22, 34–36, 43–46). For example, conG is unstructured in the absence of divalent cations (i.e., calcium) and adopts helical conformation in the presence of divalent cations representing a characteristic metal-dependent helical transition in many conantokin peptides. The metal dependent helical transition in conG is attributed to Gla residues chelating calcium by tetravalent interaction, thereby restricting the conformation of the peptide and favoring helix formation (47).
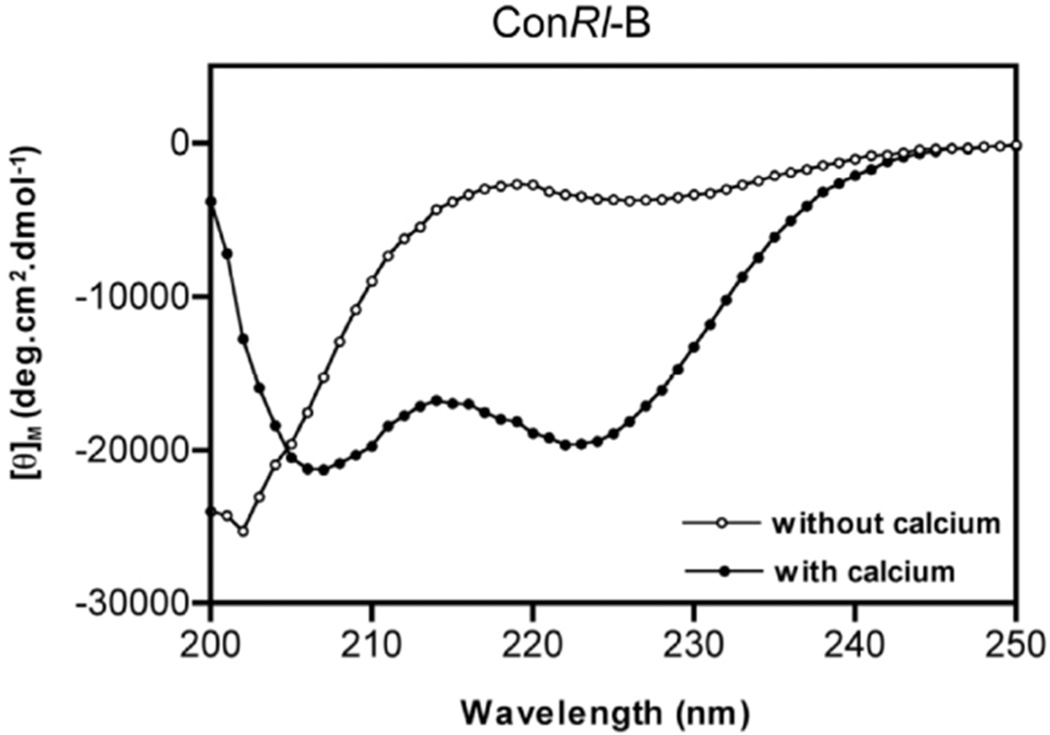
Given the sequence similarities and presence of an identical number and distribution of Gla residues in conRl-B compared to that of conG, we hypothesized that ConRl-B was structurally similar to conG. We employed circular dichroism spectroscopy to study the effect of divalent cations in inducing the helical conformation to conRl-B. Figure 5 shows CD spectra of conRl-B in the presence and absence of Ca2. +. ConRl-B is unstructured in the absence of calcium and adopts a helical conformation in the presence of calcium, a feature similar to that of conG. The estimated helical content of conRl-B in the presence of calcium is 59% (Con-G is 57%) (34). The percent of helical transition induced by calcium in conRl-B is 49% and that of conG is 44% (34). CD spectra of conRl-B[O10P] in the presence and absence of calcium (Figure S3) show that this analog is unstructured in the absence of calcium and adopts a helical conformation in the presence of calcium, similar to conRl-B. The estimated helical content of ConRl-B[O10P]-B in the presence of calcium is 51% and percent of helical transition induced by calcium is 40%. Comparison of the CD spectra of conRl-B and conRl-B[O10P]-B suggest that they have similar helical content in the absence of calcium.
Figure 5.
Circular dichroism spectra of ConRl-B. Spectra were recorded with (or) without 2mM CaCl2 containing 10 mM HEPES buffer at pH 7.0 and shown is an average spectra obtained from five independent scans (n=5). The dual minima at 208 and 222nm, in the presence of calcium, suggest that ConRl-B adopts helical conformation. Estimated percentage of helicity of peptide in the absence of calcium is 10% and in the presence of calcium is 59%.
CONCLUSION
We describe the characterization of a novel NMDA antagonist that is highly selective for NMDA receptors containing NR2B subunits and exhibits anticonvulsant activity. Two novel sequence features, the presence of a positively charged Lys residue in position 8 and Hyp in position 10 contribute to the potency and selectivity of this peptide. Prior to this report, ConG has been regarded as the most NR2B selective member of the conantokin superfamily (17, 34); however, some reports suggest that conG is more broadly selective (48, 49). ConG is reported to have biphasic effects and at least two binding sites on NR2A receptor subtypes (50). Some differences in conG pharmacology have also been attributed to variations in NR1 splicing, in particular exon 5 (i.e., (40)). In this work, we have assessed conRl-B for potency towards all four NR2 subunits in combination with either NR1a or NR1b. In all cases, conRl-B maintains high a high degree of selectivity for NR2B. Thus, conRl-B is an important subtype pharmacological tool for dissecting the role of NMDARs in the nervous system.
Supplementary Material
ACKNOWLEDGEMENTS
We greatly appreciate long-time support of HSC DNA/Peptide Synthesis and Mass Spectrometry Cores. We would like to thank the reviewers for suggesting synthesis and characterization of additional SAR analogs of conRl-B.
This work was supported by a program project GM48677 from the National Institute of General Medical Sciences. KHG acknowledges support from the INSPIRE Faculty Fellowship. HSW acknowledges support from N01-NS-4-2359.
Footnotes
Conflict of Interest disclosure: GB and HSW are scientific cofounders of NeuroAdjuvants, Inc.
SUPLEMENTAL INFORMATION
Selected examples of antagonists for NMDARs containing NR2B subunit (Table S1), purity, HPLC retention times and mass spectrometry results for conantokins studied in this work (Table S2), concentration-response curve of ConRl-C (Figure S2) and ConRl-B[O10P] (Figure S4) on the four different NR2 subunits of NMDA receptor separately co-expressed with NR1-2b in Xenopus oocytes, circular dichroism spectroscopy of ConRl-B[O10P] (Figure S3). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Chenard BL, Menniti FS. Antagonists selective for NMDA receptors containing the NR2B subunit. Current pharmaceutical design. 1999;5:381–404. [PubMed] [Google Scholar]
- 2.Chizh BA, Headley PM, Tzschentke TM. NMDA receptor antagonists as analgesics: focus on the NR2B subtype. Trends in pharmacological sciences. 2001;22:636–642. doi: 10.1016/s0165-6147(00)01863-0. [DOI] [PubMed] [Google Scholar]
- 3.Fischer G, Mutel V, Trube G, Malherbe P, Kew JN, Mohacsi E, Heitz MP, Kemp JA. Ro 25-6981, a highly potent and selective blocker of N-methyl-D-aspartate receptors containing the NR2B subunit. Characterization in vitro. The Journal of pharmacology and experimental therapeutics. 1997;283:1285–1292. [PubMed] [Google Scholar]
- 4.Gill R, Alanine A, Bourson A, Buttelmann B, Fischer G, Heitz MP, Kew JN, Levet-Trafit B, Lorez HP, Malherbe P, Miss MT, Mutel V, Pinard E, Roever S, Schmitt M, Trube G, Wybrecht R, Wyler R, Kemp JA. Pharmacological characterization of Ro 63-1908 (1-[2-(4-hydroxy-phenoxy)-ethyl]-4-(4-methyl-benzyl)-piperidin-4-ol), a novel subtype-selective N-methyl-D-aspartate antagonist. The Journal of pharmacology and experimental therapeutics. 2002;302:940–948. doi: 10.1124/jpet.102.034322. [DOI] [PubMed] [Google Scholar]
- 5.Mosley CA, Acker TM, Hansen KB, Mullasseril P, Andersen KT, Le P, Vellano KM, Brauner-Osborne H, Liotta DC, Traynelis SF. Quinazolin-4-one derivatives: A novel class of noncompetitive NR2C/D subunit-selective N-methyl-D-aspartate receptor antagonists. Journal of medicinal chemistry. 53:5476–5490. doi: 10.1021/jm100027p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mosley CA, Myers SJ, Murray EE, Santangelo R, Tahirovic YA, Kurtkaya N, Mullasseril P, Yuan H, Lyuboslavsky P, Le P, Wilson LJ, Yepes M, Dingledine R, Traynelis SF, Liotta DC. Synthesis, structural activity-relationships, and biological evaluation of novel amide-based allosteric binding site antagonists in NR1A/NR2B N-methyl-D-aspartate receptors. Bioorganic & medicinal chemistry. 2009;17:6463–6480. doi: 10.1016/j.bmc.2009.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa BM, Feng B, Tsintsadze TS, Morley RM, Irvine MW, Tsintsadze V, Lozovaya NA, Jane DE, Monaghan DT. N-methyl-D-aspartate (NMDA) receptor NR2 subunit selectivity of a series of novel piperazine-2,3-dicarboxylate derivatives: preferential blockade of extrasynaptic NMDA receptors in the rat hippocampal CA3-CA1 synapse. The Journal of pharmacology and experimental therapeutics. 2009;331:618–626. doi: 10.1124/jpet.109.156752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng B, Tse HW, Skifter DA, Morley R, Jane DE, Monaghan DT. Structure-activity analysis of a novel NR2C/NR2D–preferring NMDA receptor antagonist: 1-(phenanthrene-2-carbonyl) piperazine-2,3-dicarboxylic acid. British journal of pharmacology. 2004;141:508–516. doi: 10.1038/sj.bjp.0705644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinarsky L, Feng B, Skifter DA, Morley RM, Sherman S, Jane DE, Monaghan DT. Identification of subunit- and antagonist-specific amino acid residues in the N-Methyl-D-aspartate receptor glutamate-binding pocket. The Journal of pharmacology and experimental therapeutics. 2005;313:1066–1074. doi: 10.1124/jpet.104.082990. [DOI] [PubMed] [Google Scholar]
- 10.Morley RM, Tse HW, Feng B, Miller JC, Monaghan DT, Jane DE. Synthesis and pharmacology of N1-substituted piperazine-2,3-dicarboxylic acid derivatives acting as NMDA receptor antagonists. Journal of medicinal chemistry. 2005;48:2627–2637. doi: 10.1021/jm0492498. [DOI] [PubMed] [Google Scholar]
- 11.Acklin P, Allgeier H, Auberson YP, Bischoff S, Ofner S, Sauer D, Schmutz M. 5-Aminomethylquinoxaline-2,3-diones, Part III: Arylamide derivatives as highly potent and selective glycine-site NMDA receptor antagonists. Bioorganic & medicinal chemistry letters. 1998;8:493–498. doi: 10.1016/s0960-894x(98)00055-9. [DOI] [PubMed] [Google Scholar]
- 12.Ametamey SM, Kokic M, Carrey-Remy N, Blauenstein P, Willmann M, Bischoff S, Schmutz M, Schubiger PA, Auberson YP. Synthesis, radiolabelling and biological characterization of (D)-7-iodo-N-(1-phosphonoethyl)-5-aminomethylquinoxaline-2,3-dione, a glycine-binding site antagonist of NMDA receptors. Bioorganic & medicinal chemistry letters. 2000;10:75–78. doi: 10.1016/s0960-894x(99)00576-4. [DOI] [PubMed] [Google Scholar]
- 13.Auberson YP, Acklin P, Bischoff S, Moretti R, Ofner S, Schmutz M, Veenstra SJ. N-phosphonoalkyl-5-aminomethylquinoxaline-2,3-diones: in vivo active AMPA and NMDA(glycine) antagonists. Bioorganic & medicinal chemistry letters. 1999;9:249–254. doi: 10.1016/s0960-894x(98)00720-3. [DOI] [PubMed] [Google Scholar]
- 14.Auberson YP, Allgeier H, Bischoff S, Lingenhoehl K, Moretti R, Schmutz M. 5-Phosphonomethylquinoxalinediones as competitive NMDA receptor antagonists with a preference for the human 1A/2A, rather than 1A/2B receptor composition. Bioorganic & medicinal chemistry letters. 2002;12:1099–1102. doi: 10.1016/s0960-894x(02)00074-4. [DOI] [PubMed] [Google Scholar]
- 15.Layer RT, Wagstaff JD, White HS. Conantokins: peptide antagonists of NMDA receptors. Current medicinal chemistry. 2004;11:3073–3084. doi: 10.2174/0929867043363901. [DOI] [PubMed] [Google Scholar]
- 16.Prorok M, Castellino FJ. The molecular basis of conantokin antagonism of NMDA receptor function. Curr Drug Targets. 2007;8:633–642. doi: 10.2174/138945007780618481. [DOI] [PubMed] [Google Scholar]
- 17.Donevan SD, McCabe RT. Conantokin G is an NR2B-selective competitive antagonist of N-methyl-D-aspartate receptors. Molecular pharmacology. 2000;58:614–623. doi: 10.1124/mol.58.3.614. [DOI] [PubMed] [Google Scholar]
- 18.Sheng Z, Dai Q, Prorok M, Castellino FJ. Subtype-selective antagonism of N-methyl-D-aspartate receptor ion channels by synthetic conantokin peptides. Neuropharmacology. 2007;53:145–156. doi: 10.1016/j.neuropharm.2007.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheng Z, Liang Z, Geiger JH, Prorok M, Castellino FJ. The selectivity of conantokin-G for ion channel inhibition of NR2B subunit-containing NMDA receptors is regulated by amino acid residues in the S2 region of NR2B. Neuropharmacology. 2009;57:127–136. doi: 10.1016/j.neuropharm.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheng Z, Prorok M, Castellino FJ. Specific determinants of conantokins that dictate their selectivity for the NR2B subunit of N-methyl-D-aspartate receptors. Neuroscience. 2010;170:703–710. doi: 10.1016/j.neuroscience.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Twede VD, Miljanich G, Olivera BM, Bulaj G. Neuroprotective and cardioprotective conopeptides: an emerging class of drug leads. Current opinion in drug discovery & development. 2009;12:231–239. [PMC free article] [PubMed] [Google Scholar]
- 22.Twede VD, Teichert RW, Walker CS, Gruszczynski P, Kazmierkiewicz R, Bulaj G, Olivera BM. Conantokin-Br from Conus brettinghami and selectivity determinants for the NR2D subunit of the NMDA receptor. Biochemistry. 2009;48:4063–4073. doi: 10.1021/bi802259a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao C, Huang Y, Dong M, Hu J, Hou S, Castellino FJ, Prorok M, Dai Q. NR2B-selective conantokin peptide inhibitors of the NMDA receptor display enhanced antinociceptive properties compared to non-selective conantokins. Neuropeptides. 2008;42:601–609. doi: 10.1016/j.npep.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malmberg AB, Gilbert H, McCabe RT, Basbaum AI. Powerful antinociceptive effects of the cone snail venom-derived subtype-selective NMDA receptor antagonists conantokins G and T. Pain. 2003;101:109–116. doi: 10.1016/s0304-3959(02)00303-2. [DOI] [PubMed] [Google Scholar]
- 25.Hama A, Sagen J. Antinociceptive effects of the marine snail peptides conantokin-G and conotoxin MVIIA alone and in combination in rat models of pain. Neuropharmacology. 2009;56:556–563. doi: 10.1016/j.neuropharm.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barton ME, White HS, Wilcox KS. The effect of CGX-1007 and CI-1041, novel NMDA receptor antagonists, on NMDA receptor-mediated EPSCs. Epilepsy research. 2004;59:13–24. doi: 10.1016/j.eplepsyres.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 27.Bialer M. New antiepileptic drugs currently in clinical trials: is there a strategy in their development? Therapeutic drug monitoring. 2002;24:85–90. doi: 10.1097/00007691-200202000-00015. [DOI] [PubMed] [Google Scholar]
- 28.Bialer M, Johannessen SI, Kupferberg HJ, Levy RH, Loiseau P, Perucca E. Progress report on new antiepileptic drugs: a summary of the Sixth Eilat Conference (EILAT VI) Epilepsy research. 2002;51:31–71. doi: 10.1016/s0920-1211(02)00106-7. [DOI] [PubMed] [Google Scholar]
- 29.Han TS, Teichert RW, Olivera BM, Bulaj G. Conus Venoms -A Rich Source of Peptide-Based Therapeutics. Current pharmaceutical design. 2008;14:2462–2479. doi: 10.2174/138161208785777469. [DOI] [PubMed] [Google Scholar]
- 30.Olivera BM. Conus peptides: biodiversity-based discovery and exogenomics. The Journal of biological chemistry. 2006;281:31173–31177. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- 31.Teichert RW, Olivera BM. Natural products and ion channel pharmacology. Future medicinal chemistry. 2:731–744. doi: 10.4155/fmc.10.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santos AD, McIntosh JM, Hillyard DR, Cruz LJ, Olivera BM. The A-superfamily of conotoxins: structural and functional divergence. The Journal of biological chemistry. 2004;279:17596–17606. doi: 10.1074/jbc.M309654200. [DOI] [PubMed] [Google Scholar]
- 33.Bulaj G. Integrating the discovery pipeline for novel compounds targeting ion channels. Current opinion in chemical biology. 2008;cr12:441–447. doi: 10.1016/j.cbpa.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teichert RW, Jimenez EC, Twede V, Watkins M, Hollmann M, Bulaj G, Olivera BM. Novel Conantokins from Conus parius Venom Are Specific Antagonists of N-Methyl-D-aspartate Receptors. The Journal of biological chemistry. 2007;282:36905–36913. doi: 10.1074/jbc.M706611200. [DOI] [PubMed] [Google Scholar]
- 35.Gowd KH, Twede V, Watkins M, Krishnanb KS, Teichert RW, Bulaj G, Olivera BM. Conantokin-P, an Unusual Conantokin with a Long Disulfide Loop. Toxicon in press. 2008 doi: 10.1016/j.toxicon.2008.04.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gowd KH, Watkins M, Twede VD, Bulaj GW, Olivera BM. Characterization of conantokin Rl-A: molecular phylogeny as structure/function study. J Pept Sci. 16:375–382. doi: 10.1002/psc.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laurie DJ, Seeburg PH. Ligand affinities at recombinant N-methyl-D-aspartate receptors depend on subunit composition. European journal of pharmacology. 1994;268:335–345. doi: 10.1016/0922-4106(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 38.Laurie DJ, Seeburg PH. Regional and developmental heterogeneity in splicing of the rat brain NMDAR1 mRNA. J Neurosci. 1994;14:3180–3194. doi: 10.1523/JNEUROSCI.14-05-03180.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klein RC, Prorok M, Galdzicki Z, Castellino FJ. The amino acid residue at sequence position 5 in the conantokin peptides partially governs subunit-selective antagonism of recombinant N-methyl-D-aspartate receptors. The Journal of biological chemistry. 2001;276:26860–26867. doi: 10.1074/jbc.M102428200. [DOI] [PubMed] [Google Scholar]
- 40.Klein RC, Warder SE, Galdzicki Z, Castellino FJ, Prorok M. Kinetic and mechanistic characterization of NMDA receptor antagonism by replacement and truncation variants of the conantokin peptides. Neuropharmacology. 2001;41:801–810. doi: 10.1016/s0028-3908(01)00119-8. [DOI] [PubMed] [Google Scholar]
- 41.Bulaj G, Green BR, Lee HK, Robertson CR, White K, Zhang L, Sochanska M, Flynn SP, Scholl EA, Pruess TH, Smith MD, White HS. Design, synthesis, and characterization of high-affinity, systemically-active galanin analogues with potent anticonvulsant activities. Journal of medicinal chemistry. 2008;51:8038–8047. doi: 10.1021/jm801088x. [DOI] [PubMed] [Google Scholar]
- 42.Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, Cui J, Tu S, Sevarino KA, Nakanishi N, Tong G, Lipton SA, Zhang D. Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature. 2002;415:793–798. doi: 10.1038/nature715. [DOI] [PubMed] [Google Scholar]
- 43.Prorok M, Warder SE, Blandl T, Castellino FJ. Calcium binding properties of synthetic gamma-carboxyglutamic acid-containing marine cone snail “sleeper” peptides, conantokin-G and conantokin-T. Biochemistry. 1996;35:16528–16534. doi: 10.1021/bi9621122. [DOI] [PubMed] [Google Scholar]
- 44.Lin CH, Chan FC, Hwang JK, Lyu PC. Calcium binding mode of gamma-carboxyglutamic acids in conantokins. Protein engineering. 1999;12:589–595. doi: 10.1093/protein/12.7.589. [DOI] [PubMed] [Google Scholar]
- 45.Rigby AC, Baleja JD, Li L, Pedersen LG, Furie BC, Furie B. Role of gamma-carboxyglutamic acid in the calcium-induced structural transition of conantokin G, a conotoxin from the marine snail Conus geographus. Biochemistry. 1997;36:15677–15684. doi: 10.1021/bi9718550. [DOI] [PubMed] [Google Scholar]
- 46.Chen Z, Blandl T, Prorok M, Warder SE, Li L, Zhu Y, Pedersen LG, Ni F, Castellino FJ. Conformational changes in conantokin-G induced upon binding of calcium and magnesium as revealed by NMR structural analysis. The Journal of biological chemistry. 1998;273:16248–16258. doi: 10.1074/jbc.273.26.16248. [DOI] [PubMed] [Google Scholar]
- 47.Cnudde SE, Prorok M, Dai Q, Castellino FJ, Geiger JH. The crystal structures of the calcium-bound con-G and con-T[K7gamma] dimeric peptides demonstrate a metal-dependent helix-forming motif. Journal of the American Chemical Society. 2007;129:1586–1593. doi: 10.1021/ja065722q. [DOI] [PubMed] [Google Scholar]
- 48.Wittekindt B, Malany S, Schemm R, Otvos L, Maccecchini ML, Laube B, Betz H. Point mutations identify the glutamate binding pocket of the N-methyl-D-aspartate receptor as major site of conantokin-G inhibition. Neuropharmacology. 2001;41:753–761. doi: 10.1016/s0028-3908(01)00112-5. [DOI] [PubMed] [Google Scholar]
- 49.Alex AB, Saunders GW, Dalpe-Charron A, Reilly CA, Wilcox KS. CGX-1007 prevents excitotoxic cell death via actions at multiple types of NMDA receptors. Neurotoxicology. 32:392–399. doi: 10.1016/j.neuro.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ragnarsson L, Yasuda T, Lewis RJ, Dodd PR, Adams DJ. NMDA receptor subunit-dependent modulation by conantokin-G and Ala7-conantokin-G. Journal of neurochemistry. 2006;96:283–291. doi: 10.1111/j.1471-4159.2005.03574.x. [DOI] [PubMed] [Google Scholar]
- 51.McIntosh JM, Olivera BM, Cruz LJ, Gray WR. Gamma-carboxyglutamate in a neuroactive toxin. The Journal of biological chemistry. 1984;259:14343–14346. [PubMed] [Google Scholar]
- 52.Haack JA, Rivier J, Parks TN, Mena EE, Cruz LJ, Olivera BM. Conantokin-T. A gamma-carboxyglutamate containing peptide with N-methyl-d-aspartate antagonist activity. The Journal of biological chemistry. 1990;265:6025–6029. [PubMed] [Google Scholar]
- 53.White HS, McCabe RT, Armstrong H, Donevan SD, Cruz LJ, Abogadie FC, Torres J, Rivier JE, Paarmann I, Hollmann M, Olivera BM. In vitro and in vivo characterization of conantokin-R, a selective NMDA receptor antagonist isolated from the venom of the fish-hunting snail Conus radiatus. The Journal of pharmacology and experimental therapeutics. 2000;292:425–432. [PubMed] [Google Scholar]
- 54.Jimenez EC, Donevan S, Walker C, Zhou LM, Nielsen J, Cruz LJ, Armstrong H, White HS, Olivera BM. Conantokin-L, a new NMDA receptor antagonist: determinants for anticonvulsant potency. Epilepsy research. 2002;51:73–80. doi: 10.1016/s0920-1211(02)00101-8. [DOI] [PubMed] [Google Scholar]
- 55.Gowd KH, Twede V, Watkins M, Krishnan KS, Teichert RW, Bulaj G, Olivera BM. Conantokin-P, an unusual conantokin with a long disulfide loop. Toxicon. 2008;52:203–213. doi: 10.1016/j.toxicon.2008.04.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.