Summary
Mitochondrial fatty acid synthesis (mtFAS) shares acetyl-CoA with the Krebs cycle as a common substrate and is required for the production of octanoic acid (C8) precursors of lipoic acid (LA) in mitochondria. MtFAS is a conserved pathway essential for respiration. In a genetic screen in Saccharomyces cerevisiae designed to further elucidate the physiological role of mtFAS, we isolated mutants with defects in mitochondrial post-translational gene expression processes, indicating a novel link to mitochondrial gene expression and respiratory chain biogenesis. In our ensuing analysis, we show that mtFAS, but not lipoylation per se, is required for respiratory competence. We demonstrate that mtFAS is required for mRNA splicing, mitochondrial translation and respiratory complex assembly, and provide evidence that not LA per se, but fatty acids longer than C8 play a role in these processes. We also show that mtFAS- and LA-deficient strains suffer from a mild heme deficiency that may contribute to the respiratory complex assembly defect. Based on our data and previously published information, we propose a model implicating mtFAS as a sensor for mitochondrial acetyl-CoA availability and a coordinator of nuclear and mitochondrial gene expression by adapting the mitochondrial compartment to changes in the metabolic status of the cell.
Keywords: mitochondrial-nuclear coordination, mitochondrial gene expression, heme, lipoic acid, mitochondrial fatty acid synthesis
Introduction
The presence of the mitochondrial compartment is one of the hallmark features of eukaryotic cells, and there are grounds to argue that the acquisition of an ancestor of α-proteobacteria (Sagan, 1967) was a defining moment of the genesis of eukaryotes (van der Giezen, 2009). Many of the proteins and processes residing in mitochondria still bear witness to the prokaryotic ancestry of these organelles.
As a consequence of the endosymbiotic relationship between the prokaryotic resident and its host, a large fraction of the bacterial genome was ultimately lost or transferred to the nucleus. However, there appears to be a necessity to maintain a core group of genes encoding proteins and several RNA species within the mitochondrial genome (mtDNA) (Gray et al., 1999; Wallace, 2007). Mitochondria have retained the ability to express these genes, devoting a large fraction of the mitochondrial proteome to this task. In the yeast Saccharomyces cerevisiae, eight proteins as well as tRNAs, rRNAs and the RNA subunit of mitochondrial RNase P are still synthesized within mitochondria. With the exception of Var1, a mitoribosomal subunit, all of the proteins encoded by the yeast mitochondrial genome are integral membrane proteins that are core subunits of the oxidative phosphorylation system (OXPHOS) complexes. They assemble with nuclear-encoded subunits to form functional respiratory chain complexes III (cytochrome bc1 complex) and IV (cytochrome c oxidase; COX) and the F1FO ATP synthase or complex V. Regulation of proper subunit stoichiometry is necessary to prevent accumulation of unassembled surplus protein subunits. In mitochondrial enzyme complex assembly, this process requires coordinate expression of components from two different genomes. In no small part, gene expression in mitochondria is governed by the nucleus, which encodes the vast majority of factors involved in this process. How the coordination of gene expression of both genomes is achieved is still not well understood.
Transcription of numerous nuclear genes encoding mitochondrial proteins is regulated by the availability of heme and oxygen and can be downregulated by glucose through repressor-mediated histone deacetylation (Schuller, 2003; Turcotte et al., 2010). There is little regulation of mitochondrial transcription, although its rate is known to increase a few fold during a shift to respiration (Amiott et al., 2006a; Amiott et al., 2006b). In contrast to the elaborate transcriptional control of the nuclear genes, production of mtDNA-encoded proteins is regulated predominantly post-transcriptionally. Regulation involves factors required for RNA processing, stability, translation, and complex assembly. The regulation of mitochondrial protein synthesis by the action of mRNA-specific translational activators in yeast has been a subject of intense study during the past two decades (Herrmann et al., 2013).
For example, it has been postulated that COX1 mRNA is bound by Mss51 and Pet309, which activate translation from this template. Subsequently, Mss51 binds to the protein product and forms a stable Cox1 subassembly complex with other complex IV chaperones such as Cox14, Cox25/Coa3 and the Hsp70 chaperone Ssc1 (Perez-Martinez et al., 2003; Barrientos et al., 2004; Fontanesi et al., 2010; Mick et al., 2010; Fontanesi et al., 2011). The translational activator Mss51 is only released from this complex and then recycled when Cox1 interacts with its assembly partners, thus setting up a feedback translational regulatory loop that prevents gross overproduction of the subunit (Barrientos et al., 2004; Fontanesi et al., 2010).
If the regulation of mitochondrial gene expression was solely dependent on a linear pathway of heme and oxygen activation of nuclear genes encoding mitochondrial post-transcriptional effectors, the process would probably suffer from a delay in the synthesis of mtDNA-encoded subunits of the respiratory complexes compared to the production of nuclear-encoded subunits.
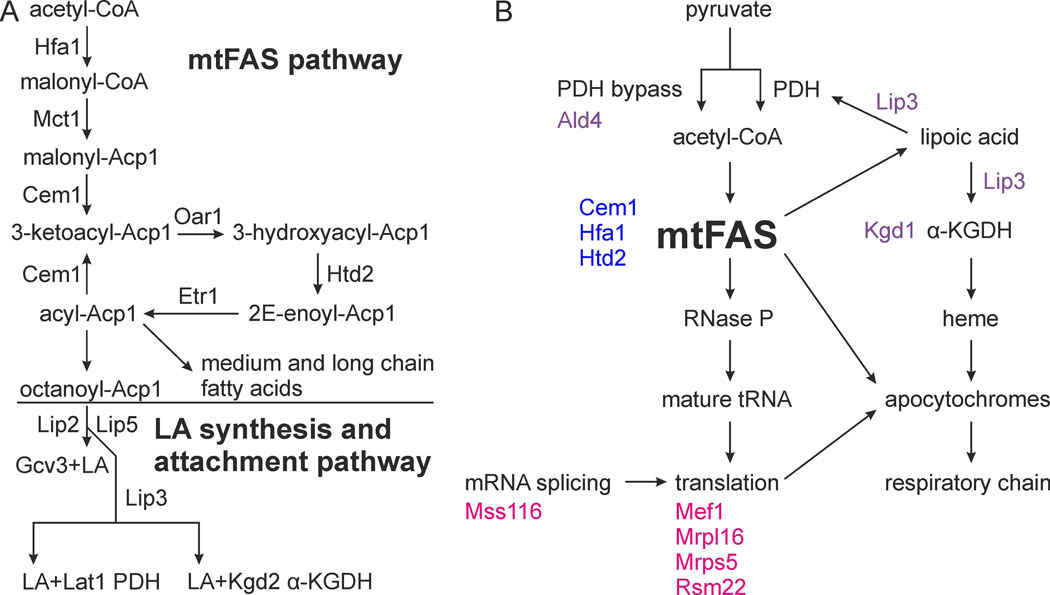
Mitochondrial fatty acid synthesis (mtFAS) is a conserved pathway essential for respiration in yeast. Deletion of mtFAS components results in small, rudimentary mitochondria, while overexpression of enzyme components of the pathway results in a dramatic enlargement of the mitochondrial compartment (Torkko et al., 2001; Kastaniotis et al., 2004). MtFAS has been implicated as the sole mitochondrial source of octanoic acid in eukaryotes. This product is required as a precursor for the synthesis of the lipoic acid (LA) cofactor that is indispensable for the function of several mitochondrial enzyme complexes including: pyruvate dehydrogenase (PDH), α-ketoglutarate dehydrogenase (α-KGDH) and the glycine cleavage system (GCS) (Fig. 1A) (Schonauer et al., 2009; Hiltunen et al., 2010). A deletion of any gene in the mtFAS and LA attachment pathways results in respiratory deficiency.
Fig. 1.
Mitochondrial fatty acid synthesis (mtFAS)/ lipoic acid (LA) synthesis and attachment pathways and synthetic mutants identified in the screen. (A) Schematic depiction of the mtFAS and LA synthesis and attachment pathways. (B) Graphic presentation of the results of the synthetic petite screen. Mutated factors are color-coded to indicate functional groups. Blue: mtFAS enzymes, magenta: post-transcriptional gene expression related proteins, purple: mutations related to LA attachment (Lip3), LA-dependent complexes (Kgd1) or the pyruvate dehydrogenase complex (PDH) bypass (Ald4).
A central metabolite for mitochondrial function is acetyl-CoA, the substrate of the Krebs cycle. Acetyl-CoA is also the substrate for ketogenesis in mammals and for mtFAS in all eukaryotes. In this work, we show that a fatty acid product of the mtFAS pathway that is longer than octanoic acid (C8) is required for mitochondrial biogenesis on multiple levels, and we present a model implicating mtFAS as a hub for mitochondrial sensing of cellular metabolic status and control of important mitochondrial post-transcriptional gene expression events.
Results
Synthetic petite interactions
To investigate possible roles of mtFAS in mitochondrial function other than supplying octanoic acid for LA synthesis (Fig. 1A), we screened for mutations causing a synthetic respiratory-deficient “petite” phenotype in combination with a compromised step in mtFAS. We have previously shown that a plasmid-borne copy of the Escherichia coli fabI gene, encoding a bacterial FAS type II enoyl reductase, fused to a yeast mitochondrial targeting sequence and expressed from a heterologous promoter (CTA1), is capable of rescuing respiratory growth of an etr1Δ strain (Torkko et al., 2001). For the work presented here, the yeast ETR1 ORF was replaced in the yeast genome by fabI under transcriptional control of ETR1 regulatory sequences. The strains (isogenic a and α) carrying this construct (“the FabI strains”) are respiratory competent but display diminished growth on glycerol and a 50 % reduction in lipoic acid content (277 ± 78 ng lipoic acid g−1 wet weight cells compared to 536 ± 101 ng lipoic acid g−1 in wild-type and 19 ± 7 ng lipoic acid g−1 in the etr1Δ negative control strain). Unlike the wild-type W1536 5B/8B strains, the FabI strains do not shift quickly from fermentation to respiration when transferred from glucose to glycerol media (not shown) and are sensitive to the fabI-specific drug triclosan on non-fermentable media (see Supporting Information Fig. S1A and S1B). The latter observation is consistent with our previous report on triclosan inhibition of the mtFabI-plasmid complemented etr1Δ strain (Torkko et al., 2003).
We employed the FabI strains in a colony-color based assay to find ethyl methane sulfonate (EMS) generated mutants unable to lose a plasmid harboring wild-type ETR1 on respiration-requiring glycerol medium, but able to lose the plasmid on medium containing fermentable glucose as the carbon source (Kastaniotis et al., 2004). 18 synthetic petite mutations were identified in 12 different genes (Fig. 1B, Supporting Information Fig. S2, Tables S1 and S2).
The interacting genes can be divided into five different categories: (I) CEM1, HFA1 and HTD2 are members of the mtFAS pathway, (II) MEF1, MRPL16, MRPS5, MSS116 and RSM22 are essential for mitochondrial translation and post-transcriptional gene expression processes, (III) LIP3 and KGD1 are linked to LA metabolism and heme synthesis, (IV) ALD4 is a mitochondrial pyruvate bypass pathway enzyme (Boubekeur et al., 1999), and (V) ASK10 encodes a transcription factor in the oxidative stress response (Cohen et al., 2003). Isolation of mutants of category I, III and IV confirmed the specificity of our screen. The synthetic mutation in ASK10, the sole member of category V, may imply a role of this factor in expression of mtFAS or LA metabolism related genes. Future analysis should shed more light on the role of the Ask10 protein in the regulation of mtFAS.
Because the synthetic mutations were obtained in a strain compromised for enoyl reductase function, initial analysis of effects of mtFAS lesions on mitochondrial gene expression was carried out in the etr1Δ strain background. To rule out gene-specific effects, we later expanded our experiments to include the htd2Δ strain also. As it became clear that these strains displayed virtually identical phenotypes, later analyses were sometimes done with only one of the deletion strains. Triclosan inhibition experiments were only carried out with an etr1Δ control, as triclosan inhibits bacterial enoyl reductase.
Respiratory chain defects in mtFAS deficient strains
Mutants of category II were the most informative in furthering our understanding of the role of mtFAS in mitochondrial respiration. We isolated several mutations in genes involved in mitochondrial post-transcriptional gene expression processes, implicating mtFAS in the production of functional respiratory complexes. While mtFAS mutants had been shown previously to lack mitochondrial cytochromes, they had been reported to have no mitochondrial translation defects (Harington et al., 1994; Yamazoe et al., 1994). In light of isolating category II mutations affecting mitochondrial gene expression in our screen, we decided to reinvestigate the levels of cytochromes and respiratory complexes in strains with lesions in mtFAS and LA attachment processes.
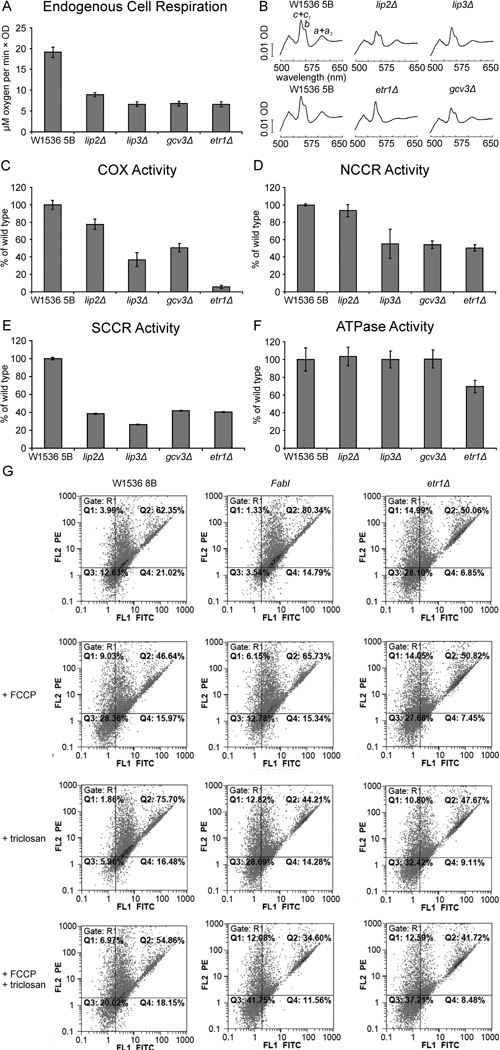
We analyzed the endogenous cell respiration rate (Fig. 2A) and determined the total mitochondrial cytochrome spectra of the knockout mutants (Fig. 2B) and found them to be diminished, which is consistent with the previously observed respiratory-deficient growth phenotype. Accordingly, respiratory chain enzyme activities also were found to be decreased. Cytochrome c oxidase (COX) (Fig. 2C, see also Supporting Information Table S3 for specific activities), NADH cytochrome c reductase (NCCR) (Fig. 2D) and succinate cytochrome c reductase (SCCR) activities (Fig. 2E) were lowered in the LA attachment deficient lip2Δ, lip3Δ and gcv3Δ strains as well as in the mtFAS deficient etr1Δ strain, suggesting a pleiotropic effect in several segments of the mitochondrial respiratory chain. However, COX activity was decreased most in the etr1Δ strain, which had a markedly more severe phenotype than the LA deficient strains (Fig. 2C). The ATP hydrolysis activity of the F1Fo ATPase was lowered only in the etr1Δ strain where it was ~70% of wild-type (Fig. 2F). Notably, it has been suggested recently that the etr1Δ strain experiences decreased mitochondrial membrane potential due to a defect in the F1Fo ATPase (Ytting et al., 2012). We examined the effect of triclosan inhibition on the mitochondrial membrane potential of wild-type W1536 8B, FabI and etr1Δ strains using FACS (fluorescence-activated cell sorting) to monitor the fluorescence signal of the membrane potential-sensitive dye JC-1 (Smiley et al., 1991). Mitochondria from wild-type cells had a high membrane potential as indicated by a high FL2 emission (Fig. 2G, for a more detailed description see Experimental Procedures). In contrast, when the proton translocator FCCP (carbonyl cyanide 4-trifluoromethoxy phenylhydrazone) was added in concert with the JC-1 dye, the FL2 emission was decreased, indicating that membrane potential-dependent mitochondrial uptake of JC-1 was inhibited. The etr1 Δ strain displayed a low FL2 emission, similar to the FCCP-treated wild-type cells, indicating a strong decrease of mitochondrial membrane potential in mtFAS deficient cells. Addition of FCCP to the etr1Δ strain, however, further decreased the FL2 emission, indicating that the membrane potential is not completely abolished in mtFAS deficient yeast strains. The FabI strain was indistinguishable from wild-type when grown in the absence of triclosan. Addition of the drug to the FabI culture gave a result similar to the etr1Δ strain, or to wild-type treated with FCCP, a phenotype suggesting a decrease in mitochondrial membrane potential. The membrane potential drops even further upon FCCP treatment, recapitulating the effect observed in the etr1Δ strain. When triclosan was added to the wild-type strain at the same concentration, it did not affect membrane potential, confirming the specificity of the triclosan effect.
Fig. 2.
Characterization of yeast strains carrying deletion mutations in genes required for lipoic acid synthesis and attachment and mitochondrial fatty acid synthesis deletion. (A) Endogenous cellular respiration, (B) cytochrome spectra, (C) cytochrome c oxidase complex (COX) activity, (D) NADH cytochrome c reductase activity (complex I + III) (NCCR) activity, (E) succinate cytochrome c reductase (complex II + III) (SCCR) activity, and (F) ATPase activity. Data are shown as the mean of three independent repetitions ± SD. (G) Mitochondrial membrane potential in etr1Δ and FabI strains inhibited by triclosan, assessed by measuring JC-1 fluorescence with FACS (fluorescence-activated cell sorting). Orange fluorescence (FL2) indicates high membrane potential, while green fluorescence (FL-1) is observed when the mitochondrial membrane potential is strongly decreased (see Experimental Procedures). FCCP (carbonyl cyanide 3- trifluoromethoxy phenylhydrazone) is an uncoupler that causes dissipation of membrane potential.
Disturbed respiratory complex assembly and translation in mtFAS and LA attachment deficient mutants
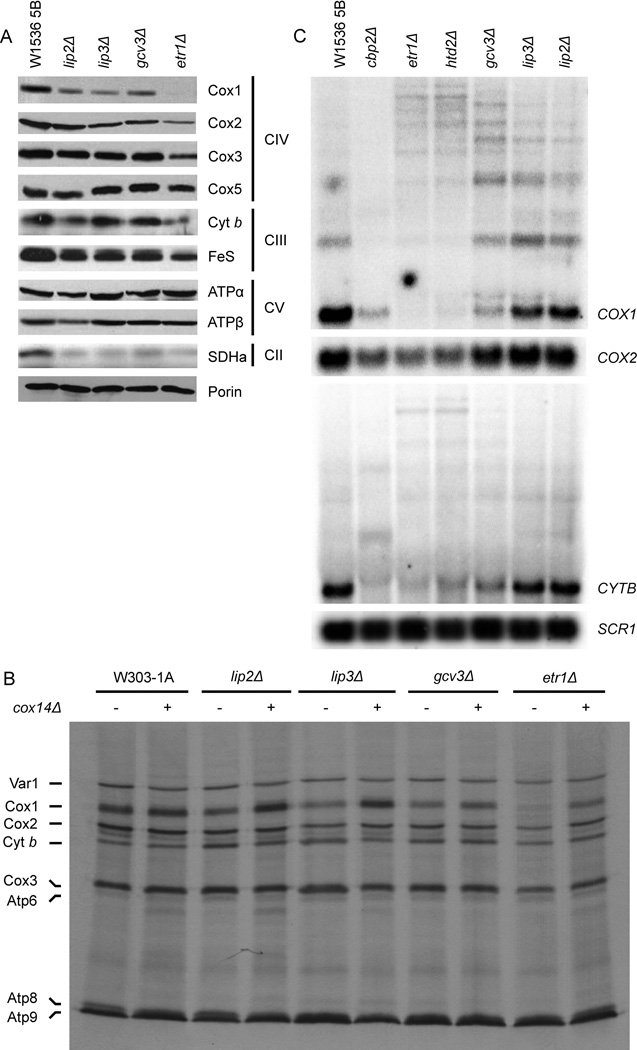
To examine if the reductions in respiration and enzyme activities were due to lowered levels of mitochondrial proteins, mitochondrial extracts (Fig. 3A) were analyzed by western blotting. Steady state levels of COX subunits were indeed specifically reduced in the etr1Δ mutant, but a much less severe phenotype was detected in the LA attachment deficient strains (gcv3Δ, lip2Δ and lip3Δ). In contrast, all the mutants showed similar levels of Cox5, a nuclear-encoded subunit known to be more stable than the mtDNA-encoded subunits when COX assembly is compromised. In addition, the steady-state levels of other components of the mitochondrial respiratory chain, such as Cyt b (a complex III or bc1 complex subunit), were lowered in some of the mutants (Fig. 3A), while the levels of SDHa (Sdh1 subunit of succinate dehydrogenase/complex II) were decreased in all mutants. The F1 subunits of ATPase accumulated at normal levels.
Fig. 3.
Mitochondrial DNA gene expression and OXPHOS enzyme subunit levels in intron-containing mitochondrial fatty acid synthesis mutant strains. (A) Steady state levels of indicated proteins by western blot analysis of isolated mitochondria. Loading control: Porin. (B) De novo translation of mtDNA-encoded proteins. (C) Processing of mitochondrial transcripts. A cpb2Δ strain was used as a non-lipoylation/non-mtFAS-defective respiratory deficient control. Total RNA extracted from the strains was hybridized with a probe complementary to COX1, COX2 and CYTB. Loading control: SCR1, the RNA subunit of the Signal Recognition Particle (SRP), a housekeeping gene.
To distinguish if decreased steady state levels of mitochondrial proteins were due to decreased synthesis or increased turnover, nascent mtDNA-encoded proteins were labeled with 35S-methionine in the presence of cycloheximide to inhibit cytoplasmic protein synthesis. MtFAS and lipoylation-defective strains were able to synthesize all mitochondrial translation products. However, all strains had attenuated levels of newly synthesized Cox1, and only the etr1Δ strain displayed a mild, more generalized, protein synthesis attenuation phenotype (Fig. 3B).
Usually, accumulation of unassembled respiratory chain enzyme subunits is limited by degradation via dedicated proteases (Langer et al., 2001). Additionally, in the case of COX, the accumulation of Cox1 is regulated translationally by a feedback mechanism involving the translational activator Mss51, which plays dual roles in COX1 mRNA translation and Cox1 chaperoning (Perez-Martinez et al., 2003; Barrientos et al., 2004; Fontanesi et al., 2010). Deletion of the COX14 gene relieves this feedback inhibition by allowing Mss51 to dissociate from unassembled Cox1 (Barrientos et al., 2004). We aimed to test whether the Cox1 synthesis attenuation observed in mtFAS mutants is due to a translational defect or is merely a result of downregulation due to defective COX assembly. For this purpose, we generated double mutants by introducing the cox14 deletion into mtFAS and LA deficient mutants. As in bona fide COX assembly mutants, deletion of the COX14 gene in these mutants resulted in a wild-type level of de novo Cox1 synthesis (Fig. 3B). These data indicate that a large part of the observed decrease in Cox1 translation is the consequence of a COX assembly defect.
Mitochondrial mRNA splicing inhibition in mtFAS and lipoylation deficient mutants
Often, even minor defects in translation of Cox1 and Cyt b result in an inhibition of splicing of the multiple introns present in their pre-mRNAs. This is due to the need to translate intron-encoded maturases for intron excision (Mittelmeier et al., 1995). To investigate whether the decrease in translation efficiency affected splicing, RNAs isolated from mtFAS and lipoylation deficient strains were probed for the COX1 and CYTB transcripts to assess transcript abundance. Our results clearly showed mRNA splicing defects of these transcripts (Fig. 3C). While the overall severity of the defect varied with the growth conditions, the splicing deficiency was consistently more pronounced in the etr1Δ and htd2Δ strains, which displayed identical phenotypes. Splicing was less severely affected in the lipoylation deficient strains. In addition to these observations, there is a noticeable decrease in COX2 transcript in the mtFAS deficient strains and the cbp2Δ control strain, but not in strains with specific lipoylation defects. Cbp2 is required for the removal of the CYTB group I intron bI5 (McGraw et al., 1983) and the cbp2Δ deletion mutant, like etr1Δ and htd2Δ strains, suffers from a severe respiratory chain defect.
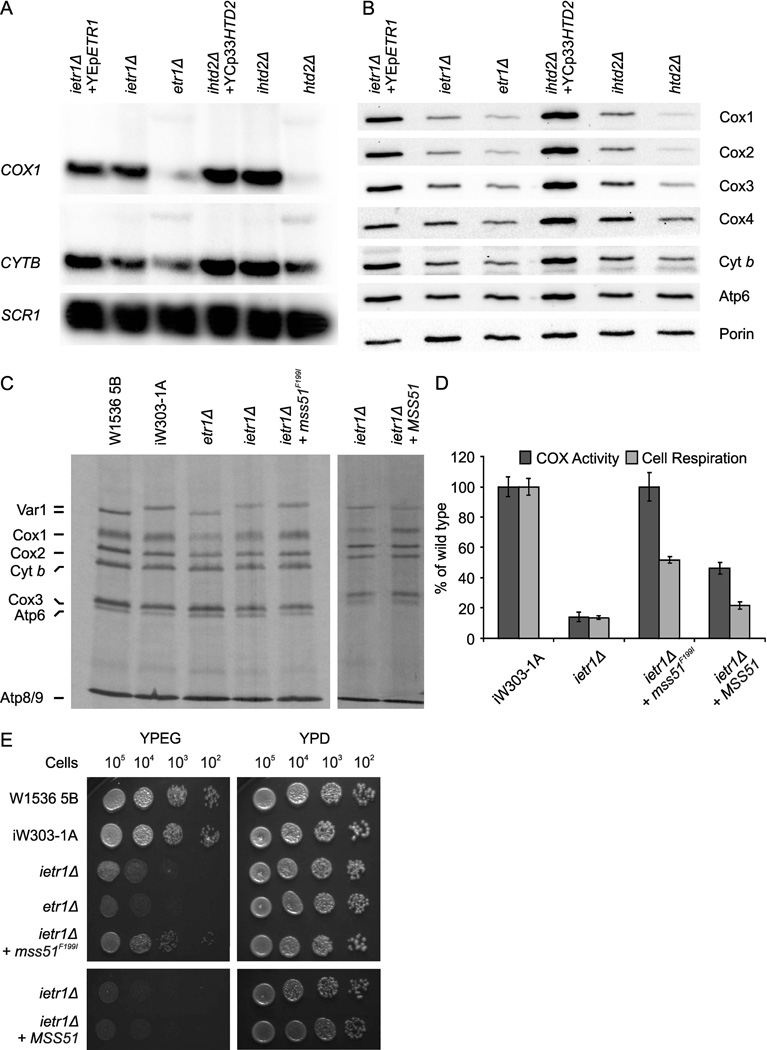
To test if the lack of mature transcripts contributed to the lack of mtDNA-encoded proteins, we generated mtFAS deletion strains harboring mitochondria with an intronless genome in the W1536 5B background (ietr1Δ and ihtd2Δ) (Fig. 4A). We indeed detected reproducible improvement of Cox1 steady-state levels in the intronless mtFAS deficient strains (Fig. 4B), as well as a mild improvement of de novo Cox1 synthesis (Fig. 4C). A slightly larger variant of Var1 was present in all strains carrying the intronless mitochondrial genome in comparison to the W303 derivatives otherwise used in this work. This is due to the presence of a different VAR1 allele in the mitochondrial genome of kar1 MCC109I (α) strain (Ellis et al., 2004), which was used as the intronless mtDNA donor in the generation of the intronless strains.
Fig. 4.
Mitochondrial gene expression, OXPHOS enzyme subunit levels, COX activity, respiration and growth of intronless mtFAS mutant strains. (A) Processing of mitochondrial transcripts in mitochondrial fatty acid synthesis deletion strains with or without mitochondrial introns. Total RNA extracted from the strains was hybridized with a probe complementary to COX1, COX2 or CYTB. Loading control: SCR1. (B) Analysis of the steady state levels of indicated proteins from mitochondrial extracts by western blot. Loading control: Porin. (C) De novo protein synthesis in intronless and control strains. (D) Cytochrome oxidase activity measurements and cell respiration in intron-containing and intronless W1536 5B etr1Δ strains and the ietr1Δ strain expressing one extra copy of either mss51F199I or wild-type MSS51. Endogenous cell respiration was measured polarographically. Data are shown as the mean of three independent repetitions ± SD. (E) Dilution series testing for growth of strains analyzed in panels C and D on non-fermentable and fermentable carbon sources.
To evaluate the impact of the COX1 mRNA splicing defect on the COX deficient phenotype in etr1 mutant yeast, we examined de novo translation and COX activities in intron-containing and intron-less strains. We detected a mild increase in both Cox1 synthesis (Fig. 4C) and COX activity (from ~ 4 to 13% of wild-type (Fig. 4D, see also Supporting Information Table S4 for specific activities) in ietr1Δ. To further assess the contribution of COX1 mRNA translational regulation to the COX deficient phenotype in etr1 mutant yeast, ietr1Δ was transformed with a plasmid expressing either wild-type or a functional mutant variant of the COX1 mRNA translational activator Mss51 (mss51F199I). We previously reported that a single copy of the mutant allele mss51F199I or additional copies of wild-type MSS51 partially suppress the respiratory defect caused by mutations in SHY1, a COX assembly factor, by increasing Cox1 synthesis (Barrientos et al., 2002). In most COX-deficient strains, mutant or additional copies of MSS51 do not suppress the COX assembly defect, but significantly increase Cox1 synthesis via bypassing the negative feedback regulation (Barrientos et al., 2004; Fontanesi et al., 2010). The restoration of Cox1 synthesis by excess Mss51 or by mss51F199I is due to its increased intrinsic capacity to avoid being trapped in a complex containing newly synthesized Cox1 when COX assembly is defective (Fontanesi et al., 2010). More recently, we have reported that Mss51 is a heme binding protein able to sense heme B levels to promote Cox1 synthesis and assembly (Soto et al., 2012). Notably, mss51F199I can perform its functions in the absence of heme (Soto et al., 2012) and thus it is ideal to test whether Cox1 translation is down-regulated in the absence of Etr1. Our results show that the additional heme-dependent wild-type Mss51 significantly restores Cox1 synthesis and increases COX activity (to 46% of wild type) and respiration (to 20%) in ietr1Δ. However, this level was insufficient to enhance respiratory growth on rich glycerol plates (Fig. 4E). In contrast, the heme-independent mss51F199I allele fully restored Cox1 synthesis and COX activity in ietr1Δ resulting in an increase of respiration to 50% of the wild-type value and a partial rescue of respiratory growth on plates (Fig. 4E).
Lipoylation defects and PDH bypass
Yeast mitochondria and probably also the mitochondria of higher eukaryotes exclusively utilize internally synthesized LA (Hiltunen et al., 2010). Since the level of LA in the FabI strain was only ~50% of that in wild-type, we expected that the synthetic petite screen would identify factors involved in LA metabolism. Lip2, Lip3, Lip5 and Gcv3 are all required for the synthesis and attachment of LA to subunits of PDH, α-KGDH, and GCS (see Fig. 1A). Thus, isolation of the lip3 and kgd1 mutants of category III as well as the ald4-1 mutation, which disables the PDH bypass, provides further evidence for the specificity of our screen.
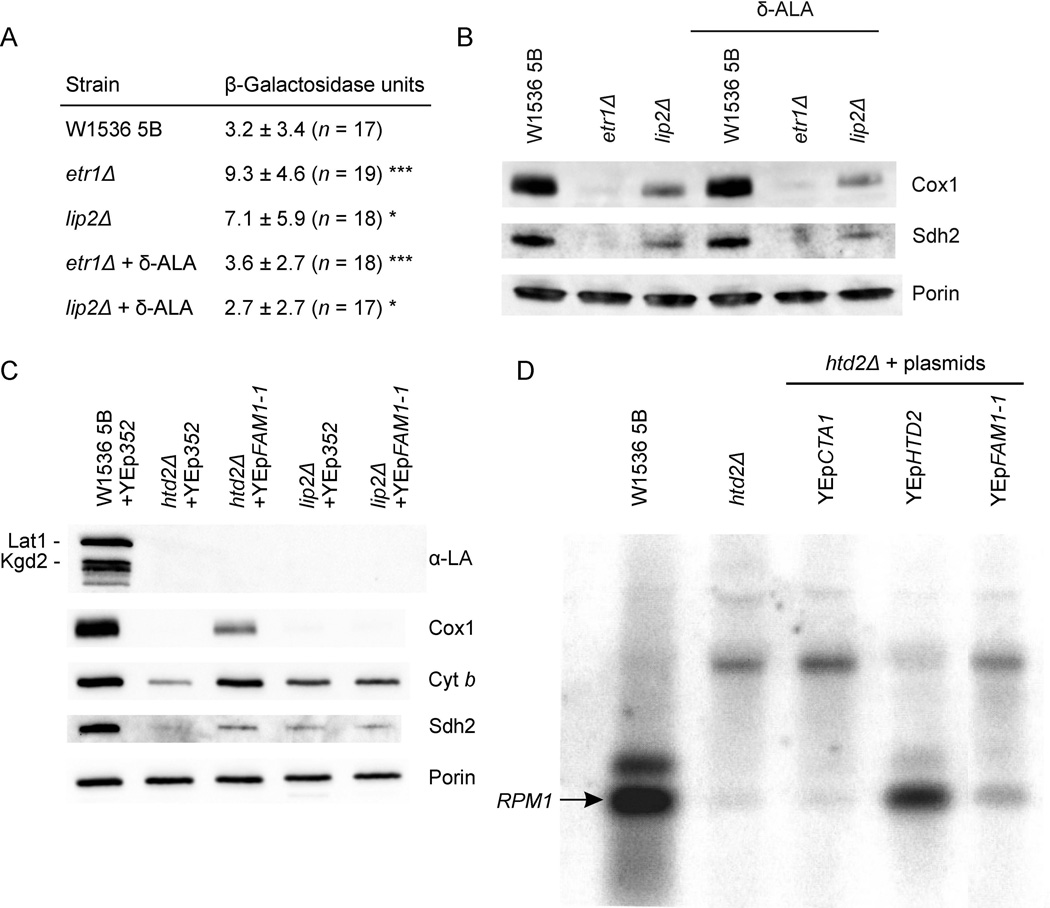
Succinyl-CoA generated by α-KGDH is required in mitochondria along with glycine in the first step of heme synthesis, which is the formation of δ-aminolevulenic acid (δ-ALA). A heme deficiency may be one of the causes of the observed cytochrome assembly defect. To assess heme levels, we measured β-galactosidase activity in yeast strains transformed with a plasmid harboring a fusion of the heme-repressed ANB1 promoter to lacZ (Deckert et al., 1998). Levels of β-galactosidase were increased about threefold in the fatty acid- or LA-deficient strains (Fig. 5A), a change so small that it could not be clearly detected by northern blotting (supplementary Fig. S3). This de-repression was suppressed by the addition of δ-ALA to the growth media (Fig. 5A). Addition of δ-ALA to the deficient strains, however, did not result in improvement of the steady state levels of respiratory complex proteins (Fig. 5B). Full de-repression increases β-galactosidase expression from this reporter by 100 to 200-fold or more (Deckert et al., 1998). Hence our observation is consistent with a mild heme deficiency in strains devoid of LA.
Fig. 5.
Effect of δ-aminolevulenic acid (δ-ALA) supplementation and FAM1-1 overexpression on the OXPHOS phenotypes of mtFAS and LA attachment mutant strains. (A) β-Galactosidase assay. Medium was supplemented with 100 µg ml−1 δ-ALA. Asterisk indicates statistical significance determined by Student's t test (***p < 0.0001, *p < 0.05). Data are represented as the mean of at least 17 measurements ± SD. (B) Supplementation with δ-ALA does not improve accumulation of respiratory complex subunits. Western blot analysis of Cox1 and Sdh2 steady state levels in mitochondria from wild-type, mtFAS and LA attachment-deficient cells grown in the presence or absence of 400 µg ml−1 δ-ALA. Loading control: Porin. (C) Western blot analyses of the steady state levels of the indicated proteins in control and mtFAS or LA attachment deficient strains overexpressing FAM1-1. Lat1 and Kgd2 are the lipoylated E2 subunits of pyruvate dehydrogenase complex and α-ketoglutarate dehydrogenase complex, respectively. Loading control: Porin. (D) Processing of the RPM1 RNA subunit of RNase P is improved in mtFAS deficient strains overexpressing FAM1-1. Total RNA extracted from the indicated strains was hybridized with a probe complementary to RPM1. The arrow indicates mature RPM1.
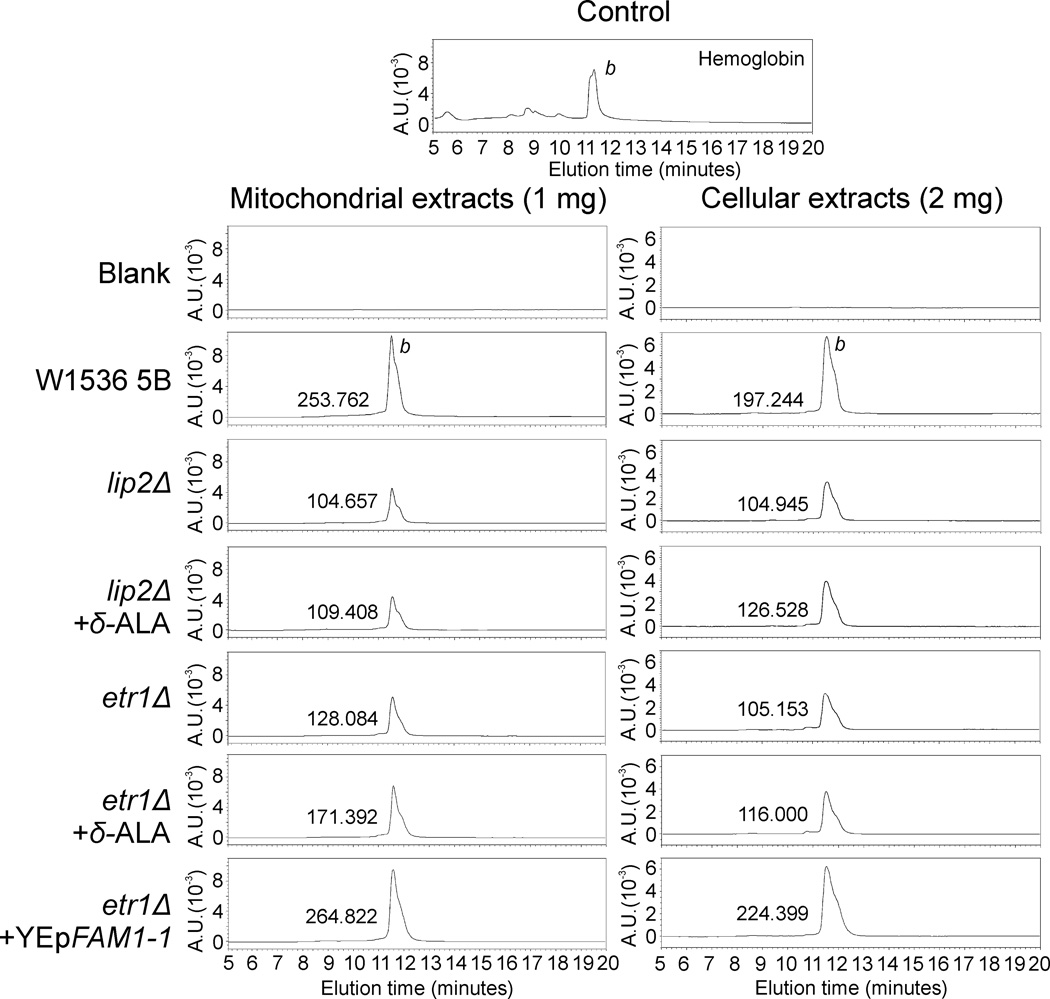
The ANB1-lacZ construct assesses “subjective” cellular heme levels, i.e. the levels that the cell senses as sufficient or deficient in terms of its needs. In order to better understand the role of heme in the mtFAS-deficient context, we tested absolute heme content in mtFAS mutants and the effect of δ-ALA supplementation. We treated etr1 and lip2 mutant strains with 100 µg/ml δ-ALA. This concentration was chosen because it was enough to complement the growth defect of a hem1 mutant strain (Haslam et al., 1979) and restored ANB1-lacZ repression in our reporter construct assay (Fig. 5A). As shown in the Figure 6, heme B content is reduced in etr1 and lip2 mutant mitochondria and whole cells. δ-ALA supplementation induced a mild but consistent increase in heme B content in whole cell extracts from etr1 and lip2 mutant strains, and a considerable increase in mitochondrial heme B only in etr1Δ. Intriguingly, the suppressor FAM1-1 (see next paragraph) completely restored heme levels to wild-type values in the etr1 mutant strain.
Fig. 6.
Effect of δ-aminolevulenic acid (δ-ALA) supplementation and FAM1-1 overexpression on heme content in mitochondrial fatty acid synthesis and lipoic acid attachment mutant strains. Hemes were extracted from isolated mitochondria (left panel) or spheroplasts (right panel) and analyzed by HPLC on a reverse phase C18 column. Hemoglobin was used to calibrate the column. The peaks corresponding to heme B were quantified by calculating the areas under the peaks and expressed in µV*sec. The peaks corresponding to heme O and heme A were basically undetectable at the represented scale and have been omitted from the figure.
Suppression of respiratory deficiency of mtFAS deficient yeast strains by FAM1-1 does not improve lipoylation status
The FAM1-1 suppressor mutation (Harington et al., 1994) partially rescues the respiratory-deficient phenotype of all mutants carrying deletions of genes encoding mtFAS enzymes (Kastaniotis et al., 2004). The mitochondrially mislocalized peroxisomal Faa2 acetyl-CoA ligase encoded by FAM1-1 has been proposed to activate free fatty acids in mitochondria, making them available to the organelle to substitute for the lack of endogenously synthesized fatty acids. Faa2 accepts substrates ranging from C7:0 – C17:0 (Knoll et al., 1994). Hence, we investigated if the suppressor allowed lipoylation of LA-dependent mitochondrial enzyme complexes. As presented in Fig. 5C, no lipoylation was detectable in these yeast strains. In contrast, there was a marked increase in the steady state levels of respiratory complex proteins Cox1, Cyt b and Sdh2, suggesting that a fatty acid longer than eight carbons is responsible for the improvement in respiratory ability in mtFAS-deficient strains. We also found by northern analysis that FAM1-1 exerted a mild suppression effect on the processing defect of RPM1, the mitochondrial RNA subunit of RNase P (Fig. 5D), which had been previously reported to be severely impaired in mtFAS-deficient strains (Schonauer et al., 2008).
Discussion
In response to a shortage of glucose in conjunction with the availability of non-fermentable carbon sources, yeast mitochondrial biogenesis is ramped up to allow the cells to cope with the changed environmental conditions. The results we present here indicate that mtFAS is required for biogenesis of respiratory-competent mitochondria at multiple levels.
Our screen for mutations causing a synthetic respiratory-deficient “petite” phenotype in combination with a compromised step in mtFAS allowed us to identified 18 mutants, most of which would not have been found in a “genome-wide screen” using the deletion strain collection. Our ensuing analyses clearly indicate a connection of mtFAS products to mitochondrial gene expression processes. Specifically, several post-transcriptional events are disturbed in mtFAS mutants. (i) The steady-state levels of COX1 and CYTB mRNA and the encoded proteins are markedly decreased in intron-containing strains because splicing of the corresponding mRNA precursors is severely retarded. Poor translation of intron-encoded maturases needed for intron excision is likely to be at least in part due to lower levels of mature tRNAs, as we have previously reported that mtFAS deletion strains are deficient in processing of the RPM1 RNA subunit of mitochondrial RNase P (Schonauer et al., 2008). (ii) Lack of accumulation of Cox1 in an intronless strain is due to a specific COX assembly defect. We have dissected the respective contributions of COX1 mRNA splicing, translation and assembly on the COX-deficient phenotype of etr1Δ yeast. When these processes were taken out of the equation by using intronless strains plus expressing an additional mss51F199I allele in the intronless ietr1Δ mutant strain, Cox1 synthesis and COX activity were fully recovered. Remarkably, the COX-sufficient strain only partially recovered respiratory growth competence, suggesting that mtFAS is required for mitochondrial functions other than COX biogenesis.
To unveil the primary cause of the respiratory-deficient phenotype in strains with defects in mtFAS and LA attachment, we pursued the characterization of suppressor strains. In general, LA attachment deficient yeast strains suffer from a milder phenotype than mtFAS mutant strains. Strikingly, our results indicate that mtFAS deficient strains suppressed by FAM1-1 are completely devoid of lipoylated Lat1, Kgd2 and Gcv3 but show an increase in the abundance of respiratory complex proteins and, as previously demonstrated (Kastaniotis et al., 2004), regain the ability to grow on medium requiring respiration. In addition, it has been reported previously that FAM1-1 restores the cytochrome spectrum of a cem1 mutant strain lacking a functional condensing enzyme (Harington et al., 1994). The absence of lipoylation in the respiratory-competent FAM1-1-suppressed mutants, in conjunction with the reported substrate preference of Fam1/Faa2, suggests that the physiologically active mtFAS product is longer than eight carbons. Hence, our data demonstrates that a mtFAS product other than octanoic acid or LA is required for respiration.
Strains with defects in mtFAS and LA attachment apparently suffer from mild heme deficiency, a factor that could explain at least part of their respiratory-deficient phenotype. Our β-galactosidase reporter results indicate that mtFAS- or lipoylation-deficient strains are sensing a mild but reproducible shortage of heme, an important cofactor for several mitochondrial respiratory complexes, which is consistent with an earlier observation of a heme deficiency in α-KGDH mutants (McCammon et al., 2003). Northern blotting for the ANB1 message confirms that the heme deficiency sensed by the cell is minuscule, at least under the growth conditions we used. However, analyses of steady-state heme levels in these strains tell a different story. Heme B is reduced by approximately 50% in both the etr1Δ and lip2Δ strains. The considerable increase of mitochondrial heme B in etr1Δ mitochondria upon addition of δ-ALA to the growth media may indicate that mitochondria in this strain background suffer more from the heme deficiency than does the rest of the cell. The full restoration of heme levels in the FAM1-1 suppressed strain is striking. However, since FAM1-1 does not restore lipoylation of α-KGDH to any detectable level, and since the heme levels in the α-KGDH-deficient lip2Δ strain do not respond much to δ-ALA supplementation, it is unlikely that the increase in steady-state cellular heme levels is due to restored α-KGDH activity. In part, this observation may result from the restoration of the respiratory chain, which ensures the retention of more heme in the cell.
Our data indicate that at least in the case of the COX deficiency observed in etr1Δ mutant yeast, COX1 mRNA splicing and Cox1 translational regulation play a role in attenuating the levels of Cox1 synthesis, and that when Cox1 biogenesis is enhanced in intronless ietr1Δ by an additional MSS51 or mss51F199I allele, COX assembly and function are restored. Importantly, the restoration of COX by MSS51 was only to ~50% of wild-type and had a minor impact on cell respiration and respiratory growth, whereas the mss51F199I allele fully restored COX activity and cellular respiration to ~50% of wild-type. We have recently shown that Mss51 function is modulated by heme B binding (Soto et al., 2012). Furthermore, restored Cox1 synthesis is also known to promote synthesis of heme A, the prosthetic group of COX (Barros et al., 2002). The possibility that mtFAS could either directly or indirectly affect the ability of wild-type Mss51 to sense heme, even when the cofactor is in abundance, warrants future investigations.
Superficially, FAM1-1 -and mss51F199I -mediated suppression are similar in their effect on respiration and growth of mtFAS deficient strains on a non-fermentable carbon source. Our data indicate however, that FAM1-1 suppression improves steady state levels of several different respiratory complex proteins, while MSS51 is a COX1- specific translation activator. Although we have not yet rigorously tested the possibility of mss51F199I effects on the expression of components of other complexes, we find such an activity not very likely. Instead, we suggest that a major contributing factor responsible for the respiratory deficiency of mtFAS mutants is a COX defect, and repair of this defect allows for improved respiration. Unless a role of mss51F199I in restoration of lipoylation is invoked, suppression of mtFAS defects by this allele is also further evidence that lipoylation of mitochondrial proteins is not required for respiration per se. Further analyses will clarify these hypotheses.
What is the connection between mtFAS and energy metabolism? The Krebs cycle and mtFAS share acetyl-CoA as their substrate. Thus, the mtFAS pathway is ideally suited to act as the master regulator of respiratory growth via synthesis of LA precursors and fatty acids required for post-transcriptional gene expression. As such, it would act as the sensor for the nutritional status of the cell by connecting acetyl-CoA availability to PDH and α-KGDH activities as well as to gene expression in mitochondria. Rather than adapting to change in nutritional status by waiting for increases in nuclear-encoded gene products, mitochondria would receive a direct metabolic signal. This would render the organelles poised to receive incoming derepressed nuclear-encoded respiratory chain gene products that will assemble with proteins synthesized in mitochondria. Prior activation of PDH and α-KGDH by lipoylation would allow for faster adaptation to a changing environment.
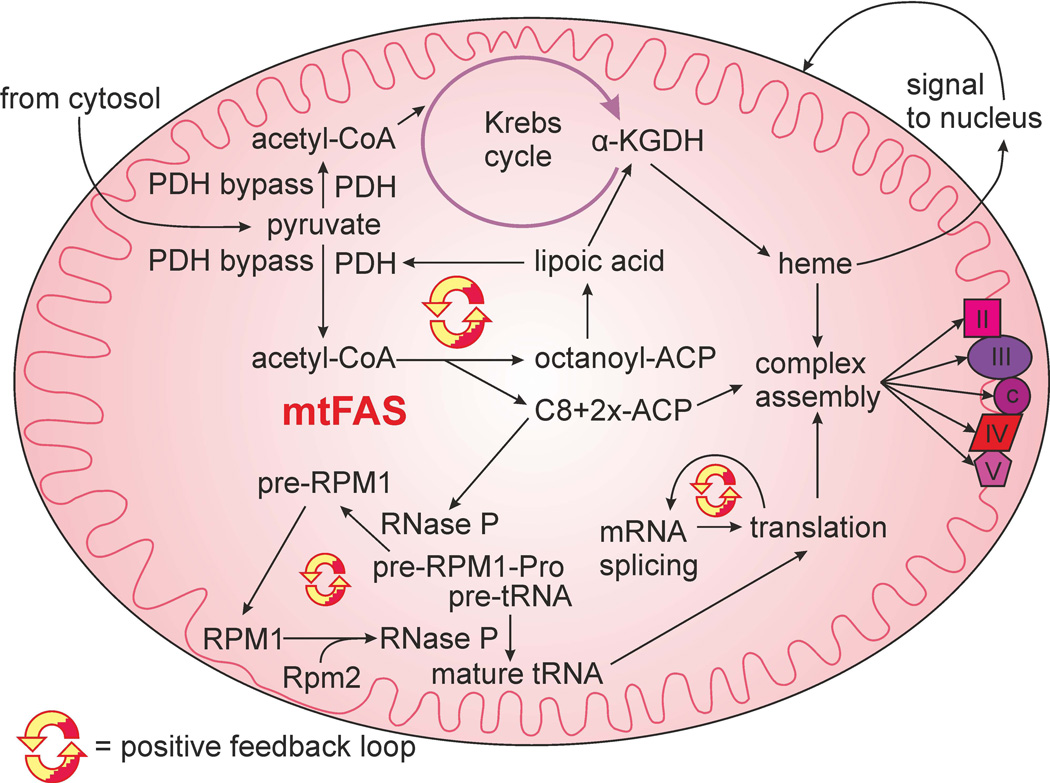
We propose a model wherein mtFAS is part of an intricate regulatory circuit, which acts as a hub and coordinator of mitochondrial biogenesis in yeast (Fig. 7). A number of these connections are conserved in higher eukaryotes (Feng et al., 2009; Mayr et al., 2011; Smith et al., 2012).
Fig. 7.
Simplified model of mtFAS-dependent regulatory circuits controlling mitochondrial gene expression. MtFAS is needed for octanoyl-ACP production, the sole octanoate source for lipoic acid (LA) synthesis. The pyruvate dehydrogenase complex (PDH) activity is part of a positive feedback loop (red and yellow circular arrow loop) with LA production and acetyl-CoA substrate production. The Krebs cycle and α-ketoglutarate dehydrogenase complex (α-KGDH) provide reducing power, succinate and precursors for heme biosynthesis. The arrows represent the flow of material and the requirement of products of the mtFAS pathway in LA synthesis, RPM1 RNA processing and respiratory complex assembly. RPM1 must be processed to assemble fully active RNase P for processing of the tRNA-containing RPM1 precursor RNA (second proposed positive feedback loop). Translation is needed for mRNA splicing, possibly resulting in mtFAS signal amplification (third proposed feedback loop). Some subunits of the cytochrome c reductase complex (III), cytochrome c oxidase complex (IV) and F1Fo ATP synthase (V) are translated in the mitochondria. The succinate dehydrogenase complex (II), as well as complexes III, IV and cytochrome c (c), require heme for their assembly and function.
Several modules of this proposed circuit may constitute positive feedback loops. The PDH complex is a source for the acetyl-CoA substrate of mtFAS and hence LA synthesis. The essential requirement of LA in the function of the PDH complex constitutes the clearest positive feedback loop (Schonauer et al., 2009). The proposed regulatory circuit suggests that lower levels of the acetyl-CoA substrate for mtFAS cause the respiratory chain defects detected in LA-deficient mutants.
A second positive feedback module may be represented by the RNase P maturation process (Schonauer et al., 2009), as RNase P requires mature RPM1 RNA for its activity, and one of the precursor RPM1 processing steps requires RNase P activity itself. This would feed forward into translation, as the 5´ cleavage step in tRNA maturation is carried out by RNase P.
Thirdly, impaired COX complex assembly, which blocks Cox1 translation initiation, contributes to a backup in the splicing of the COX1 mRNA precursor. Our model implies a dual role for heme in this regulatory circuit, as a redox cofactor in respiratory enzymes such as COX, and as a signaling molecule. In response to oxygen availability, heme levels are sensed in the nucleus and in mitochondria. Heme acts as a ligand to activate the major aerobic transcriptional activators Hap1 and the Hap2/3/4 complex in the nucleus (Zitomer et al., 1992) as well as the translational activation function of Mss51 in mitochondria (Soto et al., 2012). In our model, heme serves to integrate information of both oxygen and acetyl-CoA substrate availability.
It has been shown that the mtFAS pathway is not required for the expression, assembly or function of respiratory complexes per se (Merz et al., 2009). In the absence of the mitochondrial pyruvate importer or in strains lacking functional PDH, the level of mitochondrial protein complex lipoylation corresponds to the availability of ketogenic amino acids in the media (Herzig et al., 2012), demonstrating a possible acetyl-CoA dosage response of mtFAS. Also, our isolation of synthetic petite mutants with the FabI strain, which is not detectably translation deficient, indicates a response of mitochondrial gene expression to mtFAS output. In addition, the suppression of the COX1 translation defect and full restoration of COX activity by the heme-independent mss51F199I in the ietr1Δ mutant is consistent with a bypass of a defect in a regulatory step of COX expression by this mutant allele in the mtFAS-deficient strain. A role for mtFAS as a mitochondrial acetyl-CoA sensor is also consistent with the strict conservation of mtFAS and LA synthesis and the metabolic isolation of these processes in the organelle.
There is some evidence that this regulatory circuit may be conserved in higher eukaryotes. In a mouse mtFAS conditional knockout model, the animals suffer from a premature aging phenotype characteristic for mice with mitochondrial dysfunction (Smith et al., 2012). Remarkably, apart from LA deficiency, the authors observed lower levels of all respiratory complexes, particularly complexes II and IV. Additionally, a patient with a defect in LA synthase was reported to display various neuromuscular symptoms, defects in mitochondrial energy metabolism and acute respiratory chain deficiency, including a marked decrease in complex II activity (Mayr et al., 2011). We attribute this defect to a feedback effect of low lipoylation on mtFAS. MtFAS patients, yet to be identified, would be expected to display a rather similar phenotype.
None of our results points to a primary involvement of membrane phospholipids in the phenotypes caused by mtFAS deficiency. Ultimately, the precise identity and destination of mtFAS products and how they exert their effects on mitochondrial biogenesis will have to be determined by metabolic labeling studies. In conclusion, our model provides a framework to understand how mitochondria in single cell and more complex eukaryotes obtain information about the cellular metabolic status independent of nuclear signals.
Experimental Procedures
Strains, media and growth conditions
For strains used in this study see Table 1. Yeast cells were grown on either rich YP (1% yeast extract, 2% peptone) with 2% glucose YPD, 2% galactose YPGal, 2% ethanol, 3% glycerol YPEG, YPD supplemented with 300 µg ml−1 of geneticin, synthetic complete (SIGMA-Aldrich, Helsinki, Finland) with 2% glucose (SCD), 3% glycerol (SCG) or SCD and SCG media lacking one or more nutrients, WO-Gal (2% galactose, 0.67% yeast nitrogen base), WO-EG (2% ethanol, 3% glycerol, 0.67% yeast nitrogen base). 100–400 µg ml−1 δ-aminolevulinic acid (δ-ALA) was supplemented in media buffered at pH 5.5. In some cases media was supplemented with triclosan (Dermatologica Widmer, Helsinki, Finland). Glycerol sectoring medium has been described previously (Kastaniotis et al., 2004). Solid media contained 2% agar. Yeast was grown at 30°C with vigorous shaking for liquid cultures.
Table 1.
Strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| S. cerevisiae | ||
| MCC109I |
MATαade2-101, ura3-52, kar1-1, intronless mtDNA |
(Ellis et al., 2004) |
| W303-1A |
MATaade2-1, his3-1,15; leu2-2,112; trp1-1, ura3-1 |
R. Rothstein (Columbia University, New York, NY) |
| iW303-1A | intronless mtDNA | This study |
| W303 cox14Δ |
MATα, ade2-1, his3-1,15; leu2-2,112; trp1-1, ura3-1, yml129c::TRP1 |
(Barrientos et al., 2004) |
| cox14Δ, lip2Δ | yml129c::TRP1, ylr239c, kanMX4 | This study |
| cox14Δ, lip3Δ | yml129c::TRP1, yjl046w, kanMX4 | This study |
| cox14Δ, gcv3Δ | yml129c::TRP1, yal044c, kanMX4 | This study |
| cox14Δ, etr1Δ | yml129c::TRP1, ybr026c, kanMX4 | This study |
| W1536 5B |
MATaade2Δ, ade3Δ, can1-100, his3-11,15; leu2-3,112; trp1-1, ura3-1 |
(Kastaniotis et al., 2004) |
| ald4Δ | yor374w, kanMX4 | This study |
| cbp2Δ | yhl038c, kanMX4 | This study |
| cem1Δ | yer061c, kanMX4 | This study |
| etr1Δ | ybr026c, kanMX4 | This study |
| etr1Δ ρ0 | ybr026c, kanMX4, ρ0 | This study |
| ietr1Δ | ybr026c, kanMX4, intronless mtDNA | This study |
| ietr1Δ MSS51 | ybr026ckanMX4, intronless mtDNA, integrated MSS51 |
This study |
| ietr1Δ mss51F199I |
ybr026ckanMX4, intronless mtDNA, integrated mss51F199I |
This study |
| FabI | ybr026c, kanMX4, mtFabI | This study |
| FabI ald4Δ | ybr026c, kanMX4, mtFabI, yor374w, kanMX4 | This study |
| FabI mrps5Δ | ybr026c, kanMX4, mtFabI, ybr251w, kanMX4 | This study |
| gcv3Δ | yal044c, kanMX4 | This study |
| htd2Δ | yhr067w, kanMX4 | This study |
| htd2Δ ρ0 | yhr067w, kanMX4, ρ0 | This study |
| ihtd2Δ | yhr067w, kanMX4, intronless mtDNA | This study |
| lip2Δ | ylr239c, kanMX4 | This study |
| lip3Δ | yjl046w, kanMX4 | This study |
| mrps5Δ | ybr251w, kanMX4 | This study |
| W1536 8B |
MATα, ade2Δ, ade3Δ, can1-100, his3-11,15; leu2-3,112; trp1-1, ura3-1 |
(Kastaniotis et al., 2004) |
| etr1Δ | ybr026c, kanMX4 | This study |
| FabI | ybr026c, kanMX4, mtFabI | This study |
Plasmids
Plasmids YCplac22, YCplac33 and YCplac111 (Gietz et al., 1988), pYE352-CTA1 (Filppula et al., 1995), pYE352::YBR026c and pYE352::mtFabI (Torkko et al., 2001), the HeAl library, pTSV30A, pYE352-YHR067w and pYEFAM1-1 (Kastaniotis et al., 2004), YCp(33)AZSU -lacZ (Deckert et al., 1998) and the Lacroute library (Harington et al., 1993) and plasmids for genomic integration of MSS51 and mss51F199I (Barrientos et al., 2002) have been described. Plasmid YEp352 was generated by digesting plasmid pYE352-CTA1 with EcoRI and then religating the gapped plasmid to remove the CTA1 insert. For clarity, pYE352-CTA1, pYE352::YBR026c, pYE352-YHR067w and pYEFAM1-1 will be called YEpCTA1, YEpETR1, YEpHTD2 and YEpFAM1-1, respectively.
Manipulations of DNA and plasmid constructions were carried out using standard techniques (Ausubel et al., 1989). All new constructs were verified by sequencing. The ETR1 gene was amplified from yeast genomic DNA and cloned into vectors pTSV30A and YCplac22 to create plasmids pTSV30ETR1 and YCp22ETR1 expressing the ETR1 gene from its native regulatory sequences. The construct containing the fusion of the sequence encoding the COQ3 mitochondrial targeting sequence fused to the E. coli fabI gene was amplified from pYE352::mtFabI and the regulatory region of the ETR1 gene was amplified from the pTSV30ETR1 plasmid. These PCR products were cloned into vector YCplac22 to create plasmid YCp22mtFabI harboring the chimeric mtFabI gene under control of the ETR1 promoter and the CTA1 terminator. The ALD4, ald4-1, HTD2, MRPS5 and mrps5-1 genes were cloned into vectors YCplac33 or YCplac111 to create plasmids YCp33ALD4, YCp33ald4-1, YCp33HTD2, YCp33MRPS5, YCp111MRPS5 and YCp111mrps5-1 expressing all of the genes from their native regulatory sequences.
Disruption of open reading frames
Each open reading frame was deleted by homologous recombination with the kanMX4 cassettes amplified from the deletion strains of the EUROSCARF collection (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/) using primers specific to the 5′ upstream and 3′ downstream regions of the genes. The PCR products were purified and transformed into the wild-type strains using the lithium acetate method (Gietz et al., 2002). Proper integration of the constructs was verified by PCR and complementation. Double mutants were obtained from crosses of the respective single mutants and sporulation of the heterozygous diploids.
Generation of the etr1::mtFabI (FabI) strains
The mtFabI knock-in DNA was amplified from the YCp22mtFabI plasmid with 435 base pairs of the ETR1 promoter region on the 5’ end and a 3’ tail with 40 base pairs homologous to ETR1 terminator sequence. After selection of the respiratory competent transformants on YPG plates integration of the mtFabI construct into the ETR1 locus of W1536 5B (a) and 8B (α) etr1Δ strains was tested by sensitivity to geneticin and triclosan. Both of the strains were verified by PCR.
Generation of rho0 strains
W1536 5B (a) etr1Δ and htd2Δ strains were treated with ethidium bromide to induce loss of mitochondrial DNA (Ellis et al., 1999). 1 ml of an overnight YPD culture grown to approximately optical density (OD) at 600 nanometers of 4 were collected, washed with 5 ml Sorenson’s solution (60 mM NaPi, 140 mM KPi) and resuspended in 2 ml Sorenson’s solution with 10 µg ml−1 or 20 µg ml−1 ethidium bromide and shaken for 2 or 4 hours. After washing with Sorenson’s solution, the cells were suspended in YPD and plated on YPD plates for single colonies. The ρ0 genotype was verified by staining candidates in buffered 250 ng ml−1 DAPI (Thermo Scientific) (0.01 M MgCl2, 0.01 M Tris pH 8.0) solution and inspection by fluorescence microscopy (Olympus BX51).
Generation of the intronless strains
The intronless mitochondrial genome was transferred from the kar1 MCC109I (α) cytosolic donor strain (Ellis et al., 2004), to W1536 5B (a) etr1Δ and htd2Δ ρ0 strains harboring plasmids YEpETR1 and YCp33HTD2, respectively, to allow for selection on SCG. Saturated SCD-ura cultures of both strains were mixed with an YPD culture of MCC109I and incubated 3 hours without shaking and an additional 3 hours with mild shaking after adding fresh SCD-ura. Cultures were diluted and plated on SCD-ura. Candidates unable to grow on SCD-trp and growing on SCG were chosen for further tests. The intronless genotype was confirmed by PCR. Intronless strains are designated by the cytosolic receptor strain, preceded by lower case “i”, as in (Ellis et al., 2004).
Generation of W1536 5B ietr1Δmss51F199I and ietr1ΔMSS51
W1536 5B ietr1Δ mss51F199I and ietr1Δ MSS51 were constructed as previously described (Soto et al., 2012).
Growth curve, FACS analysis and spotting assay
For the growth curves and FACS analysis, cells were inoculated at an OD of 0.2 and grown on SCD medium for 72 hours. Triclosan was added to the samples at the 5 hour time point at 1 µg ml−1 concentration (growth curve) or 0.5 µg ml−1 (FACS) as indicated. Samples for FACS were harvested after the 72 hour growth period.
For spotting assays, strains were grown to logarithmic growth phase, harvested and adjusted to an OD of 0.5. A dilution series of undiluted, 1:10, 1:100 and 1:1000 was made and 2 µl of cells of each dilution were spotted on SCD and SCG plates and grown at 30°C for 2 and 5 days, respectively.
Lipoic acid analysis
Analysis of lipoic acid in yeast has been described before (Brody et al., 1997; Schonauer et al., 2009).
Yeast mitochondrial membrane potential analysis
Cells were harvested after the diauxic shift (OD of ~ 4.0) and subsequently treated with JC-1, a cationic, lipophilic dye used for the visualization of mitochondrial membrane potential (Smiley et al., 1991). The JC-1 lipophilic cation dye dye (5',6,6'-tetrachloro-1,1',3,3'-tetraethylbenzimidazolocarbocyanine iodide) has been shown to form so-called J-aggregates at high concentrations, resulting in a shift of absorption and fluorescence maxima. JC-1 uptake into mitochondria, where the molecules aggregate due to a concentration increase relative to the cytosol, is membrane potential dependent. Aggregated, mitochondrially-imported JC-1 emits orange fluorescence (FL2, 590 nm) while unaggregated JC-1 shows green fluorescence (FL1, 520 nm). The graphs were divided into quadrants at the x (2) y (2) to facilitate analysis. Fractions of cell populations present in the quadrants are indicated (Q1-Q4). 10 µM FCCP (carbonyl cyanide 3- trifluoromethoxy phenylhydrazone) added at the same time as JC-1 was used as the negative control. The triclosan concentration in the triclosan–supplemented media was 0.5 µg ml−1. Staining was performed following the SIGMA Mitochondria Staining Kit protocol (SIGMA-Aldrich, St. Louis, MO, USA). The fluorescence signal was detected using a Pertec fluorescence spectrophotometer equipped with a 20 mW solid state 488 nm laser (Partek, Münster, Germany), following the MitoProbe™ JC-1 Assay Kit for Flow Cytometry (M34152) protocol. Data analysis was performed by FlowMax software from Partek.
β-Galactosidase assays
Strains W1536 5B and derivatives thereof, transformed with plasmids YCp(33)AZ (Deckert et al., 1998) and YCplac33 as a control, were inoculated in 2 ml SCD-Ura medium and grown over night. The next day, these cultures were used to inoculate fresh cultures to an OD of 0.2, and these fresh cultures grown to OD of ~ 0.5, when the cells were harvested for β-galactosidase assays using the permeabilized cell assay as described (Adams et al., 1997).
Heme content determination
Mitochondria were isolated from cells grown for 16 hours in rich galactose media (YPGal) supplemented, when indicated, with 100 µg/ml δ-aminolevulinic acid (δ-ALA), as previously described (Haslam et al., 1979). Spheroplasts were obtained by zymolyase (5 mg/ml) digestion from cells grown under the same conditions. Hemes were extracted from mitochondria (1 mg) and spheroplasts (2 mg) with 250 mM HCl acetone and 50% acetonitrile and analyzed by HPLC with a C18 column as described (Soto et al., 2012).
Statistical Analysis
All experiments were done at least in triplicate. For the enzymatic and polarographic assays, data are presented as means ± SD. The values obtained for wild-type and mutants strains were compared by the t-Student test. P < 0.05 was considered significant. For quantification of western blot and radioactive signals, the images were digitalized and densitometry performed using the histogram function of the Adobe Photoshop program. The values measured in three independent assays did not differ by more than 5%.
Mitochondrial isolation for western blot analysis
Mitochondria were prepared from strains grown in media containing 2% galactose or 2% glucose as described (Meisinger et al., 2000) and (Herrmann et al., 1994).
Synthetic petite screen
The colony sectoring screen has been described (Kastaniotis et al., 2004). W1536 5B and 8B FabI strains carrying pTSV30ETR1 plasmid were subjected to EMS mutagenesis, plated on glycerol sectoring media and grown at 30°C for 10 days and at 22°C for 7 days. Non-sectoring red colonies were isolated and retested to confirm the phenotype. Mutants that depended on the presence of the pTSV30ETR1 plasmid on SCG but not on SCD were tested with YCp22ETR1, YCp22mtFabI and YEpCTA1 (negative control) for ability to grow on SCG to rule out mutations in the mtFabI replacement cassette and with YEpFAM1-1 to identify possible mutations in mtFAS pathway members (Kastaniotis et al., 2004).
We were unable to completely resolve complementation groups in complementation analysis, presumably because of a propensity of the mutants to lose mitochondrial DNA. Mutations were identified by genomic DNA library complementation of the respiratory growth defect. All these candidate genes also complemented the mutants when present on single copy plasmids.
For some of the mutants, we chose to confirm the synthetic petite phenotype in a strain background that had not undergone EMS treatment. The synthetic petite phenotypes of both mrps5-1 (Fig. S2) and ald4-1 (data not shown) were reproduced in the respective knockout strains carrying a plasmid harbouring the respective mutant allele in the FabI genetic background.
Western blot analysis
Equivalent amounts of total mitochondrial proteins were separated by SDS-PAGE (12% gel) in order to quantify the steady-state levels of mitochondrial respiratory chain enzymes and individual subunits. Proteins were transferred to nitrocellulose membrane and decorated with the indicated antisera. Anti- Porin antibody was used as loading control.
Characterization of yeast mitochondrial respiratory chain
Endogenous cell respiration was assayed in whole cells in the presence of galactose using a Clark type polarographic oxygen electrode from Hansatech Instruments (Norfolk, UK) at 24°C as described (Barrientos et al., 2002). Mitochondria prepared from the different strains were used for spectrophotometric assays carried out at 24°C. KCN-sensitive cytochrome c oxidase (COX) activity, antimycin A-sensitive NADH cytochrome c reductase (NCCR) activity and antimycin A-sensitive succinate cytochrome c reductase (SCCR) were assayed with 50 µg of mitochondria permeabilized with 0.5% sodium deoxycholate, as described (Barrientos et al., 2002). Briefly, COX activity was measured by following the oxidation of 50 µM reduced cytochrome c at 550 nm in a buffer containing 20 mM KPi (pH 7.4). The addition of 0.3 mM KCN inhibited the reaction. NCCR and SCCR activities were measured by following the reduction of oxidized 50 µM cytochrome c at 550 nm, using respectively 0.4 mM NADH or 10 mM succinate as the electron donor in a buffer containing 20 mM KPi (pH 7.4) and 2 mM EDTA. The addition of 0.4 µM antimycin A inhibited both reactions. ATPase activity was assayed by measuring release of inorganic phosphate from ATP at 37°C in the presence and absence of oligomycin (King, 1932).
Mitochondrial cytochrome spectra
Isolated mitochondria were extracted at a protein concentration of 5 mg/ml in 20 mM Tris-HCl, pH 7.5, 1 M KCl, 1% sodium deoxycholate, conditions that quantitatively solubilize all mitochondrial cytochromes (Tzagoloff et al., 1975). Samples of the extract were either oxidized with ferricyanide or reduced with sodium dithionite and the difference spectra were measured at room temperature using a UV-2401PC Shimadzu spectrophotometer.
In vivo mitochondrial protein synthesis
Mitochondrial gene products were labeled with 35S-methionine (7 mCi mmol−1, PerkinElmer) in whole cells at 30°C for 10 minutes pulse in the presence of 0.2 mg ml−1 cycloheximide. Equivalent amount of total cellular proteins were separated by SDS-PAGE on a 17.5% polyacrylamide gel, transferred to a nitrocellulose membrane and exposed to Kodak X-OMAT X-ray film.
Northern blot analysis
Northern blot analysis was done as described previously (Schonauer et al., 2008). For anti-sense oligonucleotide probes see Supporting Information Table S5.
Supplementary Material
Acknowledgements
We thank the technical assistance of Catherine Trivigno.
We thank Richard Zitomer for strains and plasmids, Steve Hanes and François Lacroute for the terrific genomic multicopy yeast libraries and Geneviève Dujardin for the anti-Cyt b antibody. We are also grateful to Telsa Mittelmeier for valuable comments on the manuscript and thank Dermatologica Widmer for the generous gift of the drug triclosan. This work was supported by the Academy of Finland and the Sigrid Juselius Foundation, the US National Institutes of Health (NIH-RO1 GM071775-06) and the Muscular Dystrophy Association.
References
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in yeast genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Amiott EA, Jaehning JA. Sensitivity of the yeast mitochondrial RNA polymerase to +1 and +2 initiating nucleotides. J Biol Chem. 2006a;281:34982–34988. doi: 10.1074/jbc.M608638200. [DOI] [PubMed] [Google Scholar]
- Amiott EA, Jaehning JA. Mitochondrial transcription is regulated via an ATP "sensing" mechanism that couples RNA abundance to respiration. Mol Cell. 2006b;22:329–338. doi: 10.1016/j.molcel.2006.03.031. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent RE, Kingston DD, Moore JG, Seidman JA, editors. Current protocols in molecular biology. New York, NY: John Wiley and Sons, Inc; 1989. Current protocols in molecular biology. [Google Scholar]
- Barrientos A, Zambrano A, Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in saccharomyces cerevisiae. EMBO J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Korr D, Tzagoloff A. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of leigh's syndrome. EMBO J. 2002;21:43–52. doi: 10.1093/emboj/21.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros MH, Tzagoloff A. Regulation of the heme A biosynthetic pathway in saccharomyces cerevisiae. FEBS Lett. 2002;516:119–123. doi: 10.1016/s0014-5793(02)02514-0. [DOI] [PubMed] [Google Scholar]
- Boubekeur S, Bunoust O, Camougrand N, Castroviejo M, Rigoulet M, Guerin B. A mitochondrial pyruvate dehydrogenase bypass in the yeast saccharomyces cerevisiae. J Biol Chem. 1999;274:21044–21048. doi: 10.1074/jbc.274.30.21044. [DOI] [PubMed] [Google Scholar]
- Brody S, Oh C, Hoja U, Schweizer E. Mitochondrial acyl carrier protein is involved in lipoic acid synthesis in Saccharomyces cerevisiae. FEBS Lett. 1997;408:217–220. doi: 10.1016/s0014-5793(97)00428-6. [DOI] [PubMed] [Google Scholar]
- Cohen TJ, Lee K, Rutkowski LH, Strich R. Ask10p mediates the oxidative stress-induced destruction of the saccharomyces cerevisiae C-type cyclin Ume3p/Srb11p. Eukaryot Cell. 2003;2:962–970. doi: 10.1128/EC.2.5.962-970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert J, Torres AM, Hwang SM, Kastaniotis AJ, Zitomer RS. The anatomy of a hypoxic operator in saccharomyces cerevisiae. Genetics. 1998;150:1429–1441. doi: 10.1093/genetics/150.4.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis TP, Helfenbein KG, Tzagoloff A, Dieckmann CL. Aep3p stabilizes the mitochondrial bicistronic mRNA encoding subunits 6 and 8 of the H+-translocating ATP synthase of saccharomyces cerevisiae. J Biol Chem. 2004;279:15728–15733. doi: 10.1074/jbc.M314162200. [DOI] [PubMed] [Google Scholar]
- Ellis TP, Lukins HB, Nagley P, Corner BE. Suppression of a nuclear aep2 mutation in saccharomyces cerevisiae by a base substitution in the 5'-untranslated region of the mitochondrial oli1 gene encoding subunit 9 of ATP synthase. Genetics. 1999;151:1353–1363. doi: 10.1093/genetics/151.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Witkowski A, Smith S. Down-regulation of mitochondrial acyl carrier protein in mammalian cells compromises protein lipoylation and respiratory complex I and results in cell death. J Biol Chem. 2009;284:11436–11445. doi: 10.1074/jbc.M806991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filppula SA, Sormunen RT, Hartig A, Kunau WH, Hiltunen JK. Changing stereochemistry for a metabolic pathway in vivo. experiments with the peroxisomal beta-oxidation in yeast. J Biol Chem. 1995;270:27453–27457. doi: 10.1074/jbc.270.46.27453. [DOI] [PubMed] [Google Scholar]
- Fontanesi F, Clemente P, Barrientos A. Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in saccharomyces cerevisiae. J Biol Chem. 2011;286:555–566. doi: 10.1074/jbc.M110.188805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi F, Soto IC, Horn D, Barrientos A. Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol Cell Biol. 2010;30:245–259. doi: 10.1128/MCB.00983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Harington A, Schwarz E, Slonimski PP, Herbert CJ. Subcellular relocalization of a long-chain fatty acid CoA ligase by a suppressor mutation alleviates a respiration deficiency in saccharomyces cerevisiae. EMBO J. 1994;13:5531–5538. doi: 10.1002/j.1460-2075.1994.tb06890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harington A, Herbert CJ, Tung B, Getz GS, Slonimski PP. Identification of a new nuclear gene (CEM1) encoding a protein homologous to a beta-keto-acyl synthase which is essential for mitochondrial respiration in saccharomyces cerevisiae. Mol Microbiol. 1993;9:545–555. doi: 10.1111/j.1365-2958.1993.tb01715.x. [DOI] [PubMed] [Google Scholar]
- Haslam JM, Astin AM. The use of heme-deficient mutants to investigate mitochondrial function and biogenesis in yeast. Methods Enzymol. 1979;56:558–568. doi: 10.1016/0076-6879(79)56054-6. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Woellhaf MW, Bonnefoy N. Control of protein synthesis in yeast mitochondria: The concept of translational activators. Biochim Biophys Acta. 2013;1833:286–294. doi: 10.1016/j.bbamcr.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Stuart RA, Craig EA, Neupert W. Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J Cell Biol. 1994;127:893–902. doi: 10.1083/jcb.127.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Raemy E, Montessuit S, Veuthey JL, Zamboni N, Westermann B, et al. Identification and functional expression of the mitochondrial pyruvate carrier. Science. 2012;337:93–96. doi: 10.1126/science.1218530. [DOI] [PubMed] [Google Scholar]
- Hiltunen JK, Autio KJ, Schonauer MS, Kursu VA, Dieckmann CL, Kastaniotis AJ. Mitochondrial fatty acid synthesis and respiration. Biochim Biophys Acta. 2010;1797:1195–1202. doi: 10.1016/j.bbabio.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Kastaniotis AJ, Autio KJ, Sormunen RT, Hiltunen JK. Htd2p/Yhr067p is a yeast 3-hydroxyacyl-ACP dehydratase essential for mitochondrial function and morphology. Mol Microbiol. 2004;53:1407–1421. doi: 10.1111/j.1365-2958.2004.04191.x. [DOI] [PubMed] [Google Scholar]
- King EJ. The colorimetric determination of phosphorus. Biochem J. 1932;26:292–297. doi: 10.1042/bj0260292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll LJ, Johnson DR, Gordon JI. Biochemical studies of three saccharomyces cerevisiae acyl-CoA synthetases, Faa1p, Faa2p, and Faa3p. J Biol Chem. 1994;269:16348–16356. [PubMed] [Google Scholar]
- Langer T, Kaser M, Klanner C, Leonhard K. AAA proteases of mitochondria: Quality control of membrane proteins and regulatory functions during mitochondrial biogenesis. Biochem Soc Trans. 2001;29:431–436. doi: 10.1042/bst0290431. [DOI] [PubMed] [Google Scholar]
- Mayr JA, Zimmermann FA, Fauth C, Bergheim C, Meierhofer D, Radmayr D, et al. Lipoic acid synthetase deficiency causes neonatal-onset epilepsy, defective mitochondrial energy metabolism, and glycine elevation. Am J Hum Genet. 2011;89:792–797. doi: 10.1016/j.ajhg.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCammon MT, Epstein CB, Przybyla-Zawislak B, McAlister-Henn L, Butow RA. Global transcription analysis of krebs tricarboxylic acid cycle mutants reveals an alternating pattern of gene expression and effects on hypoxic and oxidative genes. Mol Biol Cell. 2003;14:958–972. doi: 10.1091/mbc.E02-07-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw P, Tzagoloff A. Assembly of the mitochondrial membrane system. characterization of a yeast nuclear gene involved in the processing of the cytochrome b pre-mRNA. J Biol Chem. 1983;258:9459–9468. [PubMed] [Google Scholar]
- Meisinger C, Sommer T, Pfanner N. Purification of saccharomcyes cerevisiae mitochondria devoid of microsomal and cytosolic contaminations. Anal Biochem. 2000;287:339–342. doi: 10.1006/abio.2000.4868. [DOI] [PubMed] [Google Scholar]
- Merz S, Westermann B. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in saccharomyces cerevisiae. Genome Biol. 2009;10:R95. doi: 10.1186/gb-2009-10-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Vukotic M, Piechura H, Meyer HE, Warscheid B, Deckers M, Rehling P. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J Cell Biol. 2010;191:141–154. doi: 10.1083/jcb.201007026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelmeier TM, Dieckmann CL. In vivo analysis of sequences required for translation of cytochrome b transcripts in yeast mitochondria. Mol Cell Biol. 1995;15:780–789. doi: 10.1128/mcb.15.2.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martinez X, Broadley SA, Fox TD. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–5961. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14:255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- Schonauer MS, Kastaniotis AJ, Hiltunen JK, Dieckmann CL. Intersection of RNA processing and the type II fatty acid synthesis pathway in yeast mitochondria. Mol Cell Biol. 2008;28:6646–6657. doi: 10.1128/MCB.01162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonauer MS, Kastaniotis AJ, Kursu VA, Hiltunen JK, Dieckmann CL. Lipoic acid synthesis and attachment in yeast mitochondria. J Biol Chem. 2009;284:23234–23242. doi: 10.1074/jbc.M109.015594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller HJ. Transcriptional control of nonfermentative metabolism in the yeast saccharomyces cerevisiae. Curr Genet. 2003;43:139–160. doi: 10.1007/s00294-003-0381-8. [DOI] [PubMed] [Google Scholar]
- Smiley ST, Reers M, Mottola-Hartshorn C, Lin M, Chen A, Smith TW, et al. Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc Natl Acad Sci U S A. 1991;88:3671–3675. doi: 10.1073/pnas.88.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Witkowski A, Moghul A, Yoshinaga Y, Nefedov M, de Jong P, et al. Compromised mitochondrial fatty acid synthesis in transgenic mice results in defective protein lipoylation and energy disequilibrium. PLoS One. 2012;7:e47196. doi: 10.1371/journal.pone.0047196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto IC, Fontanesi F, Myers RS, Hamel P, Barrientos A. A heme-sensing mechanism in the translational regulation of mitochondrial cytochrome c oxidase biogenesis. Cell Metab. 2012;16:801–813. doi: 10.1016/j.cmet.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkko JM, Koivuranta KT, Kastaniotis AJ, Airenne TT, Glumoff T, Ilves M, et al. Candida tropicalis expresses two mitochondrial 2-enoyl thioester reductases that are able to form both homodimers and heterodimers. J Biol Chem. 2003;278:41213–41220. doi: 10.1074/jbc.M307664200. [DOI] [PubMed] [Google Scholar]
- Torkko JM, Koivuranta KT, Miinalainen IJ, Yagi AI, Schmitz W, Kastaniotis AJ, et al. Candida tropicalis Etr1p and saccharomyces cerevisiae Ybr026p (Mrf1'p), 2-enoyl thioester reductases essential for mitochondrial respiratory competence. Mol Cell Biol. 2001;21:6243–6253. doi: 10.1128/MCB.21.18.6243-6253.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte B, Liang XB, Robert F, Soontorngun N. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 2010;10:2–13. doi: 10.1111/j.1567-1364.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A, Akai A, Needleman RB, Zulch G. Assembly of the mitochondrial membrane system. cytoplasmic mutants of saccharomyces cerevisiae with lesions in enzymes of the respiratory chain and in the mitochondrial ATPase. J Biol Chem. 1975;250:8236–8242. [PubMed] [Google Scholar]
- van der Giezen M. Hydrogenosomes and mitosomes: Conservation and evolution of functions. J Eukaryot Microbiol. 2009;56:221–231. doi: 10.1111/j.1550-7408.2009.00407.x. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Why do we still have a maternally inherited mitochondrial DNA? insights from evolutionary medicine. Annu Rev Biochem. 2007;76:781–821. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- Yamazoe M, Shirahige K, Rashid MB, Kaneko Y, Nakayama T, Ogasawara N, Yoshikawa H. A protein which binds preferentially to single-stranded core sequence of autonomously replicating sequence is essential for respiratory function in mitochondrial of saccharomyces cerevisiae. J Biol Chem. 1994;269:15244–15252. [PubMed] [Google Scholar]
- Ytting CK, Fuglsang AT, Hiltunen JK, Kastaniotis AJ, Ozalp VC, Nielsen LJ, Olsen LF. Measurements of intracellular ATP provide new insight into the regulation of glycolysis in the yeast saccharomyces cerevisiae. Integr Biol (Camb) 2012;4:99–107. doi: 10.1039/c1ib00108f. [DOI] [PubMed] [Google Scholar]
- Zitomer RS, Lowry CV. Regulation of gene expression by oxygen in saccharomyces cerevisiae. Microbiol Rev. 1992;56:1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.