Abstract
Background
Epidemiological studies have shown that consumption of a high-fat diet (HFD) increases the risk of developing breast cancer (BC). Studies in rodents have shown HFD causes changes in the genetic programming of the maturing mammary gland (MG) increasing the susceptibility of developing the disease. Less is known about how HFD induced genes impact BC development. HFD exposure two weeks before conception to six weeks of age was previously shown to dramatically change MG gene expression in 10 week old mice. Therefore, we investigated these differentially expressed HFD-induced genes for their expression in BC using the NKI 295 breast tumor dataset.
Results
To examine the potential role of HFD induced genes in BC, we first investigated whether these HFD-induced genes in mouse MGs were differentially expressed in different types of human BC. Of the 28 HFD induced genes that were differentially expressed between BC subtypes in the NKI set, 79% were significantly higher in basal-like BC. Next, we analyzed whether HFD induced genes were associated with BC prognosis utilizing gene expression and survival data for each HFD induced gene from the NKI data and constructed Kaplan Meier survival plots. Significantly, 93% of the prognosis associated genes (13/14) were associated with poor prognosis (P = 0.002). Kaplan Meier analysis with 249 non-basal-like BC found that all but one of the genes examined were still significantly associated with poor prognosis. Furthermore, gene set enrichment analysis (GSEA) with HFD microarray data revealed that invasive BC genes where enriched in HFD samples that also had lost expression of luminal genes.
Conclusions
HFD exposed mouse MGs maintain differential expression of genes that are found highly expressed in basal-like breast cancer. These HFD-induced genes associate with poor survival in numerous BC subtypes, making them more likely to directly impact prognosis. Furthermore, HFD exposure leads to a loss in the expression of luminal genes and a gain in expression of mesenchymal and BC invasion genes in MGs. Collectively, our study suggests that HFD exposure during development induces genes associated with poor prognosis, thus identifying how HFD diet may regulate BC development.
Electronic supplementary material
The online version of this article (doi:10.1186/1756-0500-7-543) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, Basal-like breast cancer, High fat diet, Poor prognosis, Mammary gland development, Diet-induced genes
Background
Recent evidence suggests that dietary content is one causal lifestyle factor that may contribute to breast cancer (BC) development. Studies focusing on migrant populations have shown that women migrating from low (Asian, Latin American) to high BC incidence rate countries (United States and other Western countries) acquire a higher risk for BC similar to those in the new country [1–6]. These studies suggest that dietary changes might be a strong contributing factor to the increased incidence rates observed in these migrating populations. A study analyzing the effects of dietary patterns and the risk of BC in women of different ethnicities concluded that women consuming a Western diet had a higher risk of BC [7]. The Western diet consists of foods primarily high in fat. A higher intake of dietary fat has been shown to increase BC risk in adults [8, 9].
A high-fat diet (HFD) may also determine the type of BC that will develop translating to a more precise treatment strategy increasing patient survival. Breast cancer tumors have been previously classified into subtypes based on their unique molecular/genetic expression profiles that correlate with phenotypic characteristics and clinical outcome [10–13]. The expression of estrogen receptor (ER) is a distinguishing marker between these tumors subtypes in combination with other molecular cues. Luminal subtype A (luminal A) and luminal subtype B (luminal B) are tumors that have high expression levels of ER (ER+) and are associated with a favorable prognosis. ERBB2 is overexpressed in HER2 positive breast cancers whereas basal epithelial-like (basal-like) tumors are ER negative (ER-). HER2 and basal-like tumors are associated with a poor prognosis [10, 11, 13, 14]. Strikingly, the five year survival rate for patients with luminal A is 90% compared to as low as a 30% five year survival rate for patients with HER2 and basal-like BC tumors [14–16]. This difference is due, in part, to the difficulty in predicting a clinical course, tumor stage at diagnosis and availability of targeted therapy [14, 17]. Obesity significantly increases the risk of developing basal-like BC in premenopausal women, predicting a poor outcome in these individuals [18]. Commensurately, obese individuals are much less likely to develop luminal BC. Therefore, identifying the impact a HFD has on the type of BC a women develops may be critical in identifying the disease early to help predict a clinical course of action that would increase that patient’s survival.
The impact that HFD has on developing BC in humans has yet to be fully elucidated, in part, due to the limited number of human studies (and ability) to adequately assess the effect diet has on genetic regulation of the developing mammary gland (MG). Studies using rodent models have provided the most compelling evidence linking developmental exposure to a HFD to breast carcinogenesis [19–21]. These studies culminate to identify several theories to how consumption of a HFD during pre and/or postnatal development may cause BC including genetic reprogramming of the MG [22–25]. However, it is unclear as to how HFD might contribute to the development of specific types of BC correlating to the severity of the disease and patient survival. Thus, identifying MG HFD-induced transcriptional programs that are pertinent to specific types of human BC could have a tremendous impact on understanding the etiology of diet induced BC.
The purpose of this study was to examine genes induced by exposure to a HFD during MG development in the mouse for the role they may play in human BC. Luitjen et. al. [25] previously identified genes that were differentially expressed in 10 week old MGs after developmental HFD exposure. Strikingly, these changes were sustained even after HFD exposure, suggesting a long-term effect. To investigate how HFD may influence the development of BC, we examined the experimentally identified HFD-induced genes from the study by Luitjen et al. [25] in human breast tumors. The expression of HFD- induced genes was examined in BC by utilizing the widely studied NKI BC dataset from the Netherlands Cancer Institute, which contains gene expression and patient follow up/survival data from 295 women between the ages of 26 and 53 [10]. The NKI BC data set had been previously classified into five breast cancer subtypes [10]. Using Kaplan Meier analysis and gene set enrichment analysis (GSEA) we found that HFD induced genes were associated with poor prognosis and invasive BC. We also found that HFD leads to a loss in luminal gene expression and a gain in basal-like gene expression. Our data shows for the first time that HFD-induced genes are highly expressed in BCs that are of the basal-like subtype and are associated with poor prognosis.
Methods
Collection of HFD-induced genes and gene expression analysis in BC subtypes
Microarray data of differentially expressed genes was obtained from a study analyzing the effects of early diet on the genetic programming of the MG in wild-type mice [25]. As described previously [25], FVB wild-type mice were fed a HFD diet consisting of either a 24% high fat content of either n-6 (corn oils) or n-3 (flaxseed oil) PUFAs (polyunsaturated fatty acids) from two weeks prior conception to 6 weeks of age. Control animals were fed standard rodent chow (5% fat) during the same period. Gene expression profiling of the MG was performed at 10 weeks of age. Luitjen et al. [25] utilized the data from both types of fat (n-3 and n-6) in ANOVA analysis to identify differentially expressed genes. We examined the top one hundred differentially expressed genes that were induced by HFD with a p-value of 0.0001 or less based on the Luitjen ANOVA analysis for representation in the NKI dataset. The NKI dataset contains gene expression and survival data for 295 human breast tumors; these samples have also been previously classified based on breast cancer subtypes (basal-like = 46 tumors; HER2 positive = 49 tumors; luminal A = 88 tumors; luminal B = 81 tumors; normal-like = 31 tumors) [10, 11]. Our analysis focused on the HFD-induced genes found in the NKI data set (41 total). The average expression of each HFD-induced gene was determined for each BC sub-type. In order to determine whether HFD genes were differentially expressed between breast cancer subtypes, we performed the two tiered, type two T-test to determine whether each gene was expressed signficantly different between the basal-like subtype and the other sub-types of BC (HER2, luminal A, luminal B and normal-like). The T-test gave the same p-value as one way ANOVA, however a T-test anaylsis was used given we were only examining a single factor between two groups at a time. The false discovery rate (FDR) was calculated for differentially expressed genes to correct for multiple hypothesis testing. Changes in gene expression between subtypes that had an FDR of 0.01 or less were considered to be signifcant. It is improtant to note that the data sets utilized in this article were from experiemnts previously approved by their respective institutial ethics commitees and follow their national legeslation [10, 25].
Survival analysis
A Kaplan Meier survival analysis for each HFD induced gene was performed using the NKI dataset and MedCalc software (Mariakerke, Belgium) for each gene to be analyzed. The association with prognosis of human breast cancer for each gene was obtained from this analysis: poor prognosis; good prognosis; no association. 295 breast tumors were divided into two equal-sized groups based on the expression of each gene; group one represented low expression of the gene and group two represented high expression of the gene. All availabe time points were utilized. P-values of less than 0.05 were considered significant associations. Based on the Chang et al. [11] classification of the NKI database, we utilized 249 breast cancer samples for the non-basal like breast tumor analysis.
Additional statistical analysis of poor prognosis genes
HFD-induced genes that were associated with poor prognosis were analyzed as a group for overall differential expression between BC subtypes using NKI data. We performed ONE-WAY ANOVA with a Dunnett’s post test to determine the significance of mean gene expression between BC subtypes using the basal-like subtype as the reference group. Probability values of less than 0.05 were considered significant. The PC IBM INSTAT and PRISM software were used for the statistical analyses and graphs, respectively (GraphPad, San Diego, CA, USA).
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed using the Broad Institute platform [26, 27]. Samples were analyzed with weighted, Signal2Noise default settings. Microarray gene expression data from Luitjen et al. was grouped based on diet (control low fat diet or HFD samples) [25]. Both types of HFD (n-3 and n-6 as indicated) were independently studied by GSEA with similar results. (Additional file 1: Table S1) contains the list of 361 breast cancer gene sets from the Molecular Signatures database, Broad Institute, that were utilized for analysis. The luminal gene sets utilized in GSEA are in (Additional file 2: Table S2).
Results
HFD during MG development induces basal-like BC genes
Murine MGs exposed to HFD during development (two weeks before conception to 6 weeks post birth) had dramatic changes in gene expression at ten weeks of age [25]. In order to investigate how HFD may influence BC development, we examined genes from the Luitjen et al. study that were induced by HFD with a p-value of 0.0001 or less for expression in BC subtypes by utilizing data for these genes in the NKI BC dataset. Gene expression signatures classify breast cancer into 5 basic subtypes: basal-like, HER2, luminal A, luminal B and normal-like. The basal-like subtype is associated with the worst prognosis whereas the luminal A is associated with the best prognosis [12, 13]. The average expression of each HFD induced gene was determined for each BC subtype (Table 1). Changes with an FDR of 0.01 or less were considered significantly different. Twenty-eight out of the 41 HFD induced genes showed statistically different levels of gene expression between the luminal A (associated with good prognosis) and basal-like (associated with poor prognosis) subtypes of BC; 79% of these genes were significantly higher in basal-like BC compared to luminal A BC. Therefore, HFD exposed glands preferentially maintain induced expression of genes that are found highly expressed in basal-like BC.
Table 1.
Expression of HFD-induced genes in breast cancer subtypes and association with breast cancer prognosis
| Gene a | Basal-like b | HER2 c | Luminal A d | Luminal B e | Normal-like f | Survival association g |
|---|---|---|---|---|---|---|
| MMP12 | 0.17 | -0.13* | -0.24* | -0.19* | -0.33* | Poor prognosis |
| GPNMB | -0.01 | 0.00 | -0.18* | -0.06 | -0.11 | Poor prognosis |
| CTSL | 0.07 | 0.04 | -0.16* | -0.03* | -0.18* | Poor prognosis |
| ITGAX | 0.00 | 0.07* | 0.04 | 0.05 | 0.05 | - |
| LILRB4 | 0.10 | 0.02 | -0.15* | -0.04* | -0.22* | Poor prognosis |
| TM7SF1 | -0.18 | 0.05* | 0.05* | 0.01* | -0.03* | - |
| DNMT3A | 0.09 | 0.03 | -0.08* | 0.00* | -0.10* | Poor prognosis |
| TYROBP | 0.06 | 0.01 | -0.05* | -0.01 | -0.13* | - |
| DIO1 | -0.21 | -0.18 | -0.04* | -0.01* | -0.07* | - |
| SLC11A1 | 0.14 | 0.01* | -0.10* | -0.03* | -0.15* | Poor prognosis |
| GALNS | 0.05 | 0.03* | 0.01 | 0.03 | 0.01 | - |
| NCF2 | 0.11 | 0.01* | -0.15* | -0.04* | -0.16* | Poor prognosis |
| PTPNS1 | 0.23 | 0.00* | -0.15* | -0.08* | -0.14* | Poor prognosis |
| LGALS3 | -0.07 | 0.04* | -0.03 | -0.01* | 0.04* | - |
| MTHFS | -0.02 | 0.00 | 0.02 | 0.05* | -0.07 | - |
| IL1RN | 0.00 | -0.17* | -0.13* | -0.11 | -0.05 | - |
| CTSS | 0.12 | -0.04* | -0.17* | -0.06* | -0.20* | - |
| WBSCR5 | 0.03 | 0.01 | 0.00 | 0.00 | -0.06* | - |
| PRKCD | -0.05 | 0.02* | 0.03* | 0.04* | -0.02 | - |
| CAPG | 0.09 | -0.03* | -0.06* | 0.01* | -0.07* | - |
| CD68 | 0.06 | 0.09 | -0.02* | 0.02 | -0.01* | - |
| ITGB2 | 0.11 | 0.04 | -0.09* | -0.03* | -0.11* | Poor prognosis |
| CSTB | 0.18 | 0.02* | -0.04* | -0.03* | -0.07* | Poor prognosis |
| ATF3 | 0.01 | -0.13* | -0.12* | -0.09* | 0.01 | - |
| CSF2RA | 0.01 | -0.01 | -0.02 | -0.01 | -0.11* | - |
| WFS1 | -0.20 | 0.00* | 0.03* | 0.06* | -0.02* | - |
| PPGB1 | 0.00 | 0.07* | -0.05* | 0.02 | -0.07* | Poor prognosis |
| ADAM8 | 0.12 | 0.02* | -0.09* | -0.06* | -0.25* | Poor prognosis |
| CCRL2 | 0.06 | 0.07 | -0.01 | 0.02 | -0.06* | - |
| VTN | 0.00 | 0.00 | 0.01 | 0.01 | 0.06 | - |
| RGS1 | 0.04 | 0.01 | -0.03* | -0.04* | -0.13* | - |
| FCGR3 | 0.09 | 0.06 | -0.03* | 0.03* | -0.08* | Poor prognosis |
| EMR1 | 0.06 | -0.03 | 0.01 | 0.00* | 0.03 | - |
| ARPC2 | 0.06 | 0.03 | -0.06* | -0.03* | -0.03* | - |
| ESD | -0.04 | -0.04 | 0.02* | -0.01 | 0.08* | Good prognosis |
| VAMP4 | 0.06 | 0.05 | 0.05 | 0.05 | -0.03 | - |
| CDKN1C | -0.05 | -0.05 | -0.05 | -0.12 | 0.13* | - |
| HEXB | -0.04 | 0.00 | -0.02 | -0.02 | -0.03 | - |
| NAGLU | -0.01 | 0.05* | 0.02 | 0.04* | 0.12* | - |
| KCNH2 | 0.00 | -0.29* | -0.21* | -0.15* | -0.32* | - |
| MSTF1 | 0.02 | 0.03 | 0.04* | 0.04* | 0.04 | - |
aHFD induced genes (Luitjen et al.) that were present in the NKI human breast cancer data set as described in the Methods. b-fAverage gene expression among 5 different human breast cancer subtypes for these genes (Tumors: basal-like n = 46; HER2 n = 49; luminal A n = 88; luminal B n = 81; normal-like n = 31). gTotal of 13 genes associated with poor prognosis and 1 gene associated with good prognosis based on Kaplan Meier survival analysis for the indicated gene with 295 BC samples from the NKI dataset as described in the Materials and Methods. The plots for the genes that were significantly associated with prognosis are depicted in Figure 1 of this manuscript. *Gene expression statistically different with respect to basal-like subtype (False discovery Rate [FDR] < 0.01).
HFD induced genes are strongly associated with poor prognosis in BC
In order to examine whether HFD induced genes were associated with BC prognosis, we performed Kaplan Meier survival analysis using HFD induced genes from 10-week old mammary fat pads and the NKI dataset, as described in the Methods. A total of fourteen genes out of forty-one were associated with prognosis in BC (p < 0.05). Only one HFD-induced gene was associated with good prognosis; the high expression of Esterase D (ESD), showed a higher survival probability when compared to the low gene expression (Figure 1, Table 1). Significantly, 93% of the prognosis associated genes (13 out of 14 HFD-induced genes) were associated with poor prognosis; the high expression of these genes showed a lower survival probability when compared to their low expression (Figure 1, Table 1) (P = 0.002). The AmiGO database [28] was utilized to identify gene ontologies that were associated with the HFD-induced poor prognosis genes. We found that the poor prognosis genes were associated with cell adhesion, proteolysis, immunological response, ion transport and DNA methylation (Table 2). Altogether, HFD-induced genes during mammary development are highly associated with poor survival in BC.
Figure 1.
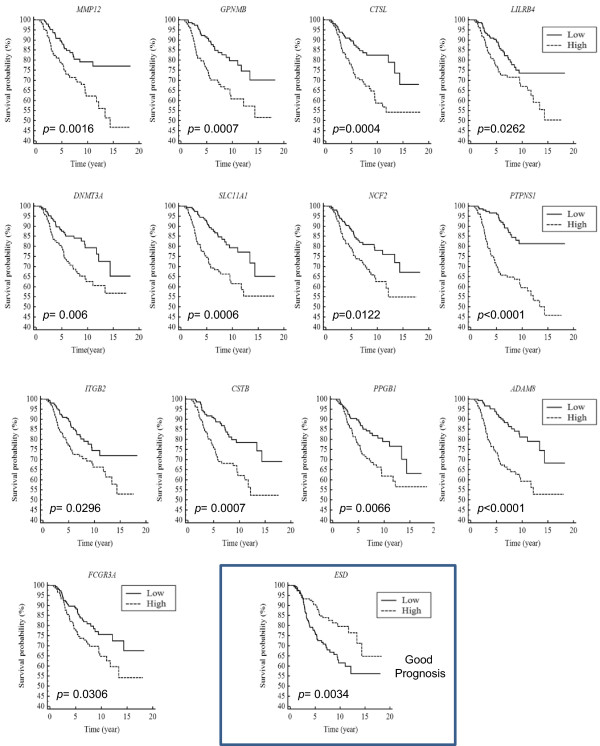
HFD-induced genes are strongly associated with poor BC prognosis. HFD induced genes from mouse MGs were examined for an association with human BC prognosis using Kaplan Meier survival analysis. Gene expression and survival data was obtained from the NKI (Netherlands Cancer Institute) dataset for each HFD induced gene. 295 breast tumors were divided into two equal-sized groups based on the expression of the indicated gene; one group represented low expression of the gene and the other group represented high expression of the gene. Depicted are 14 out of the 41 analyzed HFD induced genes; only these 14 genes showed significant association with breast cancer prognosis. 13 out of 14 HFD induced genes were significantly associated with a poor prognosis.
Table 2.
Cellular processes of genes associated with poor prognosis
| Gene | Description | GO reference | Gene ontology |
|---|---|---|---|
| Mmp12 | Matrix metallopeptidase 12 | GO:0006508 | Proteolysis |
| Gpnmb | Glycoprotein (transmembrane) | GO: 0007155 | Cell adhesion |
| Ctsl | Cathepsin L | GO:0006508 | Proteolysis |
| Lilrb4 | Leukocyte immunoglobulin-like receptor, subfamily B, member 4 | GO: 0045671 | Negative regulation of osteoclast differentiation |
| Dnmt3a | DNA (cytosine-5-)-methyltransferase 3 alpha | GO:0006306 | DNA methylation |
| Slc11a1 | Solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 | GO:0070839 | Divalent metal ion export |
| Ncf2 | Neutrophil cytosolic factor 2 | GO:0016175 | Superoxide-generating NADPH oxidase activity |
| Ptpns1 | Signal-regulatory protein alpha | GO: 0007155 | Cell adhesion |
| Itgb2 | Integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | GO:0007155 | Cell adhesion |
| Cstb | Cystatin B (stefin B) | GO:0010466 | Negative regulation of peptidase activity |
| Ppgb1 | Cathepsin A | GO: 0006508 | Proteolysis |
| Adam8 | ADAM metallopeptidase domain 8 | GO: 0006508 | Proteolysis |
| Fcgr3 | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | GO:0001788 | Antibody-dependent cellular cytotoxicity |
Gene ontologies from the AMIGO database were obtained for HFD induced genes that were found to be associated with poor prognosis.
The strong association between HFD-induced genes and poor prognosis in human breast cancer prompted us to further investigate this phenomena. We first examined the expression distribution of these genes in the 295 breast cancer tumors by plotting the expression of individual genes for each breast tumor sample, (Additional file 3: Figure S1) and (Additional file 4: Figure S2). One can see that there is a high expression of each poor prognosis associated gene in basal-like breast cancer samples whereas there is lower expression in luminal A breast cancers. Each of the poor prognosis genes is significantly less well expressed in luminal BC compared to basal-like BC, Table 1. Genes associated with poor prognosis were analyzed as a group for differential expression in human breast cancer subtypes. The average expression of these genes showed significant statistical difference in HER2, luminal A, luminal B, and normal-like subtypes with respect to the basal-like subtype (ANOVA p < 0.01, Figure 2). Of note, the differences in expression were more striking between basal-like BC samples and luminal A, luminal B and normal-like tumors, whereas a less dramatic, but still significant difference was found between basal-like tumors and HER2 positive tumors. This result is consistent with the fact that HER2 positive cancer are associated with poor prognosis as well [12, 14].
Figure 2.
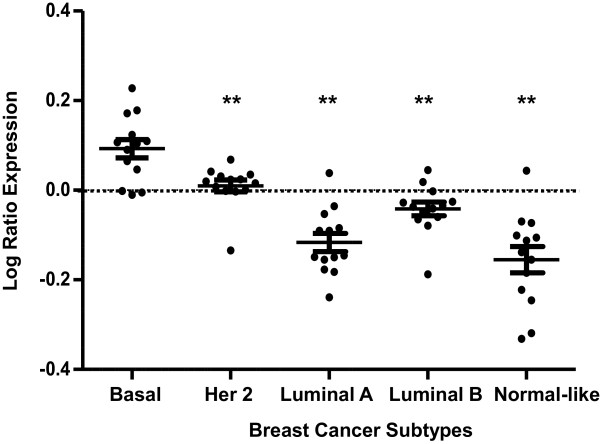
Poor prognosis HFD genes are highly expressed in the basal-like subtype of BC. We calculated the mean (±SEM) expression of each poor prognosis associated gene (from Figure 1) for each BC subtype using NKI gene expression data. Poor prognosis genes were significantly more highly expressed in the basal-like subtype compared to the other subtypes (Her2, luminal A, luminal B, and normal-like subtypes) based on ANOVA with Dunnett’s post-test using the basal-like subtype as the reference group. **p < 0.01.
Although it is clear that HFD-induced genes are more strongly expressed in basal-like breast cancer, we were interested to examine whether these genes were associated with prognosis in other subtypes of breast cancer. To investigate this, we performed Kaplan Meier analysis with 249 non-basal-like NKI samples and found that all but 1 (92%) of the genes examined (Matrix metalloproteinase 12, MMP12) were still significantly associated with poor prognosis (Table 3). Therefore, HFD-induced genes associate with poor survival in a broad range of human breast cancer subtypes, making them likely to directly impact prognosis and have an influence on survival in many types of breast cancer.
Table 3.
Genes associated with poor prognosis in non-basal-like breast cancer
| Gene | Association | P-value |
|---|---|---|
| GPNMB | Poor prognosis | P = 0.0011 |
| CTSL | Poor prognosis | P = 0.0022 |
| LILRB4 | Poor prognosis | P = 0.0274 |
| DNMT3A | Poor prognosis | P = 0.0246 |
| SLC11A1 | Poor prognosis | P = 0.0461 |
| NCF2 | Poor prognosis | P = 0.0011 |
| PTPNS1 | Poor prognosis | P = 0.0008 |
| ITGB2 | Poor prognosis | P = 0.0013* |
| CSTB | Poor prognosis | P = 0.0332 |
| PPGB | Poor prognosis | P = 0.0009 |
| ADAM8 | Poor prognosis | P = 0.0007 |
| FCGR3 | Poor prognosis | P = 0.0053 |
| MMP12 | - | P = 0.1092 |
Kaplan Meier analysis was performed with all non-basal breast cancer samples (249) from the NKI dataset and the HFD genes that were found to be associated with prognosis (from Figure 1 and Additional file 1: Table S1). *5 year Kaplan Meier curve was used instead of all time points for ITGB2. Strikingly, 12 out of 13 genes were still strongly associated with poor prognosis in non-basal breast cancer samples.
HFD exposure induces BC mesenchymal and invasion genes
HFD exposure during development in mice leads to dramatic changes in mammary fat pad gene expression. These changes strongly suggest that the breast tissue exposed to HFD are developmentally altered and may sustainably express programs that parallel those found in certain types of BC. To investigate this, we performed GSEA with 361 curated BC gene sets from the Broad Institute Molecular Signatures Database. Each of the 361 examined gene sets contained genes that were previously determined to be differentially expressed under defined condition in breast cancer. In GSEA, Kolmogorov-Smirnov-style statistics are employed to investigate whether gene sets are more highly expressed (enriched) between two conditions in microarray analyses. We compared microarray gene expression data from control low fat MGs to data from HFD MGs to determine whether any of the BC gene sets were significantly enriched in HFD samples. We found that genes associated with BC invasion were significantly enriched in HFD samples, Figure 3 and Table 4. Enrichment plots are depicted in Figure 3 in which genes are ranked based on their association with given phenotypes, in this case the phenotypes are HFD (on the right) or control diet (on the left). The lines underneath the enrichment plot depict genes that are contained on the gene list being investigated. One can see a striking enrichment of invasion genes in HFD treated samples, Figure 3. Therefore, HFD leads to an increase in the expression of genes involved in BC invasion.
Figure 3.
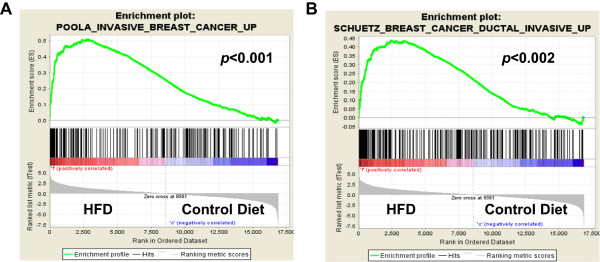
BC invasion genes are enriched in HFD microarray samples. Gene set enrichment analysis (GSEA) was performed with microarray data from 10 week old mice treated with HFD (labeled F) or control diet (labeled C) and 361 curated genes sets from the Molecular Signatures Database, Broad Institute; enrichment plots for two of the analyzed gene sets are shown. In these analyses, genes are ranked based on their association with the HFD phenotype or the control phenotype. Genes most strongly associated with HFD would be on the far right. Enrichment scores (ES) are calculated using a weighted Kolmogorov-Smirnov-style statistic. Lines underneath the graph depict the genes that are contained in the investigated gene set. A-B, genes associated with invasion in BC were highly associated with HFD samples. P values are indicated on plots.
Table 4.
Breast cancer gene sets that are enriched in HFD microarray samples
| GENE SET NAME | SIZE | ES | NES | NOM |
|---|---|---|---|---|
| p-value | ||||
| SMID_BREAST_CANCER_NORMAL_LIKE_UP | 348 | 0.50 | 2.13 | <0.001 |
| POOLA_INVASIVE_BREAST_CANCER_UP | 215 | 0.51 | 1.96 | <0.001 |
| BERTUCCI_MEDULLARY_VS_DUCTAL_BREAST_CANCER_UP | 142 | 0.46 | 1.91 | <0.001 |
| FINAK_BREAST_CANCER_SDPP_SIGNATURE | 21 | 0.65 | 1.87 | <0.001 |
| FINETTI_BREAST_CANCER_KINOME_GREEN | 15 | 0.75 | 1.86 | <0.001 |
| SCHUETZ_BREAST_CANCER_DUCTAL_INVASIVE_UP | 266 | 0.44 | 1.77 | <0.001 |
| SMID_BREAST_CANCER_LUMINAL_B_DN | 412 | 0.36 | 1.69 | <0.001 |
| JOHNSTONE_PARVB_TARGETS_3_UP | 306 | 0.34 | 1.64 | <0.001 |
Gene set enrichment analysis was preformed with microarray data from high fat and low fat diet as described in the Materials and Methods and 361 curated breast cancer gene sets on the Molecular Signatures Database, Broad Institute. In GSEA enrichment scores (ES) are calculated for each gene set using a Kolmogorov-Smirnov-style statistic. The ES indicates how strongly associated a gene set is with a given phenotype or not. GSEA generates nominal p-values (NOM p-value) using on a phenotype based permutation comparing ES with a null distribution. Normalized enrichment scores (NES) are adjusted enrichment scores based on the number of genes in each examined set. Gene sets with a nominal P-value (NOM p-value) of 0.001 or less and a normalized enrichment score (NES) of 1.5 or greater are shown. These data suggest that common genes are expressed in mammary fat pads from mice treated with HFD and invasive breast cancer.
To further examine HFD mediated changes in gene expression in the MG with regard to BC development, we performed GSEA with 53 luminal BC gene sets from the Molecular Signatures database, Broad Institute. We found that genes that are normally down-regulated in the luminal subtype of BC (and are highly expressed in mesenchymal BC) were enriched in HFD treated samples, suggesting that these mice may have altered breast development to a more basal phenotype (Figure 4 A-B, Table 5). Enrichment plots for analysis with gene sets that are down in luminal BC and high in mesenchymal BC (Charafe_luminal_versus_mesenchymal_DN) indicate a clear increase in mesenchymal BC genes upon HFD treatment, Figure 4. Furthermore, we found that stromal genes from poor prognosis BC (gene-set FINAK_BREAST_CANCER_SDPP_SIGNATURE, SDPP is an abbreviation for stroma-derived prognostic predictor of BC disease outcome), were strongly associated with HFD MGs with a Normalized Enrichment Score (NES) of 1.87, p = <0.001 (Table 4). One of the top enriched genes in our GSEA was the mesenchymal marker Vimentin. We assessed the average Vimentin expression in control MGs versus HFD fed MGs using the Luitjen et al. [25] microarray dataset and found that HFD exposed MGs expressed significantly higher levels of the basal-like BC marker Vimentin. The average expression of Vimentin was strikingly tenfold higher with n-3 HFD and 20 fold higher with n-6 HFD (Figure 4C). In sum, HFD exposure leads to a loss in the expression of luminal genes and a gain in expression of mesenchymal and BC invasion genes in MGs.
Figure 4.
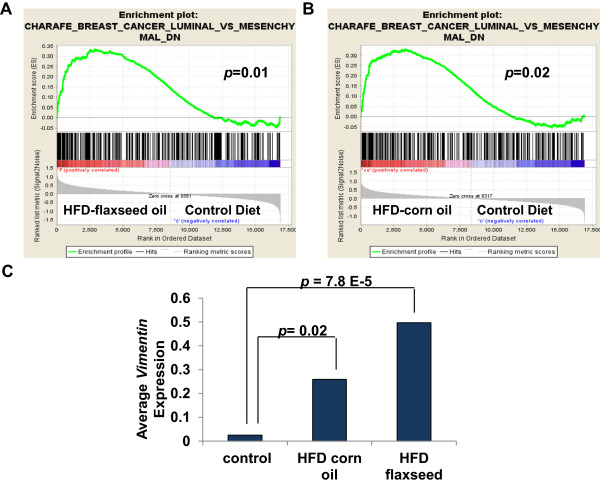
BC Mesenchymal genes are enriched in HFD microarray samples. GSEA was performed with microarray data from 10 week old mice that were fed HFD (labeled F for flaxseed or CO for corn oil) or normal diet (labeled C for control) and 361 curated gene sets from the Molecular Signatures Database, Broad Institute; enrichment plots are shown for the Charafe_Breast_Cancer_Luminal_VS_Mesenchymal_DN geneset. A-B, genes associated with mesenchymal characteristics in BC were significantly associated with the HFD samples. C, the average expression of the mesenchymal marker Vimentin was calculated for HFD (corn oil or flaxseed) and control samples using data from the Luitjen et al. microarray data set. Vimentin was strikingly expressed 10–20 fold higher in HFD treated MG compared to control MGs. P values are indicated on graphs.
Table 5.
HFD leads to a loss in the expression of luminal genes
| GENE SET NAME | Size | ES | NES | NOM |
|---|---|---|---|---|
| p-value | ||||
| SMID_BREAST_CANCER_NORMAL_LIKE_UP | 348 | 0.50 | 2.09 | <0.001 |
| BERTUCCI_MEDULLARY_VS_DUCTAL_BREAST_CANCER_UP | 142 | 0.47 | 1.95 | <0.001 |
| FINETTI_BREAST_CANCER_KINOME_GREEN | 15 | 0.75 | 1.84 | 0.01 |
| SMID_BREAST_CANCER_LUMINAL_B_DN | 412 | 0.37 | 1.70 | <0.001 |
| FARMER_BREAST_CANCER_CLUSTER_1 | 25 | 0.66 | 1.70 | 0.01 |
| CHARAFE_BREAST_CANCER_LUMINAL_VS_MESENCHYMAL_DN | 325 | 0.33 | 1.53 | 0.01 |
| HOWLIN_PUBERTAL_MAMMARY_GLAND | 60 | 0.40 | 1.50 | 0.03 |
Gene set enrichment analysis was performed with 53 gene sets containing luminal genes in breast cancer denoted (form the Molecular Signatures Database, Broad Institute). In GSEA enrichment scores (ES) are calculated for each gene set using a Kolmogorov-Smirnov-style statistic. The ES indicates how strongly associated a gene set is with a given phenotype or not. GSEA generates nominal p-values (NOM p-value) using on a phenotype based permutation comparing ES with a null distribution. Normalized enrichment scores (NES) are adjusted enrichment scores based on the number of genes in each examined set. We found that gene sets with down-regulation of luminal genes were enriched in the HFD samples.
Discussion
BC is a heterogeneous disease. Identifying dietary factors that contribute to the development of different BC subtypes is important for the prevention and treatment of the disease. A HFD has been shown to increase BC risk in humans [8, 9] and several studies using rodent models have shown that exposure to a HFD in utero and/or during postnatal development significantly increase carcinogen stimulated MG tumorigenesis [19–21]. However, the developmental processes in the MG impacted by a HFD that mediate the heterogeneity of BC are unclear. Recently, work by Luitjen et al. [25] revealed that HFD exposure during MG development leads to sustained changes in gene expression. In the present study, we found that genes induced by a HFD in the mouse developing MG were strongly expressed in the human basal-like subtype of BC, suggesting that HFD exposed MGs may have abnormal basal-like characteristics. In line with this, we found increased expression of mesenchymal genes by GSEA including the well described marker Vimentin in HFD gland samples. Our analysis reveals that HFD exposure during development leads to an increase in basal-like characteristics in MGs along with a concomitant loss of luminal gene expression (Figures 3–4), which may promote an increase in basal-like BC development. This pattern of gene expression parallels the increase in basal-like BC/decrease in luminal BC that is found in obese women [18]. It remains to be determined as to which HFD exposed cells express the basal-like genes and whether the MG architecture is dramatically altered in a sustained manner in these models and in humans. Nonetheless, HFD profoundly affects MG gene expression in a sustained way, inducing genes that are expressed in basal-like BC.
Here we show that of the HFD-induced genes that were significantly associated with prognosis in BC, a striking 93% was significantly associated with poor prognosis. All but one these genes were still strongly associated with poor prognosis in the non-basal-like set of BCs (249 tumors examined). Therefore, the association between HFD-induced genes and prognosis in BC is highly compelling and spans many subtypes. While some skepticism could be drawn due to the relevance of cross-species analysis, a strong body of evidence has demonstrated the parallels between rodent and human MG structures, MG development and carcinogenesis [29–33]. Furthermore, mouse MG tumors have been shown to replicate the diversity of human BCs including tumor initiation, hormone dependency and histopathology across multiple MG endpoints [31–33]. Thus, our data suggests a model in which HFD impacts MG development to promote poor prognosis cancer.
Interestingly, we provide evidence that a HFD, regardless of content (n-3 or n-6 PUFAs), may preferentially promote basal-like BC. Epidemiological evidence supports the idea that a HFD increases BC risk. Whether this risk is a consequence of overall percentage of fat in an individual’s diet or type of PUFAs in the diet has long been debated. Numerous rodent studies have investigated the dietary impact of (n-3) and (n-6) PUFAs on BC risk. While rodent studies support the notion that diets high in (n-6) PUFAs increase breast carcinogenesis [34–37], diets primarily consisting of n-3 PUFAs have been suggested to protect against BC [38, 39]. However, Hilakivi-Clarke et al. [22], showed that prepubertal rats exposed to a high (n-3) PUFA diet resulted in key biological changes within the MG reflecting an increased susceptibility to BC. Furthermore, studies in adult rats have shown that exposure to a diet high in (n-3) PUFAs do not inhibit carcinogen induced mammary tumorigenesis, but may promote it [40, 41]. While the timing of exposure could play a factor (fetal, adolescence, adult), human studies have yet to provide conclusive evidence that diets consisting of primarily (n-3) or (n-6) PUFAs play a profound role in increasing or decreasing BC risk. [42–44]. Thus, the influence of a HFD on BC development maybe more a consequence of the amount fat consumed regardless of the source and our data supports this hypothesis.
Conclusions
Given the striking association between HFD-induced genes during development and prognosis in BC, it is imperative to examine these phenomena in humans. The greatest strides in decreasing cancer mortality have been made by prevention as with the link between smoking and lung cancer. It will be important to determine whether developmental HFD exposure as in this study and/or exposure in adults lead to the induction of poor prognosis genes. Also, it will be critical to determine whether these changes are permanent or require the presence of HFD or are maintained for only a certain period of time. It will also be crucial to determine how any of the maintained changes in gene expression are propagated. Based on the HFD mediated induction of DNA (cytosine-5-)-methyltransferase 3 alpha (DNMT3A), perhaps an epigenetic change is responsible for these sustained shifts in gene expression. DNMT3A functions in de novo methylation, is important in development and altered expression levels have been found in several different types of human cancers [45]. If the link between poor prognosis in breast cancer and developmental HFD exposure bare out in humans, efforts can be made to prevent such exposures. In addition, if this link holds true in human breast cancer, it may be possible to identify women who are more susceptible to poor prognosis cancers based on the expression of these newly identified HFD-induced basal-like characteristics such as the mesenchymal marker Vimentin. Importantly, vimentin has been shown to play a significant role in the epithelial-mesenchymal transition (EMT) process in BC [46, 47]. EMT is a cellular reprograming process that changes the shape of epithelial cells to exhibit a more motile mesenchymal phenotype (reviewed in [48]). In vitro Vimentin over expression in human BC cells has been shown to contribute to this process, thus increasing BC cell motility and invasive properties [47]. In human breast cancer samples, vimentin expression is found in high-grade ductal carcinomas with low ER expression levels [49]. Thus, identifying early changes in vimentin expression in breast epithelium due to HFD exposure might be used to predict clinical outcomes translating to better preventive treatment strategies.
In sum, we show that HFD induces the expression of genes that are associated with poor prognosis in BC. HFD exposed MGs showed high expression of the mesenchymal marker Vimentin as well as a loss in luminal markers. We propose that developmental HFD exposure leads to sustained changes in gene expression that promote the development of poor prognosis cancers including basal-like BC.
Availability of supporting data
The data supporting the results of the article is included within the article and in supplementary materials.
Electronic supplementary material
Additional file 1: 361 breast cancer gene sets that were utilized in GSEA. This excel file details the 361 breast cancer gene sets from the Molecular Signatures Database that were utilized for gene set enrichment analysis with the HFD microarray data. NAME denotes the gene set name, original size denotes the number of genes in the curated gene set, after restricting to data set denotes the number of genes that were found on both the gene list and the HFD microarray data set, and status denotes whether the gene list was included in the analysis or not. A small number of gene lists were not included in the analysis, if there were not enough genes. (PDF 279 KB)
Additional file 2: 53 luminal breast cancer gene sets that were utilized in GSEA. This excel file details the 53 breast cancer gene sets from the Molecular Signatures Database that were utilized for gene set enrichment analysis with the HFD microarray data. NAME denotes the set name, original size denotes the number of genes in the curated gene set, after restricting to data set denotes the number of genes that were found on both the gene list and the HFD microarray data set, and status denotes whether the gene list was included in the analysis or not. A small number of gene lists were not included in the analysis, if there were not enough genes. (PDF 184 KB)
Additional file 3: Poor prognosis associated HFD-induced genes MMP12, GPNMB, CTSL and LILRB4 are highly expressed in basal-like BC. Depicted are histograms with log ration expression values for the indicated gene for each tumor (from the 295 NKI breast tumor dataset). Tumors are grouped together based on subtype. These subtypes are indicated at the top of the figure: Basal-like tumors are 1–46, HER2 are 47–95, Luminal A are 96–183, Luminal B are 184–264 and Normal-like tumors are 265–295. (PDF 326 KB)
Additional file 4: Poor prognosis associated HFD-induced genes DNMT3A, SLC11A1, NCF2 and PTPNS1 are highly expressed in basal-like BC. Depicted are histograms with log ration expression values for the indicated gene for each tumor (from the 295 NKI breast tumor dataset). Tumors are grouped together based on subtype. These subtypes are indicated at the top of the figure: Basal-like tumors are 1–46, HER2 are 47–95, Luminal A are 96–183, Luminal B are 184–264 and Normal-like tumors are 265–295. (PDF 335 KB)
Acknowledgements
RCMCh was supported by an undergraduate training grant from the Howard Hughes Medical Institute Precollege and Undergraduate Science Education Program- award number 52007568. RKD was supported by a Faculty Research Council (FRC) grant from the University of Texas-Pan American (UTPA). The content of this publication is the responsibility of the authors and does not represent the views of the HHMI.
Abbreviations
- BC
Breast cancer
- HFD
High fat diet
- ER
Estrogen receptor
- ERBB2/HER2
Epidermal growth factor receptor 2
- MG
Mammary gland
- GSEA
Gene set enrichment analysis
- PUFAs
Polyunsaturated fatty acids
- FDR
False discovery rate
- ESD
Esterase D
- MMPR
Matrix metalloproteinase 12
- DNMT3A
DNA (cytosine-5-)-methyltransferase 3 alpha
- EMT
Epithelial-mesenchymal transition
- NCI
Netherlands cancer institute
- NOM
Normal p-values
- ES
Enrichment scores
- NES
Normalized enrichment scores.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RKD conceived and designed the study. RCMCh and MK participated in the design of the study and conducted the experimental analysis. All authors participated in the analysis of data and review of the manuscript. RCMCh did the primary writing of this manuscript, but all authors contributed in this regard. All authors read and approved the final manuscript.
Contributor Information
Raquel C Martinez-Chacin, Email: raquelcmch@gmail.com.
Megan Keniry, Email: keniryme@utpa.edu.
Robert K Dearth, Email: dearthrk@utpa.edu.
References
- 1.Ziegler RG, Hoover RN, Pike MC, Hildesheim A, Nomura AM, West DW, Wu-Williams AH, Kolonel LN, Horn-Ross PL, Rosenthal JF, Hyer MB. Migration patterns and breast cancer risk in Asian-American women. J Natl Cancer Inst. 1993;85:1819–1827. doi: 10.1093/jnci/85.22.1819. [DOI] [PubMed] [Google Scholar]
- 2.Thomas D, Karagas M. Migrant Studies. In: Schottenfeld D, Fraumeni JF, editors. Cancer Epidemiology and Prevention. 3. New York: Oxford University Press; 1996. pp. 236–254. [Google Scholar]
- 3.McCredie M. Cancer epidemiology in migrant populations. Recent Results Cancer Res. 1998;154:298–305. doi: 10.1007/978-3-642-46870-4_21. [DOI] [PubMed] [Google Scholar]
- 4.Deapen D, Liu L, Perkins C, Bernstein L, Ross RK. Rapidly rising breast cancer incidence rates among Asian-American women. Int J Cancer. 2002;99:747–750. doi: 10.1002/ijc.10415. [DOI] [PubMed] [Google Scholar]
- 5.Pike MC, Kolonel LN, Henderson BE, Wilkens LR, Hankin JH, Feigelson HS, Wan PC, Stram DO, Nomura AM. Breast cancer in a multiethnic cohort in Hawaii and Los Angeles: risk factor-adjusted incidence in Japanese equals and in Hawaiians exceeds that in whites. Canc Epidemiol Biomarker Prev. 2002;11:795–800. [PubMed] [Google Scholar]
- 6.John EM, Phipps AI, Davis A, Koo J. Migration history, acculturation, and breast cancer risk in Hispanic women. Canc Epidemiol Biomarker Prev. 2005;14:2905–2913. doi: 10.1158/1055-9965.EPI-05-0483. [DOI] [PubMed] [Google Scholar]
- 7.Murtaugh MA, Sweeney C, Giuliano AR, Herrick JS, Hines L, Byers T, Baumgartner KB, Slattery ML. Diet patterns and breast cancer risk in Hispanic and non-Hispanic white women: the Four-Corners Breast Cancer Study. Am J Clin Nutr. 2008;87:978–984. doi: 10.1093/ajcn/87.4.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd NF, Stone J, Vogt KN, Connelly BS, Martin LJ, Minkin S. Dietary fat and breast cancer risk revisited: a meta-analysis of the published literature. Br J Cancer. 2003;89:1672–1685. doi: 10.1038/sj.bjc.6601314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiebaut AC, Kipnis V, Chang SC, Subar AF, Thompson FE, Rosenberg PS, Hollenbeck AR, Leitzmann M, Schatzkin A. Dietary fat and postmenopausal invasive breast cancer in the National Institutes of Health-AARP Diet and Health Study cohort. J Natl Cancer Inst. 2007;99:451–462. doi: 10.1093/jnci/djk094. [DOI] [PubMed] [Google Scholar]
- 10.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 11.Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, Dai H, He YD, van't Veer LJ, Bartelink H, van de Rijn M, Brown PO, van de Vijver MJ. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci USA. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC, Lashkari D, Shalon D, Brown PO, Botstein D. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 14.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffry SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale A. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated Observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;14:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carey LA. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;21:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 17.Rutqvist LE, Johansson H. Long-term follow-up of the randomized stockholm trial on adjuvant tamoxifen among postmenopausal patients with early stage breast cancer. Acta Oncologica. 2007;2:133–145. doi: 10.1080/02841860601034834. [DOI] [PubMed] [Google Scholar]
- 18.Turkoz FP, Solak M, Petekkaya I, Keskin O, Kertmen N, Sarici F, Arik Z, Babacan T, Ozisik Y, Altundag K. The prognostic impact of obesity on molecular subtypes of breast cancer in premenopausal women. J BUON. 2013;18:335–341. [PubMed] [Google Scholar]
- 19.Hilakivi-Clarke L, Clarke R, Onojafe I, Raygada M, Cho E, Lippman M. A maternal diet high in n-6 polyunsaturated fats alters mammary gland development, puberty onset, and breast cancer risk among female rat offspring. Proc Natl Acad Sci USA. 1997;94:9372–9377. doi: 10.1073/pnas.94.17.9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilakivi-Clarke L, Clarke R, Lippman M. The influence of maternal diet on breast cancer risk among female offspring. Nutrition. 1999;15:392–401. doi: 10.1016/S0899-9007(99)00029-5. [DOI] [PubMed] [Google Scholar]
- 21.Moral R, Escrich R, Solanas M, Vela E, Costa I, de Villa MC, Escrich E. Diets high in corn oil or extra-virgin olive oil provided from weaning advance sexual maturation and differentially modify susceptibility to mammary carcinogenesis in female rats. Nutr Cancer. 2011;63:410–420. doi: 10.1080/01635581.2011.535956. [DOI] [PubMed] [Google Scholar]
- 22.Hilakivi-Clarke L, Olivo SE, Shajahan A, Khan G, Zhu Y, Zwart A, Cho E, Clarke R. Mechanisms mediating the effects of prepubertal (n-3) polyunsaturated fatty acid diet on breast cancer risk in rats. J Nutr. 2005;135:2946–2952. doi: 10.1093/jn/135.12.2946S. [DOI] [PubMed] [Google Scholar]
- 23.Olivo-Marston SE, Zhu Y, Lee RY, Cabanes A, Khan G, Zwart A, Wang Y, Clarke R, Hilakivi-Clarke L. Gene signaling pathways mediating the opposite effects of prepubertal low-fat and high-fat n-3 polyunsaturated fatty acid diets on mammary cancer risk. Cancer Prev Res (Phila) 2008;1:532–545. doi: 10.1158/1940-6207.CAPR-08-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medvedovic M, Gear R, Freudenberg JM, Schneider J, Bornschein R, Yan M, Mistry MJ, Hendrix H, Karyala S, Halbleib D, Heffelfinger D, Clegg DJ, Anderson MW. Influence of fatty acid diets on gene expression in rat mammary epithelial cells. Physiol Genomics. 2009;38:80–88. doi: 10.1152/physiolgenomics.00007.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luijten M, Singh AV, Bastian CA, Westerman A, Pisano MM, Pennings JL, Verhoef A, Green ML, Piersma AH, de Vries A, Knudsen TB. Lasting effects on body weight and mammary gland gene expression in female mice upon early life exposure to n-3 but not n-6 high-fat diets. PLoS One. 2003;8:e55603. doi: 10.1371/journal.pone.0055603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudland PS, Barraclough R, Fernig DG, Smith JA. Growth and differentiation of the normal mammary gland and its tumours. Biochem Soc Symp. 1998;63:1–20. [PubMed] [Google Scholar]
- 30.Medina D. Chemical carcinogenesis of rat and mouse mammary glands. Breast Dis. 2007;28:63–68. doi: 10.3233/bd-2007-28107. [DOI] [PubMed] [Google Scholar]
- 31.Russo J, Russo IH. Development of the human breast. Maturitas. 2004;49:2–15. doi: 10.1016/j.maturitas.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 32.Russo J, Russo I. Molecular basis of breast cancer. New York: Springer-Verlag; 2004. [Google Scholar]
- 33.Singh M, McGinley JN, Thompson HJ. A comparison of the histopathology of premalignant and malignant mammary gland lesions induced in sexually immature rats with those occurring in the human. Lab Invest. 2000;80:221–231. doi: 10.1038/labinvest.3780025. [DOI] [PubMed] [Google Scholar]
- 34.Carroll KK. Dietary fats and cancer. Am J Clin Nutr. 1991;53:1064S–1067S. doi: 10.1093/ajcn/53.4.1064S. [DOI] [PubMed] [Google Scholar]
- 35.Fay MP, Freedman LS, Clifford CK, Midthune DN. Effect of different types and amounts of fat on the development of mammary tumors in rodents: a review. Cancer Res. 1997;57:3979–3988. [PubMed] [Google Scholar]
- 36.Hopkins GJ, Carroll KK. Relationship between amount and type of dietary fat in promotion of mammary carcinogenesis induced by 7,12-dimethylbenz[a]anthracene. J Natl Cancer Inst. 1979;72:1009–1012. [PubMed] [Google Scholar]
- 37.Rose DP, Connolly JM. Effects of dietary omega-3 fatty acids on human breast cancer growth and metastases in nude mice. J Natl Cancer Inst. 1993;85:1743–1747. doi: 10.1093/jnci/85.21.1743. [DOI] [PubMed] [Google Scholar]
- 38.Maillard V, Bougnoux P, Ferrari P, Jourdan ML, Pinault M, Lavillonniere F, Body G, Le Floch O, Chajes V. N-3 and N-6 fatty acids in breast adipose tissue and relative risk of breast cancer in a case–control study in Tours, France. Int J Cancer. 2002;98:78–83. doi: 10.1002/ijc.10130. [DOI] [PubMed] [Google Scholar]
- 39.Kaizer L, Boyd NF, Kriukov V, Tritchler D. Fish consumption and breast cancer risk: an ecological study. Nutr Cancer. 1989;12:61–68. doi: 10.1080/01635588909514002. [DOI] [PubMed] [Google Scholar]
- 40.Sasaki T, Kobayashi Y, Shimizu J, Wada M, In'nami S, Kanke Y, Takita T. Effects of dietary n-3-to-n-6 polyunsaturated fatty acid ratio on mammary carcinogenesis in rats. Nutr Cancer. 1998;30:137–143. doi: 10.1080/01635589809514653. [DOI] [PubMed] [Google Scholar]
- 41.Cohen LA, Chen-Backlund JY, Sepkovic DW, Sugie S. Effect of varying proportions of dietary menhaden and corn oil on experimental rat mammary tumor promotion. Lipids. 1993;28:449–456. doi: 10.1007/BF02535944. [DOI] [PubMed] [Google Scholar]
- 42.MacLennan M, Ma DW. Role of dietary fatty acids in mammary gland development and breast cancer. Breast Cancer Res. 2010;12:211. doi: 10.1186/bcr2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunter DJ, Spiegelman D, Adami HO, Beeson L, van den Brandt PA, Folsom AR, Fraser GE, Goldbohm RA, Graham S, Howe GR, Kushi LH, Marshall JR, McDermott A, Miller AB, Speizer FE, Wolk A, Yaun S, Willett W. Cohort studies of fat intake and the risk of breast cancer–a pooled analysis. N Engl J Med. 1996;334:356–361. doi: 10.1056/NEJM199602083340603. [DOI] [PubMed] [Google Scholar]
- 44.Smith-Warner SA, Spiegelman D, Adami HO, Beeson WL, van den Brandt PA, Folsom AR, Fraser GE, Freudenheim JL, Goldbohm RA, Graham S, Kushi LH, Miller AB, Rohan TE, Speizer FE, Toniolo P, Willett WC, Wolk A, Zeleniuch-Jacquotte A, Hunter DJ. Types of dietary fat and breast cancer: a pooled analysis of cohort studies. Int J Cancer. 2001;92:767–774. doi: 10.1002/1097-0215(20010601)92:5<767::AID-IJC1247>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 45.Kim MS, Kim YR, Yoo NJ, Lee SH. Mutational analysis of DNMT3A gene in acute leukemias and common solid cancers. APMIS. 2013;121:85–94. doi: 10.1111/j.1600-0463.2012.02940.x. [DOI] [PubMed] [Google Scholar]
- 46.Vuoriluoto K, Haugen H, Kiviluoto S, Mpindi JP, Nevo J, Gjerdrum C, Tiron C, Lorens JB, Ivaska J. Vimentin regulates EMT induction by Slug and ocogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene. 2011;12:1436–1448. doi: 10.1038/onc.2010.509. [DOI] [PubMed] [Google Scholar]
- 47.Korsching E, Packeisen J, Liedtke C, Hungermann D, Wulfing P, van Diest PJ, Brandt B, Boecker W, Buerger H. The origin of vimentin expression in invasive breast cancer: epithelial-mesenchymal transition, myoepithelial histogenesis or histogenesis from progenitor cells with bilinear differentiation potential? J Pathol. 2005;206:451–457. doi: 10.1002/path.1797. [DOI] [PubMed] [Google Scholar]
- 48.Thiery JP. Epithelial-mesenchymal transitions in tumor progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 49.Domagala W, Lasota J, Bartkowiak J, Weber K, Osborn M. Vimentin contributes to human mammary epithelial cell migration. J Cell Sci. 1999;112(Pt 24):4615–4625. doi: 10.1242/jcs.112.24.4615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: 361 breast cancer gene sets that were utilized in GSEA. This excel file details the 361 breast cancer gene sets from the Molecular Signatures Database that were utilized for gene set enrichment analysis with the HFD microarray data. NAME denotes the gene set name, original size denotes the number of genes in the curated gene set, after restricting to data set denotes the number of genes that were found on both the gene list and the HFD microarray data set, and status denotes whether the gene list was included in the analysis or not. A small number of gene lists were not included in the analysis, if there were not enough genes. (PDF 279 KB)
Additional file 2: 53 luminal breast cancer gene sets that were utilized in GSEA. This excel file details the 53 breast cancer gene sets from the Molecular Signatures Database that were utilized for gene set enrichment analysis with the HFD microarray data. NAME denotes the set name, original size denotes the number of genes in the curated gene set, after restricting to data set denotes the number of genes that were found on both the gene list and the HFD microarray data set, and status denotes whether the gene list was included in the analysis or not. A small number of gene lists were not included in the analysis, if there were not enough genes. (PDF 184 KB)
Additional file 3: Poor prognosis associated HFD-induced genes MMP12, GPNMB, CTSL and LILRB4 are highly expressed in basal-like BC. Depicted are histograms with log ration expression values for the indicated gene for each tumor (from the 295 NKI breast tumor dataset). Tumors are grouped together based on subtype. These subtypes are indicated at the top of the figure: Basal-like tumors are 1–46, HER2 are 47–95, Luminal A are 96–183, Luminal B are 184–264 and Normal-like tumors are 265–295. (PDF 326 KB)
Additional file 4: Poor prognosis associated HFD-induced genes DNMT3A, SLC11A1, NCF2 and PTPNS1 are highly expressed in basal-like BC. Depicted are histograms with log ration expression values for the indicated gene for each tumor (from the 295 NKI breast tumor dataset). Tumors are grouped together based on subtype. These subtypes are indicated at the top of the figure: Basal-like tumors are 1–46, HER2 are 47–95, Luminal A are 96–183, Luminal B are 184–264 and Normal-like tumors are 265–295. (PDF 335 KB)


