Abstract
Marked sequence variation in the mtDNA control region has been observed in human single CD34+ cells, which persist in vivo and are present also in differentiated hematopoietic cells. In this study, we analyzed 5071 single CD34+ cells from 49 individuals (including 31 maternally related members from four families and 18 unrelated donors) in order to determine the mutation spectrum within the mtDNA control region in single cells, as related to aging and family genetic background. Many highly mutated sites among family members were hypervariable sites in the mtDNA control region. Further, CD34+ cells from members of the same family also shared several unique mtDNA variants, suggesting pedigree-specific occurrence of these variants. Overall age-related accumulation of mtDNA mutations in CD34+ cells varied in different families, suggesting a specific accumulation pattern, which might be modulated by family genetic background. Our current findings have implications for the occurrence of mtDNA mutations in hematopoietic stem cells and progenitors.
Keywords: mtDNA, HSC, Somatic mutation, aging, genetic background, family
Introduction
Mitochondria play a central role in energy metabolism, in which mitochondrial oxidative phosphorylation is an extremely efficient system to produce energy required for the body (Wallace et al., 2010). Although reactive oxygen species (ROS) are a natural byproduct during the normal oxidative phosphorylation process, increased production of ROS and decline of mitochondrial respiratory capacity is observed during aging (Finkel and Holbrook, 2000; Vendelbo and Nair, 2011). Oxidation of cell components by ROS affects the function of various tissues and organs with age (Finkel and Holbrook, 2000; Vendelbo and Nair, 2011). Under the mitochondrial theory of aging, aging is a vicious cycle between elevated ROS and mitochondrial dysfunction leading to mtDNA damage and age-related cell death or senescence (Harman, 1972; Nagley and Wei, 1998; Trifunovic, 2006). This theory has been broadly supported by previous reports of accumulation of mtDNA mutations with aging (Greaves and Turnbull, 2009; Trifunovic, 2006), as well as molecular genetic and evolutionary studies (Agarwal and Sohal, 1996; Barja, 2004; Beckman and Ames, 1998; Finkel and Holbrook, 2000).
In our previous studies, we employed mtDNA sequence analysis of single hematopoietic cells to assess aging effects on the accumulation of mutations (Shin et al., 2004; Yao et al., 2007c). In particular, we found that human hematopoietic stem cells (HSCs) and progenitors, as represented by CD34+ cell surface staining, showed striking intercellular and intracellular mtDNA sequence variation (Ogasawara et al., 2005; Yao et al., 2007b; Yao et al., 2007d). Some of these sequence variations in single CD34+ cells persisted in vivo over time (Yao et al., 2007b) and were present in CD34+ cell progeny, circulating white blood cells (Ogasawara et al., 2005). In transplanted patients, CD34+ cells from the donor could be tracked by their distinctive mtDNA mutations as patients became full myeloid chimeras (Yao et al., 2007b). mtDNA mutation analysis of murine HSCs and progenitors showed age-related accumulation of mutations, dependent on the nuclear genetic background (Yao et al., 2007c). However, as we also reported (Yao et al., 2007b), a preliminary analysis of human mtDNA variation in individual CD34+ cells from the healthy adults (aged from 25 to 65 years) revealed no obvious correlation between the mtDNA mutation number and the age of the individual, and the mutation levels in CD34+ cells from the adult samples could be seemingly classified into two groups defined by 6.5–12.8 haplotypes per 100 cells and 29.7–42.6 haplotypes per 100 cells. These results are reminiscent of potential modulation from the nuclear genetic background as observed in mice (Yao et al., 2007c). In one intriguing finding, based on comparison of CD34+ cells from the maternally related donors and recipients, a very small portion of CD34+ cells shared certain nucleotide substitutions that appeared to be exclusive to the family (Yao et al., 2007b). The nature of these “family-specific” mutations in HSCs and progenitors from siblings, perhaps transmitted with the mother’s mitochondria, or occurring somatically in parallel, or otherwise modulated by the nuclear genetic background, remained to be validated and clarified.
In the current work, we have analyzed 5071 single CD34+ cells from 49 persons. Among them, 32 individuals are newly collected from four families, in age ranging from 13 to 89 years, and three samples are cord blood. We had two aims: (1) to examine the mutation spectra of the noncoding mtDNA control region in single CD34+ cells from maternally related individuals and to characterize the family-specific occurrence of mtDNA mutations; (2) to discern an aging effect on mutation accumulation in the mtDNA control region in single CD34+ cells. With an enlarged sample size and members from a family, we were able to obtain a comprehensive characterization of the mtDNA mutation spectrum, providing some insight into age, family genetic background and mtDNA mutations in human cells.
Materials and Methods
Subjects
A total of 32 healthy individuals from four families were recruited for this study (Table 1 and Fig. 1). For each individual, peripheral blood (PB, 8–10 mL) was collected in a heparinized tube and mononuclear cells (MNCs) were isolated by Ficoll density gradient centrifugation after overnight shipping and delivery. For donors in Family B living in Italy (N = 11), the same amount of PB was collected and processed locally within 24 h of phlebotomy for MNC separation, freezing, and storage; frozen MNCs were shipped to our laboratory at Bethesda on dry ice and then transferred to liquid nitrogen until they were subjected to single cell sorting. A control group was composed of 18 unrelated individuals: three cord blood (CB) samples and one member (C-2) from Family C (this study); 14 healthy individuals reported in our previous studies (Ogasawara et al., 2005; Yao et al., 2007b; Yao et al., 2007d). A total of 20 samples (1952 CD34+ cells) were obtained from these 18 unrelated individuals as two of the 14 reported healthy individuals were sampled twice at different time points (Yao et al., 2007b). All donors gave informed consent, and this study was approved by the institutional review board of the National Heart, Lung, and Blood Institute.
Table 1.
The level of mtDNA sequence variation in single CD34+ cell populations from 49 healthy donors with different ages
| Sample | Age | Sex | No. of cells | No. of haplotypes | No. of nonaggregate haploytpes by nucleotide substitutions | No. of cells with nonaggregate sequences | No. of cells with nonaggregate sequences defined by substitution | Reference |
|---|---|---|---|---|---|---|---|---|
| A-1 | 85 | F | 91 | 50 | 43 | 52 | 43 | This study |
| A-2 | 55 | M | 92 | 35 | 28 | 40 | 28 | This study |
| A-3 | 59 | F | 93 | 25 | 20 | 38 | 22 | This study |
| A-4 | 35 | M | 93 | 13 | 8 | 36 | 10 | This study |
| A-5 | 30 | M | 93 | 27 | 21 | 40 | 21 | This study |
| A-6 | 25 | F | 95 | 12 | 5 | 32 | 14 | This study |
| B-1 | 83 | M | 95 | 39 | 33 | 74 | 33 | This study |
| B-2 | 81 | M | 94 | 42 | 36 | 73 | 36 | This study |
| B-3 | 89 | F | 86 | 48 | 42 | 71 | 42 | This study |
| B-4 | 79 | F | 96 | 35 | 29 | 74 | 29 | This study |
| B-5 | 87 | F | 95 | 49 | 40 | 70 | 41 | This study |
| B-6 | 60 | F | 95 | 47 | 38 | 73 | 41 | This study |
| B-7 | 47 | M | 95 | 38 | 29 | 73 | 29 | This study |
| B-8 | 51 | M | 94 | 35 | 28 | 64 | 29 | This study |
| B-9 | 54 | F | 94 | 52 | 43 | 74 | 43 | This study |
| B-10 | 57 | F | 93 | 37 | 30 | 70 | 30 | This study |
| B-11 | 27 | M | 91 | 35 | 27 | 69 | 27 | This study |
| B-12 | 21 | M | 185 | 76 | 70 | 160 | 90 | This study |
| B-13 | 20 | M | 191 | 77 | 70 | 132 | 70 | This study |
| B-14 | 13 | M | 96 | 54 | 43 | 85 | 43 | This study |
| B-15 | 32 | F | 96 | 50 | 41 | 76 | 42 | This study |
| B-16 | 28 | F | 96 | 31 | 23 | 71 | 23 | This study |
| C-1 | 80 | F | 93 | 51 | 45 | 61 | 46 | This study |
| C-2 | 79 | M | 94 | 45 | 37 | 65 | 37 | This study |
| C-3 | 76 | F | 96 | 17 | 15 | 19 | 17 | This study |
| C-4 | 46 | F | 96 | 48 | 43 | 56 | 44 | This study |
| C-5 | 49 | M | 112 | 14 | 13 | 13 | 13 | This study |
| C-6 | 46 | F | 95 | 8 | 5 | 7 | 5 | This study |
| C-7 | 18 | M | 94 | 29 | 24 | 41 | 25 | This study |
| C-8 | 21 | M | 92 | 16 | 14 | 17 | 14 | This study |
| D-1 | 65 | F | 96 | 37 | 32 | 39 | 34 | This study |
| D-2 | 42 | M | 96 | 21 | 19 | 20 | 19 | This study |
| Donor 1 | 48 | F | 85 | 14 | 8 | 32 | 10 | Ogasawara et al., 2005 |
| Donor 2 | 44 | M | 92 | 16 | 11 | 32 | 11 | Ogasawara et al., 2005 |
| Donor 3 | 36 | M | 93 | 12 | 6 | 34 | 11 | Ogasawara et al., 2005 |
| Donor 4 | 55 | M | 93 | 14 | 8 | 41 | 23 | Ogasawara et al., 2005 |
| Donor 5 | 35 | M | 93 | 13 | 10 | 34 | 11 | Ogasawara et al., 2005 |
| Donor 6 | 35 | M | 95 | 15 | 10 | 40 | 10 | Yao et al., 2007d |
| Donor 7 | 32 | M | 93 | 40 | 35 | 53 | 35 | Yao et al., 2007d |
| Donor 8 | 25 | M | 93 | 38 | 35 | 56 | 35 | Yao et al., 2007d |
| Donor 9 | 57 | M | 93 | 31 | 28 | 38 | 35 | Yao et al., 2007b; 2007d |
| Donor 10 | 39 | M | 94 | 17 | 12 | 43 | 12 | Yao et al., 2007b; 2007d |
| QUIDC-3-14-5 | 65 | F | 91 | 39 | 34 | 48 | 39 | Yao et al., 2007b |
| HERJC-2-16-5 | 48 | F | 93 | 40 | 31 | 73 | 31 | Yao et al., 2007b |
| CABO-2-5-98 | 34 | M | 91 | 32 | 27 | 57 | 27 | Yao et al., 2007b |
| CABO-1-31-6 | 42 | M | 94 | 43 | 37 | 64 | 37 | Yao et al., 2007b |
| KIRGC-12-10-2 | 53 | F | 94 | 43 | 41 | 43 | 41 | Yao et al., 2007b |
| KIRGC-2-2-6 | 57 | F | 96 | 39 | 38 | 39 | 39 | Yao et al., 2007b |
| CB-1 | 0 | - | 93 | 6 | 4 | 7 | 4 | This study |
| CB-2 | 0 | - | 187 | 30 | 23 | 85 | 29 | This study |
| CB-3 | 0 | - | 95 | 18 | 15 | 49 | 16 | This study |
| Total | - | - | 5071 | 1693 | 1408 | 2753 | 1496 | - |
Note – Donors 9 and 10 that were reported in Yao et al. (2007d) refer to BURRC-9-23-4 and GREHC-2-10-5 in Yao et al. (2007b), respectively. We counted the number of haplotypes (including the aggregate mtDNA haplotype [which has the highest frequency in a population of cells and bears the consensus sequence; Table S3] and nonaggregate haplotypes that differed from the aggregate sequence by nucleotide substitutions or indels [insertions and deletions] or both) and the number of nonaggregate haplotypes that differed from the aggregate type only by nucleotide substitutions, based on the mutations detected in a population of single CD34+ cells from each donor (Table S4). As the nonaggregate haplotype may be shared by several single cells, which reflects the clonal expansion of the clone, we then counted the number of cells harboring the nonaggregate haplotypes or nonaggregate haplotypes by nucleotide substitutions for each donor.
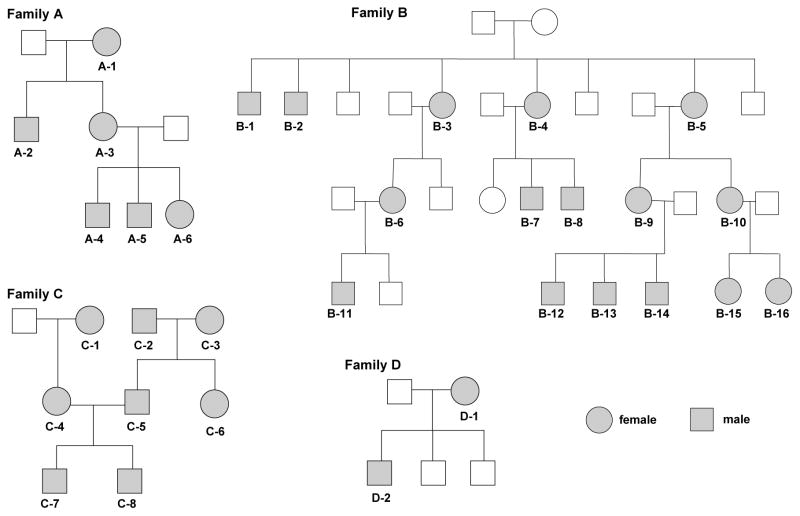
Figure 1.
Pedigrees of the four families analyzed in this study. Members of four families (A, B, C, and D) were recruited for examination of mtDNA control region sequence in single CD34+ cells. Individuals who provided blood samples are marked in grey; ages are listed in Table 1.
Single CD34+ cell sorting and PCR amplification
Single CD34+ cells were isolated and analyzed using the same procedure described in our previous studies (Ogasawara et al., 2005; Yao et al., 2007b; Yao et al., 2007d). In brief, after thawing and washing with phosphate buffered saline (PBS), cells were suspended in 100 μL of PBS containing 0.5% bovine serum albumin (BSA) and incubated with anti-CD34 phycoerythrin (PE)-conjugated monoclonal antibody (BD Bioscience, San Jose, CA) for 30 minutes at 4°C. Cells were then washed, resuspended in 600 μL of PBS with 0.5% BSA, and added 4μL of 7-amino-actinomycin D (7-AAD). Cell sorting was performed on the MoFlo Legacy high-performance cell sorter (Dako-Cytomation, Ft Collins, CO), using 100 mW of the 488 nm line of an argon laser (I-90, Coherent, Palo Alto, CA) for excitation. Forward scatter was the triggering parameter (Fig. S3). Single cell deposition was accomplished using the CyClone Automated Cloner (Dako-Cytomation, Ft Collins, CO) in a 0.5 single drop mode with gating, based on forward scatter and fluorescence. Single CD34+ cells were sorted into each well of an optical 96-well reaction plate containing 50 μL of lysis buffer (10 mM Tris-HCl [pH 8.0], 50 mM KCl, 100 μg/mL Proteinase K, and 1% Triton X-100). The plate was incubated at 56°C for 30 minutes, followed by an additional incubation at 96°C for 8 minutes.
Two-step nested PCR was used to amplify the entire mtDNA control region in single CD34+ cells, following the same PCR condition as reported previously (Yao et al., 2007b; Yao et al., 2007d). In brief, the first PCR was performed in a total volume of 30 μL, which contains 400 μM of each dNTP, 1 unit of TaKaRa LA Taq™ (which has proof reading activity, with a relative error rate of 0.16 as compared to conventional Taq DNA polymerase; Takara Bio. Inc.), 0.5 μM of each forward and reverse outer primer (L15594: 5’-CGCCTACACAATTCTCCGATC -3’ and H901: 5’-ACTTGGGTTAATCGTGTGACC -3’), and 5 μL of cell lysates. PCR amplification was run on the GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA) with the following thermal conditions: one denaturation cycle of 94°C for 3 minutes; 35 amplification cycles of 94°C for 30 seconds, 52°C for 40 seconds and 72°C for 1 minute with a 5-second increase per cycle; and a full extension cycle of 72°C for 10 minutes. The second PCR was performed in 50 μL of reaction mixture containing 400 μM of each dNTP, 2 units of TaKaRa LA Taq™, 0.5 μM of each forward and reverse inner primers (L15990: 5’ – TTAACTCCACCATTAGCACC -3’ and H650: 5’ – GAAAGGCTAGGACCAAACCTA -3’), and 5 μL of first PCR product under the same amplification conditions as the first PCR but with an extension time of 90 seconds at 72°C per cycle. Second PCR products were purified and then sequenced by using the BigDye Terminator v3.1 Cycle Sequencing Kit on the 3100 DNA Sequencer (Applied Biosystems). The same sequencing primers described previously (Yao et al., 2007b; Yao et al., 2007d) were used to determine the entire mtDNA control region sequence.
Identification of mtDNA sequence variation and statistical analysis
Germline sequence variants in the consensus /aggregate sequences of single CD34+ cells from each subject were scored relative to the revised Cambridge Reference Sequence (rCRS) (Andrews et al., 1999). Sequencing electrophoregram of each single cell was proof-read, as reported previously (Yao et al., 2007b; Yao et al., 2007d), and mutation was scored by referring to the aggregate sequence of the donor. Cells with contamination were recognized using the same strategy as described in our previous study (Yao et al., 2007a). In brief, if the mtDNA sequence of a single cell from a particular subject did not contain or had heteroplasmy at all of the variants detected in the aggregate sequence for that subject, it was regarded as being contaminated. The overall contamination rate was very low (around 0.4% of total single cells) in this study and exclusion of these contaminated cells did not influence the result. We scored for heteroplasmy (co-existence of wild-type and mutant alleles) of certain mtDNA variants, based on the electrophoregram in which mutant alleles were present at >10% of the highest peak. As nearly all identified mtDNA mutations were heteroplasmic in cells (with a mutant ratio varying from >10% to 100%; Fig. S2), we counted the mutation regardless of its exact level of heteroplasmy. Length variation of the poly C-tract in region 303–309 was scored by direct counting of the base shift of T at nucleotide 310. A length mutation of the AC repeat in region 515–524 in the third hypervariable segment was also scored on the basis of the sequencing electrophoregram. But, a length variation of the poly C-tract in region 16184–16193, caused by variant 16189T>C, was not considered in this study because most of them were heteroplasmic of multiple poly C-tracts (> 10 Cs) and could not be conservatively counted according to sequencing chromatographs.
As the majority of single cells from each donor shared the same consensus mtDNA sequence (which we defined as the aggregate haplotype), we define a cell with a unique haplotype if this cell differed from the aggregate haplotype by containing extra nucleotide alteration(s). We used the number of haplotypes in a population of 100 cells from each sample as an index to compare frequencies of mtDNA sequence variation among different samples. This index reflects the total number of mutations that occurred or retained within a given number of cells and not the frequency of the cell clone harboring certain mutation(s) (Yao et al., 2007b; Yao et al., 2007d). As polynucleotide length mutations have different mutation mechanisms compared to nucleotide substitutions, we mainly focused on analysis of nucleotide substitutions, as represented by the number of haplotypes defined by nucleotide substitutions per 100 cells in our analysis. An aging effect on an mtDNA variation level in a cell population was evaluated by the linear regression analysis using the GraphPad Prism 4.0.
Results
Data quality control
Single cell analysis has a very high sensitivity to detect mtDNA mutations present at low levels. However, this technique also poses challenges, especially to eliminate artifacts caused by multiple-step PCR and contamination (Reeve et al., 2009; Yao et al., 2007a). We benefited from our previous experience of single cell analysis (Ogasawara et al., 2005; Yao et al., 2007b; Yao et al., 2007d), and we believe that our strategy was sufficiently stringent to maintain data reliability. First, we used the same high fidelity TaKaRa LA Taq™ DNA polymerase for PCR and negative controls during the amplification. Application of the same reagents for all samples should yield similar levels of background sequence variation inevitably arising from PCR amplification, and should not affect the overall pattern of mutation rate. Second, we were able to consistently reproduce the majority of mutations (~ 80%) in single cells among duplicates in independent amplifications of single cells from donors D-2 and B-11. Moreover, the levels of mtDNA mutation in each single cell population of the duplicates from the same donor were similar. Finally, we employed the same phylogenetic approach as described for single cell analysis (Yao et al., 2007a) in order to identify and eliminate potential cases of contamination.
Aging effect on the accumulation of mtDNA variations in single CD34+ cells
A total of 5071 single CD34+ cells sorted from 51 PB samples collected from 49 healthy donors with age ranging from 0–89 years were (re-)analyzed in this study (Table 1). Among them, 32 individuals were from four families (6 from Family A, 16 from Family B, 8 from Family C, and 2 from Family D). Pedigrees of the families and selected family members are illustrated in Figure 1. The other 17 individuals were as follows: 14 healthy individuals (two donors were sampled twice at different time points (Yao et al., 2007b)) reported in our previous studies (Yao et al., 2007b; Yao et al., 2007d); three CB samples also were included due to the paucity of young individuals in our families (only one donor < 18 years old; Table 1). When variation levels of the mtDNA control region (the number of haplotypes defined by nucleotide substitutions per 100 cells) were plotted against ages of individual donors, we observed a roughly age-related accumulation pattern of mtDNA sequence variations in single CD34+ cells, although the correlation was moderate (r2 = 0.19, P = 0.001; Table 1, Fig. 2A and Table S1). We also performed the analysis using just the adult samples, to exclude potential bias of three CB samples and one young donor (age 13), a similar result was observed (r2 = 0.17, P = 0.004; Table S1). As different families had different number of donors, we included “Family” as a control variable, but the result was not changed (r = 0.40, P = 0.004; Table S1). We obtained same results when we considered the number of total haplotypes (defined by nucleotide substitutions and insertions/deletions) per 100 single cells (Table S2). Overall, a high frequency of mtDNA mutation was not always present in CD34+ cells from donors older than 75 years, as compared to donors younger than 75 years old or even much younger donors less than 30 years old. This pattern may reflect the heterogeneity within and the dynamics of CD34+ cell proliferation in vivo, and we cannot exclude a bias due to sampling of comparatively few younger donors. The high level of mtDNA variation in CD34+ cells from CB or donors with young age seemed to suggest that there is not necessarily a low mutation rate at young age and mtDNA mutations accrue in CD34+ cells not only after birth.
Figure 2.
Number of mtDNA control region haplotypes in a population of CD34+ cells. Haplotype frequencies were defined by the number of haplotypes (characterized by nucleotide substitutions)/100 single CD34+ cells from unrelated healthy individuals and family members. (A) All 51 samples were derived from 49 healthy donors in the age range of 0–89, in which two donors were sampled twice at different time points (Yao et al., 2007b). Among them, 32 individuals were newly recruited from four families (6 from family A, 16 from Family B, 8 from Family C, and 2 from Family D). The other 17 individuals consisted of previously reported 14 healthy donors (Ogasawara et al., 2005; Yao et al., 2007b; Yao et al., 2007d) and three cord blood donors (this study). The number of haplotypes in each donor was shown relative to the donors’ age: (B) 6 members of Family A, (C) 16 members of Family B, and (D) 4 members (C-1, C-4, C-7 and C-8) of Family C.
Family-specific pattern for age-dependent accumulation of mtDNA variations in single CD34+ cells
Our previous work demonstrated that the level of mtDNA sequence heterogeneity in murine HSCs and progenitors was modulated by the nuclear genetic background (Yao et al., 2007c), and we speculated that this might also be observable in human. Although the best samples to address this hypothesis should be monozygotes, maternally related members from the same family may be helpful. Plots of mtDNA mutation levels against ages in the families with at least 5 members displayed remarkably difference between families: there was a significant correlation between the age and the mtDNA mutation level in Family A (r2 = 0.79; P = 0.018; Fig. 2B and Fig. S1), whereas the mutation levels in the 16 donors in Family B (r2 = 0.04; P = 0.485) showed no clear tendency for age-dependent accumulation of mtDNA mutations in single CD34+ cells, mainly because younger donors had surprisingly high levels of mtDNA mutations (Fig. 2C). In four maternally related members in Family C, an age-related pattern was evident but not statistically significant (r2 = 0.76; P = 0.128) (Fig. 2D). The results indicated that the accumulation of mtDNA mutations in single CD34+ cells was indeed influenced by the family genetic backgrounds of families, as the accumulation rate differed in various pedigrees. Note that our observation for families A and C was based on limited number of donors, and larger cohorts will be necessary to further validate our result.
Family-specific variants or recurrent mutations in single CD34+ cells?
The age-related accumulation of specific variants, such as C150T, A189G, T408A, and T414G, has been reported in leukocytes, muscle tissues, and fibroblasts (Michikawa et al., 1999; Wang et al., 2001; Zhang et al., 2003). Some of these variants are highly hypervariable (e.g. C150T and A189G), according to the estimation of the available worldwide populations (Soares et al., 2009; Stoneking, 2000), but other sites appear to be less mutable and may be tissue or cell-specific. In our previous experiments aimed to trace donor CD34+ clones in a recipient after transplantation, we found that a small portion of CD34+ cells from the sibling samples shared certain mtDNA variants, suggesting potential family-specific variants if not recurrent mutations (Yao et al., 2007b).
To further examine this pattern for a potentially high prevalence of somatic mutations in CD34+ cells, we first compiled the mtDNA control region mutation spectrum of 1952 CD34+ cells from 20 PB samples collected from 18 unrelated individuals (Table 1 and Fig. 3A): 16 samples from 14 healthy donors reported previously (Ogasawara et al., 2005; Yao et al., 2007b; Yao et al., 2007d); three CB samples; one sample from a member (C-2) of Family C. The mutation pattern from these 18 unrelated individuals was then compared with those from four different families (A, B, C, and D). An occurrence of each variant (in heteroplasmic or homoplasmic status) was scored according to its presence in a donor, but the variant frequency in a cell population from the donor was neglected. The top five highly mutational sites were 146, 16129, 204, 16311, and 16131 in these 18 unrelated individuals (Fig. 3A and Fig. S2). Except for 16131, the positions were all listed as hypervariable sites in the mutational hotspot list compiled by Soares et al. (2009) on the basis of 2196 complete mtDNA genomes from worldwide populations (Fig. 3B). We also identified several variable sites, such as 42, 64, 16038, and 16086, which had a modest or low rate in the worldwide human mtDNA population data, but they had a relatively high frequency in the data from the 18 unrelated individuals (Fig. 3A). Further studies are required to address why these variants are so highly prevalent in CD34+ cells.
Figure 3.
Mutational hot spots of the mtDNA control region in single CD34+ cells and in reported complete mtDNAs from worldwide populations. Mutational hot spots were scored, according to their occurrences in the respective individual, but with disregarding their frequencies in a cell population from the individual. As the number of donors in each family varied, we arbitrarily regarded a certain site as a mutational hot spot if mutations hit the site at least in 3 donors in 18 unrelated healthy individuals (A), 2 donors in Family A (N=6) (C), and 4 donors in Family B (N=16) (D). The 18 unrelated healthy individuals included one member (C-2) of Family C in addition to 17 unrelated donors (14 reported healthy individuals (Ogasawara et al., 2005; Yao et al., 2007b; Yao et al., 2007d) and three cord blood donors). The mutational hot spots in worldwide mtDNAs are listed (B) when mutations appear at least 30 times in the global tree of 2196 complete mtDNA sequences (Soares et al., 2009).
Figures 3C and 3D represent the highest mutational hits in the maternally related members from Families A and B, respectively. In Family A, a total of 15 sites had mutation hits at least twice, among which 16192 and 204 were most frequent, and the other 12 sites showed a similar mutational frequency. Family B displayed a total of 16 sites with mutation hits at least four times, among which 146, 204, 16129, and 16131 were the top four, followed by sites 16093, 16150, and 16187. Of interest, the top four sites (146, 204, 16129, and 16131) in Family B matched four of the top five hypervariable sites (146, 16129, 204, 16311, and 16131) present in the 18 unrelated individuals (Fig. 3A). In particular, the occurrence of variation at site 16131 ranked fourth in both Family B and the 18 unrelated individuals, suggesting that this variant might be more prone to undergo somatic change in single CD34+ cells.
We also observed a relatively high frequency of apparently family-unique variation at sites 16192, 16079, 215 and 217 in members from Family A (Fig. 3C and Fig. S1) and at sites 16150, 16187 and other sites in members from Family B (Fig. 3D). These mutations were present at either low frequency or were absent in 1952 single CD34+ cells from the 18 unrelated individuals and in the complete mtDNA database from worldwide populations. Although the frequency number of a certain variant may be biased, due to the limited number of donors in each family, we inferred unique mutation distribution patterns in single CD34+ cells from different families.
Discussion
There is accumulating evidence that mtDNA mutations play important roles in human disease and in physiologic aging (Taylor and Turnbull, 2005; Wallace et al., 2010) and mtDNA mutations are common in normal and tumor tissues (Greaves and Turnbull, 2009; He et al., 2010; Yao et al., 2007d). The aging of hematopoietic stem cells (HSCs) is complex and multifactorial (Sahin and Depinho, 2010), accompanying with age-dependent accumulation of mtDNA mutations (Shin et al., 2004). However, whether accumulated mtDNA mutations in HSCs and progenitors affects their developmental or function has not sufficiently resolved (Sahin and Depinho, 2010; Shin et al., 2004; Yao et al., 2007c). Recently, Norddahl and coworkers (2011) demonstrated that hematopoietic defects in mice carrying a proofreading-defective mtDNA polymerase is reminiscent of premature HSC aging. They further showed that rapid accumulation of mtDNA mutations had little functional effect on the HSC pool, but would cause distinct differentiation blocks for downstream progenitor cells (Norddahl et al., 2011).
In the current work, there was a tendency for age-associated mutation accumulation in human CD34+ cells, but the mutation levels in certain young donors was not necessarily low (Fig. 2 and Table 1), suggesting that CD34+ cells accumulated mutations at all stages of growth. Furthermore, age-related mutation accumulation seemed to be affected by family genetic background, varying substantially from one family to another. Of note, various mutational hotspots in the maternally related individuals (Family A or B) were also observed as hypervariable mutations in single cell populations from the 18 unrelated individuals and in 2196 complete mtDNAs from the worldwide populations (Soares et al., 2009). Therefore, it seems probable that highly frequent variants/mutations in single CD34+ cells do not occur randomly. Forster et al. (2002) analyzed the mtDNA mutations in 248 pedigrees living in the high-radiation peninsular and in nearby low-radiation islands in Kerala, India, to characterize radiation-associated mtDNA mutations; they surmised that radioactive conditions significantly accelerated mtDNA mutations in evolutionarily hot spots between mothers and their offspring. Our current study of mutational hotspots in single CD34+ cells is consistent with their observation.
In Drosophila, mtDNA haplotypes’ influence on the longevity has varied on different nuclear backgrounds, suggesting the mtDNA effects could be masked or exaggerated by significant interactions with the nuclear allelic variation (Rand et al., 2006). In humans, there also appeared to be an effect of inherited mtDNA variants on longevity, dependent on the population, probably due to different nuclear genetic background (Alexe et al., 2007; Dato et al., 2004; Santoro et al., 2006). Therefore, aging and longevity may be regulated by nuclear gene variants associated with inherited and somatic mtDNA variations/mutations in nuclear-mitochondrial interaction (Santoro et al., 2006; Vendelbo and Nair, 2011). As more than 98% of the mitochondria’s proteins are encoded by nuclear genes, and coordinated expression of nuclear and mitochondrial genes is indispensable for mitochondrial function (Calvo and Mootha, 2010), it is highly plausible that nuclear-mitochondrial epistatic interactions in different families account for the diverse pattern of mtDNA mutation accumulation in single CD34+ cells. The lack of aging-related accumulation of mtDNA mutation in Family B, but not in families A and C, was simply caused by an elevated level of mtDNA mutation in donors at young age (Fig. 2). Does Family B harbor a “mutagenic background” that accounts for their high load of mutations? Apparently, we have no clear answer to this question based on the current observation. As CD34+ cells are very heterogeneous and replenish during hematopoiesis (Doulatov et al., 2012), it is possible that mutation level will finally reach a platform due to the balance of accumulation and differentiation/disappearance of mutated cells in those donors with a “mutagenic background”, irrespective of age. Considering the results of functional assays of HSCs and progenitors with mtDNA mutations (Yao et al., 2007c), we would expect no tight connection between mutation frequency and functional consequence in the mutated CD34+ cells, or in the hematopoietic systems of these young individuals with a high level of mtDNA mutation.
In this study, besides the high hits for the evolutionarily hypervariable sites, we also noticed that certain mtDNA variants were shared by family members, pedigree-specific occurrence of mtDNA variants. This observation was unexpected but confirmed our previous speculation for a family-specific occurrence of mtDNA variants among siblings (Yao et al., 2007b). We hypothesized that this variation was shaped by family genetic background. Indeed, others have recently shown that somatic point mutations in the mtDNA control region are influenced by genetic background and associated with healthy aging (Rose et al., 2010). How this seemingly family-specific mtDNA mutation occurs and accumulates and whether it is restricted to CD34+ cells but not other tissues are unknown. As the family differences for mutation accumulation and presence of family-specific variants are obvious in this study, one likely explanation for this observation is the maternal inheritance of low-level variants or maternal influence on occurrence of low-level variants. Direct experimental data is needed to confirm this speculation. Nonetheless, our observations contribute to an inference of complexity in the mechanism of mtDNA mutations in single cells. The pedigree-specific mtDNA variants could have potential forensic consequences and individual patterns of mutations could help to identify individuals instead of matri-lineages (Salas et al., 2007).
The mtDNA control region plays a crucial role in regulating mtDNA replication and transcription (Falkenberg et al., 2007; Shadel and Clayton, 1997). Deleterious mutations in this region would be expected to have functional consequences (Bi et al., 2010). We do not know the exact role of the mutations that were frequently observed in the mtDNA control region of CD34+ cells. As some of these highly mutated sites, such as 146, 204 and 16129, are also hypervariable in the general populations, indicating that they are unlikely to be of deleterious significance. However, the accumulation of these mutations is unlikely to be simple byproduct of oxidative damage, as we observed high frequencies of various mutations at both evolutionarily hypervariable sites and seemingly family-specific sites. For instance, the frequency of variant A215G is relatively high in pedigrees from a high radiation area (Forster et al., 2002), but this variant also was present in two out of six individuals in Family A. The age-associated variant T414G (Michikawa et al., 1999) was only found in two CD34+ cells from a 48-year old healthy donor in our previous study (Yao et al., 2007b) and in one 79-year old donor (C-2) in this study. Further work is required to clarify putative functional effects of these variants on mtDNA replication and mtDNA copy number within a cell (Falkenberg et al., 2007; Shadel and Clayton, 1997).
Our current study has some limitations. First, due to the relatively low number of subjects per family and limited number of families analyzed, the family-related patterns as we observed here need to be further confirmed. Second, we did not perform any functional characterizations for those CD34+ cells with and without mtDNA mutations, to show the potential effect of mtDNA variants. It remains unknown whether there is a family-specific mutation accumulation in mtDNA coding region in CD34+ cells. Further study should be carried out to answer these questions.
In summary, we provide, to the best of our knowledge, the mutation spectra of the human mtDNA control region in CD34+ hematopoietic cells from a large cohort of maternally related and unrelated individuals. The results in general support age-related accumulation of mtDNA mutations, but further discerned a family-specific rate of mtDNA mutation accumulation and mutational sites. The exact biological significance of accumulation of aging-related and family-specific mtDNA mutations in CD34+ cells awaits future study.
Supplementary Material
Highlights.
Many mtDNA highly mutated sites in family members are evolutionarily hypervariable
CD34+ cells from members of the same family shared family-specific mtDNA variants
Age-related accumulation of CD34+ cell mtDNA mutations varied in different families
Acknowledgments
This research was supported (in part) by the Intramural Research Program of the NIH, NHLBI. Y.-G.Y was supported by the Ministry of Science and Technology of China (2011CB910900), Yunnan Province (2009CI119), and Natural Science Foundation of China (30925021).
We thank our blood donors for participating in this study and Thomas M. Herndon and Carol Boss for help with sample collection. This research was supported (in part) by the Intramural Research Program of the NIH, NHLBI. Y.-G.Y was supported by the Ministry of Science and Technology of China (2011CB910900), Yunnan Province (2009CI119), and Natural Science Foundation of China (30925021).
Footnotes
Authorship Contributions
Y.-G.Y. designed research, performed research, analyzed data and wrote the paper. S.K. designed research and wrote the paper. X.F., L.S. and J.P.M. performed research. G.T. contributed vital samples. N.S.Y. designed research and wrote the paper.
Disclosure of Conflicts of Interest
The authors declared no conflict of interest.
Supplementary data to this article can be found online at xxxx.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal S, Sohal RS. Relationship between susceptibility to protein oxidation, aging, and maximum life span potential of different species. Exp Gerontol. 1996;31:365–372. doi: 10.1016/0531-5565(95)02039-x. [DOI] [PubMed] [Google Scholar]
- Alexe G, Fuku N, Bilal E, Ueno H, Nishigaki Y, Fujita Y, Ito M, Arai Y, Hirose N, Bhanot G, Tanaka M. Enrichment of longevity phenotype in mtDNA haplogroups D4b2b, D4a, and D5 in the Japanese population. Hum Genet. 2007;121:347–356. doi: 10.1007/s00439-007-0330-6. [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Barja G. Free radicals and aging. Trends Neurosci. 2004;27:595–600. doi: 10.1016/j.tins.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- Bi R, Zhang AM, Zhang W, Kong QP, Wu BL, Yang XH, Wang D, Zou Y, Zhang YP, Yao YG. The acquisition of an inheritable 50-bp deletion in the human mtDNA control region does not affect the mtDNA copy number in peripheral blood cells. Hum Mutat. 2010;31:538–543. doi: 10.1002/humu.21220. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dato S, Passarino G, Rose G, Altomare K, Bellizzi D, Mari V, Feraco E, Franceschi C, De Benedictis G. Association of the mitochondrial DNA haplogroup J with longevity is population specific. Eur J Hum Genet. 2004;12:1080–1082. doi: 10.1038/sj.ejhg.5201278. [DOI] [PubMed] [Google Scholar]
- Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10:120–136. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Forster L, Forster P, Lutz-Bonengel S, Willkomm H, Brinkmann B. Natural radioactivity and human mitochondrial DNA mutations. Proc Natl Acad Sci U S A. 2002;99:13950–13954. doi: 10.1073/pnas.202400499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves LC, Turnbull DM. Mitochondrial DNA mutations and ageing. Biochim Biophys Acta. 2009;1790:1015–1020. doi: 10.1016/j.bbagen.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA, Jr, Kinzler KW, Vogelstein B, Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science. 1999;286:774–779. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- Nagley P, Wei YH. Ageing and mammalian mitochondrial genetics. Trends Genet. 1998;14:513–517. doi: 10.1016/s0168-9525(98)01580-7. [DOI] [PubMed] [Google Scholar]
- Norddahl GL, Pronk CJ, Wahlestedt M, Sten G, Nygren JM, Ugale A, Sigvardsson M, Bryder D. Accumulating mitochondrial DNA mutations drive premature hematopoietic aging phenotypes distinct from physiological stem cell aging. Cell Stem Cell. 2011;8:499–510. doi: 10.1016/j.stem.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Ogasawara Y, Nakayama K, Tarnowka M, McCoy JP, Jr, Kajigaya S, Levin BC, Young NS. Mitochondrial DNA spectra of single human CD34+ cells, T cells, B cells, and granulocytes. Blood. 2005;106:3271–3284. doi: 10.1182/blood-2005-01-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Fry A, Sheldahl L. Nuclear-mitochondrial epistasis and drosophila aging: introgression of Drosophila simulans mtDNA modifies longevity in D. melanogaster nuclear backgrounds. Genetics. 2006;172:329–341. doi: 10.1534/genetics.105.046698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve AK, Krishnan KJ, Taylor G, Elson JL, Bender A, Taylor RW, Morris CM, Turnbull DM. The low abundance of clonally expanded mitochondrial DNA point mutations in aged substantia nigra neurons. Aging Cell. 2009;8:496–498. doi: 10.1111/j.1474-9726.2009.00492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G, Romeo G, Dato S, Crocco P, Bruni AC, Hervonen A, Majamaa K, Sevini F, Franceschi C, Passarino G. Somatic point mutations in mtDNA control region are influenced by genetic background and associated with healthy aging: a GEHA study. PLoS ONE. 2010;5:e13395. doi: 10.1371/journal.pone.0013395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A, Bandelt HJ, Macaulay V, Richards MB. Phylogeographic investigations: the role of trees in forensic genetics. Forensic Sci Int. 2007;168:1–13. doi: 10.1016/j.forsciint.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Santoro A, Salvioli S, Raule N, Capri M, Sevini F, Valensin S, Monti D, Bellizzi D, Passarino G, Rose G, De Benedictis G, Franceschi C. Mitochondrial DNA involvement in human longevity. Biochim Biophys Acta. 2006;1757:1388–1399. doi: 10.1016/j.bbabio.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- Shin MG, Kajigaya S, McCoy JP, Jr, Levin BC, Young NS. Marked mitochondrial DNA sequence heterogeneity in single CD34+ cell clones from normal adult bone marrow. Blood. 2004;103:553–561. doi: 10.1182/blood-2003-05-1724. [DOI] [PubMed] [Google Scholar]
- Soares P, Ermini L, Thomson N, Mormina M, Rito T, Röhl A, Salas A, Oppenheimer S, Macaulay V, Richards MB. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneking M. Hypervariable sites in the mtDNA control region are mutational hotspots. Am J Hum Genet. 2000;67:1029–1032. doi: 10.1086/303092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A. Mitochondrial DNA and ageing. Biochim Biophys Acta. 2006;1757:611–617. doi: 10.1016/j.bbabio.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Vendelbo MH, Nair KS. Mitochondrial longevity pathways. Biochim Biophys Acta. 2011;1813:634–644. doi: 10.1016/j.bbamcr.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Michikawa Y, Mallidis C, Bai Y, Woodhouse L, Yarasheski KE, Miller CA, Askanas V, Engel WK, Bhasin S, Attardi G. Muscle-specific mutations accumulate with aging in critical human mtDNA control sites for replication. Proc Natl Acad Sci U S A. 2001;98:4022–4027. doi: 10.1073/pnas.061013598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YG, Bandelt HJ, Young NS. External contamination in single cell mtDNA analysis. PLoS ONE. 2007a;2:e681. doi: 10.1371/journal.pone.0000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YG, Childs RW, Kajigaya S, McCoy JP, Jr, Young NS. Mitochondrial DNA sequence heterogeneity of single CD34+ cells after nonmyeloablative allogeneic stem cell transplantation. Stem Cells. 2007b;25:2670–2676. doi: 10.1634/stemcells.2007-0269. [DOI] [PubMed] [Google Scholar]
- Yao YG, Ellison FM, McCoy JP, Chen J, Young NS. Age-dependent accumulation of mtDNA mutations in murine hematopoietic stem cells is modulated by the nuclear genetic background. Hum Mol Genet. 2007c;16:286–294. doi: 10.1093/hmg/ddl457. [DOI] [PubMed] [Google Scholar]
- Yao YG, Ogasawara Y, Kajigaya S, Molldrem JJ, Falcão RP, Pintão MC, McCoy JP, Jr, Rizzatti EG, Young NS. Mitochondrial DNA sequence variation in single cells from leukemia patients. Blood. 2007d;109:756–762. doi: 10.1182/blood-2006-01-011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Asin-Cayuela J, Fish J, Michikawa Y, Bonafe M, Olivieri F, Passarino G, De Benedictis G, Franceschi C, Attardi G. Strikingly higher frequency in centenarians and twins of mtDNA mutation causing remodeling of replication origin in leukocytes. Proc Natl Acad Sci U S A. 2003;100:1116–1121. doi: 10.1073/pnas.242719399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.