Abstract
The identification of resting state networks (RSNs) and the quantification of their functional connectivity in resting-state fMRI (rfMRI) are seriously hindered by the presence of artefacts, many of which overlap spatially or spectrally with RSNs. Moreover, recent developments in fMRI acquisition yield data with higher spatial and temporal resolutions, but may increase artefacts both spatially and/or temporally. Hence the correct identification and removal of non-neural fluctuations is crucial, especially in accelerated acquisitions. In this paper we investigate the effectiveness of three data-driven cleaning procedures, compare standard against higher (spatial and temporal) resolution accelerated fMRI acquisitions, and investigate the combined effect of different acquisitions and different cleanup approaches. We applied single-subject independent component analysis (ICA), followed by automatic component classification with FMRIB’s ICA-based X-noiseifier (FIX) to identify artefactual components. We then compared two first-level (within-subject) cleaning approaches for removing those artefacts and motion-related fluctuations from the data. The effectiveness of the cleaning procedures were assessed using timeseries (amplitude and spectra), network matrix and spatial map analyses. For timeseries and network analyses we also tested the effect of a second-level cleaning (informed by group-level analysis). Comparing these approaches, the preferable balance between noise removal and signal loss was achieved by regressing out of the data the full space of motion-related fluctuations and only the unique variance of the artefactual ICA components. Using similar analyses, we also investigated the effects of different cleaning approaches on data from different acquisition sequences. With the optimal cleaning procedures, functional connectivity results from accelerated data were statistically comparable or significantly better than the standard (unaccelerated) acquisition, and, crucially, with higher spatial and temporal resolution. Moreover, we were able to perform higher dimensionality ICA decompositions with the accelerated data, which is very valuable for detailed network analyses.
Keywords: functional magnetic resonance imaging (fMRI), resting-state, artefact removal, functional connectivity, multiband acceleration
1. Introduction
Resting-state functional magnetic resonance imaging (rfMRI) is a widespread and powerful technique for investigating the functional connectivity of the human brain. This form of connectivity is measured by evaluating the similarity between different voxels’ timeseries, given the evidence that the spontaneous timeseries from functionally related brain regions are correlated (Biswal et al., 1995). Several resting state networks (RSNs) have been identified and changes in their connectivity have been observed in a variety of diseases (for a review, see Cole et al., 2010). One major problem with rfMRI, however, is the presence of artefacts, many of which share some spatial or spectral overlap with RSNs and affect their correct identification and quantification of their connectivity. Unlike task-fMRI, where there is prior knowledge about the temporal signal of interest, in rfMRI any non-neural activity shared across multiple voxels can yield misleading results.
Spatially extended artefacts can be caused by the MRI hardware/software (e.g., timing instabilities), or more frequently, they are caused by physiology and subject motion (Murphy et al., 2013). For example, it is well known that even relatively small amounts of head motion represent a significant confound for rfMRI network identification (Van Dijk et al., 2012; Power et al., 2012; Satterthwaite et al., 2012). Other confounds relate to physiological noise, including cardiac and respiratory cycles (Birn et al., 2006; Shmueli et al., 2007), which occur at relatively high frequency (≈ 1 Hz and ≈ 0.3 Hz, respectively) but are generally aliased into lower frequencies at standard TR (2–3 s) (Lowe et al., 1998). Vascular tone also represents a source of physiological noise, generating low-frequency oscillations (<0.1 Hz) in absence of stimulus (Aalkjaer et al., 2011). Only by removing such confounds is it possible to obtain reliable functional connectivity measures. Moreover, due to the increasing use of high field MRI, hardware innovations, and the introduction of innovative accelerated sequences (Moeller et al., 2010; Feinberg et al., 2010; Setsompop et al., 2012), we are able to obtain data with higher SNR and/or spatial/temporal resolution, but this may come at the price of an associated increase in certain structured artefacts. It is therefore highly desirable to develop an effective method for artefact identification and removal that retains as much neuronally-related signal as possible.
Current noise removal methods for individual resting dataset cleanup can be divided into two main categories (see Murphy et al., 2013 for a detailed review): those that use external physiological recordings and those that are data-driven. In the former category, with techniques like retrospective image correction (RETROICOR – Glover et al., 2000), low-order Fourier series are fit to the image data based on the time of each image acquisition relative to the phase of the cardiac and respiratory cycles. This approach has been extended to include the regression of low-frequency changes in heart rate (Shmueli et al., 2007), while Birn and colleagues (2006) developed a method to remove the variance of respiration-induced changes from the data through the regression of the respiration volume per time (RVT).
However, physiological monitoring data are often not available, and are not expected to relate to all common forms of artefact; hence, several methods have been proposed to estimate and remove the artefacts using only the fMRI data itself. The simplest approach is to apply temporal filtering (e.g., a band-pass filter keeping frequencies 0.01–0.08 Hz) that removes the primary cardiac and respiration frequencies if the TR is short enough, but not their aliased lower-frequency components with more standard TR. Moreover, the removal of high frequency signals through bandpass filtering may remove signal that contributes to resting state networks (Niazy et al., 2011). Another standard pre-processing step for fMRI analyses is rigid-body head motion correction. Usually, all the volumes are aligned to a reference volume, with the volume-to-volume movement of the head described by three translations and three rotations. However, even with perfect geometric correction of head motion, spin history effects result in residual motion-related artefacts; it is common to attempt to remove these from the data using a multiple linear regression, with the confound regressors derived from the estimated motion parameters. However, it has been shown (Power et al., 2012; Satterthwaite et al., 2013) that such approaches are often not capable of completely removing the effects of motion. Power and colleagues (Power et al., 2012) described a technique called “scrubbing” to deal with remaining artefacts: volumes (time points) affected by excessive motion are simply excluded from the functional connectivity analyses. In another study (Satterthwaite et al., 2013) the authors proposed an improved pre-processing procedure by removing a higher number of motion-derived regressors (24 regressors, derived from the 6 motion parameters, their temporal derivatives, and the squares of the 12 resulting regressors). Further nuisance regressors can be derived from the resting data itself. Under the assumption that any process that affects all brain voxels is unrelated to the neural activity, global signal regression removes the global mean signal computed across all voxels in the brain (Desjardins et al., 2001; Greicius et al., 2003). However, it has been demonstrated (Murphy et al., 2009; Saad et al., 2012) that the global regression process also introduces spurious anti-correlations that are difficult to interpret. Some studies (Popa et al., 2009; de Pasquale et al., 2010) indicate that the global signal can include a significant amount of neural activity; therefore, many argue that its removal should be avoided. As BOLD signal related to neural activity should be predominantly in the grey matter, an alternative method is to regress out of the timeseries derived from just the white matter and/or cerebrospinal fluid (CSF) voxels (Weissenbacher et al., 2009). Confound removal can also be performed through a modification of the acquisition sequence. The dual echo approach proposed by Bright and Murphy (Bright and Murphy, 2013) consists of simultaneous acquisitions of a short echo time (TE) and a BOLD-weighted (standard TE) fMRI data, followed by voxelwise regression of the short TE data from the BOLD-weighted data, to remove noise variance. A related method uses a multi-echo approach that allows separation of BOLD and non-BOLD signal components based on TE-dependence (Kundu et al., 2012). Multi-echo data at 3 TEs were acquired and fed into independent component analysis. Components were analyzed for the degree to which their signal changes fit models for relaxation-rate (R2*) and initial signal intensity (when the TE=0) change, and summary scores were developed to characterize each component as BOLD-like or not BOLD-like. These scores clearly differentiated BOLD-like RSN components from non BOLD-like components (related to motion, pulsatility, and other nuisance effects), and non BOLD-like component timecourses were used as noise regressors to improve seed-based correlation mapping.
Independent component analysis (ICA) (McKeown et al., 1998) h as proven to be a successful technique for detecting consistent spatial components and separating signal from noise. ICA decomposes the 4D (space X time) data into multiple components, each described by a single 3D spatial map and an associated timecourse. Ideally, some components purely reflect BOLD signal, and others purely reflect artefactual processes. If the latter can be identified, then they can be subtracted from (or regressed out of) the data. Identification of artefactual components by hand is time-consuming, operator dependent, and requires expert knowledge about signal and noise fluctuations’ spatial and temporal characteristics. Thus, there have been several approaches proposed that attempt to automate ICA-based denoising using different strategies to classify the components as signal or noise. To mention a few examples in addition to the work of Kundu et al. (2012): Thomas et al., (2002) identify the noise components to remove using an unsupervised algorithm that examines the Fourier decomposition of the time series obtained after principal components analysis or ICA; Kochiyama and colleagues (2005) proposed an automatic solution for removing the effects of task-related motion, characterising the non-artefactual ICs by virtue of their task-related signal changes; Perlbarg et al. (2007) remove signal fluctuations that match known spatial patterns of physiological noise; similarly, Beall and Lowe (2007) estimated cardiac and respiratory fluctuations from resting state data with temporal ICA and generated spatial weight matrices applicable to other resting data.
The presence of multiple distinct kinds of artefacts in rfMRI data requires the identification and removal of a wide range of component types (i.e., having potentially quite varied artefactual spatial and/or temporal characteristics). Therefore, Tohka et al. (2008) proposed a richer set of spatial and temporal features that capture a wider range of ICs’ characteristics, while De Martino et al., (2007) used a representation of the components in a multidimensional space of descriptive measures (IC-fingerprints), which are then used to classify the components by feeding the features into a support vector machine. The “features” are quantities derived from the ICA spatial maps and/or timecourses; for example, one feature might be the fraction of the supra-threshold spatial map overlaying grey matter, and another might be the fraction of power in the timeseries spectrum lying above 0.05Hz. A set of distinct features can be fed into a trained multivariate classifier in order to attempt to classify each ICA component as “good” or “bad”.
We recently developed FMRIB’s ICA-based Xnoiseifier (FIX), which is an automated approach (once trained) for cleaning fMRI data of various types of artefact (Salimi-Khorshidi et al., 2014). The general cleaning procedure in FIX, as with some of the above-mentioned methods, consists of several steps: spatial ICA, component-wise feature extraction, classifier training (using expert-hand-labelled data), components’ classification (i.e., predicting components’ likelihood of being signal vs. noise, in new data) and denoising (removal of the artefactual components). The first four steps are extensively described in Salimi-Khorshidi et al., 2014: the ICA step is performed with MELODIC (Beckmann and Smith, 2004); over 180 spatial and temporal features are extracted for each ICA component; the training and prediction phases are performed with a hierarchical fusion of multiple classifiers (K-nearest neighbour, support vector machines and decision trees). The performance of FIX in component classification against manual labelling (the “gold standard” for component classification) has been assessed on various rfMRI datasets, achieving over 99% classification accuracy on the best datasets, and around 95% accuracy on more “standard” acquisitions.
In this paper, we investigated different approaches for carrying out the last step of the denoising procedure – the removal of the nuisance components identified by the classifier, resulting in “cleaned” fMRI data. This is not trivial, as the noise components can share variance with components containing RSN signal. For this reason, the first aim of the present work was to compare several cleaning approaches in the framework of the FIX strategy and find a recommended procedure for noise removal, in order to clean the fMRI data of artefacts, while minimising the loss of signal. To achieve this, we tested the efficacy of the different cleaning procedures through timeseries, network and spatial map RSN analyses, feeding uncleaned and cleaned datasets into both low- and high-dimensional group-level ICA to identify resting-state networks (see sections 2.5 and 3 for details).
Recently, multiband echo planar imaging has proven to be very powerful for obtaining fMRI data (Moeller et al., 2010), with improved temporal and/or spatial resolution, and improvements in sensitivity to detection of RSNs (Feinberg et al., 2010). Slice accelerated EPI has seen rapid adoption in the neuroimaging community, for example, in the Human Connectome Project and the most recent Thousand Connectomes datasets from the Nathan Kline Institute. The second aim of this study was to further evaluate the potential of the multiband accelerated EPI sequence, and to investigate the effect of combining different acquisition pulse sequences with different artefact cleaning approaches (the accelerated multiband acquisitions might well give different results for different cleanup approaches, compared with standard EPI). Therefore, all evaluations were performed on two large datasets from one set of subjects - a “Standard” EPI sequence and a multiband accelerated EPI sequence, demonstrating the efficacy of FIX cleaning on both acquisitions via investigation of RSN timeseries, correlation networks and spatial maps (see sections 2.5 and 3 for details). Partly as a result of this study, public releases of HCP rfMRI data are being cleaned using FIX using the “soft” cleanup approach described below (Smith et al., 2013).
2. Methods
2.1. Subjects and MRI Data Acquisition
Data from 76 subjects, participants in the Whitehall II MRI study (ages 69.1 ± 5.8 years, M/F = 52/24), were acquired using a 3T Siemens Verio MRI scanner with a 32-channel head coil. All subjects gave written informed consent to participate in the study. The following MRI sequences were acquired in fixed order for all subjects:
3D high-resolution T1-weighted MR images were acquired using a MEMPRAGE sequence (TR = 2530 ms, TE = 1.79/3.65/5.51/7.37 ms, flip angle = 7°, field of view = 256 mm, voxel dimension = 1 mm isotropic, acquisition time = ~6 min);
Multiband accelerated rfMRI acquisition (hereafter referred as MB6): single-shot EPI T2*-weighted images (TR = 1300 ms, multiband factor MB = 6, PEshift=FOV/3, TE = 40 ms, Flip Angle = 66°, voxel dimension = 2 mm isotropic, whole brain, acquisition time = 10 min for a total of 460 time points), developed partly for the Human Connectome Project (Moeller et al., 2010; Feinberg et al., 2010; Setsompop et al. 2012; Xu et al., 2013);
Standard rfMRI acquisition: single-shot EPI T2*-weighted images (TR = 3000 ms, TE = 30 ms, Flip angle = 90°, voxel dimension = 3 mm isotropic, whole brain, acquisition time = 10 min for a total of 200 time points);
Field maps were acquired to reduce EPI distortion due to magnetic field inhomogeneity (TR = 400 ms, TE = 5.19/7.65 ms, flip angle = 60°, field of view = 258 mm, voxel dimension = 3 mm isotropic, acquisition time = ~1 min).
2.2. rfMRI Data Pre-processing
The same pre-processing was performed on both Standard and MB6 rfMRI data using FSL (Smith et al., 2004; Jenkinson et al., 2012). Each rfMRI dataset was corrected for head motion using MCFLIRT (Jenkinson et al., 2002) and then corrected for EPI distortions using FMRIB’s Utility for Geometrically Unwarping EPIs (FUGUE), which performs the unwarping of the EPI images based on fieldmap data. Non-brain tissue was removed with BET (Smith, 2002) and data were high-pass temporal filtered to remove slow drifts (cutoff period ~ 100.0 s). Data were not spatially smoothed, in order to make it easier to compare and interpret the effect of the different acquisitions’ resolutions (e.g., on SNR and apparent spatial detail). Each 4D pre-processed dataset was then fed into MELODIC (Multivariate Exploratory Linear Optimised Decomposition of Independent Components – Beckmann and Smith, 2004) to perform within-subject spatial-ICA with automatic dimensionality estimation (we explain below how the ICA outputs were then used). The 76 subjects’ datasets were then randomly split into two age-matched subsets: 23 subjects were used as the training dataset for FIX, and 53 as the test dataset. The rationale for this group size difference is because we judged 23 subjects to be sufficient to train FIX well and provide robust group-level ICA decomposition for the templates (see section 2.3), reducing the manual intervention as much as possible. Moreover, many of the final evaluations depend on cross-subject variance estimations, and hence the size of the second group is arguably more important to our quantifications than the size of the first. Subsequent analysis steps are summarized in Fig. 1.
Fig. 1.

Graphical illustration of overall evaluation.
2.3. Group-ICA RSN templates creation
Next we applied ICA-based artefact removal for each subjects’ data in the training dataset. Every ICA component from every single-subject dataset (from both MB6 and Standard acquisitions) was hand-classified into signal or noise, looking first at the thresholded spatial map and then at the temporal power spectrum and at the timecourse. For more detail and examples regarding how the hand-classification decisions were made see Salimi-Khorshidi et al., 2014. Briefly, the typical characteristics of signal components are:
spatial maps with well-defined grey matter clusters;
spatial pattern attributable to the RSNs’ patterns described in literature (Beckmann 2005; De Luca 2006; Rytty et al., 2013);
predominantly low-frequency (<0.1 Hz) power spectra;
while noise components can have:
spatial overlap with white matter, CSF, or blood vessels;
signal localised at the edges of the brain (motion) or in areas of signal drop (susceptibility);
spatial maps with irregular/not well-defined clusters;
non-predominantly low-frequency (<0.1 Hz) power spectra;
spikes in the timeseries.
Next, the full space of all artefact timeseries and motion parameters (as opposed to the partial space, where the non-artefact timeseries are also included in the regression; see section 2.4 for further details on this distinction) was regressed out of the 4D pre-processed data, to achieve “aggressive” artefact removal. The rationale for applying more aggressive artefact regression was that we wanted to obtain the cleanest possible group-level ICA maps from the training dataset, to be used as a reference of “true signal”. The aggressive within-subject-cleaning approach maximizes the noise removal at the expense of potential loss of some RSN-related signal in some subjects, which is compensated for by utilizing multiple subjects for the group-ICA-based template generation. We registered subjects’ cleaned rfMRI 4D data to their high-resolution structural image using FLIRT linear registration, enhanced with brain-boundary-registration (BBR Greve and Fischl, 2009), and then to MNI152 standard space via application of the nonlinear FNIRT tool applied to the structural image (Andersson et al., 2007a, b). All 4D rfMRI datasets were resampled to 2 × 2 × 2 mm3 resolution in the final MNI152 space.
We then performed (separately for Standard and MB6 datasets) group-ICA using MELODIC. The group ICA was performed at two dimensionalities (d): with 30 dimensions for a more “conventional” ICA analysis, and with 100 to achieve a more fine-detailed functional parcellation of the data (this is useful for more detailed network analyses). These four template sets of group-average maps were used as reference templates in subsequent analyses performed on the test datasets. The templates’ components were manually classified as RSNs or artefacts based on previous knowledge of the RSNs’ patterns described in literature (Beckmann 2005; De Luca 2006; Rytty et al., 2013), and following the same rules used for single-subject manual labelling described in Salimi-Khorshidi et al., 2014 and (Smith et al., 2013), (including study of the mean cross-subject temporal power spectrum for each component), and further double-checked by two experts. (Although ICA-based cleanup was applied to each separate dataset, some artefactual components can still emerge at the group-level. For example, low-level artefactual processes that are too weak to be identified by single-session ICA may be consistent across subjects and hence appear more strongly at the group level).
2.4. Automated classification and cleanup procedures with FIX
In addition to generating the group-ICA template maps, the hand-labelled components from the training dataset were also used to train the FIX (FMRIB’s ICA-based X-noiseifier) component classifier (Salimi-Khorshidi et al., 2014). We created one training dataset for the Standard data and extended a pre-existing training dataset for MB6 (though our classification accuracy results were extremely similar for MB6, if we only used this current study’s MB6 training dataset). We evaluated FIX’s classification accuracy via leave-one-out bootstrap testing (see Salimi-Khorshidi et al., 2014, for details).
The single-subject ICA components of the test dataset were then automatically classified into signal and noise using FIX. As the ground truth of signal and noise is unknown, from now on we will refer to “signal” and “noise” components identified in the test dataset as defined by the FIX classification (purely for convenience of notation). For the cleanup procedure we used both the noise components’ timeseries and 24 motion-estimation confound timeseries (the six rigid-body parameter timeseries, their backwards-looking temporal derivatives, and the squares of all twelve resulting regressors) (Satterthwaite et al., 2013). The 24 motion confound timeseries then had the same temporal highpass filtering applied to them that had been applied to the data.
We evaluated two different cleaning procedures: an “aggressive” cleanup, and a “soft” cleanup.
The “aggressive” approach consisted of regression of the full space of all artefacts (noise components) and the motion confounds out of the 4D pre-processed data (Y), using:
| (1) |
where C is the matrix of artefact and motion timeseries (C=[Cmotion ICA(bad)]) and pinv() is the matrix pseudo-inverse (for example, as estimated via pinv(C)=(CTC)−1CT). With this method, the contribution of the motion and the artefacts is fully removed from the data. In general, the signal and noise ICA components are not completely orthogonal; this approach removes all shared variance between the two (hence the term “aggressive”).
The “soft”, less aggressive, approach consisted of three steps. First, we regressed out the full space of the motion confounds (Cmotion) from both the data and from all the (“good” and “bad”) ICA component timeseries (ICA), in order to fully (aggressively) remove the effect of motion from the data (resulting in Ym) and the ICA timeseries (resulting in ICAm):
| (2) |
| (3) |
Second, we estimated the contribution of both good and bad components ( ) via multiple regression of the data against all (motion-cleaned) ICA timeseries, in order to be able to identify the unique variance of the artefacts (Eq 4):
| (4) |
Finally, using this, the unique contribution of the bad components was removed from the data, utilising only the bad ICA components’ timeseries and regression coefficients:
| (5) |
Hence we obtained 6 different test datasets: uncleaned, softly-cleaned and aggressively-cleaned data for MB6 and Standard acquisitions. For each dataset, the raw temporal-SNR image was formed for each subject, eroded to exclude brain-edge effects, and the median SNR value was calculated as a first measure of the cleaning effect (note that the temporal “noise” in the SNR here includes valid RSN-related fluctuations). All subjects’ rfMRI test data were then resampled into 2 mm MN I152 space, as carried out on the training datasets.
2.5. Dual-Regression and analyses
In order to evaluate sensitivity and consistency of RSNs estimated from the test datasets, we applied dual-regression (described below) of the training-dataset group-level template spatial maps into each test dataset. This resulted in test-dataset subject-specific spatial maps corresponding to the template maps, and associated subject-specific timeseries.
The four sets of group-ICA templates were used (separately from each other, i.e. in four different analyses) in the first stage of dual regression (Filippini et al., 2009) as sets of spatial regressors. For each version of each single-subject rfMRI test dataset (created as described above), the full set of d template spatial maps was regressed into the single-subject data (as a spatial regression), the output being d single-subject timeseries (separately for each subject in the test dataset). These subject-level timeseries (from each of the 53 test dataset subjects) were then used to perform timeseries (temporal standard deviation to represent the timeseries amplitude, and temporal power spectra to analyze the frequency content) and correlation-based network analyses (see section 3.4–3.6 for details).
As the group ICA contained some artefactual components, we introduced a further timeseries cleanup (“Nets cleanup”), where the timeseries corresponding to the artefactual group-level components manually identified in the templates (see section 2.3) were regressed out of the remaining RSNs’ timeseries using Eq.6:
| (6) |
In the case of unknown group-level components (not artefacts but also not clearly RSNs), these were simply discarded for the purposes of the Nets cleanup evaluations, and not regressed out of the good components. Timeseries and network analyses were evaluated only using timeseries derived from good template components, with 6 different combinations of first-level (no cleaning, soft, aggressive) and timeseries (with vs. without Nets) cleaning. For the temporal analyses we calculated the mean amplitude of the timeseries for each component (section 3.4) and the mean power spectrum across subjects and components (section 3.5). For the network analysis, d × d full correlation, partial correlation and L1-norm regularised partial correlation matrices (Smith et al., 2011) were estimated using the set of RSN timeseries (section 3.6). The temporal characteristics and network matrices were estimated separately for each subject.
Finally, the second stage of dual regression was carried out for each subject, resulting in subject-specific spatial maps corresponding to the template maps. This was achieved by regressing the subject-specific rfMRI datasets against the set of d subject-specific timeseries as estimated in stage 1. We calculated two sets of test-dataset cross-subject group-level maps (one using mixed-effects (ME) statistics and one using approximate fixed-effects (FE) statistics), and compared them against the template maps, under the assumption that the more a group RSN map reflects the corresponding template map, the more the cleaning approach (or the acquisition sequence) correctly identifies the true signal. To do this, we calculated the spatial correlation coefficients between the group maps and the templates, (as well as several measures of sensitivity and specificity), which were compared between different cleaning procedures and acquisitions. All d components were used in the two dual-regression stages, but only the non-artefactual components were then used when comparing the test-dataset maps against the training-dataset template maps (see section 3.7 for details).
In the following section we describe the different comparison analyses and present the comparison results. Section 4 then includes a broader discussion of the results.
3. Results
3.1. Single-subject independent component classification
The single-subject ICA decompositions, performed on all 76 subjects, found 69.8 ± 14.9 components (per dataset) with the Standard sequence data, and 124.9 ± 25. 7 with MB6 sequence data, as judged by the MELODIC automatic dimensionality estimation (Beckmann and Smith, 2004). Of these, the artefact components (which were manually identified on the 23-subject training/template dataset) were more than 85% of all components (60.9 ± 14.9, i.e., 87.7%, for Standard and 111.5 ± 24.0, i.e., 88%, for MB6). On the remaining 53 subjects, FIX automatically identified 57.4 ± 15.8 (82%) artefactual components on Standard data and 108.1 ± 29.8 (87%) on MB6 data, in line with the proportion of artefacts found with hand labelling. FIX performed with higher classification accuracy on MB6 data, with an overall accuracy of 98% for MB6 and 95% for Standard (see Salimi-Khorshidi et al., 2014, for details).
3.2. Temporal SNR results
Based on the expectation that the cleaning procedure should decrease fluctuation of the signal around its mean, we first compared the temporal “SNR” for each cleanup and sequence. For each rfMRI run (both for Standard and MB6 sequences), a temporal-SNR image was generated after motion correction and highpass filtering; the “noise” here includes RSN-related fluctuations, which therefore places an upper limit on the possible SNR. The SNR image was eroded by 3 voxels at the brain edge to avoid edge effects, and the median SNR value across voxels was computed. This process was repeated after soft and aggressive cleanup. The results were then compared across cleaning approaches and between sequences by means of two-tailed paired t-tests.
The boxplots in Fig. 2 show the distributions of the median (across space) SNR value over the 53 subjects for the two acquisitions with different cleaning options. The raw SNR results (Fig. 2.A) show that the cleaning procedure significantly increases the SNR (p<0.01), while, when comparing the two sequences, the combined decrease in voxel volume and the EPI acceleration in MB6 results in significant lower SNR (p<0.01) (due more to the former than the latter effect (Smith et al., 2013)). However, if the increased number of timepoints is taken into account (Fig. 2.B), the statistical power for simple analyses applied to MB6 data is seen to be comparable to those from the standard acquisition. This is of great value because it means that the increase in statistical power due to the acceleration counters the loss in SNR caused by the increase in spatial resolution. Of course, these SNR results do have somewhat limited meaningfulness, because of the fact that the “noise” in the “SNR” includes both interesting signal fluctuations as well as various sources of noise.
Fig. 2.
Temporal SNR estimation for various cleaning procedures and acquisition protocols. The boxplots show the distribution across 53 subjects. From the raw SNR results (2.A), it is clear that the cleaning procedure increases the SNR, while the reduced voxel volume and EPI acceleration decreases it. However, taking into account the increased number of timepoints (2.B), the statistical power for simple analyses applied to MB6 data is seen to be comparable to those from the standard acquisition. This is of great value because it means that the increase in statistical power due to the acceleration counters the loss in SNR caused by the increase in spatial resolution.
3.3. Group ICA components and dual regression - summary
As described above, group ICA was performed at two dimensionalities (d=30 and 100). Based on visual inspection (3 experts in agreement with each other) of each component’s spatial map and (mean cross-subject) temporal power spectrum, 28/30 and 58/100 group-ICA components were judged to be non-artefactual in the MB6 templates, and 19/30 and 43/100 in the Standard ones. We identified 1/30 and 7/100 components as unknown (i.e., components which could not be unambiguously identified as good or bad) in the MB6 templates and 1/30 and 17/100 in the Standard ones. For each dimensionality, the difference in the proportions of good and bad components between the two sequences was tested with a binomial test. We found that the proportion of good components found in the templates was significantly higher for MB6 (p<0.05), and the proportion of unknown components significantly lower (except for the case of only having 1/30 unknown component for both acquisitions with the d=30 analyses). These results strongly suggest that MB6 data can be more effectively cleaned of artefacts by FIX, both at low and high group-ICA dimensionality.
As we were interested in comparing both different cleaning procedures and the two acquisitions, we performed 12 sets of dual regressions (two sequences X two dimensionalities X three cleanings). However, noise removal and high dimensional dual-regression was only fully possible with MB6, because, with Standard data, the temporal degrees of freedom after the cleaning procedure were not sufficient to perform the full dual-regression with d=100. For this reason, subsequent comparisons among different cleaning approaches were performed on Standard d=30, MB6 d=30 and MB6 d=100, while the comparisons between the two acquisitions was performed at d=30. Where possible and meaningful we included and discussed the results obtained on Standard d=100. For each dataset, we obtained subject-level timeseries (first stage of dual regression) and spatial maps (second stage), which were analysed to test the effect of the different cleaning procedures and the differences between the two acquisitions. More specifically, the output of the first stage of dual regression (subject-level timeseries of the 53 test subjects) was used to perform timeseries (amplitude and power spectra) and network analyses, while the output of the second stage was entered into spatial map analyses.
3.4. Timeseries’ amplitude analysis
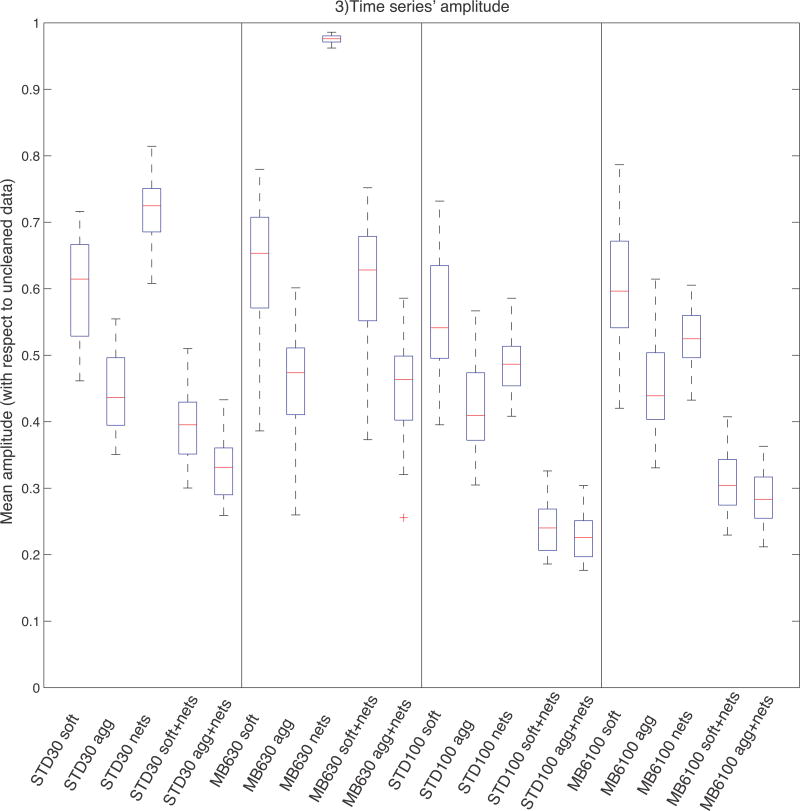
Given the hypothesis that the cleaning procedure should decrease the fluctuation of the subject-level timeseries with respect to the uncleaned data, we compared the timeseries’ amplitude across cleanings and sequences. This measure was obtained by scaling each timeseries’ standard deviation (for each subject and component) by the standard deviation of the corresponding uncleaned timeseries. Unlike the SNR analysis, where the normalisation factor was the corresponding mean signal, here the amplitudes are normalised by the uncleaned data amplitude. The boxplots in Fig. 3 show the distribution of amplitudes across components (with the scaled amplitude for a given component averaged across subjects) obtained with different cleaning approaches. We directly compared these amplitude values through two-tailed paired t-tests and the results obtained with the two sequences using two-tailed independent t-tests, and the results are shown in Supplemental Tab. S1. With all protocols, each cleaning step (both at 1st and 2nd level) significantly reduced the timeseries amplitude (p<0.01). Regarding 1st level cleanup, the largest difference was observed between uncleaned and cleaned data (almost 50% amplitude reduction), while the difference between soft and aggressive approach is less strong, although statistically significant (p<0.01). Nets cleanup significantly reduces the amplitude, especially if the data have not been cleaned at 1st level. On MB6 d=30 data the effect of nets cleanup was statistically significant but limited in effect size, because only one artefactual component was removed.
Fig. 3.
Timeseries amplitudes. This measure was obtained by scaling (the standard deviation of the) single-subject timeseries associated with each group-level map by the standard deviation of the corresponding uncleaned timeseries. The boxplots show the distribution of amplitudes across components (each component is first averaged across subjects). All cleanup approaches decrease the amplitude; the amplitude is higher with MB6 sequence than with Standard. STD=Standard sequence; uncl=uncleaned; soft=FIX soft cleaning; agg=FIX aggressive cleaning; nets=Nets cleaning.
As with the SNR results, these results have limited interpretability because it is not known what the balance is between remaining noise and signal contributions to the amplitudes; nevertheless, the results are useful indicators of how much variance is being removed in the various cases. For example, if there was almost no variance being removed by a given method, further investigations would be unnecessary.
Regarding the comparison between the two acquisitions (see Supplemental Tab. S2), amplitude with MB6 was always higher than Standard: this difference was significant only on nets cleanup results at low dimensionality, and for all cleaning options at high dimensionality. This means that a slightly smaller fraction of the temporal variance is removed by the cleaning of the MB6 data.
3.5. Timeseries’ power spectra
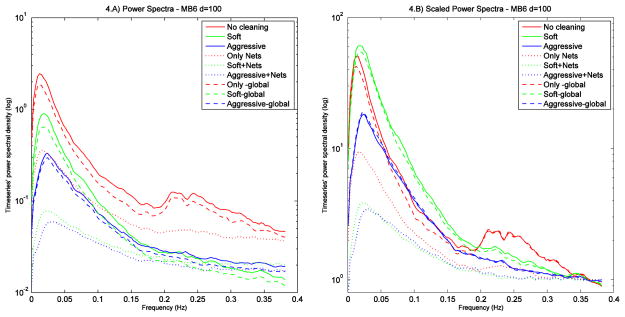
In order to evaluate the impact of the cleaning procedures at different frequencies, timeseries’ power spectra were also generated after scaling each time series (for each subject and component) by the standard deviation of the corresponding uncleaned timeseries. The mean spectra were obtained by averaging the spectra across subjects and calculating the median across components. With this qualitative analysis we compared: no cleaning, soft cleanup and aggressive cleanup for first-level (FIX) cleaning; no cleaning, nets cleanup and global signal removal, for second-level (“Nets” timeseries) cleaning. As global signal removal is frequently used in the literature (Fox et al., 2009), we included this option when comparing our different cleaning approaches, calculating the global signal as the mean timeseries across all components (good and bad) for each subject. Fig. 4.A shows the power spectra at each cleaning step for the MB6 sequence at high dimensionality (results were similar for other protocols, see Supplemental Fig. S1). As expected, uncleaned data have the highest power both at low frequency (LF, predominantly signal) and at high frequency (HF, predominantly noise, both artefactual and thermal). After soft cleanup the power is more reduced at HF than at LF, while aggressive cleanup causes the biggest power reduction both at LF and HF. Regarding the effect of second-level cleaning, there is not much additional reduction after the removal of the global signal (slightly lower power both at LF and HF), while Nets cleanup caused a large additional reduction.
Fig. 4.
Temporal power spectra (4.A) for different cleaning approaches, obtained from scaled timeseries (i.e., each normalised by the amplitude of the corresponding uncleaned timeseries), averaging the spectra across subjects and then calculating median spectra across components. Uncleaned data have the highest power both at low and high frequency; however, after normalising for power at the highest frequencies (last 0.8% of the spectrum width, where the content of thermal noise is higher than the content in signal) (4. B), it is clear that with soft cleanup we obtained the highest contrast-to-noise ratio. Results are shown for MB6 data, at d=100 (y axis in logarithmic scale).
In order to obtain a different measure of contrast-to-noise ratio (CNR), we scaled each power spectrum (Fig. 4.B) according to the amount of thermal noise. We assumed the thermal noise level to be approximated by the mean value of the power spectra at the highest frequencies (last 0.8% of the spectrum width), where the spectral curve has (or nearly has) asymptoted, and where the content of thermal noise is higher than the content in signal (Cordes et al., 2002; Triantafyllou et al., 2005). After this normalisation, it is clear that the highest CNR was obtained with soft cleaning (green line). This cleaning approach is also the most affected by nets cleanup, probably because nets cleanup is not effective if the first-level cleaning has not been performed (red dotted line), while most of the noise has already been removed with aggressive cleanup (blue line).
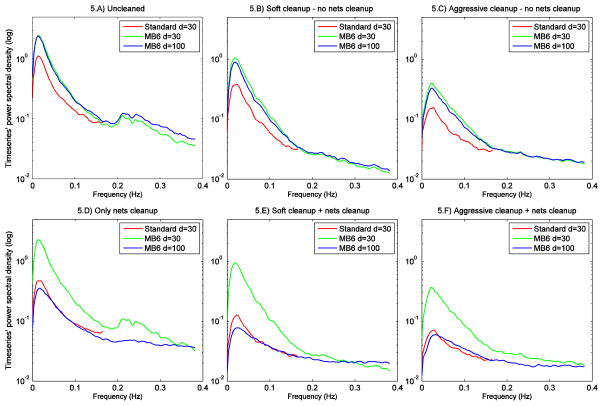
In Fig. 5 we present related comparisons, but now concentrating on comparing the power spectra (not normalised for high frequency power) across the different protocols for the different cleaning options. The distinct artefact peak that was seen in non-cleaned MB6 data at ~0.25 Hz is not apparent in Standard data power spectra because of temporal aliasing of physiological artefacts; the lower temporal resolution (TR=3s) does not allow the capturing of such fluctuations cleanly, so the effect of physiological artefacts is mixed in with the true underlying fluctuations at other frequencies. At d=30, the LF peak (dominated by RSN signal) is always higher for MB6 than Standard, and the difference is increased with both first and second level cleaning. The large differences between MB6 d=30 and d=100 after nets cleanup arise because of the different number of bad components removed from each dataset (1 bad component for MB6 d=30 and 35 bad components for MB6 d=100). From these figures we can also observe that Nets cleanup is not as effective as FIX: in fact, comparing the spectra obtained with the two methods (Fig. 5 B vs 5 D), we can see that the HF peak is always lower using FIX. Regarding the effect on LF, for MB6 d=100 data the LF peak is higher with FIX, suggesting that FIX retains more RSNs-related signal than Nets cleaning. On the contrary, for MB6 d=30 the LF peak is higher with Nets cleaning. However, it must be taken into account that, as only one component is removed with Nets cleanup, the spectra is almost identical to the one from uncleaned data (Fig. 5 A), indicating that FIX still offers the best balance between noise removal and signal loss. No significant differences at LF peak were observable on Standard d=30.
Fig. 5.
Temporal power spectra of the different protocols for each cleaning option. The spectra were obtained by averaging the single subjects’ spectra (from timeseries scaled using the amplitude of uncleaned data) and calculating median spectra across components, without normalisation for high frequency power. Y axis in logarithmic scale.
3.6. Network analyses
Network analysis was performed by estimating correlations between all pairs of timeseries (good components only). Full correlation, partial correlation, and L1-norm regularised partial correlation (“ICOV” - obtained by regularising the inverse of the covariance matrix) were used for the computation of network matrices. Full correlation evaluates the similarity between two timeseries directly, reflecting both direct and indirect functional connections, while partial correlation evaluates the similarity between two timeseries after regressing out all other timeseries, and should emphasize direct functional connections, rather than indirect (Marrelec et al., 2006; Smith et al., 2011; Smith, 2012). The L1-norm regularised partial correlation (regularised ICOV) shrinks entries that are close to zero more than those that are not (Friedman et al., 2008). In our analyses we used a regularisation-controlling parameter lambda=0.1. In this way we were able to estimate regularised partial correlation network matrices also for Standard data at high dimensionality d=100, for which the degrees of freedom were not sufficient to estimate unregularised partial correlation. Correlation matrices were transformed into z-scores using the Fisher transform (including an empirical correction for temporal autocorrelation using FSLNets1) to improve normality.
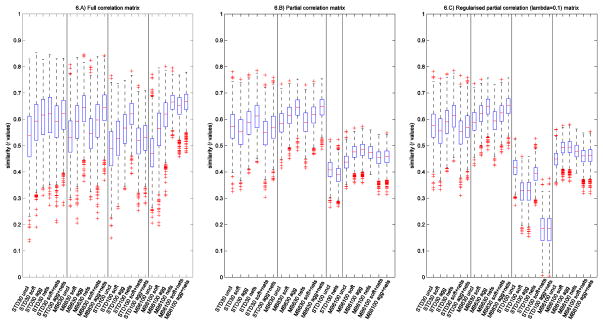
The effect of the different cleaning approaches was evaluated by comparing how similar the network matrices are across subjects. In a homogeneous group of healthy controls, effective cleaning should increase the networks’ consistency (similarity) across subjects (Smith et al., 2005). This analysis was performed by calculating the correlation coefficient between the two network matrices (unwrapped into long vectors for the purpose of correlating the networks against each other) for each pair of subjects, giving a global index of network similarity across subjects; see Fig. 6. These values were then compared across acquisitions with two-tailed paired t-tests, and the results are shown in supplemental Tab. S3 and Tab. S4.
Fig. 6.
Networks’ similarity across subjects. The boxplots show the correlation coefficients (full correlation, partial correlation, and regularised ICOV) between network matrices (unwrapped into a vector of network matrix edges) for all pairs of subjects, with different cleaning steps and for different protocols. ICOV= L1-regularised partial correlation; STD=Standard sequence; uncl=uncleaned; soft=FIX soft cleaning; agg=FIX aggressive cleaning; nets=Nets cleaning.
It can be observed that FIX cleanup significantly improved the similarity across subjects in almost all cases. Aggressive cleaning generally led to higher similarity than soft, when considering full correlation. As full correlation is more influenced by the presence of any shared signal/noise it therefore benefits more from more aggressive noise removal. This difference is lower for partial correlation and regularised ICOV, particularly for the more detailed network modelling (d=100) (which is probably the set of results likely to be of highest general interest). In this case almost all of the improvement in similarity is already achieved with soft cleaning. When using partial correlation, Nets cleanup caused a significant reduction in timeseries amplitude, but in most of the evaluations, had very little effect on network matrix similarity between subjects. Nets cleanup increases the similarity with full correlation at d = 100, but (for Standard) not in combination with 1st level cleaning. However, it must be taken into account that the results shown in this study were obtained removing the contribution of the templates’ bad components, that were obtained with aggressively FIX-cleaned training data (as they were used as the reference of “true signal” and “true noise”). In this case the good and bad group components were clearly identifiable in the templates. However, if FIX cleanup is not performed, the group ICA results themselves would have been less clean, and this would affect the results of Nets cleanup, making this approach possibly less effective than in this study.
Comparing the two sequences (d=30), with MB6 we obtained higher similarity than with Standard, especially with partial correlation and regularised ICOV. However, when moving to higher parcellation (d=100) the superiority of MB6 is evident on full correlation results and, even if the regularisation allows the estimation of the network matrix, the degrees of freedom are not sufficient to achieve successful cleanup and (regularised or unregularised) partial correlation network modelling with the Standard data.
Finally, as the cleaning procedure should enhance not only the consistency across subjects but also the discriminability regarding classifications of interest, we tested if it was possible to predict the subjects’ age from the network matrices using multiple regression. We used the (d × d) correlation values between timeseries for each subjects as design matrix columns (features), pre-selecting the strongest features (inside the leave-one-out loop, the features were ranked according their correlation with age and the 25% of all correlations having the highest age-correlation values were used to predict the age of the subject excluded from the loop), and performed leave-one-out and permutation testing using FSLNets. As shown in Table 1, the predicted age was significantly correlated (pcorr<0.05) with the actual age only with MB6 and soft cleanup on high dimensional (d=100) partial correlation and regularised ICOV matrices.
Table 1.
Prediction of subjects’ age with multiple regression.
| Protocol | correlation matrix | p value
|
|||||
|---|---|---|---|---|---|---|---|
| uncleaned | FIXsoft | FIXagg | Nets | FIXsoft+Nets | FIXagg+Nets | ||
| MB6 d=100 |
full | 0.180 | 0.953 | 0.974 | 0.114 | 0.833 | 0.835 |
| partial | 0.739 | 0.048 | 0.076 | 0.111 | 0.342 | 0.272 | |
| ICOV 0.1 | 0.767 | 0.046 | 0.052 | 0.128 | 0.362 | 0.258 | |
|
| |||||||
| Standard d=100 |
full | 0.091 | 0.163 | 0.295 | 0.108 | 0.398 | 0.616 |
| partial | 0.928 | - | - | 0.115 | - | - | |
| ICOV 0.1 | 0.936 | 0.259 | 0.075 | 0.097 | 0.273 | 0.300 | |
|
| |||||||
| MB6 d=30 |
full | 0.252 | 0.301 | 0.709 | 0.144 | 0.484 | 0.774 |
| partial | 0.543 | 0.215 | 0.368 | 0.415 | 0.305 | 0.423 | |
| ICOV 0.1 | 0.520 | 0.286 | 0.419 | 0.408 | 0.322 | 0.483 | |
|
| |||||||
| Standard d=30 |
full | 0.660 | 0.342 | 0.515 | 0.298 | 0.340 | 0.408 |
| partial | 0.350 | 0.248 | 0.673 | 0.399 | 0.637 | 0.536 | |
| ICOV 0.1 | 0.357 | 0.328 | 0.708 | 0.412 | 0.660 | 0.521 | |
Significant results (pcorr<0.05) are highlighted in bold.
To summarise the most important/relevant2 results from these tests: networks are more reproducible across subjects when using the multiband acquisition, and in this case, when carrying out more detailed network modelling (higher dimensionality) with partial correlation, soft cleanup and aggressive cleanup gave good (and similar) results.
3.7. Spatial maps analyses
The output of the second stage of dual regression (subject-level RSN spatial maps for the 53 test dataset subjects) was used for spatial map analyses. For mixed effect (ME) cross-subject analysis, group maps were obtained by performing a one-sample t-test on all subjects’ spatial maps, for each component, calculating the corresponding z-statistic map and applying a mixture model correction to ensure comparable null distributions in different tests (Beckmann and Smith, 2004). For fixed effects (FE) analysis the single subject z maps were mixture model corrected, averaged and multiplied by the square root of Nsubjects to obtain valid z-statistics.
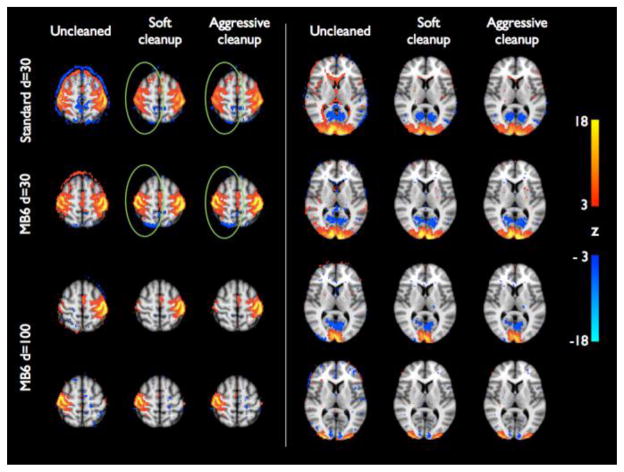
In Fig. 7 we show example group maps derived from the Standard data, 30-dimensional group ICA, MB6 d=30, and MB6 d=100 using the corresponding training-dataset templates for dual regression (dual regression stage 2 could not be run at high dimensionality for the cleaned Standard data, because it did not have enough timepoints). These components (sensory-motor and lateral visual networks) are shown without and with soft or aggressive cleanup. The effects of the cleanup are quite strong in these components and the cortical signal is more focal in the cleaned data. At low dimensionality, these RSNs show similar spatial patterns in Standard and MB6 data, but the signal is stronger with MB6 (especially in the right sensory-motor network). With high dimensionality group ICA decomposition, these RSNs are split into multiple components, allowing a more detailed analysis of network connectivity.
Fig. 7.
Group-level z-statistic maps of two RSNs (sensory-motor network and visual areas), derived from Standard (d=30) and MB6 (d=30 and d=100) datasets using the corresponding training data templates, without and with soft or aggressive FIX cleanup. Individual subjects’ z-statistic maps were mixture model corrected and combined using fixed-effects averaging. Group maps are thresholded at abs(z)>3 (red-yellow colour coding for positive z values, blue-light blue for negative ones). The effect of the cleaning is quite strong in terms of noise removal and more focal signal (as highlighted with the ring around the right sensory-motor network). With high dimensionality the RSNs are split into multiple components, allowing a more detailed analysis of network connectivity.
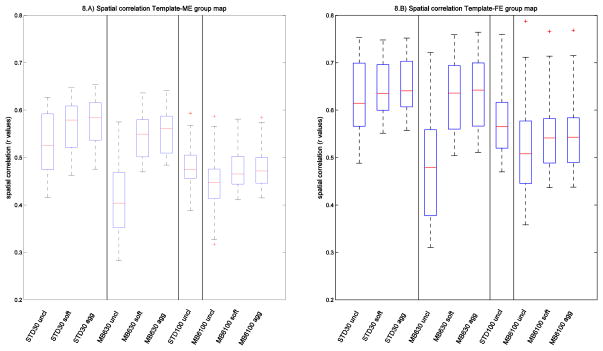
Under the hypothesis that similarity between a group map and the corresponding template map reflects the ability of the cleanup (or acquisition method) to improve correct identification of the true signal, we calculated the spatial correlation between the group maps (ME and FE) and the templates, as a quantitative measure to describe the ability of the cleaning approach and/or the acquisition to correctly detect RSNs. We then compared the correlation values across cleanings with two-tailed paired t-tests and across sequences with two-tailed unpaired t-tests.
The results reported in Figure 8 and supplemental Tables S5 and S6 show that the cleaning significantly increases the similarity between the group maps and the template. Soft and aggressive cleanup are quite similar to each other; although they are statistically significantly different, the difference in mean correlation coefficient is less than 0.01. When comparing the two acquisition methods on uncleaned data, the correlation values obtained with Standard data are higher than MB6. This is mainly due to the lower spatial resolution (and higher SNR) of the standard sequence; Feinberg et al (2010) already demonstrated that slice acceleration (hence TR reduction), while keeping spatial resolution fixed, increases RSN spatial maps’ z-statistics. After cleaning, the difference between MB6 and standard sequence ME maps at d=30 is no longer statistically significant, demonstrating the additional benefit of the combination of acceleration and cleaning, besides the feasibility of high dimensionality ICA analyses.
Fig. 8.
Spatial correlations. The boxplots show the distributions across components of the correlation coefficients between the group maps (obtained with ME and FE statistics) and the corresponding templates, for different cleaning approaches and for different acquisition protocols. STD=Standard sequence; uncl=uncleaned; soft=FIX soft cleaning; agg=FIX aggressive cleaning.
Finally, in order to further quantitate the spatial map results, we calculated five measures of sensitivity and specificity, taking as reference (“true signal”) the template maps derived from the training dataset, to compare the effect of cleaning and the differences between the two sequences (see Supplemental material). As shown in supplemental Table S7, the cleaning procedure allows to better identify the RSNs (higher sensitivity) and their specific activation pattern, while if uncleaned, the data contain false positives that lead to noise outside the RSNs (lower specificity) and overestimated values within RSNs. Regarding the comparison between Standard and MB6 at low dimensionality (d=30), we obtained comparable results between the two sequences (see supplemental Table S8): this further underline the ability of MB6 sequence to provide not only higher spatial and temporal resolution, but also a more truthful detection of the activations, especially within small clusters.
4. Discussion
FMRIB’s ICA-based Xnoiseifier (FIX) is a fully automatic solution (once trained) for cleaning fMRI data of various types of noise. The cleaning procedure with FIX consists of four major operations: spatial ICA, classifier training, component classification (noise detection), and denoising. The first three steps have been extensively described in Salimi-Khorshidi et al., 2014, demonstrating FIX’s ability to classify the independent components into signal or noise with high accuracy. Here we investigated the final stage of the cleaning procedure by carrying out detailed spatial and temporal analysis in order to successfully remove the noise components while preserving as much signal as possible.
In this study we evaluated the efficacy of FIX’s automatic denoising step, testing two different first-level (single subject) cleaning approaches (aggressive and soft) for removing the artefactual components from the fMRI timeseries data, previously identified in the classification step. We compared their effect on both temporal and spatial rfMRI analyses: RSNs’ timeseries (amplitude and power spectra), network matrices (full correlation, partial correlation and L1-norm regularised partial correlation) and spatial maps. In this way we were able to investigate the balance between artefact removal and signal reduction. All these evaluations were performed on two datasets from the same set of 76 subjects: a Standard EPI acquisition and a multiband slice accelerated EPI acquisition (MB6) (Moeller et al. 2010, Feinberg et al., 2010; Setsompop et al., 2012) developed partly for the Human Connectome Project.
Our results showed the efficacy of artefact removal, which proved to be important for reliable temporal and spatial rfMRI analyses. If an artefact is not cleaned at the single subject level and its spatial pattern is overlapping one of the RSNs, it will in general influence the single-subject RSNs’ timeseries, i.e. the output of the first stage of dual regression. Consequently, the non-cleaned timeseries will have higher amplitude with respect to the cleaned data, often with high frequency confounds visible in the power spectra. The presence of shared noise will in general also produce less consistent network matrices across subjects. During the second step of dual regression, the noise contained in the timeseries corrupts the RSN maps. This leads to noisy subject-level z-maps, and affects any following group-level analyses, reducing the ability to detect specific activation patterns within the RSNs (resulting here in lower spatial correlation with the training data templates). This problem does not just affect rfMRI analyses utilising group-ICA followed by dual-regression, but would also affect “back-projection” ICA results and seed-based resting-state correlation maps, for similar reasons.
Comparing the two cleaning approaches (soft vs aggressive), we obtained similar results within the spatial maps both for spatial correlation and sensitivity / specificity measures. Also the network analysis results were generally comparable, especially at high dimensionality. However, regarding mean amplitudes and power spectra analyses, we observed that the reduction in the mean standard deviation (timeseries amplitude) after soft cleaning was caused by a more selective removal of high frequency power, which increases the contrast-to-noise ratio. With aggressive cleaning, the significant reduction in mean amplitude, strongly affects the low frequency peak, causing a significant signal loss. For this reason we would in general suggest the use of the soft cleaning approach, which consists of: 1) removing the full space of the 24 motion parameters (Satterthwaite et al., 2013) from the data and the ICA timeseries; 2) estimating the contribution (spatial regression coefficients) of both good and bad components, in order to identify the unique variance of the artefacts; 3) subtracting the contribution of the bad components from the data (the outer product of their timeseries and spatial regression coefficients, summed over noise components). In this way we achieve a good balance between noise removal and signal loss. The overall level of artefact removal is significant, as shown in Figs 3–5; see also supplemental movie files showing the effect of FIX ICA-based artefact removal on resting-state data acquired with Standard (Movie 1) and MB6 sequences (Movie 2).
We also tested the effect of Nets cleanup, a cleaning option for timeseries and network analyses, applied within-subject to the timeseries output by dual-regression stage 1, with one timeseries for each group-ICA component. This is carried out by regressing the timeseries corresponding to the group-level artefact components out of those corresponding to the non-artefact components (and then discarding the former), the alternative being simply to discard the artefact-related timeseries, and keep the non-artefact timeseries unaltered. However, despite the improvement observed on amplitude and network similarity results, this approach proved to be quite aggressive and to cause a significant amplitude reduction both at low and high frequencies (and not selectively at high frequency as FIX-based cleanup does), as demonstrated with the power spectra analysis. Moreover, if FIX cleanup is not performed, the group ICA used for the Nets cleanup would have contained a greater number of artefact components, possibly making this approach less effective than seen in this study.
The rationale behind partial correlation might be taken to imply that bad timeseries should be included when estimating the partial correlation (so that the increased specificity and interpretability of any node-node “direct” connection can be fully realised); given this logic, one could therefore argue that Nets cleanup should be applied if node timeseries are to be removed before estimation of network matrices (because this will then be equivalent to including the bad node timeseries when estimating the partial correlation).
A detailed analysis of the effects of global signal regression was beyond the scope of this work, as resting-state research is increasingly focusing on network matrices estimated with methods related to partial correlation, and spatial maps derived from multiple temporal regression. Indeed, standard partial correlation analyses cannot be carried out after global signal regression, as the correlation matrix is no longer full rank and it is not possible to invert it. Similarly, the generation of RSN spatial maps via dual regression (i.e., against RSN timeseries) is a multiple regression, so global signal regression is largely irrelevant. However, we believe it was an interesting result that global signal removal has almost no effect on power spectra because it equally affects both low and high frequency, resulting in a non-specific amplitude loss, and on its own provides a poor level of cleanup.
The results discussed so far (the relative merits of different cleanup options) were similar for both the Standard and MB6 data, demonstrating the efficacy of the cleaning procedure on two quite different EPI acquisitions. The only difference in the cleaning approach for the two datasets was the training dataset used for the FIX classification which was tailored to each specific acquisition. The use of a good training dataset is important because it allows FIX to optimize the classification training for the kind of data from a specific study.
Once successfully cleaned, the second aim of this work was to directly compare the data from the Standard and MB6 acquisitions using the same sets of analysis. As multiband EPI has proven to be powerful for obtaining sub-second (or close) whole brain images, reducing the acquisition time and/or increasing spatial resolution (Moeller et al., 2010; Feinberg et al., 2010), we wanted to further investigate the potential of this sequence for identifying the activation patterns of RSNs and detecting their functional connectivity.
The results suggest that the use of multiband EPI is advantageous for rfMRI analysis for several reasons. First, the increased quantity (voxels and/or timepoints) with the slice accelerated acquisition results in a better FIX classification accuracy (98% for MB6 versus 95% for Standard, with leave-one-out testing). We obtained, on average, 8.6 good single-subject ICs for the Standard sequence and 15.2 for MB6. Thus, per subject, on average, 0.43 good components are classified as bad in the Standard and only 0.3 good components are misclassified with MB6 (i.e., one might describe this by saying that on average one subject out of three has a single good component misclassified, and the other two subjects have none). Second, a considerably higher proportion of non-artefactual group-ICA components was identified in the MB6 dataset than with the Standard dataset, suggesting more successful ICA-based cleanup of MB data (even when driven by hand-labelling of the ICA components). Third, the MB accelerated data allowed a more detailed timeseries and network analyses through higher dimensionality decomposition, which was not achievable with the Standard sequence because of its lower temporal degrees of freedom. Fourth, despite comparable mean amplitudes of the timeseries, the spectra after cleaning showed considerably less structured artefact (i.e., deviation from the expected clean 1/f-like spectrum), and networks are more reproducible across subjects with MB6. Finally, the results of spatial map analyses (spatial correlation and sensitivity and specificity measures) were similar between the two sequences, notwithstanding much higher static image SNR in the lower-resolution Standard data. Arguably, a limitation of this study is the lack of a direct comparison between the two sequences at the same resolution, to test the pure effect of acceleration. However, this has already been evaluated by Feinberg et al (2010), showing significant statistical advantages of slice acceleration (with up to a 60% increase in z statistics), with all acquisitions held at 3×3×3mm. Hence, in our study, given the large number of comparisons planned (with different combinations of various cleanup approaches, and interactions of these with the two acquisitions), we wanted to make a targeted comparison, given what is already known from the Feinberg et al. results – a comparison between a typical EPI acquisition, against an optimized alternative using multiband accelerated EPI, where improvements to both spatial and temporal resolution have been balanced against each other.
The fixed acquisition order of the two rfMRI sequences could also represent a possible confound (although both runs follow on from a previous “resting” run, in the sense that the first follows on from the structural scan of reasonable duration). However, it has been demonstrated that rfMRI connectivity measures obtained with ICA and dual regression have moderate to high both short- and long-term test-retest reliability (Zuo et al., 2010). Moreover, in a recent pilot study (Smith et al., 2013) on data acquired in different sessions and in randomised order across subjects, multiband EPI was shown to give higher performances with respect to non-accelerated EPI, in agreement with our findings. For these reasons we believe that our results are not significantly influenced by the acquisition order.
Our results have been informative for analysis developments in the Human Connectome Project: rfMRI are acquired with the same spatial resolution as our MB6 data, and with even greater temporal resolution (MB8, 2×2×2mm, 0.72s; Smith et al., 2013); FIX is now in use as part of the default HCP analysis pipeline, and FIX-cleaned data is the recommended version of the resting-state fMRI data that is publicly available already – over 200 subjects’ worth of hour-long datasets having been released to date.
In this work we used a dataset of healthy (albeit older) controls in order to evaluate the effect of cleaning and acquisition protocols on the identification of RSNs; we also demonstrated that the combination of the accelerated acquisition and the optimized cleaning (FIX soft) enhance not only the consistency across subjects but also the discriminability with respect to a variable of interest (subjects’ age). An interesting future development could be the study of the impact of the cleaning procedure on the between-group discriminability regarding other classifications of interest (controls vs patients, or correlation with behavioural or cognitive indices etc.)(Tian et al., 2013).
In conclusion, we have demonstrated that, by combining an accurate ICA component classifier with an effective approach for noise removal, we are able to remove artefacts automatically and with confidence that we are not removing significant amounts of non-artefact signal. Moreover, with multiband accelerated sequences and effective cleaning, we can perform higher dimensionality decompositions and more detailed RSN analyses than with a standard EPI acquisition.
Supplementary Material
Highlights.
Artefact removal via single-subject ICA and automatic classification is investigated
Different approaches are compared using temporal, network and spatial analysis
Effective cleaning is achieved by removing the unique variance of artefacts
Multiband gives comparable/improved sensitivity and higher spatiotemporal resolution
Accelerated acquisition allows for more detailed network analyses
Acknowledgments
Funding: NIH 1U54MH091657-01 P30-NS057091 P41-RR08079/EB015894, UK MRC G1001354 (Whitehall II MRI Sub-study); The HDH Wills 1965 Charitable Trust (NF, AM). CES and CEM were supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre based at Oxford University Hospitals NHS Trust and University of Oxford. We thank: Charlotte Allan, Abda Mahmood, Amanda Pipkin, Anja Topiwala for recruiting and assessing participants; Radiographers (Caroline Young, Michael Sanders, Jon Campbell) for acquiring scans; Mika Kivimäki, Archana Singh-Manoux for giving access to Whitehall II data, and John R Geddes for applying for funding.
Footnotes
We emphasize here the importance of higher dimensional network modelling using partial correlation because we consider this to be the most interesting/useful general approach for network modelling (compared with the other analyses being tested here).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aalkjaer C, Boedtkjer D, Matchkov V. Vasomotion - what is currently thought? Acta Physiologica (Oxford, England) 2011;202(3):253–269. doi: 10.1111/j.1748-1716.2011.02320.x. [DOI] [PubMed] [Google Scholar]
- Andersson JLR, Jenkinson M, Smith S. Non-linear registration aka spatial normalisation. FMRIB Technial Report TR07JA2 2007 [Google Scholar]
- Andersson JLR, Jenkinson M, Smith S. Non-linear optimisation. FMRIB Technical Report TR07JA1 2007 [Google Scholar]
- Beall EB, Lowe MJ. Isolating physiologic noise sources with independently determined spatial measures. Neuroimage. 2007;37(4):1286–1300. doi: 10.1016/j.neuroimage.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Transactions on Medical Imaging. 2004;23(2):137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences. 2005;360(1457):1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM, Diamond JB, Smith MA, Bandettini PA. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. Neuroimage. 2006;31(4):1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magnetic Resonance in Medicine: Official Journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 1995;34(4):537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Bright MG, Murphy K. Removing motion and physiological artifacts from intrinsic BOLD fluctuations using short echo data. Neuroimage. 2013;64:526–537. doi: 10.1016/j.neuroimage.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DM, Smith SM, Beckmann CF. Advances and pitfalls in the analysis and interpretation of resting-state FMRI data. Frontiers in Systems Neuroscience. 2010;4:8. doi: 10.3389/fnsys.2010.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes D, Haughton V, Carew JD, Arfanakis K, Maravilla K. Hierarchical clustering to measure connectivity in fMRI resting-state data. Magnetic Resonance Imaging. 2002;20(4):305–317. doi: 10.1016/s0730-725x(02)00503-9. [DOI] [PubMed] [Google Scholar]
- De Luca M, Beckmann CF, De Stefano N, Matthews PM, Smith SM. fMRI resting state networks define distinct modes of long-distance interactions in the human brain. Neuroimage. 2006;29(4):1359–1367. doi: 10.1016/j.neuroimage.2005.08.035. [DOI] [PubMed] [Google Scholar]
- De Martino F, Gentile F, Esposito F, Balsi M, Di Salle F, Goebel R, Formisano E. Classification of fMRI independent components using IC-fingerprints and support vector machine classifiers. Neuroimage. 2007;34(1):177–194. doi: 10.1016/j.neuroimage.2006.08.041. [DOI] [PubMed] [Google Scholar]
- de Pasquale F, Della Penna S, Snyder AZ, Lewis C, Mantini D, Marzetti L, Belardinelli P, Ciancetta L, Pizzella V, Romani GL, et al. Temporal dynamics of spontaneous MEG activity in brain networks. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(13):6040–6045. doi: 10.1073/pnas.0913863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins AE, Kiehl KA, Liddle PF. Removal of confounding effects of global signal in functional MRI analyses. Neuroimage. 2001;13(4):751–758. doi: 10.1006/nimg.2000.0719. [DOI] [PubMed] [Google Scholar]
- Feinberg DA, Moeller S, Smith SM, Auerbach E, Ramanna S, Gunther M, Glasser MF, Miller KL, Ugurbil K, Yacoub E. Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PloS One. 2010;5(12):e15710. doi: 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, Matthews PM, Beckmann CF, Mackay CE. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(17):7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Zhang D, Snyder AZ, Raichle ME. The global signal and observed anticorrelated resting state brain networks. Journal of Neurophysiology. 2009;101(6):3270–3283. doi: 10.1152/jn.90777.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics (Oxford, England) 2008;9(3):432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magnetic Resonance in Medicine: Official Journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2000;44(1):162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: A network analysis of the default mode hypothesis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(1):253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve DN, Fischl B. Accurate and robust brain image alignment using boundary-based registration. Neuroimage. 2009;48(1):63–72. doi: 10.1016/j.neuroimage.2009.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. Neuroimage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Kochiyama T, Morita T, Okada T, Yonekura Y, Matsumura M, Sadato N. Removing the effects of task-related motion using independent-component analysis. Neuroimage. 2005;25(3):802–814. doi: 10.1016/j.neuroimage.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Kundu P, Inati SJ, Evans JW, Luh WM, Bandettini PA. Differentiating BOLD and non-BOLD signals in fMRI time series using multi-echo EPI. Neuroimage. 2012;60(3):1759–1770. doi: 10.1016/j.neuroimage.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe MJ, Mock BJ, Sorenson JA. Functional connectivity in single and multislice echoplanar imaging using resting-state fluctuations. Neuroimage. 1998;7(2):119–132. doi: 10.1006/nimg.1997.0315. [DOI] [PubMed] [Google Scholar]
- Marrelec G, Krainik A, Duffau H, Pelegrini-Issac M, Lehericy S, Doyon J, Benali H. Partial correlation for functional brain interactivity investigation in functional MRI. Neuroimage. 2006;32(1):228–237. doi: 10.1016/j.neuroimage.2005.12.057. [DOI] [PubMed] [Google Scholar]
- McKeown MJ, Makeig S, Brown GG, Jung TP, Kindermann SS, Bell AJ, Sejnowski TJ. Analysis of fMRI data by blind separation into independent spatial components. Human Brain Mapping. 1998;6(3):160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Ugurbil K. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magnetic Resonance in Medicine: Official Journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2010;63(5):1144–1153. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Bandettini PA. Resting-state fMRI confounds and cleanup. Neuroimage. 2013;80:349–359. doi: 10.1016/j.neuroimage.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. The impact of global signal regression on resting state correlations: Are anti-correlated networks introduced? Neuroimage. 2009;44(3):893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niazy RK, Xie J, Miller K, Beckmann CF, Smith SM. Spectral characteristics of resting state networks. Progress in Brain Research. 2011;193:259–276. doi: 10.1016/B978-0-444-53839-0.00017-X. [DOI] [PubMed] [Google Scholar]
- Perlbarg V, Bellec P, Anton JL, Pelegrini-Issac M, Doyon J, Benali H. CORSICA: Correction of structured noise in fMRI by automatic identification of ICA components. Magnetic Resonance Imaging. 2007;25(1):35–46. doi: 10.1016/j.mri.2006.09.042. [DOI] [PubMed] [Google Scholar]
- Popa D, Popescu AT, Pare D. Contrasting activity profile of two distributed cortical networks as a function of attentional demands. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2009;29(4):1191–1201. doi: 10.1523/JNEUROSCI.4867-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59(3):2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytty R, Nikkinen J, Paavola L, Abou Elseoud A, Moilanen V, Visuri A, Tervonen O, Renton AE, Traynor BJ, Kiviniemi V, et al. GroupICA dual regression analysis of resting state networks in a behavioral variant of frontotemporal dementia. Frontiers in Human Neuroscience. 2013;7:461. doi: 10.3389/fnhum.2013.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad ZS, Gotts SJ, Murphy K, Chen G, Jo HJ, Martin A, Cox RW. Trouble at rest: How correlation patterns and group differences become distorted after global signal regression. Brain Connectivity. 2012;2(1):25–32. doi: 10.1089/brain.2012.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi-Khorshidi G, Douaud G, Beckmann CF, Glasser M, Griffanti L, Smith SM. Automatic denoising of functional MRI data: Combining independent component analysis and hierarchical fusion of classifiers. Neuroimage. 2014;90:449–468. doi: 10.1016/j.neuroimage.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Wolf DH, Loughead J, Ruparel K, Elliott MA, Hakonarson H, Gur RC, Gur RE. Impact of in-scanner head motion on multiple measures of functional connectivity: Relevance for studies of neurodevelopment in youth. Neuroimage. 2012;60(1):623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Elliott MA, Gerraty RT, Ruparel K, Loughead J, Calkins ME, Eickhoff SB, Hakonarson H, Gur RC, Gur RE, et al. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage. 2013;64:240–256. doi: 10.1016/j.neuroimage.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsompop K, Gagoski BA, Polimeni JR, Witzel T, Wedeen VJ, Wald LL. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magnetic Resonance in Medicine: Official Journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2012;67(5):1210–1224. doi: 10.1002/mrm.23097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmueli K, van Gelderen P, de Zwart JA, Horovitz SG, Fukunaga M, Jansma JM, Duyn JH. Low-frequency fluctuations in the cardiac rate as a source of variance in the resting-state fMRI BOLD signal. Neuroimage. 2007;38(2):306–320. doi: 10.1016/j.neuroimage.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM. The future of FMRI connectivity. Neuroimage. 2012;62(2):1257–1266. doi: 10.1016/j.neuroimage.2012.01.022. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Human Brain Mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, Ramsey JD, Woolrich MW. Network modelling methods for FMRI. Neuroimage. 2011;54(2):875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- Smith SM, Beckmann CF, Ramnani N, Woolrich MW, Bannister PR, Jenkinson M, Matthews PM, McGonigle DJ. Variability in fMRI: A re-examination of inter-session differences. Human Brain Mapping. 2005;24(3):248–257. doi: 10.1002/hbm.20080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–19. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Smith SM, Beckmann CF, Andersson J, Auerbach EJ, Bijsterbosch J, Douaud G, Duff E, Feinberg DA, Griffanti L, Harms MP, et al. Resting-state fMRI in the human connectome project. Neuroimage. 2013;80:144–168. doi: 10.1016/j.neuroimage.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CG, Harshman RA, Menon RS. Noise Reduction in BOLD-Based fMRI Using Component Analysis. Neuroimage. 2002;17:1521–1537. doi: 10.1006/nimg.2002.1200. [DOI] [PubMed] [Google Scholar]
- Tian L, Kong Y, Ren J, Varoquaux G, Zang Y, Smith SM. Spatial vs. temporal features in ICA of resting-state fMRI - A quantitative and qualitative investigation in the context of response inhibition. PloS One. 2013;8(6):e66572. doi: 10.1371/journal.pone.0066572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohka J, Foerde K, Aron AR, Tom SM, Toga AW, Poldrack RA. Automatic independent component labeling for artifact removal in fMRI. Neuroimage. 2008;39(3):1227–1245. doi: 10.1016/j.neuroimage.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafyllou C, Hoge RD, Krueger G, Wiggins CJ, Potthast A, Wiggins GC, Wald LL. Comparison of physiological noise at 1.5 T, 3 T and 7 T and optimization of fMRI acquisition parameters. Neuroimage. 2005;26(1):243–250. doi: 10.1016/j.neuroimage.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Van Dijk KR, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012;59(1):431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenbacher A, Kasess C, Gerstl F, Lanzenberger R, Moser E, Windischberger C. Correlations and anticorrelations in resting-state functional connectivity MRI: A quantitative comparison of preprocessing strategies. Neuroimage. 2009;47(4):1408–1416. doi: 10.1016/j.neuroimage.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Xu J, Moeller S, Auerbach EJ, Strupp J, Smith SM, Feinberg DA, Yacoub E, Ugurbil K. Evaluation of slice accelerations using multiband echo planar imaging at 3T. Neuroimage. 2013;83:991–1001. doi: 10.1016/j.neuroimage.2013.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Kelly C, Adelstein JS, Klein DF, Castellanos FX, Milham MP. Reliable intrinsic connectivity networks: test-retest evaluation using ICA and dual regression approach. Neuroimage. 2010;49(3):2163–2177. doi: 10.1016/j.neuroimage.2009.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.