Abstract
During somatic hypermutation (SHM), activation-induced deaminase (AID) mutates deoxycytidine on single-stranded DNA (ssDNA) generated by the transcription machinery, but the detailed mechanism remains unclear. Here we report a higher abundance of RNA polymerase II (Pol II) at the immunoglobulin heavy chain variable (Igh-V) region compared to the constant region and partially transcribed Igh RNAs, suggesting a slower Pol II progression at Igh-V that could result in some early/premature transcription termination after prolonged pausing/stalling of Pol II. Knocking down RNA exosome complexes, which could decrease premature transcription termination, leads to decreased SHM. Knocking down Spt5, which can augment premature transcription termination, leads to increases in both SHM and the abundance of ssDNA substrates. Collectively, our data support the model that, following the reduction of Pol II progression (pausing or stalling) at the Igh-V, additional steps such as premature transcription termination are involved in providing ssDNA substrates for AID during SHM.
Introduction
While antibodies are responsible for the humoral immune response, those antibodies encoded by germ-line sequences often do not have sufficient affinity or specificity to provide full protection against diverse pathogens. To deal with this, B cells have developed mechanisms to somatically hypermutate (SHM) the V regions of their germ-line encoded heavy and light chain antibody genes. Most mutations are accumulated in the complementarity-determining regions (CDRs) of antibodies to refine their antigen contacting surfaces. B cells also diversify antibody function by carrying out class-switch recombination (CSR), which allows different antigen binding sites to be expressed with different constant regions. Together, SHM and CSR provide us with antibodies that can interact with all antigens, are distributed throughout the body and mediate effective humoral immune protection1.
Both SHM and CSR require the endogenous mutagenic enzyme AID2, 3. As a result of intensive study over the last 13 years, it is now known that AID: 1) deaminates dC to dU and creates point mutations, abasic sites and G:U mismatches4; 2) recruits error prone DNA repair processes that further contribute to the diversification of mutation spectra by introducing mutations at A/T sites5; 3) requires transcription of the targeted genomic region during both SHM and CSR6; and 4) utilizes ssDNA as its biochemical substrate7. The fact that AID binds and mutates only ssDNA substrates is puzzling because ssDNA is a transient and short-lived form of DNA that is mostly covered by protein complexes during transcription or replication. It is thus unclear exactly how ssDNA is generated and made accessible for AID during SHM or CSR and whether the two different processes share the same mechanism(s). When carrying out SHM and CSR in B cells, AID also occasionally introduces mutations at genomic loci other than antibody genes and these “off-target” mutations are responsible for many B cell malignancies8.
Since transcription is necessary for SHM and CSR, AID mediated mutagenesis must be coordinated with transcriptional machinery9. In fact, Pol II10, Spt511 and RNA polymerase associate factor (PAF1)12 have all been reported to interact with AID directly and to affect CSR. However, even though transcription is always accompanied by ssDNA around the synthesis center of the Pol II complex—the structure referred as a transcription bubble, the conventional transcription cycle of initiation, elongation and termination is unlikely to supply ssDNA substrates for AID for the following reasons. First, recent structural studies suggest that ssDNA created in the transcription bubble is almost fully covered by either the Pol II complex itself or co-factors like the DRB sensitivity-inducing factor (DSIF, composed of Spt4 and Spt5 proteins)13. Such coverage is probably present throughout elongation and even during transcription termination triggered by poly-A signals14. Second, whereas transcription initiation involves TFIIH mediated dsDNA melting at promoter regions before a complete Pol II complex is assembled15, AID may be excluded from those regions because a high density of transcription factors normally cover promoters. This is consistent with the fact that AID preferentially targets the Igh-V coding region but spares regions that are within ~200bp immediately after the transcription start site (TSS)4, 16, 17.
Nevertheless, several processes accompanying or resulting from transcription have been proposed to help expose ssDNA for AID targeting. These processes include: 1) R-loops resulting from special features in the DNA sequence at Ig-h switch regions but not Igh-V regions18, 19; 2) negative DNA supercoils occurring at the trailing end of transcription bubbles20; and 3) paused Pol II complexes that are frequently present proximal to the TSS regions21. It is noteworthy that pausing only represents one of the three possible states of stalled Pol II complexes and the other two scenarios include backtracking Pol II due to transcription errors and early terminating Pol II due to the failure of error correction22. While DNA supercoils generate AID accessible regions on dsDNA plasmids in vitro23 and paused Pol II complexes play essential roles in CSR11, 24, 25, the source(s) of ssDNA substrates for AID during SHM of the V region in vivo remains to be established.
Premature transcription termination refers to the process when Pol II is released in the middle of a transcribing gene independent of a poly-A termination signal. It is a plausible way of exposing ssDNA around the Pol II catalytic center for two reasons. First, during premature transcriptional termination, the release of Pol II complex from template DNA may not be coordinated with the transcriptional termination complexes, which could leave the melted dsDNA unprotected. Second, partial transcripts resulting from premature transcription termination negatively regulate Pol II progression specifically around the termination site26, which could in turn maintain ssDNA levels at V regions. While detailed mechanisms remain unclear, it is well established that DSIF complex component Spt5 is essential for Pol II processivity; and the loss of Spt5 leads to an increase in premature transcription termination in yeast and in mammalian cells27, 28, 29. On the other hand, a sustained level of existing premature transcription termination requires the RNA exosome through a newly identified feedback mechanism26. Based on these considerations we tested the role of premature transcription termination in SHM by manipulating the levels of Spt5 and the RNA exosome in the mutating human B cell line, Ramos. Our data suggest that, in addition to Pol II pausing, there is a second set of events like the premature transcription termination process which B cells may indeed “hijack” to supply ssDNA substrates for AID to mutate at immunoglobulin V regions during SHM.
Results
Reduced progression of Pol II at Igh-V region in Ramos cells
Since Pol II abundance at specific regions of a transcribing gene is widely used to assess local progression efficiency, we first examined the distribution of Pol II complexes at the actively transcribing Igh-V locus in Ramos cells. The human Burkitt’s lymphoma derived Ramos cell line has many characteristics of a germinal center B cell including the constitutive capacity to undergo SHM and a pattern of AID induced mutations at G:C sites that is similar to what has been observed in vivo30. However, analysis of Pol II abundance in wild type Ramos cells is challenging because the size of the hypermutating V region (~400 bp) is at the resolution limit of the chromatin immunoprecipitation (ChIP) analysis. To circumvent this problem, we expanded the AID targeting region in Ramos by introducing a mCherry fluorescent protein coding sequence into the 2nd exon of the Igh-V locus. Using recombinase-mediated cassette exchange (RMCE)31, we replaced the endogenous Ramos V region with an in-frame fusion of the mCherry gene and the endogenous V region (Fig 1a) to provide a ~1.2kb target for AID as well as a reporter for AID induced mutations. We confirmed that the entire fusion V exon was able to be targeted by AID mediated SHM (see below). Using the ChIP assay, we detected on the average ~2–3 times more Pol II at the 2nd exon of Igh-V—the 1.2kb hypermutating mCherry/Igh-V fusion region—than at other areas of the gene, i.e. the promoter, the downstream intronic enhancer Eμ and the constant region (Fig 1b). The relatively higher abundance of Pol II was quite consistent amongst the different parts of the mCherry/Igh-V fusion (c-f) compared to the relatively lower abundance within the downstream Eμ-Cμ region (h-k) (Fig 1c). This significant but moderate increase in Pol II occupancy in the mCherry/Igh-V region suggested a slowing-down of Pol II progression. We also conducted ChIP analysis for Pol II phosphorylated at serine 5 of the C-terminal repeat domain (Pol IIS5P), a form of Pol II that is enriched during transcription initiation and gradually decreases throughout elongation. We found that Pol IIS5P occupancy stayed high from the promoter region until ~2.5kb downstream of TSS (Fig 1d). This suggested that Pol II started to transit from initiation phase to elongation phase immediately before Eμ.
Figure 1. Abundance of Pol II complexes at Igh-V region in Ramos cells.
a) The structure of the Igh-V region in the reporter Ramos cell line is depicted with grey bars indicating the positions of each primer set used in the following figures and rectangular boxes representing coding exons; b) Chromatin immunoprecipitation (ChIP) was conducted on reporter cells expressing the AID-ER fusion protein but without 4-OHT treatment using anti-total Pol II, quantified by real-time PCR and normalized to the level at Cμ region after subtraction of IgG background. Average total-Pol II occupancy at Igh-V region and downstream regions were compared using student T test; c) The abundance of Pol II at each region shown in a; d) ChIP was conducted on reporter cells using anti-Pol II S5P was quantified by real-time PCR. Data were then normalized to signals at the Cμ region after subtraction of IgG background. Data represent the average of two independent experiments and error bars represent SD.
Spt5 knockdown affects Pol II occupancy and ssDNA patches
Reduced progression of Pol II complexes suggested the existence of stalled Pol II at Igh-V regions. Stalled Pol II complexes could represent a paused Pol II, a backtracking Pol II or an early terminating Pol II22. To address whether the stalled Pol II contributes to SHM and if so, which of these three processes is responsible, we chose to manipulate the level of the DSIF complex component Spt5. This is because: 1) Spt5 mediates proximal TSS pausing of the RNA polymerase32; and 2) it positively maintains Pol II processivity during elongation27, 28, 29. Therefore, a decrease in Spt5 level is expected to reduce Pol II pausing but facilitate premature Pol II termination.
Of five shRNA constructs against human Spt5 (SUPT5H) tested in Ramos cells, two (# 4 and 6) achieved ~50% knockdown at both mRNA and protein levels (Fig 2a, b, supplementary Fig. 1). Spt5 knockdown (Spt5KD) cells and cells transduced with a control short hairpin RNA (Ctrl-shRNA) expressed similar amounts of steady-state IgH and AID mRNA (Fig 2c). We also found that, as expected, Spt5 abundance at the IgH region was consistently decreased in Spt5KD cells and, to a more variable extent, the total Pol II complex abundance decreased accordingly in two independent experiments (Fig 2d).
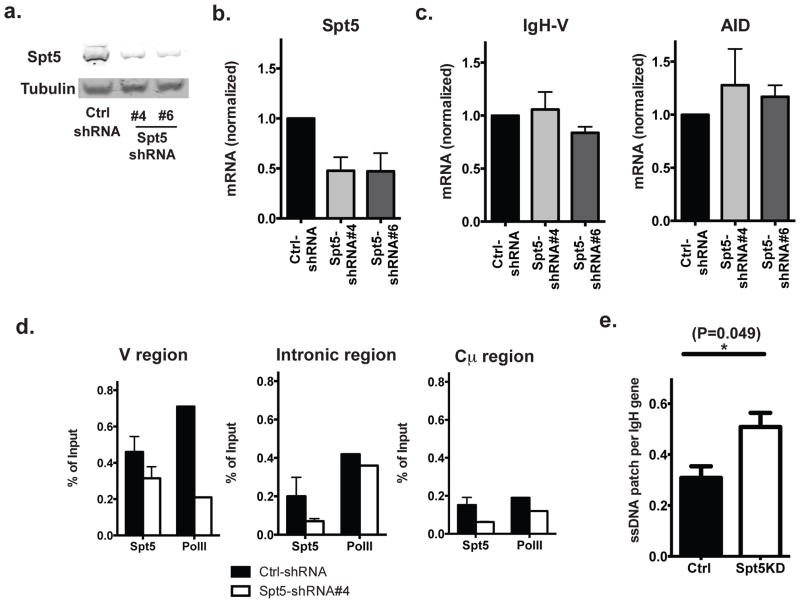
Figure 2. Decrease in the cellular Spt5 level is associated with more ssDNA patches.
a) Western blot; and b) quantitative PCR analysis of Spt5 protein and mRNA level (normalized by cyclophilin B expression) in cells transduced with indicated shRNA(s); c) Quantitative PCR analysis of mRNA levels of IgH and AID in cells either transduced with control shRNA constructs or indicated shRNA against Spt5; d) ChIP was conducted for Spt 5 and Pol II on cells either transduced with control (Ctrl) shRNA or shRNA #4 against Spt5 and ChIP products were quantified by quantitative PCR at the indicated region of Igh gene. e) Abundance of exposed ssDNA patches was quantified using native bisulfite conversion method (see Methods) followed by DNA sequencing. Data represent 4 independent experiments for part a and 2 independent experiments for part d (the error bars in this case are the variation in the duplicate PCRs in that experiment wherever applicable). Part b and c represent the average of at least 3 independent experiments. Part e represents a compiled analysis from two independent experiments using t-test and error bars represent the SD.
Transient patches of ssDNA in crosslinked chromatin can be detected by treating cross-linked nuclei with bisulfite reagents under non-denaturing conditions33, 34. In this assay, the bisulfite treatment only converts dCs on exposed ssDNA to dUs but cannot convert dCs that are either hybridized with other nucleic acids (dsDNA or RNA:DNA hybrids) or covered by proteins. Using this bisulfite method, we found significantly more non-protected ssDNA patches at the Igh-V region in Spt5KD cells than in Ctrl-shRNA transduced cells (Fig 2e), suggesting that the decrease in cellular Spt5 levels was associated with more potential AID substrates.
Spt5 knock down is associated with an increase in SHM
We first estimated the change of SHM rate upon Spt5 reduction in an IgM− Ramos sub-clone that expressed endogenous levels of AID and had a nonsense mutation in the native V region. We used the IgM gene reversion assay established previously35 (see Methods) and found that the ~50% reduction in the Spt5 level was accompanied by a significant increase in the SHM rate (5.46x10−5 vs. 7.52x10−5 per nucleotide per generation (p<0.05)) (Fig 3a).
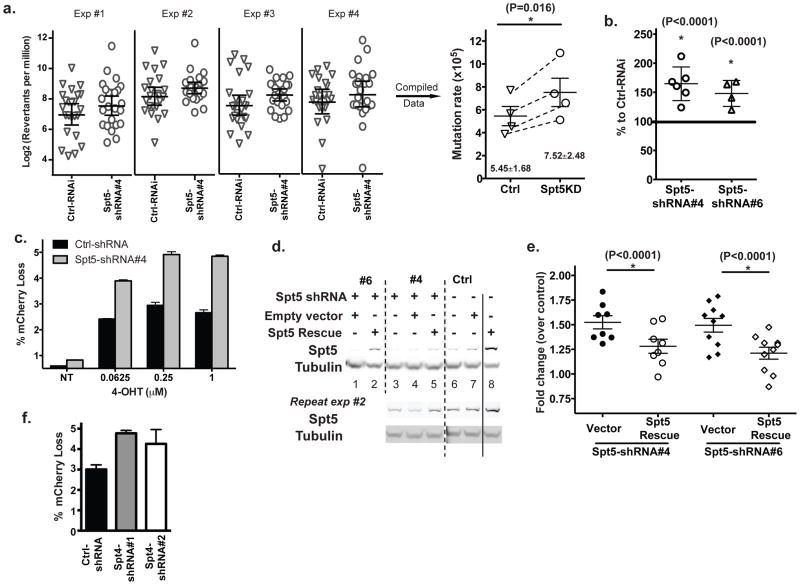
Figure 3. Decrease in cellular Spt5 leads to an increase in SHM.
a) Mutation rates of wild type Ramos cells transduced with either control shRNA or shRNA #4 against Spt5 were estimated using the reversion assay. In each of 4 independent experiments, reversion of 24 clones transduced with either the Ctrl or the #4 Spt5 shRNA was compared (shown in the left panel) and rates of mutation calculated from each experiment were represented by corresponding symbols in the right panel. Statistical analysis was conducted on data compiled from the 4 independent experiments; b) Mutation frequency assessed by the reporter cell line as percentage of cells that had lost fluorescence upon 4-OHT induction. Data was normalized to control shRNA-transduced cells; c) Indicated amounts of 4-OHT were used to induce AID mediated mutation as assessed by the loss of fluorescence (NT refers as cells without 4-OHT induction). Data were a representative result of two independent experiments with each condition done in triplicate; d) An exogenous Spt5 coding construct with partial resistance to shRNA (7 out of 21 nucleotides mutated with no change in amino acid sequence) was introduced into cells followed by the transduction of indicated shRNA against Spt5. Cells with the exogenous Spt5 rescue construct showed substantial resistance to shRNA mediated knockdown of Spt5 (lane 2, 5 vs lane 1, 3). Western blots from two independent experiments are shown. Dash and solid lines were used to help distinguish different experimental groups; e) Restoring the Spt5 level in Spt5KD cells reduced the SHM frequency; f) Augmented SHM in cells transduced with shRNA against the Spt4 gene. Compiled data are shown in a, b, e, and f with each symbol representing an independent set of experiments. Paired t-test (panel a & e) or z-test (panel b) was used for the statistical analysis and error bars represent the SD. Representative data of at least two independent experiments were shown in c, d and g.
We then used an independent Ramos cell line (“reporter line” described in Fig 1a and the Methods) that carries the mCherry/VH4-34 fusion at Igh-V locus and expresses AID-ER fusion protein) to confirm the effect of knocking-down Spt5 on SHM. With this reporter line, the mutation rate can then be quantitatively assessed based on the percentage of cells that lose their fluorescence upon 4-hydroxy-tamoxifen (4-OHT) induction of the nuclear localization of AID36. Consistent with the reversion assay, ~50%–60% more cells lost their fluorescence under the Spt5KD condition than the control cells after 7 days of induction (Fig 3b). While the SHM level reached a plateau with the induction concentration of ~0.25 μM 4-OHT in both Spt5KD and control cells, Spt5KD cells mutated the Igh-V region more efficiently at a lower concentration of inducer (0.0625 μM) than control cells did at the maximum induction level (Fig 3c). These data indicated that the increased level of SHM in Spt5KD cells was not due to the excessive level of active AID molecules in nucleus. Because of the significant increase in experimental efficiency using the reporter line instead of the reversion assay in analyzing the efficiency of SHM, we chose to perform all further mutation analyses on the reporter platform.
We next used an shRNA resistant form (see Methods) of Spt5 to rescue the effect phenotypically. The exogenous Spt5 rescue construct restored Spt5 levels in the knockdown cells close to normal levels (lanes 2 & 5 vs. 6 in Fig 3d, supplementary Fig. 2). Consistent with the regulatory role of Spt5 on SHM, Spt5KD cells (with either the #4 or #6 shRNA construct) significantly reduced their mutation frequencies (P<0.0001) when cellular Spt5 levels were largely restored (Fig 3e). These data confirmed that the reduced level of Spt5 was the cause of the observed increase in SHM in the Spt5-specific shRNA transduced cells.
Since the DSIF complex is composed of Spt5 and Spt4 molecules, we also tested whether reducing the Spt4 level in cells would have an impact on SHM similar to Spt5KD. Indeed, a similar increase in SHM was observed when endogenous Spt4 level was reduced in the reporter Ramos cells (Fig 3f). Nevertheless, the experiment needs to be interpreted with caution because Spt4 is also a stabilizing factor for Spt537 and the effect of Spt4KD can at least partially be caused by the reduction in the level of general DSIF complexes.
Impact of Spt5 knockdown on the characteristics of SHM
In the previous section, we used both the reversion of a nonsense mutation in the native Ramos V region and the loss of fluorescence of a reporter in the endogenous Igh-V locus to show that the knock down of Spt5 resulted in a 50%–60% increase in V region mutation. To provide an independent measure of the frequency of mutation per mutated Igh-V gene and to examine the characteristics of the additional mutations, we went on to examine the sequences of the Igh-V fusion genes in mutated Ramos reporter cells by Sanger sequencing. Since in Ramos cells only a small % of the V regions undergo mutation, we sequenced only those cells that mutated their Igh-V gene sufficiently to cause a loss of fluorescence. Most of the mutations in both the control and the knockdown cells were in AID hot spots. Reduction of Spt5 did not cause a statistically significant change in the distribution of mutations in either the mCherry or the 4–34 part of the fusion V region (Fig 4a). However, the average number of mutations harbored by each mutated Igh-V gene was consistently (but not statistically significantly) increased by about 25% based on 3 independent experiments (Fig 4b). This provides independent evidence that a decrease in Spt5 leads to not only more V regions being targeted for mutation but also more mutations accumulating in a particular mutated Igh-V gene. While these are two different ways of quantifying mutation, the overall impact was estimated by multiplying the ~1.6 fold increase in frequency (based on the fluorescence reporter assay) by the 1.25 increase in mutations per V region, which resulted in a ~2 fold overall increase of AID induced mutations due to the knock down of Spt5. The magnitude of this increase is similar to what can be achieved by artificially introducing a termination signal at the immunoglobulin light chain V region in chicken DT40 cells38. Spt5 levels did not influence the strand distribution of mutations (mutation preference on the template or the non- template strand) because the incidence of mutations from C or from G was similar (~50%) in both Spt5KD and Ctrl-shRNA transduced cells (Fig 4c). Interestingly, the frequency of G:C transversions in Spt5KD cells was consistently and significantly higher than control cells (Fig 4d), suggesting that Spt5 might suppress base excision repair (BER) or the lack of Spt5 might expose docking sites that facilitate the recruitment of BER machinery. Nevertheless, a difference of UNG recruitment is not likely to be the main reason for our observed increase in SHM upon Spt5 reduction because: 1) UNG mediates only one of the two pathways in the second phase of SHM and accounts for both transversion mutations and error free repair4; 2) UNG promotes both SHM and CSR while reduction of Spt5 leads to opposite effects on SHM and CSR (see discussion); and 3) changes in the availability of UNG does not influence overall mutation frequency39. Hence, the increase of the overall SHM rate most likely resulted from the initial increase of the frequency of deamination. Overall, these data suggest that reduction in the Spt5 level promotes SHM in a general manner without any spatial preference.
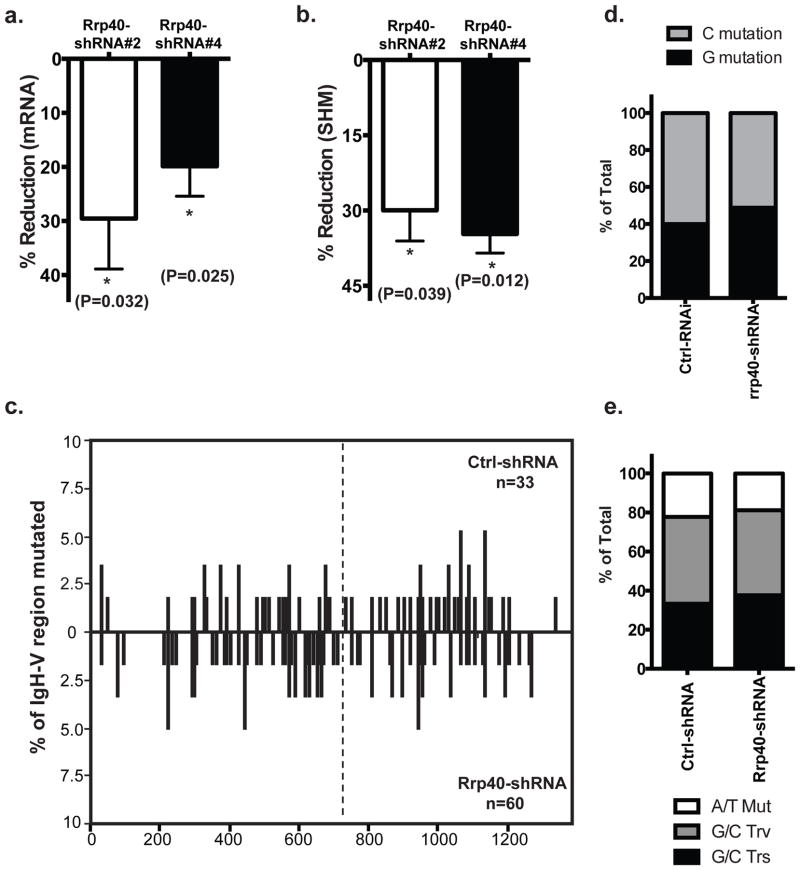
Figure 4. Impact of Spt5KD on the characteristics of SHM.
a) Distribution of mutations in the fusion V region (see Fig 1) from cells either transduced with Ctrl-shRNA or shRNA against Spt5 were analyzed by SHMTool (http://scb.aecom.yu.edu/cgi-bin/p1). The illustrated distribution is calculated from one representative sequencing experiment. The vertical dashed line at 720 base pairs marks the border between the mCherry and endogenous 4–34 V regions; b) Mutations per Igh-V gene, c) G or C mutation frequency and d) mutation types were summarized in indicated cell types from two independent sequencing experiments. In total, 45 Igh-V sequences were sequenced for Ctrl-shRNA transduced cells and 54 for Spt5KD cells. 2 by 2 contingency table analysis were used for statistical analysis in b, c and d.
RNA exosome and premature transcription termination in SHM
The finding that a decrease in Spt5 levels is accompanied by an increase in the rate of SHM suggested that premature transcription termination might be associated with and could contribute to SHM. To explore this possibility further, we knocked down the RNA exosome complex because: 1) the RNA exosome helps to induce premature termination of transcription26; and 2) the exosome core components facilitate SHM in a biochemical assay40 in vitro, though this has not been established in vivo. The RNA exosome complex is composed of two ribonuclease units and a nine sub-unit core including Rrp40 (EXOSC3)41, 42, a molecule that has a profound impact in AID mediated mutation activity in vitro40. Therefore, we tested the role of Rrp40 in SHM using Ramos cells.
The maximum reduction we achieved from five shRNAs against the exosome core component Rrp40 was ~30% at mRNA level (Fig 5a). This is probably because an extensive decrease in its level is cytotoxic considering the essential role of RNA exosome for ribosomal RNA processing43. However, based on data from two independent shRNA constructs, this small decrease of Rrp40 level was sufficient to reduce the frequency of SHM by ~30–40% (P<0.01) (Fig 5b). The Rrp40 knockdown did not significantly change the overall distribution of mutations in either the mCherry or the endogenous V region (Fig 5c). In addition, there was no increase in the numbers of mutations on the non-template strand compared to the template strand (Fig 5d), nor did we find any change in mutation pattern resulting from the reduced Rrp40 level (Fig 5e). This indicated that the RNA exosome complexes could promote SHM in a general way rather than solely through the suggested nascent RNA degradation mechanism that would help to preferentially expose template ssDNA40. Our data, however, cannot rule out the possibility that the latter mechanism may also play an important role in SHM because our limited ability to reduce RNA exosome levels might have precluded us from observing such an effect.
Figure 5. RNA exosome and SHM.
Knockdown efficiency of Rrp40 was assessed by quantitative PCR and percentage of reduction was calculated comparing to Ctrl-shRNA transduced cells; b) SHM rate was assessed in Rrp40 knockdown cells and Ctrl-shRNA transduced cells in the Ramos reporter cells and the reduction of SHM level was calculated accordingly. Combined data from three independent experiments were used to detect significant reduction associated with a reduced level of cellular Rrp40; c) Distribution of mutations in cells either transduced with control shRNA or shRNA against Rrp40 were analyzed by SHMTool. Data represent the compiling analysis from two independent sequencing experiments; d) G or C mutation frequency and e) mutation type were summarized in cells under the indicated conditions. A total of 33 Igh-V sequences were sequenced from Ctrl-shRNA transduced cells and 60 from Rrp40KD cells. A compiled analysis of 3 independent experiments was conducted in panels a and b using t-test and error bars represent the SD. 2 by 2 contingency table analysis were used for statistical analysis in d and e.
Premature transcription termination likely occurs at Igh-V
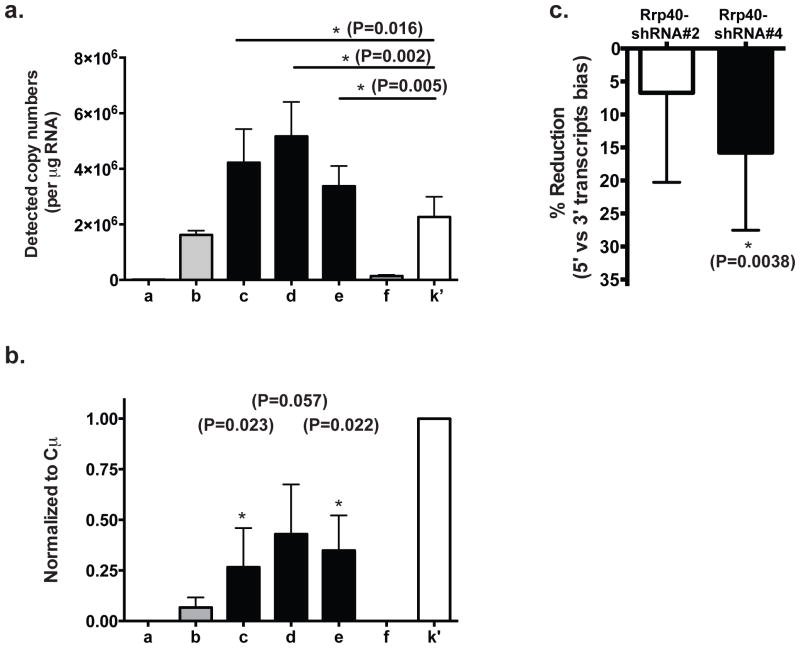
Overall, our experiments confirmed the positive role of RNA exosome core component Rrp40 in optimal SHM. Together with the observed increase in SHM upon Spt5 reduction, they provided strong support for a positive role of premature transcription termination in SHM. We therefore tested whether premature transcription termination events happened on the Igh gene. Early transcription termination events should result in a higher abundance of transcripts containing only the 5′-end of the gene compared to transcripts containing both 5′- and 3′-ends. Such an imbalance could serve as a surrogate indicator for premature transcription termination and could be estimated by quantifying the absolute copy number of RNA transcripts containing various portions of the gene in total RNA. To do this, we constructed a plasmid harboring a single copy of each region to be investigated. Using this plasmid and real-time quantitative PCR, we generated individual standard curves of the copy numbers (estimated by the molecular weight of the plasmid) against the Ct (cycle threshold) values for each region of interest. These standard curves were then used to determine the absolute quantity of RNA transcripts containing the designated region in total RNA. This quantification method worked effectively, as exemplified by its ability to distinguish the intron region f because of its low abundance (~2–4%) compared to the mature full length Igh mRNA (Fig 6a). In support of our hypothesis, we observed ~2 fold more RNA transcripts containing the Igh variable region (Igh-V fusion) than those containing the constant region (Cμ) in Ramos cells (Fig 6a) when cDNAs were synthesized using random hexamers. Partial Igh transcripts containing only the 5′ end of the Igh gene were not polyadenylated (Fig 6b) since this bias was not observed when cDNAs were synthesized using poly-T oligonucleotides (the lower signals detected at the 5′ end of the mRNA compared to the constant region were likely due to the limitation in the enzymatic processivity of the reverse transcriptases). Consistent with the important role of RNA exosome in premature transcription termination, the reduction in Rrp40 levels reduced the bias of higher 5′-only transcripts over the full-length mRNA (Fig 6c). These findings suggest that premature transcription termination does occur naturally at the Igh-V region with its level positively correlating with SHM in B cells.
Figure 6. Premature transcription termination likely occurs at Igh-V region.
a) Absolute quantitation of transcript abundance containing each indicated gene region was conducted by real-time PCR using the corresponding primer sets. Ct value was used to calculate the copy number from a standard curve generated in parallel using the control plasmid template (see Experimental Procedures). Data represent the average of four independent experiments (error bar represents SD) and paired t-test is used in statistical analysis. b) RNA quantification analysis similar to that in part (a) was performed on cDNA from poly-T mediated reverse transcription templates. Data represent the average of three independent experiments. T-test was employed to detect any significant reduction. c) The bias of abundance in transcripts containing only 5′-end vs full-length were computed as a ratio of the copy number containing 5′-coding region over transcripts containing Cμ from either Rrp40 knockdown cells or Ctrl-shRNA transduced cells. The reduction of bias was then calculated by comparing Rrp40KD cells to Ctrl-shRNA transduced cells. Data represented the average of all three regions detected by quantitative PCR at the 2nd exon of Igh gene (region c, d e indicated in Fig 1a) from three independent experiments. Panel a and b represent a compiled analysis of 3 independent experiments with error bars represent the SD.
Discussion
As Spt5 is essential for optimal CSR11, 44 and interacts with AID11, it is surprising that a reduction in Spt5 levels facilitates SHM. However, this discrepancy can be an indication of the distinct sources of ssDNA substrates for AID during SHM and CSR. Whereas R-loops are the major source of ssDNA during CSR, we propose here (see Fig 7) that premature transcription termination provides ssDNA substrates for AID during SHM. Since mutation rates exceeding 10−3 per base per generation will introduce too many nonsense mutations for efficient affinity maturation45, the complex role of Spt5 in SHM may have evolved to achieve optimal antibody diversification. Moreover, it is noteworthy that Spt5 seems to also influence the non-homologous end joining (NHEJ) and homologous recombination (HR) processes44, both of which are important for CSR but largely dispensable for SHM. Thus, dissimilar roles of Spt5 in SHM and CSR probably reflect the distinct molecular mechanisms involved in these two processes.
Figure 7. Model of the premature transcription termination as a source of ssDNA AID substrates.
The progression of Pol II reduces at the mutating Igh-V region, which leads to accumulation of stalling Pol II complexes (a, b). Most stalled Pol II complexes return to elongation after Pol II co-factor Spt5 recruits P-TEFb complexes that release pausing by phosphorylating C-terminal domains of both Spt5 and Pol II (c). However, non-resolvable pausing occurs stochastically, leading to premature transcription termination. Those early-terminated Pol II complexes leave unwound DNA region open with non-template ssDNA strand ready for AID targeting immediately (d). RPA complexes are then recruited to cover exposed non-template ssDNA while RNA:DNA hybrids are processed likely through nascent RNA removal process of transcription termination, which leaves single strand template DNA sensitive to AID (e). Partial transcripts are processed by the exosome complex, likely providing short RNA to in turn reduce Pol II progression at Igh-V and sustain the premature termination level there (f).
Genome-wide Pol II occupancy studies have revealed a large number of genes with stalled Pol II complexes in the transcribing regions. Although the fate of these stalled Pol II complexes remains under debate, one possibility among several others (like pausing or stalling), is that at least some of them will eventually lose all processivity and terminate transcription prematurely21, 22. Our RNA transcript analysis identified partially transcribed Igh genes in B cells (Fig 6). Although a bias towards 5′-region containing transcripts could result from preferential 3′-degradation of mRNA, it more likely reflects a role of premature transcription termination in SHM since: 1) Pol II is enriched in Igh-V region at a similar level as that reported around transcription termination sites (Fig 1)21, 22; 2) a reduction in Spt5 leads to an increase in SHM and in the frequency of ssDNA patches at Igh-V region (Fig 2e, 3); 3) the RNA exosome is required for optimal SHM (Fig 5); and 4) insertion of a transcription termination signal at the Igh-V region in DT40 cells results in increases of both Pol II accumulation and SHM upstream of the termination signal38. Together these findings support the idea that premature transcription termination contributes to SHM by supplying ssDNA substrates for AID. This premature transcription termination at the Igh-V region could be the consequence of frequent stalling (slow progression) and the stochastic loss of Pol II processivity physiologically, which could expose unwound DNA templates for AID to mediate SHM (Fig 7). Consistent with this idea, NEDD4, an E3-ubiquitinase that can mediate the degradation of unresolvable stalling Pol II, has been found at AID targeted regions during CSR46. This confirms that Pol II complexes can indeed “leave” their template in the middle of transcriptional process, which may provide accessibility for AID to the single-stranded transcription bubble. Interestingly, this model is also consistent with the observation that hypermutating dark zone germinal center B cells actually reduced their surface Ig level and that could be due to an increase in premature transcription termination at hypermutating V regions47.
Two different processes have recently been proposed to provide ssDNA substrates for AID. First, paused Pol II complexes correlated with the generation of R-loop structures at IgH switch regions, which positively contributes to CSR18, 19. If paused Pol II complexes themselves are also sufficient to facilitate SHM, the reduction in DSIF (Spt5/Spt4 complex) level should lead to a decrease in both Pol II pausing and SHM. On the contrary, we observed an increase in SHM when the Spt5 level was reduced in Ramos B cells. Since the reduction of cellular Spt5 is known to decrease Pol II processivity and hence promote premature transcription termination27, 28, 29, these observations suggested that premature transcription termination could be playing an important role in SHM. Second, accumulation of DNA supercoils around the transcribing Pol II complex has also been proposed as a source of AID substrates23, 34. AID can mutate supercoiled DNA in vitro and reduction of the DNA supercoil relieving factor topoisomerase I results in an increase in SHM48. However, supercoiled DNA should expose both strands of a DNA molecule symmetrically, but this remains to be examined by the ssDNA patch analysis by deeper sequencing for the V region in vivo33, 34.
Our model (Fig 7) of the premature transcription termination process as a source of ssDNA for AID during SHM suggests that: 1) the hypersensitive mutation region is similar to the size of a transcription bubble (14–18 base pairs); 2) any factors that reduce Pol II processivity such as a decreased level of elongation factors or increased abnormal DNA structures like supercoils will facilitate SHM48; 3) an artificial increase of Pol II complex termination by adding termination signals at V region promotes SHM38; and 4) template and non-template ssDNA are not likely exposed to AID simultaneously. We also (Fig 7) hypothesize that upon premature termination, unwound non-template ssDNA is exposed while template strand may remain hybridized with nascent RNA. Later, non-template ssDNA in the transcription bubble gets protected by RPA while RNA:DNA hybrids in the same transcription bubble will be processed through an RNA removal mechanism during termination49 to expose the template strand ssDNA for AID to target. Such sequential exposure of ssDNA for AID targeting is probably important for keeping the frequency of DNA double strand breaks low (<~10%) at the Igh-V regions during SHM30.
How AID targets Ig loci with high specificity in B cells remains a central question in B cell biology. Although our data indicate that the frequency of AID substrates strongly influence the mutation frequency in a SHM competent cell, the occurrence of ssDNA at the Igh-V region is probably not sufficient for eliciting the SHM process. In fact, the premature transcription termination process is likely to be determined by DNA elements independent of AID and SHM. This notion is consistent with recent observations that neither the accumulation of Pol II at the switch regions24 nor the ssDNA frequency33 at Igh-V region is dependent on AID. Moreover, the level of Pol II accumulation at the Ig-L locus in DT40 cells is independent of either the surrounding cis-elements capable of promoting SHM or the presence of AID50. Thus, although we have elucidated a novel mechanism utilized by B cells to provide ssDNA substrate for AID during SHM, the susceptibility of the variable region to SHM and the specificity of AID targeting to those regions in B cells are probably subject to several more levels of control rather than a simple interaction of the enzyme and its substrates. It is however interesting to investigate in the future what source(s) of ssDNA is used in off-target mutation sites of AID and whether they share similar mechanisms with the Igh-V region.
Methods
Cell lines and antibodies
The wild type human Burkitt’s lymphoma cell line Ramos has been described30, 35. The modified reporter Ramos cell line was established by replacing the endogenous 4–34 Igh-V region with mCherry-4–34 fusion fragment (Fig1a) using recombinase-mediated cassette change (RMCE). Briefly, a LoxP flanked hygromycin resistance gene was integrated into endogenous Igh-V locus by homologous recombination. The mcherry reporter cassette was then knocked into the Igh-V locus through Cre mediated recombination31. The Ramos clone used to construct this reporter line was preselected to have an undetectable level of AID protein and confirmed not to undergo SHM. Cells containing mCherry-Igh-V fusion were then transfected with AID-ER fusion protein 36 and clones were selected based on their capacity for undergoing SHM in a 4-OHT (Sigma-Aldrich) inducible manner. Cells were considered to have undergone SHM when they reduced their mCherry fluorescence based on flow cytometry analysis. Antibodies used in this study were anti-Pol II C-terminal repeats (Abcam), antiPol II C-terminal repeats (Serine 5 phosphorylated) (Abcam), anti-Spt5 (Santa Cruz) (1:250 dilution), anti-tubulin (Sigma-Aldrich) (1:2500 dilution).
Real-time PCR quantitation
In ChIP experiments, ChIP DNA precipitated with a specific antibody (1ug per reaction) was analyzed with the ΔΔCt method using SYBR Green PCR Master Mix (Life Technology or KAPA Biosystems) and was normalized to input DNA. PCR efficiency was estimated using serial dilutions and the signal was adjusted accordingly. In all experiments, rabbit anti-rat polyclonal antibody was used as the negative control and the ChIP signal from it was subtracted from each sample before further analysis. For absolute quantification of RNA transcripts, a single plasmid containing a single copy of each region of interest was made and the copy number was estimated using the molecular weight calculated based on the size of the plasmid. The plasmid was then used as a standard in the standard curve method of quantitative PCR and the copy number was calculated by ViiA 7 real-time PCR software (Life Technology). Primers used in the study are listed in Supplementary Table 1.
Lentiviral transduction and shRNA
Lentiviral particles containing designated shRNA were prepared by shRNA Core facility at Albert Einstein College of Medicine. All the shRNA constructs were obtained from the human TRC library (Thermo Scientific) with sequences listed in Supplementary Table 2. Control shRNA (Ctrl-shRNA) is the SHC002 construct from Sigma-Aldrich. Ramos cells were transduced with ~3:1 multiplicity of infection (MOI) and were subjected to Puromycin (Gibco, Life Technology) selection for 7–9 days. Successful knockdown of targeted genes was verified by real-time PCR and in the case of Spt5, western analysis as well. An shRNA resistant form of human Spt5 was created by replacing 7 of the 21 nucleotides of the shRNA targeting sequence but keeping amino acid sequence untouched.
Mutation Analysis
To obtain the mutation pattern, cells that had lost their mCherry fluorescence were sorted by flow cytometry to extract their genomic DNA (Qiagen). The mCherry-Igh-V fusion region was amplified using PfuTurbo (Agilent) cloned into the sequencing vector and Sanger sequenced in both directions to cover the whole ~1.3kb region. Sequencing data were then aligned by ClustalW2 and analyzed using SHMTool
ssDNA detection by Bisulfite treatment
In situ bisulfite treatment of cross-linked cellular nuclei under non-denaturing conditions was conducted to detect ssDNA patches that were natively exposed in chromatin33, 34. The final concentration of bisulfite was reduced to ~2 M to improve DNA recovery and a new KAPA HiFi Uracil+ kit (KAPA Biosystems) was used in the DNA amplification step to improve the accuracy of the analysis. The whole fusion of mcherry and Ramos Igh-V was amplified, cloned into vector and sequenced by Sanger sequencing.
Estimation of mutation rate by IgM reversion assay
Individual clones of IgM negative Ramos cells expressing the endogenous VH4-34 heavy chain V region bearing a stop-codon35 were allowed to accumulate mutations for three weeks. Any mutations that change the stop-codon into a sense codon will allow its descendants to become IgM positive. The frequency of those reverted cells was examined by flow cytometry. After analyzing sufficient numbers of clones, a mutation rate at that specific site could be estimated by maximal likelihood method51.
Statistical analysis
All statistical analyses were conducted using Prism 6 software. Throughout the data, * represented that the statistical significance was achieved and P value was illustrated correspondingly. Error bars represent standard deviation (SD) among independent experiments. In the cases where multiple experiments were compiled together, a paired Student’s T-test was used.
Supplementary Material
Acknowledgments
We thank Drs. Barbara Birshtein, Hilda Ye, Jonathan Warner and Richard Chahwan for their critical comments on the project. We thank the core facilities at Albert Einstein College for their technical support on Flow Cytometry and shRNA viral particle preparation. Funding for this work was provided by National Institutes of Health grants R01 CA072649, R01 CA102705 and an NCI/CFAR Pilot Project from P30CA013330 to M.D. Scharff. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing financial interests:
The authors have no competing financial interests.
Contributions:
X.W. and M.F. performed the experiments. S.K. and L.W. provided essential reagents and technical supports. X. W. and M.D.S designed the study and wrote the manuscript.
References
- 1.Victora GD, Nussenzweig MC. Germinal centers. Annual review of immunology. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 2.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 3.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102(5):565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 4.Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, et al. The biochemistry of somatic hypermutation. Annual review of immunology. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 5.Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Molecular cell. 2004;16(2):163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annual review of immunology. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(7):4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141(1):27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kenter AL. AID targeting is dependent on RNA polymerase II pausing. Seminars in immunology. 2012;24(4):281–286. doi: 10.1016/j.smim.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nambu Y, Sugai M, Gonda H, Lee CG, Katakai T, Agata Y, et al. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302(5653):2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- 11.Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143(1):122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willmann KL, Milosevic S, Pauklin S, Schmitz KM, Rangam G, Simon MT, et al. A role for the RNA pol II-associated PAF complex in AID-induced immune diversification. The Journal of experimental medicine. 2012;209(11):2099–2111. doi: 10.1084/jem.20112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. The EMBO journal. 2011;30(7):1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes & development. 2012;26(19):2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouzine F, Wojtowicz D, Yamane A, Resch W, Kieffer-Kwon KR, Bandle R, et al. Global regulation of promoter melting in naive lymphocytes. Cell. 2013;153(5):988–999. doi: 10.1016/j.cell.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longerich S, Tanaka A, Bozek G, Nicolae D, Storb U. The very 5′ end and the constant region of Ig genes are spared from somatic mutation because AID does not access these regions. The Journal of experimental medicine. 2005;202(10):1443–1454. doi: 10.1084/jem.20051604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woo CJ, Martin A, Scharff MD. Induction of somatic hypermutation is associated with modifications in immunoglobulin variable region chromatin. Immunity. 2003;19(4):479–489. doi: 10.1016/s1074-7613(03)00261-9. [DOI] [PubMed] [Google Scholar]
- 18.Zarrin AA, Alt FW, Chaudhuri J, Stokes N, Kaushal D, Du Pasquier L, et al. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nature immunology. 2004;5(12):1275–1281. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]
- 19.Huang FT, Yu K, Balter BB, Selsing E, Oruc Z, Khamlichi AA, et al. Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Smu class switch region. Molecular and cellular biology. 2007;27(16):5921–5932. doi: 10.1128/MCB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 21.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322(5909):1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature reviews Genetics. 2012;13(10):720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(35):12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. The Journal of experimental medicine. 2009;206(8):1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajagopal D, Maul RW, Ghosh A, Chakraborty T, Khamlichi AA, Sen R, et al. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. The Journal of experimental medicine. 2009;206(6):1237–1244. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagschal A, Rousset E, Basavarajaiah P, Contreras X, Harwig A, Laurent-Chabalier S, et al. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell. 2012;150(6):1147–1157. doi: 10.1016/j.cell.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Molecular cell. 2005;17(6):831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 28.Wu-Baer F, Lane WS, Gaynor RB. Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. Journal of molecular biology. 1998;277(2):179–197. doi: 10.1006/jmbi.1997.1601. [DOI] [PubMed] [Google Scholar]
- 29.Bourgeois CF, Kim YK, Churcher MJ, West MJ, Karn J. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Molecular and cellular biology. 2002;22(4):1079–1093. doi: 10.1128/MCB.22.4.1079-1093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sale JE, Neuberger MS. TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity. 1998;9(6):859–869. doi: 10.1016/s1074-7613(00)80651-2. [DOI] [PubMed] [Google Scholar]
- 31.Baughn LB, Kalis SL, MacCarthy T, Wei L, Fan M, Bergman A, et al. Recombinase-mediated cassette exchange as a novel method to study somatic hypermutation in Ramos cells. mBio. 2011;2(5) doi: 10.1128/mBio.00186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaguchi Y, Shibata H, Handa H. Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochimica et biophysica acta. 2013;1829(1):98–104. doi: 10.1016/j.bbagrm.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Ronai D, Iglesias-Ussel MD, Fan M, Li Z, Martin A, Scharff MD. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. The Journal of experimental medicine. 2007;204(1):181–190. doi: 10.1084/jem.20062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsa JY, Ramachandran S, Zaheen A, Nepal RM, Kapelnikov A, Belcheva A, et al. Negative supercoiling creates single-stranded patches of DNA that are substrates for AID-mediated mutagenesis. PLoS genetics. 2012;8(2):e1002518. doi: 10.1371/journal.pgen.1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W, Bardwell PD, Woo CJ, Poltoratsky V, Scharff MD, Martin A. Clonal instability of V region hypermutation in the Ramos Burkitt’s lymphoma cell line. International immunology. 2001;13(9):1175–1184. doi: 10.1093/intimm/13.9.1175. [DOI] [PubMed] [Google Scholar]
- 36.Doi T, Kinoshita K, Ikegawa M, Muramatsu M, Honjo T. De novo protein synthesis is required for the activation-induced cytidine deaminase function in class-switch recombination. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(5):2634–2638. doi: 10.1073/pnas.0437710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding B, LeJeune D, Li S. The C-terminal repeat domain of Spt5 plays an important role in suppression of Rad26-independent transcription coupled repair. The Journal of biological chemistry. 2010;285(8):5317–5326. doi: 10.1074/jbc.M109.082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kodgire P, Mukkawar P, Ratnam S, Martin TE, Storb U. Changes in RNA polymerase II progression influence somatic hypermutation of Ig-related genes by AID. The Journal of experimental medicine. 2013;210(7):1481–1492. doi: 10.1084/jem.20121523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451(7180):841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 40.Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, Eccleston J, et al. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144(3):353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lykke-Andersen S, Brodersen DE, Jensen TH. Origins and activities of the eukaryotic exosome. Journal of cell science. 2009;122(Pt 10):1487–1494. doi: 10.1242/jcs.047399. [DOI] [PubMed] [Google Scholar]
- 42.Houseley J, Tollervey D. The nuclear RNA surveillance machinery: the link between ncRNAs and genome structure in budding yeast? Biochimica et biophysica acta. 2008;1779(4):239–246. doi: 10.1016/j.bbagrm.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 43.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic acids research. 2000;28(8):1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanlie A, Begum NA, Akiyama H, Honjo T. The DSIF subunits Spt4 and Spt5 have distinct roles at various phases of immunoglobulin class switch recombination. PLoS genetics. 2012;8(4):e1002675. doi: 10.1371/journal.pgen.1002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381(6585):751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 46.Sun J, Keim CD, Wang J, Kazadi D, Oliver PM, Rabadan R, et al. E3-ubiquitin ligase Nedd4 determines the fate of AID-associated RNA polymerase II in B cells. Genes & development. 2013;27(16):1821–1833. doi: 10.1101/gad.210211.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacLennan IC. Germinal centers. Annual review of immunology. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 48.Kobayashi M, Sabouri Z, Sabouri S, Kitawaki Y, Pommier Y, Abe T, et al. Decrease in topoisomerase I is responsible for activation-induced cytidine deaminase (AID)-dependent somatic hypermutation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(48):19305–19310. doi: 10.1073/pnas.1114522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuehner JN, Pearson EL, Moore C. Unravelling the means to an end: RNA polymerase II transcription termination. Nature reviews Molecular cell biology. 2011;12(5):283–294. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kohler KM, McDonald JJ, Duke JL, Arakawa H, Tan S, Kleinstein SH, et al. Identification of core DNA elements that target somatic hypermutation. Journal of immunology. 2012;189(11):5314–5326. doi: 10.4049/jimmunol.1202082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hall BM, Ma CX, Liang P, Singh KK. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 2009;25(12):1564–1565. doi: 10.1093/bioinformatics/btp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.