Abstract
Tau is an intrinsically disordered protein (IDP) implicated in Alzheimer’s disease. Recently, tau proteins were discovered to be able to catalyze self-acetylation, which may promote its pathological aggregation. Understanding the paradox of tau’s random-like conformations, aggregation propensity, and enzymatic activity are challenging questions. We characterized the atomic structures of two truncated tau constructs, K18 and K19, consisting of, respectively, only the four- and three-repeats of tau protein, providing structural insights into tau’s paradox. Extensive 4.8 μs replica-exchange molecular dynamics simulations of the tau proteins achieved quantitative correlation with experimental Cα chemical shifts. Our results revealed (1) dynamically ordered conformations with close lysine–cysteine distances essential for tau self-acetylation and (2) high β-sheet content and large hydrophobic surface exposure for the two critical hexapeptides (275VQIINK280 and 306VQIVYK311), crucial for tau aggregation. Together, they illuminate tau’s perplexing behavior of how its disordered state can accomplish both roles.
Alzheimer’s disease (AD) is characterized by the coexistence of the intracellular neurofibrillary tangles of tau and the extracellular senile plaques of amyloid-β proteins.1 Tau proteins have two major isoforms with either three or four microtubule (MT) binding repeats (3R or 4R).2 Both isoforms are found in the fibrils deposited in the brain.3 K18 and K19 are the truncated constructs of the 4R and 3R tau, consisting of, respectively, only the four (residues 244–372) and three (with repeat-two being deleted) repeats. They can form amyloid fibrils with cross-β structure, which constitutes the core of the paired helical filaments (PHFs) of tau.4−7 A number of biophysical and spectroscopic studies revealed that tau belongs to the category of intrinsically disordered proteins (IDPs),8−11 which lack a folded structure. In solution, the monomeric tau appears as a random polymer,12,13 although some regions exhibit a preference for helical or β-strand conformations.14 However, it was recently found that tau possesses intrinsic enzymatic activity capable of catalyzing self-acetylation mediated by a pair of catalytic cysteine residues residing within the MT-binding domain.15 The acetylation of tau was shown to be able to inhibit its function and promote its pathological aggregation.15−17 The paradox of tau proteins’ random-like conformations, aggregation propensity, and enzymatic activity poses several fundamental questions; among these is how the conformational ensembles of IDPs are related to their function and aggregation.18 Although IDPs appear to contrast the structure–function paradigm that the protein’s function is determined by its unique single folded structure, their multiple distinct conformations are optimized by evolution and exploited for function.19,20 Thus, an atomic-level characterization of the conformational ensemble of IDPs is of great importance for understanding their biological activity and pathological aggregation.
An earlier study using small-angle X-ray scattering reported that each individual domain of tau has random coil features.13 Solution NMR data revealed tau’s structural diversity and intricate network of transient long-range contacts.10 Single-molecule Förster resonance energy transfer suggested that different tau domains display distinct conformational properties that are strongly correlated with their degree of disorder and that relate to their roles in aggregation.21,22 Recently, by selecting an ensemble from a large pool of statistical coil conformers and mapping the conformational ensemble at the residue level from NMR data, Ozenne et al. identified enhanced populations of turn and helical regions in K18.23 Although these experimental studies greatly improved our understanding of the conformational properties of tau proteins and hinted at local structures, an atomistic detailed characterization of the conformations adopted by tau proteins and their long-range structural arrangements have yet to emerge from experimental/computational studies.
Here, we explore the conformational ensemble of K18 and K19 monomers by conducting two 4.8 μs replica-exchange molecular dynamics (REMD) simulations24,25 using the GROMACS-4.5.3 software package.26 We chose the all-atom CHARMM27 force field with CMAP corrections,27 in accord with our recent combined simulation and experimental studies on K18 and K19 oligomers.28,29 A previous study on the evaluation of eight different force fields showed that four force fields including CHARMM27 provide a reasonably accurate description of the native states of two small proteins with α-helical and β-sheet structures,30 close to the ensembles that were reconstructed to fit the experimental data.31 Twenty-four different starting states (see Figure S1, Supporting Information) were used for each REMD simulation. The simulations were conducted in the NPT ensemble using 48 replicas, 100 ns per replica, at temperatures exponentially spaced between 310 and 430 K. The last 70 ns of trajectories at T = 310 K were used in the analysis. Details about the REMD simulations, the initial state modeling, and analysis methods are given in the Supporting Information.
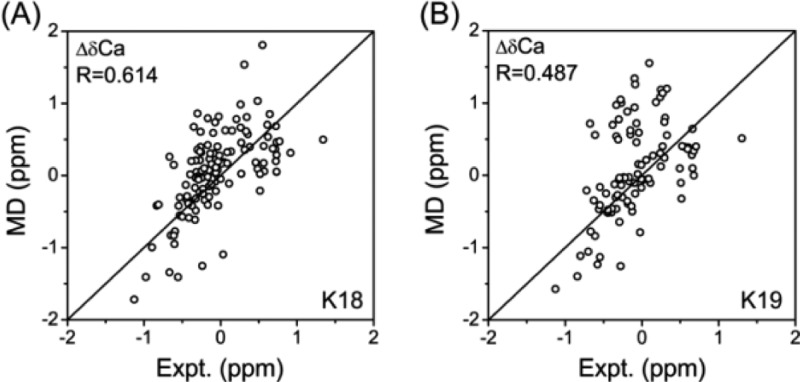
REMD simulations with explicit water have been shown to be able to reveal the conformational organization of small disordered histone tails,32 indicating their potential to sample the conformational space of IDPs. K18 and K19 consist of 130 and 99 amino acid (aa) residues, respectively. We note that large sizes of systems such as K18 and K19 may need simulation time longer than 100 ns to reach equilibrium. However, long REMD simulations are almost computationally prohibitive. For our 48 replica REMD simulations of K18/K19 in explicit solvent, 1.2 ns was obtained per day using 192 cores on the Biowulf PC/Linux cluster at the NIH for the REMD run of both K18 and K19. Our studies represent the longest REMD simulations in explicit water for such large protein systems. Figure S2 (Supporting Information) shows that the initially disordered conformations can evolve into structured conformations with high helix and β-sheet contents, indicating that K18 and K19 are not trapped in local energy minima. Figures S3 and S4 and Table S1 (Supporting Information) demonstrate that the probability of secondary structure content, secondary structure percentages of each residue, the numbers of clusters, the probability density function (PDF) of the radius of gyration (Rg), and the solvent-accessible surface area (SASA) of K18 and K19 within two independent time intervals (30–65 and 65–100 ns) are quite similar, indicating convergence of the REMD simulations. The number of clusters within the two time windows for both K18 and K19 decreased by a small amount (Table S1, Supporting Information), even though the secondary structures from the two time windows overlap very well (Figure S4, Supporting Information), indicating that random structures are decreasing and more structured conformations evolve in the simulations. We also compared our calculated secondary chemical shifts (SCSs) of Cα atoms with those from previous NMR experiments.8 Our predicted and experimentally measured Cα SCSs show a Pearson correlation coefficient of 0.614 for K18 and 0.487 for K19 (Figure 1), indicating that our simulation results are consistent with those from previous experiments.8 The correlations are encouraging for such large proteins with 130 aa’s for K18 and 99 aa’s for K19 in explicit water. A recent REMD study of a small 20 residue IDP fragment produced Cα SCSs having a correlation coefficient of 0.73 with experimental values.33 The calculated Cα chemical shifts from recent REMD simulations on α-synuclein multimers have a correlation coefficient of 0.991 with experimental values.34 We also achieved excellent agreement with experimental Cα chemical shifts with a correlation coefficient of 0.989 for K18 and 0.984 for K19 (Figure S5, Supporting Information). These calculated Cα chemical shifts indicate that the present simulations sample an ensemble of configurations that is consistent with low-resolution NMR experimental data, but we cannot exclude the possibility that some other low free energies may exist.
Figure 1.
Scatter plots comparing experimental (Expt.) and SPARTA-predicted (MD) SCSs of the Cα atom for (A) K18 and (B) K19. The Pearson correlation coefficients (R) between experimental and MD-generated SCSs are indicated.
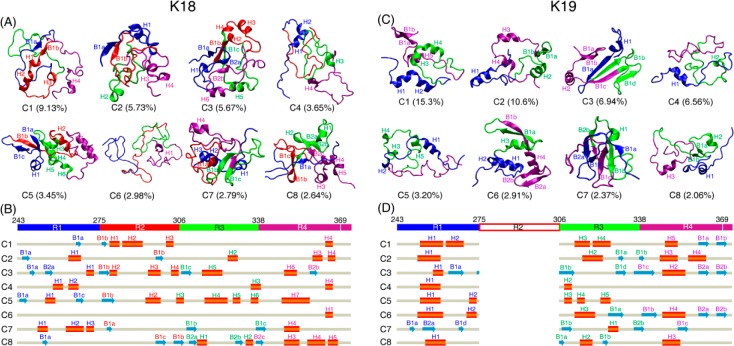
The atomic structures of K18 and K19 monomers at T = 310 K were characterized using RMSD-based cluster analysis of the conformational ensemble. The representative structures of the top eight most-populated clusters for K18 and K19 (Figure 2) revealed both the ordered and disordered nature of the two proteins. The calculated smallest Cα-RMSD values of the first eight clusters with one of the 24 initial states for both K18 and K19 (Table S2, Supporting Information) show that 7/6 out of the 8 clusters of K18/K19 have the smallest Cα-RMSD of larger than 1.1 nm, indicating that the simulation results are not biased by the initial states. Common structural properties of K18 and K19 were observed, with the middle part of each repeat favoring helical structure and the two terminal regions of each repeat preferring β-sheet structure. Each repeat has a similar β–α–β-turn structural pattern. Differences in structural properties were also seen. In K18, R4 has a higher helix probability than the other three repeats, while in K19, R1 and R4 both have higher helix probabilities than R3 (Figure 2 and Figure S6, Supporting Information). Overall, K19 has a higher helix probability than K18. The distribution of secondary structures in both K18 and K19 is similar to those obtained by previous NMR characterization.8,23
Figure 2.
Structural analysis of K18 and K19 monomers in aqueous solution at 310 K. Representative conformations for the top eight most-populated clusters along with their corresponding probabilities for K18 (A) and K19 (C). Secondary structures are displayed in new-cartoon style, with different colors representing different repeats, blue for R1, red for R2, green for R3, and purple for R4 and the last four residues after R4. For each structure, helices are indicated with H1, H2, ..., and β-sheets are labeled with B1, B2, ...; β-strands in the same sheet are labeled with Bna, Bnb, ... (n = 1, 2, ...). Two adjacent β-strands use the neighboring letters in the alphabet. Sequence views of the eight clusters for K18 (B) and K19 (D). The aa residue numbering is based on the full-length 441 aa tau protein. The β-strand is shown with blue arrow and the helix with the red cylinder. Each helix/β-strand is labeled with the same label as used in (A) and (C).
A previous study using a Bayesian weighting (BW) algorithm, which is based on techniques from Bayesian statistics, reported that the ordered parameters of K18 should be at least 1 order of magnitude higher than that estimated from random coil models, providing evidence for residual structure in K18.35 Consistently, we found that in K18, the percentages of conformers with at least 50 and 40% residues adopting α-helix and β-sheet structures are 0.0 and 4.8%, respectively. In K19, these percentages are 1.6 and 37.6%, respectively.
The global intramolecular interactions were examined from the residue–residue and side-chain–side-chain (SC–SC) contact probability maps (Figure S7A,B, Supporting Information). Distinct differences are observed between K18 and K19. In K18, repeat R1 has strong interaction with R2, and R3 has strong interaction with R4, while there are almost no interactions between R1/R2 and R3/R4, consistent with previous NMR studies on the tau protein at single-residue resolution.10 In contrast, in K19, R1 interacts with both R3 and R4. These results indicate that the domain–domain interactions are not completely random in K18 and K19. The spatial distributions of α-helix/β-sheet in K18 and K19 (Figure S7C,D, Supporting Information) also provide evidence for both the ordered and disordered nature of tau proteins.
We found that helical structures are mainly located in the middle region of each repeat, that is, 250MPDLKNVKSKI260 in R1, 280KKLDLSNVQSK290 in R2, 315LSKVTSKCGSL325 in R3, and 345DFKDRVQSKIG355 in R4. The atomic-level structures of the most populated helix in each repeat are given in Figure S8 (Supporting Information). As seen from this figure, all of the helical conformations are amphipathic helical structures. The solvent-exposed hydrophobic surface may facilitate aggregation. As the net charges for the helix in each repeat are positive, it is very likely that the helical motifs are responsible for binding and stabilizing the negatively charged MT. We also found that in K18 and K19, the populations of lysine in the helical structure are, respectively, 16.1 and 28.4% versus 7.9 and 13.8% in the full-length isoform. A nucleated assembly mechanism suggested that dimerization is one of the rate-limiting steps for PHF formation.36 Our results may suggest a mechanism for polyanion-induced PHF formation where the polyanions (e.g., heparin) interact with the positively charged lysines in the helical structures, linking two tau monomers and promoting dimerization.
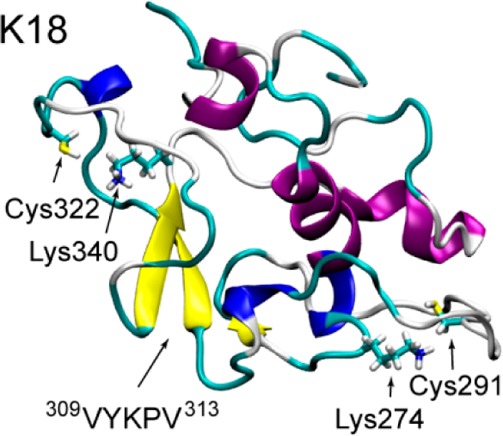
Recently, it was reported that tau isoforms, including K18 and K19, possess intrinsic enzymatic activity capable of catalyzing self-acetylation mediated by a pair of catalytic cysteine residues residing within the MT-binding domain.15 Cysteine-containing sequences 283DLSNVQSKCGS293 in R2 and 314DLSKVTSKCGS324 in R3 prefer α-helical structures (Figure S6, Supporting Information), and they are highly similar to the catalytic regions in MYST-family acetyltransferase Esa1 and Tip60.15 Enzymatic activity requires structural stability and conformational dynamics.37 Although there are ample data relating to the intrinsically disordered feature of tau, this is not the case for the structured features. Figure S8 (Supporting Information) illustrates that the locations of cysteine residue C322 in K18 and K19 have relatively stable helix environments, which are essential to catalyze a chemical reaction.
We then check the distance distribution between lysine and cysteine residues to see if they are close enough as in acetyltransferase.38 In the crystal structure of yeast Esa1, the Cα–Cα distance between Lys262 and Cys304 is 8.4 Å.38 In the K18 conformational ensemble, there are peaks between 5 and 10 Å in the lysine–cysteine distance distribution curve (Figure 3A). The structural details can be seen by representative structures of C5 and C7 in Figure 3B. In the C5 structure, the Lys274 sits on an α-helix, and a nearby Cys291 is located at the end of another α-helix. Lysine and cysteine residues in C7 are in close contact. As seen from Figure 3B, both Cys291 and Cys322 have nearby lysines (Lys274 and Lys340), which are similar to what is observed in the yeast Esa1 acetyltransferase.38 The close contact between lysine and cysteine residues in K18 may facilitate the self-acetylation activity of tau protein. These data provide atomic-level evidence for the close lysine–cysteine contact critical for the self-acetylation activity of tau proteins.
Figure 3.
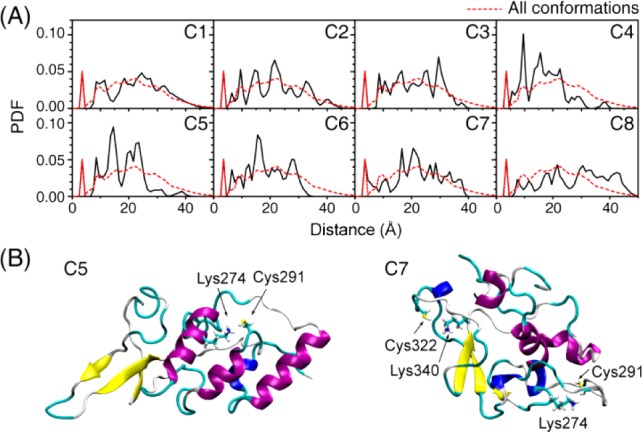
Minimum distance distributions between lysine and cysteine residues in K18 indicate that the structured conformational ensembles of K18 could enable tau’s acetyltransferase activity mediated by cysteines.
It was reported that short β-strand stretches often cooperatively trigger fibril formation in long amyloidogenic sequences.39 The probability distribution of β-strand length in K18 and K19 proteins (Figure S9, Supporting Information) shows that the most populated β-strand lengths are in the range of 2–5 residues, with longer β-strands in K19 than those in K18. The two critical hexapeptides (275VQIINK280 and 306VQIVYK311) are the core part in the parallel β-structures in the tau PHFs.40,41 Previous studies on the aggregation of these two hexapeptides and a longer peptide (273GKVQIINKKLDL284) reported that these peptides can form β-sheet-rich oligomers42,43 and fibrils44 with the β-strand in parallel alignment. Secondary structure analysis of the conformational ensemble of K18 and K19 shows that the hexapeptides 264ENLKHQ269 in R1, 275VQIINK280 and 295DNIKHV300 in R2 (K19 does not have R2), 306VQIVYK311 and 326GNIHHK331 in R3, and 337VEVKSE342 in R4 have a preference to adopt β-sheet structures (Figure 2B,D and Figure S6, Supporting Information). To examine the extent of solvent exposure of these β-sheet-rich regions, we plot in Figure 4 the free-energy landscape (FEL) projected on two reaction coordinates, the hydrophobic and hydrophilic SASAs of each hexapeptide. As can be seen in Figure 4, in K18, 275VQIINK280 in R2 and 306VQIVYK311 in R3 have larger SASAs than those for other hexapeptides. In K19, 306VQIVYK311 in R3 has the largest hydrophobic SASA. These results indicate that 275VQIINK280 in R2 and 306VQIVYK311 in R3 of K18 and 306VQIVYK311 in R3 of K19 are likely the aggregation centers, serving as a hydrophobic β-sheet core and facilitating the formation of PHFs, consistent with the current view that considers 275VQIINK280 and 306VQIVYK311 as the PHF cores.
Figure 4.
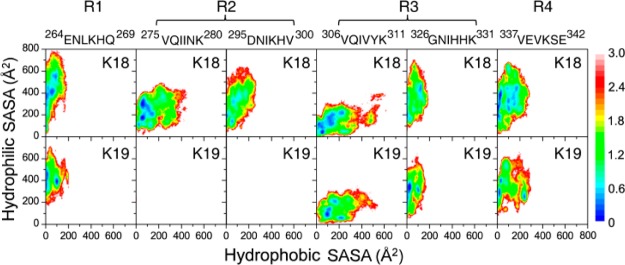
FEL of each β-sheet-rich hexapeptide in K18 and K19 as a function of hydrophilic and hydrophobic SASAs.
K18 and K19 are highly charged proteins. There are 21 positively charged residues and 11 negatively charged residues in K18 and 16 positively charged residues and 9 negatively charged residues in K19. Thus, it is important to investigate the salt bridge distributions in the monomers. The histograms of the number of salt bridges with respect to the number of intervening residues (NI) (Figure S10, Supporting Information) show that most of the salt bridges are formed between sequentially close residues in both K18 and K19 (NI ≤ 11). The average number of intrarepeat and inter-repeat salt bridges (Table S3, Supporting Information) demonstrates that most of the local salt bridges (NI ≤ 11) in both isoforms are formed within R4, correlated with the high propensity of helical structures. In K18, the average numbers of inter-repeat salt bridges of R1–R2 and R3–R4 are larger than those for other repeat pairs, consistent with the contact probability map of K18 showing two well-separated contact regions in Figure S7 (Supporting Information), R1–R2 and R3–R4. In K19, the average number of inter-repeat salt bridges between R1 and R4 is much larger than that in R1–R3 and R3–R4. In the fibrils, the inter-repeat contacts are between R1–R2, R2–R3, and R3–R4 for K18 and R1–R3, R1–R4, and R3–R4 for K19.29 Thus, the addition of the K18 monomer to the fibril does not require large domain reorganization, while K19 needs to break the R1–R4 contact for polymerization. Experimentally, K19 is slower to form amyloid fibrils in the absence of heparin catalysis.28 The reorganization of salt bridges could be one of the factors to slow down the fibrillation of K19.
In summary, our results revealed that the conformational ensemble of K18/K19 is a mixture of disordered and ordered structures. The highly polymorphic ordered structures are only transiently stable (i.e., low population), which is challenging to characterize experimentally. The ordered structures provide the structural basis for the intrinsic acetyltransferase enzymatic activity. The locations of cysteine residues in K18 and K19 have relatively stable environments, with the lysine and cysteine residues sufficiently close to enable the catalytic reaction. The conformational ensemble with both disordered and ordered structures also offers insights into tau aggregation. The β-sheet structures, preformed in the monomeric states, possibly provide seeds for the very early stage of aggregation; especially the two critical hexapeptides (275VQIINK280 and 306VQIVYK311), with large hydrophobic SASAs, are likely to be the seeding nucleus. These results not only point to a structural motif allowing tau to function as an acetyltransferase but also provide significant insights into the molecular mechanism of tau pathological aggregation.
Acknowledgments
B.M and R.N. acknowledge the financial support from NCI, NIH, under Contract Number HHSN261200800001E. G.W. acknowledges the financial support from the NSF of China (Grant No.: 91227102 and 11274075). Simulations were performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the NIH and National High Performance Computing Center of Fudan University.
Supporting Information Available
The details of initial state modeling, REMD simulations, analysis methods, and some details of the results. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This paper was published ASAP on August 22, 2014. The Supporting Information was updated. The revised paper was reposted on August 26, 2014.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Hardy J. A.; Higgins G. A. Alzheimer’s Disease: The Amyloid Cascade Hypothesis. Science 1992, 256, 184–185. [DOI] [PubMed] [Google Scholar]
- Buee L.; Bussiere T.; Buee-Scherrer V.; Delacourte A.; Hof P. R. Tau Protein Isoforms, Phosphorylation and Role in Neurodegenerative Disorders. Brain Res. Rev. 2000, 33, 95–130. [DOI] [PubMed] [Google Scholar]
- Goedert M.; Spillantini M. G.; Cairns N. J.; Crowther R. A. Tau Proteins of Alzheimer Paired Helical Filaments: Abnormal Phosphorylation of All Six Brain Isoforms. Neuron 1992, 8, 159–168. [DOI] [PubMed] [Google Scholar]
- Barghorn S.; Mandelkow E. Toward a Unified Scheme for the Aggregation of Tau into Alzheimer Paired Helical Filaments. Biochemistry 2002, 41, 14885–14896. [DOI] [PubMed] [Google Scholar]
- Siddiqua A.; Margittai M. Three- and Four-Repeat Tau Coassemble into Heterogeneous Filaments: An Implication for Alzheimer Disease. J. Biol. Chem. 2010, 285, 37920–37926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqua A.; Luo Y.; Meyer V.; Swanson M. A.; Yu X.; Wei G.; Zheng J.; Eaton G. R.; Ma B.; Nussinov R.; et al. Conformational Basis for Asymmetric Seeding Barrier in Filaments of Three- and Four-Repeat Tau. J. Am. Chem. Soc. 2012, 134, 10271–10278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daebel V.; Chinnathambi S.; Biernat J.; Schwalbe M.; Habenstein B.; Loquet A.; Akoury E.; Tepper K.; Muller H.; Baldus M.; et al. β-Sheet Core of Tau Paired Helical Filaments Revealed by Solid-State NMR. J. Am. Chem. Soc. 2012, 134, 13982–13989. [DOI] [PubMed] [Google Scholar]
- Mukrasch M. D.; Biernat J.; von Bergen M.; Griesinger C.; Mandelkow E.; Zweckstetter M. Sites of Tau Important for Aggregation Populate β-Structure and Bind to Microtubules and Polyanions. J. Biol. Chem. 2005, 280, 24978–24986. [DOI] [PubMed] [Google Scholar]
- Jeganathan S.; von Bergen M.; Mandelkow E. M.; Mandelkow E. The Natively Unfolded Character of Tau and Its Aggregation to Alzheimer-Like Paired Helical Filaments. Biochemistry 2008, 47, 10526–10539. [DOI] [PubMed] [Google Scholar]
- Mukrasch M. D.; Bibow S.; Korukottu J.; Jeganathan S.; Biernat J.; Griesinger C.; Mandelkow E.; Zweckstetter M. Structural Polymorphism of 441-Residue Tau at Single Residue Resolution. PLoS Biol. 2009, 7, e1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalbe M.; Ozenne V.; Bibow S.; Jaremko M.; Jaremko L.; Gajda M.; Jensen M. R.; Biernat J.; Becker S.; Mandelkow E.; et al. Predictive Atomic Resolution Descriptions of Intrinsically Disordered hTau40 and α-Synuclein in Solution from NMR and Small Angle Scattering. Structure 2014, 22, 238–249. [DOI] [PubMed] [Google Scholar]
- Schweers O.; Schonbrunn-Hanebeck E.; Marx A.; Mandelkow E. Structural Studies of Tau Protein and Alzheimer Paired Helical Filaments Show No Evidence for β-Structure. J. Biol. Chem. 1994, 269, 24290–24297. [PubMed] [Google Scholar]
- Mylonas E.; Hascher A.; Bernado P.; Blackledge M.; Mandelkow E.; Svergun D. I. Domain Conformation of Tau Protein Studied by Solution Small-Angle X-ray Scattering. Biochemistry 2008, 47, 10345–10353. [DOI] [PubMed] [Google Scholar]
- Eliezer D.; Barre P.; Kobaslija M.; Chan D.; Li X.; Heend L. Residual Structure in the Repeat Domain of Tau: Echoes of Microtubule Binding and Paired Helical Filament Formation. Biochemistry 2005, 44, 1026–1036. [DOI] [PubMed] [Google Scholar]
- Cohen T. J.; Friedmann D.; Hwang A. W.; Marmorstein R.; Lee V. M. The Microtubule-Associated Tau Protein Has Intrinsic Acetyltransferase Activity. Nat. Struct. Mol. Biol. 2013, 20, 756–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S. W.; Cho S. H.; Zhou Y.; Schroeder S.; Haroutunian V.; Seeley W. W.; Huang E. J.; Shen Y.; Masliah E.; Mukherjee C.; et al. Acetylation of Tau Inhibits Its Degradation and Contributes to Tauopathy. Neuron 2010, 67, 953–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T. J.; Guo J. L.; Hurtado D. E.; Kwong L. K.; Mills I. P.; Trojanowski J. Q.; Lee V. M. The Acetylation of Tau Inhibits Its Function and Promotes Pathological Tau Aggregation. Nat. Commun. 2011, 2, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.; Stultz C. M. Finding Order within Disorder: Elucidating the Structure of Proteins Associated with Neurodegenerative Disease. Future Med. Chem. 2009, 1, 467–482. [DOI] [PubMed] [Google Scholar]
- Tsai C. J.; Ma B.; Sham Y. Y.; Kumar S.; Nussinov R. Structured Disorder and Conformational Selection. Proteins: Struct., Funct., Bioinf. 2001, 44, 418–427. [DOI] [PubMed] [Google Scholar]
- Dunker A. K.; Cortese M. S.; Romero P.; Iakoucheva L. M.; Uversky V. N. Flexible Nets. The Roles of Intrinsic Disorder in Protein Interaction Networks. FEBS J. 2005, 272, 5129–5148. [DOI] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S.; Rhoades E. Identification of an Aggregation-Prone Structure of Tau. J. Am. Chem. Soc. 2012, 134, 16607–16613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath A.; Sammalkorpi M.; DeWitt D. C.; Trexler A. J.; Elbaum-Garfinkle S.; O’Hern C. S.; Rhoades E. The Conformational Ensembles of α-Synuclein and Tau: Combining Single-Molecule FRET and Simulations. Biophys. J. 2012, 103, 1940–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozenne V.; Schneider R.; Yao M.; Huang J. R.; Salmon L.; Zweckstetter M.; Jensen M. R.; Blackledge M. Mapping the Potential Energy Landscape of Intrinsically Disordered Proteins at Amino Acid Resolution. J. Am. Chem. Soc. 2012, 134, 15138–15148. [DOI] [PubMed] [Google Scholar]
- Sugita Y.; Okamoto Y. Replica-Exchange Molecular Dynamics Method for Protein Folding. Chem. Phys. Lett. 1999, 314, 141–151. [Google Scholar]
- Hansmann U. H. E.; Okamoto Y. Prediction of Peptide Conformation by Multicanonical Algorithm: New Approach to the Multiple-Minima Problem. J. Comput. Chem. 1993, 14, 1333–1338. [Google Scholar]
- Hess B.; Kutzner C.; van der Spoel D.; Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [DOI] [PubMed] [Google Scholar]
- Bjelkmar P. r.; Larsson P.; Cuendet M. A.; Hess B.; Lindahl E. Implementation of the CHARMM Force Field in GROMACS: Analysis of Protein Stability Effects from Correction Maps, Virtual Interaction Sites, and Water Models. J. Chem. Theory Comput. 2010, 6, 459–466. [DOI] [PubMed] [Google Scholar]
- Luo Y.; Dinkel P.; Yu X.; Margittai M.; Zheng J.; Nussinov R.; Wei G.; Ma B. Molecular Insights into the Reversible Formation of Tau Protein Fibrils. Chem. Commun. (Cambridge, U.K.) 2013, 49, 3582–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.; Luo Y.; Dinkel P.; Zheng J.; Wei G.; Margittai M.; Nussinov R.; Ma B. Cross-Seeding and Conformational Selection between Three- and Four-Repeat Human Tau Proteins. J. Biol. Chem. 2012, 287, 14950–14959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen K.; Maragakis P.; Piana S.; Eastwood M. P.; Dror R. O.; Shaw D. E. Systematic Validation of Protein Force Fields against Experimental Data. PLoS One 2012, 7, e32131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange O. F.; Lakomek N. A.; Fares C.; Schroder G. F.; Walter K. F.; Becker S.; Meiler J.; Grubmuller H.; Griesinger C.; de Groot B. L. Recognition Dynamics up to Microseconds Revealed from an RDC-Derived Ubiquitin Ensemble in Solution. Science 2008, 320, 1471–1475. [DOI] [PubMed] [Google Scholar]
- Potoyan D. A.; Papoian G. A. Energy Landscape Analyses of Disordered Histone Tails Reveal Special Organization of Their Conformational Dynamics. J. Am. Chem. Soc. 2011, 133, 7405–7415. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Chu X.; Longhi S.; Roche P.; Han W.; Wang E.; Wang J. Multiscaled Exploration of Coupled Folding and Binding of an Intrinsically Disordered Molecular Recognition Element in Measles Virus Nucleoprotein. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, E3743–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurry T.; Ullman O.; Fisher C. K.; Perovic I.; Pochapsky T.; Stultz C. M. The Dynamic Structure of α-Synuclein Multimers. J. Am. Chem. Soc. 2013, 135, 3865–3872. [DOI] [PubMed] [Google Scholar]
- Fisher C. K.; Stultz C. M. Protein Structure along the Order–Disorder Continuum. J. Am. Chem. Soc. 2011, 133, 10022–10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedhoff P.; von Bergen M.; Mandelkow E. M.; Davies P.; Mandelkow E. A Nucleated Assembly Mechanism of Alzheimer Paired Helical Filaments. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 15712–15717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B.; Nussinov R. Enzyme Dynamics Point to Stepwise Conformational Selection in Catalysis. Curr. Opin. Chem. Biol. 2010, 14, 652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H.; Rossetto D.; Mellert H.; Dang W.; Srinivasan M.; Johnson J.; Hodawadekar S.; Ding E. C.; Speicher K.; Abshiru N.; et al. MYST Protein Acetyltransferase Activity Requires Active Site Lysine Autoacetylation. EMBO J. 2012, 31, 58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L.; Cui W.; He Z.; Shi X.; Feng K.; Ma B.; Cai Y. D. Cooperativity among Short Amyloid Stretches in Long Amyloidogenic Sequences. PLoS One 2012, 7, e39369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bergen M.; Friedhoff P.; Biernat J.; Heberle J.; Mandelkow E. M.; Mandelkow E. Assembly of Tau Protein into Alzheimer Paired Helical Filaments Depends on a Local Sequence Motif (306VQIVYK311) Forming Beta Structure. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 5129–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bergen M.; Barghorn S.; Li L.; Marx A.; Biernat J.; Mandelkow E. M.; Mandelkow E. Mutations of Tau Protein in Frontotemporal Dementia Promote Aggregation of Paired Helical Filaments by Enhancing Local β-Structure. J. Biol. Chem. 2001, 276, 48165–48174. [DOI] [PubMed] [Google Scholar]
- Li D. W.; Mohanty S.; Irback A.; Huo S. Formation and Growth of Oligomers: A Monte Carlo Study of an Amyloid Tau Fragment. PLoS Comput. Biol. 2008, 4, e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larini L.; Gessel M. M.; LaPointe N. E.; Do T. D.; Bowers M. T.; Feinstein S. C.; Shea J. E. Initiation of Assembly of Tau(273–284) and Its DK280 Mutant: An Experimental and Computational Study. Phys. Chem. Chem. Phys. 2013, 15, 8916–8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goux W. J.; Kopplin L.; Nguyen A. D.; Leak K.; Rutkofsky M.; Shanmuganandam V. D.; Sharma D.; Inouye H.; Kirschner D. A. The Formation of Straight and Twisted Filaments from Short Tau Peptides. J. Biol. Chem. 2004, 279, 26868–26875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.