Abstract
Rearrangements, or gene fusions, involving the ETS family of transcription factors are common driving events in both prostate cancer and Ewing’s sarcoma. These rearrangements result in pathogenic expression of the ETS genes, and trigger activation of transcriptional programs enriched for invasion and other oncogenic features. While ETS gene fusions represent intriguing therapeutic targets, transcription factors, such as those comprising the ETS family, have been notoriously difficult to target. Recently, preclinical studies have demonstrated an association between ETS gene fusions and components of the DNA damage response pathway, such as poly (ADP)-ribose polymerase 1 (PARP1), the catalytic subunit of DNA protein kinase (DNA-PK), and histone deactylase 1 (HDAC1), and have suggested that ETS fusions may confer sensitivity to inhibitors of these DNA repair proteins. In this review, we discuss the role of ETS fusions in cancer, the preclinical rationale for targeting ETS fusions with inhibitors of PARP1, DNAPK, and HDAC1, as well as ongoing clinical trials targeting ETS gene fusions.
BACKGROUND
ETS transcription factors are aberrantly expressed in several cancers including prostate cancer (1), the Ewing’s sarcoma family of tumors (2), melanoma (3), secretory breast carcinoma (4), acute lymphoblastic leukemia (5), gastrointestinal stromal tumors (6) and rare cases of acute myeloid leukemia (7). The ETS family consists of 28 unique genes (reviewed in (8)), of which ERG, FLI1 and ETV1 are the most frequently deregulated in cancer. Prostate cancer frequently harbors rearrangements of ETS genes, in which ERG (50% of all prostate cancers) and ETV1 (5%) are fused to the androgen-regulated promoter and 5′ untranslated region of the TMPRSS2 gene (1, 9). This creates an androgen-regulated TMPRSS2-ETS fusion transcript that encodes a nearly full-length ETS transcription factor (Figure 1). In addition, almost all Ewing’s sarcomas contain an ETS rearrangement, including EWS-FLI1 (~90%) or EWS-ERG (~5-10%) gene fusions, which encode a chimeric protein notable for several features, including: 1) provision of an activation domain (from the EWS gene) to the ETS fusion and 2) replacement of the N-terminus of the ETS protein by an RNA binding domain from the EWS protein that enhances post-transcriptional splicing of ETS target genes (10) (Figure 1).
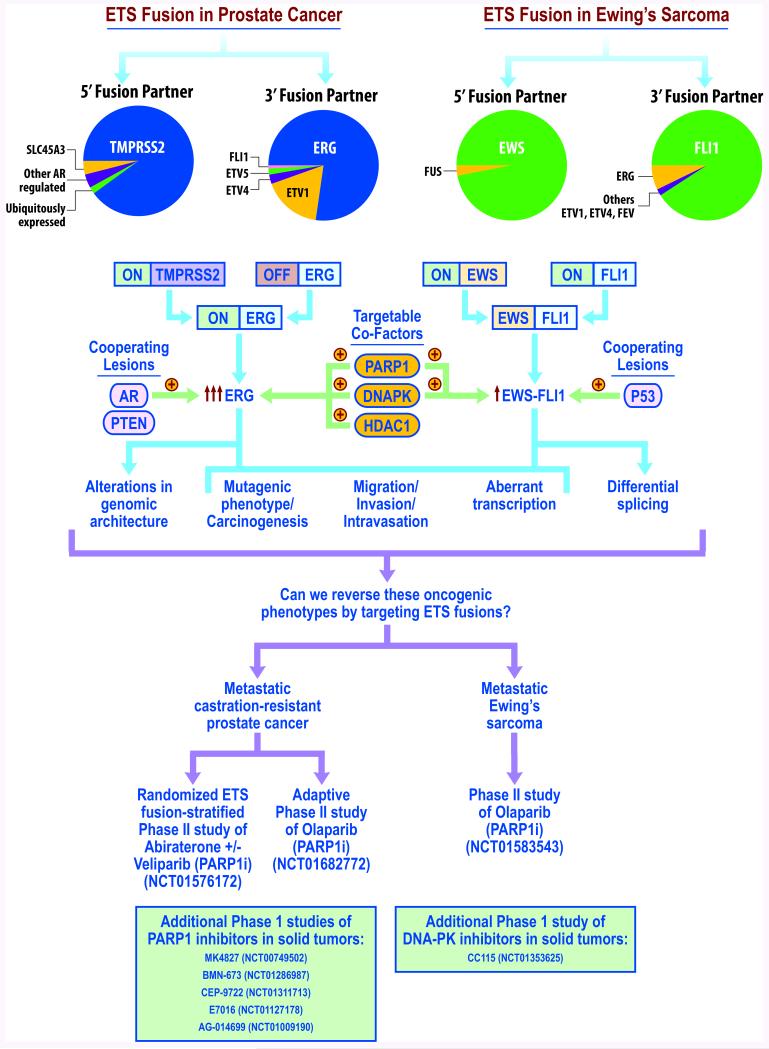
Figure 1. Overview of the role of ETS fusions in cancer and ongoing clinical trials targeting these fusions.
ETS gene fusions are common driving events in both prostate cancer and the Ewing’s sarcoma. The prevalence of different 5′ and 3′ fusion partners is depicted via pie chart. Formation of these fusions results in an aberrant transcriptional program enriched in invasion as well as induction of DNA breaks, consistent with a mutator phenotype. ETS fusion-mediated tumorigenesis has been demonstrated to be accelerated in the presence of cooperating lesions, such as PTEN loss and AR overexpression in prostate cancer, and P53 alterations in Ewing’s sarcoma. Recent preclinical studies have identified targetable co-factors, such as PARP1, DNAPK, and HDAC1, and inhibition of these co-factors has conferred preferential sensitivity to ETS-positive malignancies in preclinical models. While there are a series of phase I studies of PARP1, DNAPK, or HDAC1 inhibitors in patient populations including ERG fusion-positive malignancies, ongoing phase II studies have focused on PARP1 inhibition as a strategy to target ETS fusion-positive disease, including a randomized study in which patients are stratified based on ETS fusion status (NCT01576172).
Both prostate cancer and Ewing’s sarcoma ETS genomic rearrangements are thought to occur early in malignant progression. For example, TMPRSS2-ERG fusions are observed during the transition from high-grade prostatic intraepithelial neoplasia (PIN) lesions to invasive carcinoma (9, 11) and are formed at high frequency in androgen-stimulated cell lines under genotoxic stress (12-14). However, mice genetically engineered to express androgen-regulated ERG or ETV1 develop prostatic intraepithelial neoplasia-like lesions, but do not progress to frank carcinoma (9, 11, 15-17). This suggests that complete ETS-mediated transformation may require additional collaborating mutations and while this spectrum is only beginning to emerge (18-20), it is clear that ERG accelerates prostate carcinogenesis following loss of a highly recurrent prostate cancer tumor suppressor protein called PTEN or in the context of overexpression of the androgen receptor (15-17). Interestingly, TMPRSS2-ERG overexpression leads to increased self-renewal over multiple plating generations in Sca-1hi/EpCAM+ basal/progenitor cells isolated from genetically engineered mice (21) suggesting a role for ETS fusions in prostate cancer progenitor populations. In contrast to prostate cancer, the cells from which Ewing’s sarcoma are derived are still unknown, limiting the interpretation of genetic mouse models. Despite this impediment, EWS-FLI1 overexpression has been shown to induce leukemic phenotypes when expressed in hematoepoetic stem cells (22), to induce skeletal disruption when expressed in mesenchymal progenitors using a PRX1 promoter (23), and to accelerate tumor formation in conjunction with TP53 deletion (23).
Consistent with their role in prostate cancer and Ewing’s sarcoma progression, ETS transcription factors drive downstream signaling pathways with a number of functional consequences. RNA interference-mediated disruption of either TMPRSS2-ERG or EWS-FLI1 expression inhibits cell proliferation, invasion, metastasis and xenograft growth of prostate cancer or Ewing’s sarcoma cell line models that harbor the respective fusions (24-26). Accordingly, the transcriptional program driven by overexpression of ETS gene fusions is enriched for invasion and metastasis-associated gene signatures (1, 27, 28). Recently, our group found that both prostate cancer and Ewing’s sarcoma ETS gene fusions induce DNA double strand breaks (25, 26). This suggests that ETS gene fusions may drive a mutator phenotype and cause increased genomic instability in some cells.
Given the pathogenic roles of ETS fusions in the progression of both prostate cancer and Ewing’s sarcoma, ETS fusion products represent intriguing potential therapeutic targets. However, transcription factors, such as the ETS family, have been notoriously difficult to target (29). Potential strategies for targeting ETS fusion genes include therapies directed at the gene promoter, the RNA transcript, the fusion product itself, co-regulators of the fusion product, other collaborating lesions, and downstream targets of the fusion. While each of these strategies holds promise, this review focuses on agents available to patients or currently in clinical trials, leading to an emphasis on therapies directed at the androgen-responsive promoter (in prostate cancer) or against co-regulators of the fusion product.
CLINICAL-TRANSLATIONAL ADVANCES
Targeting the promoter of ETS fusions
The fact that the predominant ETS fusions in prostate cancer contain an androgen-responsive promoter (1, 24, 30, 31) provides a strong rationale for treating fusion-positive prostate malignancies with approaches directed against the androgen signaling axis. However, retrospective analyses of clinical samples have not consistently supported the theory that ETS fusion-positive prostate cancers should be preferentially sensitive to androgen deprivation therapy or anti-androgen approaches. In the context of castration-sensitive disease, while data from a radical prostatectomy series suggests that ETS fusion status predicts for response to adjuvant androgen deprivation therapy (32), results from other series have suggested that there is no association between ETS fusion status and response in patients managed with either definitive or adjuvant androgen deprivation therapy or anti-androgen therapy (33, 34). This discrepancy between studies may stem from the inherent issues associated with retrospective biomarker studies, such that imbalances between comparison groups in prognostic factors which are not fully taken into account. Alternatively, ETS fusions may simply not predict for response to androgen deprivation therapy in this setting, as all castration-sensitive disease may be similarly responsive to androgen deprivation therapy initially. Regardless, this relationship should be evaluated in prospective studies involving larger numbers of patients.
Following upfront androgen deprivation therapy, many patients will relapse with castration-resistant prostate cancer. The restoration of androgen signaling (35) and TMPRSS2-ERG expression (36) in castration-resistant disease provides a foundation for the hypothesis that ETS-positive castration-resistant prostate cancer may be preferentially responsive to next-generation anti-androgen therapy, such as abiraterone acetate. Abiraterone blocks androgen synthesis by inhibiting the enzyme cytochrome P450 17 alpha-hydroxysteroid dehydrogenase (37) and has improved clinical outcomes for patients with castration-resistant disease in large phase III clinical trials (38, 39). Using patient specimens from smaller phase I/II studies of metastatic patients treated with abiraterone, Attard et al found that the presence of the predominant ETS fusion, the TMPRSS2:ERG rearrangement, in circulating tumor cells (CTCs) correlated with prostate-specific antigen (PSA) response (40). In this study, 38% of patients with ERG fusion-positive CTCs had a >90% decline in PSA level with abiraterone, compared to 7% of patients with ERG-fusion-negative CTCs (40). In contrast, Danila et al found that TMPRSS2:ERG status in CTCs did not associate with response to abiraterone (41). As with the castration-sensitive setting, these discrepancies raise additional questions, such as whether ETS fusion status in the CTCs accurately reflects fusion status in the metastatic lesions. To address these questions, a multi-institutional randomized phase II clinical trial (clinicaltrials.gov identifier: NCT01576172) was initiated by our group at the University of Michigan with the objective of assessing several key questions, including the relationships between ETS fusion status and the response to anti-androgen therapy. Specifically, this trial, which requires biopsy of metastatic prostate cancer lesions for enrollment, prospectively stratifies patients by ETS fusion status in biopsies prior to randomization to treatment, which includes an arm consisting of abiraterone alone. This trial represents one of the first biomarker-driven trials in prostate cancer, and in comprehensively assessing ETS status in metastases, the primary tumor, circulating blood RNA, and CTCs, the study design should provide more definitive answers regarding whether ETS fusion-positive castration-resistant prostate cancer can be preferentially targeted with a standard next-generation anti-androgen.
Targeting the activity of ETS fusion products
Given the uncertainty on whether anti-androgen therapies can preferentially target ETS-positive prostate cancers, it is clear that better ETS-directed therapies need to be developed. While transcription factors themselves have conventionally been considered poor druggable targets (29), targeting co-factors necessary for functioning of the ETS gene fusion products may represent a more viable strategy. To date, the most promising co-factors, based on available clinical agents, for inhibiting ETS fusion activity include the enzymes poly (ADP)-ribose polymerase 1 (PARP1), the catalytic subunit of DNA protein kinase (DNA-PK), and histone deactylase 1 (HDAC1) (Figure 1).
PARP1 inhibition as a therapeutic approach for ETS-rearranged malignancies
Our group previously discovered an interaction between the TMPRSS2-ERG gene fusion product and PARP1, a protein involved in DNA damage response (26). We mapped the interaction to the conserved ETS DNA binding domain of ERG and demonstrated that PARP1 also interacts with ETV1, EWS-ERG and EWS-FLI1 (25, 26). Preclinical experiments demonstrated that ETS transcription factor activity was dependent on PARP1 expression and that inhibition of PARP1 could potentiate ETS-induced DNA damage leading to a long term loss of cell viability (25, 26). Overexpression of either TMPRSS2-ERG or EWS-FLI1 was sufficient to make cell line xenografts sensitive to PARP1 inhibition, indicative of a synthetic phenotype. These findings led to the hypothesis that ETS rearranged tumors are sensitive to PARP1 inhibition. To test this hypothesis, we completed 11 different cell line or primary tumor xenografts, and of these, only the 6 xenografts with an ETS rearrangement were sensitive to PARP1 inhibition (3 TMPRSS2-ERG, 1 ETV1 rearranged, 2 EWS-FLI1) (25, 26). Subsequent independent validation of the finding that the EWS-FLI1 fusion is associated with response to PARP1 inhibitors was performed via a high-throughput screen of 639 cell lines against 130 drugs under clinical or preclinical evaluation (42); this screen likely could not detect similar associations with prostate cancer ETS fusions as it included only one cell line harboring such a fusion.
The clinical use of PARP1 inhibitors has gained momentum secondary to previous preclinical reports demonstrating that cancers with impaired HR such as BRCA1/2-deficient cancers were extremely sensitive to PARP1 inhibition (43, 44). These studies proposed that PARP1 inhibitors cause replication forks to collapse leading to increased DNA damage, which goes unrepaired in the absence of HR, and early clinical studies have suggested that the PARP1 inhibitor olaparib has activity on the context of BRCA mutant cancers (45). Of interest, in preclinical studies, ETS fusion-positive xenografts were as sensitive to olaparib as a naturally BRCA1-deficient breast cancer xenograft (26), further strengthening the rationale to assess this biomarker-therapy combination clinically.
PARP1 inhibitors are now being actively evaluated in the clinic for both ETS-rearranged metastatic prostate cancer and Ewing’s sarcoma. Several of these trials are depicted in Figure 1. NCT01576172, the multi-institutional phase II trial described earlier, stratifies castration-resistant prostate cancer patients prospectively by ETS fusion status and randomizes them to abiraterone acetate alone versus abiraterone acetate combined with the PARP1 inhibitor veliparib (ABT-888). In addition to assessing the potential relationship between ETS fusion status and outcomes following abiraterone treatment, this trial also aims to prospectively determine if ETS status can predict for response to the addition of PARP1 inhibition to anti-androgen therapy. Other PARP1 inhibitors being assessed as monotherapy specifically in castration-resistant disease include olaparib (AZD-2281/KU-0059436) (Phase II, clinicaltrials.gov identifier NCT01682772) and niraparib (MK-4827) (Phase I expansion in prostate cancer, NCT00749502). The phase II olaparib study is interesting in design, as it employs a two stage scheme; the first stage is designed to screen for potential biomarkers of response to PARP1 inhibition, and the second is an expansion cohort enriched in identified biomarkers from the first stage (personal communication, J. De Bono). Initial results from the phase I niraparib study were recently reported(46); analysis of archival tumor samples from 18 patients with metastatic castration-resistant prostate cancer did not demonstrate an association between ETS fusion status and response to therapy. While these findings should be confirmed in larger studies with biopsies obtained immediately prior to treatment initiation, they do raise the issue that the response to PARP1 inhibitors is likely multifactorial in nature. The results from the prospective biomarker-stratified phase II studies described above will more conclusively determine whether the ETS-PARP1 association seen in vitro will hold up clinically.
Outside of prostate cancer, olaparib has been assessed as monotherapy in a Phase II trial for patients with recurrent or metastatic Ewing’s sarcoma (NCT01583543). As only 4 out of the initial 12 patients achieved stable disease (6-18 weeks) and none achieved partial or complete response, further accrual to this trial has been discontinued(47). Because molecular diagnosis was not required for this study, it is unclear whether the results from this trial stem from biologic or pharmacologic factors. In addition to this study, several Phase I studies including PARP1 inhibitors are currently underway or near completion for patients with any solid tumor; these include the agents BMN-673(NCT01286987), CEP-9722 (NCT01311713), E7016 (NCT01127178), and rucaparib (AG-014699/PF-01367338) (NCT01009190).
Several issues need to be addressed when assessing PARP1 inhibitors as a strategy for targeting ETS fusion-positive malignancies. One major concern is which PARP1 inhibitor will be most efficacious in this context. While some of these studies may appear redundant in clinical context, it is clear that not all PARP1 inhibitors behave similarly. A recent study suggests that PARP inhibitors differ markedly in their ability to cause cytoxicity by trapping PARP1 and PARP2 enzymes at damaged DNA, a difference which does not correlate with the catalytic inhibitory properties for each agent (48). This suggests that certain PARP1 inhibitors may be more effective than others for treating ETS-positive cancers; however, to address this concern, more investigation is needed into the mechanism by which PARP1 inhibitors are cytotoxic in the context of ETS rearrangements. A second issue is whether PARP1 inhibitors are best administered as a monotherapy or in combination with other potentiating agents. While initial PARP1 inhibitor trials employed monotherapy approaches, several PARP1 inhibitor combination studies have been completed in both the Phase I and II trial settings using various chemotherapeutics for other malignancies (reviewed (49)). Many of these have integrated alkylating agents due to the observation that PARP1−/− mice are extremely sensitive to this class of therapeutics (49), while others use topoisomerase inhibitors. For example, ABT-888 has been shown to enhance the effects of topotecan in adults with refractory solid tumors or lymphomas (50). Notably, our group demonstrated that olaparib and temozolomide significantly reduced tumor volumes in a TMPRSS2-ERG rearranged prostate cancer cell line xenograft and completely regressed tumors in an EWS-FLI1 rearranged Ewing’s sarcoma cell line xenograft (25, 26). These results suggest that the combination of temozolamide with a PARP inhibitor would be worthwhile to assess clinically for Ewing’s sarcoma; in fact, two ongoing phase I studies (NCT01858168 and NCT02044120) are exploring this regimen in patients with this disease.
DNA-PK inhibition as a treatment strategy for ETS-rearranged malignancies
As an alternative to PARP1 inhibition, blocking the activity of the DNA repair protein DNA-PK represents another potential strategy for targeting ETS fusion-positive cancers. Our group has previously demonstrated that the catalytic subunit of DNA-PK also physically interacts with ETS fusion products, such as ERG, ETV1, EWS-ERG and EWS-FLI1 (25, 26). In vitro studies demonstrated that DNA-PK expression and activity were necessary for ETS transcriptional activity, and pharmacologic inhibition or genetic knockdown of DNA-PK could also potentiate ETS-induced DNA damage (25, 26). While no clinical-grade DNA-PK inhibitor was available at the time of these initial studies, CC-115, a potent dual inhibitor of both DNA-PK and mTOR, has since been developed and is now in evaluation in a Phase I study in solid tumors (Figure 1). mTOR is a key effector in the phosphoinositide 3-kinase (PI3K) pathway. Given the recently demonstrated cross-talk between androgen receptor signaling and the PI3K pathway in PTEN-deficient prostate cancers (51), the associations between ERG fusions and PTEN deletion in prostate cancer (52), and the cooperativity between ERG overexpression and PTEN loss in carcinogenesis (16, 17), dual targeting of DNA-PK and MTOR has intriguing potential in ETS-positive prostate cancer. In addition, since mTOR inhibition downregulates the EWS-FLI1 protein and has been demonstrated to synergize with antisense oligonucleotides against EWS-FLI1 (reviewed in (53)), the dual activity of CC-115 provides theoretical advantages in the treatment of Ewing’s sarcoma as well.
HDAC1 inhibition as a treatment approach for ETS fusion-positive cancers
Another potential strategy for targeting ETS fusions is via inhibition of HDAC1, an enzyme which modifies histones and drives epigenetic gene regulation via transcriptional co-repression (reviewed in (54)). Previous studies have demonstrated that overexpression of ERG results in a gene expression signature notable for upregulation of HDAC1 and downregulation of its targets (55), and that HDAC1 indirectly interacts with ERG via the ERG-associated protein ESET (56, 57). In addition, inhibition of HDAC1 repressed ERG and was preferentially sensitive in ERG-positive cell lines (58). However, translation of these findings to the clinic have been less promising, as two phase II studies of HDAC inhibitors in CRPC yielded disappointing results; one of these studies, assessing the agent vorinostat, demonstrated significant toxicities which limited efficacy assessment, and the second, assessing the agent romidespin, demonstrated minimal antitumor activity (59, 60). Several other trials assessing HDAC inhibitors, as mono- or combination therapy in CRPC, are still ongoing (clinicaltrials.gov identifiers (NCT01075308, NCT00878436, and NCT01174199).
CONCLUSIONS
While ETS fusions were discovered several years ago, and are important preclinically in several aspects of prostate cancer initiation and progression, targeting of ETS fusions remains a work in progress. While recent advances have been made in the preclinical space of targeting ETS fusions with clinically available agents, such as inhibitors of PARP1, DNAPK, and HDAC1, these findings need to be validated in clinical trials. Of these agents, the studies targeting ETS with PARP1 inhibitors are furthest along in development, and should yield results within the next few years.
Acknowledgments
We thank Karen Giles for critically reading the manuscript and Steven Kronenberg and Vishal Kothari for graphic art support.
Grant Support This work is supported in part by a Physician Research Training Award from the Department of Defense (F.Y. Feng), an American Cancer Society Research Professor Award (A.M. Chinnaiyan), a Burroughs Welcome Foundation Award in Clinical Translational Research (A.M. Chinnaiyan), the Doris Duke Charitable Foundation Clinical Scientist Award (A.M. Chinnaiyan), NIH funding (R01CA132874 andP50CA69568; to A.M. Chinnaiyan), the Early Detection Research Network (UO1 CA113913; to A.M. Chinnaiyan), the Department of Defense (PC094231; to A.M. Chinnaiyan), and grants from the Prostate Cancer Foundation (F.Y. Feng, J.C. Brenner, and A.M. Chinnaiyan).
Footnotes
Disclosure of Potential Conflicts of Interest
F.Y. Feng reports receiving a commercial research grant from Celgene, speakers bureau honoraria from Ventana, and is a consultant/advisory board member for Medivation/Astellas. J.C. Brenner reports receiving royalties from the University of Michigan for its intellectual property on the use of PARP inhibitors in ETS-positive cancers, which is licensed to Hologic. A.M. Chinnaiyan is a consultant/advisory board member for Celgene, Hologic, and Oncofusion Therapeutics; reports receiving royalties from the University of Michigan for its intellectual property on the use of ETS fusions in prostate cancer and PARP inhibitors in ETS-positive cancers, which is licensed to Hologic; and has ownership interest in Oncofusion Therapeutics. No potential conflicts of interest were disclosed by the other author.
References
- 1.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 2.de Alava E, Kawai A, Healey JH, Fligman I, Meyers PA, Huvos AG, et al. EWS-FLI1 fusion transcript structure is an independent determinant of prognosis in Ewing’s sarcoma. J Clin Oncol. 1998;16:1248–55. doi: 10.1200/JCO.1998.16.4.1248. [DOI] [PubMed] [Google Scholar]
- 3.Jane-Valbuena J, Widlund HR, Perner S, Johnson LA, Dibner AC, Lin WM, et al. An oncogenic role for ETV1 in melanoma. Cancer Res. 2010;70:2075–84. doi: 10.1158/0008-5472.CAN-09-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–76. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 5.Shurtleff SA, Buijs A, Behm FG, Rubnitz JE, Raimondi SC, Hancock ML, et al. TEL/AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis. Leukemia. 1995;9:1985–9. [PubMed] [Google Scholar]
- 6.Chi P, Chen Y, Zhang L, Guo X, Wongvipat J, Shamu T, et al. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature. 2010;467:849–53. doi: 10.1038/nature09409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimizu K, Ichikawa H, Tojo A, Kaneko Y, Maseki N, Hayashi Y, et al. An ets-related gene, ERG, is rearranged in human myeloid leukemia with t(16;21) chromosomal translocation. Proc Natl Acad Sci U S A. 1993;90:10280–4. doi: 10.1073/pnas.90.21.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem. 2011;80:437–71. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–9. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 10.Knoop LL, Baker SJ. EWS/FLI alters 5′-splice site selection. J Biol Chem. 2001;276:22317–22. doi: 10.1074/jbc.M008950200. [DOI] [PubMed] [Google Scholar]
- 11.Klezovitch O, Risk M, Coleman I, Lucas JM, Null M, True LD, et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci U S A. 2008;105:2105–10. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–83. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42:668–75. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zong Y, Xin L, Goldstein AS, Lawson DA, Teitell MA, Witte ON. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc Natl Acad Sci U S A. 2009;106:12465–70. doi: 10.1073/pnas.0905931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casey OM, Fang L, Hynes PG, Abou-Kheir WG, Martin PL, Tillman HS, et al. TMPRSS2- Driven ERG Expression In Vivo Increases Self-Renewal and Maintains Expression in a Castration Resistant Subpopulation. PLoS One. 2012;7:e41668. doi: 10.1371/journal.pone.0041668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torchia EC, Boyd K, Rehg JE, Qu C, Baker SJ. EWS/FLI-1 induces rapid onset of myeloid/erythroid leukemia in mice. Mol Cell Biol. 2007;27:7918–34. doi: 10.1128/MCB.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin PP, Pandey MK, Jin F, Xiong S, Deavers M, Parant JM, et al. EWS-FLI1 induces developmental abnormalities and accelerates sarcoma formation in a transgenic mouse model. Cancer Res. 2008;68:8968–75. doi: 10.1158/0008-5472.CAN-08-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–88. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenner JC, Feng FY, Han S, Patel S, Goyal SV, Bou-Maroun LM, et al. PARP-1 inhibition as a targeted strategy to treat Ewing’s sarcoma. Cancer Res. 2012;72:1608–13. doi: 10.1158/0008-5472.CAN-11-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–78. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helgeson BE, Tomlins SA, Shah N, Laxman B, Cao Q, Prensner JR, et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68:73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- 28.Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 29.Darnell JE., Jr. Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–9. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 30.Bonaccorsi L, Nesi G, Nuti F, Paglierani M, Krausz C, Masieri L, et al. Persistence of expression of the TMPRSS2:ERG fusion gene after pre-surgery androgen ablation may be associated with early prostate specific antigen relapse of prostate cancer: preliminary results. J Endocrinol Invest. 2009;32:590–6. doi: 10.1007/BF03346514. [DOI] [PubMed] [Google Scholar]
- 31.Mertz KD, Setlur SR, Dhanasekaran SM, Demichelis F, Perner S, Tomlins S, et al. Molecular characterization of TMPRSS2-ERG gene fusion in the NCI-H660 prostate cancer cell line: a new perspective for an old model. Neoplasia. 2007;9:200–6. doi: 10.1593/neo.07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karnes RJ, Cheville JC, Ida CM, Sebo TJ, Nair AA, Tang H, et al. The ability of biomarkers to predict systemic progression in men with high-risk prostate cancer treated surgically is dependent on ERG status. Cancer Res. 2010;70:8994–9002. doi: 10.1158/0008-5472.CAN-10-1358. [DOI] [PubMed] [Google Scholar]
- 33.Leinonen KA, Tolonen TT, Bracken H, Stenman UH, Tammela TL, Saramaki OR, et al. Association of SPINK1 expression and TMPRSS2:ERG fusion with prognosis in endocrine-treated prostate cancer. Clin Cancer Res. 2010;16:2845–51. doi: 10.1158/1078-0432.CCR-09-2505. [DOI] [PubMed] [Google Scholar]
- 34.Boormans JL, Hermans KG, Made AC, van Leenders GJ, Wildhagen MF, Collette L, et al. Expression of the androgen-regulated fusion gene TMPRSS2-ERG does not predict response to endocrine treatment in hormone-naive, node-positive prostate cancer. Eur Urol. 2010;57:830–5. doi: 10.1016/j.eururo.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Attard G, Reid AH, Olmos D, de Bono JS. Antitumor activity with CYP17 blockade indicates that castration-resistant prostate cancer frequently remains hormone driven. Cancer Res. 2009;69:4937–40. doi: 10.1158/0008-5472.CAN-08-4531. [DOI] [PubMed] [Google Scholar]
- 36.Cai C, Wang H, Xu Y, Chen S, Balk SP. Reactivation of androgen receptor-regulated TMPRSS2:ERG gene expression in castration-resistant prostate cancer. Cancer Res. 2009;69:6027–32. doi: 10.1158/0008-5472.CAN-09-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Attard G, Reid AH, A’Hern R, Parker C, Oommen NB, Folkerd E, et al. Selective inhibition of CYP17 with abiraterone acetate is highly active in the treatment of castration-resistant prostate cancer. J Clin Oncol. 2009;27:3742–8. doi: 10.1200/JCO.2008.20.0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2012 doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Attard G, Swennenhuis JF, Olmos D, Reid AH, Vickers E, A’Hern R, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–8. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 41.Danila DC, Anand A, Sung CC, Heller G, Leversha MA, Cao L, et al. TMPRSS2-ERG status in circulating tumor cells as a predictive biomarker of sensitivity in castration-resistant prostate cancer patients treated with abiraterone acetate. Eur Urol. 2011;60:897–904. doi: 10.1016/j.eururo.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 44.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 45.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 46.Sandhu SK, Schelman WR, Wilding G, Moreno V, Baird RD, Miranda S, et al. The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet Oncol. 2013;14:882–92. doi: 10.1016/S1470-2045(13)70240-7. [DOI] [PubMed] [Google Scholar]
- 47.Choy E, Butrynski J, Harmon D, Morgan J, George S, Wagner A, et al. Translation of preclinical predictive sensitivity of Ewing sarcoma to PARP inhibition: phase II study of olaparib in adult patients with recurrent/metastatic Ewing sarcoma following failure of prior chemotherapy [abstract]; Proceedings of the 104th Annual Meeting of the American Association for Cancer Research; Washington, DC; AACR. 2013. [Google Scholar]
- 48.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–99. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Do K, Chen A. Molecular Pathways: Targeting PARP in Cancer Treatment. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kummar S, Chen A, Ji J, Zhang Y, Reid JM, Ames M, et al. Phase I study of PARP inhibitor ABT-888 in combination with topotecan in adults with refractory solid tumors and lymphomas. Cancer Res. 2011;71:5626–34. doi: 10.1158/0008-5472.CAN-11-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009;22:1083–93. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balamuth NJ, Womer RB. Ewing’s sarcoma. Lancet Oncol. 2010;11:184–92. doi: 10.1016/S1470-2045(09)70286-4. [DOI] [PubMed] [Google Scholar]
- 54.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 55.Iljin K, Wolf M, Edgren H, Gupta S, Kilpinen S, Skotheim RI, et al. TMPRSS2 fusions with oncogenic ETS factors in prostate cancer involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer Res. 2006;66:10242–6. doi: 10.1158/0008-5472.CAN-06-1986. [DOI] [PubMed] [Google Scholar]
- 56.Yang L, Mei Q, Zielinska-Kwiatkowska A, Matsui Y, Blackburn ML, Benedetti D, et al. An ERG (ets-related gene)-associated histone methyltransferase interacts with histone deacetylases 1/2 and transcription co-repressors mSin3A/B. Biochem J. 2003;369:651–7. doi: 10.1042/BJ20020854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang L, Xia L, Wu DY, Wang H, Chansky HA, Schubach WH, et al. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene. 2002;21:148–52. doi: 10.1038/sj.onc.1204998. [DOI] [PubMed] [Google Scholar]
- 58.Bjorkman M, Iljin K, Halonen P, Sara H, Kaivanto E, Nees M, et al. Defining the molecular action of HDAC inhibitors and synergism with androgen deprivation in ERG-positive prostate cancer. Int J Cancer. 2008;123:2774–81. doi: 10.1002/ijc.23885. [DOI] [PubMed] [Google Scholar]
- 59.Molife LR, Attard G, Fong PC, Karavasilis V, Reid AH, Patterson S, et al. Phase II, two-stage, single-arm trial of the histone deacetylase inhibitor (HDACi) romidepsin in metastatic castration-resistant prostate cancer (CRPC) Ann Oncol. 2010;21:109–13. doi: 10.1093/annonc/mdp270. [DOI] [PubMed] [Google Scholar]
- 60.Bradley D, Rathkopf D, Dunn R, Stadler WM, Liu G, Smith DC, et al. Vorinostat in advanced prostate cancer patients progressing on prior chemotherapy (National Cancer Institute Trial 6862): trial results and interleukin-6 analysis: a study by the Department of Defense Prostate Cancer Clinical Trial Consortium and University of Chicago Phase 2 Consortium. Cancer. 2009;115:5541–9. doi: 10.1002/cncr.24597. [DOI] [PMC free article] [PubMed] [Google Scholar]