To the editor
One approach to improve risk prognostication in patients with Multiple Myeloma (MM) is to use new technologies to stratify patients based on distinct outcomes1. Outcome prediction for most patients is based on the International Staging System (ISS) and the presence or absence of specific fluorescent in-situ hybridization (FISH) abnormalities. Additional biomarkers are necessary to improve precision, and there is evidence that small non-coding RNAs, microRNAs (miRNAs), are involved in MM pathogenesis2–4, but the predictive role of circulating miRNAs remains to be fully evaluated. To address this question, we measured serum miRNA levels of a large cohort of newly diagnosed MM patients that were uniformly treated and followed, correlating miRNA levels with clinical outcome to test their prognostic impact in a multivariate model.
Patients with previously untreated transplant ineligible MM enrolled on a Gruppo Italiano Malattie EMatologiche dell’Adulto (GIMEMA) phase III clinical trial and with material available for analysis (n=288) were studied for serum levels of miRNAs5. Patients received Velcade-Melphalan-Prednisone (VMP) or Velcade-Melphalan-Prednisone-Thalidomide followed by maintenance with Velcade-Thalidomide (VMPT-VT) through the parent clinical trial (NCT#01063179).
RNA was isolated from the serum of 54 randomly selected MM patients, analyzed by NanoString, and only those miRNAs consistently expressed over the background threshold in at least 20% of samples were selected. These miRNAs were measured by qRT-PCR analyses using the 2^-dct methods6, 7. In addition, a composite indicator of high risk genetic features was generated and evaluated in these analyses, where it was positive if the patient had any of the following: del17, t(4;14), t(14;16); otherwise, they were classified as standard risk.
Univariate Cox regression models were used to assess the influence of continuous expression levels for the miRNAs in relation to PFS and OS. We tested the proportional hazards assumption for a Cox regression model fit for each of the covariates8. In the presence of non-proportional hazards, weighted estimation in the Cox regression model was used to provide unbiased average hazard ratio estimates9. Any miRNA, clinical, and/or molecular covariates that were significant at the p<0.10 level were brought forward for evaluation in the multivariable setting.
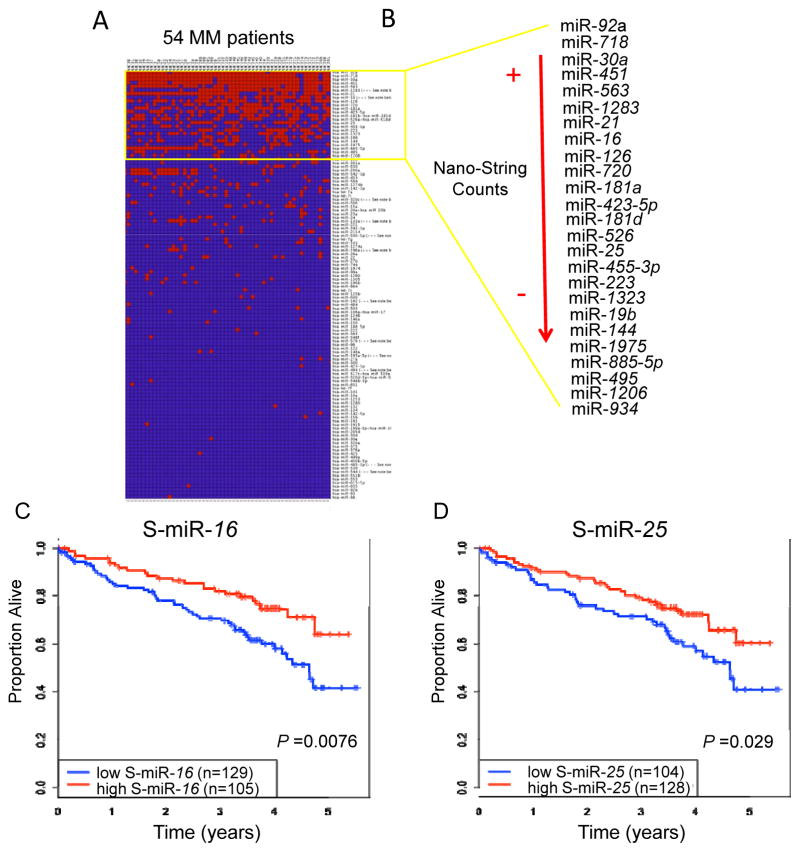
To identify circulating miRNAs with potential clinical impact, we analyzed by Nanostring the serum samples of a discovery set of 54 newly diagnosed MM patients randomly selected for analysis from 288 patients enrolled on a randomized phase III study of VMP versus VMPT-VT. (Fig. A1, Supplementary Information Table A1). Out of the 800 miRNAs evaluated, only 25 were detectable (≥100 counts) in at least 20% of the patients (Figure. 1A,B). The expression of these miRNAs were then measured in a validation set of 234 patients by qRT-PCR. Of the 25 serum miRNAs (S-miRNAs), only 10 (S-miRs-92a, 21, 30a, 720, 451, 223, 126, 19b, 25 and S-miR-16) were validated to be differentially expressed among the patients in the validation set (Supplementary Information Table A2). These 10 miRNAs were not significantly associated with clinical or molecular characteristics at diagnosis (Supplementary Information Table A3). One exception was for chromosome 13q14 deletion (Del13), which was more frequently found in patients with low S-miR-16 (62% vs. 38%, p=0.003) and S-miR-19b (64% vs. 41%, p=0.01) level (Supplementary Information Fig. A3). In addition, some of the continuous miRNA levels were significantly correlated with age, hemoglobin and creatinine (Supplementary Information Table A3); however, these were weak associations (|r|<0.22) and scatter-plots reflected weak if any relationships with these markers (Supplementary Information Fig. A4).
Figure 1.
(A) Supervised clustering analysis representative of the 800 miRNAs assessed using NanoString nCounter Technology and analyzed in 54 MM patients. Red cells indicate high expression and blue cells indicate low expression (B) list of the 25 miRNAs identified to be consistently expressed in at least 20% of MM samples analyzed. Overall survival (OS) for low vs. high expressors log2 S-miR-16 (C) and S-miR-25 (D) expression value were used to build OS curves. The median value of S-miR-16 and S-miR-25 for the healthy normal subjects was used as the cutoff expression value.
To assess prognostic impact, miRNA levels were evaluated as continuous variables. Of the 10 miRNAs, only S-miR-25 was significantly associated with both PFS (p=0.034) and OS (p=0.0005) in the univariate setting; S-miR-16 and S-miR30a were only associated with OS (p=0.008 and p=0.016 respectively). Patients with higher levels of S-miR-25 (HR 0.81; p=0.0005), S-miR-16 (HR 0.87; p=0.008), and S-miR-30a (HR 0.86, p=0.016) had longer OS than those with lower expression of these miRNAs (Table 1). Only S-miR-25 was significantly associated with PFS duration, where higher S-miR-25 expression was associated with better PFS duration (HR=0.92; p=0.034). A borderline significance for PFS and OS was also seen with S-miR-720, but was not statistically significant in multivariable models.
Table 1.
Significant and shared characteristics from univariate analyses for PFS and OS
| Factor or Marker | Progression-free survival (PFS) | Overall survival (OS) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariable | Univariate | Multivariable | |||||||||
| HR | 95% CI | p-value | HR | 95% CI | p-value | HR | 95% CI | p-value | HR | 95% CI | p-value | |
| VMPT (vs. VMP)1 | 0.67 | 0.49,0.92 | 0.013 | -- | -- | -- | 0.83 | 0.52,1.31 | 0.41 | -- | -- | -- |
| ISS stage2 | 1.25 | 0.99,1.58 | 0.06 | 1.41 | 1.05,1.89 | 0.022 | 1.42 | 1.02,1.98 | 0.039 | 1.34 | 0.82,2.18 | 0.24 |
| Creatinine | 1.30 | 0.92,1.83 | 0.14 | -- | -- | -- | 1.89 | 1.07,3.34 | 0.03 | 1.003 | 0.28,3.57 | 0.997 |
| Male (vs. female) | 1.03 | 0.75,1.42 | 0.84 | -- | -- | -- | 1.80 | 1.13,2.88 | 0.014 | 2.29 | 1.05,4.98 | 0.037 |
| Del172 | 2.12 | 1.25,3.59 | 0.005 | 2.29 | 1.17,4.50 | 0.016 | 2.13 | 1.13,4.03 | 0.02 | 1.30 | 0.51,3.30 | 0.59 |
| Del13 | 1.24 | 0.87,1.77 | 0.23 | -- | -- | -- | 1.76 | 1.05,2.96 | 0.03 | 3.44 | 1.63,7.27 | 0.001 |
| miR-163 | 0.94 | 0.88,1.02 | 0.13 | -- | -- | -- | 0.87 | 0.79,0.97 | 0.008 | -- | -- | -- |
| miR-25 | 0.92 | 0.84,0.99 | 0.034 | 0.99 | 0.88,1.10 | 0.80 | 0.81 | 0.72,0.91 | 0.0005 | 0.76 | 0.62,0.94 | 0.013 |
| miR-30a4 | 0.97 | 0.90,1.05 | 0.49 | -- | -- | -- | 0.86 | 0.77,0.97 | 0.016 | 1.03 | 0.90,1.19 | 0.66 |
| miR-7204 | 0.92 | 0.85,1.01 | 0.077 | 0.89 | 0.79,1.01 | 0.077 | 0.87 | 0.76,1.01 | 0.06 | 0.91 | 0.77,1.07 | 0.26 |
In the multivariable analyses, the Cox regression models were stratified on treatment arm to accommodate any inherent differences in PFS or OS attributable to treatment arm
Failed assumptions of proportional hazards and adjusted for non-proportionality of hazards either by hypothesis test or by graphical interpretation of the Shoenfeld residuals
High positive correlation and issues of strong multicollinearity between miR-25 and miR-16 necessitated only including miR-25 in the model, which was the stronger of the two (r = 0.75, p<0.000001)
Only failed proportional hazards assumption for OS, but not for PFS
ISS=International Staging System for multiple myeloma, ranging from 1 to 3; for this univariate model, ISS was treated as an ordinal variable. All serum miRs were log-transformed continuous measures. Hazard ratio >1 indicates that those with this characteristic are at increased risk of the event; i.e. they tend to have shorter progression free survival (PFS) and overall survival (OS).
There was a statistically significant relationship between the presence of deletion 17 on FISH and both PFS (HR 2.12, p=0.005) and OS (HR 2.13, p=0.02). We noted that serum creatinine was highly significant for OS in the univariate setting (p=0.008). We converted serum creatinine levels to the more clinically meaningful estimated glomerular filtration rate (GFR) using the MDRD equation and thus adjusting for age, gender, and race. GFR was not significantly associated with OS (p=0.15). Of interest, univariate analyses for OS showed a strong gender effect (HR 1.8, p=0.014), where women had a survival advantage over men in this cohort. However, upon further evaluation, we uncovered that there was also a very significant relationship between gender and serum creatinine levels (p<0.0001). This multicollinearity adds complexity to model building; despite this collinearity, both gender and serum creatinine were included in the multivariable model. Although not as “normalized” as the GFR measure we did not want to miss an effect of serum creatinine on OS in these patients.
To assess the independent prognostic impact of the miRNAs on PFS and OS duration, we evaluated several continuous serum miRNA markers in multivariable models that also adjusted for clinical and molecular factors of interest. When we evaluated S-miR-25 and S-miR-720 in relation to PFS when adjusting for the other identified significant factors (treatment arm, ISS stage, and del17), it was no longer significant for PFS (p=0.80). While not significant, S-miR-720 did maintain at least borderline significance for PFS (p=0.077).
In looking at OS, both S-miR-16 and S-miR-25 were each significantly associated with OS in the univariate setting. However, these two miRNA markers were also highly correlated (r = 0.75; p<0.00001); therefore, we carried forward only S-miR-25 to the multivariable model since it had a greater observed effect and proved to provide a better fit in the models than when S-miR-16 was included. miR-25 was also significantly correlated with miR-30a and miR-720 (Table A5), but these relationships were not as pronounced. To assess and identify a stable model, we evaluated the models with and without these correlated miRNA markers. Even when we included these additional miRNA markers in the model in addition to the clinical and molecular markers, S-miR-25 retained its significance as a prognostic factor for OS whereas the others did not (Table 1).
Overall, multivariable models for PFS and OS showed that serum miRNA markers had a significant or at least borderline significant role in determining prognosis. This association was more pronounced when looking at OS, where S-miR-25 remained a significant and relevant factor (p=0.013) even when adjusting for Del17, Del13, ISS stage, treatment arm, and even creatinine and gender (Table 1). The multivariable model for PFS had fewer significant prognostic factors overall, although S-miR-720 still retained at least borderline significance (p=0.077) even when ISS stage and Del17 were included in the model (Table 1). The impact of S-miR-25 on OS can be readily seen when these levels are dichotomized as high vs. low (using a median cutpoint from S-miR-25 levels of healthy normals; HR=1.66, p=0.029, Figure 1C).
In this analysis we found that gender and creatinine were both significant in the univariate setting. While the importance of gender has been rarely reported10, we discovered that creatinine and gender were collinear and MDRD, which depends on both gender and creatinine, was not an adequate composite variable. Creatinine is also a biomarker of more than renal function in this elderly population, varying by age, gender, weight, and often reflects muscle mass11.
Circulating miRNAs in MM are poorly understood. Their origin (cancer or normal cell), transit (actively secreted or derived at the time of apoptosis), and most influential location (within PBMCs, extracellular vesicles, or free floating) are all unknown. One report of circulating miRNA in MM identified let-7e and miR-744 as critical to differentiate from normal as well as correlate with overall survival, but this was based on a discovery set of just 13 patients12. A study of PBMCs from only 5 MM patients found numerous differences compared to 5 healthy controls13, but the small sample size limits generalizability.
We compared intracellular miR-16 and -25 expression in 32 paired samples of serum and PCs obtained patients with a new myeloma diagnosis, but there was no correlation, suggesting that circulating S-miR-16 and S-miR-25 do not reflect the miRNA content of myeloma cells in the marrow. By studying the biological significance of circulating miRNAs we found that miR-25 and miR-16 are encapsulated in extracellular vesicles but these miRNAs do not regulate their target genes post-transcriptionally, suggesting a different mechanism of regulation14. Additional work will be required to elucidate the mechanism of action of miR-25 and miR-16 in the extracellular compartment.
In conclusion, we show that circulating miR-16 and 25 are independent prognosticators in newly diagnosed MM of OS more so than PFS. We aim to validate these biomarkers in order to use them in combination with the ISS to better risk stratify MM patients.
Supplementary Material
Acknowledgments
We thank Dr Leif Bergsagel for careful reading of the manuscript and for helpful scientific discussion. We thank P. Fadda for the NanoString data (Nucleic Acid Shared Resource, the Ohio State University Comprehensive Cancer Center) and J. Corry for technical support. We also thank Octavia Bowers for the administrative support. Funding provided by: Kimmel (FP), Multiple Myeloma Opportunities for Research & Education (MMORE) (FP), Programma di Rilevante Interesse Nazionale 2009, Ricerca Sanitaria sulle Malattie Rare 2008. This work was supported by the Ohio State University Pelotonia Fellowship Program (R.A.) and Pelotonia Idea Grant (F.P.)
Footnotes
None of the authors have a relevant conflict of interest to report.
Supplementary information is available at Leukemia’s website.
Authorship roles: AR, FP: Designed the study, analyzed the data, performed the experiments and wrote the manuscript. TT, VD, JC, AS, JG: Performed the experiments. SG: performed statistical analysis, analyzed the data, wrote the manuscript. GM, CCH: designed the study, analyzed the data and wrote the manuscript. LC: analyzed Nanostring data. OP, BS, IG, RD, GS, PM, UG, RR, BG, CV, MV, BM, AP recruited the patients and designed study. BW, DMB, YAE, JCB: support the analysis of clinical data. All the authors have contributed substantially to the analysis and interpretation of data, revising the manuscript for important content, and final approval.
References
- 1.Kumar SK, Uno H, Jacobus SJ, Van Wier SA, Ahmann GJ, Henderson KJ, et al. Impact of gene expression profiling-based risk stratification in patients with myeloma receiving initial therapy with lenalidomide and dexamethasone. Blood. 2011 Oct 20;118(16):4359–4362. doi: 10.1182/blood-2011-03-342089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2008 Sep 2;105(35):12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lionetti M, Agnelli L, Mosca L, Fabris S, Andronache A, Todoerti K, et al. Integrative high-resolution microarray analysis of human myeloma cell lines reveals deregulated miRNA expression associated with allelic imbalances and gene expression profiles. Genes Chromosomes Cancer. 2009 Jun;48(6):521–531. doi: 10.1002/gcc.20660. [DOI] [PubMed] [Google Scholar]
- 4.Lionetti M, Biasiolo M, Agnelli L, Todoerti K, Mosca L, Fabris S, et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood. 2009 Dec 10;114(25):e20–26. doi: 10.1182/blood-2009-08-237495. [DOI] [PubMed] [Google Scholar]
- 5.Palumbo A, Bringhen S, Rossi D, Cavalli M, Larocca A, Ria R, et al. Bortezomib-Melphalan-Prednisone-Thalidomide Followed by Maintenance With Bortezomib-Thalidomide Compared With Bortezomib-Melphalan-Prednisone for Initial Treatment of Multiple Myeloma: A Randomized Controlled Trial. Journal of Clinical Oncology 2010. 2010 Dec 1;28(34):5101–5109. doi: 10.1200/JCO.2010.29.8216. [DOI] [PubMed] [Google Scholar]
- 6.Pichiorri F, Okumura H, Nakamura T, Garrison PN, Gasparini P, Suh SS, et al. Correlation of fragile histidine triad (Fhit) protein structural features with effector interactions and biological functions. J Biol Chem. 2009 Jan 9;284(2):1040–1049. doi: 10.1074/jbc.M806638200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pichiorri F, Palmieri D, De Luca L, Consiglio J, You J, Rocci A, et al. In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation. The Journal of experimental medicine. 2013 May 6;210(5):951–968. doi: 10.1084/jem.20120950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grambsch PM, Therneau TM. Proportional hazards Tests and Diagnostics Based on Weighted Residuals. Biometrika. 1994;81(3):515–526. [Google Scholar]
- 9.Schemper M, Wakounig S, Heinze G. The estimation of average hazard ratios by weighted Cox regression. Statistics in medicine. 2009 Aug 30;28(19):2473–2489. doi: 10.1002/sim.3623. [DOI] [PubMed] [Google Scholar]
- 10.Boyd KD, Ross FM, Chiecchio L, Dagrada G, Konn ZJ, Tapper WJ, et al. Gender disparities in the tumor genetics and clinical outcome of multiple myeloma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2011 Aug;20(8):1703–1707. doi: 10.1158/1055-9965.EPI-11-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung SH, Yang DH, Ahn JS, Lee SS, Ahn SY, Kim YK, et al. Decreased body mass index is associated with poor prognosis in patients with multiple myeloma. Annals of hematology. 2013 Dec 6; doi: 10.1007/s00277-013-1977-9. [DOI] [PubMed] [Google Scholar]
- 12.Kubiczkova L, Kryukov F, Slaby O, Dementyeva E, Jarkovsky J, Nekvindova J, et al. Circulating serum microRNAs as novel diagnostic and prognostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. Haematologica. 2013 Nov 15; doi: 10.3324/haematol.2013.093500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campo S, Allegra A, D’Ascola A, Alonci A, Scuruchi M, Russo S, et al. MiRNome expression is deregulated in the peripheral lymphoid compartment of multiple myeloma. British journal of haematology. 2014 Mar 12; doi: 10.1111/bjh.12828. [DOI] [PubMed] [Google Scholar]
- 14.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proceedings of the National Academy of Sciences of the United States of America. 2012 Jul 31;109(31):E2110–2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.