Abstract
Epithelial–mesenchymal transition (EMT) has been thought to occur during early embryogenesis, and also the differentiation process of human embryonic stem (hES) cells. Spontaneous differentiation is sometimes observed at the peripheral of the hES cell colonies in conventional culture conditions, indicating that EMT occurs in hES cell culture. However, the triggering mechanism of EMT is not yet fully understood. The balance between self-renewal and differentiation of human pluripotent stem (hPS) cells is controlled by various signal pathways, including the fibroblast growth factor (FGF)-2. However, FGF-2 has a complex role for self-renewal of hES cells. FGF-2 activates phosphatidylinositol-3 kinase/AKT, mitogen-activated protein kinase/extracellular signal-regulated kinase-1/2 kinase, and also protein kinase C (PKC). Here, we showed that a PKC rapidly induced an early growth response protein-1 (EGR-1) in hES cells, which was followed by upregulation of EMT-related genes. Before the induction of EMT-related genes, EGR-1 was translocated into the nucleus, and then bound directly to the promoter region of SNAIL, which is a master regulator of EMT. SNAIL expression was attenuated by knockdown of EGR-1, but upregulated by ectopic expression of EGR-1. EGR-1 as the downstream signal of PKC might play a key role in EMT initiation during early differentiation of hES cells. This study would lead to a more robust understanding of the mechanisms underlying the balance between self-renewal and initiation of differentiation in hPS cells.
Introduction
The balance between self-renewal and differentiation of human pluripotent stem (hPS) cells, including embryonic stem (hES) and induced pluripotent stem cells, is controlled by various signal pathways [1–8], including transforming growth factor-β/activin A/Nodal [9–11], sphingosine-1-phosphate/platelet-derived growth factor [12], insulin growth factor (IGF)/insulin [13], and fibroblast growth factor-2 (FGF-2) [14–17]. In particular, FGF-2 signaling appears indispensable to maintain the undifferentiated state of hPS cells [18–20]. However, FGF-2 has a complex role for self-renewal of hES cells [21]. FGF-2 activates phosphatidylinositol-3 kinase (PI3K)/AKT and mitogen-activated protein kinase/extracellular signal-regulated kinase-1/2 (ERK-1/2) kinase. The PI3K/AKT pathway plays important roles in the proliferation and survival of hPS cells [22–24]. On the other hand, the ERK-1/2 pathway has reported to have diverse roles in hPS cells, such as self-renewal, cell attachment, and cell differentiation [2,25–27]. FGF-2 also induces inactive phosphorylation of glycogen synthase kinase-3 (GSK-3) [1,3,4,28] of which function has been reported to be controversial for self-renewal of hPS cells [29,30]. Moreover, we have recently reported that FGF-2 induces protein kinase C (PKC), which leads to phosphorylation of GSK-3β [4]. An activator of PKC, phorbol 12-myristate 13-acetate (PMA) changes the sharp-edged, flat, and tightly packed colonies of hPS cells to scattered cells, indicating that PKC induces the epithelial–mesenchymal transition (EMT) in hPS cells [4]. Consistent with our findings, Thomson's group reported that activation of PKC is associated with EMT, leading the hPS cell differentiation into extraembryonic endodermal (ExEn) cells [5]. However, the downstream of the PKC pathway in regulating the EMT initiation is not yet understood.
In early embryogenesis, the EMT plays crucial roles in the differentiation of multiple tissues and organs and the formation of a body plan [31]. During the EMT process, E-cadherin is downregulated. Several transcription factors have been implicated in this repression, including SNAIL, SLUG, and TWIST1 [32–37]. Actually, in hPS cell culture, spontaneous differentiation is sometimes observed with the expression of SNAIL and VIMENTIN at the peripheral of the hPS cell colonies in conventional culture conditions, indicating that EMT occurs in hPS cell culture [38–41].
In this study, we investigated molecules involved in the cascade of PKC-induced EMT in hES cells using the minimal growth factor-defined culture medium hESF9, which includes FGF-2 as the sole growth factor to remove the effects of other growth factors on EMT [17]. The results showed that the early growth response protein-1 (EGR-1; also known as NGFI-A, KROX-24, ZIF268, or TIS8) was induced by a PKC activator, PMA, and bound to the regulatory region of SNAIL, resulting in the upregulation of SNAIL and EMT-related genes. PMA-induced SNAIL expression was attenuated by knockdown of EGR-1, whereas ectopic expression of EGR-1 induced EMT-related genes expression. These results indicated that a downstream effector of PKC signaling, EGR-1, contributed to the induction of EMT in hES cell differentiation. This study would lead to a more robust understanding of the mechanisms underlying the balance between self-renewal and initiation of differentiation in hPS cells.
Materials and Methods
Cell culture
The hES cell line, H9 [19,42] (WA09, WISC Bank; WiCell Research Institute), was routinely maintained as previously described [19]. For the experiment, the cells were seeded on a six-well plate (BD Falcon) coated with bovine fibronectin (FN; Sigma; 2 μg/cm2) in the hESF9 medium [17] consisting of the ESF basal medium (CSTI) [43] without 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid supplemented with l-ascorbic acid-2-phophate (Wako), 2-mercaptoethanol, 2-ethanolamine, sodium selenite, insulin, transferrin, oleic acid conjugated with bovine fatty acid-free albumin, heparan sulfate sodium salt (all from Sigma), and FGF-2 (Katayama Kagaku Kogyo Ltd.). PMA dissolved in dimethyl sulfoxide (DMSO) was added into the medium at a final concentration of 10 nM (containing a final concentration of 0.1% DMSO). The experiments using hES cells were performed following the Guidelines for utilization of hES cells of the Ministry of Education, Culture, Sports, Science and Technology of Japan with the approval by the institutional research ethics committee.
Immunocytochemistry
Immunocytochemistry was performed as previously described [4]. The image analysis was performed by IN Cell Analyzer 2000 and IN Cell Developer Toolbox software (GE Healthcare). The primary and secondary antibodies used are listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/scd).
Real-time quantitative reverse transcription–polymerase chain reaction
Real-time quantitative reverse transcription–polymerase chain reaction (qRT-PCR) and real-time quantitative PCR (qPCR) were performed based on the SYBR Green gene expression technology in a 7300 Real Time PCR System (Applied Biosystems), according to the manufacturer's instructions. Specific primers used are listed in Supplementary Table S2.
DNA microarray
DNA microarray analysis was performed using the whole human genome DNA microarray 4x44K kit (ver.2.0) and a microarray scanner G2565BA (Agilent) according to the manufacturer's instructions (Agilent). The signal intensity data produced for each of the spots were analyzed using feature extraction (Agilent) and GeneSpring GX software (Agilent).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed using the ChIP-IT Express kit (Active Motif) according to the manufacturer's instructions. Chromatin was precipitated with EGR-1 antibodies (Cell Signaling Technology) or H3K9ac antibodies (MAB Institute). The immunoprecipitated DNA samples were analyzed by qPCR. The SNAIL promoter was amplified with the primer pairs listed in Supplementary Table S2.
Construction of EGR-1 expression vector
The EGR-1 expression vector was constructed as follows. The EGR1-2A-eGFP fragment coding EGR-1 (NM_001964.2), a self-cleaving 2A peptide [44], and the enhanced green fluorescent protein (eGFP) were synthesized by the GeneArt gene synthesis service (Life Technologies). The synthesized fragment was inserted into the XhoI and NotI sites of the episomal pEBmulti-Hyg plasmid (Wako Pure Chemical Industries) to create the pEGR1-2A-eGFP plasmid. To generate the peGFP control vector, the pEGR1-2A-eGFP plasmid was digested with XhoI and SalI restriction enzymes, and the EGR1 fragment was removed from the pEGR1-2A-eGFP plasmid. The plasmids were verified by sequencing.
Transfection
Before transfection, the hES cells were incubated with ROCK inhibitor Y-27632 (10 μM) for 1 h and dissociated into single cells by a cell dissociation solution, TrypLE™ Select (Life Technologies). Pellets of 1×106 cells were mixed with 2 μg of the EGR-1 expression plasmid (pEB-EGR1-2A-eGFP) or control plasmid (peGFP) in 100 μL of Neon R buffer solution (Life Technologies). The cell suspension was transferred to a cuvette and electroporated using a Neon Transfection system (Life Technologies) with program (voltage 1,050, width 30, and pulses 2) according to the manufacturer's protocol. The electroporated cells were plated onto a six-well plate coated with FN in the hESF9 medium supplemented with ROCK inhibitor (10 μM). The eGFP-positive cells were selected on hygromycin B (200 μg/mL) for 6 days in the hESF9 medium.
Transfections with siRNA
Transfections with siRNA targeting human EGR-1 (SMARTpool ON-TARGETplus, L-006526-00) or nontargeting control siRNA (ON-TARGETplus Non-targeting Pool, D-001810-10) were performed using Dharmafect1 (Dharmacon) as previously described [4]. Total RNAs or proteins were extracted for analysis 72 h after the fast transfection.
Western blot analyses
Western blot analyses were performed as described previously [4]. The protein was separated by 12.5% sodium dodeyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore). The membranes were reacted with primary antibodies, peroxidase-conjugated secondary antibodies, and ECL Plus reagent (GE Healthcare). Protein bands were visualized using the LAS-4000 imager (Fujifilm). The primary antibodies used are listed in the Supporting Information Table S2.
Cell imaging analysis
The images of eGFP-positive cells in culture were captured in a cell imaging system, BioStation CT (Nikon Instruments, Inc.) at 37°C 10% CO2. The images were analyzed by a software CL-Quant (Nikon Instruments, Inc.).
Results
EMT induction of hES cells by PMA
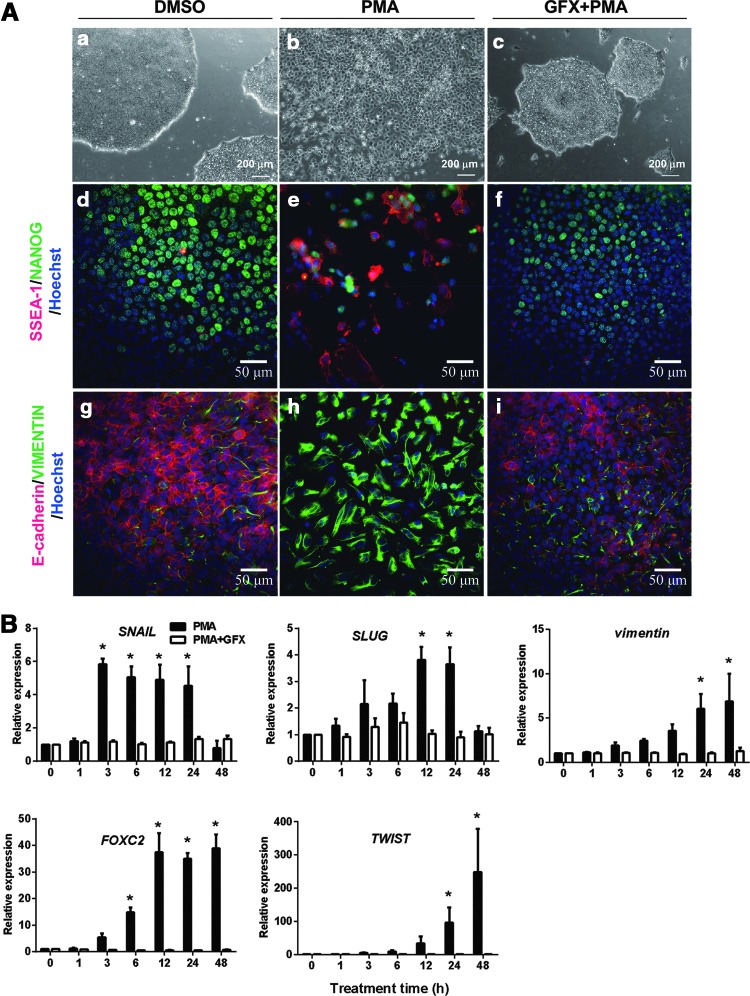
To confirm whether a PKC activator, PMA, induces EMT in hES cells, PMA was added into the culture of H9 hES cells grown in the defined culture conditions. The colony morphology of H9 cells was changed within 24 h after PMA addition. Compared with the control (Fig. 1A[a]), the packed colonies of undifferentiated H9 hES cells came loose and flatter cells scattered out (Fig. 1A[b]). PMA addition together with the PKC inhibitor GFX did not change the colony morphology of H9 hES cells (Fig. 1A[c]). The scattered cells were positive for SSEA-1 and VIMENTIN, and negative for NANOG or E-cadherin, and vice versa for the cells treated with PMA together with GFX (Fig. 1A[d–i]). These results indicated that the activation of PKC induced the EMT process.
FIG. 1.
Phorbol 12-myristate 13-acetate (PMA)-induced epithelial–mesenchymal transition (EMT) process in human embryonic stem (hES) cells. (A) Phase-contrast images and immunostaining of H9 hES cells 24 h after addition of PMA (10 nM) with or without GFX (5 μM) in the defined medium hESF9. (a–c) Phase-contrast images of H9 cells. (a) The cells in the presence of dimethyl sulfoxide (DMSO) (control). (b) The cells treated with PMA. (c) The cells treated with PMA and GFX. Scale bars=200 μm. (d–f) Immunostaining for NANOG (green) and SSEA-1 (red) of the cells with DMSO (d), PMA (e), and PMA and GFX (f). (g–i) Immunostaining for E-cadherin (red) and VIMENTIN (green) of the cells with DMSO (g), PMA (h), and PMA and GFX (i). Nuclei were stained with Hoechst 33342 (blue). Scale bars=50 μm. (B) Quantitative reverse transcription–polymerase chain reaction (qRT-PCR) analysis of the EMT-related gene expression. The cells were treated with PMA (black bars) or PMA in the presence of GFX (white bars) in the hESF9 medium for 48 h. Expression levels were all normalized against GAPDH. The relative expression levels of each gene were calculated from the undifferentiated H9 hES cells untreated with PMA or PMA together with GFX. The data are represented as mean±standard error (SE, n=3). *P<0.05.
To determine whether PMA promotes the expression of EMT-related genes or lineage-specific cell differentiation-associated genes, H9 hES cells were cultured in the presence of PMA with or without GFX for 48 h, and then for 4 more days after changing to a fresh medium without PMA or GFX (Supplementary Fig. S1A). qRT-PCR analysis showed that the expression of SNAIL rapidly increased 3 h after PMA treatment (Fig. 1B). After the SNAIL expression, the expression of SLUG, VIMENTIN, FOXC2, and TWIST significantly increased in a time-dependent manner in the cells treated with PMA. After the gene expression, the expression of an ExEn marker gene SOX7 and endoderm marker genes SOX17, EOMES, GATA4, and GATA6 was significantly increased in the cells (Supplementary Fig. S1B, C). On the contrary, the expression of a primitive streak marker gene GSC (Supplementary Fig. S1C) or ectoderm genes MSI1, SOX9, MAP2, Nestin, and PAX3 was not induced by PMA (Supplementary Fig. S1D). PMA addition together with GFX did not change the expression profile of EMT-related genes or cell differentiation marker genes. These results confirmed that the activation of PKC induced EMT-related genes and led to ExEn differentiation.
The expression of EGR-1
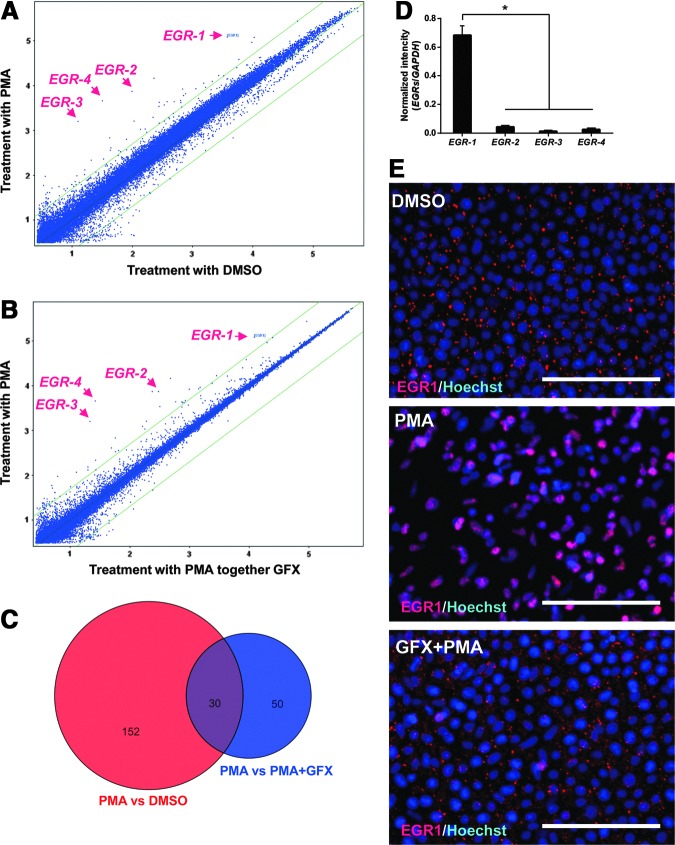
To determine PKC downstream molecules involved in the EMT initiation, differences in gene expression profiles among the cells treated with DMSO (control), PMA, or PMA together with GFX were analyzed by using DNA microarrays (Fig. 2A, B). The cells were treated with PMA for 1 h, in which time period, the SNAIL expression was before increase as shown in Fig. 1B. We previously reported that GFX, which is a selective inhibitor of PKC-α, β, γ, δ, and ζ negated PMA-induced differentiation of hPS cells, whereas Gö6976, which is a selective inhibitor of PKC-α, β, and γ isoforms could not counteract the effect of PMA [4]. Therefore, among genes influenced by PMA (fold change ≥5.0), genes of which expression were different between the cells treated PMA or together with GFX (fold change ≥5.0) were considered to be involved in EMT (Fig. 2C). Accordingly, the analysis nominated 26 activated genes and 2 repressed genes as the candidates for PKC downstream genes (Supplementary Table S3). Among them, we found that a zinc finger transcription factor EGR gene family members, EGR-1, EGR-2, EGR-3, and EGR-4 were increased in the PMA-treated cells. The expression levels of EGR-1 were significantly higher in the EGR gene family members (Fig. 2D). Therefore, we focused on EGR-1, which is reported to be involved in the EMT process in epithelial cells [45,46].
FIG. 2.
Early growth response protein-1 (EGR-1) increases in the hES cells treated with PMA. (A) DNA microarray analysis of H9 hES cells treated with or without PMA for 1 h. (B) DNA microarray analysis of H9 hES cells treated with PMA or PMA together with GFX for 1 h. (C) Venn diagram of probes (purple) shared by the PMA-influenced probes (red) and GFX-influenced probes (blue) in DNA microarray analysis. Values in Venn diagram indicated the number of probes changed (fold change ≥5.0) by DNA microarray analysis at (A) and (B). (D) The fluorescence intensity of each spot of EGR genes in the microarray was normalized against the fluorescence intensity of GAPDH gene. The data are represented as mean±standard error (SE, n=3). *P<0.05. (E) Immunostaining for EGR-1 (Alexa fluor 647; red) in H9 hES cells treated with PMA (10 nM) or PMA together with GFX (5 μM). Nuclei were stained with Hoechst 33342 (blue). Scale bars=50 μm.
To confirm the expression of EGR-1 in the cells treated with PMA, immunocytochemistry analysis was performed using the anti-EGR-1 antibody (Fig. 2E). The result indicated EGR-1 protein expression in the cells 1 h after PMA treatment and accumulation of EGR-1 in the nucleus. EGR-1 protein expression was at low levels in the cells treated with PMA together with GFX. From these results, it was confirmed that EGR-1 expression was induced by PMA in hES cells.
Relation between EGR-1 and SNAIL
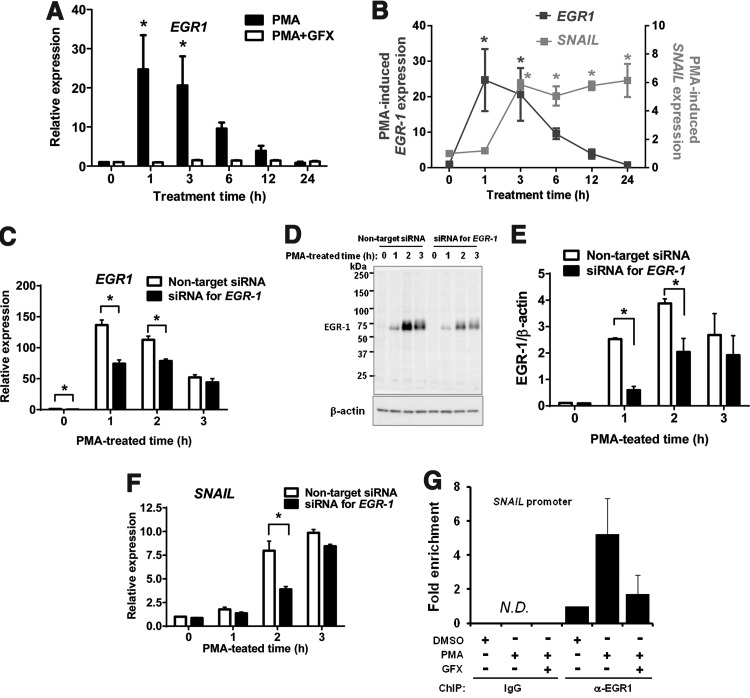
To investigate the induced expression level of EGR-1 in the cells treated with PMA, qRT-PCR analysis was performed. The results showed that the EGR-1 expression peaked at 1 h after PMA treatment (Fig. 3A) and the induction of SNAIL expression peaked at 3 h after PMA treatment (Fig. 3B).
FIG. 3.
PMA-induced EGR-1 expression in hES cells. (A) qRT-PCR analysis of EGR-1 expression in the cells treated with PMA (black bars) or PMA together with GFX (white bars) for 24 h. (B) qRT-PCR analysis of EGR-1 (black square) or SNAIL (gray square) expression in the cells treated with PMA for 24 h. Expression levels were all normalized against GAPDH. The relative expression levels of each gene were calculated from the undifferentiated H9 hES cells untreated with PMA or PMA together with GFX. The data are represented as mean±SE (n=3). *P<0.05. (C) qRT-PCR analysis of PMA-induced EGR-1 expression in the cells transfected with siRNA targeting EGR-1 (black bars) or nontarget control (white bars). Cells were treated with PMA for 3 h. Expression levels were all normalized against GAPDH. The relative expression levels of EGR-1 were calculated from the nontarget siRNA-transfected H9 hES cells. The data are represented as mean±SE (n=3). *P<0.05. (D) Western blot analysis of PMA-induced EGR-1 expression in the cells transfected with siRNA targeting EGR-1 or nontarget control. Cells were treated with PMA for 3 h. (E) Quantification of the western blot result at (D). Cells were transfected with siRNA targeting EGR-1 (black bars) or nontarget control (white bars). The data are represented as mean±SE (n=3). *P<0.05. (F) qRT-PCR analysis of PMA-induced SNAIL expression in the cells transfected with siRNA targeting EGR-1 (black bars) or nontarget control (white bars). Cells were treated with PMA for 3 h. Expression levels were all normalized against GAPDH. The relative expression levels of SNAIL were calculated from the nontarget siRNA-transfected H9 hES cells. The data are represented as mean±SE (n=3). *P<0.05. (G) Chromatin immunoprecipitation (ChIP) assay. The cells were treated with PMA (10 nM) or PMA together with GFX (5 μM) for 1 h. N.D., not detected.
From these data above, it was predicted that the PMA-induced EGR-1 might directly regulate the induction of SNAIL. To determine the relationship between EGR-1 and SNAIL, the cells were transfected with siRNA targeting EGR-1. When the cells were treated with PMA for 3 h, the expression of PMA-induced EGR-1 mRNA (Fig. 3C) and its protein levels (Fig. 3D, E) were significantly reduced at 1–2 h PMA treatment time, and then SNAIL expression (Fig. 3F) was significantly reduced at 2 h PMA treatment time, in the cells transfected with siRNA of EGR-1 compared with those in the cells transfected with nontarget siRNA. Then, we searched the ChIP-on-chip data for EGR-1, provided by the FANTOM 4 database (http://fantom.gsc.riken.jp/4/) [47]. Data showed that EGR-1 binds to the region of the SNAIL promoter in human THP-1 cells (Supplementary Fig. S2A). Thus, ChIP analysis was performed to determine whether EGR-1 could actually bind to the promoter region of SNAIL, which contains the putative EGR-1 binding site in hES H9 cells (Supplementary Fig. S2B). The result showed that EGR-1 bound to the promoter region of SNAIL, and that the amounts of immunoprecipitated DNA with the EGR-1 antibody was increased by PMA treatment, but decreased by PMA treatment together with GFX (Fig. 3G). It is reported that EGR-1 is colocalized with histone H3 lysine 9 acetylation (H3K9ac) sites, which is tightly associated with the transcription start sites of genes [47]. Thus, H3K9ac is important for EGR-1 target site selection for gene activation. To confirm H3K9ac modification around the EGR-1 binding site in the SNAIL promoter, ChIP analysis was performed using the anti-H3K9ac antibody (Supplementary Fig. S2C). The result indicated that the EGR-1 binding region in the SNAIL promoter was colocalized with the H3K9ac site. These above results demonstrated that the PMA-induced EGR-1 was directly bound to the SNAIL promoter region and it resulted in the expression of SNAIL, suggesting that EGR-1 may play a role in EMT initiation during early differentiation of hES cells.
EMT induction by ectopic EGR-1
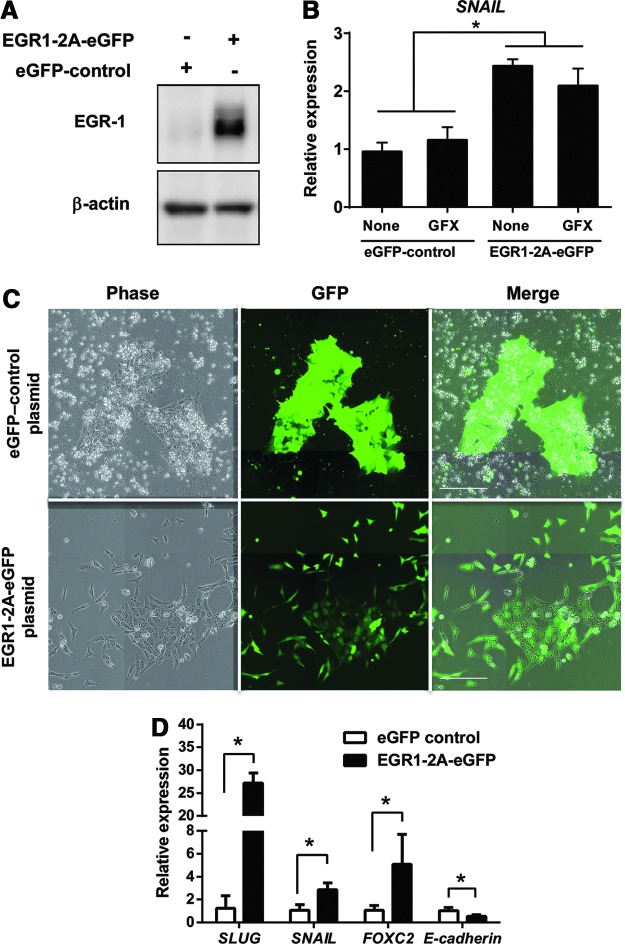
To confirm the function of EGR-1 as an EMT inducer in hES cells, an episomal vector carrying a CAG promoter-driven EGR1-2A-eGFP was transiently transfected into H9 cells. The expression of ectopic EGR-1 in H9 hES cells 24 h after the transfection was confirmed by western blotting analysis using the specific anti-EGR-1 antibody (Fig. 4A). The expression of SNAIL was significantly increased in the cells expressing ectopic EGR-1 with or without GF109203X (GFX) (Fig. 4B). Furthermore, the cells expressing ectopic EGR-1-2A-eGFP were selected on hygromycin B for 6 days, colonies became loose and scattered flatter cells. Control cell colonies maintained undifferentiated morphology (Fig. 4C). The result by qRT-PCR showed that the expression of EMT-related genes SNAIL, SLUG, and FOXC2 were significantly upregulated in the cells expressing ectopic EGR-1, but the expression of E-cadherin was significantly downregulated (Fig. 4D). These results indicated that the expression of EGR-1 induced EMT in hES cells.
FIG. 4.
EMT-related gene expression by ectopic EGR-1. (A) Western blot analysis of ectopic EGR-1 expression in the cells transfected with EGR1-2A-eGFP or with enhanced green fluorescent protein (eGFP) control plasmid. Transiently ectopic EGR-1 expression was confirmed by the specific EGR-1 antibody at 24 h after transfection. (B) qRT-PCR analysis of SNAIL in the cells expressing EGR1-2A-eGFP or eGFP alone in the presence or absence of GFX. Expression levels of the SNAIL gene were all normalized against GAPDH. The data are represented as mean±SE (n=3). *P<0.05. (C) Phase-contrast and fluorescence images of H9 hES cells transfected with pEGR1-2A-eGFP or peGFP control plasmid. The EGR1-2A-eGFP or eGFP alone expressing cells were selected on hygromycin B (200 μg/mL) in the hESF9 medium for 6 days. Scale bars=200 μm. (D) qRT-PCR analysis of the EMT-related genes. Expression levels of genes in the cells expressing EGR1-2A-eGFP (black bars) or eGFP alone (white bars) were all normalized against GAPDH. The data are represented as mean±SE (n=3). *P<0.05.
Discussion
In this study, we showed that PMA rapidly induced the expression of EGR-1 and accumulation of EGR-1 into the nucleus, which resulted in the expression of SNAIL in hES cells. PMA-induced SNAIL expression was attenuated by knockdown of EGR-1, whereas ectopic EGR-1 expression induced EMT-related genes expression, resulting in the induction of the EMT process in hES cells. Furthermore, EGR-1 may function in hES-specific lineage cell differentiation. The ChIP-on-ChIP database of EGR-1, provided by the FANTOM 4 database (http://fantom.gsc.riken.jp/4/) [47], showed that EGR-1 binds not only to the region of SNAIL promoter, but also to the promoter and intragenic regions of a number of genes. For example, EGR-1 binding sites are present in the promoter region of EMT-related genes; E-cadherin, TWIST, VIMENTIN, and MMP2, and extraembryonic and/or definitive endodermal cell lineage-associated genes; EOMES, GATA6, SOX7, FOXA2, and CXCR4 in human THP-1 cells (Supplementary Fig. S3). These findings are consistent with our results that PMA induced EMT and also cell differentiation into the extraembryonic endoderm in hPS cells.
The previous study reported that the SNAIL protein is phosphorylated by GSK-3β, and then the phosphorylated SNAIL protein binds to the promoter region of E-cadherin and downregulates the transcript of E-cadherin, leading to the EMT process [48]. These findings implied that the function of SNAIL protein induced by EGR-1 might be regulated by GSK-3β in the nucleus [48]. Although GSK-3β is generally considered to be a cytoplasmic protein, previous reports suggest that GSK-3β also functions in the nucleus [49–51]. It means that Wnt signaling promotes EMT in hPS cells. We have previously reported that phosphorylation of GSK-3β is induced by FGF-2-activated PKC and that this process might be related to hPS cell differentiation [4]. This is consistent with previous studies that canonical Wnt signaling does not appear to promote stem cell maintenance [52,53]. However, this conclusion contradicts the findings of previous studies, which demonstrated that canonical Wnt signaling supports self-renewal of stem cells [6,38]. A recent study has shown the dual function of Wnt signaling in hES cells, suggesting that the pathways of self-renewal or differentiation are dependent on the presence of hES cell supporting factors [7,30,54,55]. From these studies, GSK-3 has emerged as an important regulator of undifferentiated state in hPS cells.
GSK-3β has been reported to be controlled by various growth factors, including insulin, IGF, FGF-2, and WNT3A [4,22,29,56,57]. We reported that in hPS cells, FGF-2-induced GSK-3β phosphorylation is inhibited in the presence of the PKC inhibitor, resulting in the inhibition of the PKC/GSK-3β signaling pathway to support the maintenance of an undifferentiated state [4]. From the findings above, it is implied that FGF-2 induces the expression of EGR-1 through the PKC signaling pathway, EGR-1 induced the SNAIL protein, and then PKC/GSK-3β activates SNAIL, leading to the EMT process [48] (Fig. 5). However, if FGF-2 continues to amplify the EMT process, hPS cells should rapidly differentiate. Inhibitory signaling related to the dual function of GSK-3β might exist in hPS cell culture and it can balance the FGF-2-induced EMT process to maintain the undifferentiated state although the PKC inhibitor can artificially stabilize the balance between undifferentiated state and cell differentiation. Further investigation of mechanisms involved in the regulation of PKC/EGR1/SNAIL and PKC/GSK-3β activity should be performed in future.
FIG. 5.
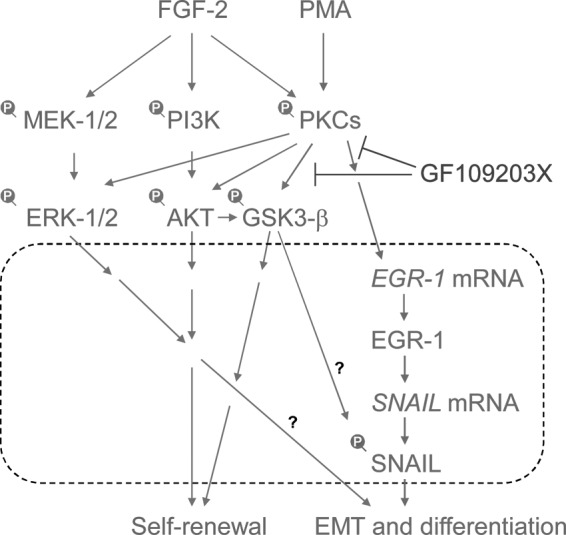
A model of EMT-triggering pathways using protein kinase C (PKC) in fibroblast growth factor (FGF-2) signaling. PMA induces PKCs, and then induces EGR-1. EGR-1 directly induces the expression of SNAIL mRNA, leading to the EMT process [48]. GF109203X, a selective inhibitor of PKCs, negated the PKC-induced EGR-1/SNAIL expression, EMT process, and differentiation of human pluripotent stem cells. PKC-EGR-1-SNAIL pathways promote EMT and cell differentiation. Dot line box indicates the process in the nucleus.
This study clarified that PKC activation induced the EMT process through the EGR-1/SNAIL pathway suggesting that EGR-1 plays a role in the differentiation of hES cells. This finding would lead to a better understanding of the precise mechanism regulating the balance between undifferentiated state and cell differentiation in hPS cells.
Supplementary Material
Acknowledgments
This study was supported by grants-in-aid from the Ministry of Health, Labor and Welfare of Japan to M.K.F., the Ministry of Education, Culture, Sports, Science and Technology of Japan to M.K.F. and M.K., and the New Energy and Industrial Technology Development Organization (NEDO) of Japan to M.K.F. We thank Hiroko Matsumura, Ayaka Fujiki, Naoko Ueda, Yujung Liu, Daiki Tateyama, and Minako Okamura for excellent technical support, and Takayuki Fukuda and Kana Yanagihara for technical support and discussion.
Author Disclosure Statement
All the authors have read and approved the article, and hereby declare that none of them has any competing interest.
References
- 1.Dreesen O. and Brivanlou AH. (2007). Signaling pathways in cancer and embryonic stem cells. Stem Cell Rev 3:7–17 [DOI] [PubMed] [Google Scholar]
- 2.Na J, Furue MK. and Andrews PW. (2010). Inhibition of ERK1/2 prevents neural and mesendodermal differentiation and promotes human embryonic stem cell self-renewal. Stem Cell Res 5:157–169 [DOI] [PubMed] [Google Scholar]
- 3.Armstrong L, Hughes O, Yung S, Hyslop L, Stewart R, Wappler I, Peters H, Walter T, Stojkovic P, et al. (2006). The role of PI3K/AKT, MAPK/ERK and NFκβ signalling in the maintenance of human embryonic stem cell pluripotency and viability highlighted by transcriptional profiling and functional analysis. Hum Mol Genet 15:1894–1913 [DOI] [PubMed] [Google Scholar]
- 4.Kinehara M, Kawamura S, Tateyama D, Suga M, Matsumura H, Mimura S, Hirayama N, Hirata M, Uchio-Yamada K, et al. (2013). Protein kinase C regulates human pluripotent stem cell self-renewal. PLoS One 8:e54122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng X, Zhang J, Smuga-Otto K, Tian S, Yu J, Stewart R. and Thomson JA. (2012). Protein kinase C mediated extraembryonic endoderm differentiation of human embryonic stem cells. Stem Cells 30:461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato N, Meijer L, Skaltsounis L, Greengard P. and Brivanlou AH. (2004). Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med 10:55–63 [DOI] [PubMed] [Google Scholar]
- 7.Dravid G, Ye Z, Hammond H, Chen G, Pyle A, Donovan P, Yu X. and Cheng L. (2005). Defining the role of Wnt/β-catenin signaling in the survival, proliferation, and self-renewal of human embryonic stem cells. Stem Cells 23:1489–1501 [DOI] [PubMed] [Google Scholar]
- 8.Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, et al. (2011). BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 9:144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vallier L, Reynolds D. and Pedersen RA. (2004). Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev Biol 275:403–421 [DOI] [PubMed] [Google Scholar]
- 10.Vallier L, Alexander M. and Pedersen RA. (2005). Activin/nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci 118:4495–4509 [DOI] [PubMed] [Google Scholar]
- 11.James D, Levine AJ, Besser D. and Hemmati-Brivanlou A. (2005). TGFβ/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development 132:1273–1282 [DOI] [PubMed] [Google Scholar]
- 12.Pebay A, Wong RC, Pitson SM, Wolvetang EJ, Peh GS, Filipczyk A, Koh KL, Tellis I, Nguyen LT. and Pera MF. (2005). Essential roles of sphingosine-1-phosphate and platelet-derived growth factor in the maintenance of human embryonic stem cells. Stem Cells 23:1541–1548 [DOI] [PubMed] [Google Scholar]
- 13.Bendall SC, Stewart MH, Menendez P, George D, Vijayaragavan K, Werbowetski-Ogilvie T, Ramos-Mejia V, Rouleau A, Yang J, et al. (2007). IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature 448:1015–1021 [DOI] [PubMed] [Google Scholar]
- 14.Dvorak P, Dvorakova D, Koskova S, Vodinska M, Najvirtova M, Krekac D. and Hampl A. (2005). Expression and potential role of fibroblast growth factor 2 and its receptors in human embryonic stem cells. Stem Cells 23:1200–1211 [DOI] [PubMed] [Google Scholar]
- 15.Avery S, Inniss K. and Moore H. (2006). The regulation of self-renewal in human embryonic stem cells. Stem Cells Dev 15:729–740 [DOI] [PubMed] [Google Scholar]
- 16.Ding VM, Boersema PJ, Foong LY, Preisinger C, Koh G, Natarajan S, Lee DY, Boekhorst J, Snel B, et al. (2011). Tyrosine phosphorylation profiling in FGF-2 stimulated human embryonic stem cells. PLoS One 6:e17538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furue MK, Na J, Jackson JP, Okamoto T, Jones M, Baker D, Hata R, Moore HD, Sato JD. and Andrews PW. (2008). Heparin promotes the growth of human embryonic stem cells in a defined serum-free medium. Proc Natl Acad Sci U S A 105:13409–13414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman LM. and Carpenter MK. (2005). Characterization and culture of human embryonic stem cells. Nat Biotechnol 23:699–708 [DOI] [PubMed] [Google Scholar]
- 19.Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, Itskovitz-Eldor J. and Thomson JA. (2000). Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol 227:271–278 [DOI] [PubMed] [Google Scholar]
- 20.Xu RH, Peck RM, Li DS, Feng X, Ludwig T. and Thomson JA. (2005). Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat Methods 2:185–190 [DOI] [PubMed] [Google Scholar]
- 21.Eiselleova L, Matulka K, Kriz V, Kunova M, Schmidtova Z, Neradil J, Tichy B, Dvorakova D, Pospisilova S, Hampl A. and Dvorak P. (2009). A complex role for FGF-2 in self-renewal, survival, and adhesion of human embryonic stem cells. Stem Cells 27:1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh AM, Reynolds D, Cliff T, Ohtsuka S, Mattheyses AL, Sun Y, Menendez L, Kulik M. and Dalton S. (2012). Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell 10:312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Downward J. (2004). PI 3-kinase, Akt and cell survival. Semin Cell Dev Biol 15:177–182 [DOI] [PubMed] [Google Scholar]
- 24.Ohgushi M, Matsumura M, Eiraku M, Murakami K, Aramaki T, Nishiyama A, Muguruma K, Nakano T, Suga H, et al. (2010). Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell 7:225–239 [DOI] [PubMed] [Google Scholar]
- 25.Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP. and Thomson JA. (2002). BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol 20:1261–1264 [DOI] [PubMed] [Google Scholar]
- 26.Pera MF, Andrade J, Houssami S, Reubinoff B, Trounson A, Stanley EG, Ward-van Oostwaard D. and Mummery C. (2004). Regulation of human embryonic stem cell differentiation by BMP-2 and its antagonist noggin. J Cell Sci 117:1269–1280 [DOI] [PubMed] [Google Scholar]
- 27.Li J, Wang G, Wang C, Zhao Y, Zhang H, Tan Z, Song Z, Ding M. and Deng H. (2007). MEK/ERK signaling contributes to the maintenance of human embryonic stem cell self-renewal. Differentiation 75:299–307 [DOI] [PubMed] [Google Scholar]
- 28.Schlessinger J. (2004). Common and distinct elements in cellular signaling via EGF and FGF receptors. Science 306:1506–1507 [DOI] [PubMed] [Google Scholar]
- 29.Ding VM, Ling L, Natarajan S, Yap MG, Cool SM. and Choo AB. (2010). FGF-2 modulates Wnt signaling in undifferentiated hESC and iPS cells through activated PI3-K/GSK3β signaling. J Cell Physiol 225:417–428 [DOI] [PubMed] [Google Scholar]
- 30.Blauwkamp TA, Nigam S, Ardehali R, Weissman IL. and Nusse R. (2012). Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat Commun 3:1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thiery JP, Acloque H, Huang RY. and Nieto MA. (2009). Epithelial-mesenchymal transitions in development and disease. Cell 139:871–890 [DOI] [PubMed] [Google Scholar]
- 32.Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M. and Cano A. (2003). The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci 116:499–511 [DOI] [PubMed] [Google Scholar]
- 33.Kang Y. and Massague J. (2004). Epithelial-mesenchymal transitions: twist in development and metastasis. Cell 118:277–279 [DOI] [PubMed] [Google Scholar]
- 34.Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D. and van Roy F. (2001). The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell 7:1267–1278 [DOI] [PubMed] [Google Scholar]
- 35.Perez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, Nieto MA. and Cano A. (2001). A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J Biol Chem 276:27424–27431 [DOI] [PubMed] [Google Scholar]
- 36.Grooteclaes ML. and Frisch SM. (2000). Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene 19:3823–3828 [DOI] [PubMed] [Google Scholar]
- 37.Mani SA, Yang J, Brooks M, Schwaninger G, Zhou A, Miura N, Kutok JL, Hartwell K, Richardson AL. and Weinberg RA. (2007). Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci U S A 104:10069–10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ullmann U, Gilles C, De Rycke M, Van de Velde H, Sermon K. and Liebaers I. (2008). GSK-3-specific inhibitor-supplemented hESC medium prevents the epithelial-mesenchymal transition process and the up-regulation of matrix metalloproteinases in hESCs cultured in feeder-free conditions. Mol Hum Reprod 14:169–179 [DOI] [PubMed] [Google Scholar]
- 39.Ullmann U, In't Veld P, Gilles C, Sermon K, De Rycke M, Van de Velde H, Van Steirteghem A. and Liebaers I. (2007). Epithelial-mesenchymal transition process in human embryonic stem cells cultured in feeder-free conditions. Mol Hum Reprod 13:21–32 [DOI] [PubMed] [Google Scholar]
- 40.Van Hoof D, Braam SR, Dormeyer W, Ward-van Oostwaard D, Heck AJ, Krijgsveld J. and Mummery CL. (2008). Feeder-free monolayer cultures of human embryonic stem cells express an epithelial plasma membrane protein profile. Stem Cells 26:2777–2781 [DOI] [PubMed] [Google Scholar]
- 41.Eastham AM, Spencer H, Soncin F, Ritson S, Merry CL, Stern PL. and Ward CM. (2007). Epithelial-mesenchymal transition events during human embryonic stem cell differentiation. Cancer Res 67:11254–11262 [DOI] [PubMed] [Google Scholar]
- 42.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS. and Jones JM. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147 [DOI] [PubMed] [Google Scholar]
- 43.Furue M, Okamoto T, Hayashi Y, Okochi H, Fujimoto M, Myoishi Y, Abe T, Ohnuma K, Sato GH, Asashima M. and Sato JD. (2005). Leukemia inhibitory factor as an anti-apoptotic mitogen for pluripotent mouse embryonic stem cells in a serum-free medium without feeder cells. In Vitro Cell Dev Biol Anim 41:19–28 [DOI] [PubMed] [Google Scholar]
- 44.Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA. and Choi SY. (2011). High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6:e18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grotegut S, von Schweinitz D, Christofori G. and Lehembre F. (2006). Hepatocyte growth factor induces cell scattering through MAPK/Egr-1-mediated upregulation of Snail. EMBO J 25:3534–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuo PL, Chen YH, Chen TC, Shen KH. and Hsu YL. (2011). CXCL5/ENA78 increased cell migration and epithelial-to-mesenchymal transition of hormone-independent prostate cancer by early growth response-1/snail signaling pathway. J Cell Physiol 226:1224–1231 [DOI] [PubMed] [Google Scholar]
- 47.Kubosaki A, Tomaru Y, Tagami M, Arner E, Miura H, Suzuki T, Suzuki M, Suzuki H. and Hayashizaki Y. (2009). Genome-wide investigation of in vivo EGR-1 binding sites in monocytic differentiation. Genome Biol 10:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M. and Hung MC. (2004). Dual regulation of Snail by GSK-3β-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol 6:931–940 [DOI] [PubMed] [Google Scholar]
- 49.Bechard M. and Dalton S. (2009). Subcellular localization of glycogen synthase kinase 3β controls embryonic stem cell self-renewal. Mol Cell Biol 29:2092–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bechard M, Trost R, Singh AM. and Dalton S. (2012). Frat is a phosphatidylinositol 3-kinase/Akt-regulated determinant of glycogen synthase kinase 3β subcellular localization in pluripotent cells. Mol Cell Biol 32:288–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh AM, Bechard M, Smith K. and Dalton S. (2012). Reconciling the different roles of Gsk3β in “naive”. and “primed”. pluripotent stem cells. Cell Cycle 11:2991–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davidson KC, Adams AM, Goodson JM, McDonald CE, Potter JC, Berndt JD, Biechele TL, Taylor RJ. and Moon RT. (2012). Wnt/β-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proc Natl Acad Sci U S A 109:4485–4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sumi T, Tsuneyoshi N, Nakatsuji N. and Suemori H. (2008). Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/β-catenin, Activin/Nodal and BMP signaling. Development 135:2969–2979 [DOI] [PubMed] [Google Scholar]
- 54.Cai L, Ye Z, Zhou BY, Mali P, Zhou C. and Cheng L. (2007). Promoting human embryonic stem cell renewal or differentiation by modulating Wnt signal and culture conditions. Cell Res 17:62–72 [DOI] [PubMed] [Google Scholar]
- 55.Villa-Diaz LG, Pacut C, Slawny NA, Ding J, O'Shea KS. and Smith GD. (2009). Analysis of the factors that limit the ability of feeder cells to maintain the undifferentiated state of human embryonic stem cells. Stem Cells Dev 18:641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaidanovich-Beilin O. and Woodgett JR. (2011). GSK-3: functional insights from cell biology and animal models. Front Mol Neurosci 4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cohen P. and Frame S. (2001). The renaissance of GSK3. Nat Rev Mol Cell Biol 2:769–776 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.