Background: How antimicrobial histones participate in invertebrate defense was still unclear.
Results: Upon injury or infection, oyster immune cells release antimicrobial histones and extracellular DNA traps in a ROS-dependent manner.
Conclusion: DNA traps are involved in the defense of Lophotrochozoa. Their mechanistic bases are shared with vertebrates.
Significance: This is a novel mechanism in the evolutionary conserved invertebrate immune arsenal.
Keywords: Antimicrobial Peptide (AMP), DNA, Innate Immunity, Invertebrate, Reactive Oxygen Species (ROS), NET, Mollusk
Abstract
Although antimicrobial histones have been isolated from multiple metazoan species, their role in host defense has long remained unanswered. We found here that the hemocytes of the oyster Crassostrea gigas release antimicrobial H1-like and H5-like histones in response to tissue damage and infection. These antimicrobial histones were shown to be associated with extracellular DNA networks released by hemocytes, the circulating immune cells of invertebrates, in response to immune challenge. The hemocyte-released DNA was found to surround and entangle vibrios. This defense mechanism is reminiscent of the neutrophil extracellular traps (ETs) recently described in vertebrates. Importantly, oyster ETs were evidenced in vivo in hemocyte-infiltrated interstitial tissues surrounding wounds, whereas they were absent from tissues of unchallenged oysters. Consistently, antimicrobial histones were found to accumulate in oyster tissues following injury or infection with vibrios. Finally, oyster ET formation was highly dependent on the production of reactive oxygen species by hemocytes. This shows that ET formation relies on common cellular and molecular mechanisms from vertebrates to invertebrates. Altogether, our data reveal that ET formation is a defense mechanism triggered by infection and tissue damage, which is shared by relatively distant species suggesting either evolutionary conservation or convergent evolution within Bilateria.
Introduction
Histones play an essential role in the organization and architecture of the chromatin, and their post-translational modifications are essential to gene regulation (1). Since 1942 (2), histones have been shown to display antimicrobial activities against bacteria, fungi, viruses, and protozoa (3). In Protostomia, antimicrobial histones have been isolated from the shrimp Litopenaeus vannamei (4), the scallop Chlamys farreri (5), the abalone Haliotis discus discus (6), and recently the oyster Crassostrea virginica (7, 8). However, the mechanisms facilitating histone release, which is a prerequisite for their antimicrobial activities on potential pathogens, has long remained unidentified.
In 2004, a new antimicrobial mechanism relying on the release, by mammalian neutrophils, of extracellular DNA-carrying histones and granular antimicrobial proteins bound to the decondensed nucleic acids was uncovered (9). More recently, those extracellular traps (ETs)5 have been observed to form massively in infected tissues by intravital microscopy, demonstrating further their role in host defense (10). ETs can be released in response to bacteria, fungi, parasites, and viruses (9, 11, 12), to microbe-associated molecular patterns such as lipopolysaccharide (LPS), and to host inflammatory signals associated with tissue damage such as interleukin-8 (9) and tumor necrosis factor (13). ETs were reported to entrap bacteria, fungi, and parasites (9, 11, 12) and to kill them by their content in antimicrobial peptides/proteins including histones, bactericidal permeability-increasing proteins, and hydrolases (9, 12, 14–16). However, some bacteria such as Streptococcus pneumoniae, Mycobacterium tuberculosis, and group A Streptococci are found entrapped into ETs without being killed (17, 18). ETs can then play two important roles in the control of infections, first by entrapping microbes and preventing their dissemination, and second by concentrating antimicrobials and potentially killing microbes (10, 16, 19).
Although ETs have been well studied in vertebrates (Deuterostomia) over the past decade, studies on invertebrates have remained sparse and limited to arthropods (Ecdysozoa, Protostomia). In 2008, a first report suggested that extracellular nucleic acids enhance immunity and induce hemolymph coagulation in Galleria mellonella (20). More recently, an in vitro study showed that hemocytes from the shrimp L. vannamei release ETs able to entrap bacteria upon challenge with LPS, phorbol myristate acetate (PMA), or bacteria (21). To the best of our knowledge, clear evidences of ETs formation in vivo and characterization of the underlying mechanisms have not been reported yet in any invertebrate.
Here we performed a comprehensive study on DNA extracellular traps in the defense of a lophotrochozoan, the oyster Crassostrea gigas. Our work reveals that C. gigas hemocytes form ETs associated with antimicrobial histones both in vitro and in vivo, in response to infections and tissue damage, and that these ETs can entrap bacteria. By using a quantitative approach, we also show that similar to vertebrate neutrophils, oyster hemocytes require the production of reactive oxygen species to release ETs. Altogether, our data reveal that C. gigas, a lophotrochozoan, uses ET formation as defense mechanism that can be triggered by infection and tissue damage. From this study, this defense mechanism is shared by distant species among the main branches of the Bilateria.
EXPERIMENTAL PROCEDURES
Cationic Protein Extraction from C. gigas Tissues
C. gigas adult oysters were carved in the dorsal side of the shell with a small notch and acclimated for 5 days in seawater tanks. Then, 16 oysters were challenged by injection in the adductor muscle of 100 μl of Vibrio tasmaniensis LGP32 (1 × 107 CFU/oyster), an oyster pathogen (22) recently assigned to V. tasmaniensis within the Splendidus clade (23). After 24 h, gills were dissected, frozen at −80 °C, and ground to fine powder. Gill powder was resuspended in 5% acetic acid and a mixture of protease inhibitors (Sigma). After sonication, proteins were acid-extracted for 3 h at 4 °C and centrifuged twice at 13,000 × g, 4 °C, 30 min. pH was adjusted to 6.8 before the addition of a cation exchange resin (CM Macro-Prep, Bio-Rad). After overnight incubation at 4 °C, the resin was washed with 25 mm ammonium acetate, pH 6.8, and proteins were eluted twice with 1% TFA in ultrapure water.
Purification of Antimicrobial Proteins
Cation exchange extracted proteins were fractionated on a C18 reversed-phase HPLC column (UP5ODB 25QS, 5 μm, 250 × 2.0 mm, Interchim) using a linear gradient of 0% to 80% acetonitrile in 0.05% trifluoroacetic acid (TFA) over 90 min at a flow rate of 0.7 ml/min. Fractions were dried under vacuum, dissolved in ultrapure water, and tested for antibacterial activity against Staphylococcus aureus SG511. Antimicrobial fractions were purified by a second step of reversed-phase HPLC (X-bridge BEH130, 4.6 mm x 150 mm, Waters) using a biphasic gradient of 0–26% and 26–46% acetonitrile in 0.05% TFA over 5 and 80 min at a flow rate of 0.25 ml/min. Fractions were dried under vacuum, dissolved in ultrapure water, and tested for antimicrobial activity.
Protein Identification
Purity of active fractions was assessed by MALDI-TOF-MS, while sequences were obtained by nano-LC-MS/MS after digestion with trypsin or V8 endopeptidase. LC-MS/MS spectra were analyzed using the automated Mascot algorithm (Matrix Science Ltd., London, UK), and homology searches of the purified protein sequences were performed using a Basic Local Alignment Search Tool (BLAST) search on the National Center for Biotechnology Information (NCBI) server (www.ncbi.nlm.nih.gov/BLAST). Results were validated by the software IRMa (Mascot Results Interpretation). Sequence alignment was performed with the ClustalW2 tool of European Bioinformatics Institute server (EBI).
Antimicrobial Assays
Antibacterial activity of HPLC fractions was assayed against the Gram-positive Micrococcus lysodeikticus Collection de l'institut Pasteur (CIP) 5345, Bacillus megaterium CIP 66.20, S. aureus SG 511, as well as the Gram-negative Escherichia coli SBS363 and V. tasmaniensis LGP32. Minimum inhibitory concentrations (MICs) were determined in poor broth (1% Bacto-Tryptone, 0.5% NaCl w/v, pH 7.5) medium by the liquid growth inhibition assay as described previously (24). Poor broth was supplemented with 2.9% NaCl for the marine V. tasmaniensis. Incubation was performed for 18 h under shaking (150 rpm) at 30 °C for M. lysodeikticus and B. megaterium, at 37 °C for S. aureus and E. coli, and at 20 °C for V. tasmaniensis. Growth was monitored by optical density at 620 nm on a microplate reader infinite M200 (Tecan).
Induction of Extracellular Traps
Hemolymph withdrawn from the oyster pericardial cavity was kept on ice before plating hemocytes on 13-mm glass coverslips in 24-well culture plates at 2.5 × 105 cells/cm2. One h after plating, ET formation was induced by adding V. tasmaniensis LGP32, V. tasmaniensis LMG20012T, Brevibacterium stationis CIP 101282, or Zymosan particles to hemocytes at a multiplicity of infection of 50:1. Plates were centrifuged for 5 min at 500 × g to synchronize binding and further incubated at 20 °C for 30 min, 1 h, or 2 h. To assess the involvement of reactive oxygen species (ROS) in ET formation, hemocytes were pretreated with 10 μm diphenylene iodonium chloride (DPI, Sigma) for 1 h before microbial challenge. For microscopy analyses of living cells, 0.5 μm Sytox Green nucleic acid stain (Molecular Probes) was added to hemocytes. Live imaging was performed on an Axiovert 200M Zeiss inverted microscope. For other experiments, cells were fixed with 4% paraformaldehyde and then permeabilized with 0.01% Triton X-100 for 10 min and stained with 1.25 μg/ml DAPI (Sigma) and either 16.5 nm phalloidin-Alexa Fluor 488 (Molecular Probes) or 2 μg/ml wheat germ agglutinin (WGA)-TRITC (Sigma). Immunostaining was performed with a rabbit anti-H5-like histone antibody generated against the NH2-TPKPAKAKKAAKPKKPASHC-COOH peptide conjugated to Keyhole limpet hemocyanin (KLH) (Proteogenix). Fixed and permeabilized hemocytes were first incubated for 45 min in 50 mm NH4Cl and then for 20 min in PBS containing 5% BSA. Then, 20 μg/ml antibody dissolved in PBS containing 5% BSA was added to coverslips and incubated for 1 h. After three washes in PBS, 10 μg/ml anti-rabbit secondary antibody coupled to Alexa Fluor 488 was added and incubated for 1 h. Coverslips were then washed, stained with DAPI, and mounted with fluorescent mounting medium (DAKO).
Monitoring of Reactive Oxygen Species Production
Hemocytes freshly withdrawn from oysters were plated on a 96-well plate at a density of 6 × 105 cells/cm2. After 1 h of incubation at 17 °C to let the cells settle down, the wells were washed with sterile seawater (SSW) and incubated for 1 h in SSW supplemented with 1 μm luminol (Sigma). Then, zymosan particles (at a multiplicity of infection of 50:1) or PMA (at a final concentration of 1 μm, Sigma) were quickly added, and the plate was immediately placed into a microplate reader infinite M200 (Tecan) to quantify the luminescence emission every 2 min for 2 h. To inhibit ROS production, 10 μm DPI was added to hemocytes 1 h before the addition of ROS inducers (PMA or zymosan).
Histology
Whole oysters were fixed with Davidson's fixative for 42 h. After dissection, muscles and gills were embedded in paraffin. Histological sections and hematoxylin-eosin staining were performed at the technical platform of RHEM (Réseau d'Histologie Expérimentale de Montpellier UMS3426 CNRS, US9 INSERM, University Montpellier 1 and 2). After rehydratation, histological sections were permeabilized with 0.01% Triton X-100 for 10 min, washed three times in PBS, and stained with 0.25 μg/ml DAPI for 1 h. After three washes in PBS, coverslips were mounted over histological sections with fluorescent mounting medium (DAKO).
Image Acquisition and Extranuclear Histone Quantification
Histological sections stained with DAPI; coverslips labeled with DAPI, WGA-TRITC, and phalloidin were observed with 40× or 63× objectives, and images were captured using a Leica TCS SPE confocal scanning laser microscope. Extranuclear histones were quantified on hemocytes immunostained with anti-H5-like histone antibody and counterstained with DAPI using a method adapted from Brinkmann et al. (25). Briefly, for every conditions, 80 images were taken randomly over the entire surface of the coverslip (covering more than a thousand hemocytes) using a 40× objective on a Zeiss Axio Imager upright fluorescence microscope equipped with an AxioCam MRm 2 digital microscope camera. The image files were then analyzed with FIJI software (26). For every image, the DAPI-stained nuclei of hemocytes were counted. The total area revealed by anti-H5-like histone staining and the total area revealed by DNA staining DAPI-staining were measured in μm2. The area occupied by extranuclear histones only was obtained by subtracting the DAPI-stained area to the anti-H5-like histone stained area. The area of extranuclear histones was finally divided by the number of counted hemocytes (nuclei) to normalize on hemocyte density (μm2/cell).
RESULTS
Antimicrobials Accumulate in Oyster Gills 24 h after Infection or Injury
Antimicrobial peptides/proteins were isolated from gills of oysters 24 h after an intramuscular injection of V. tasmaniensis LGP32 (LGP32) or SSW. For that, the dissected gills were subjected to acid extraction, cation exchange chromatography, and reversed-phase HPLC. Gills of non-injected oysters were used as a negative control. Two absorbance peaks strongly increased upon injection (Fig. 1). The corresponding HPLC fractions were the only fractions showing antimicrobial activity against S. aureus SG511 (Fig. 1, Tables 1 and 2). Two were isolated from LGP32-injected oysters (LGP32–36 and LGP32–37), and one was isolated from SSW-injected oysters (SSW36). No antimicrobial activity could be recorded in any fractions isolated from non-injected oysters.
FIGURE 1.
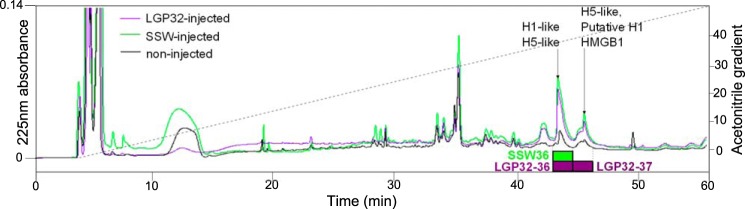
Antimicrobials accumulate in oyster gills 24 h after infection or injury. Reversed-phase HPLC was performed on gill extracts from non-injected oysters (black line), SSW-injected oysters (green line), and V. tasmaniensis LGP32-injected (LGP32-injected) oysters (purple line) using a 0–80% acetonitrile gradient (dotted line) developed over 90 min, on a UP5ODB-25QS column. Absorbance at 225 nm (continuous lines) showed an increase in intensity for the fractions eluted at 36% of acetonitrile in LGP32-injected (LGP32–36) and SSW-injected oysters (SSW36) and for the fraction eluted at 37% of acetonitrile in LGP32-injected oysters (LGP32–37). Purple and green oblongs show antimicrobial fractions in LGP32-injected and SSW-injected oysters, respectively. LGP32–36, LGP32–37, and SSW36 were the only fractions showing antimicrobial activity against S. aureus SG511. The molecules found by LC-MS/MS in active fractions are displayed with arrows.
TABLE 1.
MICs of native H1-like histone (GenBank EKC17653)
| MIC | |
|---|---|
| μm | |
| M. lysodeikticus CIP 53.45 | 0.7 |
| B. megaterium CIP 66.20 | 0.18 |
| S. aureus SG511 | 0.7 |
| E. coli SBS363 | 0.7 |
| V. tasmaniensis LGP32 | >0.7 |
TABLE 2.
Estimated MIC of the LGP32–36 HPLC fraction
| MICa | |
|---|---|
| μm | |
| M. lysodeikticus CIP 53.45 | 0.35 |
| B. megaterium CIP 66.20 | 0.35 |
| S. aureus SG511 | 0.7 |
| E. coli SBS363 | 0.7 |
| V. tasmaniensis LGP32 | >0.7 |
a MICs were determined in μg/ml and converted into μm considering an average molecular mass of 20176 Da (Fig. 2).
After a second reversed-phase HPLC step, the active fractions were analyzed by MALDI-TOF mass spectrometry (Fig. 2). The SSW36 fraction contained one single molecule with a measured molecular mass of 20,748.7 Da. The LGP32–36 fraction contained two molecules: a major one of 19,602.8 Da and a minor one of 20,749.7 Da, similar to that found in the SSW36 fraction. Finally, the LGP32–37 fraction contained one single molecule of 20,625.8 Da (Fig. 2).
FIGURE 2.
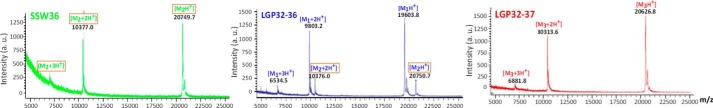
Mass spectrometry analysis of antimicrobial HPLC fractions. MALDI-TOF spectra were acquired on the antimicrobial fractions issued from two rounds of reversed-phase HPLC. The SSW36 fraction (green) is pure, showing single-, double-, and triple-charged ions of one single molecule (molecular mass M2) at MH+ = 20,749.7 Da. The LGP32–36 fraction (blue) contains two molecular species, showing ions of one major molecule (molecular mass M1) at MH+ = 19,603.8 Da and ions of a minor molecule at MH+ = 20,750.7 Da also found in SSW36. The LGP32–37 fraction (red) is pure, showing ions of one single molecule (molecular mass M3) at MH+ = 20,626.8 Da. a. u., arbitrary units.
H1- and H5-like Histones Are the Major Antimicrobials of Challenged Oyster Gills
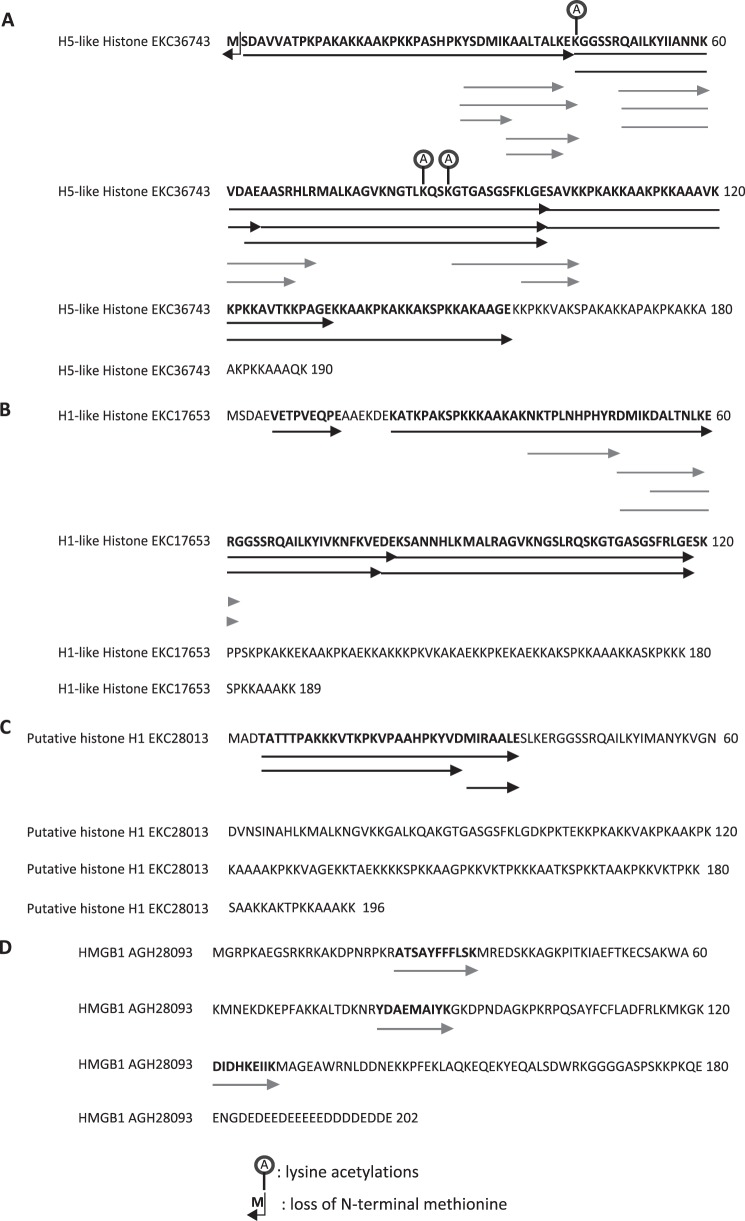
The SSW36, LGP32–36, and LGP32-37 antimicrobial fractions were analyzed further by LC-MS/MS after trypsin and endopeptidase V8 digestion. Histones were found in all three fractions (Fig. 1). An H5-like histone (GenBankTM EKC36743) present in the three fractions was identified with a coverage of 81.6% (Fig. 3A). From the molecular mass measured by MALDI-TOF on the LGP32–36 fraction (19,602.8 Da) (Fig. 2) and from the modifications observed by MS/MS sequencing (Fig. 3), it could be deduced that it lacks its N-terminal methionine (−131 Da) and carries one lysine acetylation (+42 Da) (calculated mass = 19,601.8 Da). An H1-like histone (GenBank EKC17653) only found in LGP32–36 and SSW-36 was identified with a coverage of 57.1% (Fig. 3B). This lower coverage prevented its post-translational status to be established accurately. Finally, traces of a putative histone H1 (GenBank EKC28013) and an high mobility group 1 domain protein (HMGB1, GenBank AGH28093) were found in the LGP32–37 fraction (Figs. 1 and 3, C and D). Interestingly, this accumulation of antimicrobial histones in oyster tissues in response to damage/infection (Fig. 1) correlated with hemocyte infiltration in gill tissue (Fig. 4). This suggests that H1-like and H5-like antimicrobial histones could be brought by hemocytes.
FIGURE 3.
Alignment of LC-MS/MS sequenced peptides with the full-length sequences of H1- and H5-like histones and HMGB1. A–D, H1-like histone (A), H5-like histone (B), putative H1-like histone (C), and HMGB1 (D) full sequences were aligned with LC-MS/MS peptides. Black arrows indicate peptides obtained after endopeptidase V8 digestion, and gray arrows indicate for peptides obtained after trypsin digestion. Sites of acetylation observed by LC-MS/MS are displayed above the sequences.
FIGURE 4.
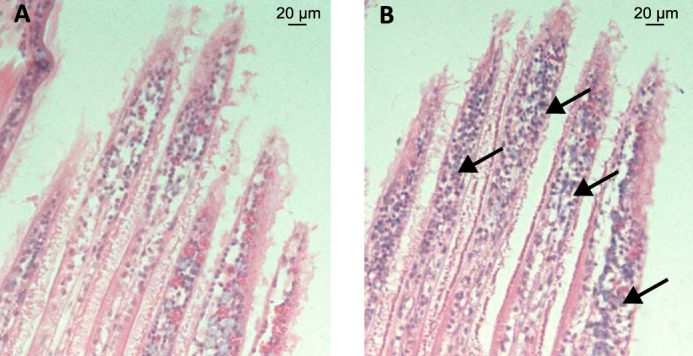
Hemocyte infiltration in gills of LGP32-infected oysters. A and B, histological sections of gills of unchallenged oyster (A) and of oysters infected for 24 h with V. tasmaniensis LGP32 (B). Hematoxylin-eosin staining shows a major hemocyte infiltration in gills of oyster infected with V. tasmaniensis LGP32.
Oyster Histones Present Potent Antibacterial Activity
Due to minute amounts of native proteins purified to homogeneity from oyster tissues, only the H1-like histone (isolated from SSW36) was tested for antimicrobial activity in a low range of concentrations (0–0.7 μm). Potent activities were observed against M. lysodeikticus CIP 53.45, S. aureus SG511, E. coli SBS363, and B. megaterium CIP 66.20 with MICs < 0.7 μm but not against V. tasmaniensis LGP32 (Table 1). The MICs estimated for LGP32–36, which contains the H5-like histone together with low amounts of H1-like histone, were also in the low micromolar range against most of the tested strains (Table 2).
Oyster Hemocytes Release DNA Extracellular Traps in Response to Bacterial Challenge
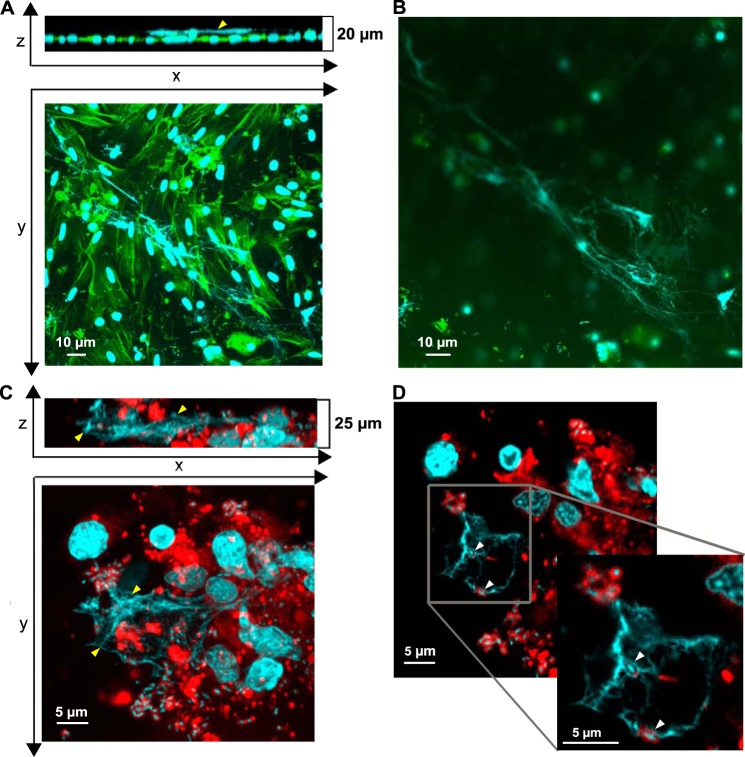
To identify the defense mechanism by which histones are released in oyster tissues, oyster hemocytes were challenged with LGP32 in vitro. The cell-impermeant nucleic acid dye, Sytox Green, added to the primary cultures revealed clusters of permeabilized cells and extracellular DNA only 45 min after LGP32 challenge, whereas most of the hemocytes remained unstained (Fig. 5, C and D). In control hemocytes, permeabilized cells were rarely detectable without any visible extracellular DNA (Fig. 5, A and B). To get higher imaging resolution of the extracellular DNA structures, confocal microscopy was performed. Extracellular networks of DNA filaments were observed 1 h after LGP32 challenge. They were found to extend 5–10 μm above the hemocyte monolayer whose actin cytoskeleton was stained with fluorescent phalloidin (Fig. 6, A and B). To determine whether vibrios could be entrapped into the extracellular DNA, confocal three-dimensional imaging was performed at a higher magnification after staining the bacterial cell wall with the WGA-TRITC lectin. Numerous vibrios were found entrapped in these extracellular DNA networks as shown on a representative confocal section (0.2-μm thickness) (Fig. 6, C and D) and in the reconstituted z-stack containing all the focal planes (Online supplement 1). Altogether, these data show that (i) C. gigas hemocytes can form large DNA ETs upon challenge with V. tasmaniensis LPG32, and (ii) vibrios are entrapped into these ETs.
FIGURE 5.
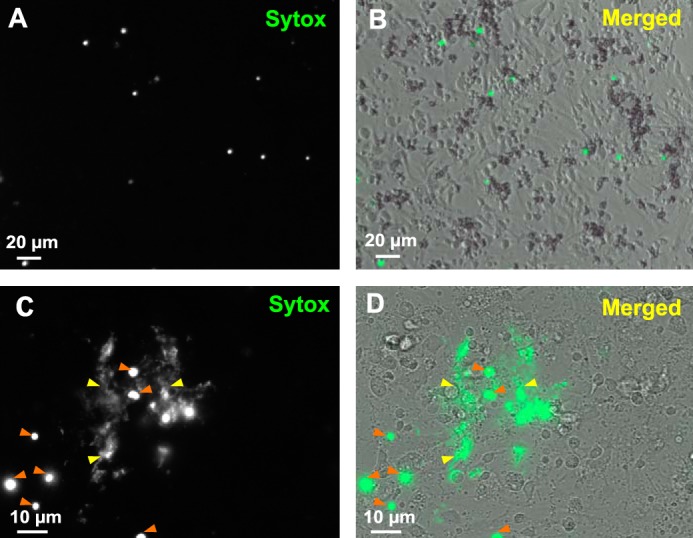
Living oyster hemocytes release extracellular DNA upon bacterial challenge. A–D, cultures of unchallenged hemocytes (A and B) and hemocytes challenged for 45 min with V. tasmaniensis LGP32 (C and D) were stained with Sytox Green and observed by epifluorescence microscopy to reveal extracellular DNA and dead cell nuclei. B and D, bright field images acquired to visualize total cells were merged to epifluorescence images. C and D, extracellular DNA networks (yellow arrowheads) were observed in areas where cells are dead (orange arrowheads) after V. tasmaniensis LGP32 challenge only.
FIGURE 6.
Oyster hemocytes release DNA extracellular traps in which bacteria are entrapped. A and B, DNA ETs released by oyster hemocytes challenged for 1 h with V. tasmaniensis LGP32. Confocal microscopy images were acquired after staining of DNA with DAPI (blue) and filamentous actin with phalloidin-Alexa Fluor 448 (green). A, projections of a z stack and a y stack made from 20 confocal sections. An extracellular DNA ET can be observed extending from 5 to 10 μm above the layer of adherent hemocytes (yellow arrowhead). B, an upper confocal section depicting the longest ET observed in the preparation. C and D, bacteria entrapped in ETs of hemocytes challenged for 1 h with V. tasmaniensis LGP32. Confocal microscopy images were acquired after staining of DNA with DAPI (blue) and cellular membranes with WGA-TRITC (red). C, projections of a z stack and a y stack made from 125 confocal sections. ETs stained with DAPI (yellow arrowheads) were observed in areas with a high bacterial density and extending up to 10 μm above adherent hemocytes. D, bacteria were found entrapped in ETs (white arrowheads), as revealed by their blue nucleic acid staining (DAPI) and red membrane staining (WGA-TRITC) on one representative confocal section of 0.2-μm thickness. See Online supplement 1 for full z-stack.
Release of H5-like Histones Is Associated with ET Formation and Dependent on the Production of ROS
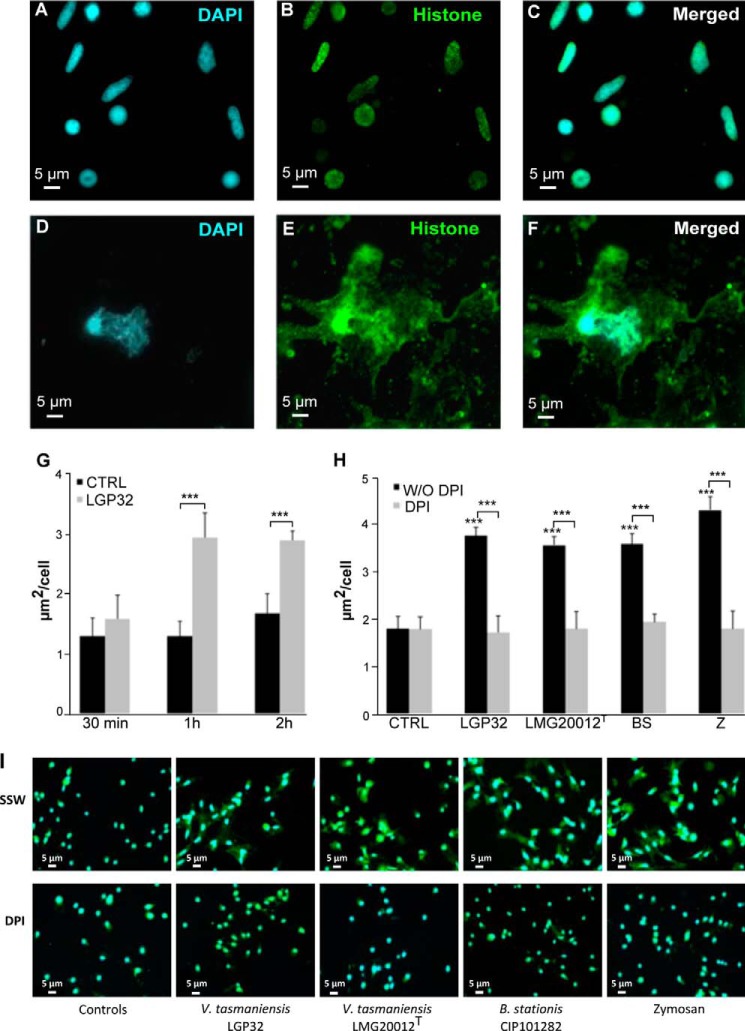
To determine whether the antimicrobial histones isolated from gills of infected/wounded oysters could be released during ET formation, a polyclonal antibody was raised against an N-terminal peptide designed on the sequence of the H5-like histone. Immunostaining was performed on both control and LGP32-challenged hemocytes. As expected, in control hemocytes, H5-like histones were found only within hemocyte nuclei (Fig. 7, A–C and I). Conversely, in LGP32-challenged hemocytes, a significant amount of H5-like histones was found to be extranuclear both in the cytosol and in the extracellular space surrounding ETs, showing that H5-like histones are released along with DNA during ET formation (Fig. 7, D–F and I). Thanks to the H5-like histone immunostaining, we were able to develop a method by automated image analyses to quantify the release of H5-like histone by oyster hemocytes, which is indicative of ET formation (see “Experimental Procedures”). In control hemocytes, the amounts of extranuclear histones remained stable over time (Fig. 7G). In accordance with our previous observations (Figs. 5 and 6), a significant nuclear release of H5-like histones was measured in LGP32-challenged hemocytes as soon as 1 h after challenge with a 2-fold increase in the average size of extranuclear histone areas (p < 0.001, Fig. 7G). There was no significant difference in the amount of extranuclear histones between 1 and 2 h, indicating that the release of H5-like histone and ET formation was rapid and synchronized for most hemocytes capable of forming ETs in our experimental conditions.
FIGURE 7.
ET release is triggered by different microbes and depends on ROS production. A–F, H5-like histone immunostaining of ETs. Epifluorescence microscopy images were acquired after staining of DNA with DAPI (blue, A and D) and immunostaining of C. gigas H5-like histones (green, B and E). In unchallenged hemocytes (A–C), H5-like histones co-localized with nuclear DNA (merge in C). In hemocytes challenged for 1 h with V. tasmaniensis LGP32 (D–F), H5-like histones formed large extranuclear areas around ETs (merge in F). G, time course of ET formation in response to challenge with V. tasmaniensis LGP32. Extranuclear histone areas (μm2/cells) were similar to unchallenged hemocytes (controls (CTRL), black) at 30 min. A 2-fold increase was observed 1 and 2 h after V. tasmaniensis LGP32 challenge (V. tasmaniensis, light gray). Averages and standard deviations were calculated from two independent experiments. ***, p < 0.001 (Student's t test). H, inhibition of ET formation upon blocking of ROS production. V. tasmaniensis LGP32 (LGP32), V. tasmaniensis LMG20012T (LMG20012T), B. stationis CIP101282 (BS), or zymosan (Z) triggered similar ET formation (black) after a 1-h challenge as indicated by extranuclear histone quantification. In all challenge conditions, DPI treatment (light gray) was sufficient to inhibit release of extranuclear histone down to levels of unchallenged hemocytes. Averages and standard deviations were calculated from three independent experiments. ***, p < 0.001 (Student's t test). I, the most representative photographs of the hemocyte response to microbial challenge in the presence/absence of DPI. Epifluorescence microscopy images were acquired after staining of DNA with DAPI (blue) and immunostaining of C. gigas H5-like histones (green). The extracellular histones observed in non-treated samples (SSW) are absent in DPI-treated samples. Sets of eighty similar images were used for extranuclear histone quantification.
To investigate the diversity of microbial challenges that could trigger ET formation, different bacterial strains and microbe-associated molecular patterns were tested including the Gram-negative Vibrio tasmaniensis LMG20012T and LGP32, the Gram-positive B. stationis CIP 101282, and zymosan particles used as a yeast surrogate. All of them induced a massive release of H5-like histones indicative of ET formation without significant difference between microbial challenges (Fig. 7, H and I). As neutrophil ET formation in mammals has been shown to depend on the production of ROS (27), a ROS production inhibitor (DPI) was used in our assays. Remarkably, DPI treatment was sufficient to completely inhibit the release of extranuclear H5-like histones, whose quantities remained as low as in unchallenged hemocytes for any kind of microbial challenge (Fig. 7, H and I).
To determine whether ROS production is sufficient to induce ET formation by oyster hemocytes, we used PMA, a well described inducer of oxidative burst and ET formation in mammalian neutrophils (27). Surprisingly, in contrast to zymosan, PMA did not induce ET formation nor the release of extranuclear H5-like histones by hemocytes (Fig. 8A). This correlated with a lack of ROS production in PMA-stimulated hemocytes, as determined by chemiluminescence (Fig. 8B). On the contrary, zymosan induced a strong oxidative burst in hemocytes, which correlated with the induction of ET formation and the release of extranuclear H5-like histones (Fig. 8, A and B). Supporting the hypothesis of a ROS-dependent process, DPI was sufficient to inhibit zymosan effects, i.e. ROS production (Fig. 8B) and ET formation (Fig. 7). Taken together, these results show that ET formation in C. gigas hemocytes can be triggered by a broad diversity of microbial agents and depends strongly on ROS production.
FIGURE 8.
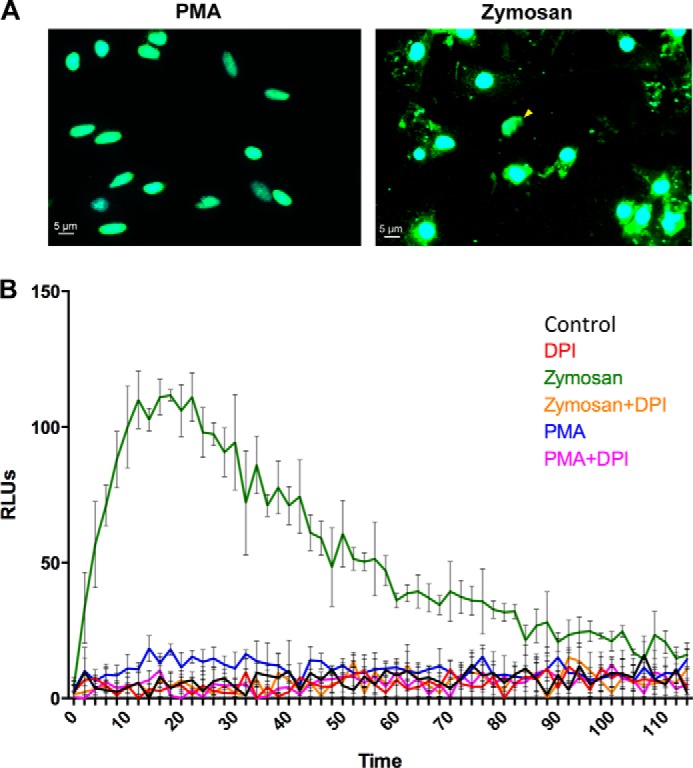
Zymosan but not PMA induces ROS production and ET formation by oyster hemocytes. A, representative photographs of the hemocyte response to stimulation with PMA or zymosan. Epifluorescence microscopy images were acquired after staining of DNA with DAPI (blue) and immunostaining of C. gigas H5-like histones (green). The extranuclear histones and ETs observed in zymosan-stimulated samples are absent in PMA-stimulated samples. B, ROS production was monitored in control hemocytes (black line) and after hemocyte stimulation with PMA (blue line) or zymosan (green line). The inhibition of ROS production by DPI treatment was monitored in control hemocytes (red line), PMA-stimulated hemocytes (pink line), and zymosan-stimulated hemocytes (orange line). Results are expressed in relative luminescence units (RLUs) indicative of luminol oxidation. Averages and standard deviations were calculated from three independent experiments.
ETs Are Observed in Vivo in Infected or Wounded Tissues
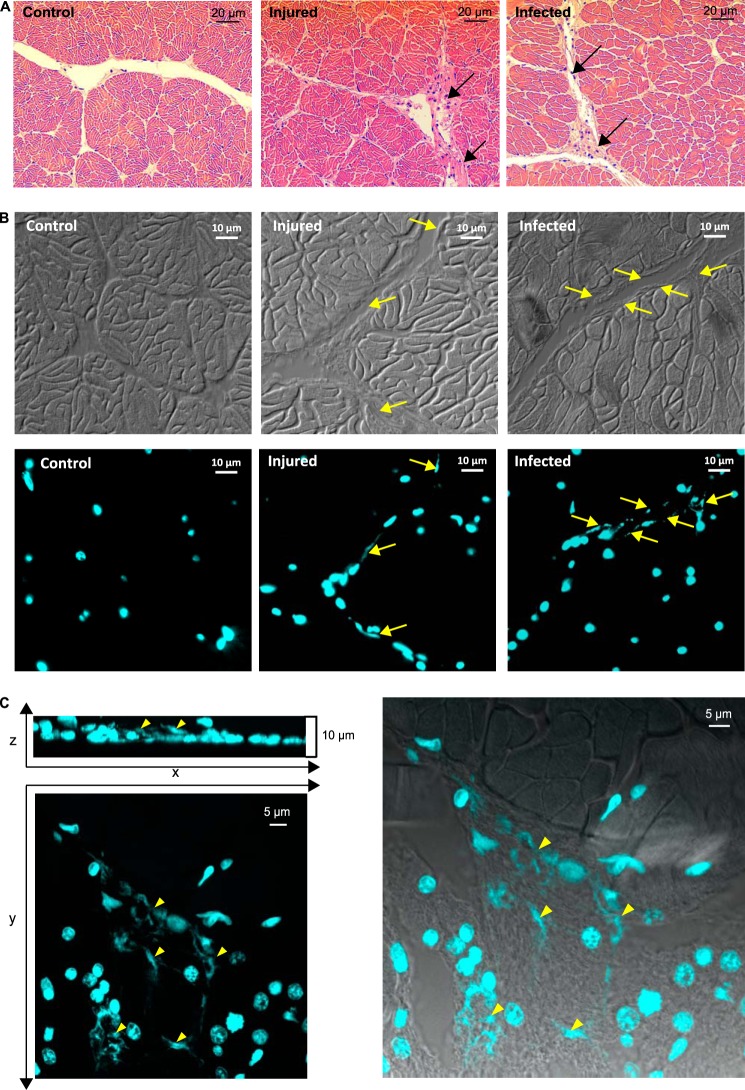
Finally, we assessed whether ETs could form in vivo. Oysters were either challenged with a sublethal injection of V. tasmaniensis LGP32 or injured by a sterile puncture in the adductor muscle. Massive hemocyte infiltrations were observed in gills (Fig. 4) and in the adductor muscles (Fig. 9A). In gills, the high hemocyte density precluded the imaging resolution needed to observe extracellular DNA filaments. In adductor muscles, cell infiltrates were found within interstitial tissue between fascicles of muscle fibers at the wound periphery in both LGP32-challenged and injured oysters 1 day after the challenge (Fig. 9, A and B). More importantly, DNA filaments were observed in these hemocyte-infiltrated regions, whereas only intact cell nuclei were observed in control animals after DAPI staining (Fig. 9C). Confocal microscopy was performed on a thick tissue section to get a better three-dimensional reconstruction of the areas containing extracellular DNA. Images confirmed the presence of ETs between fascicles of muscle fibers, and in the z-projected image, intact cell nuclei were observed above and below the ET areas, indicating that the DNA was not released as a result of any nuclei damaging during the preparation of histological sections (Fig. 9C). Taken together, these data showed that infiltrating hemocytes can release ETs in vivo in response to infection or tissue damage.
FIGURE 9.
ETs are observed in vivo in muscles of infected or injured oysters. A, hematoxylin-eosin stained adductor muscles of unchallenged (Control), V. tasmaniensis LGP32-infected, and injured oysters. Infiltrating hemocytes are found between muscle fascicles after injury (black arrows, middle panel) or after infection (black arrows, right panel). B, epifluorescence microscopy observation of DAPI-stained ETs within histological sections of muscle infiltrated with hemocytes. Contrast phase images are shown below epifluorescence images. ETs are observed in cell-infiltrated interstitial tissues between fascicles of muscle fibers after injury (yellow arrows, middle panel) or after V. tasmaniensis LGP32 infection (yellow arrows, right panel). No ETs are observed in muscle of control animals (left panel). C, confocal microscopy observation of DAPI-stained ETs within a thick histological section of muscle infiltrated with hemocytes. Projections of a z stack and a y stack display several ETs (yellow arrowheads, left panel). Intact cell nuclei are observed above and below ETs (z projection, left panel). Superposition of the phase confocal section and y projection (right panel) shows that ETs located in cell-infiltrated interstitial tissues between fascicles of muscle fibers.
DISCUSSION
Results showed that antimicrobial histones accumulate together with extracellular DNA in oyster tissues in response to infection and injury. Tissues of non-injected oysters were devoid of such antimicrobial histones (Fig. 1), indicating that histone accumulation in tissues was induced by the challenges. The major antimicrobial histones isolated here were H5-like and H1-like histones. Antimicrobial H2B and H4 histones have also been isolated from another oyster species, C. virginica (7) (8). Similar to our observations, protein levels of H4 histone strongly increased in hemocyte lysates and extracellular hemolymph of C. virginica oysters infected with Perkinsus marinus (8). Therefore, accumulation of histones in oyster tissues could be a common response to infection and injury.
The antimicrobial histones isolated from oyster gills showed a broad spectrum of antimicrobial activities with MICs as low as 0.18 μm against Bacillus megaterium CIP 66.20 for native H1-like histone (Table 1). Together with oyster defensins (24), the H1-like histone is therefore one of the most potent antimicrobials of C. gigas described so far. No activity could be recorded against the oyster pathogen V. tasmaniensis LGP32, whereas the H2B and H4 histones from C. virginica were reported to be active against vibrios (7, 8). This may be due to the intrinsic resistance of V. tasmaniensis LGP32 to antimicrobials (28) or to the low range of concentrations tested (0–0.7 μm). Because tissues of C. gigas oysters are poor in antimicrobial peptides/proteins (29), the isolation of histones as the only antimicrobials found in gills of challenged oysters strongly argues in favor of their role in the oyster antimicrobial defense.
In vitro, antimicrobial histones were rapidly released (in less than 1 h) by oyster hemocytes together with extracellular DNA when exposed to diverse microbial agents (Figs. 5–8 and Online supplement 1). Importantly, this phenomenon was also observed in vivo (Fig. 9). Indeed, both LGP32-infected and injured oysters showed a massive hemocyte infiltration and release of extracellular DNA in tissues surrounding the site of injury (Fig. 9). These DNA structures are reminiscent of the neutrophil ETs well studied in vertebrates such as mammals, birds, and fish over the past decade (15, 16, 30). In the present work, bacteria were found entrapped into such extracellular DNA networks (Fig. 6 and Online supplement 1). This strongly suggests that, as in vertebrates, oyster ETs participate in host defense by capturing large numbers of microbes and preventing their dissemination (16). Surrounding and entangling of bacteria in DNA or peptide networks are indeed increasingly recognized as conserved mechanisms of antimicrobial defense (31). Moreover, as shown in some vertebrates, the antimicrobial properties of the ET-associated antimicrobials including histones (Table 1) and their concentration on ETs could also contribute to kill the entrapped microorganisms (10, 16, 19).
The formation of oyster ETs was shown here to be dependent on ROS production by hemocytes (Fig. 7H), which results from NADPH-oxidase activity and/or mitochondrial respiration (32). Indeed, we observed a strong positive correlation between ROS production and ET formation (Figs. 7 and 8). This result is particularly important because ROS play a central role in initiating ET formation in vertebrates (27). However, unlike in vertebrates, PMA failed to trigger the oxidative burst and the formation of ETs by oyster hemocytes under our experimental conditions. Along with ROS, damage-associated molecular patterns (DAMPs) released by injured tissues (33) likely regulate ET formation in oysters. Indeed, we first showed that an aseptic injury is sufficient to induce accumulation of histones and extracellular DNA release in vivo. Second, an HMG domain protein group 1 (HMGB1) was identified in the LGP32–37 HPLC fraction, issued from LGP32-infected oysters (Fig. 1). HMGB1 is a DAMP of intracellular origin well known in vertebrates (34) but also in C. gigas, in which it enhances the Rel-dependent NF-κB activation (35). Importantly, HMGB1 was recently shown to promote neutrophil ET formation through TLR-4 activation (36). Therefore, the ROS-dependent production of ETs by oyster hemocytes can be triggered by microbial agents and potentially by DAMPs released in the extracellular milieu in response to injury or cell lysis.
One limitation of the present study is that the hemocyte subset producing ETs has not been identified. We believe that ETs are produced by certain hemocyte subsets only because numerous intact cells are observed in ET-containing regions in both in vivo and in vitro microscopy observations (Figs. 5 and 8). In vertebrates, ETs are formed mostly by neutrophils (9, 15, 16). However, the accurate determination of ET-forming hemocytes remains challenging in oysters, hemocytes subsets being still mainly classified based on morphological features rather than molecular markers (37).
In conclusion, the present study shows that oysters use the release of DNA extracellular traps and antimicrobial histones as part of their immune defense. Therefore, this defense mechanism is shared by relatively distant species belonging to the different branches of the Bilateria, not only the Deuterostomia (mammals, birds, fishes) and the Ecdysozoa (shrimp) but also the Lophotrochozoa (oyster). The identification of ROS as a secondary messenger required for ET formation in oysters supports the evolutionary conservation within Bilateria of an important immune strategy without ruling out the hypothesis of a convergent evolution between phylogenetically distant species.
Supplementary Material
Acknowledgments
We are grateful to Agnès Vergnes and Marc Leroy for technical assistance. We thank the Montpellier RIO Imaging platform for access to confocal and epifluorescence microscopy, the “Réseau d'Histologie Expérimentale de Montpellier” histology facility for histology expertise and tissue processing, and the mass spectrometry platform of the “Institut des Biomolécules Max Mousseron” for MALDI-TOF-MS.
This work was supported by grants from the Agence Nationale de la Recherche (ANR) (Vibriogen project, Blanc SVSE7 2011) and the Languedoc-Roussillon region (REVARESP project, Chercheur(se) d'avenir 2009).

This article contains Online supplement 1.
- ET
- extracellular trap
- ROS
- reactive oxygen species
- PMA
- phorbol myristate acetate
- MIC
- minimum inhibitory concentration
- DPI
- diphenylene iodonium chloride
- WGA
- wheat germ agglutinin
- TRITC
- tetramethylrhodamine isothiocyanate
- SSW
- sterile seawater
- DAMP
- damage-associated molecular pattern
- HMG
- high mobility group
- CIP
- Collection de l'institut Pasteur.
REFERENCES
- 1. Parseghian M. H., Luhrs K. A. (2006) Beyond the walls of the nucleus: the role of histones in cellular signaling and innate immunity. Biochem. Cell Biol. 84, 589–604 [DOI] [PubMed] [Google Scholar]
- 2. Miller B. F., Abrams R., Dorfman A., Klein M. (1942) Antibacterial properties of protamine and histone. Science 96, 428–430 [DOI] [PubMed] [Google Scholar]
- 3. Kawasaki H., Iwamuro S. (2008) Potential roles of histones in host defense as antimicrobial agents. Infect. Disord. Drug. Targets 8, 195–205 [DOI] [PubMed] [Google Scholar]
- 4. Patat S. A., Carnegie R. B., Kingsbury C., Gross P. S., Chapman R., Schey K. L. (2004) Antimicrobial activity of histones from hemocytes of the Pacific white shrimp. Eur. J. Biochem. 271, 4825–4833 [DOI] [PubMed] [Google Scholar]
- 5. Li C., Song L., Zhao J., Zhu L., Zou H., Zhang H., Wang H., Cai Z. (2007) Preliminary study on a potential antibacterial peptide derived from histone H2A in hemocytes of scallop Chlamys farreri. Fish Shellfish Immunol. 22, 663–672 [DOI] [PubMed] [Google Scholar]
- 6. De Zoysa M., Nikapitiya C., Whang I., Lee J. S., Lee J. (2009) Abhisin: a potential antimicrobial peptide derived from histone H2A of disk abalone (Haliotis discus discus). Fish Shellfish Immunol. 27, 639–646 [DOI] [PubMed] [Google Scholar]
- 7. Seo J. K., Stephenson J., Noga E. J. (2011) Multiple antibacterial histone H2B proteins are expressed in tissues of American oyster. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 158, 223–229 [DOI] [PubMed] [Google Scholar]
- 8. Dorrington T., Villamil L., Gómez-chiarri M. (2011) Upregulation in response to infection and antibacterial activity of oyster histone H4. Fish Shellfish Immunol. 30, 94–101 [DOI] [PubMed] [Google Scholar]
- 9. Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D. S., Weinrauch Y., Zychlinsky A. (2004) Neutrophil extracellular traps kill bacteria. Science 303, 1532–1535 [DOI] [PubMed] [Google Scholar]
- 10. Yipp B. G., Petri B., Salina D., Jenne C. N., Scott B. N., Zbytnuik L. D., Pittman K., Asaduzzaman M., Wu K., Meijndert H. C., Malawista S. E., de Boisfleury Chevance A., Zhang K., Conly J., Kubes P. (2012) Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat. Med. 18, 1386–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jenne C. N., Wong C. H., Zemp F. J., McDonald B., Rahman M. M., Forsyth P. A., McFadden G., Kubes P. (2013) Neutrophils recruited to sites of infection protect from virus challenge by releasing neutrophil extracellular traps. Cell Host. Microbe. 13, 169–180 [DOI] [PubMed] [Google Scholar]
- 12. Guimarães-Costa A. B., Nascimento M. T., Wardini A. B., Pinto-da-Silva L. H., Saraiva E. M. (2012) ETosis: a microbicidal mechanism beyond cell death. J. Parasitol. Res. 2012, 929743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y., Li M., Stadler S., Correll S., Li P., Wang D., Hayama R., Leonelli L., Han H., Grigoryev S. A., Allis C. D., Coonrod S. A. (2009) Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J. Cell Biol. 184, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Urban C. F., Reichard U., Brinkmann V., Zychlinsky A. (2006) Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell Microbiol. 8, 668–676 [DOI] [PubMed] [Google Scholar]
- 15. Brinkmann V., Zychlinsky A. (2007) Beneficial suicide: why neutrophils die to make NETs. Nat. Rev. Microbiol. 5, 577–582 [DOI] [PubMed] [Google Scholar]
- 16. Papayannopoulos V., Zychlinsky A. (2009) NETs: a new strategy for using old weapons. Trends Immunol. 30, 513–521 [DOI] [PubMed] [Google Scholar]
- 17. Beiter K., Wartha F., Albiger B., Normark S., Zychlinsky A., Henriques-Normark B. (2006) An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr. Biol. 16, 401–407 [DOI] [PubMed] [Google Scholar]
- 18. Ramos-Kichik V., Mondragón-Flores R., Mondragón-Castelán M., Gonzalez-Pozos S., Muñiz-Hernandez S., Rojas-Espinosa O., Chacón-Salinas R., Estrada-Parra S., Estrada-García I. (2009) Neutrophil extracellular traps are induced by Mycobacterium tuberculosis. Tuberculosis (Edinb.) 89, 29–37 [DOI] [PubMed] [Google Scholar]
- 19. McDonald B., Urrutia R., Yipp B. G., Jenne C. N., Kubes P. (2012) Intravascular neutrophil extracellular traps capture bacteria from the bloodstream during sepsis. Cell Host. Microbe 12, 324–333 [DOI] [PubMed] [Google Scholar]
- 20. Altincicek B., Stötzel S., Wygrecka M., Preissner K. T., Vilcinskas A. (2008) Host-derived extracellular nucleic acids enhance innate immune responses, induce coagulation, and prolong survival upon infection in insects. J. Immunol. 181, 2705–2712 [DOI] [PubMed] [Google Scholar]
- 21. Ng T. H., Chang S. H., Wu M. H., Wang H. C. (2013) Shrimp hemocytes release extracellular traps that kill bacteria. Dev. Comp. Immunol. 41, 644–651 [DOI] [PubMed] [Google Scholar]
- 22. Gay M., Renault T., Pons A. M., Le Roux F. (2004) Two vibrio splendidus related strains collaborate to kill Crassostrea gigas: taxonomy and host alterations. Dis. Aquat. Organ 62, 65–74 [DOI] [PubMed] [Google Scholar]
- 23. Sawabe T., Ogura Y., Matsumura Y., Feng G., Amin A. R., Mino S., Nakagawa S., Sawabe T., Kumar R., Fukui Y., Satomi M., Matsushima R., Thompson F. L., Gomez-Gil B., Christen R., Maruyama F., Kurokawa K., Hayashi T. (2013) Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio. tritonius sp. nov. Front Microbiol. 4, 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmitt P., Wilmes M., Pugnière M., Aumelas A., Bachère E., Sahl H. G., Schneider T., Destoumieux-Garzón D. (2010) Insight into invertebrate defensin mechanism of action: oyster defensins inhibit peptidoglycan biosynthesis by binding to lipid II. J. Biol. Chem. 285, 29208–29216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brinkmann V., Goosmann C., Kühn L. I., Zychlinsky A. (2012) Automatic quantification of in vitro NET formation. Front Immunol. 3, 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fuchs T. A., Abed U., Goosmann C., Hurwitz R., Schulze I., Wahn V., Weinrauch Y., Brinkmann V., Zychlinsky A. (2007) Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 176, 231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Duperthuy M., Binesse J., Le Roux F., Romestand B., Caro A., Got P., Givaudan A., Mazel D., Bachère E., Destoumieux-Garzón D. (2010) The major outer membrane protein OmpU of Vibrio splendidus contributes to host antimicrobial peptide resistance and is required for virulence in the oyster Crassostrea gigas. Environ Microbiol. 12, 951–963 [DOI] [PubMed] [Google Scholar]
- 29. Schmitt P., Rosa R. D., Duperthuy M., de Lorgeril J., Bachère E., Destoumieux-Garzón D. (2012) The antimicrobial defense of the Pacific oyster, Crassostrea gigas: how diversity may compensate for scarcity in the regulation of resident/pathogenic microflora. Front Microbiol. 3, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Palić D., Ostojić J., Andreasen C. B., Roth J. A. (2007) Fish cast NETs: neutrophil extracellular traps are released from fish neutrophils. Dev. Comp. Immunol. 31, 805–816 [DOI] [PubMed] [Google Scholar]
- 31. Chu H., Pazgier M., Jung G., Nuccio S. P., Castillo P. A., de Jong M. F., Winter M. G., Winter S. E., Wehkamp J., Shen B., Salzman N. H., Underwood M. A., Tsolis R. M., Young G. M., Lu W., Lehrer R. I., Bäumler A. J., Bevins C. L. (2012) Human α-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 337, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Donaghy L., Kraffe E., Le Goïc N., Lambert C., Volety A. K., Soudant P. (2012) Reactive oxygen species in unstimulated hemocytes of the Pacific oyster Crassostrea gigas: a mitochondrial involvement. PLoS One 7, e46594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matzinger P. (1994) Tolerance, danger, and the extended family. Annu. Rev. Immunol. 12, 991–1045 [DOI] [PubMed] [Google Scholar]
- 34. Rubartelli A., Lotze M. T. (2007) Inside, outside, upside down: damage-associated molecular-pattern molecules (DAMPs) and redox. Trends Immunol. 28, 429–436 [DOI] [PubMed] [Google Scholar]
- 35. Li J., Zhang Y., Xiang Z., Xiao S., Yu F., Yu Z. (2013) High mobility group box 1 can enhance NF-κB activation and act as a pro-inflammatory molecule in the Pacific oyster, Crassostrea gigas. Fish Shellfish Immunol. 35, 63–70 [DOI] [PubMed] [Google Scholar]
- 36. Tadie J. M., Bae H. B., Jiang S., Park D. W., Bell C. P., Yang H., Pittet J. F., Tracey K., Thannickal V. J., Abraham E., Zmijewski J. W. (2013) HMGB1 promotes neutrophil extracellular trap formation through interactions with Toll-like receptor 4. Am. J. Physiol. Lung Cell Mol. Physiol. 304, L342–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bachère E., Gueguen Y., Gonzalez M., de Lorgeril J., Garnier J., Romestand B. (2004) Insights into the anti-microbial defense of marine invertebrates: the penaeid shrimps and the oyster Crassostrea gigas. Immunol. Rev. 198, 149–168 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.